Abstract
Objective: The repurposing of electronic health records (EHRs) can improve clinical and genetic research for rare diseases. However, significant information in rare disease EHRs is embedded in the narrative reports, which contain many negated clinical signs and family medical history. This paper presents a method to detect family history and negation in narrative reports and evaluates its impact on selecting populations from a clinical data warehouse (CDW).
Materials and Methods: We developed a pipeline to process 1.6 million reports from multiple sources. This pipeline is part of the load process of the Necker Hospital CDW.
Results: We identified patients with “Lupus and diarrhea,” “Crohn’s and diabetes,” and “NPHP1” from the CDW. The overall precision, recall, specificity, and F-measure were 0.85, 0.98, 0.93, and 0.91, respectively.
Conclusion: The proposed method generates a highly accurate identification of cases from a CDW of rare disease EHRs.
Keywords: data warehouse, search engine, natural language processing, rare diseases, electronic health records
INTRODUCTION
Secondary use of electronic health records (EHRs) for research purposes requires information retrieval tools to make structured data and information contained within clinical text accessible. Narrative clinical reports allow flexibility of expression and representation of elaborate clinical entities, including clinical signs, patient history, and family history.1–7
In order to facilitate this needed capacity for data exploration at our institution (Necker Enfants Malades and Imagine Institute), we have designed and deployed Dr Warehouse®, a full-text clinical data warehouse (CDW) for cohort identification and data extraction. Necker/Imagine specializes in rare diseases. A detailed description of the symptoms, including negative findings, is documented in the patient record, as it is required to lead to an accurate diagnosis. Therefore, it is crucial to include mechanisms for detecting negation in text. A narrative rare-disease patient report includes a large amount of detailed information from 3 generations of relatives. Exploiting this information and distinguishing between family history and patient information is crucial not only for monogenic disorders (e.g., NPHP1 mutation, responsible for steroid resistant nephrotic syndrome) but also for all diseases with a known hereditary component (e.g., Crohn’s disease and lupus).8,9 Moreover, patients who suffer from 1 autoimmune disease are more likely to present other autoimmune disorders. Genomewide association studies have identified several genes that might be associated with increased susceptibility to diabetes and Crohn’s disease.10 It is therefore crucial to (1) identify if a patient has only 1 disease or both and (2) detect whether the condition affects other members of the family. Finally, some diseases can affect multiple organs, and clinical presentation often includes a wide array of symptoms; for example, lupus enteritis is suspected when a patient presents with abdominal pain, diarrhea, and vomiting.11 Given the low incidence (only 5% of lupus patients have diarrhea,12 and the incidence of lupus is about 5.5/100 000 persons13) and nonspecific clinical findings, it is important to identify those cases.
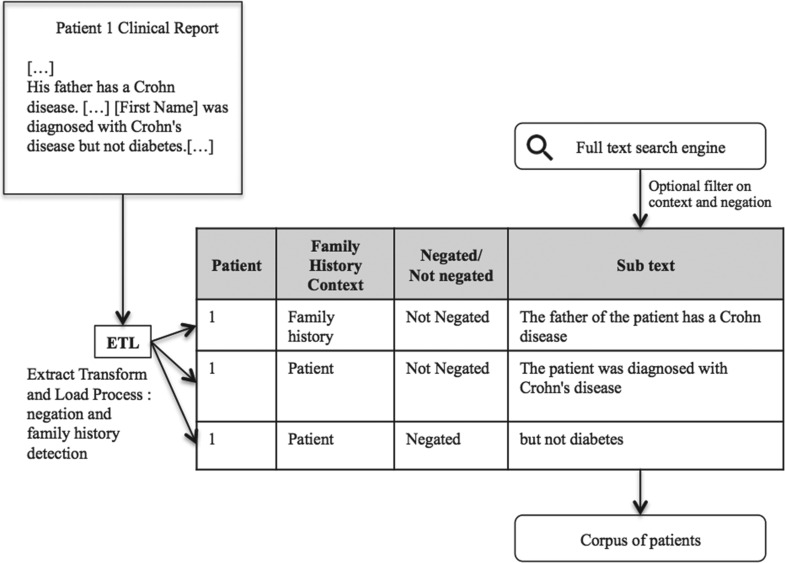
We developed a method that detects the negated segments and the segments related to the family history in the narrative reports. This method is part of the Extract-Transform-Load process of the CDW (Figure 1). We evaluated the benefits of using this method to distinguish between true positives (TPs) and false positives (FPs) in 3 corpora of rare disease patient records related, respectively, to Crohn’s disease, lupus, and NPHP1 from the CDW.
Figure 1.
Illustration of the objective of our method and its context.
Related Work
Negation detection
Most of the published NLP methods are based on concept extraction and their assertion classifications.14–17 The most common algorithm for negation detection is NegEx.18 It is based on regular expressions targeted before and after a concept in a document to determine whether a concept is negated. NegEx has been ported and evaluated on clinical texts in French and has shown good performance (a recall of 85% and a precision of 89%) on cardiology notes.19,20
In 2006, Goryachev et al.21 evaluated NegEx and 3 other methods of negation detection. Among them, NegExpander was developed by Aronow et al.22 and was based on NegEx, with extension to conjunctive phrases and to all Unified Medical Language System (UMLS) terms. The 2 other methods were based on machine-learning classifiers using Weka machine-learning software and a naïve Bayes classifier associated with a support vector machine classifier. Goryachev concluded that NegEx was the most effective.
Family history detection
As for family history, most authors have developed methods that extract family information from specific or focused sections in clinical reports. Friedlin and McDonald23 focused on sentences identified by titles with a family history–type phrase in the admission notes. They used the Regenstrief data eXtraction tool to associate 12 diseases to family histories in these sentences, with a precision and recall of 97% and 96%, respectively. Goryachev et al.24 also assumed that the medical narrative reports were structured in sections. Inside each section identified using section headings, the entities were extracted and categorized using the UMLS semantic types. The authors developed a set of rules to associate the finding with a family member or with the patient. Their pipeline achieved 97.2% sensitivity and 99.7% specificity in detecting the family history findings. Lewis et al.25 examined the entire text, considering that a patient’s family history may be spread throughout the patient’s clinical records and is mostly recorded in the clinical notes. They used a Stanford NLP Parser to detect dependency between the disease and the family relationship with a precision of 61% and a recall of 51%.
The ConText algorithm developed by Chapman et al.26 is based on the NegEx approach; it is a regular-expression–based algorithm that searches for trigger terms preceding or following the indexed clinical conditions to determine if clinical conditions mentioned in the clinical reports are negated, hypothetical, historical, or experienced by someone other than the patient. Chapman et al. showed that it was relevant to use only regular expressions to detect these contextual values to classify the concepts extracted from the text.
The main limitations of these approaches are the restriction to preidentified concepts and/or the use of semistructured reports (reports with explicit sections).
Our aim is to extract subtexts from each original patient record and classify them into 4 categories: patient–not negated/patient–negated/family history–not negated/family history–negated. These subtexts would be integrated through the Extract-Transform-Load process in the CDW to improve the performance of the full-text search engine.
Methods
Overview
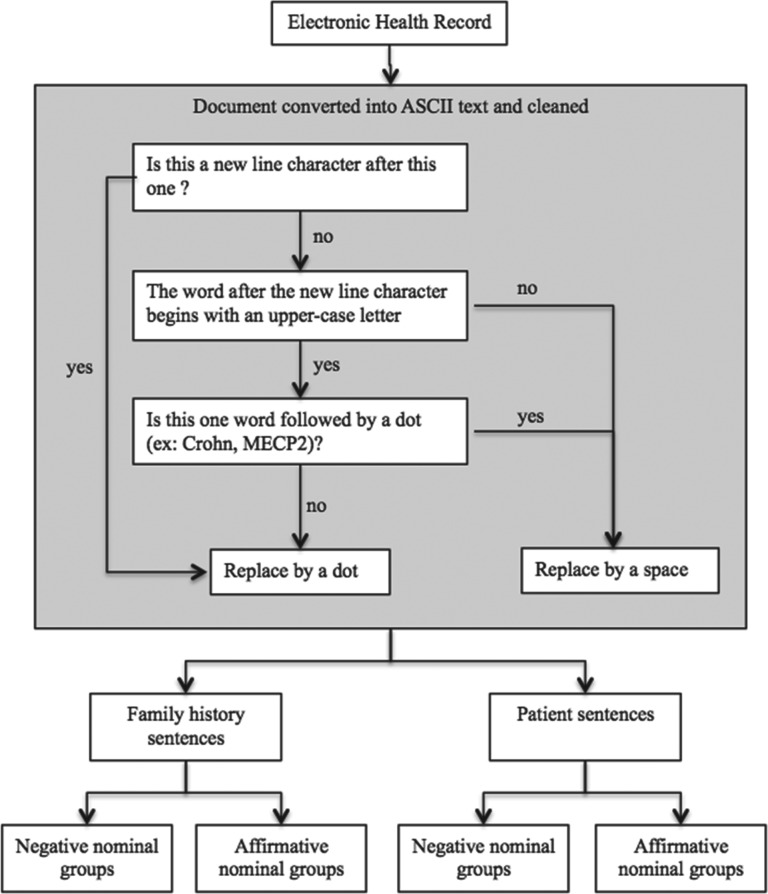
We built a pipeline (Figure 2) to process the medical records in 3 steps: cleaning text, detecting context (patient data/family history), and detecting negation in each context. This pipeline was designed to process unstructured reports and free text from multiple sources and authors.
Figure 2.
Overview of the pipeline.
Text processing
The first step consisted of converting all the Word® or PDF (Portable Document Format) documents to ASCII (American Standard Code for Information Interchange), then cleaning the documents from erroneous newline characters generated by the automated conversion. To determine whether to replace a newline character, we developed a decision tree based on regular expression rules to choose the correct character replacement for each newline character (Figure 2).
Family history detection
We listed all the French words related to family relationships (Supplementary Appendix 1). The algorithm takes into account the age of the patient at the date of the document. For example, when the patient is under 18 years old, it does not consider the words “son” and “daughter” in the list because this is the parent referring to their child, who is actually the patient. Moreover, we added several expressions that were meaningful for family history such as “family history,” “paternal ancestry,” and “maternal ancestry.”
Each sentence in which a term of the above list matched was classified as belonging to the family history context; otherwise, the sentence was classified in the patient context.
Negation detection
We built a corpus of 3900 narrative documents by querying Dr Warehouse® for 4 diseases: terminal renal failure, autism, Rett Syndrome, and Currarino.
We used this corpus to build (Table 1):
a list of French regular expression rules to split the sentence into propositions or nominal groups (Supplementary Appendix 2).
a list of negated regular expressions to determine whether a proposition had a negative meaning.
a list of exclusion rules for the double negatives such as “we cannot exclude” or “without any doubt.”
Table 1.
List of negated regular expressions and exclusion rules
| Negated expressions | Exclusion rules | |
|---|---|---|
| 1 | /[^a-z]pas\s[a-z]*\s*d/i |
|
| 2 | /\sn(e |')(l[ae] |l')?[a-z]+ pas[^a-z]/i |
|
| 3 | /[^a-z]sans\s/i |
|
| 4 | /aucun/ |
|
| 5 | /\selimine/i |
|
| 6 | /\sinfirme/i |
|
| 7 | /[^a-z]exclu[e]?[s]?[^a-z]/i |
|
| 8 | /[^a-z]jamais\s[a-z]*\s*d/i | |
| 9 | /[^a-z]ni\s/i | |
| 10 | /oriente pas vers/i | |
| 11 | /:\s*non[^a-z]/i | |
| 12 | /^\s*non[^a-z]+$/i | |
| 13 | /:\s*aucun/i | |
| 14 | /:\s*exclu/i | |
| 15 | /:\s*absent/i | |
| 16 | /:\s*inconnu/i | |
| 17 | /absence/i | |
| 18 | /absent/i | |
| 19 | /\sne pas\s/i | |
| 20 | /\snegati.*/i | |
| 21 | /[^a-z]normale?s?\s/i | /pas\s+[^a-z]normale?s?\s/i |
| 22 | /[^a-z]normaux\s/i | /pas\s+[^a-z]normaux\s/i |
Suggested English equivalent: do not (1, 2, 19), without (3), no/none (4, 11, 12, 13), eliminate (5, 6), exclude/exclusion (7, 14), never (8), neither (9), not lead to (10), absent/absence (15, 17, 18), unknown (16), negative/negation (20), normal (21, 22).
The algorithm classifies the expression of normality as negative information, e.g., “gene MECP2 normal” is equivalent to “MECP2 not mutated.” We decided to keep hypothetical diagnostic and “research for” as affirmative information as long as no final diagnosis had been confirmed. Based on these rules, the algorithm classifies each proposition and nominal group as negative or non-negative.
CDW integration
Dr Warehouse® relies on Oracle® 11 g, and the search engine is based on the Oracle® Text module. For a given document, all of the triplets {subtext, context, negation} are stored in Dr Warehouse®. Context is either “patient” or “family history.”
Evaluation
To evaluate the impact of negation and family history detection on the performance of the search engine, we used the pipeline on the entire CDW. The textual documents were de-identified using an internally developed algorithm based on name, first name, birth date, address, and hospital ID. To protect confidentiality, authorized internal staff at the Necker Hospital conducted the study.
We evaluated the system on 3 corpora extracted from Dr Warehouse® at Necker/Imagine Institute by querying the whole CDW, which contains 1.6 million EHRs for 350 000 individual patients (January 2016). The corpora corresponded to 3 clinical use cases, namely “Crohn’s and diabetes,” “lupus and diarrhea,” and “NPHP1.” For each of them, we counted the number of TPs, FPs, true negatives (TN), and false negatives (FNs) without any filtering (i.e., before applying our algorithm), with family history detection only, with negation detection only, and with both detection algorithms. We evaluated the FNs regarding negation and family history detection only, as it was not possible to calculate them for the nonfiltered queries. We defined TPs as patients with all terms present in their EHRs in a non-negative expression and with no family history expression. The TN patients were defined as having all the terms but at least 1 in a negative expression or in a family history context.
Two persons independently evaluated the system and the Kappa score was calculated. In case of discordance, a consensus was reached. Four metrics were used to assess the performance of our algorithm: recall, precision, specificity, and F-measure.
Results
The overall interevaluator agreement measured by the Kappa coefficient was 0.98. Table 2 displays the results for each use case. Before applying our algorithm, 145 patients (262 documents) were classified as “Lupus and diarrhea,” 173 patients (269 documents) “Crohn’s and diabetes,” and 32 patients (95 documents) “NPHP1.” This corresponds to a total of 626 heterogeneous documents distributed on several clinical services (Supplementary Appendix 3) and several types of records (Supplementary Appendix 4). For the 3 use cases, negation detection and family history filters provided, separately, an increased precision and F-measure. The combination of both filters further improved the precision score and F-measure. The precision increased from 0.37 to 0.91 for “Lupus and diarrhea,” from 0.13 to 0.73 for “Crohn’s and diabetes,” and from 0.66 to 0.84 for “NPHP1.” For “lupus and diarrhea” and “Crohn’s and diabetes,” the recall decreased to, respectively, 0.98 and 0.96 with the filters. For “NPHP1,” the recall remained equal to 1.
Table 2.
Precision, recall, specificity and F-measure for each use case and for pooled data
| Lupus and diarrhea | TP | FP | TN | FN | Precision | Recall | Specificity | F-measure |
|---|---|---|---|---|---|---|---|---|
| No filtering | 53 | 92 | 0 | 0 | 0.37 | 1.00 | 0.00 | 0.54 |
| Family history | 53 | 41 | 51 | 0 | 0.56 | 1.00 | 0.55 | 0.72 |
| Negation | 52 | 39 | 53 | 1 | 0.57 | 0.98 | 0.58 | 0.72 |
| Family history and negation | 52 | 5 | 87 | 1 | 0.91 | 0.98 | 0.95 | 0.95 |
| Crohn’s and diabetes | ||||||||
| No filtering | 23 | 150 | 0 | 0 | 0.13 | 1.00 | 0.00 | 0.23 |
| Family history | 22 | 25 | 125 | 1 | 0.47 | 0.96 | 0.83 | 0.63 |
| Negation | 23 | 107 | 43 | 0 | 0.18 | 1.00 | 0.29 | 0.30 |
| Family history and negation | 22 | 8 | 142 | 1 | 0.73 | 0.96 | 0.95 | 0.83 |
| NPHP1 | ||||||||
| No filtering | 21 | 11 | 0 | 0 | 0.66 | 1.00 | 0.00 | 0.79 |
| Family history | 21 | 11 | 0 | 0 | 0.66 | 1.00 | 0.00 | 0.79 |
| Negation | 21 | 4 | 7 | 0 | 0.84 | 1.00 | 0.64 | 0.91 |
| Family history and negation | 21 | 4 | 7 | 0 | 0.84 | 1.00 | 0.64 | 0.91 |
| Total | ||||||||
| No filter | 97 | 253 | 0 | 0 | 0.28 | 1.00 | 0.00 | 0.43 |
| Family history | 96 | 77 | 176 | 1 | 0.55 | 0.99 | 0.70 | 0.71 |
| Negation | 96 | 150 | 103 | 1 | 0.39 | 0.99 | 0.41 | 0.56 |
| Family history and negation | 95 | 17 | 236 | 2 | 0.85 | 0.98 | 0.93 | 0.91 |
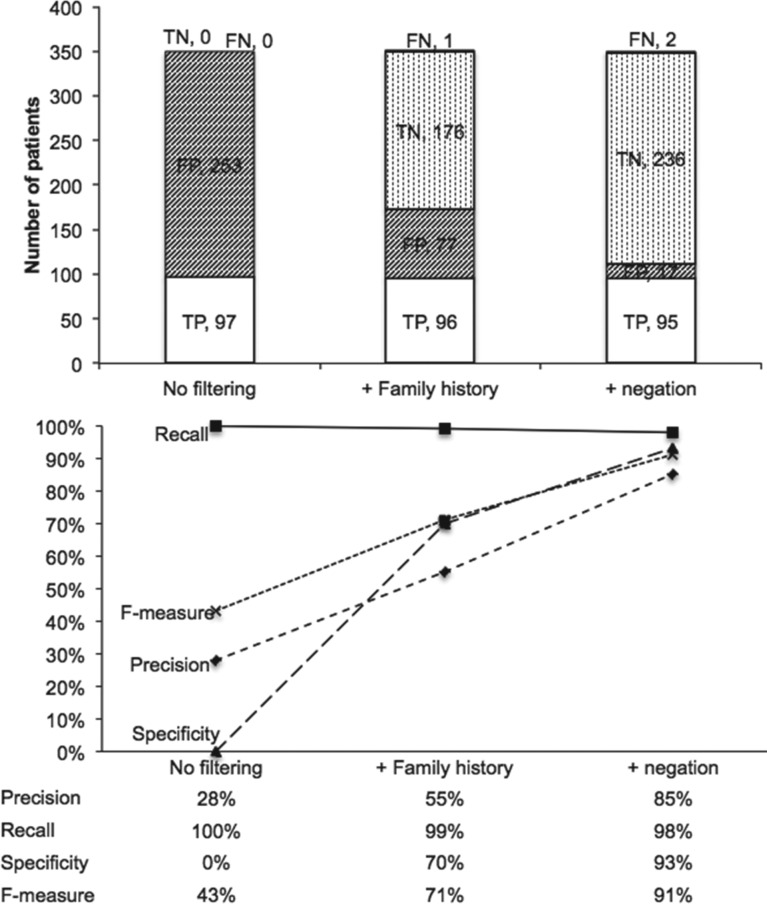
The overall precision, recall, specificity, and F-measure for the pooled data were 0.85, 0.98, 0.93, and 0.91, respectively (Table 2). In addition, only 2 patients were erroneously excluded out of the 95 TPs by the algorithm.
While negative propositions were more often present in emergency reports, with 22% of the propositions classified as negative, family history sentences were more present in consultation reports (Table 3).
Table 3.
For each report type, number of propositions (prop) with negative regex over the total number of propositions, and the number of sentences with family regex over the total number of sentences
| Report type | Nb of prop with negative regex over the total nb of prop (%) | Nb of sentences with family regex over the total nb of sentences (%) |
|---|---|---|
| Consultation reports | 2659/17296 (15) | 683/9346 (7) |
| Day hospitalization reports | 1413/11253 (13) | 280/7448 (4) |
| Emergency | 32/144 (22) | 2/90 (2) |
| Hospitalization reports | 9262/69861 (13) | 1074/45773 (2) |
| Letter | 1618/13200 (12) | 196/8077 (2) |
| Operative reports | 26/267 (10) | 4/182 (2) |
The regex negative expressions most useful in excluding FP patients were rules 1, 2, and 9, with, respectively, 64, 16, and 24 FP patients detected. The only FN patient induced was due to the rule “absence of” (Supplementary Appendix 5).
The processing (combining negation and family history) took an average of 18 ms per document. The queries used to build the corpora took, respectively, 3 s for “lupus and diarrhea,” 1 s for “Crohn’s and diabetes,” and 1 s for “NPHP1” on a server with 32 Go RAM and 8 cores 2.4 Ghz.
Discussion
The aim of the present study was to develop an automated tool to filter negated expressions and family context from clinical narrative reports. The results indicate that use of the automated approach is feasible and dramatically decreases the rate of FPs: 71% before filtering vs 15% using our algorithm. Our method achieved very good overall precision (0.85), recall (0.98), specificity (0.93), and F-measure (0.91) and also for each use case (Figure 3). One of the strengths of this study is that these results were obtained on a comprehensive corpus that contains both inpatient and outpatient reports.
Figure 3.
Both the number of patients retrieved and the accuracy (represented by precision, recall, specificity, and F-measure) are presented without filtering, with family history detection and with both detection (family history and negation).
Limitations and perspectives
Three major causes explain the remaining FPs (17 patients):
Seven were due to an incorrect split of a sentence in nominal groups. This error is due to undeleted erroneous newline characters.
Five were misclassified because a diagnosis was disscussed but was ultimately deferred until a later time.
Our system does not resolve issues of coreference (5 cases), which is the task of finding all expressions that refer to the same entity in a text, i.e., “The father is Caucasian. He has Crohn’s disease.” This type of limitation is also found in the other systems mentioned in the Related work section.24 There is ongoing research on computational methods for coreference resolution that may be implemented in our system in the future.27,28
Regarding the evaluation of certainty, we decided to use only 2 modalities: non-negative and negative. A third level should be added to our scale to consider suspicion.
Comparison to other works
Harkema et al.26 applied ConText to 6 types of clinical reports, e.g., Surgical Pathology, in which the conditions experienced by someone other than the patient were very rarely found. The number of occurrences of “other experiencer” occurred only 5 times in all the reports in their evaluation set; i.e., it was strictly 0% for radiology reports, surgical pathology, and operative procedures. Conversely, 24% of our report set corresponding to rare disease patients contained mention of some condition experienced by their family members.
Tanushi highlighted the lack of regular expressions representing double negation or normality in Negex.29 Similarly, we did not find any of them in the csv file displayed by Chapman.19 Noticeably, we identified 497 occurrences of double negatives with “we cannot exclude” and 7831 occurrences of “without any doubt” in the CDW. We proposed a list of expressions to take into account potential misclassification due to double negation and normality.
Deleger developed an algorithm to detect negation of medical problems in cardiology notes in French.20 Their algorithm focused on negation detection and their method was based on concept classification (F-measure 0.87). Our algorithm combines detection of family history and negation, and was applied to a large CDW of 1.6 million EHRs. The algorithm is part of a full-text search engine with the objective of classifying patients as cases or not (F-measure 0.91).
Conclusion
We developed an integrated pipeline to enable negation and family history context detection in the full-text search engine of a document-oriented CDW. The automated method achieved an overall F-measure of 0.91. The method is generalizable, and can be adapted to English and other languages.
The tool is available at https://github.com/imagine-bdd/DrWH-negation
Funding
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Competing Interests
The authors have no competing interests to declare.
Contributors
Substantial contributions to the conception or design of the work, or the acquisition, analysis, or interpretation of data for the work: Conception of the work: NG, AB. Acquisition of data: NG, VB. Interpretation of data: NG, AN, RS, AB. Evaluation: RS, AB. Drafting the work or revising it critically for important intellectual content: Drafting the work: NG, AN, AB. Revising the work critically for important intellectual content: NG, AN, VB, RS, AB. Final approval of the version to be published: NG, AN, VB, RS, AB. Agreement to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved: NG, AN, VB, RS, AB.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
Supplementary Material
REFERENCES
- 1. Murphy SN, Mendis ME, Berkowitz DA et al. Integration of clinical and genetic data in the i2b2 architecture. AMIA Annu Symp Proc. 2006;2006:1040. [PMC free article] [PubMed] [Google Scholar]
- 2. Hebbring SJ, Rastegar-Mojarad M, Ye Z et al. Application of clinical text data for phenome-wide association studies (PheWASs). Bioinformatics. 2015;31(12):1981–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cuggia M, Garcelon N, Campillo-Gimenez B et al. Roogle: an information retrieval engine for clinical data warehouse. Stud Health Technol Inform. 2011;169:584–8. [PubMed] [Google Scholar]
- 4. Huan Mo, William K Thompson, Luke V Rasmussen et al. Desiderata for computable representations of electronic health records-driven phenotype algorithms. J Am Med Inform Assoc. 2015;22(6):1220–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Denny JC. Chapter 13: Mining electronic health records in the genomics era. PLoS Comput Biol 2012;8(12):e1002823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cuggia M, Bayat S, Garcelon N et al. A full-text information retrieval system for an epidemiological registry. Stud Health Technol Inform. 2010;160(Pt 1):491–5. [PubMed] [Google Scholar]
- 7. Escudié JB, Rance B. Reviewing 741 patients records in two hours with FASTVISU. Proc AMIA Symp. 2015; 2015:553–9. [PMC free article] [PubMed] [Google Scholar]
- 8. Chapman WW, Bridewell W, Hanbury P et al. Evaluation of negation phrases in narrative clinical reports. Proc AMIA Symp. 2001;2001:105–9. [PMC free article] [PubMed] [Google Scholar]
- 9. Chu D, Dowling JN, Chapman WW. Evaluating the effectiveness of four contextual features in classifying annotated clinical conditions in emergency department reports. AMIA Annu Symp Proc. 2006;2006:141–5. [PMC free article] [PubMed] [Google Scholar]
- 10. Sharp RC, Abdulrahim M, Naser ES, Naser SA. Genetic Variations of PTPN2 and PTPN22: role in the pathogenesis of type 1 diabetes and Crohn’s disease. Front Cell Infect Microbiol. 2015;5:95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sran S, Sran M, Patel N, Anand P. Lupus enteritis as an initial presentation of systemic lupus erythematosus. Case Rep Gastrointest Med. 2014;2014:962735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sultan SM, Ioannou Y, Isenberg DA. A review of gastrointestinal manifestations of systemic lupus erythematosus. Rheumatology (Oxford). 1999; 38(10):917–32. [DOI] [PubMed] [Google Scholar]
- 13. Somers EC, Marder W, Cagnoli P et al. Population-based incidence and prevalence of systemic lupus erythematosus: the Michigan Lupus Epidemiology and Surveillance program. Arthritis Rheumatol. 2014; 66(2):369–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Savova GK, Masanz JJ, Ogren PV et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc. 2010;17(5):507–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. South BR, Phansalkar S, Swaminathan AD et al. Adaptation of the NegEx algorithm to Veterans Affairs electronic text notes for detection of influenza-like illness (ILI). AMIA Annu Symp Proc. 2007;2007:1118. [PubMed] [Google Scholar]
- 16. Skeppstedt M. Negation detection in Swedish clinical text: an adaption of NegEx to Swedish. J Biomed Semantics. 2011;2 (Suppl 3):S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. King B, Wang L, Provalov I et al. Cengage learning at TREC 2011 Medical Track. In National Institute of Standards and Technology Proc TREC. 2011. Maryland. [Google Scholar]
- 18. Chapman W, Bridewell W, Hanbury P et al. A simple algorithm for identifying negated findings and diseases in discharge summaries. J Biomed Inform. 2001;34(5):301–10. [DOI] [PubMed] [Google Scholar]
- 19. Chapman WW, Hillert D, Velupillai S et al. Extending the NegEx lexicon for multiple languages. Stud Health Technol Inform. 2013;192:677–81. [PMC free article] [PubMed] [Google Scholar]
- 20. Deléger L, Grouin C. Detecting negation of medical problems in French clinical notes. InProc the 2nd ACM SIGHIT International Health Informatics Symposium (IHI ‘12). New York, NY, USA: ACM; 2012: 697–702. [Google Scholar]
- 21. Goryachev S, Sordo M, Zeng QT et al. Implementation and evaluation of four different methods of negation detection. Boston, MA, DSG. 2006. Boston, Maryland; Decision Systems Group Technical Report. [Google Scholar]
- 22. Aronow DB, Fangfang F, Croft WB. Ad hoc classification of radiology reports. J Am Med Inform Assoc. 1999; 6(5):393–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Friedlin J, McDonald CJ. Using a natural language processing system to extract and code family history data from admission reports. AMIA Annu Symp Proc. 2006;2006:925. [PMC free article] [PubMed] [Google Scholar]
- 24. Goryachev S, Kim H, Zeng-Treitler Q. Identification and extraction of family history information from clinical reports. AMIA Annu Symp Proc. 2008; 2008:247–51. [PMC free article] [PubMed] [Google Scholar]
- 25. Lewis N, Gruhl D, Yang H. Extracting family history diagnosis from clinical texts. Int Conf Bioinform Comput Biol. New Orleans, Louisiana. 2011: 128–133. [Google Scholar]
- 26. Chapman W, Chu D, Dowling JN. ConText: an algorithm for identifying contextual features from clinical text . InProc Workshop on BioNLP 2007: Biological, Translational, and Clinical Language Processing (BioNLP ‘07). USA, PA: Association for Computational Linguistics, Stroudsburg; 2007: 81–88. [Google Scholar]
- 27. Kim Y, Riloff E, Gilbert N. The taming of reconcile as a biomedical conference resolver. ACL. Workshop BioNLP- Shared task;Proceedings of BioNLP Shared Task 2011 Workshop, Portland, Oregon, USA, Association for Computational Linguistics June 2011;2011:89–93. [Google Scholar]
- 28. Zheng J, Chapman W, Miller T et al. A system for coreference resolution for the clinical narrative. Am Med Inform Assoc. 2012;19(4):660–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tanushi H, Dalianis H, Duneld M et al. Negation scope delimitation in clinical text using three approaches: NegEx, PyConTextNLP and SynNeg. In: Oepen S, Hagen K, Bondi Johannessen J, eds. Proc 19th NODALIDA, NEALT Linköping, Sweden: Linköping University Electronic Press; 2013:387–97. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.