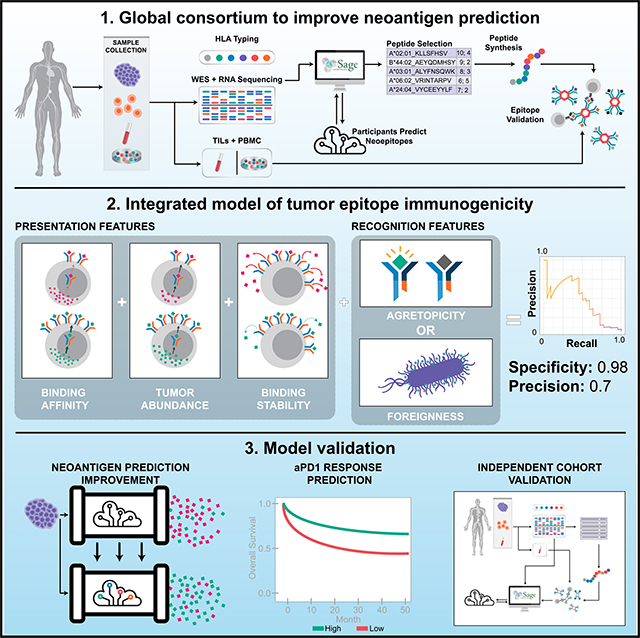
SUMMARY
Many approaches to identify therapeutically relevant neoantigens couple tumor sequencing with bioinformatic algorithms and inferred rules of tumor epitope immunogenicity. However, there are no reference data to compare these approaches, and the parameters governing tumor epitope immunogenicity remain unclear. Here, we assembled a global consortium wherein each participant predicted immunogenic epitopes from shared tumor sequencing data. 608 epitopes were subsequently assessed for T cell binding in patient-matched samples. By integrating peptide features associated with presentation and recognition, we developed a model of tumor epitope immunogenicity that filtered out 98% of non-immunogenic peptides with a precision above 0.70. Pipelines prioritizing model features had superior performance, and pipeline alterations leveraging them improved prediction performance. These findings were validated in an independent cohort of 310 epitopes prioritized from tumor sequencing data and assessed for T cell binding. This data resource enables identification of parameters underlying effective anti-tumor immunity and is available to the research community (https://www.synapse.org/#!Synapse:syn21048999).
In Brief
Genomic tumor sequencing data with matched measurements of tumor epitope immunogenicity allows for insights into the governing parameters of epitope immunogenicity and generation of models for effective neoantigen prediction.
Graphical Abstract
INTRODUCTION
Somatic alterations are a hallmark of cancer (Hanahan and Weinberg, 2011). These alterations can result in the generation of mutated peptide fragments that, when presented on class I major histocompatibility complex (MHC I) molecules, elicit a protective anti-tumor immune response. Such mutant peptides, called “neoantigens,” are hypothesized to comprise an important class of tumor antigens that drive anti-tumor immunity (Schumacher and Schreiber, 2015; Tran et al., 2017; Yarchoan et al., 2017).
Neoantigens have long been viewed as promising therapeutic targets because they are tumor-specific and are not subject to either pre-existing immune tolerance or likely to generate autoimmunity. Neoantigen vaccines have been shown to induce tumor rejection of mice bearing transplanted sarcomas (Gubin et al., 2014). In humans, neoantigen vaccine studies have shown the ability to generate neoantigen-specific T cells in melanoma and glioblastoma (Carreno et al., 2015; Keskin et al., 2019) and are supportive of an ability of these therapies to be protective against tumor recurrence in melanoma (Ott et al., 2017) and to induce vaccine-related tumor regression in melanoma (Sahin et al., 2017). Adoptive transfer of autologous T cell products containing high fractions of neoantigen-specific T cells has generated tumor regression in a range of cancer types (Tran et al., 2014, 2016; Yee et al., 2002; Zacharakis et al., 2018) and has further shown the ability to mediate durable complete regression in a substantial fraction of patients with metastatic melanoma (Goff et al., 2016; Rosenberg et al., 2011). Moreover, in these neoantigen-targeting therapies, documented cases of off-target immune response against the wild-type non-mutated peptide are exceptionally rare (Schumacher et al., 2019; Strønen et al., 2016; Tran et al., 2017) corroborating studies in animal models showing no evidence of cross-reactivity between mutant peptides and wild-type peptides (Alspach et al., 2019; Gubin et al., 2014, 2015; Kreiter et al., 2015; Matsushita et al., 2012). Neoantigen-targeting therapies are thus viewed as a safe and effective approach to generate anti-tumor immune responses (Sahin and Türeci, 2018; Yamamoto et al., 2019).
The ability to accurately and reproducibly identify neoantigens capable of eliciting a tumor-specific immune response from available sample material is paramount for the success of these therapeutic approaches and is still early in development (Garcia-Garijo et al., 2019; Hacohen et al., 2013; Nature Biotechnology, 2017; Vitiello and Zanetti, 2017; Yadav et al., 2014). Epitope immunogenicity is dependent on a complex chain of events, including variant expression, peptide processing, transport and presentation, and ultimately generation of a T cell response. Predicting which somatic alterations will generate immunogenic peptides relies on in silico algorithms that leverage advances in next generation sequencing (NGS), associated bioinformatic tools (Finotello et al., 2019; Fritsch et al., 2014; Richters et al., 2019; Schumacher and Hacohen, 2016), and inferred rules governing epitope immunogenicity. These rules typically incorporate the predicted affinity of that epitope to the specific HLA alleles associated with the subject (Hoof et al., 2009; O’Donnell et al., 2018) as well as filters and ranking criteria derived from a priori knowledge. To date, proposed rules have been derived from a heterogeneous set of studies using a range of validation techniques and models (Cohen et al., 2015; Hundal et al., 2016; Łuksza et al., 2017; Rajasagi et al., 2014; Richman et al., 2019) and may be further confounded by inherent biases present in the single pipeline used to generate candidate epitopes. Although crowdsourced biomedical research efforts (Saez-Rodriguez et al., 2016) such as the Dialogue on Reverse Engineering Assessment and Methods (DREAM) and the Critical Assessment of Genome Interpretation (CAGI), have made substantial contributions to improving methods for tumor subclonal reconstruction (Salcedo et al., 2020), predicting phenotype from exome sequencing (Daneshjou et al., 2017), multi-target drug identification (Schlessinger et al., 2017), identifying the effect of CDKN2A variants (Carraro et al., 2017), and others (Guinney et al., 2017; Hoskins et al., 2017; Keller et al., 2017; Kreimer et al., 2017), no such effort for neoepitope prediction has been undertaken. To date, there does not yet exist a comprehensive, unbiased resource with which the key parameters governing tumor epitope immunogenicity can be systematically compared. Such a resource would further enable comparison between different pipelines to improve neoantigen prediction.
Here, we report on the results obtained by the Tumor Neoantigen Selection Alliance (TESLA), a global community-based initiative seeking to understand tumor epitope immunogenicity, improve neoantigen prediction, and ultimately provide a broadly accessible reference dataset for effective benchmarking. We identify key parameters governing tumor epitope immunogenicity by analyzing predictions made by multiple independent pipelines on a common set of tumor samples and using a centralized set of validation experiments. Data from TESLA are available for download and serve as an open benchmark to accelerate the development of neoantigen-based therapies (https://www.synapse.org/#!Synapse:syn21048999).
RESULTS
Consortium Structure
TESLA brings together teams of researchers from academia, industry, and non-profit groups to compare their approaches to neoantigen prediction through three main principles: (1) all teams are provided the same genomic information derived from subject samples: tumor/normal whole exome sequencing (WES), tumor RNA-sequencing, and clinical-grade HLA typing (STAR Methods); (2) teams use these data to generate neoepitope predictions and return a ranked list of neoepitopes predicted to bind to the relevant MHC class I molecules (pMHC) and elicit an immune response; and (3) a subset of highly ranked predicted pMHC from each team are tested in vitro to determine MHC binding and peptide immunogenicity, the latter determined via the detection of pMHC-restricted T cells in subject-matched peripheral blood mononuclear cells (PBMCs) and/or tumor infiltrating T lymphocytes (TILs) (Figure 1A). A more complete description of TESLA, including data collected, can be found in the STAR Methods and Table S1, and QC metrics on the WES and RNA samples can be found in Table S2.
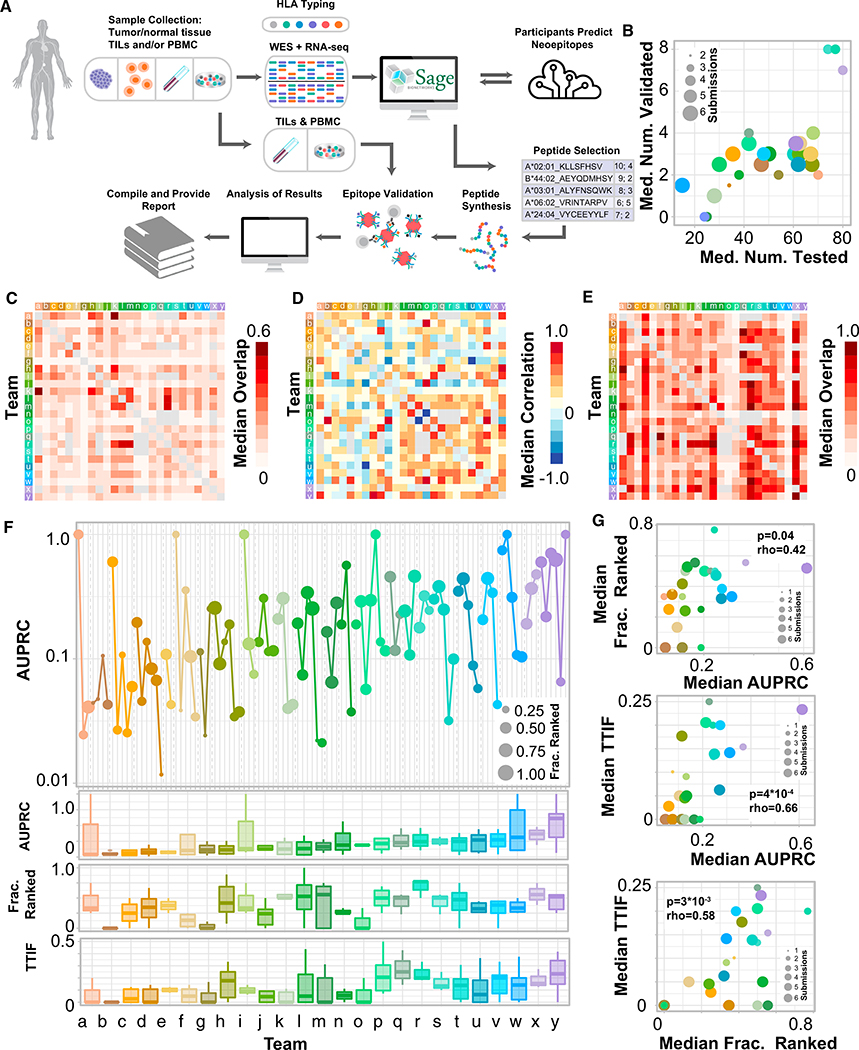
Figure 1. Overview of TESLA Team Performance.
(A) Schematic of TESLA.
(B) Scatterplot of median number of peptides tested for immunogenicity (x axis) versus median number of peptides with validated immunogenicity (y axis). Dot size is the number of patients a given team submitted for.
(C) Heatmap of the median overlap (described in STAR Methods) in the top 100 predicted pMHC for each pair of teams.
(D) Heatmap of the Spearman correlation of rank between overlapping pMHC for each pair of teams.
(E) Heatmap of the median overlap between the top 100 predicted pMHC from one team (y axis) versus the entire submitted set of pMHC (ranked and unranked) of another (x axis).
(F) AUPRC for each submission for each team. Dot size represents the fraction ranked for that particular submission. Bottom: boxplot of AUPRC, FR, and TTIF for each team, aggregated over all submissions.
(G) Scatterplots of median AUPRC versus median FR (top), median TTIF versus median AUPRC (center), and median TTIF versus median FR (bottom) for each team. Dot size represents the number of evaluable submission from that team included in the calculation. Rho, spearman rho; AUPRC, area under the precision recall curve; FR, fraction ranked; TTIF, top twenty immunogenic fractions.
TESLA Participation and Immunogenicity Validation Results
Samples from six subjects were analyzed: 3 from subjects with metastatic melanoma and 3 from subjects with non-small cell lung cancer (NSCLC) (Table S3). 28 unique teams submitted ranked neoantigen predictions on these samples; predictions from 3 teams were excluded from further analyses due to an insufficient number of subjects included in their prediction (at least 2), and subsequent analyses were performed on the predictions from 25 teams. Teams submissions ranged from 7–81,904 ranked pMHC per tumor sample (median: 204). From these submitted predictions, 608 peptides selected from among the top-ranked peptides from all groups (median, 97/subject; range, 73–144, see Table S4 for complete list of tested peptides) were tested for immunogenicity by pMHC multimer-based assays and 37 (6%) of those were found to be immunogenic, a validation rate similar to what has previously been reported (Yadav et al., 2014). Each TESLA team had a median of 51 of their submitted peptides tested of which on median 3 (6%) were immunogenic (Figure 1B).
Prediction Overlap
To understand the extent to which the predictions differed between teams, we calculated the overlap of the top 100 ranked pMHC for each pair of TESLA teams (see STAR Methods). The median overlap is shown in Figure 1C. The overlap between teams was low (less than 20%) in the majority of the cases. The median overlap between teams was 13% with a maximum overlap of 62%. Next, we looked at the Spearman correlation of pMHC rank between teams which was also, on median, low (rho less than 0.3) in the majority of cases (median correlation: 0.18) (shown in Figure 1D). Finally, we looked at the overlap between the top-100 ranked pMHC for one team with the entire list (ranked and unranked) of another (Figure 1E). Median overlap in this setting was substantially larger (32%), suggesting that the lack of consensus in prediction by TESLA participants is driven in part by differences in epitope filtering and/or ranking. Neoantigen prediction pipelines in TESLA also showed substantial diversity in the features they used, suggesting one possible origin of the observed diversity in rankings (Figure S1).
Identification and Ranking of Immunogenic Peptides
To assess the performance of the different pipelines, we calculated three metrics (described in STAR Methods): (1) “AUPRC,” the area under the precision recall curve, which measures the ability of a team to rank immunogenic pMHC ahead of the peptides for which T cell responses were not identified; (2) “fraction ranked” (FR), which measures the fraction of immunogenic pMHC for a given subject that a team included in their top-100 pMHC, capturing the ability of a team to identify immunogenic peptides; and (3) “top-20 immunogenic fraction” (TTIF), which is a surrogate metric to approximate the fraction of peptides that would be immunogenic in a therapeutic application (metrics shown in (Figure 1F; all performance metrics for each submission given in Table S5). AUPRC and FR showed a weak yet significant correlation (Figure 1G, top; rho = 0.42, p = 0.04, n = 25), whereas TTIF was more strongly correlated with both metrics (Figure 1G, center and bottom; rho = 0.66, p = 4*10−4 and rho = 0.58, p = 3*10−3, respectively), suggesting that the ability to rank and identify immunogenic peptides are related but distinct characteristics of a neoepitope prediction pipeline, and both are strongly associated with the ability to effectively produce an efficacious set of peptides for a therapeutic. One allele had a significantly higher validation rate than any other (A*03:01) but the values of AUPRC, FR, and TTIF were found to be overall robust to the presence of this allele (Figure S2). Taken together, these results suggest that there are substantial differences among TESLA participants in their ability to identify and rank immunogenic pMHC and highlight the diversity of predictions used to generate epitope candidates in TESLA.
Pipeline Combination Improves Prediction Performance
Every team identified and ranked at least one of the confirmed immunogenic epitopes in their top 100 (across all patients), however, substantial differences were observed in the number and ranking of these epitopes (Figure 2A). In particular, no team included more than 20 of the 37 immunogenic peptides in their top 100, and several of the identified immunogenic peptides were predicted by only a small subset of teams. Thus, substantial prediction diversity was observed, even for highly performing teams.
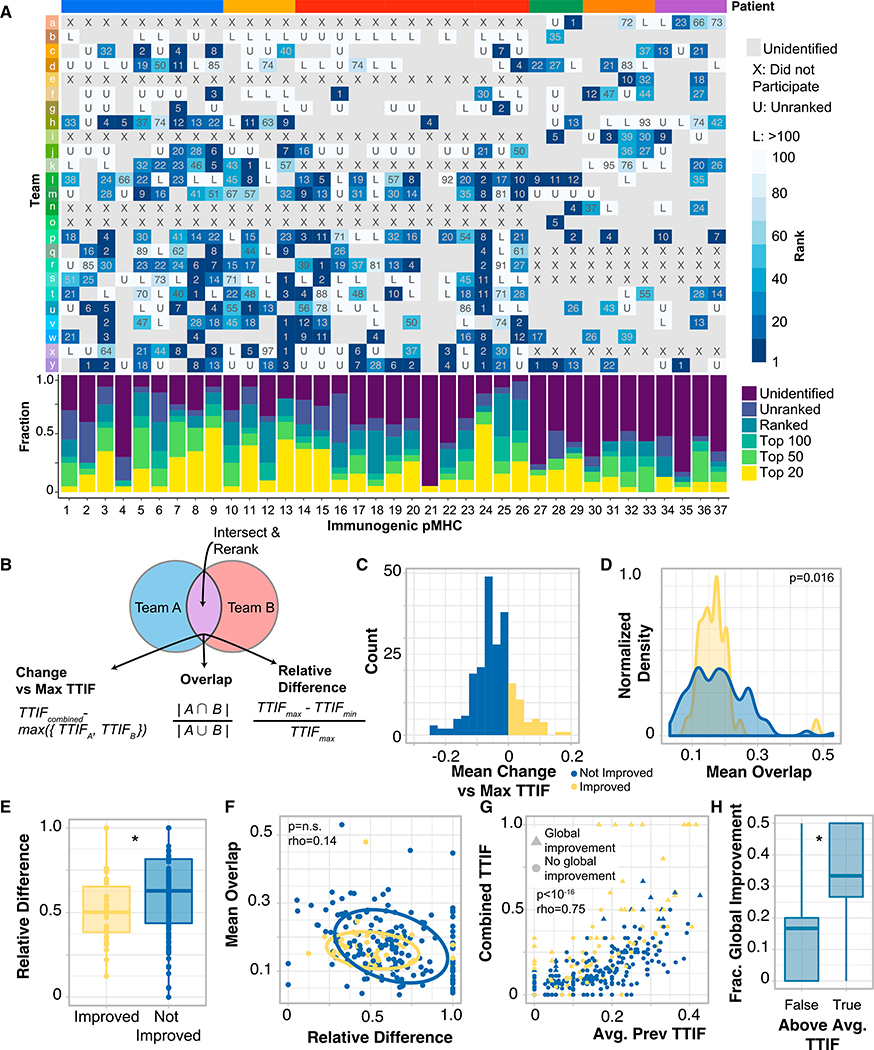
Figure 2. Rational Combination of Neoantigen Predictions Improve Prediction Performance.
(A) Overview of each pMHC across all of TESLA that validated in a multimer-based assay. For each pMHC, the rank of that pMHC for a given team is shown. Places where a team did not identify a particular pMHC in any of their submissions are shown in gray. Bottom: for each peptide, the fraction of time that peptide was ranked in different ranking groups (left).
(B) Schematic of prediction combination method along with metric calculation.
(C) Histogram of average change in combined TTIF. Yellow, combination improves prediction on average; blue, combination is detrimental to prediction on average.
(D) Density plot of average overlap, stratified by improvement status. Distributional difference assessed by Kolmogorov-Smirnov test.
(E) Boxplot of relative difference stratified by improvement status. *p < 0.05.
(F) Scatterplot of average overlap by relative difference. Ellipses represent best fit at one standard deviation. Rho, Spearman rho.
(G) Combined TTIF by the average of the two initial TTIF values, by team pair and patient. Color represented improvement status (as in C), and shape represents if a particular value was a global improvement (larger than all previous TTIF values for that patient, triangle) or not (circle). (F) Fraction of predictions that are global improvements stratified by whether average team-pair TTIF is above or below the median. *p < 0.05.
Another area where substantial prediction diversity is observed is in somatic variant calling. In that field, researchers have developed strategies to rationally combine predictions from different callers to improve specificity while maintaining sensitivity (Callari et al., 2017; Łuksza et al., 2017). We hypothesized that a similar approach could be beneficial for neoantigen prediction and sought to identify the criteria under which the combination of a pair of pipelines would be most likely to improve neoantigen prediction. For each pair of pipelines that provided predictions on a given sample, we combined those predictions by intersecting them and re-ranking them by the average rank of the remaining pMHC (Figure 2B). For a given pair of pipelines, the change in predictive ability was captured as difference from max TTIF of the two inputted predictions on the same subject. Finally, for each pipeline pair, we defined two metrics: overlap and relative difference. Briefly, overlap captures the extent to which pipeline predictions are similar, whereas relative difference captures the difference in TTIF between each pipeline pair, for a given subject (depicted in Figure 2B; STAR Methods). 19% of pipeline combinations demonstrated improved performance (Figure 2C). Pipeline pairs with improved performance had a significantly more constrained distribution of overlap values (Figure 2D, p = 0.016, Kolmogorov-Smirnov test) and a significantly smaller relative difference (Figure 2E, p = 0.015, Mann-Whitney U), and these two metrics were independent (Figure 2F). Larger average TTIF values for pairs of pipelines were positively correlated with the combined TTIF values (Figure 2G, rho = 0.75, p < 10−16, Spearman correlation), and pairs of pipelines with improved performance were significantly more likely to produce combined TTIF values that were larger than all previously observed TTIF values from single pipelines for the same patient (OR = 6.06, p = 4*10−9, termed “global improvement”). Finally, improved team pairs with above-median TTIF over all patients were more likely to produce prediction results that were global improvements (Figure 2F, p = 0.017, Mann-Whitney U). Together, these results suggest an approach to improving neoantigen prediction that does not rely on developing a pipeline with best-in-class performance: identify two pipelines with consistently strong performances that produce distinct predictions and combine those predictions using an intersect and rank approach.
Presentation Features of Immunogenic Peptides
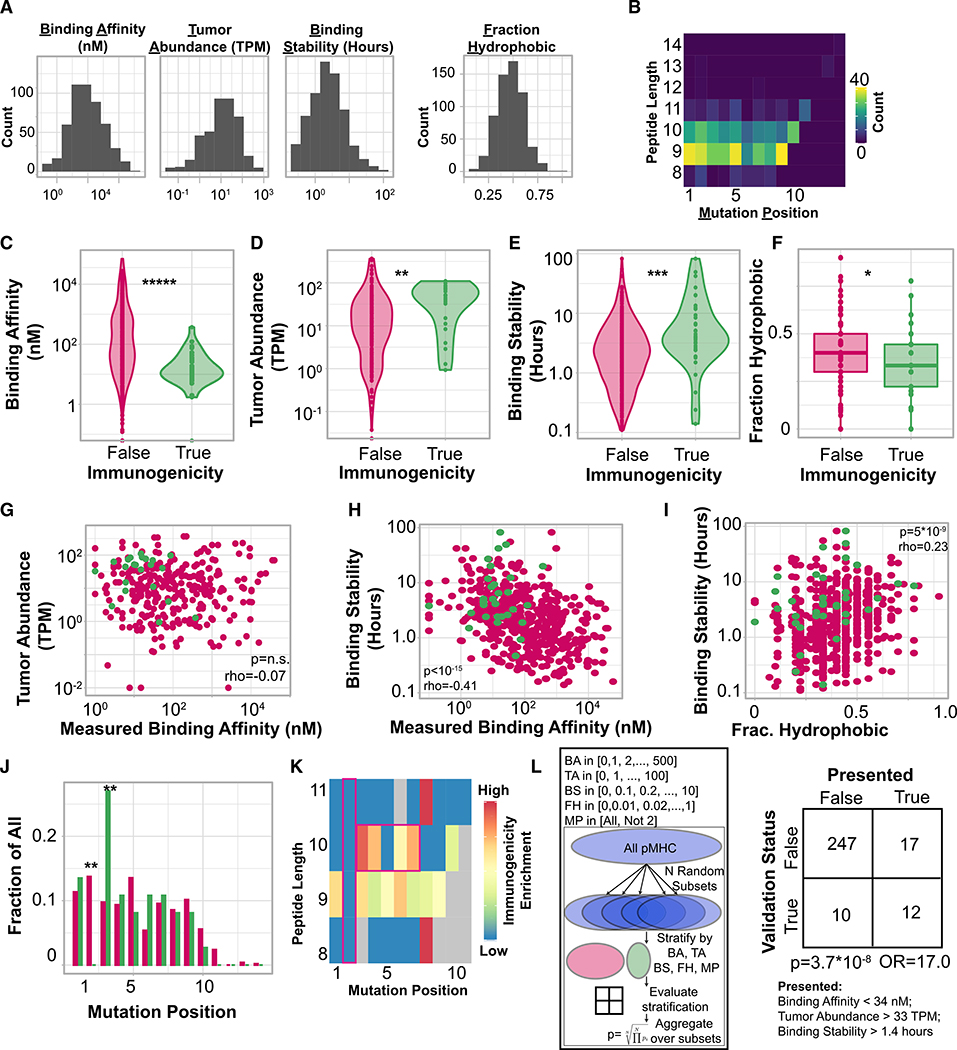
We sought to characterize the features of peptides that were associated with the presence of detectable antigen-specific T cells compared to those that were not. The current model of epitope immunogenicity proposes that for a peptide to elicit an anti-tumor T cell response, it must first be presented by an MHC I allele, and subsequently be recognized as foreign by a T cell (Schreiber et al., 2011). As such, we first considered “presentation features” hypothesized to be associated with effective antigen presentation: MHC binding affinity, expression of the originating gene (“tumor abundance”), expected duration of peptide-MHC interaction (“binding stability”), peptide hydrophobicity, and mutation position (Chowell et al., 2015; Duan et al., 2014; Hundal et al., 2016; Łuksza et al., 2017; Rasmussen et al., 2016). Peptide features are included in Table S4, and their calculation is described in STAR Methods. Overall, a wide range of values for each peptide presentation feature (Figure 3A) and mutation position (Figure 3B) was observed. We next assessed if these characteristics differed between immunogenic and non-immunogenic pMHC. Immunogenic pMHC had significantly stronger measured binding affinity (Figure 3C; p = 4*10−6, Mann-Whitney U), significantly higher tumor abundance (Figure 3D; p = 0.01, Mann-Whitney U), significantly higher binding stability (Figure 3E, p = 1.4*10−4, Mann-Whitney U), and were significantly less hydrophobic (Figure 3F, p = 0.04, Mann-Whitney U) than non-immunogenic tested pMHC. As seen previously (Abelin et al., 2017), pMHC binding affinity was not correlated with tumor abundance, and high tumor abundance did not compensate for weak binding affinity in immunogenic pMHC (Figure 3G, Spearman rho). However, pMHC binding affinity was significantly inversely correlated with binding stability (Figure 3H, p < 10−15, Spearman rho), and binding stability was positively correlated with the peptide hydrophobicity (Figure 3I, p = 5*10−9, Spearman rho), although this association was less pronounced in immunogenic peptides. Finally, in this dataset, immunogenic peptides were never derived from mutations that changed the second amino acid in a peptide (p = 0.006; Fisher exact test), although this was the most commonly predicted mutation location, while they were much more likely to be derived from mutations to the third amino acid (p = 0.003; Fisher exact test; Figure 3J). We note that the second amino acid is the anchor residue for peptides of multiple lengths (Chowell et al., 2015). Furthermore, immunogenic peptides of 10 amino-acid length were found to be enriched in mutations in the presented residues of the peptides (position 3 to 7), while no position-dependent enrichment was found in 9-mers peptides (Figure 3K). Overall, the majority of tested peptides were 9- and 10-mers, and no association was found between peptide length and immunogenicity (Figure S3).
Figure 3. Presentation Features Associated with Peptide Immunogenicity.
(A) Histogram of each feature considered.
(B) Heatmap of peptide length compared to mutation position.
(C) Violin plot of binding affinity stratified by peptide immunogenicity. *****p < 10−5, Mann-Whitney U test.
(D) Violin plot of tumor abundance stratified by peptide immunogenicity. **p < 0.01, Mann-Whitney U test.
(E) Violin plot of binding stability stratified by peptide immunogenicity. ***p < 0.001, Mann-Whitney U test.
(F) Boxplot of peptide hydrophobicity fraction stratified by peptide immunogenicity. *p < 0.05, Mann-Whitney U test.
(G) Scatterplot of binding affinity compared to tumor abundance. Correlation: Spearman rho.
(H) Scatterplot of binding affinity compared to binding stability. Correlation: Spearman rho.
(I) Scatterplot of binding stability compared to hydrophobicity fraction. Correlation: Spearman rho.
(J) Barplot of mutation position, normalized to each subset (immunogenic/non-immunogenic) separately. **p < 0.01, Fisher’s exact test.
(K) Length-dependent enrichment of mutational position. Enrichment calculated as odds ratio from Fisher’s exact test. Gray denotes pairs that did not occur in our dataset.
(L) Schematic of cross-validation scheme to select feature and threshold set. BA, binding affinity; TA, tumor abundance; BS, binding stability; FH, fraction hydrophobic; MP, mutation position. Right: contingency table using the optimal stratification parameters (below). p, Fisher’s exact test.
See also Figures S2, S3, S4, and S5 and Table S4.
Although each feature considered above was found to be associated with peptide immunogenicity, interdependence between features makes it challenging to identify an optimal threshold set to differentiate immunogenic versus non-immunogenic peptides. To overcome this, we developed a repeated-random-subsample-based method to identify a set of features and associated threshold values that stratify immunogenic peptides (strategy depicted in Figure 3L, described in STAR Methods). We applied this method to our peptide set to identify optimal thresholds for each presentation feature (i.e., binding affinity, tumor abundance, binding stability, fraction hydrophobic, and mutational position)—shown in Figure 3L. 286 (out of the 608) peptides had measurements of all 5 of these variables, and it is this set we analyzed. Using our approach, we identified a set of thresholds on these variables that filtered out 93% of non-immunogenic peptides while maintaining 55% of immunogenic peptides (Figure 3L, p = 3.7*10−8, Fisher’s exact test). This threshold set is composed of binding affinity less than 34 nM, tumor abundance greater than 33 TPM, and binding stability greater than 1.4 h. Because each of the variables considered here is associated with robust, durable peptide presentation, we term any peptide that passes all filters “presented,” and a total of 29 peptides are included in this group (shown in Figure 3L). Notably, several of the thresholds identified by our model, including thresholds for minimum binding affinity and tumor abundance, are substantially more stringent than proposed in existing literature (Bulik-Sullivan et al., 2018; Rajasagi et al., 2014). Neither peptide hydrophobicity nor mutational position was found to be important for optimal filtering. Of those immunogenic peptides that were filtered out, 50% (5/10) were filtered out due to low tumor abundance, whereas 40% (4/10) were filtered out by more than one filter (Figure S4). These results identify MHC binding affinity, binding stability, and tumor abundance as crucial presentation-associated parameters associated with peptide immunogenicity.
Recognition Features of Immunogenic Peptides
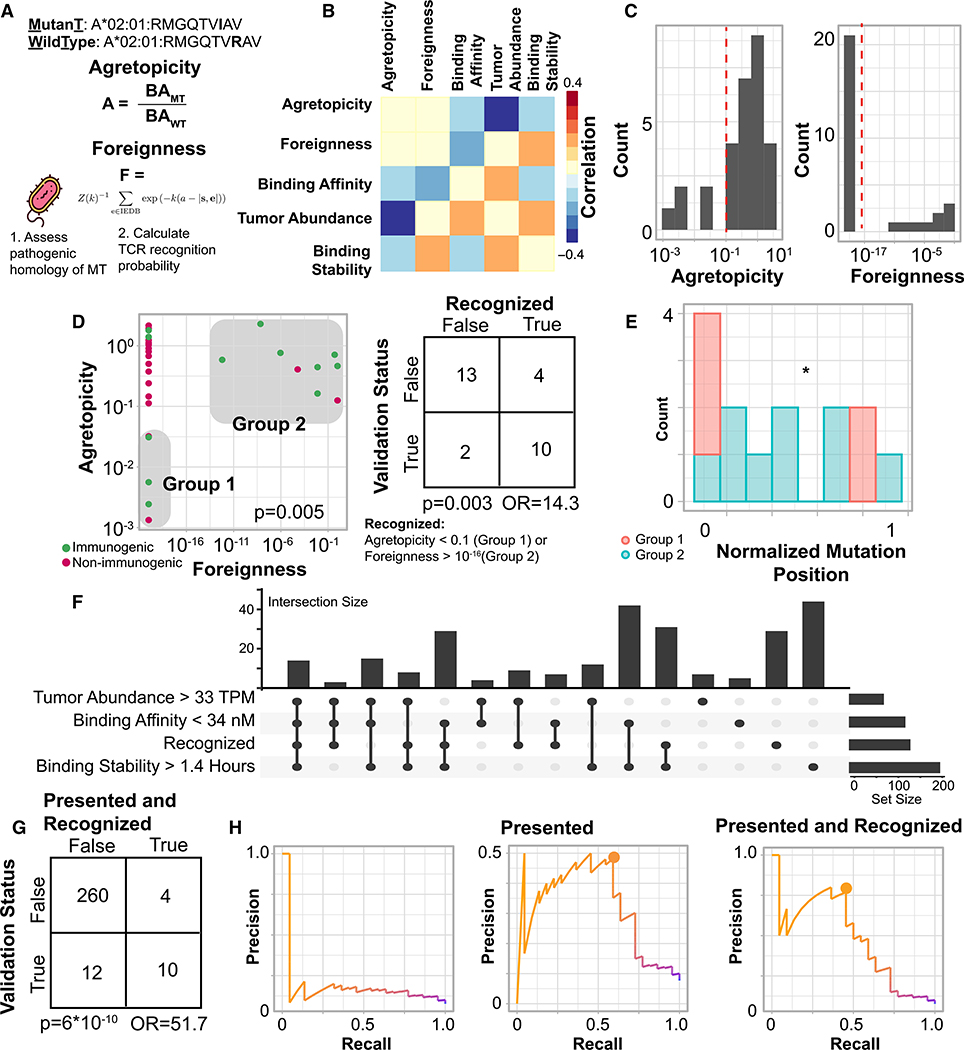
We next considered two peptide features hypothesized to be associated the absence of pre-existing tolerization: “agretopicity”—the ratio of mutant binding affinity to wild-type binding affinity (Duan et al., 2014; Ghorani et al., 2018) and “foreignness”—TCR recognition probability derived from homology to known pathogenic peptides in IEDB (Balachandran et al., 2017; Łuksza et al., 2017; Richman et al., 2019). We reasoned that these two features would be associated with immunogenicity only among peptides that were the likeliest to be presented. Thus, we considered only the 29 pMHC that passed all of the presentation-associated criteria defined in the previous section. Specifically, the pMHC in this set all had MHC binding affinity stronger than 34 nM, tumor abundance greater than 33 TPM, and binding stability greater than 1.4 h. This pMHC set was comprised of 12 immunogenic peptides and 17 non-immunogenic peptides. For these pMHC, agretopicity and foreignness were found to be independent of presentation-associated parameters (Figure 4B). Furthermore, we observed that the majority of these pMHC had agretopicity greater than 0.1 and foreignness less than 10−16 (Figure 4C), while smaller subsets were found to have values that were orders of magnitude smaller (agretopicity, “group 1”) or larger (foreignness, “group 2”). These subsets of low agretopicity or high foreignness pMHC were mutually exclusive (p = 0.005, binomial test). We term the presence of either low agretopicity or high foreignness as “recognition” and term any peptide in either group 1 or group 2 as “recognized.” Importantly, recognition is defined here as purely a property of a peptide, and is not a priori associated with the immunogenicity of that peptide in a particular patient.
Figure 4. Recognition Features Associated with Peptide Immunogenicity.
(A) Illustration of agretopicity and foreignness features.
(B) Correlation between recognition and presentation associated features. Correlation calculated with spearman rho. All correlations not significant.
(C) Histograms of agretopicity (left) and foreignness (right) among presented peptides.
(D) Scatterplot of foreignness compared to agretopicity. Color: immunogenicity. Gray boxes denote low agretopicity or high foreignness peptides. Right: contingency table comparing validation status to recognition status among presented peptides. p, Fisher’s exact test; OR, odds ratio.
(E) Barplot of mutation position by low agretopicity or high foreignness.
(F) Upset plot of all four features associated with immunogenicity. Right: total number of peptides with that feature present.
(G) Contingency table over all peptides comparing validation status to presented and recognized status. p, Fisher’s exact test; OR, odds ratio.
(H) Precision-recall curves of peptides ranked only by MHC binding affinity (left), prioritizing presented peptides (center), and prioritizing presented and recognized peptides (right). Circles represent optimal precision-recall tradeoffs.
Recognized peptides were strongly enriched in immunogenic pMHC (Figure 4D; p = 0.003, Fisher’s exact test; odds ratio, 14.3). The significance of this enrichment was found to be robust to the threshold values of binding affinity, tumor abundance and binding stability used to select the presented set (Figure S5). Among the 29 presented peptides, those in group 1 were enriched in mutations near anchor residues compared to those in group 2. (Figure 4E, p = 0.03, Fisher’s exact test). These results suggest that agretopicity and foreignness are distinct, mutually exclusive peptide features that together enrich for presented peptides more likely to generate a T cell response.
Our results highlight 4 distinct, independent pMHC features—strong binding affinity, high tumor abundance, high binding stability, and peptide recognition—that when all present, greatly enrich for immunogenicity, and the overlap in these features across the entire 286 pMHC considered in this analysis is shown in Figure 4F. The presence of all four features was very strongly enriched in immunogenic peptides (Figure 4G, p = 6*10−10, Fisher’s exact test; odds ratio: 51.7), filtering out 98% of non-immunogenic peptides while preserving 45% of immunogenic ones. We finally compared the effects of the presentation and recognition filters using precision recall curves in Figure 4H. Ranking pMHC only by their measured MHC binding affinity had a uniform precision less than 20%. Ranking presented pMHC first and then ranking by MHC binding affinity had an optimal precision of ~50% with 55% recall, while ranking presented and recognized pMHC and then ranking by MHC binding affinity had an optimal precision greater than 70% with a recall of 45% (see STAR Methods). Our results support an integrative model of peptide immunogenicity that requires both MHC presentation and T cell recognition and where T cell recognition can be achieved through low peptide agretopicity or high peptide foreignness.
Submission Features Associated with Pipeline Performance
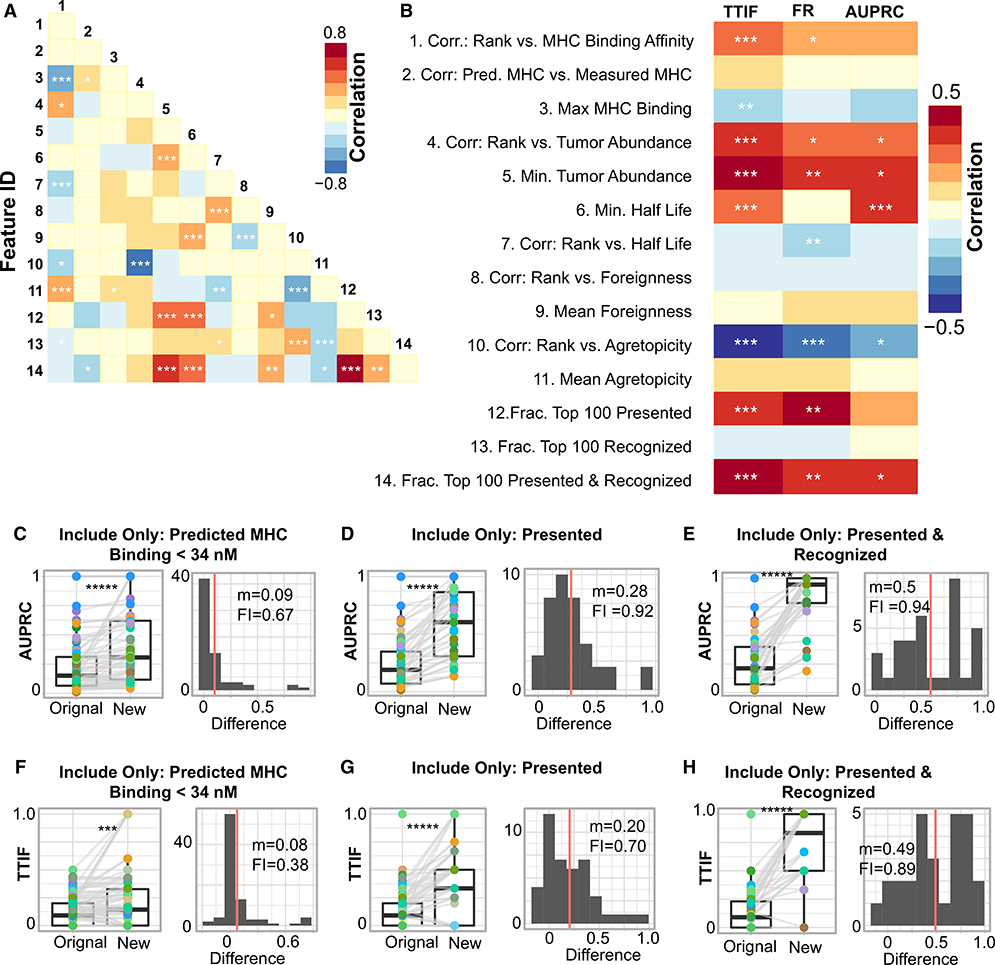
Based on these identified characteristics of tumor epitopes, we hypothesized that neoantigen prediction pipelines that prioritized these characteristics would perform better. We addressed this question by analyzing the core output of each pipeline, a ranked list of pMHC. For each submitted pMHC list, we calculated a set of 14 distinct, quantitative features, described in Table S6, termed “submission features.” In general, there was limited correlation between these values, and only 20% of possible submission feature pairs had a significant correlation (Figure 5A). Thus, this set of submission features captured numerous, distinct elements of each submission from a participants’ pipeline.
Figure 5. Directed Interventions on Submission Features Improves Neoantigen Pipeline Performance.
(A) Spearman correlation between each feature pair across all teams. Feature IDs are those in (A).
(B) Spearman correlation between 3 performance metrics (AUPRC, FR, and TTIF variability) and the 17 submission features plotted in (A) over all submissions. ***q < 0.05; **q < 0.1; *q < 0.25.
(C–H) Two pipeline performance metrics are considered (AUPRC, C–E; TTIF, F–H), and for each metric, three interventions are demonstrated. For each intervention, the boxplot (left) shows the change in the performance metrics from the original prediction to the new prediction (post intervention). Significance values are calculated using a paired Mann-Whitney U test. ***p < 0.001; *****p < 10−5. The histogram (right) shows the distribution of changes to the performance metric.
Red line, median; m, median improvement; FI, fraction improved.
We sought to identify associations between submission features and overall metrics of pipeline performance. We calculated the Spearman correlation between these 14 submission features and the three performance metrics introduced previously (TTIF, FR, and AUPRC) (Figure 5B). Numerous submission features were correlated with TTIF, FR, and AUPRC. Submissions that prioritized pMHC with stronger MHC binding affinity (feature 1), more stringently filtered out pMHC with weak MHC binding affinity (feature 3), low tumor abundance (feature 5), and low binding stability (feature 6) all performed better across numerous metrics. Conversely, submissions that explicitly prioritized peptide foreignness (features 8 and 9), agretopicity (features 10 and 11), or both (feature 13), without accounting for presentation, either had no difference in performance or performed worse. Correlation between predicted binding affinity and measured binding affinity (feature 2) was not associated with improved prediction performance. Finally, submissions with a large fraction of their top 100 pMHC presented (feature 12) or presented and recognized (feature 14) had substantially larger values of TTIF, FR, and AUPRC. Together, these results demonstrate that submissions that prioritized strong binding affinity, high tumor abundance, and high binding stability, potentially coupled with peptide recognition, had superior performance in TESLA.
Directed Interventions Improve Pipeline Performance
We identified traits of neoantigen prediction pipelines that are correlated with improved ability to rank and identify neoantigens. We next sought to understand if the implementation of these traits would improve prediction, in particular, in under-performing pipeline submissions. For AUPRC and TTIF separately, we implemented a particular intervention on the filtering and/or ranking in all submissions (Figures 5C–5H). The three interventions considered were to remove all pMHC with binding affinity greater than 34 nM (Figures 5C and 5F), remove all pMHC that do not pass the presented criteria from Figure 3 (Figures 5D and 5G), and remove all pMHC that do not pass both the presented and the recognized criteria from Figure 4 (Figures 5D and 5G). In all cases, these interventions significantly improved prediction compared to the initial submission set. Notably, although filtering on MHC improved only a fraction of submissions (AUPRC, 67%; TTIF, 38%), these values were greatly increased upon filtering for presentation and presentation and recognition. Furthermore, these interventions were rarely detrimental to prediction. Together, these results demonstrate that neoantigen prediction performance can be substantially improved through the implementation of pipeline interventions identified by TESLA. We also applied these interventions to a set of predictions made by an independent set of TESLA teams who joined the consortium after peptide selection was complete. In this setting as well, the interventions identified here significantly improved neoantigen prediction (Figure S6).
Predicted and Recognized Neoantigen Abundance Is Associated with Overall Survival to PD1 Blockade
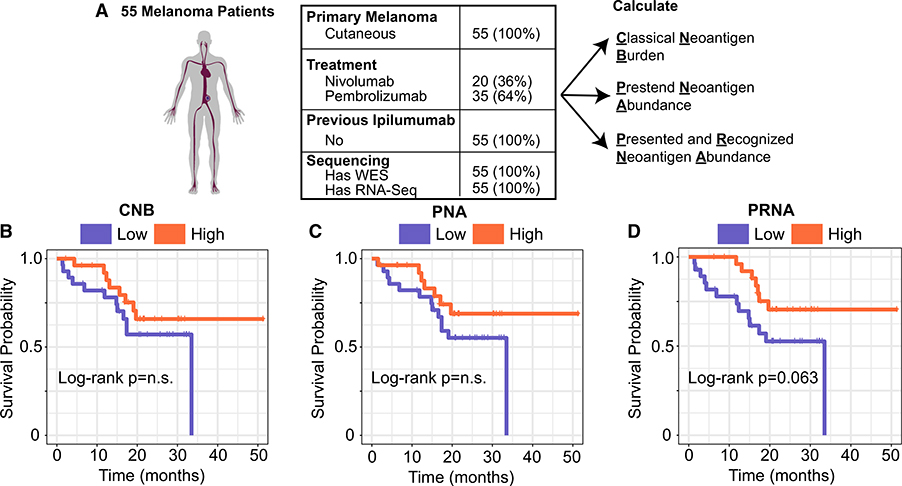
Tumor mutation burden (TMB) has been proposed as a biomarker for response to immune checkpoint inhibitors (ICI) (Samstein et al., 2019), however, its predictive value is inconsistent (Łuksza et al., 2017) and in diseases such melanoma, may be confounded with primary type (Liu et al., 2019). Based on our results above, we hypothesized that a metric that integrates relevant aspects of both tumor epitope presentation and recognition may offer improved predictive power for benefit from ICI. Thus, we developed the metric predicted and recognized neoantigen abundance (PRNA), the sum abundance of all mutations that satisfy both the “presented” criteria and the “recognized” criteria (STAR Methods). We sought to determine if this metric would outperform other metrics which do not incorporate all relevant features of tumor epitope immunogenicity, by comparing it to classical neoantigen burden (CNB, an analog of TMB that filters out only those mutations with MHC binding affinity greater than 500 nM) and to the predicted neoantigen abundance (PNA, the sum abundance of all mutations that satisfy the “presented” criteria, STAR Methods). We applied these metrics to a cohort of 55 melanoma patients homogeneous in treatment (anti-PD1), primary type (cutaneous), and treatment history (ICI-naive), who all had whole exome and RNA-sequencing of tumor biopsy samples before treatment initiation (Liu et al., 2019) (Figure 6A). On these samples, we calculated CNB, PNA, and PRNA. Stratifying this patient cohort by the median value of each metric showed no significant association with CNB or PNA with overall survival (Figures 6B and 6C, p: log rank test). However, patients with tumors with above-median PRNA values did have significantly longer overall survival than those with below-median PRNA values (Figure 6D, p = 0.063, log rank test). Furthermore, PRNA-high patients were ~50% more likely to survive longer than 2 years. These results suggest that the integrated portrait of epitope immunogenicity developed here may help improve understanding of response to ICI.
Figure 6. Predicted and Recognized Neoantigen Abundance Is Associated with Overall Survival to Anti-PD1.
(A) Patient cohort table displaying primary type, treatment, previous immunotherapies, and presence of sequencing.
(B) Kaplan-Meier plot of overall survival stratified by CNB-high/low status. p value, log rank test.
(C) Kaplan-Meier plot of overall survival stratified by PNA-high/low status. p value, log rank test.
(D) Kaplan-Meier plot of overall survival stratified by PRNA-high/low status. p value, log rank test. All high/low cutoffs were taken to be the median across the cohort.
Validation in an Independent Cohort
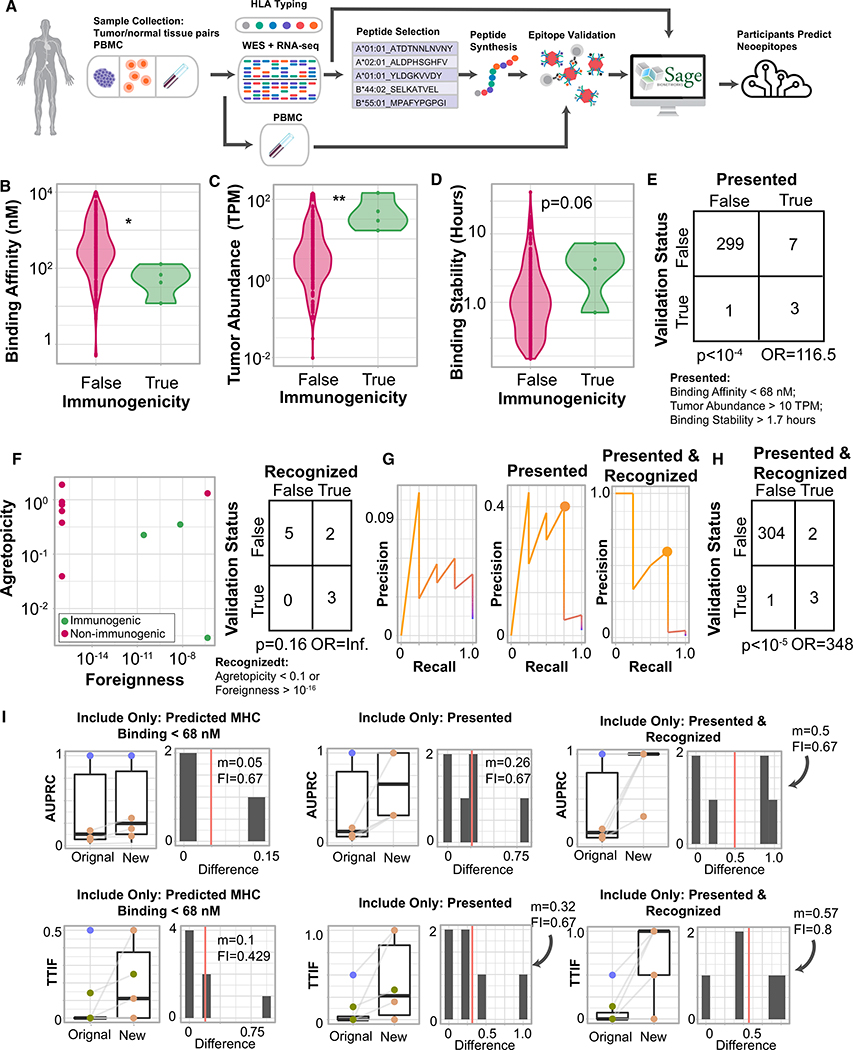
To assess whether the results identified herein are robust beyond our initial cohort, we identified an independent cohort of 3 melanoma patients with whole exome tumor-normal DNA sequencing and tumor RNA sequencing from tissue samples and for which 310 pMHC had been tested for immunogenicity using a tetramer-based assay in patient-matched PBMC samples (Figure S7; Table S7). Of those pMHC tested, 4 were found to be immunogenic (Figure 7A). In this cohort, immunogenic peptides had significantly stronger binding affinity (Figure 7B, p = 0.012, Mann-Whitney U); significantly higher tumor abundance (Figure 7C, p = 0.033, Mann-Whitney U), and longer binding stability (Figure 7D, p = 0.067, Mann-Whitney U). Applying the same repeated-random-subsample method on these peptides identified a threshold set that incorporated all three of these features with similar values to our initial cohort and that filtered out 97% of non-immunogenic peptides while preserving 75% of immunogenic ones (Figure 7E, p = 9*10−5, odds ratio: 116.5; Fisher’s exact test). Of the remaining 10 pMHC that met all of the “presented” criteria, 100% of immunogenic pMHC also met the recognition criteria (low agretopicity or high foreignness), whereas only 28% of non-immunogenic pMHC did (Figure 7F). Filtering pMHC on both presentation and recognition criteria achieved a recall of 75% and filtered out 99% of non-immunogenic peptides (Figures 7G and 7H; p = 8*10−6, odds ratio: 348, Fisher’s exact test). Finally, to assess if these parameters were associated with improved neoantigen prediction, we gathered neoepitope predictions from the DNA and RNA sequencing data from four TESLA participants. Although the predictive ability of participants on these data was low, potentially owing to reduced number of identified immunogenic peptide, interventions identical to those on the initial cohort demonstrated marked improvement in neoepitope prediction, with the large majority of submissions showing substantial improvement (Figure 7I).
Figure 7. Features Associated with Improved Neoepitope Prediction in a Validation Cohort.
(A) Schematic of the validation experiment.
(B) Violin plot of binding affinity stratified by peptide immunogenicity. **p < 10−2, Mann-Whitney U test.
(C) Violin plot of tumor abundance stratified by peptide immunogenicity. *p < 0.05, Mann-Whitney U test.
(D) Violin plot of binding stability stratified by peptide immunogenicity. p = 0.06, Mann-Whitney U test.
(E) Contingency table using optimal stratification parameters (below). p, Fisher’s exact test.
(F) Scatterplot of foreignness compared to agretopicity. Color: immunogenicity. Right: contingency table comparing validation status to recognition status among presented peptides. p, Fisher’s exact test; OR, odds ratio.
(G) Precision-recall curves of peptides ranked only by MHC binding affinity (left), prioritizing presented peptides (center), and prioritizing presented and recognized peptides (right). Circles represent optimal precision-recall tradeoffs.
(H) Contingency table over all peptides comparing validation status to presented and recognized status. p, Fisher’s exact test. OR, odds ratio.
(I) Two pipeline performance metrics are considered (AUPRC, top; TTIF, bottom), and for each metric, three interventions are demonstrated. For each intervention, the boxplot (left) shows the change in the performance metrics from the original prediction to the new prediction (post intervention). The histogram (right) shows the distribution of changes to the performance metric. Red line, median; m, median improvement; FI, fraction improved.
See also Table S7.
DISCUSSION
Effective neoantigen prediction relies on understanding the parameters governing epitope immunogenicity. By directly measuring epitope immunogenicity in patient-matched T cells, we uncover features of peptides and pMHC associated with in vivo immune recognition to construct an integrated model of tumor epitope immunogenicity. Because candidate epitopes were drawn from a broad set of prediction pipelines, we increased the diversity of tested predictions and removed potential bias associated with using predictions from only a single pipeline. The importance of the features we identify, such as agretopicity and foreignness, has previously been hypothesized from in vitro or otherwise indirect methods, and this is among the first studies demonstrating their importance in a human cancer setting. These results comprehensively characterize ~50% of immunogenic tumor epitopes: they are those tumor peptides that have strong MHC binding affinity and long half-life, are expressed highly, and have either low agretopicity or high foreignness.
This work does not address 4 further considerations about neoepitope immunogenicity. First, the peptides tested were generated from small somatic variants (SNV and/or indels); complex variant structures such as fusion proteins (Yang et al., 2019), splice-isoforms (Kahles et al., 2018; Robinson et al., 2019), aberrantly expressed introns of mutant genes (Smart et al., 2018), modified peptides (Cobbold et al., 2013), or non-proteinaceous antigens, which are currently difficult to identify using common genomic approaches, were not considered. In some cases, however, neoantigens arising from these variants may represent major classes of highly immunogenic abnormal molecules expressed in tumor cells (Smith et al., 2019). Second, we focused our validation efforts on identifying peptides associated with pre-existing T cell specificities. By definition, these would arise from tumor neoantigens that are sufficient to drive naturally occurring immune responses. However, this validation approach might overlook neoantigens whose immunogenicity could be enhanced by appropriate vaccination approaches. Third, our validation approach focused only on MHC-I restricted antigens and did not take into account the important role that MHC-II restricted antigens, and more broadly, the role CD4+ T cells, play in the development of effective immune responses to tumors (Abelin et al., 2019; Alspach et al., 2019; Kreiter et al., 2015; Ott et al., 2017). Finally, the approach we take here looks at immunogenicity of tumor antigens in patient-matched samples and does not address the rules governing epitope immunogenicity in the context of allogeneic T cells (Strønen et al., 2016). Identifying those rules may aid in the development of allogeneic neoantigen-directed adoptive T cell therapies (Bethune and Joglekar, 2017).
The analysis approach we take attempts to balance between specificity and sensitivity in the identification of neoepitopes. In some treatment settings, however, like in RNA-based vaccines or in searching for tumor-reactive TIL (Sahin et al., 2017; Yamamoto et al., 2019), specificity is not as serious a challenge, and the parameter values used here may be tuned to increase sensitivity or other metrics of interest. Furthermore, although this study identified key parameters governing epitope immunogenicity, the values of those parameters may differ based on the approach used for DNA/RNA sequencing, binding affinity prediction, and other bioinformatic steps, as well as the type of cancer. Notably, the tumor types considered here are characterized by a high mutational burden originating from exposure to UV radiation (melanoma) or cigarette smoke (NSCLC), which could influence the peptides included in this study (Hellmann et al., 2018; Samstein et al., 2019). As such, parameter values in other settings may need further calibration to optimally meet the criteria for a particular treatment setting and approach. For these reasons, it is our goal that the data assembled here be a resource for the community to identify new features that differentiate immunogenic pMHC, calibrate and tune neoepitope prediction pipelines for particular use cases, and ultimately, be used to benchmark and improve neoantigen prediction methods.
STAR★METHODS
Detailed methods are provided in the online version of this paper and include the following:
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents will be fulfilled by the Lead Contact, Daniel K. Wells (dwells@parkerici.org).
Materials Availability
Tetramers used in TESLA are available upon signature of an MTA.
Data and Code Availability
TESLA data is available here: https://www.synapse.org/#!Synapse:syn21048999. All molecular data (WES and RNA) from the melanoma cohort can be found in dbGap (accession number phs000452.v3.p1) and clinical variables can be found as supplemental information in the associated manuscript (Liu et al., 2019). Code to recreate the scoring metrics used in this study is included as a supplemental file to this manuscript.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
All clinical investigation was conducted according to the principles expressed in the Declaration of Helsinki. Written informed consent was obtained from the participants. Samples were collected under Institutional Review Board (IRB).
METHOD DETAILS
Subjects, Treatment, and Specimen Collection
The study was conducted with samples from subjects with metastatic melanoma or non-small cells lung cancer (NSCLC), previously collected and stored at UCLA or MSKCC respectively, and for which adequate matched pre-treatment control/tumor biopsy were available along with baseline and/or on-treatment matched peripheral blood mononuclear cells (PBMCs; melanoma) or tumor lysates (NSCLC). Collection and genomic analyses of the melanoma tumor biopsies, corresponding normal samples and PBMC were approved by UCLA Institutional Review Board 11–001918 and 11–003066. Collection and genomic analyses of the NSCLC tumor biopsies, corresponding normal samples and tumor lysates were approved by MSKCC Institutional Review Board 06–107. In all cases, tumor biopsies used in genetic analyses were snap-frozen by immediate immersion in liquid nitrogen for genetic analyses. PBMCs were prepared from fresh whole blood by Ficoll-Paque density gradient centrifugation and cryopreserved. Tissue lysate was prepared as described below. Additional information on the subjects with melanoma and NSCLC included in this analysis can be found in Table S3.
Tissue lysate preparation
Tumor sample lysate was prepared using the Miltenyi gentleMACS Octo Dissociator (Bergish, Germany). Briefly, tumor samples were placed in a 100mm culture dish in serum free RPML medium and minced into small pieces (1–3mm3) with sterile scalpels. Minced tumor was submerged in 10mL of disassociating solution in a C tube and subsequently incubated at 37°C using the Human Tumor Dissociation cycle “1 hour with continuous shaking” in the gentleMACS Octo Dissociator. Once complete, tumor was passed through a 100 μM sterile cell strainer (BD Falcon) on a 50mL conical tube and spun at 1500 RPM for 7 minutes in a refrigerated centrifuge. Cells were resuspended in freezing media at a concentration of approximately 3 million cells/mL and transferred into 1mL cryovial aliquots. Cells were stored at −80°C in a slow freeze container, and transferred to liquid nitrogen for long term storage.
Whole Exome Sequencing and RNA Sequencing
Both DNA and RNA were co-extracted from snap-frozen tumor biopsies using the QIAGEN AllPrep Kit according to the manufacturer’s instructions (QIAGEN, Valencia, CA). Melanin, a known inhibitor of enzymatic reactions, coprecipitates with the RNA, therefore the RNA required further purification performed as described (Lagonigro et al., 2004), modified with a RNeasy (QIAGEN, Valencia, CA) column-based clean-up to remove the additives used to bind the melanin.
IDT Exome
Automated (dual indexes) were constructed with 200–250ng of genomic DNA utilizing the KAPA HTP Library Kit (KAPA Biosystems) according to the manufacturer’s instructions. Automated libraries were performed on the SciClone NGS (Perkin Elmer) instrument targeting 200–250bp inserts. Samples were pooled in a 1:1 ratio and processed through the capture pipeline in the McDonnell Genome Institute at Washington University School of Medicine (St. Louis, MO). Libraries were pooled prior to capture hybridizations with IDT Exome Library IDT xGen Exome Research Panel that spans a 39 Mb target region (19,396 genes). Capture processing was performed following the company protocol. KAPA qPCR was used to quantify the libraries and determine the appropriate concentration to produce optimal recommended cluster density on a HiSeq4000 v2 (PE150bp) sequencing run. Sample pools were sequenced on two lanes of an Illumina HiSeq4000 v2 (PE150bp) instrument. All sequencing runs were completed according the manufacturer’s recommendations (Illumina Inc, San Diego, CA)
RNA-seq
Manual libraries (single indexes) were constructed utilizing Illumina TruSeq Stranded total RNA kit (Illumina) (100–500ng of total RNA) according to the manufacturer’s instructions. KAPA qPCR was used to quantify the libraries and determine the appropriate concentration to produce optimal recommended cluster density on a HiSeq4000 v2 (PE150bp) sequencing run. Final pool of samples were sequenced across two lanes HiSeq4000. All sequencing runs were completed according the manufacturer’s recommendations (Illumina Inc, San Diego, CA).
Human Leukocyte Antigen (HLA) typing
Clinical grade (six-digit) HLA typing was obtained for each subject from the normal DNA by Histogenetics (Ossining, NY)
Peptide Synthesis
Peptides > 90% pure by analytical HPLC were synthesized by New England Peptides (Gardner, MA).
Neoepitope Predictions
Once sequencing of samples was completed, fastq files of tumor DNA sequence, tumor RNA sequence, and normal DNA sequence were uploaded to Sage Bionetworks Synapse platform (https://synapse.org/) along with clinical grade HLA typing information. These data were made accessible to the consortium participating teams to identify neoepitopes using their respective prediction pipeline.
To enable comparison across the different pipelines, all participants were required to align sequence data to GRCh38 (Ensembl). All participants were required to submit three files for each sample for which they predicted neoepitopes:
A ranked list of neoepitopes and the HLA allele those neoepitopes were identified with
An unranked list of filtered neoepitopes and the HLA allele those neoepitopes were identified with
A list of identified variants
Additional requirements, including a detailed description of each of the above files is found in Table S1. Teams were asked to complete their predictions in 6 weeks after downloading the data.
Workflow Survey
As part of their submissions, the teams were asked to include a description of the steps included in their neoantigen prediction workflow in a numbered list in a text file. These free text workflow descriptions were manually reviewed, and a representative list of common workflow steps was compiled. To do this, each step of each workflow was added to a cumulative list if that step was deemed substantially different from other steps already present on the list. This process established a controlled vocabulary for describing those steps. The vocabulary was then organized into groups of steps performing a similar higher-level function, e.g., “variant calling,” “variant filtering” and “peptide ranking.” Then, each step in each workflow was annotated with the controlled vocabulary to make sure that each workflow was represented by the vocabulary. Finally, a workflow survey was created such that each team was asked to provide a binary response about whether their workflow included each step defined by the controlled vocabulary. The survey contained 6 high-level categories each containing 3–11 specific steps.
Process for Choosing Neoepitopes to Test in Validation Assays
For each sample used in TESLA there is capacity to test between 100–200 neoepitopes for immunogenicity. Given that each participating team submitted 5–100,000 peptides for each sample, a sub-set of peptides for validation had to be selected. To do this, we focused on ensuring that each participant would have roughly the same number and quality (ranking) of peptides validated (“fairness”) while also making sure that, if there are differences between the algorithms, we would be able to detect them (“distinguishability”). These principles were constrained by the fact that some peptides are too hydrophobic (or otherwise hard to make/use) to be used in our assays, as well as the fact that not every neoepitope can be tested in every assay due to HLA restriction requirements. In practice, we used the following guidelines to choose peptides:
To the extent possible, each participant had their top 5 neoepitopes selected, taking into account the MHC for which they are restricted
Additionally, neoepitopes that were the most recurrently ranked in the top 50 by all participants were also selected taking into consideration the amount of biological material available for testing, the demand for each assay and the MHC constraints for all four assays.
50 was chosen as a cutoff to reflect the upper bound on the current number of epitopes that can be included in a personalized therapeutic approach.
Validation Experiments
Selected peptides were evaluated in a set of experiments all together referred as ‘validation experiments’. Validation experiments include HLA binding and immunological analyses that assess the presence of existing T cells able to recognize the selected peptide.
Class I Peptide Binding Assay (‘HLA binding’)
Classical competition assays to quantitatively measure peptide binding to class I MHC molecules are based on the inhibition of binding of an allele-specific, high affinity radiolabeled peptide to purified MHC molecules, and were performed as detailed elsewhere (Sidney et al., 2013). Briefly, 0.1–1 nM of radiolabeled peptide was co-incubated at room temperature with 1 μM to 1 nM of purified MHC in the presence of a cocktail of protease inhibitors and 1 μM β2-microglobulin. Following a two-day incubation, MHC-bound radioactivity was determined by capturing MHC/peptide complexes on W6/32 (anti-HLA class I) antibody coated Lumitrac 600 plates (Greiner Bio-one, Frickenhausen, Germany), and measuring bound cpm using the TopCount (Packard Instrument Co., Meriden, CT) microscintillation counter. In the case of competitive assays, the concentration of peptide yielding 50% inhibition of the binding of the radiolabeled peptide was calculated. Under the conditions utilized, where [label] < [MHC] and IC50 ≥ [MHC], the measured IC50 values are reasonable approximations of the true Kd values. Each competitor peptide was tested at six different concentrations covering a 100,000-fold dose range, and in three or more independent experiments. As a control, the unlabeled version of the respective radiolabeled probe also was tested in each experiment.
MHC Class I Multimer Binding Assay (Flow Cytometry)
Multimer binding assays were performed independently by two groups: (A) The Immunomonitoring Laboratory [IML], Washington University in Saint Louis, USA, and (B) Netherlands Cancer Institute [NKI], Amsterdam, NL. Similar approaches were followed by both groups for multimer construction, T cell binding and detection, as described below unless mentioned otherwise.
Production of Exchanged Peptide-MHC Class I Monomers –
MHC I monomers refolded with an ultraviolet light-cleavable conditional ligand were prepared as previously described (Andersen et al., 2012; Toebes et al., 2006). In brief, recombinant HLA heavy chains and human β2-microglobulin light chain were produced in Escherichia coli (Agilent Technologies), isolated from inclusion bodies and refolded in the presence of UV-cleavable peptides. Monomers were captured by anion exchange (HiTrap Q HP, GE), biotinylated, and purified by gel filtration FPLC (A) or HPLC (B). Specific peptide-MHC I (pMHC I) complexes were then generated by UV-induced ligand exchange in a 96 well plate and the peptides’ exchange were verified by a sandwich like ELISA. In brief, pMHC I complexes loaded with UV-sensitive peptide (100 μg/mL-1) were subjected to 366 nm UV light (Camag) for 1 h at 4°C in the presence of rescue peptide (200 μM). Re-folded monomer was detected using anti- β2 microglobulin. Monomers were also exchanged with viral peptides which bind to distinct HLA alleles, included as positive and negative controls for staining (MP1 Influenza A: GILGFVFTL, P1804 EBV1: RVRAYTYSK, PB1 Influenza: VSDGGPNLY, EBV: YVLDHLIVV, 1807 EBV: RAKFKQLL, 1813 EBV: IVTDFSVIK, 1822 Influenza A: SRYWAIRTR, 1824 EBV: RRIYDLIEL, EBV6’ EBNAC3:EENLLDFVRF, and 1803 A3 influenza A: RVLSFIKGTK).
Production of Multimer Staining –
pMHC I multimers were generated using fluorescent streptavidin (SA) conjugates. For each pMHC I monomer, conjugation was performed with two fluorochromes. Mixtures were incubated for 30 min on ice. NaN3 (0.02% wt/vol) and an excess of D-biotin (26.4 μM, Sigma) was added to block residual binding sites. For T cells staining, the pMHC I multimers were collected and centrifuged for 2 minutes at 10.000 rpm, before being added to the subject PBMC or tumor lysates, and incubated at 37°C for 15 minutes. Subsequently, anti-CD45, anti-CD8, a ‘dump channel’ mix including: anti-CD4, anti-CD14, anti-CD16,and anti-CD19, and anti-CD3 was used for positive gating (Biolegend). A marker for living cells (LIVE∕DEAD® Fixable IR Dead Cell Stain Kit) was added for a 20 minute incubation on ice as previously described (Kvistborg et al., 2012). All incubations were done in the dark.
Data Acquisition and Analysis –
Before flow cytometric analysis, cells were washed twice. Data were acquired on a BD LSR Fortessa X-20 (A) or a BD Symphony A5 (B) following instrument compensation using UltraComp eBeads, and analyzed using Diva Software v8 and Flowjo v9.9.6 (A) or FlowJo v10.5.3 or earlier v10 packages (B). A given peptide is considered ‘validated’ when peptide-specific T cells are detected and confirmed in an independent staining experiment conducted with new reagents including a different set of fluorescent dyes. Example data from this workflow are shown in Figure S7.
MHC I Nanoparticle Pull-Down Assay (Microscopy)
The nanoparticle (NP) pulldown method utilizes multiple reagents that can be prepared ahead of time and assembled just prior to use and is described in detail elsewhere (Peng et al., 2019). Briefly, these include streptavidin-coated magnetic NPs (500 nm radius, Invitrogen Dynabeads MyOne T1), biotinylated neoantigen pMHC I monomers, cysteine-modified streptavidin (SAC) scaffold (Sano and Cantor, 1990), and ssDNA oligomers that are amine-modified (oligomer #1), biotinylated (oligomer #2), and dye-labeled (oligomer #3).
Oligomer #1 is appended to the cysteine site on the SAC scaffold via maleimide HyNic/S-4FB crosslinkers (TriLink BioTechnologies, LLC) to make SAC-DNA. Oligomer #2, which has hybridization regions for oligomer #1 (on SAC-DNA) and oligomer #3, is attached to the NPs to make NP-DNA. The NPs, which have up to > 105 sites for DNA attachment, were prepared according to the manufacturer’s protocol. Briefly, the NPs are mixed with oligomer #2 at a 1:20 ratio, and excess oligomers were removed by washing the NPs three times with PBS. A pMHC I monomer library element is added to SAC-DNA at a 4:1 ratio to form the pMHC I multimer-DNA. This multimer is then attached to the NP-DNA via DNA hybridization (between multimer-bound oligomer #1 and NP-bound oligomer #2). The dye-labeled oligomer #3 is then hybridized onto one of the additional sites on oligomer #2. The oligomer #2 and #3 were chosen so that a given neoantigen is associated with a unique dye color in multiplex analyses. A fully assembled neoepitopes multimer/NP reagent contains > 104 neoepitopes multimers per NP and is referred to as a pNP.
In a typical assay, four colors are used to enable 3-plex analysis. Cells are stained with CellTracker Orange CMRA (a fluorescent dye retained in live cells, ThermoFisher), and 3 sets of pNPs, each with a unique fluorophore (Alexa Fluor 488, Cy5, or Alexa Fluor 750) are used to pulldown up to 3 distinct neoantigen-specific T cell populations in parallel. Thus, for a 15-element library, 5 sets of measurements are performed, with each measurement using about 10,000 CD8+ PBMCs and 75 million pNPs (~25 million pNPs for each antigen specificity).
Live CD8+ PBMCs are sorted by FACS or negatively enriched using MACS (Miltenyi biotec). Before analysis, the sorted cells are stained with CellTracker Orange, treated with DNase, and non-selective cells are removed using a small library of irrelevant pNPs. The processed cells are then incubated with a 3-element pNP library for 15–30 min, and NP-labeled cells are then enriched by magnet pulldown. The enriched T cells are washed once with 0.5% BSA in PBS to remove unlabeled cells. The NP-labeled cells are then loaded into a hemocytometer and imaged by bright field and 4-color fluorescent microscopy. Cells that are specific for a given neoantigen are identified as those that are fluorescent in only CellTracker Orange and a single additional fluorophore from one set of pNP (typically 5–20 pNPs are labeled on each specific T cell). Dead, barely viable, or non-selective cells are found to be non-selectively labeled by multiple pNP library elements. For the experiments described here, assays were carried out in 4–5 replicates for each neoepitope pMHC I, using different vials of thawed PBMCs. Cells that are called as positive for a specific neoantigen were observed in at 2 of those runs, with multiple detection per run.
Analytic Methods
Peptide Overlap
Peptide overlap was calculated as the length of the intersection of two lists, normalized to the length of the first list.
Area Under the Precision Recall Curve (AUPRC)
AUPRC was calculated using the ‘pr.curve’ function in the PRROC package (Grau et al., 2015) with default parameters.
Fraction Ranked (FR)
Fraction ranked is calculated as the fraction of all peptides with detected immunogenicity for a particular subject that were included in the top 100 ranked pMHC by a participant. A cutoff of 100 peptides was chosen to calculate FR to normalize for differing numbers of pMHC submitted by each team. The value of 100 was chosen in particular to ensure consistency with other analyses and to be reflective of the total number of epitopes which might be considered for inclusion in a personalized therapeutic.
Top-20 Immunogenic Fraction (TTIF)
Top-20 immunogenic fraction for a particular subject and participant is calculated as the ratio of the top-20 ranked pMHC with detected immunogenicity to the total number of top-20 pMHC which were tested for immunogenicity. As TTIF was designed to assess the therapeutic efficacy of a specific peptide set, the threshold of 20 was selected since therapeutic vaccine platforms reported to date have included ~20 neoepitopes (Ott et al., 2017; Sahin et al., 2017).
Foreignness
Peptide foreignness was calculated as described previously (Łuksza et al., 2017). Briefly, candidate epitopes were aligned against the Immune Epitope Database and Analysis Resource (Vita et al., 2019), the TCR recognition probability was calculated using the multistate thermodynamic model introduced in the above. All parameters for the original approach were maintained; specifically, alignments were performed against the BLOSUM62 amino acid similarity matrix (Henikoff and Henikoff, 1992) and required to be gapless; a = 26; k = 4.87. Foreignness for all peptides tested in TESLA are provided in Table S4, and were calculated using the antigen.garnish package available here: https://github.com/immune-health/antigen.garnish
Binding Stability
Binding stability was calculated using NetMHCStabPan (Rasmussen et al., 2016) using default parameters.
Hydrophobicity Fraction
Hydrophobicity fraction was calculated as the fraction of peptide residues that were hydrophobic. Hydrophobic residues were considered to be “V,” “I,” “L,” “F,” “M,” “W,” and “C” (Barnes, 2003)
Parameter Selection via Repeated Random Subsampling
For each continuous parameter (MHC binding affinity, MHC binding stability, tumor abundance, hydrophobicity fraction) a range of values covering two orders in magnitude was generated while for binary features (mutation position) both levels were considered. For each unique parameter combination, 10 random subsets of 70% of tested peptides (with immunogenic/non-immunogenic ratio equal to the whole dataset) were selected and stratified using the given features. Immunogenicity stratification was calculated using a Fisher exact test. The parameter set with the smallest average p value over all random subsets was chosen to be the one with the best overall stratification ability.
Optimal Precision/Recall Cutoff
Optimal precision recall cutoff was chosen as the maximum of the precision/recall sum.
Predicted Neoantigen Abundance
Potential immunogenic peptides were generated using a previously generated set of mutation calls (Liu et al., 2019) and predicted MHC binding affinity was assigned using NetMHCPan4.0. Predicted neoantigen abundance was taken as the sum of the normalized transcripts per million (TPM) of the mutations which passed all “presented” filters from Figure 3L (excluding the abundance filter) – specifically, MHC binding affinity stronger than 34 nM and MHC binding stability longer than 1.4 hours, and mutational position not 2.
Predicted and Recognized Neoantigen Abundance
Potential immunogenic p eptides were generated using a previously generated set of mutation calls (Liu et al., 2019) and predicted MHC binding affinity was assigned using NetMHCPan4.0. Predicted and recognized neoantigen abundance was taken as the sum of the normalized transcripts per million (TPM) of the mutations which passed all “presented” features (excluding the abundance filter, identical to predicted neoantigen abundance) and the “recognized’ filters from Figure 4D – specifically, peptide agretopicity less than 0.1 or peptide foreignness greater than 10−16.
QUANTIFICATION AND STATISTICAL ANALYSIS
All statistical analysis was performed in base R. The statistical test used were two-sided (where applicable) and are indicated in the text and/or figure legends. The “n” for each analysis, where n could represent pMHC, number of participants, number of submissions, or similar, is indicated in the main text or in figure legends of relevant analyses. Significance was set at p < 0.05 and p values between 0.05 and 0.1 were reported in their numerical form. Non-parametric methods were used for all statistical analyses obviating any assumption of normality for any assessed metric.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| BUV805 mouse anti-human CD45 [clone HI30] | BD Bioscience | Cat#564915 |
| BUV395 mouse anti-human CD3 [clone UCHT1] | BD Bioscience | Cat#563546 |
| AlexaFluor700 mouse anti-human CD8a [clone HIT3a] | Biolegend | Cat#300919 |
| FITC mouse anti-human CD16 Antibody [Clone: 3G8] | Biolegend | Cat#302006 |
| FITC mouse anti-human CD14 Antibody [Clone: M5E2] | Biolegend | Cat#301804 |
| FITC mouse anti-human CD40 Antibody [Clone: 5C3] | Biolegend | Cat#334306 |
| FITC mouse anti-human CD4 Antibody [Clone: SK3] | Biolegend | Cat#344604 |
| FITC mouse anti-human CD19 Antibody [Clone: HIB19] | Biolegend | Cat#302206 |
| LIVE/DEAD® Fixable IR Dead Cell Stain Kit | ThermoFisher | Cat#L34975 |
| PE-CY7 Streptavidin | Life Technologies | Cat#501137616 |
| Brilliant Violet 650 Streptavidin | Biolegend | Cat#405231 |
| Brilliant Violet 605 Streptavidin | Biolegend | Cat#405229 |
| Brilliant Violet 711 Streptavidin | Biolegend | Cat#405241 |
| Brilliant Violet 421 Streptavidin | Biolegend | Cat#405225 |
| Brilliant Violet 510 Streptavidin | BD Bioscience | Cat#BDB563261 |
| APC Streptavidin | Life Technologies | Cat#SA1005 |
| PE Streptavidin | Biolegend | Cat#405204 |
| PE-CF594 Streptavidin | BD Bioscience | Cat#BDB562318 |
| W6/32 (anti-HLA class I) | ATCC | HB-95 |
| Bacterial and Virus Strains | ||
| BL21-CodonPlus(DE3)-RIPL Competent cells | Agilent Technologies | Cat#230280 |
| Biological Samples | ||
| Tumor lysates from patients with NSCLC | This paper | N/A |
| Human PBMCs from patients with metastatic melanoma | This paper | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| UltraComp eBeads Compensation Beads | Life Technologies | 01-2222-42 |
| NaN3 | Sigma Aldrich | Cat# S2002 |
| D-biotin | Sigma Aldrich | Cat# B4501 |
| HRP anti-human b2-microglobulin | BioLegend | Cat# 280303 |
| MP1 Influenza A: GILGFVFTL | Peptide 2.0 | N/A |
| P1804 EBV1: RVRAYTYSK | Peptide 2.0 | N/A |
| PB1 Influenza: VSDGGPNLY | Peptide 2.0 | N/A |
| EBV: YVLDHLIVV | Peptide 2.0 | N/A |
| 1807 EBV: RAKFKQLL | Peptide 2.0 | N/A |
| 1813 EBV: IVTDFSVIK | Peptide 2.0 | N/A |
| 1822 Influenza A: SRYWAIRTR | Peptide 2.0 | N/A |
| 1824 EBV: RRIYDLIEL | Peptide 2.0 | N/A |
| EBV6’ EBNAC3:EENLLDFVRF | Peptide 2.0 | N/A |
| 1803 A3 influenza A: RVLSFIKGTK | Peptide 2.0 | N/A |
| Putative neo-antigens for patient samples | This paper | N/A |
| Dynabeads MyOne T1 streptavidin-coated NPs (500 nm radius) | Invitrogen | Cat#65602 |
| CellTracker Orange CMRA Dye | ThermoFisher | Cat#C34551 |
| Bovine Serum Albumin (BSA) heat-shock fraction | Sigma-Aldrich | A3059 |
| Critical Commercial Assays | ||
| AllPrep DNA/RNA Kit | QIAGEN | catalog #80204 |
| RNeasy MinElute Clean-up Kit | Sigma 52365-50G | catalog #52365-50G |
| KAPA HTP Library Kit (KAPA Biosystems) | Roche | 7961901001 |
| TruSeq Stranded Total RNA with RiboZero Plus | Illumina | 20020598 |
| Deposited Data | ||
| Whole exome sequencing data (melanoma cohort) | Liu et al., 2019 | phs000452.v3.p1 |
| Transciptome data (melanoma cohort) | Liu et al., 2019 | phs000452.v3.p1 |
| Whole exome sequencing data (melanoma and NSCLC patients) | This paper | syn21048999 |
| Oligonucleotides | ||
| DNA for NP modification (ssDNA, 5′-biotin-) | Peng et al., 2019 | N/A |
| DNA for barcoding (ssDNA, 5-Cy5/Cy3/AlexaFluor488/AlexaFluor750) | Peng et al., 2019 | N/A |
| DNA for streptavidin labeling (ssDNA, 5-NH2-) | Peng et al., 2019 | N/A |
| Recombinant DNA | ||
| Plasmid containing SAC gene | Sano and Cantor, 1990 | Addgene Plasmid #17329 |
| HLA-A*02:01 plasmid | Andersen et al., 2012 | N/A |
| β-2-microglobulin plasmid | Andersen et al., 2012 | N/A |
| Software and Algorithms | ||
| FlowJo, LLC version 10.5.3 | Becton Dickinson | https://www.flowjo.com/solutions/flowjo/downloads/previous-versions |
| FacsDiva, BD software version 8 | Becton Dickinson | https://www.bdbiosciences.com/en-us/instruments/research-instruments/research-software/flow-cytometry-acquisition/facsdiva-software |
| Other | ||
| UV-lamp 366nm | CAMAG | Cat# 022.9070 |
| HiTrap Q HP | GE Healthcare | Cat# 17-1154-01 |
Highlights.
Diverse neoantigen predictions on shared genomic data from a global consortium
37 out of 608 tested peptide-MHCs are bound by patient-matched T cells
Epitope presentation and recognition characteristics predict immunogenicity
Model-based interventions improve neoantigen prediction
ACKNOWLEDGMENTS
We thank all the subjects who contributed to this study through donation of tumor and blood samples, as well as the research staff at UCLA and MSKCC for sample collection and processing. We acknowledge Olga Malkova, Diane E. Bender, Likui Yang, and Tammi Vickery for their work on MHC I multimer binding assay and nucleic acid isolation and sequencing; Jeff Bluestone, Jeff Hammerbacher, Ansuman Satpathy, and Robert Vonderheide for helpful and supportive comments; and David Liu and Eliezer van Allen for help in obtaining access to published data. TESLA was conceived collaboratively between the Parker Institute for Cancer Immunotherapy (PICI) and the Cancer Research Institute (CRI), and primary financial support came from PICI, a not-for-profit organization. Additional financial support was provided by NIH (R21 AI34127 to A.S.), an NIH training grant (GM08042 to J.M.Z.), a UCLA Tumor Immunology training grant (NIH T32CA009120), the CRI Irvington Postdoctoral Fellowship Program (to K.M.C.), and the Queen Wilhelmina Cancer Research Award (to T.N.S.).
DECLARATION OF INTERESTS
D.K.W. is a paid scientific advisor and shareholder in Immunai and receives research support from Bristol-Myers Squibb. M.M.v.B. is a stockholder and employee of BioNTech. V.M.H.-L. is an unpaid scientific advisor and holds equity in FX Biopharma. B.C.-A. has a contract grant with Kite Pharma and is a member of the Institutional Biosafety Committee (IBC) at Advarra Inc. N.H. is a stockholder in BioNTech, K.M.C. is a stockholder in Geneoscopy. J.Z. is an equity/stock holder and consultant to PACT Pharma. A.R. has received honoraria from consulting with Amgen, Bristol-Myers Squibb, Chugai, Genentech, Merck, Novartis, and Roche, is or has been a member of the scientific advisory board, and holds stock in Advaxis, Arcus Biosciences, Bioncotech Therapeutics, Compugen, CytomX, Five Prime, FLX-Bio, ImaginAb, Isoplexis, Kite-Gilead, Lutris Pharma, Merus, PACT Pharma, Rgenix, and Tango Therapeutics. M.D.H. receives research support from Bristol-Myers Squibb, has been a compensated consultant for Merck, Bristol-Myers Squibb, AstraZeneca, Genentech/Roche, Nektar, Syndax, Mirati, Shattuck Labs, Immunai, Blueprint Medicines, Achilles, and Arcus, received travel support/honoraria from AstraZeneca, Eli Lilly, and Bristol-Myers Squibb, has options from Shattuck Labs, Immunai, and Arcus, and has a patent filed by his institution related to the use of tumor mutation burden to predict response to immunotherapy (PCT/US2015/062208), which has received licensing fees from PGDx. P.K. is a consultant for Neon Therapeutics and Personalis. J.R.H. is board member and founder of Isoplexis and board member and founder of PACT. F.R. is an advisor/consultant to Equillium Bio, Good Therapeutics, SelectION, Inc., Cascade Drug Development Group, aTyr Pharma, and Lumos Pharma, and is a founder and holds equity in Sonoma Biotherapeutics. R.D.S. is a cofounder, scientific advisory board member, stockholder, and royalty recipient of Jounce Therapeutics and Neon Therapeutics and is a scientific advisory board member for A2 Biotherapeutics, BioLegend, Codiak Biosciences, Constellation Pharmaceuticals, NGM Biopharmaceuticals, and Sensei Biotherapeutics. J.S. and A.S. receive funding from BMS and Gritstone, are consultants for Turnstone, and perform fee-for-service assays for Neon. A.S. is a consultant for Gritstone. N.B. receives research funds from Novocure, Celldex, Ludwig institute, Genentech, Oncovir, Melanoma Research Alliance, Cancer Research Institute, Leukemia & Lymphoma Society, 485, NYSTEM, and Regeneron, and is on the advisory boards of Neon, Tempest, Checkpoint Sciences, Curevac, Primevax, Novartis, Array BioPharma, Roche, and Avidea. T.N.S. receives research funds from Merck KGaA, is consultant/advisory board member for Adaptive Biotechnologies, AIMM Therapeutics, Allogene Therapeutics, Merus, Neogene Therapeutics, Neon Therapeutics, Scenic Biotech, and Third Rock Ventures, and is a stockholder in AIMM Therapeutics, Allogene Therapeutics, BioNTech, Merus, Neogene Therapeutics, Scenic Biotech, and Third Rock Ventures Fund IV and V. A.R. has received honoraria from consulting with Amgen, Bristol-Myers Squibb, Chugai, Genentech, Merck, Novartis, Roche, and Sanofi, is or has been a member of the scientific advisory board, holds stock in Advaxis, Apricity, Arcus Biosciences, Bioncotech Therapeutics, Compugen, CytomX, Five Prime, FLX-Bio, ImaginAb, Isoplexis, Kite-Gilead, Lutris Pharma, Merus, PACT Pharma, Rgenix, and Tango Therapeutics, has received research funding from Agilent and from Bristol-Myers Squibb through Stand Up to Cancer (SU2C), and has received payment for licensing a patent on non-viral T cell gene editing to Arsenal. The remaining authors declare no conflicts of interest.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.cell.2020.09.015.
REFERENCES
- Abelin JG, Keskin DB, Sarkizova S, Hartigan CR, Zhang W, Sidney J, Stevens J, Lane W, Zhang GL, Eisenhaure TM, et al. (2017). Mass Spectrometry Profiling of HLA-Associated Peptidomes in Mono-allelic Cells Enables More Accurate Epitope Prediction. Immunity 46, 315–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abelin JG, Harjanto D, Malloy M, Suri P, Colson T, Goulding SP, Creech AL, Serrano LR, Nasir G, Nasrullah Y, et al. (2019). Defining HLA-II Ligand Processing and Binding Rules with Mass Spectrometry Enhances Cancer Epitope Prediction. Immunity 51, 766–779. [DOI] [PubMed] [Google Scholar]
- Alspach E, Lussier DM, Miceli AP, Kizhvatov I, DuPage M, Luoma AM, Meng W, Lichti CF, Esaulova E, Vomund AN, et al. (2019). MHC-II neoantigens shape tumour immunity and response to immunotherapy. Nature 574, 696–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen RS, Kvistborg P, Frøsig TM, Pedersen NW, Lyngaa R, Bakker AH, Shu CJ, Straten Pt., Schumacher TN, and Hadrup SR (2012). Parallel detection of antigen-specific T cell responses by combinatorial encoding of MHC multimers. Nat. Protoc 7, 891–902. [DOI] [PubMed] [Google Scholar]
- Balachandran VP, Łuksza M, Zhao JN, Makarov V, Moral JA, Remark R, Herbst B, Askan G, Bhanot U, Senbabaoglu Y, et al. ; Australian Pancreatic Cancer Genome Initiative; Garvan Institute of Medical Research; Prince of Wales Hospital; Royal North Shore Hospital; University of Glasgow; St Vincent’s Hospital; QIMR Berghofer Medical Research Institute; University of Melbourne, Centre for Cancer Research; University of Queensland, Institute for Molecular Bioscience; Bankstown Hospital; Liverpool Hospital; Royal Prince Alfred Hospital, Chris O’Brien Lifehouse; Westmead Hospital; Fremantle Hospital; St John of God Healthcare; Royal Adelaide Hospital; Flinders Medical Centre; Envoi Pathology; Princess Alexandria Hospital; Austin Hospital; Johns Hopkins Medical Institutes; ARC-Net Centre for Applied Research on Cancer (2017). Identification of unique neoantigen qualities in long-term survivors of pancreatic cancer. Nature 551, 512–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes MR (2003). Bioinformatics for Geneticists. (Wiley; ), pp. 39–70. [Google Scholar]
- Bethune MT, and Joglekar AV (2017). Personalized T cell-mediated cancer immunotherapy: progress and challenges. Curr. Opin. Biotechnol 48, 142–152. [DOI] [PubMed] [Google Scholar]
- Bulik-Sullivan B, Busby J, Palmer CD, Davis MJ, Murphy T, Clark A, Busby M, Duke F, Yang A, Young L, et al. (2018). Deep learning using tumor HLA peptide mass spectrometry datasets improves neoantigen identification. Nat. Biotechnol 37, 55. [DOI] [PubMed] [Google Scholar]
- Callari M, Sammut S-J, De Mattos-Arruda L, Bruna A, Rueda OM, Chin S-F, and Caldas C (2017). Intersect-then-combine approach: improving the performance of somatic variant calling in whole exome sequencing data using multiple aligners and callers. Genome Med 9, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carraro M, Minervini G, Giollo M, Bromberg Y, Capriotti E, Casadio R, Dunbrack R, Elefanti L, Fariselli P, Ferrari C, et al. (2017). Performance of in silico tools for the evaluation of p16INK4a (CDKN2A) variants in CAGI. Hum. Mutat 38, 1042–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreno BM, Magrini V, Becker-Hapak M, Kaabinejadian S, Hundal J, Petti AA, Ly A, Lie W-R, Hildebrand WH, Mardis ER, and Linette GP (2015). Cancer immunotherapy. A dendritic cell vaccine increases the breadth and diversity of melanoma neoantigen-specific T cells. Science 348, 803–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowell D, Krishna S, Becker PD, Cocita C, Shu J, Tan X, Greenberg PD, Klavinskis LS, Blattman JN, and Anderson KS (2015). TCR contact residue hydrophobicity is a hallmark of immunogenic CD8+ T cell epitopes. Proc. Natl. Acad. Sci. USA 112, E1754–E1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobbold M, Peña HDL, Norris A, Polefrone JM, Qian J, English AM, Cummings KL, Penny S, Turner JE, Cottine J, et al. (2013). MHC Class I–Associated Phosphopeptides Are the Targets of Memory-like Immunity in Leukemia. Sci. Transl. Med 5, 203ra125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen CJ, Gartner JJ, Horovitz-Fried M, Shamalov K, Trebska-McGowan K, Bliskovsky VV, Parkhurst MR, Ankri C, Prickett TD, Crystal JS, et al. (2015). Isolation of neoantigen-specific T cells from tumor and peripheral lymphocytes. J. Clin. Invest 125, 3981–3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daneshjou R, Wang Y, Bromberg Y, Bovo S, Martelli PL, Babbi G, Lena PD, Casadio R, Edwards M, Gifford D, et al. (2017). Working toward precision medicine: Predicting phenotypes from exomes in the Critical Assessment of Genome Interpretation (CAGI) challenges. Hum. Mutat 38, 1182–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan F, Duitama J, Al Seesi S, Ayres CM, Corcelli SA, Pawashe AP, Blanchard T, McMahon D, Sidney J, Sette A, et al. (2014). Genomic and bioinformatic profiling of mutational neoepitopes reveals new rules to predict anticancer immunogenicity. J. Exp. Med 211, 2231–2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finotello F, Rieder D, Hackl H, and Trajanoski Z (2019). Next-generation computational tools for interrogating cancer immunity. Nat. Rev. Genet 20, 724–746. [DOI] [PubMed] [Google Scholar]
- Fritsch EF, Rajasagi M, Ott PA, Brusic V, Hacohen N, and Wu CJ (2014). HLA-binding properties of tumor neoepitopes in humans. Cancer Immunol. Res 2, 522–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Garijo A, Fajardo CA, and Gros A (2019). Determinants for Neoantigen Identification. Front. Immunol 10, 1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghorani E, Rosenthal R, McGranahan N, Reading JL, Lynch M, Peggs KS, Swanton C, and Quezada SA (2018). Differential binding affinity of mutated peptides for MHC class I is a predictor of survival in advanced lung cancer and melanoma. Ann. Oncol 29, 271–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff SL, Dudley ME, Citrin DE, Somerville RP, Wunderlich JR, Danforth DN, Zlott DA, Yang JC, Sherry RM, Kammula US, et al. (2016). Randomized, Prospective Evaluation Comparing Intensity of Lympho-depletion Before Adoptive Transfer of Tumor-Infiltrating Lymphocytes for Patients With Metastatic Melanoma. J. Clin. Oncol 34, 2389–2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grau J, Grosse I, and Keilwagen J (2015). PRROC: computing and visualizing precision-recall and receiver operating characteristic curves in R. Bioinformatics 31, 2595–2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubin MM, Zhang X, Schuster H, Caron E, Ward JP, Noguchi T, Ivanova Y, Hundal J, Arthur CD, Krebber W-J, et al. (2014). Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature 515, 577–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubin MM, Artyomov MN, Mardis ER, and Schreiber RD (2015). Tumor neoantigens: building a framework for personalized cancer immunotherapy. J. Clin. Invest 125, 3413–3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guinney J, Wang T, Laajala TD, Winner KK, Bare JC, Neto EC, Khan SA, Peddinti G, Airola A, Pahikkala T, et al. ; Prostate Cancer Challenge DREAM Community (2017). Prediction of overall survival for patients with metastatic castration-resistant prostate cancer: development of a prognostic model through a crowdsourced challenge with open clinical trial data. Lancet Oncol 18, 132–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacohen N, Fritsch EF, Carter TA, Lander ES, and Wu CJ (2013). Getting personal with neoantigen-based therapeutic cancer vaccines. Cancer Immunol. Res 1, 11–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, and Weinberg RA (2011). Hallmarks of cancer: the next generation. Cell 144, 646–674. [DOI] [PubMed] [Google Scholar]
- Hellmann MD, Nathanson T, Rizvi H, Creelan BC, Sanchez-Vega F, Ahuja A, Ni A, Novik JB, Mangarin LMB, Abu-Akeel M, et al. (2018). Genomic Features of Response to Combination Immunotherapy in Patients with Advanced Non-Small-Cell Lung Cancer. Cancer Cell 33, 843–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, and Henikoff JG (1992). Amino acid substitution matrices from protein blocks. Proc. Natl. Acad. Sci. USA 89, 10915–10919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoof I, Peters B, Sidney J, Pedersen LE, Sette A, Lund O, Buus S, and Nielsen M (2009). NetMHCpan, a method for MHC class I binding prediction beyond humans. Immunogenetics 61, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins RA, Repo S, Barsky D, Andreoletti G, Moult J, and Brenner SE (2017). Reports from CAGI: The Critical Assessment of Genome Interpretation. Hum. Mutat 38, 1039–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hundal J, Carreno BM, Petti AA, Linette GP, Griffith OL, Mardis ER, and Griffith M (2016). pVAC-Seq: A genome-guided in silico approach to identifying tumor neoantigens. Genome Med 8, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahles A, Lehmann K-V, Toussaint NC, Hüser M, Stark SG, Sachsenberg T, Stegle O, Kohlbacher O, Sander C, and Rätsch G; Cancer Genome Atlas Research Network (2018). Comprehensive Analysis of Alternative Splicing Across Tumors from 8,705 Patients. Cancer Cell 34, 211–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller A, Gerkin RC, Guan Y, Dhurandhar A, Turu G, Szalai B, Mainland JD, Ihara Y, Yu CW, Wolfinger R, et al. ; DREAM Olfaction Prediction Consortium (2017). Predicting human olfactory perception from chemical features of odor molecules. Science 355, 820–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keskin DB, Anandappa AJ, Sun J, Tirosh I, Mathewson ND, Li S, Oliveira G, Giobbie-Hurder A, Felt K, Gjini E, et al. (2019). Neoantigen vaccine generates intratumoral T cell responses in phase Ib glioblastoma trial. Nature 565, 234–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreimer A, Zeng H, Edwards MD, Guo Y, Tian K, Shin S, Welch R, Wainberg M, Mohan R, Sinnott-Armstrong NA, et al. (2017). Predicting gene expression in massively parallel reporter assays: A comparative study. Hum. Mutat 38, 1240–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreiter S, Vormehr M, van de Roemer N, Diken M, Löwer M, Diekmann J, Boegel S, Schrörs B, Vascotto F, Castle JC, et al. (2015). Mutant MHC class II epitopes drive therapeutic immune responses to cancer. Nature 520, 692–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvistborg P, Shu CJ, Heemskerk B, Fankhauser M, Thrue CA, Toebes M, van Rooij N, Linnemann C, van Buuren MM, Urbanus JHM, et al. (2012). TIL therapy broadens the tumor-reactive CD8(+) T cell compartment in melanoma patients. OncoImmunology 1, 409–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagonigro MS, De Cecco L, Carninci P, Di Stasi D, Ranzani T, Rodolfo M, and Gariboldi M (2004). CTAB-urea method purifies RNA from melanin for cDNA microarray analysis. Pigment Cell Res 17, 312–315. [DOI] [PubMed] [Google Scholar]
- Liu D, Schilling B, Liu D, Sucker A, Livingstone E, Jerby-Arnon L, Zimmer L, Gutzmer R, Satzger I, Loquai C, et al. (2019). Integrative molecular and clinical modeling of clinical outcomes to PD1 blockade in patients with metastatic melanoma. Nat. Med 25, 1916–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Łuksza M, Riaz N, Makarov V, Balachandran VP, Hellmann MD, Solovyov A, Rizvi NA, Merghoub T, Levine AJ, Chan TA, et al. (2017). A neoantigen fitness model predicts tumour response to checkpoint blockade immunotherapy. Nature 551, 517–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushita H, Vesely MD, Koboldt DC, Rickert CG, Uppaluri R, Magrini VJ, Arthur CD, White JM, Chen Y-S, Shea LK, et al. (2012). Cancer exome analysis reveals a T-cell-dependent mechanism of cancer immunoediting. Nature 482, 400–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell TJ, Rubinsteyn A, Bonsack M, Riemer AB, Laserson U, and Hammerbacher J (2018). MHCflurry: Open-Source Class I MHC Binding Affinity Prediction. Cell Syst 7, 129–132. [DOI] [PubMed] [Google Scholar]
- Ott PA, Hu Z, Keskin DB, Shukla SA, Sun J, Bozym DJ, Zhang W, Luoma A, Giobbie-Hurder A, Peter L, et al. (2017). An immunogenic personal neoantigen vaccine for patients with melanoma. Nature 547, 217–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng S, Zaretsky JM, Ng AHC, Chour W, Bethune MT, Choi J, Hsu A, Holman E, Ding X, Guo K, et al. (2019). Sensitive Detection and Analysis of Neoantigen-Specific T Cell Populations from Tumors and Blood. Cell Rep 28, 2728–2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajasagi M, Shukla SA, Fritsch EF, Keskin DB, DeLuca D, Carmona E, Zhang W, Sougnez C, Cibulskis K, Sidney J, et al. (2014). Systematic identification of personal tumor-specific neoantigens in chronic lymphocytic leukemia. Blood 124, 453–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen M, Fenoy E, Harndahl M, Kristensen AB, Nielsen IK, Nielsen M, and Buus S (2016). Pan-Specific Prediction of Peptide-MHC Class I Complex Stability, a Correlate of T Cell Immunogenicity. J. Immunol 197, 1517–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richman LP, Vonderheide RH, and Rech AJ (2019). Neoantigen Dissimilarity to the Self-Proteome Predicts Immunogenicity and Response to Immune Checkpoint Blockade. Cell Syst 9, 375–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richters MM, Xia H, Campbell KM, Gillanders WE, Griffith OL, and Griffith M (2019). Best practices for bioinformatic characterization of neoantigens for clinical utility. Genome Med 11, 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson TJ, Freedman JA, Abo MA, Deveaux AE, LaCroix B, Patierno BM, George DJ, and Patierno SR (2019). Alternative RNA Splicing as a Potential Major Source of Untapped Molecular Targets in Precision Oncology and Cancer Disparities. Clin. Cancer Res 25 10.1158/1078-0432.CCR-18-2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg SA, Yang JC, Sherry RM, Kammula US, Hughes MS, Phan GQ, Citrin DE, Restifo NP, Robbins PF, Wunderlich JR, et al. (2011). Durable complete responses in heavily pretreated patients with metastatic melanoma using T-cell transfer immunotherapy. Clin. Cancer Res 17, 4550–4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez-Rodriguez J, Costello JC, Friend SH, Kellen MR, Mangravite L, Meyer P, Norman T, and Stolovitzky G (2016). Crowdsourcing biomedical research: leveraging communities as innovation engines. Nat. Rev. Genet 17, 470–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin U, and Türeci Ö (2018). Personalized vaccines for cancer immunotherapy. Science 359, 1355–1360. [DOI] [PubMed] [Google Scholar]
- Sahin U, Derhovanessian E, Miller M, Kloke B-P, Simon P, Löwer M, Bukur V, Tadmor AD, Luxemburger U, Schrörs B, et al. (2017). Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature 547, 222–226. [DOI] [PubMed] [Google Scholar]
- Salcedo A, Tarabichi M, Espiritu SMG, Deshwar AG, David M, Wilson NM, Dentro S, Wintersinger JA, Liu LY, Ko M, et al. ; DREAM SMC-Het Participants (2020). A community effort to create standards for evaluating tumor subclonal reconstruction. Nat. Biotechnol 38, 97–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samstein RM, Lee C-H, Shoushtari AN, Hellmann MD, Shen R, Janjigian YY, Barron DA, Zehir A, Jordan EJ, Omuro A, et al. (2019). Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat. Genet 51, 202–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano T, and Cantor CR (1990). Expression of a cloned streptavidin gene in Escherichia coli. Proc. Natl. Acad. Sci. USA 87, 142–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlessinger A, Abagyan R, Carlson HA, Dang KK, Guinney J, and Cagan RL (2017). Multi-targeting Drug Community Challenge. Cell Chem. Biol 24, 1434–1435. [DOI] [PubMed] [Google Scholar]
- Schreiber RD, Old LJ, and Smyth MJ (2011). Cancer immunoediting: integrating immunity’s roles in cancer suppression and promotion. Science 331, 1565–1570. [DOI] [PubMed] [Google Scholar]
- Schumacher TN, and Hacohen N (2016). Neoantigens encoded in the cancer genome. Curr. Opin. Immunol 41, 98–103. [DOI] [PubMed] [Google Scholar]
- Schumacher TN, and Schreiber RD (2015). Neoantigens in cancer immunotherapy. Science 348, 69–74. [DOI] [PubMed] [Google Scholar]
- Schumacher TN, Scheper W, and Kvistborg P (2019). Cancer Neoantigens. Annu. Rev. Immunol 37, 173–200. [DOI] [PubMed] [Google Scholar]
- Sidney J, Southwood S, Moore C, Oseroff C, Pinilla C, Grey HM, and Sette A (2013). Measurement of MHC/Peptide Interactions by Gel Filtration or Monoclonal Antibody Capture. Curr. Protoc. Immunol 100, 18.3.1–18.3.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart AC, Margolis CA, Pimentel H, He MX, Miao D, Adeegbe D, Fugmann T, Wong K-K, and Van Allen EM (2018). Intron retention is a source of neoepitopes in cancer. Nat. Biotechnol 36, 1056–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CC, Selitsky SR, Chai S, Armistead PM, Vincent BG, and Serody JS (2019). Alternative tumour-specific antigens. Nat. Rev. Cancer 19, 465–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strønen E, Toebes M, Kelderman S, van Buuren MM, Yang W, van Rooij N, Donia M, Böschen M-L, Lund-Johansen F, Olweus J, and Schumacher TN (2016). Targeting of cancer neoantigens with donor-derived T cell receptor repertoires. Science 352, 1337–1341. [DOI] [PubMed] [Google Scholar]
- Toebes M, Coccoris M, Bins A, Rodenko B, Gomez R, Nieuwkoop NJ, van de Kasteele W, Rimmelzwaan GF, Haanen JBAG, Ovaa H, and Schumacher TN (2006). Design and use of conditional MHC class I ligands. Nat. Med 12, 246–251. [DOI] [PubMed] [Google Scholar]
- Tran E, Turcotte S, Gros A, Robbins PF, Lu Y-C, Dudley ME, Wunderlich JR, Somerville RP, Hogan K, Hinrichs CS, et al. (2014). Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science 344, 641–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran E, Robbins PF, Lu Y-C, Prickett TD, Gartner JJ, Jia L, Pasetto A, Zheng Z, Ray S, Groh EM, et al. (2016). T-Cell Transfer Therapy Targeting Mutant KRAS in Cancer. N. Engl. J. Med 375, 2255–2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran E, Robbins PF, and Rosenberg SA (2017). “Final common pathway” of human cancer immunotherapy: targeting random somatic mutations. Nat. Immunol 18, 255–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vita R, Mahajan S, Overton JA, Dhanda SK, Martini S, Cantrell JR, Wheeler DK, Sette A, and Peters B (2019). The Immune Epitope Database (IEDB): 2018 update. Nucleic Acids Res 47 (D1), D339–D343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitiello A, and Zanetti M (2017). Neoantigen prediction and the need for validation. Nat. Biotechnol 35, 815–817. [DOI] [PubMed] [Google Scholar]
- Yadav M, Jhunjhunwala S, Phung QT, Lupardus P, Tanguay J, Bumbaca S, Franci C, Cheung TK, Fritsche J, Weinschenk T, et al. (2014). Predicting immunogenic tumour mutations by combining mass spectrometry and exome sequencing. Nature 515, 572–576. [DOI] [PubMed] [Google Scholar]
- Yamamoto TN, Kishton RJ, and Restifo NP (2019). Developing neoantigen-targeted T cell-based treatments for solid tumors. Nat. Med 25, 1488–1499. [DOI] [PubMed] [Google Scholar]
- Yang W, Lee K-W, Srivastava RM, Kuo F, Krishna C, Chowell D, Makarov V, Hoen D, Dalin MG, Wexler L, et al. (2019). Immunogenic neoantigens derived from gene fusions stimulate T cell responses. Nat. Med 25, 767–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarchoan M III, Johnson BA 3rd, Lutz ER, Laheru DA, and Jaffee EM (2017). Targeting neoantigens to augment antitumour immunity. Nat. Rev. Cancer 17, 209–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee C, Thompson JA, Byrd D, Riddell SR, Roche P, Celis E, and Greenberg PD (2002). Adoptive T cell therapy using antigen-specific CD8+ T cell clones for the treatment of patients with metastatic melanoma: in vivo persistence, migration, and antitumor effect of transferred T cells. Proc. Natl. Acad. Sci. USA 99, 16168–16173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharakis N, Chinnasamy H, Black M, Xu H, Lu Y-C, Zheng Z, Pasetto A, Langhan M, Shelton T, Prickett T, et al. (2018). Immune recognition of somatic mutations leading to complete durable regression in metastatic breast cancer. Nat. Med 24, 724–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Editorial (2017). The problem with neoantigen prediction. Nat. Biotechnol 35, 97. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
TESLA data is available here: https://www.synapse.org/#!Synapse:syn21048999. All molecular data (WES and RNA) from the melanoma cohort can be found in dbGap (accession number phs000452.v3.p1) and clinical variables can be found as supplemental information in the associated manuscript (Liu et al., 2019). Code to recreate the scoring metrics used in this study is included as a supplemental file to this manuscript.