Abstract
Aims and Objectives:
The aim and objective of this study is to detect invasive fungal infections (IFIs) early and with more sensitivity by the nested polymerase chain reaction (PCR) for fungus as compared to fungal culture in clinically suspected patients and also explore its correlation in reference to age, duration of symptoms, immunocompromised status, and other risk factors predisposing to IFIs.
Materials and Methods:
In this cross-sectional study, 50 suspected patients admitted in medical acute care unit/intensive care unit (ACU/ICU) of Sir Sunderlal Hospital, Banaras Hindu University, Varanasi, India, comprised the study. All cases were selected based on the predefined inclusion and exclusion criteria. A detailed history, clinical examination, and all required investigations were done in all suspected patients. Blood samples were taken for nested-PCR for fungus and culture. Nested PCR was performed on extracted DNA form samples collected from all participants under the study.
Results:
Our study comprised of 50 suspected immunocompromised patients of IFIs. Among the participants under the study, the most common risk factor was diabetes mellitus (28% cases). Nearly two-thirds (60%) of the cases were 50 years or more. Around 70% of the cases had a history of illness more than 2 weeks. Nested PCR for fungus came out to be positive in 21/50 patients (42%); however, fungal culture was positive in none. Among the admitted patient in ACU/ICU, 75% were neutropenic.
Conclusions:
IFIs are more common in immunocompromised individuals, patients with comorbidities, long history of symptoms, and elderly population. Nested PCR for fungus has a high sensitivity (as compared to the fungal culture), and also they are rapid in giving the results. Thus, nested PCR for fungus can be used in a cost-effective manner for the early and reliable diagnosis of clinically suspected IFIs.
Keywords: Febrile neutropenia, invasive fungal infection, polymerase chain reaction
Introduction
Fungi are eukaryotic organisms and infection do not occur in individual with normal immunity. The most important risk factor leading to predisposition for an IFI is the presence of a compromised immunity. The most prevalent of the reasons for this compromised immunity are the presence of febrile neutropenia, hematological malignancy, solid-organ transplantation, acquired immunodeficiency syndrome (AIDS), prolonged use of steroids, other immunemodulator drugs, and diabetes mellitus. There are multiple virulence factors which help the fungi invade the human system and evade the host-defence mechanism such as the presence of chitin in their cell wall and release of certain proteolytic enzymes among others.
Not all fungal infections are invasive. Fungi normally colonize human mucosal surfaces. When there is a breach in the immunity, fungi invade the host and cause an invasive fungal infection (IFI). The European Organization for Research and Treatment of Cancer/IFIs Cooperative Group has put up a definition for IFIs. The term IFI is used only to characterize systemic, generalized, deep seated, visceral, and severe life-threatening fungal infections, in contrast to superficial, local, benign, self-limiting fungal diseases. Definition of invasive fungal disease, as per ISHLT infectious diseases “presence of fungus in the respiratory secretions (sputum or broncho-alveolar lavage [BAL]) detected by the culture, polymerase chain reaction (PCR), or biomarker (GM/cryptococcal antigen) in the presence of symptoms, radiologic, and endobronchial changes, or presence of histological changes consistent with fungal invasion of the tissue.[1]
The diagnosis of IFIs is difficult because of the lack of specific clinical signs and symptoms until late. The difficulty associated with making a diagnosis with current diagnostic tools in developing country, obtaining infected tissue/sample required to establish a specific diagnosis, and in some cases defining the isolated agent's sensitivity to the currently available therapeutic regimen being advocated in a given patient.
In our country, there have been inadequacies in the availabilities of nonculture diagnostics, access to antifungal drugs, and local guidelines as the major gaps in the management of IFIs. These gaps are targets for improvement.
Moreover, diagnostic test available for diagnosing IFIs has not been sensitive enough to catch them. The conventional tests such as routine microscopy and fungal culture are not much sensitive and specific to diagnose. Therefore, there has been always a growing need for tests which can diagnose IFIs with much accuracy and rapidity. Molecular methods such as PCR which is not only a rapid but highly sensitive method, unlike fungal culture which can take many days to give the result, has made it a very a popular and a reliable method to diagnose IFIs.
This study includes all those patients who can be suspected to have IFIs either clinically or by routine investigations including radiological investigations and then these patients’ sample would be subjected to fungal culture and nested PCR.
Materials and Methods
This cross-sectional study was conducted among 50 patients medical acute care unit/intensive care unit (ACU/ICU) in Sir Sunder Lal Hospital, Varanasi, India, from October 2017 to June 2019. All cases were selected based on the inclusion and exclusion criteria. A detailed history, clinical examination, and all required investigations were done for all patients under the study. The present study was approved by the Institutional Ethical Committee.
Study population
Inclusion criteria
Patients with age more than 14 years with a high suspicion of fungal infection based on history, clinical parameters, and other routine investigations were included in the study.
Exclusion criteria
Patients who had already taken any antifungal agent whether prophylactically or therapeutically were excluded from the study.
Methods and procedure
Written informed consent was obtained from each patient. For patients who were too ill to be communicated, permission to enrol the patients in the study was obtained from the next of kin
Particulars that were collected at admission included age, gender, and medical history
All patients underwent a general physical examination and a systemic examination at the time of admission
Routine biochemical investigations for all patients included complete blood count, liver function tests, renal function tests, and Galactomannan assay
Whole blood samples were collected for culture and PCR
Wherever required other fluids such as urine, cerebrospinal fluid (CSF), sputum, pus, BAL, pleural tap, ascitic tap, or biopsy were collected
Computed tomography (CT) of the thorax or abdomen was performed in all patients with suspected fungal lesions either clinical and or radiological basis
Finally, clinical, radiological, microbiological, and other laboratory data were evaluated and compared in the diagnosis of IFIs.
DNA isolation from whole blood sample
Procedure
500 μl of blood sample was taken into an Eppendorf tube and mixed well
Equal volume of Tris-ethylenediaminetetraacetic acid buffer was added having a pH of 8 to the blood sample
The above mixture was incubated (in a hot water bath) at 65°C–70°C for about 60 min
Then, 50–60 μl of 10% Sodium dodecyl sulfate was added to the above mixture
2–3 μl of Proteinase K was added to the above mixture and mixed well
It was then incubated for 24 h at 37°C
5M NaCl (sodium chloride) solution was then added to the mixture
To this, 100–120 μl of cetyl trimethylammonium bromide was added and mixed well and incubated (in a hot water bath) at 65°C–70°C for 30–45 min
P: C: I (Phenol: Chloroform: Isoamyl alcohol) (25:24:1) was added to the above sample and gently mixed
Above mixture was centrifuged at 10,000 rpm. Aqueous phase was collected in a fresh Eppendorf tube. Equal volume of C: I (Chloroform: Isoamyl alcohol) (24:1) was added and mixed well
The mixture was centrifuged at 10,000 rpm. Aqueous phase was collected in a fresh Eppendorf tube
Equal volume of chilled iso-propanol was added. Turbidity appeared (i.e., DNA precipitating). It was left for 5 min at the room temperature. It was then centrifuged at 10,000 rpm, and the aqueous phase was discarded
Pellets were collected and washed with 70% ethanol (three times). It was let to dry and stored in TE buffer (pH 8).
Quantification and qualitative analyses of isolated DNA
Quantification and qualification was done with the help of nanodrop spectrophotometer.
Quality checking of DNA to carry out polymerase chain reaction
Sometimes, DNA gets degraded during storage. Hence, before proceeding to PCR, it is important to check whether DNA is intact or degraded. This was checked by Agarose gel electrophoresis. If DNA remained intact within or near the well, it meant it was not degraded but if smear formed it meant DNA had degraded.
Polymerase chain reaction amplification of target gene
At first, fungal species were identified with the help of primer internal transcribed spacer (ITS) 1 and ITS4 which are universal forward and reverse primers, respectively. Then, primers were taken that target the ITS region of rDNA, i.e., ITS1 and ITS2.
Primary cycle
Sample was subjected to initial denaturation at 95°C for 7 min. Followed by 35 cycles of denaturation at 95°C for 30 s, annealing at 55°C for 1 min, and extension was given at 72°C for 45 s and final extension of 10 min at 72°C.
Primers were ITS1 and ITS4. These amplify the partial sequence of 18s rRNA gene plus complete sequence of ITS1 region plus complete sequence of 5.8S rRNA plus complete sequence of ITS 4 region and partial sequence of 28S rRNA gene. These are universal fungal oligonucleotide primers.
Secondary cycle or (semi-nested cycle)
PCR mixture for secondary run was subjected to denaturation at 95°C for 7 min followed by 35 cycles of denaturation at 95°C for 30 s, annealing at 55°C for 1 min, and extension at 72°C for 45 s and final extension was at 72°C for 7 min. Seminested primers were ITS1 and ITS2 forward and reverse primers, respectively. These primers would amplify the region of 18S rRNA + 5.8SrDNA and 5.8S + 28S rDNA genes, respectively.
Gel electrophoresis
Agarose gel electrophoresis was performed to check the amplified segments of nucleic acid.
Statistical methods
The statistical analysis was performed using the SPSS Statistics software version 16.0. SPSS(Version 23.0;SPSS Inc., Chicago, IL, USA). The data were presented as mean ± standard deviation for parametric continuous variables and frequency with their respective percentages for the categorical variables.
Observations
In this study, 10% patients were of <40 years of age, 30% were between 41 and 50 years of age, 26% were between 51 and 60 years, whereas 34% patients were of more than 60 years of age. Thus, in our study, 60% (26% +34%) patients were of more than 50 years of age [Figure 1]. In this study, male patients outnumbered the females; males being 58%.
Figure 1.
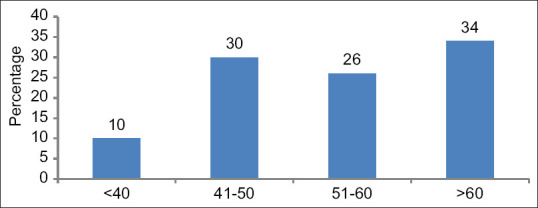
Age distribution (%) of the patients
There were 30% patients who had symptom duration between 7 and 14 days, 34% had a history between 15 and 28 days, and 36% patients had symptom duration of more than 28 days. Thus, majority of the patients i.e., 70% (36%+34%) had a history of more than 15 days [Figure 2].
Figure 2.
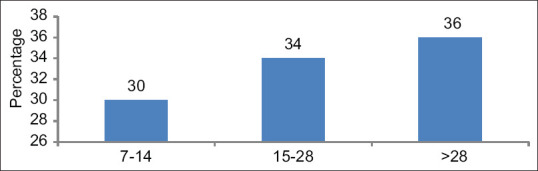
Percentage distribution according to the duration of symptoms
Among the 50 patients, most of the patients were found to be immunocompromised due to some predisposing factors most common being diabetes mellitus, medical ACU/ICU admission, and immunosuppressant therapy among others [Figure 3].
Figure 3.
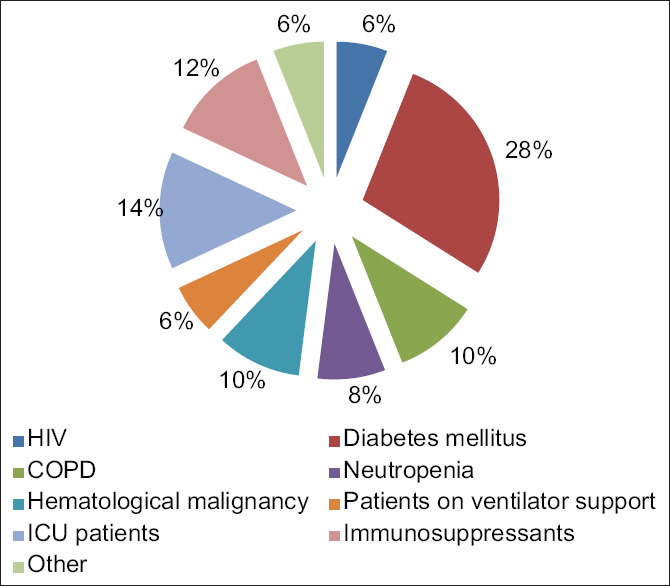
Predisposing factors and its distribution
In our study, the mean duration of symptoms among 50 cases was 28.16 ± 19.221 days. The mean age was 54.14 ± 13.86 years, whereas the mean total leukocyte count (TLC) was 21235.48 ± 72930.435 cells/μL [Table 1].
Table 1.
Clinical and laboratory parameters (n=50)
| Minimum | Maximum | Mean±SD | |
|---|---|---|---|
| Duration | 7 | 90 | 28.16±19.221 |
| Age (years) | 20 | 81 | 54.14±13.864 |
| TLC (cells/µL) | 490 | 522000 | 21235.48±72930.435 |
SD=Standard deviation, TLC=Total leukocyte count
Among 50 patients, 42% came out to be PCR positive for fungus, whereas none came out to be fungal culture positive [Table 2].
Table 2.
Frequency of nested polymerase chain reactionpositive patients for fungus (n=50)
| Fungal PCR | Frequency (%) |
|---|---|
| Negative | 29 (58.0) |
| Positive | 21 (42.0) |
| Total | 50 (100.0) |
PCR=Polymerase chain reaction
On analysis of risk factors involved in PCR positive for fungus patients (21/50), we found that diabetes mellitus was major risk factor (28%) followed by ICU admission with or without ventilator. As per expectation rate of IFI was more common among neutropenics and those admitted in ICU for other reasons [Figure 4 and Table 3].
Figure 4.
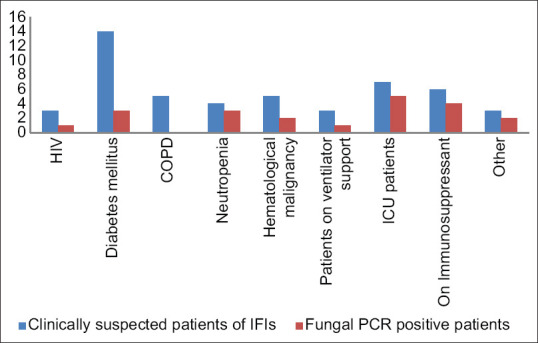
Predisposing factors and polymerase chain reaction positivity for fungus
Table 3.
Predisposing factors and polymerase chain reaction positivity for fungus
| Risk factors | Clinically suspected patients of IFIs | PCR-positive patients | Proportion of PCR-positive patients (%) within risk factor |
|---|---|---|---|
| HIV | 3 | 1 | 33.33 |
| Diabetes mellitus | 14 | 3 | 21.42 |
| COPD | 5 | 0 | 0 |
| Neutropenia | 4 | 3 | 75.0 |
| Hematological malignancy | 5 | 2 | 40.0 |
| Patients on ventilator support | 3 | 1 | 33.33 |
| ICU patients | 7 | 5 | 71.42 |
| On immunosuppressant | 6 | 4 | 66.67 |
| Others | 3 | 2 | 66.67 |
| Total | 50 | 21 | 42.0 |
COPD=Chronic obstructive pulmonary disease, ICU=Intensive care unit, PCR=Polymerase chain reaction, IFIs=Invasive fungal infections
Around 67% (28.6 + 38.1) of PCR-positive patients for fungus were of >50 years [Table 4] and 81% of the PCR-positive patients had a history of more than 15 days.
Table 4.
Age distribution among polymerase chain reaction-positive patients for fungus
| Age | n (%) |
|---|---|
| <40 | 2 (9.5) |
| 41-50 | 5 (23.8) |
| 51-60 | 6 (28.6) |
| >60 | 8 (38.1) |
| Total | 21 (100) |
Discussion
The term IFIs is used to characterize systemic, generalized, deep-seated, visceral, and severe life-threatening fungal infections. These have a very high rate of morbidity and mortality if not diagnosed early and treated promptly.[2,3]
The incidence of IFIs has been increasing over the last few decades. This is mainly because of increased number of immunocompromised population like those of AIDS, increased number of patients with hematological and solid organ malignancies, increased number of patients undergoing haematopoietic stem-cell transplantation, and solid organ transplantation.[4,5,6]
The diagnosis of IFIs has always been a problem. The conventional methods of diagnosis such as fungal culture and others have a very low sensitivity and also take a very long time in giving the results. Therefore, newer methods were needed which can diagnose these IFIs with high sensitivity and also with accuracy. With the advent of molecular methods in diagnostics, those were also applied in the field of diagnosing fungal infections. PCR for fungus is one of these newer molecular methods which have high sensitivity and rapidity in the diagnosis of IFIs.
Our study comprised of 50 patients, suspected to be having IFIs either clinically and or from routine investigations including radiological. To confirm their diagnosis, samples for fungal culture and PCR were sent to our department microbiology laboratory. As with all IFI studies climate, demographics and sample size significantly influence the results. In our study, 58% of the cases were male, 42% were female, and none was transgender. In a similar study done in 2015 by Magda et al.,[7] around 65% were male, whereas around 35% were female.
The age ranged from 20 to 81 years, whereas the mean of the age being 54.14 ± 13.86 years suggesting preponderance of IFIs in aging population, 60% of our patients were of more than 50 years of age. This is similar to the studies done by Meersseman et al.[8] and Montagna et al.[9] where maximum numbers of cases of IFIs were of older age group (60 years or more). In a study by Soliman et al. (2012),[10] patients with hematological malignancies, the incidence of IFIs was higher in younger individuals similar to our study.
Among the host factors predisposing for fungal infections, in our study, out of all the cases 28% were diabetic, 14% were ICU admitted patients, 12% were on immunosuppressants, and 10% were known cases of chronic obstructive pulmonary disease and hematological malignancies each, 8% were neutropenic, whereas 6% were on mechanical ventilation and 6% were HIV-positive patients. In 2010, Slavin et al.[11] observed that IFIs are more common with patient on prolonged (>3 weeks) corticosteroid therapy.
None of our patient samples was positive on fungal culture despite repetition. Isolation of fungus from culture as such is a cumbersome process. The sample has to be collected from sterile sites such as blood, CSF, or from the tissues. It has a very poor sensitivity and often repeated blood cultures have to be done to get a growth of the fungus. Moreover, filamentous fungi such as Aspergillus are rarely isolated from blood. Therefore, a negative culture does not exclude the possibility of having a fungal infection.[12] This is an important drawback associated with the conventional diagnostic techniques.
To overcome this problem, newer tests have been developed to ease the fungal diagnostics. Fungal PCR is one of such test which can detect fungal DNA even from nonviable cells with good sensitivity and specificity.[13] In our study, 42% of the patients came out to be fungal PCR positive. In a study done by Ribeiro et al., (2006),[14] only 35% of the patients came out to be fungal PCR positive. In a study done by Badiee et al.,[2] 17.7% patients were positive for fungal PCR. While in a study done by El-Sayed et al.,[15] around 57% patients were positive for fungal PCR. This variation in the results in various studies can be attributed to the different sample size in each study, demographics, difference in host factors, sample collection methods, and DNA extraction methods.
Fungal culture remains the gold standard for the diagnosis of IFIs, but it takes a very long time to give the result and has a poor sensitivity, while fungal PCR is a method which can be used to diagnose IFIs early and with a higher sensitivity. Now with advent NGS, the diagnosis of IFIs can be more accurate.
There is a poor agreement between fungal culture and fungal PCR in our study. Similar disagreement was also seen in the study of El-Sayed et al.[15] They explained the disparity between the results of fungal culture and PCR to the poor sensitivity of the culture method and the ability of the PCR method to detect even the DNA of the fungus which has been phagocytised by the circulating blood cells.[16]
Eighty-one percent of the PCR-positive patients were found to have a history of their clinical symptoms of more than 15 days. It will not be prophesy but reality, now this is the era of virus and fungus infections and bacterial infection has taken back seat for a while. Hence, in my opinion, any critically ill febrile patient admitted to ICU/medical ACU should all be subjected to PCR for fungus as an additional investigation for better treatment outcome and reduced the prolonged hospitalization. Further, it will be prudent that in all suspect immunocompromised febrile patient having illness of >15 days must be subjected to PCR for fungus. Unnecessary, nonjudicious prolonged use of antibiotics at periphery will not only produce antibiotic resistance but make patients more prone to fungal infections surfaced due to suppressed defence in prolonged illness.
In this era of transplant, increasing use of biologics, immune modulatory drugs molecular diagnostics for IFIs is the need of hour and should be practiced more frequently in relevant settings.[17] In IFIs considering the host characteristics, the use of novel diagnostic tools and use of currently available therapeutics will surely cut down the mortality and morbidity.[18]
Conclusions
FIs are more common in immunocompromised individuals, older individuals, patients with comorbidities, and with a prolonged history. The most common symptoms with which they present is fever and cough and also commonly have a high TLC
Nested PCR for fungus was positive in 42% of patients clinically suspected to have IFIs. Thus, PCR has a high sensitivity (compared to fungal culture) and also is rapid in giving the results. PCR positivity is increased in patients with a long history or certain subsets of patients like those with neutropenia or those who are admitted in the ICU
Further, it will be prudent that in all suspect immunocompromised febrile patient having illness of >15 days must be subjected to PCR for fungus for early and reliable diagnosis and judicious use of available therapeutic.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
References
- 1.Husain S, Mooney ML, Danziger-Isakov L, Mattner F, Singh N, Avery R, et al. A 2010 working formulation for the standardization of definitions of infections in cardiothoracic transplant recipients. J Heart Lung Transplant. 2011;30:361–74. doi: 10.1016/j.healun.2011.01.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Badiee P, Kordbacheh P, Alborzi A, Malekhoseini S, Ramzi M, Mirhendi H, et al. Study on invasive fungal infections in immunocompromised patients to present a suitable early diagnostic procedure. Int J Infect Dis. 2009;13:97–102. doi: 10.1016/j.ijid.2008.04.011. [DOI] [PubMed] [Google Scholar]
- 3.De Pauw B, Walsh TJ, Donnelly JP, Stevens DA, Edwards JE, Calandra T, et al. Revised definitions of invasive fungal disease from the European Organization for Research and Treatment of Cancer/Invasive Fungal Infections Cooperative Group and the National Institute of Allergy and Infectious Diseases Mycoses Study Group (EORTC/MSG) Consensus Group. Clin Infect Dis. 2008;46:1813–21. doi: 10.1086/588660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baddley JW. Clinical risk factors for invasive aspergillosis. Med Mycol. 2011;49(Suppl 1):S7–12. doi: 10.3109/13693786.2010.505204. [DOI] [PubMed] [Google Scholar]
- 5.Badiee P, Alborzi A, Farhoudi F. A case of Candida mediastinitis after dental extraction. J Infect Dev Ctries. 2011;5:75–8. doi: 10.3855/jidc.1086. [DOI] [PubMed] [Google Scholar]
- 6.Pagano L, Caira M, Candoni A, Offidani M, Fianchi L, Martino B, et al. The epidemiology of fungal infections in patients with hematologic malignancies: The SEIFEM-2004 study. Haematologica. 2006;91:1068–75. [PubMed] [Google Scholar]
- 7.Azab MM, Abo Taleb AF, Mohamed NA, Omran FH. Rapid diagnosis of invasive fungal infections. Int J Curr Microbiol App Sci. 2015;4:470–86. [Google Scholar]
- 8.Meersseman W, Vandecasteele SJ, Wilmer A, Verbeken E, Peterman's WE, van Wijngaerden E. Invasive aspergillosis in critically ill patients without malignancy. Am J Respir Crit Care Med. 2004;170:621–5. doi: 10.1164/rccm.200401-093OC. [DOI] [PubMed] [Google Scholar]
- 9.Montagna MT, Caggiano G, Lovero G, De Giglio O, Coretti C, Cuna T, et al. Epidemiology of invasive fungal infections in the intensive care unit: Results of a multicenter Italian survey (AURORA Project) Infection. 2013;41:645–53. doi: 10.1007/s15010-013-0432-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soliman MH, Azab MM, Abu Taleb FM, Mohamed NA. Identification of different types of fungi isolated from patients with hematologic malignancy and from their surrounding exogenous sources. Zagazig Univ Med J. 2002;18:951–61. [Google Scholar]
- 11.Slavin MA, Sorrell TC, Marriott D, Thursky KA, Nguyen Q, Ellis DH, et al. Candidaemia in adult cancer patients: Risks for fluconazole-resistant isolates and death. J Antimicrob Chemother. 2010;65:1042–51. doi: 10.1093/jac/dkq053. [DOI] [PubMed] [Google Scholar]
- 12.Badiee P, Hashemizadeh Z. Opportunistic invasive fungal infections: Diagnosis & amp; clinical management. Indian J Med Res. 2014;139:195–204. [PMC free article] [PubMed] [Google Scholar]
- 13.Chen SC, Ftnillday CL, Meyer W. A review of nucleic acid-based diagnostic tests for systemic mycosis with an emphasis on polymerase chain based assays. Med Mycol. 2001;40:333–57. doi: 10.1080/mmy.40.4.333.357. [DOI] [PubMed] [Google Scholar]
- 14.Ribeiro P, Costa F, Monteiro A, Caldas J, Silva M, Ferreira G, et al. Polymerase chain reaction screening for fungemia and/or invasive fungal infections in patients with hematologic malignancies. Support Care Cancer. 2001;14:469–74. doi: 10.1007/s00520-005-0903-7. [DOI] [PubMed] [Google Scholar]
- 15.El-Sayed ZA, Hasan ZE, Nasr RA. Real-Time PCR in the early detection of invasive fungal infection in immune-deficient infants and children. Egypt J Pediatr Allergy Immunol. 2012;10:67–74. [Google Scholar]
- 16.Ahmaad S, Khan Z, Mustafa A, Khan ZU. Semi-nested PCR for diagnosis of candidemia: Comparison with culture, antigen detection and biochemical methods for species identification. J Clin Microbiol. 2002;40:2483–89. doi: 10.1128/JCM.40.7.2483-2489.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kidd SE, Chen SCA, Meyer W, Halliday CL. A New Age in Molecular Diagnostics for Invasive Fungal Disease. Are We Ready Front Microbiol. 2020;10:2903. doi: 10.3389/fmicb.2019.02903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stohs E, Zimmer A. An approach to suspected invasive fungal infection in patients with hematologic malignancy and HCT recipients with persistent neutropenic fever despite mold-active prophylaxis. Curr Fungal Infect Rep. 2020;14:89–98. [Google Scholar]


