Abstract
Purpose:
To optimize diffusion-relaxation MRI with tensor-valued diffusion encoding for precise estimation of compartment-specific fractions, diffusivities and T2 values with a two-compartment model of white matter, and to explore the approach in vivo.
Methods:
Sampling protocols featuring different b-values (b), b-tensor shapes (bΔ), and echo times (TE) were optimized using Cramér-Rao lower bounds (CRLB). Whole-brain data were acquired in children, adults, and elderly with white matter lesions. Compartment fractions, diffusivities and T2 values were estimated in a model featuring two microstructural compartments represented by a ‘stick’ and a ‘zeppelin’.
Results:
Precise parameter estimates were enabled by sampling protocols featuring seven or more ‘shells’ with unique b/bΔ/TE-combinations. Acquisition times were approximately 15 minutes. In white matter of adults, the ‘stick’ compartment had a fraction of approximately 0.5 and, compared with the ‘zeppelin’ compartment, featured lower isotropic diffusivities (0.6 vs. 1.3 μm2/ms) but higher T2 values (85 vs. 65 ms). Children featured lower ‘stick’ fractions (0.4). White matter lesions exhibited high ‘zeppelin’ isotropic diffusivities (1.7 μm2/ms) and T2 values (150 ms).
Conclusions:
Diffusion-relaxation MRI with tensor-valued diffusion encoding expands the set of microstructure parameters that can be precisely estimated and therefore increases their specificity to biological quantities.
Keywords: tensor-valued diffusion encoding, diffusion-relaxation MRI, Fisher information, brain microstructure
1 |. INTRODUCTION
Diffusion MRI (dMRI) has gained widespread use due to its sensitivity to brain tissue microstructure (1). Methods such as diffusion tensor imaging (DTI) (2) and diffusion kurtosis imaging (DKI) (3) detect microstructural alterations in diseases such as multiple sclerosis (4), white matter lesions (5, 6), gliomas (7) and parkinsonian disorders (8) as well as in normal processes such as learning (9, 10) and maturation (11–13). Although sensitive, DTI and DKI parameters do not report on specific quantities of the tissue microstructure (14, 15). Recent research has therefore aimed to develop methods with specificity to microstructural quantities such as the axonal density.
One approach to estimate microstructural quantities from dMRI data is to use a forward model that parameterizes the signal in terms of such quantities and fit the model to acquired data (1). Such models tend to represent tissue by ‘compartments’ with specific signal contributions weighted by fractions (16–28). In white matter, water diffusion in the intra- and extra-axonal spaces are typically modelled by ‘stick’ and ‘zeppelin’ diffusion tensors, respectively. Correlations have been found between parameters of such models and microstructural quantities from histology. For example, the ‘stick’ fraction (the fraction of the ‘stick’ compartment) has been correlated with the axonal volume fraction (29) and the radial diffusivity of the ‘zeppelin’ compartment has been correlated with the myelin volume fraction (30).
While a complete description of the tissue microstructure requires many parameters, dMRI normally supports the estimation of just a few (31–33). Trying to estimate many microstructural parameters from dMRI data leads to problems with degeneracy and low precision, which has been addressed by the introduction of various model constraints. For example, compartment diffusivities may be fixed to specific values or assumed to follow given relations (16, 23, 27). Although such constraints increase precision, they introduce another problem: the estimated parameters represent microstructural quantities truthfully only when the constraints are valid. For example, interpreting the ‘stick’ fraction as a volume fraction without estimating compartment-specific T2 relaxation values is valid only if these values are equal. If they are different, as in white matter lesions, the results can be strongly misleading (28).
To estimate microstructural quantities across a wide range of conditions, model constraints should be replaced with independent information. Here we explore two approaches for acquiring such information. The first is tensor-valued diffusion encoding, which adds information by acquiring data with multiple shapes of the b-tensor (34–36). Such data can be used to estimate all parameters in the relatively unconstrained ‘standard model’ (37, 38). The second is diffusion-relaxation experiments, which adds information by acquiring diffusion data for multiple echo times (TE) (39–42). Such data can be used to estimate compartment-specific T2 values in brain tissue (28, 40). These approaches have previously been used separately but not in combination.
In this work, we investigate how to best combine tensor-valued diffusion encoding and diffusion-relaxation experiments for maximal information encoding. For this purpose, we defined sampling protocols in terms of ‘shells’ with different b-values (b), shapes of the b-tensor (bΔ) and echo times (TE). A minimally constrained two-compartment model was used to optimize protocols for minimal parameter variance based on Cramér-Rao lower bounds (CRLB) (43, 44). The importance of each independent acquisition dimension was assessed, and the performance of different sampling protocols was evaluated. The clinical feasibility of the approach was tested by using the principles learned from the optimization to design an in vivo protocol and acquire data in children, adults, and elderly with white matter lesions.
2 |. THEORY
2.1 |. A general diffusion-relaxation compartment model
We start by considering the class of models (18, 19, 29, 33, 40, 45–47) that describe the dMRI signal as an ‘isotropic convolution’ () (48) on the unit sphere between an axisymmetric microstructure kernel K and an orientation distribution function (ODF) P, according to
| Eq. 1 |
where u is the symmetry axis of the diffusion encoding and n is the symmetry axis of the kernel.
To define a general compartment model of diffusion and T2 relaxation, we consider kernels on the form
| Eq. 2 |
The experiment is described by the axisymmetric diffusion-encoding tensor (b-tensor) B with symmetry axis u (35, 36) and the echo time TE. Each compartment is described by an axisymmetric diffusion tensor Dj and a T2 value T2;j, and is weighted by a fraction fj (Σfj = 1). Assuming that the model captures the diffusion and T2 relaxation properties of all relevant compartments, the fractions theoretically represent the proton density- and T1-weighted volume fractions of those compartments.
As in Eriksson (49), we parameterize D in terms of its isotropic diffusivity DI and anisotropy DΔ, which relate to the axial (D||) and radial (D⊥) diffusivities according to DI = (D|| + 2D⊥)/3 and DΔ = (D|| – D⊥)/(D|| + 2D⊥). The double inner product B:D is given by
| Eq. 3 |
where b is the conventional b-value (50), bΔ is the shape of the b-tensor, which ranges from –0.5 (planar tensor encoding; PTE) through zero (spherical tensor encoding; STE) to unity (linear tensor encoding; LTE), and L2 is the second Legendre polynomial (49).
We simplify the convolution in Eq. 1 by expanding K and P in the spherical harmonic basis, according to
| Eq. 4 |
for order l = 0, 2, …, and degree m = –l, –l + 1, …, l. With u = (sinθ cosφ, sinθ sinφ, cosθ), the spherical harmonic basis functions are given by
| Eq. 5 |
where are the associated Legendre polynomials (m = 0 yields the regular Legendre polynomials) (48, 51). The coefficients for K (kl0) and P (plm) are obtained by inner products, according to
| Eq. 6 |
where the overbar denotes complex conjugation. Eq. 4 uses the axial symmetry of K. First, K(θ,φ) = K(θ), wherefore klm = 0 for m ≠ 0. Second, K(–θ) = K(θ), wherefore kl0 ≠ 0 only for even l (odd-order harmonics are antisymmetric). Eqs. 2–6 yield kl0 (referred to as kl), according to
| Eq. 7 |
where
| Eq. 8 |
and
| Eq. 9 |
For the first two harmonic orders, the integral evaluates to
| Eq. 10 |
and
| Eq. 11 |
Note that fitting the l = 0 term in Eq. 4 to the signal’s zeroth order coefficient 〈S|Y00〉 is equivalent to the ‘powder-averaging’ approach (52–54), and that jointly fitting the l = 2 terms to the second order coefficients 〈S|Y2m〉 (after transformation to rotational invariants) yields the ‘RotInv’ approach (33, 40, 46).
2.2 |. Two-compartment diffusion-relaxation model
We define a two-compartment diffusion-relaxation model by limiting the kernel (Eq. 2) to feature a ‘stick’ compartment (S) with DΔ;S = 1 (D⊥;S = 0) and a ‘zeppelin’ compartment (Z). The ‘stick’ property of zero radial diffusivity (16) is expected for diffusion within cylindrical structures having a radius smaller than the resolution limit (55) and has been experimentally indicated in white matter (53, 56, 57). Using Eqs. 4–11 with the harmonic expansion truncated at lmax = 2, and p00 = Y00 = 1/ for ODF normalization, yields the full signal equation
| Eq. 12 |
where m ∈ {–2, –1, 0, 1, 2}. It is a function of five scalar experiment-related parameters,
| Eq. 13 |
where θ and φ are the colatitude and longitude of the b-tensor symmetry axis, and twelve scalar model parameters,
| Eq. 14 |
where fS is the ‘stick’ fraction, DI;S and DI;Z are the isotropic diffusivities of the ‘stick’ and ‘zeppelin’, DΔ;Z is the ‘zeppelin’ shape, T2;S and T2;Z are the T2 values of the ‘stick’ and ‘zeppelin’, respectively. The requirement of a real ODF enforces conjugate symmetry, according to p2-m = (–1)m, leaving five real numbers describing it. As in Novikov (33), we use the rotational invariant Euclidean norm of the p2m coefficients to derive an ‘orientation coherence’ parameter, according to
| Eq. 15 |
The normalization ensures pl ∈ [0 1]. Since pl is rotational invariant and increases with ODF anisotropy, pl = plmax when P’ = δ(θ = 0)/(4π), where δ is the delta function. With P’ plugged into Eq. 6, Eq. 15 yields plmax = 4π, since Ylm(θ = 0) = 0 for m = 0. Using Eq. 5 and Ll(1) = 1 yields plmax = . Here, the parameter of interest is p2.
2.3 |. Parameter relations commonly used to constrain the two-compartment diffusion-relaxation model
Models adapted for conventional multi-shell dMRI acquired with a single TE tend to address degeneracy issues by constraining parameters to follow given relations. What we refer to as the ‘density assumption’ is a relation implicitly enforced when referring to compartment fractions as volume fractions without accounting for T2 relaxation (28). It assumes equal T2 values across compartments, here according to
| Eq. 16 |
With a shared compartment T2 value and data acquired for a single TE, the T2 dependence is factored into S0. Another set of relations, here referred to as ‘proportional diffusivities’, link axial diffusivities between compartments as
| Eq. 17 |
This has been used to enforce equal axial diffusivities (c = 1) (16, 23, 25) or equal isotropic diffusivities (c = 3) (27, 58). Yet another relation is the ‘tortuosity relation’, which connects the shape of the ‘zeppelin’ compartment to the ‘stick’ fraction (21, 23, 25, 59, 60), according to
| Eq. 18 |
In this work, we extend this expression to incorporate myelin. We first note that Eq. 18 was derived for diffusion around parallel cylinders where simulations showed that (61)
| Eq. 19 |
where vE is the extra-axonal volume fraction. While Eq. 18 assumes vE = 1 – fS, a more plausible relation includes myelin, according to vE = 1 – vA – vM, where vA and vM are the volume fractions of axons and myelin, respectively. These are related via the g-ratio, according to g2 = vA / (vA + vM) (62, 63). By adjusting for MR-invisible myelin, we can relate fS to vA, according to fS = vA / (1 – vM), to obtain
| Eq. 20 |
This relation could be used to update Eq. 18, which assumes g = 1 (no myelin), with g-ratio values obtained through independent means. Alternatively, it could be used to fit g using estimates of DΔ;Z and fS from the two-compartment diffusion relaxation model. However, note that this assignment is valid if and only if the compartment model is also accurate.
Finally, if the ODF is assumed to be axisymmetric, the Watson distribution is a less complex alternative to the truncated spherical harmonic expansion used here (64). It is described by mean orientation unit vector μ and concentration parameter κ with values from zero (spherical ODF) to infinity (delta function ODF). The OD parameter of Zhang (23) remaps κ to represent orientation dispersion from zero (OD = 0; κ = ∞) to complete (OD = 1; κ = 0), according to
| Eq. 21 |
2.4 |. Cramér-Rao lower bounds
Cramér-Rao lower bounds (CRLB) express the lower bound of the noise-induced variance in unbiased estimators (43, 44) and have been used previously to quantify the impact of the sampling protocol on the variance in model parameter estimates (59, 60, 65, 66). The CRLB are obtained as the diagonal elements of the inverse of the Fisher information matrix (F). For an experiment described by N samples en, assuming Gaussian noise with standard deviation σnoise, F has the elements
| Eq. 22 |
where ∂Sn/∂mi and ∂Sn/∂mj are partial derivatives of the signal model for measurement n with respect to model parameters mi and mj ∈ m (59). Here, we defined F using S, e and m from Eqs. 12, 13 and 14, respectively.
3 |. METHODS
The procedures described in this section were performed using MATLAB (R2015b, MathWorks, Natick, MA, USA) and in-house code, available at https://github.com/belampinen/lampinen_mrm_2019.
3.1 |. Protocol optimization
We optimized sampling protocols by minimizing an objective function based on the model parameters’ CRLB (from Eq. 22) converted into an optimization metric referred to as the ‘weighted parameter variance’ (VW), defined as
| Eq. 23 |
where w is a vector of weights, vC = [CRLB1, …, CRLB12], and the average is across the prior sets in Table 1, which were intended to represent white matter (A), deep gray matter (B), and white matter lesions (C). Furthermore, fTacq is an ‘efficiency factor’ that makes VW invariant to the actual acquisition time (Tacq). The weights were defined to ignore S0 and the ODF parameters, using w = 0 for these, and to bring the remaining kernel parameters to comparable scales, using w–1 values of 0.05 for fS, 0.1 μm2/ms for DI;S and DI;Z, 0.1 for DΔ;Z and 10 ms for T2;S and T2;Z. The efficiency factor was defined as fTacq = Tacq/Tacq;ref with Tacq;ref = 30 minutes and Tacq = (τfat + TEmax + τEPI /2) nsliceN, using the time for the fat saturation pulse (τfat = 5 ms), the maximal TE, the EPI readout time (τEPI = 45 ms), the number of slices (nslice = 40) and the protocol’s number of measurements (N). The final objective function was given by
| Eq. 24 |
Table 1 –
Model parameter prior sets and bounds
| Prior sets | fS | DI;S | DI;Z | DΔ;Z | T2;S | T2;Z | ODb |
|---|---|---|---|---|---|---|---|
| A | 0.45 | 0.6 | 1.3 | 0.57 | 80 | 60 | 0.20 |
| B | 0.15 | 0.3 | 0.9 | 0.40 | 75 | 55 | 0.45 |
| C | 0.40 | 0.6 | 1.7 | 0.40 | 80 | 150 | 0.35 |
| Bounds | fS | DI;S | DI;Z | DΔ;Z | T2;S | T2;Z | |
| Minimum | 0 | 0.07a | 0.2a | –0.46a | 30 | 30 | |
| Maximum | 1 | 1.33a | 4.0a | 0.86a | 300 | 1000 | |
Diffusivities are in μm2/ms and T2 values are in ms.
From enforcing D||;S, D||;Z and D⊥;Z ∈ [0.2 4.0] μm2/ms.
The parameters describing spherical harmonic coefficients, p20, Re(p21), Im(p21), Re(p22), Im(p21), are, for simplicity, represented by the corresponding OD from the Watson distribution (Eq. 21) assuming a main direction of [0 0 1].
To account for scanner limitations, the penalty factors ϕSNR and ϕgrad approximate step functions by (linearly) increasing rapidly from unity under certain conditions (see GitHub link for details). The first factor penalized SNR < 2, where the Gaussian noise assumption in Eq. 22 is particularly biased (67). The SNR was estimated based on data from Szczepankiewicz (68) assuming an approximate voxel T2 of 80 ms. The second factor penalized gradient amplitudes g > gmax (default 80 mT/m).
As in our previous work (66), we minimized Eq. 24 using the stochastic Self Organizing Migrating Algorithm (SOMA) (69). SOMA was repeated six times per optimization after which the best protocol was chosen. The optimization task was to specify b, bΔ and TE for each of a predetermined number of shells as well as the number of b-tensor symmetry axis orientations (ndir) across which to repeat these parameters. The allowed parameter values were b ∈ [0, 0.1, …, 10] ms/μm2, bΔ ∈ [–0.50, –0.45, …, 1], TE ∈ [50, 60, …, 300] ms, and ndir ∈ [6, 10, 15, 30, 45]. The directional set [θi, φi] corresponding to each ndir was selected from platonic solids (36) for simplicity and optimization performance. The lower bound of TE = 50 ms should guarantee that myelin water contributes to the non-diffusion weighted signal by less than 2%, assuming an approximate volume fraction of 0.15 with a T2 value of 15 ms in white matter (70, 71). To obtain a protocol suitable for clinical conditions, referred to as the ‘in vivo protocol’, we manually adapted a CRLB-optimized protocol to repeat a set of DTI and DKI-compatible shells using LTE (bΔ = 1) for different TE.
3.2 |. Model fitting
Fitting was performed with least-squares minimization using MATLAB and the multidimensional dMRI toolbox (72). Minimization used the lsqcurvefit function with default settings and parameter bounds from Table 1. The diffusivity bounds were enforced through penalties ensuring axial and radial diffusivities between 0.2 and 4 μm2/ms. A lower bound above zero is the ‘zeppelin’ compartment’s main characteristic and avoids representing water at the ‘dot-compartment’ limit of zero isotropic diffusivity (73–76), which is presumably negligible in healthy white matter (57, 76, 77). For T2 values, the lower bound avoids representing water presumably fully attenuated at our echo times (e.g. myelin water), and the upper bounds were considered safe assumptions for water within ‘stick-like’ structures and in tissue without major contamination with cerebrospinal fluid (CSF), respectively. Fitting was performed twice with initial guesses randomly selected from uniform distributions within the parameter bounds, after which the best fitting solution was kept. The probability of finding the global solution using two initial guesses was estimated to approximately 99.96% using in vivo data.
3.3 |. Optimization evaluation (simulations)
To assess the impact on parameter precision of the different acquisition dimensions, we optimized protocols under restrictions on b, bΔ and TE, one at a time, as well as on the number of unique combinations of these properties (shells). To assess the effect of the maximal gradient amplitude, the optimizations under b and TE restrictions were performed for gmax values of 200 mT/m and infinity in addition to 80 mT/m.
To illustrate the ability to determine the ‘stick’ fraction using different protocols, we used the normalized residual variance (NRV) (28). For a given protocol, a signal was synthesized for each prior set in Table 1 using Eq. 12, after which Eq. 12 was fitted with fS fixed to different values between zero and one. The NRV then yields a goodness-of-fit metric for each fixed fS, according to
| Eq. 25 |
where the average is across I = 10 realizations of Gaussian noise with standard deviation σnoise, Sn is the noised signal, Sn-overbar is the fitted signal, N is the protocol’s number of measurements and M is the number of free model parameters (M = 11, after fixing fS). Plotting the NRV against the fixed fS illustrates how precisely this parameter can be determined with a given protocol, which is challenging using a model with free compartment T2 values and diffusivities (28).
To explore the precision of parameter estimates under challenging model priors, we fitted Eq. 12 to signals synthesized with the in vivo protocol (Table 2, protocol II) assuming four different sets of priors for the two-compartment model, modified so that the ODF was represented by a Watson distribution rather than the truncated spherical harmonics. The three first sets of priors mimicked set A in Table 1, but with gradually increasing orientation dispersion achieved by setting OD (Eq. 21) to 0.05, 0.5, and 1. The fourth set mimicked the full orientation dispersion set but with equal compartment isotropic diffusivities (0.95 μm2/ms) and T2 values (70 ms). The simulations were performed for 200 realizations of noise from the Rice distribution to obtain distributions of estimated parameter values.
Table 2 –
Sampling protocols: an optimized protocol (I), the in vivo protocol (II), an optimized LTE-only protocol (III) and a protocol from Lampinen (28) (IV), featuring multiple TE only for low b-values.
| Protocol | Shell # | b [ms/μm2] | bΔ | TE [ms] | ndir |
|---|---|---|---|---|---|
| I | 1 | 0 | 1 | 50 | 10 |
| 2 | 0 | 1 | 90 | 10 | |
| 3 | 0.7 | 0.1 | 75 | 30 | |
| 4 | 1.9 | 1 | 70 | 30 | |
| 5 | 0 | 1 | 140 | 10 | |
| 6 | 1.2 | 0.1 | 140 | 6 | |
| 7 | 1.8 | 1 | 140 | 15 | |
| 8 | 2.1 | 1 | 140 | 45 | |
| 9 | 6.2 | 1 | 80 | 45 | |
| 10 | 3.6 | 0.15 | 105 | 45 | |
| II | 1,2,3a | 0.1,1.0,2.0 | 1 | 63 | 6,15,45 |
| 4,5,6,7b | 0.1,1.0,2.0,5.0 | 1 | 85 | 6,6,15,45 | |
| 8,9,10c | 0.1,1.0,2.0 | 1 | 130 | 30,6,30 | |
| 11,12,13d | 0.1,2.0,2.5 | 0.6 | 85 | 6,15,45 | |
| III | 1,2 | 0,0.4 | 1 | 50 | 6,45 |
| 3 | 0.9 | 1 | 60 | 15 | |
| 4 | 2.7 | 1 | 70 | 45 | |
| 5 | 8.9 | 1 | 85 | 45 | |
| 6 | 10 | 1 | 90 | 30 | |
| 7,8,9,10 | 0,2.1,2.1,2.2 | 1 | 100 | 6,30,10,10 | |
| IV | 1,2,3,4,5 | 0.1,0.5,1.0,1.5,2.0 | 1 | 106 | 6,6,10,16,30 |
| 6,7,8,9,10 | 0.1,0.5,1.0,1.5,2.0 | 0 | 106 | 6,6,10,16,30 | |
| 11,12 | 0,0.5 | 1 | 50 | 1,6 | |
| 13,14 | 0,0.5 | 1 | 85 | 1,6 | |
| 15,16 | 0,0.5 | 1 | 120 | 1,6 | |
| 17,18 | 0,0.5 | 1 | 155 | 1,6 |
The gradient waveforms used for the in vivo protocol (II) had a pause duration of 12.4 ms and durations before (δ1) and after (δ2) refocusing given by
δ1 = 16.5 ms, δ2 = 16.5 ms
δ1 = 27.5 ms, δ2 = 27.5 ms
δ1 = 50.0 ms, δ2 = 50.0 ms
δ1 = 30.0 ms, δ2 = 27.5 ms
All simulations used the same SNR estimations as described for the optimization. In the NRV simulations, acquisition time differences were accounted for according to SNR’ = SNR · (Tacq;ref/Tacq)1/2, with Tacq;ref = 30 minutes.
3.4 |. In vivo acquisition and processing
To study white and deep gray matter of the normal adult and maturing brain, we included five adult volunteers (age 31 ± 5 years; male/female = 3/2) and five children from a reading intervention study (age 6 ± 1 years; male/female = 4/1). To study age-related white matter lesions, we included data from five elderly subjects that were controls in a Parkinson’s disease study (age 76 ± 3 years; male/female = 2/3), some of which contributed data to a previous study (28). The experiments were approved by the regional ethical review board in Lund and all subjects gave informed consent.
dMRI data were acquired on a MAGNETOM Prisma 3T system (Siemens Healthcare, Erlangen, Germany), using a prototype diffusion-weighted spin-echo sequence (68). Encoding gradient waveforms were optimized to maximize the b-value for a given TE (78) and to remove concomitant field gradient effects (79). The diffusion-encoding directions were translated into gradient waveform rotations as specified in Szczepankiewicz (80). For the adults and the children, protocol II in Table 2 was used, featuring LTE, prolate (PrTE) b-tensors and multiple TE. The sampling order was volume-interleaved to distribute energy consumption and heating evenly over time (81) and to reduce potential bias from system drift (82). Total acquisition time was 15 minutes for a 2.5 mm isotropic resolution, matrix size = 88×88, 40 slices, multiband factor 2 (83), parallel imaging factor 2 (GRAPPA), partial Fourier factor 0.75, bandwidth = 1775 Hz/pixel, and ‘strong’ fat saturation. TR was 3.4 s, except for one subject with SMS (3.7 s) and another subject without SMS (6.4 s). For the elderly subjects, protocol IV in Table 2 was used, featuring LTE, STE and multiple TE for a single set of low b-values and directions, see Lampinen (28). All data were corrected for eddy-currents and subject motion using ElastiX (84) with extrapolated target volumes (85). To obtain parameter maps, Eq. 12 was fitted voxel-by-voxel to data smoothed using a Gaussian kernel with a standard deviation of 0.45 times the voxel dimensions.
3.5 |. In vivo data evaluation
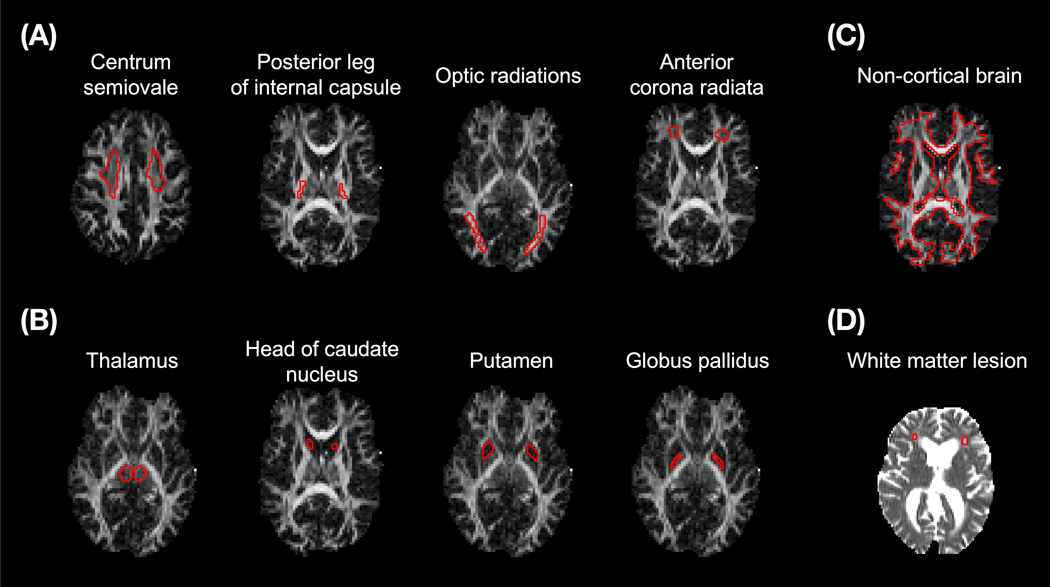
To assess parameter values from fitted maps, values were extracted from regions of interests (ROIs; Figure 1). For the adults and the children, ROIs were defined in white matter (Figure 1A), deep gray matter (Figure 1B), and the whole brain excluding the cortex and the ventricles to avoid CSF contamination (Figure 1C). For the elderly subjects, the anterior corona radiata featuring a white matter lesion was defined (Figure 1D).
Figure 1 –
Regions of interest (ROIs) are shown in red for one adult volunteer (A–C) on top of an FA map and for one elderly subject (D) on top of an MD map. The ROIs were defined on maps obtained from a DTI analysis of data from shell 1 and 2 of the in vivo protocol (II in Table 2). Non-cortical brain tissue (C) was defined by manually including deep gray matter and the brainstem within a white matter mask generated by thresholding the μFA parameter from a QTI analysis (36).
To explore whether our data supported popular or novel model constraints, we assessed different parameter relations using data obtained with our comparatively rich sampling protocol. First, we compared the predictions of some commonly enforced relations (Eqs. 16–18) with our (ROI-averaged) estimates. Second, we investigated the impact on estimated parameters from enforcing these relations by fitting the corresponding simplified models (using c = 1 in Eq. 17) to our data and comparing the results to those obtained when fitting the full model (Eq. 12). For each model, parameter values were obtained from all subjects by fitting to the ROI-averaged signals within white matter (Figure 1A and 1D), and parameter maps were obtained from one subject through voxel-by-voxel fitting within one slice of non-cortical brain (Figure 1C). For these maps, data were smoothed using a Gaussian kernel with a standard deviation of 0.75 times the voxel dimensions. Third, we tested whether enforcing these relations resulted in a significantly worse fit. Model selection maps were obtained for the non-cortical brain slice by thresholding an F-statistic at a 5% confidence level. F was calculated as the quotient between (SSR1 – SSR2)/(M2 – M1) and SSR2/(N – M2), using the sum of squared residuals (SSR) and the number of fitted parameters (M) of the simplified (1) and the full model (2) and the number of measurements N. The SSR values were obtained by fitting to unsmoothed data and, for each voxel, averaging across a 3×3 neighborhood. Fourth, we explored alternative parameter relations, including the tortuosity relation based on different g-ratios (Eq. 20).
4 |. RESULTS
4.1 |. Sampling protocols and parameter precision
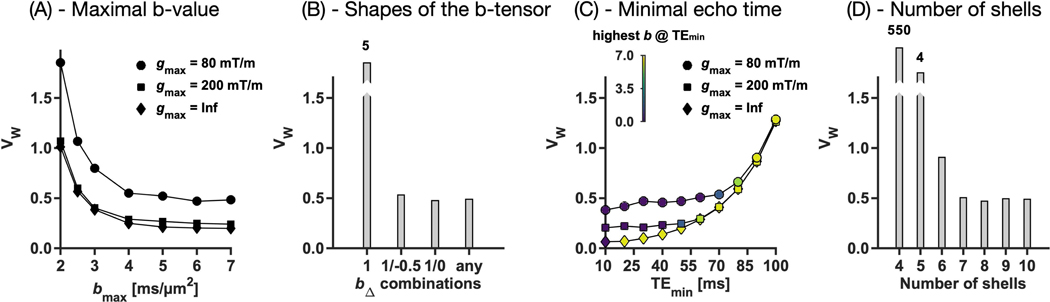
Figure 2 illustrates the impact on precision of the acquisition dimensions represented by b-value (b; A), b-tensor shape (bΔ; B) and echo time (TE; C), as well as of the number of shells (b/bΔ/TE-combinations; D). The x-axes show protocols optimized under different restrictions, and the y-axes show parameter variance by the CRLB-derived VW parameter (Eq. 23). Numerical simulations confirmed that the CRLB accurately quantified the parameter variance (Supporting Information Figure S1).
Figure 2 –
The impact of the acquisition dimensions on parameter precision. Plots show the weighted parameter variance (VW; Eq. 23; lower is better) for sampling protocols optimized under different restrictions indicated on the x-axis: maximal b-value (bmax) in panel A, shapes of the b-tensor (bΔ) in panel B, minimal echo times (TEmin ) in panel C, and number of shells in panel D. The impact on precision from bmax and TEmin was independent on the maximal gradient amplitude (gmax; indicated by shape), although stronger gradients increased precision by allowing shorter TE.
Increasing bmax improved precision up until approximately 4 ms/μm2 (Figure 2A). Acquiring data using at least two b-tensor shapes dramatically improved precision (Figure 2B). For example, using both bΔ = 1 and bΔ = –½ reduced VW to a tenth compared with using only bΔ = 1. No particular shape combinations were favored, however. Decreasing TEmin improved precision, but primarily when this was achievable for high b-values (Figure 2C). As a consequence, returns diminished below approximately 60–70 ms for gmax = 80 mT/m. Protocols with at least seven shells had the best precision and adding shells beyond the seventh had marginal impact (Figure 2D). Access to higher maximal gradient amplitudes improved precision by enabling shorter TE (Figure 2A/C). Increasing gmax from 80 to 200 mT/m reduced VW by approximately 50%.
Table 2 shows four protocols: an optimized protocol with 10 shells (I), the in vivo protocol (II), an optimized LTE-only protocol (III), and a protocol used in Lampinen (28) (IV). Protocol I is representative of optimized protocols in general. In particular, it features both low and high b-values using short TE (shells 1–4) as well as long TE (shells 5–8), an ‘ultra-high’ b-value (shell 9) and a high b-value for a non-LTE b-tensor shape (bΔ ≠ 1; shell 10). Protocol II was designed for in vivo use and also exhibited these features but was adapted to enable separate DTI and DKI analyses. Protocol III was restricted to only use LTE and featured a very high maximal b-value and comparatively short TE. Protocol IV from Lampinen (28) had a different design and acquired a range of b-values using two b-tensor shapes (bΔ = 0 and 1) for a single TE and low b-values for multiple TE.
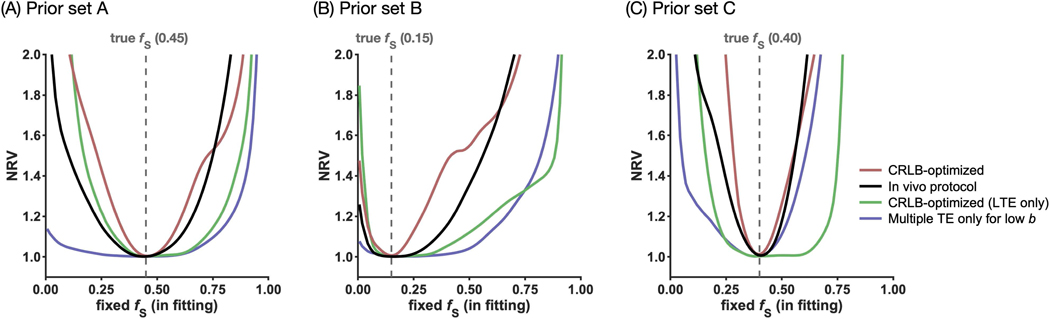
Figure 3 illustrates the ability to determine the ‘stick’ fraction using the protocols in Table 2 under the priors in Table 1. The NRV curves (Eq. 25) indicate parameter precision by the width of the minima around the true fS. For the optimized protocol (I) and the in vivo protocol (II), the minima were narrow, indicating high precision. For the LTE-only protocol (III) and the protocol acquiring multiple TE only for low b-values (IV), however, the curve minima were wide and flat, indicating multiple equivalent solutions and poor precision. The exception was a narrower minimum for protocol IV under large compartmental differences in isotropic diffusivities (Figure 3C), as expected from Lampinen (28).
Figure 3 –
Illustration of the ability to determine the ‘stick’ fraction (fS). The plot shows the NRV (normalized residual variance, Eq. 25) versus the value to which fS was fixed while fitting the other model parameters, for the protocols in Table 2 under the priors in Table 1. The width of the curve around the ground truth (dashed vertical line) indicates precision by the range of fS that yield similar goodness-of-fit values. The minima were narrow (high precision) for the CRLB-optimized protocol (I) and the in vivo protocol (II), but wide (poor precision) for the LTE-only protocol (III) and the protocol with multiple TE only for low b-values (IV). However, protocol IV achieved better precision where compartmental difference in isotropic diffusivities were large (C). CRLB-optimized protocols can be susceptible to local minima (B), although the in vivo protocol appeared robust.
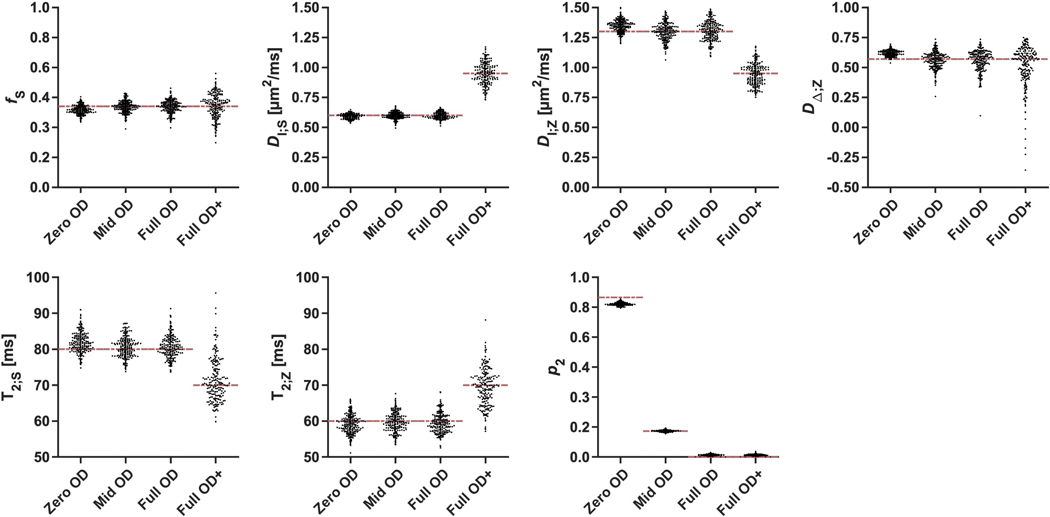
Figure 4 illustrates parameter precision using the in vivo protocol (Table 2, protocol II) under challenging model priors, showing distributions of parameter estimates from simulations. Precision was highest under no orientation dispersion (‘Zero OD’), decreased only slightly for a more realistic scenario (‘Mid OD’) and was largely retained even under full orientation dispersion (‘Full OD’). Removing both the directional information and compartmental differences in isotropic diffusivity (‘Full OD+’) substantially reduced precision, however. Precision for p2 was high under all conditions.
Figure 4 –
Parameter precision illustrated by distributions of parameter estimates (point swarms) simulated using the in vivo protocol under priors denoted on the x-axes. Precision was highest for ‘Zero OD’ and decreased slightly for the ‘Mid OD’ and the ‘Full OD’ priors. The Full OD+ priors, featuring full orientation dispersion together with equal compartment isotropic diffusivities and T2 values, had a large detrimental impact on precision.
4.2 |. In vivo results
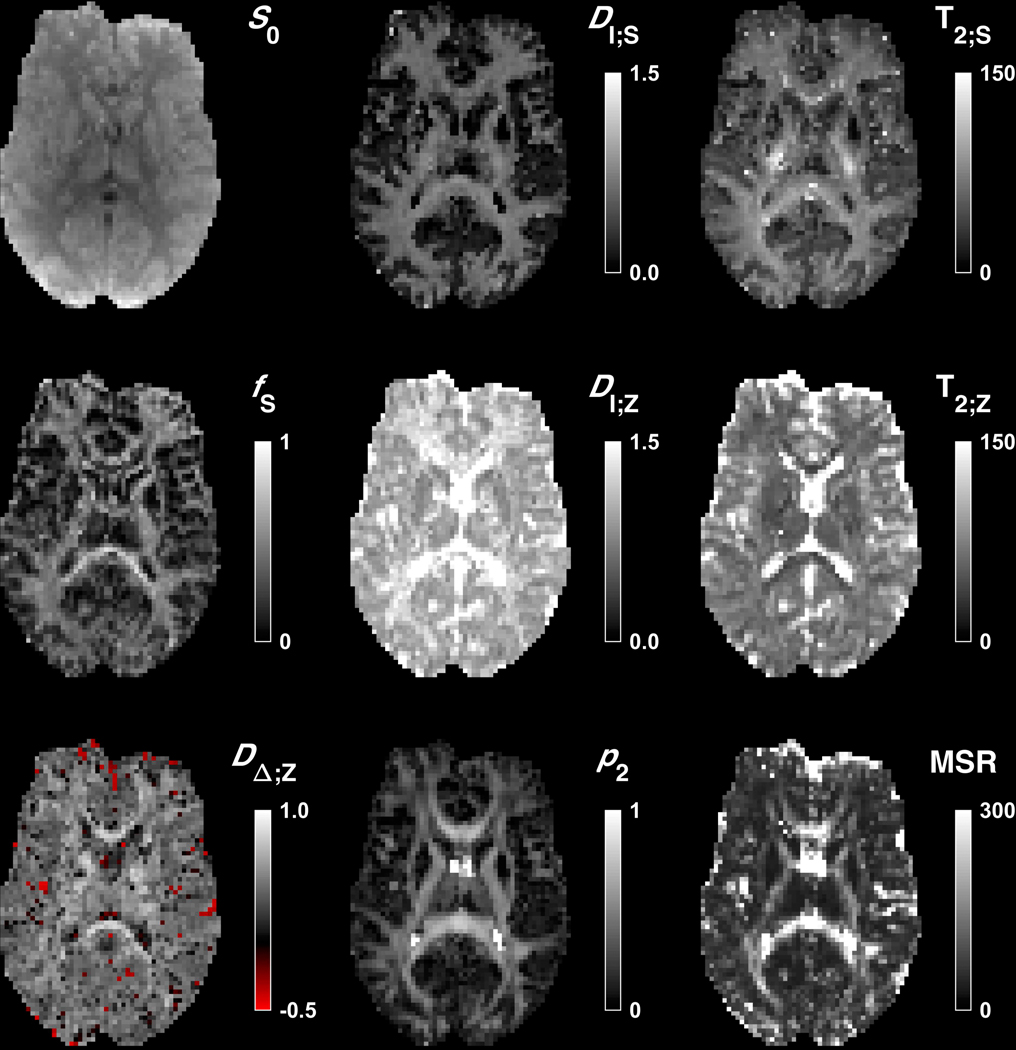
Maps of the kernel parameters are presented in Figure 5 and in Supporting Information Figure S2 together with p2 and the mean squared residuals (MSR). Parameter estimates were generally precise, indicated by the lack of visible noise in the parameter maps and confirmed by a test-retest experiment (Supporting Information Figure S3). The exception was DΔ;Z, with somewhat noisier maps. The maps of fS, DI;S, DI;Z, DΔ;Z, T2;S and p2 were brighter in white matter than in gray matter, although the DI;Z maps were brightest in the ventricles and the T2;S maps were particularly bright in the cerebrospinal tract. The T2;Z maps were relatively homogeneous but darker in iron-rich deep gray matter and brightest in the ventricles. The p2 maps were bright in coherent white matter and appeared to correlate with larger fit residuals. The median SNR in the non-cortical brain (Figure 1C) was approximately 50 at TE = 63 ms and b = 0.1 ms/μm2 based on the MSR (thus including both noise and model errors).
Figure 5 –
Maps of the kernel parameters in an adult volunteer together with p2 and mean squared residuals (MSR) from the fitting. Additional slices are shown in Supporting Information Figure S2. Isotropic diffusivities are in μm2/ms, T2 values are in ms and remaining parameters are dimensionless. All maps were masked to exclude voxels outside of the brain. In addition, the DI;S, T2;S, and p2 maps were masked to exclude voxels where fS < 0.1. The S0 maps exhibited proton density-weighting and lacked typical T2-related hypointensities. The fS (‘stick’ fraction) maps were bright in white matter and dark in deep gray matter, although somewhat brighter in the lateral thalamus. The DI;S and DI;Z (the ‘stick’ and ‘zeppelin’ isotropic diffusivities) maps were brightest in white matter, with DI;S maps being very dark in deep gray matter. The DΔ;Z (‘zeppelin’ shape) maps appeared somewhat noisy and were brightest in white matter and in orientationally coherent structures such as the corpus callosum. Negative DΔ;Z values (red) were sparse. The T2;S (‘stick’ T2 value) maps were brightest in white matter and particularly in the cerebrospinal tract. The T2;Z (‘zeppelin’ T2 value) maps were homogenous but darker in deep gray matter and brighter close to CSF. The p2 maps were smooth and resembled FA maps, being bright in white matter except in crossing-fiber regions such as the anterior corona radiata. The MSR maps were similar to p2 maps, possibly reflecting the ‘bluntness’ of the spherical harmonic ODF truncated at lmax = 2.
Parameter values are presented in Table 3 from the adults, children, and white matter lesions of the elderly subjects. In adults, white matter showed similar ‘stick’ properties across all regions with fS ≈ 0.5, DI;S ≈ 0.6 μm2/ms and T2;S ≈ 80 ms, with somewhat higher values in the internal capsule. ‘Zeppelins’ featured higher isotropic diffusivities and lower T2 values, with DI;Z ≈ 1.3 μm2/ms and T2;Z ≈ 65 ms, respectively. The properties of ‘sticks’ were similar also between deep gray matter regions, although the ‘stick’ fractions and isotropic diffusivities were lower than in white matter, with fS ≈ 0.15, DI;S ≈ 0.25 μm2/ms and T2;S ≈ 80 ms. ‘Zeppelins’ in deep gray matter featured lower isotropic diffusivities and T2 values than in white matter, with DI;Z ≈ 0.9 μm2/ms and T2;Z ≈ 50 ms. In children, most values were similar to in adults, but white matter ‘stick’ fractions were 10–30% lower and deep gray matter ‘zeppelin’ T2 values were 10–30% higher. Markedly high ‘stick’ T2 values and low ‘stick’ fractions were seen in the head of the caudate nucleus and the putamen of children, possibly due to imaging artifacts. In white matter lesions, we found elevated ‘zeppelin’ isotropic diffusivities and T2 values, with DI;Z = 1.68 μm2/ms and T2;Z = 149 ms, respectively, and a tendency towards more isotropic diffusion (DΔ;Z closer to zero). The ‘stick’ fractions and the diffusivities and T2 values of ‘sticks’ were similar to those found in adults, however. The lesion parameters also exhibited high inter-subject variance.
Table 3 –
ROI parameter values with means and inter-subject standard deviations for adults (Ad; n = 5), children (Ch, n = 5) and elderly subjects with white matter lesions (Le; n = 5).
| Non-cortical brain tissue | fS | DI;S | DI;Z | DΔ;Z | T2;S | T2;Z | p2 | D||;Z | D⊥;Z | |
|---|---|---|---|---|---|---|---|---|---|---|
| Ad | 0.40 (0.01) | 0.54 (0.03) | 1.23 (0.05) | 0.47 (0.01) | 77 (2) | 67 (2) | 0.37 (0.01) | 2.41 (0.10) | 0.64 (0.02) | |
| Ch | 0.34 (0.02) | 0.51 (0.03) | 1.22 (0.02) | 0.49 (0.01) | 81 (2) | 72 (4) | 0.40 (0.01) | 2.43 (0.06) | 0.61 (0.01) | |
| White matter | fS | DI;S | DI;Z | DΔ;Z | T2;S | T2;Z | p2 | D||;Z | D⊥;Z | |
| Centrum semiovale | Ad | 0.49 (0.02) | 0.62 (0.03) | 1.31 (0.04) | 0.46 (0.03) | 82 (3) | 62 (3) | 0.29 (0.03) | 2.54 (0.10) | 0.69 (0.05) |
| Ch | 0.46 (0.02) | 0.61 (0.05) | 1.28 (0.07) | 0.49 (0.03) | 77 (3) | 69 (7) | 0.36 (0.03) | 2.55 (0.17) | 0.64 (0.05) | |
| Posterior leg of internal capsule | Ad | 0.53 (0.03) | 0.66 (0.04) | 1.41 (0.16) | 0.63 (0.05) | 107 (8) | 65 (6) | 0.48 (0.03) | 3.20 (0.38) | 0.52 (0.10) |
| Ch | 0.41 (0.07) | 0.58 (0.05) | 1.37 (0.12) | 0.65 (0.02) | 102 (17) | 66 (3) | 0.55 (0.02) | 3.15 (0.25) | 0.48 (0.06) | |
| Optic radiations | Ad | 0.45 (0.01) | 0.62 (0.04) | 1.30 (0.08) | 0.50 (0.04) | 79 (3) | 68 (5) | 0.49 (0.04) | 2.60 (0.23) | 0.64 (0.04) |
| Ch | 0.39 (0.03) | 0.58 (0.03) | 1.30 (0.04) | 0.48 (0.02) | 74 (1) | 80 (11) | 0.51 (0.05) | 2.55 (0.05) | 0.67 (0.04) | |
| Anterior corona radiata | Ad | 0.45 (0.04) | 0.58 (0.04) | 1.36 (0.09) | 0.44 (0.03) | 69 (3) | 60 (4) | 0.30 (0.03) | 2.57 (0.23) | 0.75 (0.04) |
| Ch | 0.40 (0.03) | 0.59 (0.03) | 1.32 (0.04) | 0.46 (0.05) | 68 (3) | 61 (7) | 0.32 (0.02) | 2.56 (0.11) | 0.70 (0.08) | |
| Le | 0.44 (0.11) | 0.57 (0.04) | 1.68 (0.16) | 0.37 (0.05) | 71 (30) | 149 (29) | 0.23 (0.02) | 2.70 (0.18) | 1.05 (0.11) | |
| Deep gray matter | fS | DI;S | DI;Z | DΔ;Z | T2;S | T2;Z | p2 | D||;Z | D⊥;Z | |
| Thalamus | Ad | 0.15 (0.02) | 0.33 (0.05) | 0.99 (0.03) | 0.52 (0.03) | 97 (16) | 53 (3) | 0.31 (0.01) | 2.01 (0.10) | 0.47 (0.03) |
| Ch | 0.13 (0.01) | 0.28 (0.03) | 1.02 (0.02) | 0.50 (0.01) | 115 (17) | 64 (3) | 0.32 (0.02) | 2.06 (0.06) | 0.50 (0.02) | |
| Head of caudate nucleus | Ad | 0.14 (0.03) | 0.25 (0.05) | 0.81 (0.03) | 0.34 (0.05) | 77 (19) | 60 (3) | 0.18 (0.03) | 1.38 (0.11) | 0.53 (0.04) |
| Ch | 0.07 (0.02) | 0.29 (0.07) | 0.82 (0.02) | 0.32 (0.04) | 172 (42) | 68 (3) | 0.19 (0.03) | 1.35 (0.08) | 0.56 (0.03) | |
| Putamen | Ad | 0.10 (0.02) | 0.20 (0.02) | 0.83 (0.02) | 0.41 (0.03) | 77 (13) | 51 (3) | 0.16 (0.01) | 1.53 (0.08) | 0.49 (0.02) |
| Ch | 0.10 (0.03) | 0.27 (0.09) | 0.85 (0.04) | 0.36 (0.05) | 108 (22) | 67 (3) | 0.13 (0.01) | 1.47 (0.14) | 0.54 (0.03) | |
| Globus pallidus | Ad | 0.15 (0.04) | 0.26 (0.08) | 1.06 (0.05) | 0.54 (0.03) | 80 (16) | 41 (5) | 0.20 (0.01) | 2.23 (0.16) | 0.48 (0.01) |
| Ch | 0.15 (0.01) | 0.31 (0.05) | 1.09 (0.06) | 0.51 (0.03) | 84 (9) | 55 (2) | 0.23 (0.02) | 2.23 (0.17) | 0.52 (0.01) | |
Diffusivities are in μm2/ms and T2 values are in ms.
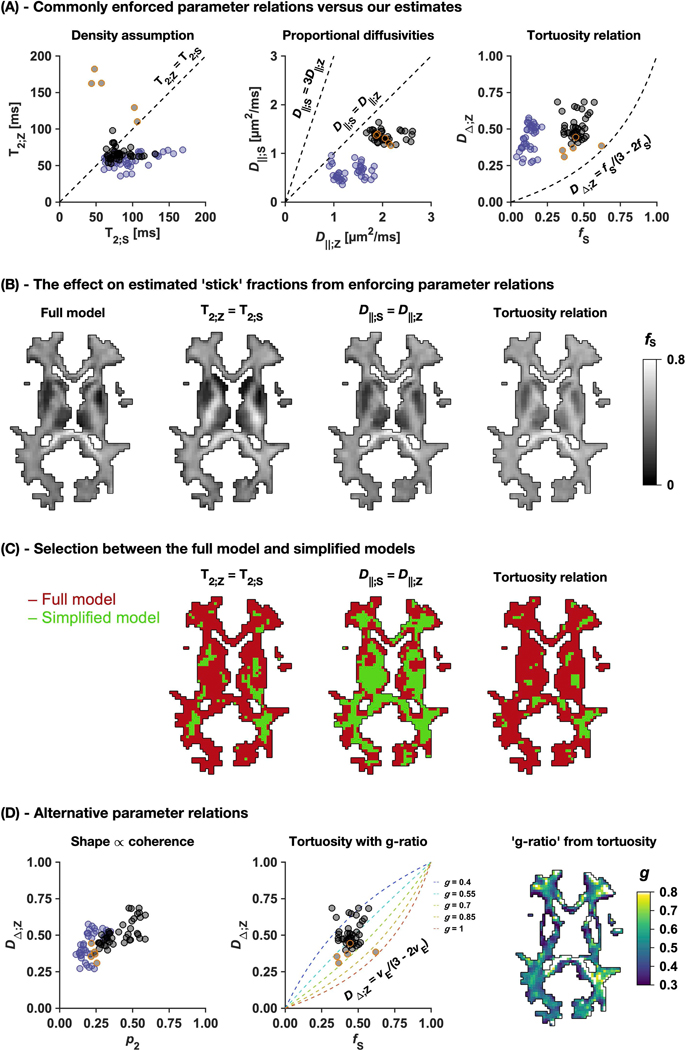
Parameter relations are explored in Figure 6. Three commonly enforced parameter relations were contradicted by our estimates (Figure 6A). The density assumption predicts equal compartment T2 values (Eq. 16) but data indicated higher T2 values for ‘sticks’ than for ‘zeppelins’ in healthy white matter and the opposite in white matter lesions. Proportional diffusivities link compartment axial diffusivities by a constant (Eq. 17) but data did not suggest a simple linear relationship between these parameters. The tortuosity relation connects the ‘zeppelin’ shape to the ‘stick’ fraction (Eq. 18), but again, observations did not agree with assumptions. Enforcing these relations yielded ‘stick’ fraction maps that were smoother but featured rather different values compared with using the full model (Eq. 12; Figure 6B). Enforcing equal T2 values resulted in different ‘stick’ fractions (fS´) that were higher in healthy white matter (fS´ – fS = ΔfS ≈ 0.05–0.15), and particularly in the cerebrospinal tract, but lower in white matter lesions (ΔfS ≈ –0.30), and thus substantially exaggerating their effect. Enforcing equal axial diffusivities resulted in ‘stick’ fractions that were similar in white matter but higher in white matter lesions (ΔfS ≈ 0.15). Enforcing the tortuosity relation resulted in higher ‘stick’ fractions both within normal (ΔfS ≈ 0.05–0.10) and lesioned (ΔfS ≈ 0.15) white matter. Using the full model yielded a significantly better fit within most regions of the healthy brain compared with enforcing equal compartment T2 values or the tortuosity relation, although enforcing equal axial diffusivities appeared acceptable in many regions (Figure 6C).
Figure 6 –
Evaluation of model simplification through enforcing parameter relations. Panel A shows commonly enforced relations (dashed lines) together with our estimates (dots). The estimates are ROI-averages from adults and children in white matter regions (black) and deep gray matter regions (blue), and from elderly subjects in white matter lesions (orange-bordered black). Panel B compares ‘stick’ fraction maps obtained either using the full model or simplified models enforcing different parameter relations. Panel C shows model selection maps (F-statistics thresholded at a level corresponding to a significance level of 5%, uncorrected for multiple comparisons) between the full model and the simplified models in panel B. Panel D shows alternative parameter relations that may be compatible with data, including the tortuosity relation allowing a variable g-ratio (Eq. 20) together with a fitted ‘g-ratio’ map.
Data suggested some alternative parameter relations (Figure 6D). The ‘zeppelin’ shape appeared positively related to the orientation coherence in white matter. Furthermore, allowing different g-ratios (Eq. 20) may explain variation unaccounted for by the ‘ordinary’ tortuosity relation (g = 1; Eq. 18). Finally, a tortuosity-based ‘g-ratio’ map is shown within white matter from fitting g using Eq. 20 and our estimates of fS and DΔ;Z. Although noisy, the map featured a median of approximately 0.5, which is close to the value of 0.6 where conduction velocities are theoretically optimized (86). The shown data points are available at https://github.com/belampinen/lampinen_mrm_2019/tree/master/estimates.
5 |. DISCUSSION
In this paper, we show what is required of a diffusion-relaxation sampling protocol to enable precise estimation of compartment-specific fractions, diffusivities and T2 values, and present a 15-minute whole-brain imaging protocol. Three protocol features were critical to parameter precision. First, at least seven unique b/bΔ/TE-combinations (shells) were required for robust estimation of seven kernel parameters (counting S0; Figure 2D). Using fewer shells resulted in parameters becoming undetermined under high orientation dispersion (starting with DΔ;Z for six shells; Supporting Information Figures S4 and S5). Second, using at least two shapes of the b-tensor substantially enhanced precision (Figure 2B). This was expected, because the two-compartment ‘standard model’ is degenerate for data acquired with a single b-tensor shape (LTE) (32, 33, 37, 38), even when using very high b-values (33, 87) or multiple echo times (40). With multiple b-tensor shapes, this degeneracy is resolved (37, 38). Third, multiple echo times for high b-values were required to distinguish the T2 values of compartments with different anisotropy but similar isotropic diffusivities (Figure 3A/B) (28, 40). Protocols with these three features allowed estimation of seven kernel parameters, compared with three kernel parameters allowed by contemporary approaches using multi-shell dMRI (27, 28, 31). The estimates were robust to full orientation dispersion (Figure 4; Supporting Information Figure S4), where the ‘standard model’ is degenerate even for data acquired with multiple shapes of the b-tensor (37, 38). This confirms that diffusion-relaxation experiments improve precision in diffusion parameters, at least in the presence of compartmental differences in T2 values (88).
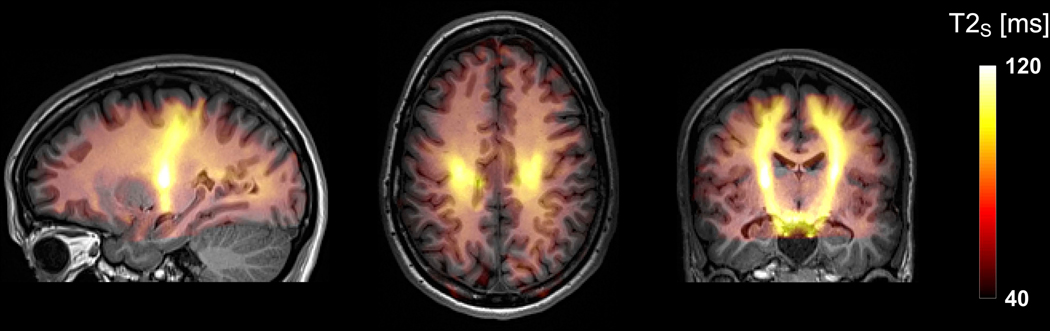
In vivo parameter maps were smooth, indicating precise estimates (Figure 5), which invites a tentative interpretation of the parameters in terms of microstructure. In white matter, the ‘stick’ fractions close to 50% are consistent with estimates from similar approaches (40), and are aligned with axonal volume fractions from histology (30, 71, 89) if corrected for the space occupied by MR-invisible myelin, according to fS = vA / (1 – vM). In children, white matter ‘stick’ fractions were lower than in adults (Table 3), potentially reflecting incomplete maturation (90). The ‘stick’ axial diffusivities were low (D||;S ≈ 1.8 μm2/ms; Table 3) in comparison with estimates for intra-axonal water obtained using planar signal filtering (D|| ≈ 2.25 μm2/ms) (91). Potential explanations include the comparatively longer TE used in Dhital (91), if longer axonal T2 are related to higher axial diffusivities. The ‘stick’ axial diffusivities were low also compared with those of ‘zeppelins’ (D||;Z ≈ 2.7 μm2/ms), possibly indicating obstruction of intra-axonal diffusion from, for example, variation in axonal caliber (92). The axial diffusivities were more similar between compartments in previous studies (33, 40), possibly due to model differences and undetected bias in those or the present study. The ‘zeppelin’ shape assumed negative values only sparsely and incoherently (Figure 5), indicating an absence of ‘oblate’ diffusion tensors in healthy brain. The ‘stick’ T2 values (Table 3) are largely consistent with previous observations (40, 93), including high values in the internal capsule. Higher T2 values within the more anisotropic (stick) compartment is also consistent with previous findings both in the human brain (40, 93, 94) and in ex vivo animal nervous tissue (95–98), possibly reflecting a reduction of ‘zeppelin’ T2 from exchange between the extracellular space and myelin water (99). Additionally, we found that T2;S yielded a plausible segmentation of the cerebrospinal tract all the way from the brainstem to the motor cortex (Figure 7). This suggests that diffusion-relaxation experiments increase specificity to particular fiber bundles, which could potentially improve regional segmentation or aid separating crossing fibers in tractography (100).
Figure 7 –
Maximum intensity projections of the T2;S parameter in three planes shown on top of a coregistered T1-weighted image. The result yielded a plausible segmentation of the cerebrospinal tract all the way from the brainstem to close to the motor cortex.
In gray matter, ‘stick’ fractions between 10 and 20% are consistent with previous findings (28, 33). However, the findings are not consistent with interpreting ‘sticks’ as neurites, because the combined (neurite) density of axons and dendrites is approximately 60% according to histology (101–103). We hypothesize, as in Lampinen (28), that this discrepancy is due to dendrites not exhibiting stick-like diffusion, due to, for example, exchange with soma or between short segments with different orientations. Thus, the model may be too simplistic for gray matter.
In white matter lesions, the increased ‘zeppelin’ isotropic diffusivities and T2 values are consistent with demyelination enlarging the extracellular space (104). Disambiguating between demyelination and axonal loss requires either data with independent sensitivity to myelin or the use of assumptions, however. By the tortuosity assumption extended to variable g-ratios (Eq. 20), and excluding the outlier lesion in Fig. 6, our results would suggest similar axonal volume fractions (vA ≈ 0.2) but lower myelin volume fractions (vM ≈ 0.4 versus 0.5) in lesions versus in the healthy anterior corona radiata, which is in line with the results of Coelho (105).
This study had limitations. First, for efficiency reasons, the optimization used a small set of priors (Table 1). One may be concerned that the choice of priors could have a strong effect on the protocol and in some way bias the fitting procedure. However, using different priors revealed similar sampling requirements and no bias towards particular solutions was seen in the fitting. Second, CLRB-optimized protocols can be susceptible to local minima (Figure 3A/B). However, these can be avoided by using multiple random initial guesses in the fitting, and the manually adjusted in vivo protocol appeared robust. Third, we did not address reproducibility. Future studies should investigate the impact on parameter values from changing the sampling protocol. Fourth, the gradient waveforms used to generate linear and prolate tensor encoding differed with respect to time-dependent diffusion encoding, which may lead to bias (106, 107). Such time dependence is only expected for very short encoding times (108), however. For the longer encoding times used in this study, we have not been able to detect time dependence in the healthy brain (20, 28). Fifth, a spherical harmonic expansion truncated at lmax = 2 may be insufficient for representing sharp ODFs, as indicated by the fit being worst in regions with high orientation coherence (Figure 5). However, comparing our estimates with those obtained using the ‘RotInv’ approach suggests that this is not likely to bias the kernel parameters (Supporting Information Figure S6). Finally, even the relatively unconstrained model used here is simplistic compared with the complexity of neural tissue and does not account for myelin (71), soma (109), the microvasculature (50), glial cells (110) or CSF (111), wherefore parameters should be interpreted with care. Preliminary results indicate that our present data would support an additional free parameter to include a free water compartment (Supporting Information Figure S7), and that bias from neglecting free water is small but non-negligible also within non-cortical brain, possibly indicating contributions from perivascular spaces (112). This should be further investigated and validated in future studies using the b/bΔ/TE measurement space to drive models with higher capacity.
5 |. CONCLUSIONS
Optimized diffusion-relaxation sampling enabled precise and efficient whole-brain estimation of compartment-specific fractions, diffusivities and T2 values. Acquiring data using the independent dimensions of b-value, b-tensor shape and echo time expands the set of microstructure parameters measurable with MRI, as well as the range of conditions in which they may be accurately estimated. According to our findings, the conventional approach of improving precision through enforcing parameter relations causes bias even in the healthy brain. Future studies could use diffusion-relaxation MRI with tensor-valued diffusion encoding for enhanced specificity to microstructural quantities. Our in vivo results showed that ‘stick’ fractions were lower in the white matter of children than in adults, and that white matter lesions featured changes compatible with demyelination.
Supplementary Material
FIGURE S1 Validation of the CRLB by comparing the VW metric based on variance estimates from either CRLB or simulations.
FIGURE S2 A multi-slice version of Figure 5.
FIGURE S3 A test-retest experiment on an adult volunteer, showing test and retest parameter maps, difference maps, and Bland-Altman plots.
FIGURES S4 and S5 The importance of the number of ‘shells’ versus the degree of orientation dispersion for the ability to determine all model parameters.
FIGURE S6 Comparison of parameter maps obtained by fitting Eq. 12 directly to data versus those obtained using the ‘RotInv’ approach.
FIGURE S7 Comparison of parameter maps obtained using the two-compartment model (Eq. 12) versus a model extended to include a free water compartment.
ACKNOWLEDGEMENTS
This study was supported by grants from Swedish Research Council (2016-03443), the NIH (P41EB015902 and R01MH074794), Random Walk Imaging AB (grant no. MN15), The Swedish Society for Medical Research (P15-0077) and BAGADILICO. The funding sources had no role in the design and conduct of the study; in the collection, analysis, interpretation of the data; or in the preparation, review, or approval of the manuscript. We thank Siemens Healthcare for providing access to the pulse sequence programming environment, Thomas Witzel and Daniel Park for supporting the implementation of SMS into the FWF pulse sequence, and Massachusetts General Hospital for providing the SMS source code.
Footnotes
CONFLICT OF INTEREST
MN declares ownership interests in Random Walk Imaging, and patent applications in Sweden (1250453-6 and 1250452-8), USA (61/642 594 and 61/642 589), and PCT (SE2013/050492 and SE2013/050493). MN and FS are inventors on pending patents pertaining to the methods presented herein. Remaining authors declare no conflict of interest.
REFERENCES
- 1.Alexander DC, et al. , Imaging brain microstructure with diffusion MRI: Practicality and applications. NMR in Biomedicine, 2017. [DOI] [PubMed] [Google Scholar]
- 2.Basser PJ, Mattiello J, and LeBihan D, MR diffusion tensor spectroscopy and imaging. Biophysical journal, 1994. 66(1): p. 259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jensen JH, et al. , Diffusional kurtosis imaging: The quantification of non-gaussian water diffusion by means of magnetic resonance imaging. Magnetic Resonance in Medicine, 2005. 53(6): p. 1432–1440. [DOI] [PubMed] [Google Scholar]
- 4.Werring D, et al. , The pathogenesis of lesions and normal-appearing white matter changes in multiple sclerosis: a serial diffusion MRI study. Brain, 2000. 123(8): p. 1667–1676. [DOI] [PubMed] [Google Scholar]
- 5.Ropele S, et al. , Quantitation of brain tissue changes associated with white matter hyperintensities by diffusion-weighted and magnetization transfer imaging: The LADIS (leukoaraiosis and disability in the elderly) study. Mag Reson Med, 2009. 29(2): p. 268–274. [DOI] [PubMed] [Google Scholar]
- 6.Svärd D, et al. , The effect of white matter hyperintensities on statistical analysis of diffusion tensor imaging in cognitively healthy elderly and prodromal Alzheimer’s disease. PloS one, 2017. 12(9): p. e0185239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van Cauter S, et al. , Gliomas: diffusion kurtosis MR imaging in grading. Radiology, 2012. 263(2): p. 492–501. [DOI] [PubMed] [Google Scholar]
- 8.Surova Y, et al. , Assessment of global and regional diffusion changes along white matter tracts in parkinsonian disorders by MR tractography. PloS one, 2013. 8(6): p. e66022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scholz J, et al. , Training induces changes in white-matter architecture. Nature neuroscience, 2009. 12(11): p. 1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sagi Y, et al. , Learning in the fast lane: new insights into neuroplasticity. Neuron, 2012. 73(6): p. 1195–1203. [DOI] [PubMed] [Google Scholar]
- 11.Lebel C, et al. , Microstructural maturation of the human brain from childhood to adulthood. Neuroimage, 2008. 40(3): p. 1044–1055. [DOI] [PubMed] [Google Scholar]
- 12.Cheung MM, et al. , Does diffusion kurtosis imaging lead to better neural tissue characterization? A rodent brain maturation study. NeuroImage, 2009. 45(2): p. 386–392. [DOI] [PubMed] [Google Scholar]
- 13.Löbel U, et al. , Diffusion tensor imaging: the normal evolution of ADC, RA, FA, and eigenvalues studied in multiple anatomical regions of the brain. Neuroradiology, 2009. 51(4): p. 253–263. [DOI] [PubMed] [Google Scholar]
- 14.Novikov DS, Kiselev VG, and Jespersen SN, On modeling. Magnetic resonance in medicine, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nilsson M, et al. , Imaging brain tumour microstructure. Neuroimage, 2018. 182: p. 232–250. [DOI] [PubMed] [Google Scholar]
- 16.Behrens TE, et al. , Characterization and propagation of uncertainty in diffusion-weighted MR imaging. Magnetic resonance in medicine, 2003. 50(5): p. 1077–1088. [DOI] [PubMed] [Google Scholar]
- 17.Assaf Y. and Basser PJ, Composite hindered and restricted model of diffusion (CHARMED) MR imaging of the human brain. Neuroimage, 2005. 27(1): p. 48–58. [DOI] [PubMed] [Google Scholar]
- 18.Jespersen SN, et al. , Modeling dendrite density from magnetic resonance diffusion measurements. Neuroimage, 2007. 34(4): p. 1473–1486. [DOI] [PubMed] [Google Scholar]
- 19.Kaden E, Knösche TR, and Anwander A, Parametric spherical deconvolution: inferring anatomical connectivity using diffusion MR imaging. NeuroImage, 2007. 37(2): p. 474–488. [DOI] [PubMed] [Google Scholar]
- 20.Nilsson M, et al. , On the effects of a varied diffusion time in vivo: is the diffusion in white matter restricted? Magnetic resonance imaging, 2009. 27(2): p. 176–187. [DOI] [PubMed] [Google Scholar]
- 21.Zhang H, et al. , Axon diameter mapping in the presence of orientation dispersion with diffusion MRI. Neuroimage, 2011. 56(3): p. 1301–1315. [DOI] [PubMed] [Google Scholar]
- 22.Sotiropoulos SN, Behrens TE, and Jbabdi S, Ball and rackets: inferring fiber fanning from diffusion-weighted MRI. Neuroimage, 2012. 60(2): p. 1412–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang H, et al. , NODDI: practical in vivo neurite orientation dispersion and density imaging of the human brain. Neuroimage, 2012. 61(4): p. 1000–16. [DOI] [PubMed] [Google Scholar]
- 24.White NS, et al. , Probing tissue microstructure with restriction spectrum imaging: histological and theoretical validation. Human brain mapping, 2013. 34(2): p. 327–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaden E, et al. , Multi-compartment microscopic diffusion imaging. NeuroImage, 2016. 139: p. 346–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tariq M, et al. , Bingham–NODDI: mapping anisotropic orientation dispersion of neurites using diffusion MRI. NeuroImage, 2016. 133: p. 207–223. [DOI] [PubMed] [Google Scholar]
- 27.Lampinen B, et al. , Neurite density imaging versus imaging of microscopic anisotropy in diffusion MRI: A model comparison using spherical tensor encoding. NeuroImage, 2017. 147: p. 517–531. [DOI] [PubMed] [Google Scholar]
- 28.Lampinen B, et al. , Searching for the neurite density with diffusion MRI: Challenges for biophysical modeling. Human brain mapping, 2019. 40(8): p. 2529–2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jespersen SN, et al. , Neurite density from magnetic resonance diffusion measurements at ultrahigh field: comparison with light microscopy and electron microscopy. Neuroimage, 2010. 49(1): p. 205–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jelescu IO, et al. , In vivo quantification of demyelination and recovery using compartment-specific diffusion MRI metrics validated by electron microscopy. Neuroimage, 2016. 132: p. 104–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kiselev VG and Il’yasov KA, Is the “biexponential diffusion” biexponential? Magnetic resonance in medicine, 2007. 57(3): p. 464–469. [DOI] [PubMed] [Google Scholar]
- 32.Jelescu IO, et al. , Degeneracy in model parameter estimation for multi-compartmental diffusion in neuronal tissue. NMR in Biomedicine, 2016. 29(1): p. 33–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Novikov DS, et al. , Rotationally-invariant mapping of scalar and orientational metrics of neuronal microstructure with diffusion MRI. NeuroImage, 2018. 174: p. 518–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mitra PP, Multiple wave-vector extensions of the NMR pulsed-field-gradient spin-echo diffusion measurement. Physical Review B, 1995. 51(21): p. 15074. [DOI] [PubMed] [Google Scholar]
- 35.Westin C-F, et al. Measurement tensors in diffusion MRI: generalizing the concept of diffusion encoding International Conference on Medical Image Computing and Computer-Assisted Intervention, 2014. 2014. Springer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Westin C-F, et al. , Q-space trajectory imaging for multidimensional diffusion MRI of the human brain. NeuroImage, 2016. 135: p. 345–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Coelho S, et al. , Resolving degeneracy in diffusion MRI biophysical model parameter estimation using double diffusion encoding. Mag Reson Med, 2019. 82(1): p. 395–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reisert M, Kiselev VG, and Dhital B, A unique analytical solution of the white matter standard model using linear and planar encodings. Mag Reson Med, 2019. 81(6): p. 3819–3825. [DOI] [PubMed] [Google Scholar]
- 39.Benjamini D. and Basser P, Use of marginal distributions constrained optimization (MADCO) for accelerated 2D MRI relaxometry and diffusometry. Journal of magnetic resonance, 2016. 271: p. 40–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Veraart J, Novikov DS, and Fieremans E, TE dependent Diffusion Imaging (TEdDI) distinguishes between compartmental T2 relaxation times. Neuroimage, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Almeida Martins JP and Topgaard D, Multidimensional correlation of nuclear relaxation rates and diffusion tensors for model-free investigations of heterogeneous anisotropic porous materials. Scientific reports, 2018. 8(1): p. 2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ning L, et al. , Joint RElaxation-Diffusion Imaging Moments (REDIM) to probe neurite microstructure. IEEE Trans Med Imaging, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rao CR, Information and accuracy attainable in the estimation of statistical parameters. Bulletin of the Calcutta Mathematical Society, 1945. 37(3): p. 81–91. [Google Scholar]
- 44.Cramér H, Mathematical methods of statistics. Vol. 9 1946: Princeton university press. [Google Scholar]
- 45.Tournier J-D, et al. , Direct estimation of the fiber orientation density function from diffusion-weighted MRI data using spherical deconvolution. NeuroImage, 2004. 23(3): p. 1176–1185. [DOI] [PubMed] [Google Scholar]
- 46.Reisert M, et al. , Disentangling micro from mesostructure by diffusion MRI: A Bayesian approach. 2017. 147: p. 964–975. [DOI] [PubMed] [Google Scholar]
- 47.Jespersen SN, et al. , Diffusion time dependence of microstructural parameters in fixed spinal cord. Neuroimage, 2018. 182: p. 329–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kennedy RA, Lamahewa TA, and Wei L, On azimuthally symmetric 2-sphere convolution. Digital Signal Processing, 2011. 21(5): p. 660–666. [Google Scholar]
- 49.Eriksson S, et al. , NMR diffusion-encoding with axial symmetry and variable anisotropy: Distinguishing between prolate and oblate microscopic diffusion tensors with unknown orientation distribution. The Journal of chemical physics, 2015. 142(10): p. 104201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Le Bihan D, et al. , MR imaging of intravoxel incoherent motions: application to diffusion and perfusion in neurologic disorders. Radiology, 1986. 161(2): p. 401–407. [DOI] [PubMed] [Google Scholar]
- 51.Driscoll JR and Healy DM, Computing Fourier transforms and convolutions on the 2-sphere. Advances in applied mathematics, 1994. 15(2): p. 202–250. [Google Scholar]
- 52.Jespersen SN, et al. , Orientationally invariant metrics of apparent compartment eccentricity from double pulsed field gradient diffusion experiments. NMR in Biomedicine, 2013. 26(12): p. 1647–1662. [DOI] [PubMed] [Google Scholar]
- 53.Kroenke CD, Ackerman JJ, and Yablonskiy DA, On the nature of the NAA diffusion attenuated MR signal in the central nervous system. Magnetic resonance in medicine, 2004. 52(5): p. 1052–1059. [DOI] [PubMed] [Google Scholar]
- 54.Lasič S, et al. , Microanisotropy imaging: quantification of microscopic diffusion anisotropy and orientational order parameter by diffusion MRI with magic-angle spinning of the q-vector. Frontiers in Physics, 2014. 2: p. 11. [Google Scholar]
- 55.Nilsson M, et al. , Resolution limit of cylinder diameter estimation by diffusion MRI: The impact of gradient waveform and orientation dispersion. NMR in Biomedicine, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McKinnon ET, et al. , Dependence on b-value of the direction-averaged diffusion-weighted imaging signal in brain. Magnetic resonance imaging, 2017. 36: p. 121–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Veraart J, Fieremans E, and Novikov DS, On the scaling behavior of water diffusion in human brain white matter. NeuroImage, 2019. 185: p. 379–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guerreri M, et al. , Revised NODDI model for diffusion MRI data with multiple b-tensor encodings. In Proceedings of the 26th Annual Meeting of ISMRM, Paris, France, 2018 p. 5241. 2018. [Google Scholar]
- 59.Alexander DC, A general framework for experiment design in diffusion MRI and its application in measuring direct tissue-microstructure features. Magn Reson Med, 2008. 60(2): p. 439–48. [DOI] [PubMed] [Google Scholar]
- 60.Alexander DC, et al. , Orientationally invariant indices of axon diameter and density from diffusion MRI. Neuroimage, 2010. 52(4): p. 1374–1389. [DOI] [PubMed] [Google Scholar]
- 61.Szafer A, et al. , Diffusion-weighted imaging in tissues: Theoretical models. NMR in Biomedicine, 1995. 8(7): p. 289–296. [DOI] [PubMed] [Google Scholar]
- 62.Nilsson M, et al. , The role of tissue microstructure and water exchange in biophysical modelling of diffusion in white matter. MAGMA, 2013. 26(4): p. 345–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stikov N, et al. , Bound pool fractions complement diffusion measures to describe white matter micro and macrostructure. Neuroimage, 2011. 54(2): p. 1112–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mardia KV and Jupp PE, Directional statistics. Vol. 494 2009: John Wiley & Sons. [Google Scholar]
- 65.Cercignani M. and Alexander DC, Optimal acquisition schemes for in vivo quantitative magnetization transfer MRI. Magn Reson Med, 2006. 56(4): p. 803–810. [DOI] [PubMed] [Google Scholar]
- 66.Lampinen B, et al. , Optimal experimental design for filter exchange imaging: Apparent exchange rate measurements in the healthy brain and in intracranial tumors. Magnetic resonance in medicine, 2017. 77(3): p. 1104–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gudbjartsson H. and Patz S, The Rician distribution of noisy MRI data. Magn Reson Med, 1995. 34(6): p. 910–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Szczepankiewicz F, et al. , Tensor-valued diffusion encoding for diffusional variance decomposition (DIVIDE): Technical feasibility in clinical MRI systems. PloS One, 2019. 14(3): p. e0214238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zelinka I, SOMA—self-organizing migrating algorithm, in New optimization techniques in engineering. 2004, Springer; p. 167–217. [Google Scholar]
- 70.Whittall KP, et al. , In vivo measurement of T2 distributions and water contents in normal human brain. Magnetic Resonance in Medicine, 1997. 37(1): p. 34–43. [DOI] [PubMed] [Google Scholar]
- 71.Stikov N, et al. , In vivo histology of the myelin g-ratio with magnetic resonance imaging. Neuroimage, 2015. 118: p. 397–405. [DOI] [PubMed] [Google Scholar]
- 72.Nilsson M, et al. , An open-source framework for analysis of multidimensional diffusion MRI data implemented in MATLAB. In Proceedings of the 26th Annual Meeting of ISMRM, Paris, France, 2018 p. 5355. 2018. [Google Scholar]
- 73.Panagiotaki E, et al. , Compartment models of the diffusion MR signal in brain white matter: a taxonomy and comparison. Neuroimage, 2012. 59(3): p. 2241–2254. [DOI] [PubMed] [Google Scholar]
- 74.Ferizi U, et al. , A ranking of diffusion MRI compartment models with in vivo human brain data. Magnetic resonance in medicine, 2014. 72(6): p. 1785–1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zeng Q, et al. , A Modified Tri-Exponential Model for Multi-b-value Diffusion-Weighted Imaging: A Method to Detect the Strictly Diffusion-Limited Compartment in Brain. Frontiers in neuroscience, 2018. 12: p. 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tax CM, et al. , The dot-compartment revealed? Diffusion MRI with ultra-strong gradients and spherical tensor encoding in the living human brain. NeuroImage, 2020: p. 116534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dhital B, et al. , The absence of restricted water pool in brain white matter. Neuroimage, 2018. 182: p. 398–406. [DOI] [PubMed] [Google Scholar]
- 78.Sjölund J, et al. , Constrained optimization of gradient waveforms for generalized diffusion encoding. Journal of Magnetic Resonance, 2015. 261: p. 157–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Szczepankiewicz F, Westin CF, and Nilsson M, Maxwell-compensated design of asymmetric gradient waveforms for tensor-valued diffusion encoding. Mag Reson Med, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Szczepankiewicz F, Hoge S, and Westin C-F, Linear, planar and spherical tensor-valued diffusion MRI data by free waveform encoding in healthy brain, water, oil and liquid crystals. Data in brief, 2019. 25: p. 104208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hutter J, et al. Highly efficient diffusion MRI by slice-interleaved free-waveform imaging (SIFI). In proceeding of the 26th Annual Meeting of ISMRM, Montreal, Canada, 2018. 2018. [Google Scholar]
- 82.Vos SB, et al. , The importance of correcting for signal drift in diffusion MRI. Mag Reson Med, 2017. 77(1): p. 285–299. [DOI] [PubMed] [Google Scholar]
- 83.Setsompop K, et al. , Blipped-controlled aliasing in parallel imaging for simultaneous multislice echo planar imaging with reduced g-factor penalty. Mag Reson Med, 2012. 67(5): p. 1210–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Klein S, et al. , Elastix: a toolbox for intensity-based medical image registration. Medical Imaging, IEEE Transactions on, 2010. 29(1): p. 196–205. [DOI] [PubMed] [Google Scholar]
- 85.Nilsson M, et al. , Extrapolation-Based References Improve Motion and Eddy-Current Correction of High B-Value DWI Data: Application in Parkinson’s Disease Dementia. PloS one, 2015. 10(11): p. e0141825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rushton W, A theory of the effects of fibre size in medullated nerve. The Journal of physiology, 1951. 115(1): p. 101–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fieremans E, et al. Effect of combining linear with spherical tensor encoding on estimating brain microstructural parameters. In proceedings of the 26th Annual Meeting of ISMRM, Paris, France, 2018. 2018. [Google Scholar]
- 88.Veraart J, Fieremans E, and Novikov D, Quantifying neuronal microstructure integrity with TE dependent Diffusion imaging (TEdDI). In Proceedings of the 25th Annual Meeting of ISMRM, Honolulu, HI, USA, 2017 p. 836 2017. [Google Scholar]
- 89.Xu J, et al. , Mapping mean axon diameter and axonal volume fraction by MRI using temporal diffusion spectroscopy. NeuroImage, 2014. 103: p. 10–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Paus T, et al. , Structural maturation of neural pathways in children and adolescents: in vivo study. Science, 1999. 283(5409): p. 1908–1911. [DOI] [PubMed] [Google Scholar]
- 91.Dhital B, et al. , Intra-axonal diffusivity in brain white matter. Neuroimage, 2019. 189: p. 543–550. [DOI] [PubMed] [Google Scholar]
- 92.Lee H-H, et al. , Probing axonal swelling with time dependent diffusion MRI. arXiv preprint: 12685, 2019. [Google Scholar]
- 93.McKinnon ET and Jensen JH, Measuring intra-axonal T2 in white matter with direction-averaged diffusion MRI. Mag Reson Med, 2019. 81(5): p. 2985–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.De Santis S, Assaf Y, and Jones D. The influence of t2 relaxation in measuring the restricted volume fraction in diffusion MRI. In proceedings of the 24th Scientific Meeting, International Society for Magnetic Resonance in Medicine, Singapore, 2016. 2016. [Google Scholar]
- 95.Peled S, et al. , Water diffusion, T2, and compartmentation in frog sciatic nerve. Mag Reson Med, 1999. 42(5): p. 911–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wachowicz K. and Snyder RE, Assignment of the T2 components of amphibian peripheral nerve to their microanatomical compartments. Mag Reson Med, 2002. 47(2): p. 239–245. [DOI] [PubMed] [Google Scholar]
- 97.Bonilla I. and Snyder RE, Transverse relaxation in rat optic nerve. J Magnetic Resonance in Medicine, 2007. 20(2): p. 113–120. [DOI] [PubMed] [Google Scholar]
- 98.Dortch RD, et al. , Compartment-specific enhancement of white matter and nerve ex vivo using chromium. Magnetic resonance in medicine, 2010. 64(3): p. 688–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Dortch RD, et al. , Characterizing inter-compartmental water exchange in myelinated tissue using relaxation exchange spectroscopy. Mag Reson Med, 2013. 70(5): p. 1450–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.De Santis S, et al. , Resolving relaxometry and diffusion properties within the same voxel in the presence of crossing fibres by combining inversion recovery and diffusion-weighted acquisitions. Magnetic resonance in medicine, 2016. 75(1): p. 372–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ikari K. and Hayashi M, Aging in the Neuropil of Cerebral Cortex–A Quantitative Ultrastructural Study. Psychiatry and Clinical Neurosciences, 1981. 35(4): p. 477–486. [DOI] [PubMed] [Google Scholar]
- 102.Chklovskii DB, Schikorski T, and Stevens CF, Wiring optimization in cortical circuits. Neuron, 2002. 34(3): p. 341–347. [DOI] [PubMed] [Google Scholar]
- 103.Braitenberg V. and Schüz A, Cortex: statistics and geometry of neuronal connectivity. 2013: Springer Science & Business Media. [Google Scholar]
- 104.Englund E. and Brun A, White matter changes in dementia of Alzheimer’s type: the difference in vulnerability between cell compartments. Histopathology, 1990. 16(5): p. 433–439. [DOI] [PubMed] [Google Scholar]
- 105.Coelho S, et al. , Local volume fraction distributions of axons, astrocytes, and myelin in deep subcortical white matter. NeuroImage, 2018. 179: p. 275–287. [DOI] [PubMed] [Google Scholar]
- 106.Jespersen SN, et al. , Effects of nongaussian diffusion on “isotropic diffusion” measurements: an ex-vivo microimaging and simulation study. Journal of Magnetic Resonance, 2019. 300: p. 84–94. [DOI] [PubMed] [Google Scholar]
- 107.Lundell H, et al. , Multidimensional diffusion MRI with spectrally modulated gradients reveals unprecedented microstructural detail. Scientific reports, 2019. 9(1): p. 9026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Szczepankiewicz F, et al. Is spherical diffusion encoding rotation invariant? An investigation of diffusion time dependence in the healthy brain. In proceedings for the 27th Annual Meeting of ISMRM, Montreal, Canada 2019. [Google Scholar]
- 109.Palombo M, et al. , SANDI: a compartment-based model for non-invasive apparent soma and neurite imaging by diffusion MRI. arXiv preprint:.02832, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Stanisz GJ, et al. , An analytical model of restricted diffusion in bovine optic nerve. Magnetic Resonance in Medicine, 1997. 37(1): p. 103–111. [DOI] [PubMed] [Google Scholar]
- 111.Pasternak O, et al. , Free water elimination and mapping from diffusion MRI. Mag Reson Med, 2009. 62(3): p. 717–730. [DOI] [PubMed] [Google Scholar]
- 112.Sepehrband F, et al. , Perivascular space fluid contributes to diffusion tensor imaging changes in white matter. NeuroImage, 2019. 197: p. 243–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FIGURE S1 Validation of the CRLB by comparing the VW metric based on variance estimates from either CRLB or simulations.
FIGURE S2 A multi-slice version of Figure 5.
FIGURE S3 A test-retest experiment on an adult volunteer, showing test and retest parameter maps, difference maps, and Bland-Altman plots.
FIGURES S4 and S5 The importance of the number of ‘shells’ versus the degree of orientation dispersion for the ability to determine all model parameters.
FIGURE S6 Comparison of parameter maps obtained by fitting Eq. 12 directly to data versus those obtained using the ‘RotInv’ approach.
FIGURE S7 Comparison of parameter maps obtained using the two-compartment model (Eq. 12) versus a model extended to include a free water compartment.