Erythropoietin (EPO) stimulates erythroid differentiation and maturation. Though the transcriptional regulation of EPO has been well studied, the molecular determinants of EPO secretion remain unknown. Here, we generated a HEK293T reporter cell line that provides a quantifiable and selectable readout of intracellular EPO levels and performed a genome-scale CRISPR screen that identified SURF4 as an important mediator of EPO secretion. Targeting SURF4 with multiple independent single guide RNAs (sgRNAs) resulted in intracellular accumulation and extracellular depletion of EPO.
KEYWORDS: erythropoietin, CRISPR screen, erythropoiesis, erythropoietin
ABSTRACT
Erythropoietin (EPO) stimulates erythroid differentiation and maturation. Though the transcriptional regulation of EPO has been well studied, the molecular determinants of EPO secretion remain unknown. Here, we generated a HEK293T reporter cell line that provides a quantifiable and selectable readout of intracellular EPO levels and performed a genome-scale CRISPR screen that identified SURF4 as an important mediator of EPO secretion. Targeting SURF4 with multiple independent single guide RNAs (sgRNAs) resulted in intracellular accumulation and extracellular depletion of EPO. Both of these phenotypes were rescued by expression of SURF4 cDNA. Additionally, we found that disruption of SURF4 resulted in accumulation of EPO in the endoplasmic reticulum (ER) compartment and that SURF4 and EPO physically interact. Furthermore, SURF4 disruption in Hep3B cells also caused a defect in the secretion of endogenous EPO under conditions mimicking hypoxia, ruling out an artifact of heterologous overexpression. This work demonstrates that SURF4 functions as an ER cargo receptor that mediates the efficient secretion of EPO. Our findings also suggest that modulating SURF4 may be an effective treatment for disorders of erythropoiesis that are driven by aberrant EPO levels. Finally, we show that SURF4 overexpression results in increased secretion of EPO, suggesting a new strategy for more efficient production of recombinant EPO.
INTRODUCTION
Approximately one-third of the proteins encoded by the mammalian genome are secretory proteins (1, 2). These proteins traffic from the endoplasmic reticulum (ER) to the Golgi apparatus via coat protein complex II (COPII) vesicles before reaching their final destinations: endosomes, lysosomes, plasma membrane, or extracellular space. COPII vesicles have an inner coat composed of SAR1 and SEC23-SEC24 heterodimers and an outer coat composed of SEC13-SEC31 heterotetramers (3). Though transmembrane cargo proteins may directly interact with COPII components, the physical barrier created by the ER membrane prevents direct interaction between soluble cargos and the COPII coat. Therefore, soluble cargos either passively flow into COPII vesicles (bulk flow) or are captured in COPII vesicles through recognition by intermediary receptors or adapters (cargo capture) (4).
Support for receptor-mediated cargo capture arose from early electron microscopy studies and in vitro assays of cargo packaging in COPII vesicles, which demonstrated efficient selection and concentration of cargos into COPII vesicles, as well as physical interactions between soluble cargos and COPII components (4–9). Subsequent studies uncovered LMAN1 as the first ER cargo receptor that mediates ER export of soluble cargos in mammals (10–12). LMAN1, together with its adapter MCFD2, forms a complex that is required for the efficient secretion of coagulation factors V and VIII, cathepsins, and alpha1-antitrypsin (12–16). While a handful of additional interactions between soluble cargos and ER receptors have been described in mammals (4, 9, 17), the extent to which bulk flow and cargo capture contribute to recruitment of proteins in COPII vesicles is unclear. This is primarily due to the fact that ER cargo receptors that are necessary for the efficient secretion of the majority of soluble cargos remain unidentified.
Erythropoietin (EPO) is a glycoprotein that is produced predominantly by specialized kidney peritubular fibroblasts and secreted into the plasma (18–21). EPO binds to its receptor expressed on erythroid precursors and promotes cell survival, proliferation, and differentiation (22–24). EPO plays a crucial role in the regulation of red blood cell production (erythropoiesis). Failure to make sufficient amounts of EPO, as seen in the setting of chronic kidney disease, results in anemia. In contrast, supraphysiological EPO levels, as seen in the context of EPO-secreting tumors, result in polycythemia. Though the transcriptional regulation of EPO production has been well studied (25–30), the molecular basis of EPO trafficking remains poorly understood.
In this study, in an effort to identify proteins involved in EPO secretion, we developed a genome-scale CRISPR/Cas9 knockout screen that provides a quantifiable and selectable readout of intracellular EPO levels. This screen, followed by several validation experiments, identified the ER cargo receptor SURF4 as a key mediator of efficient EPO secretion. These findings suggest that modulation of SURF4 in the EPO-producing cells could provide a novel strategy for altering plasma EPO levels and therefore regulating erythropoiesis. Additionally, this report suggests a novel strategy for improving the efficiency of recombinant EPO production.
RESULTS
Generation of a reporter cell line that allows for a quantifiable and selectable readout of intracellular EPO levels.
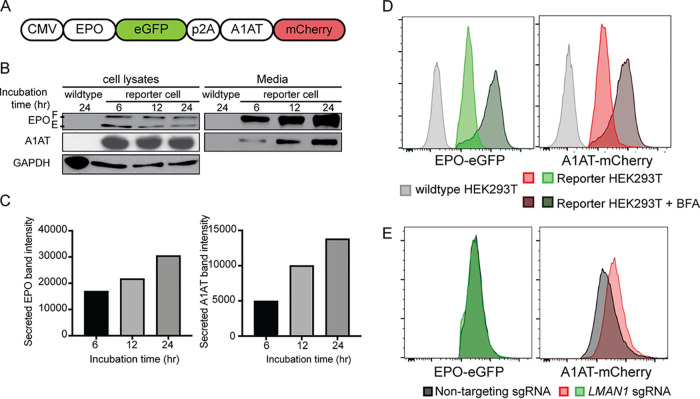
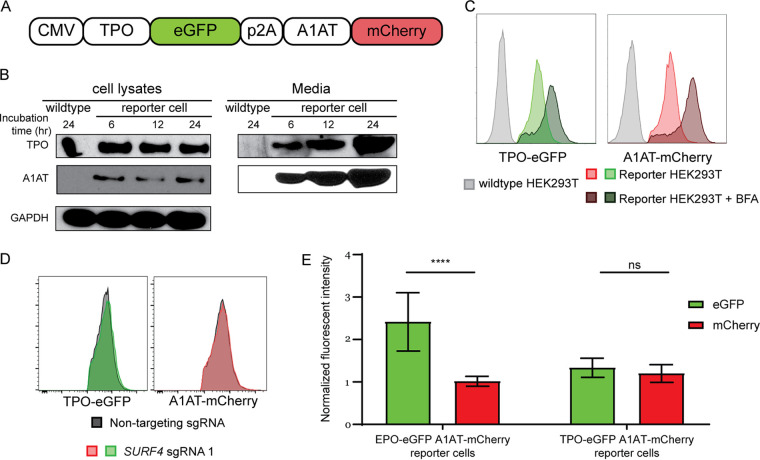
To identify proteins that regulate the intracellular trafficking of EPO, we developed a genome-scale functional screen that provides a quantifiable and selectable readout of intracellular EPO accumulation. Therefore, we generated a reporter HEK293T cell line stably expressing enhanced green fluorescent protein (eGFP)-tagged EPO (EPO-eGFP) and, as an internal control, mCherry-tagged alpha1-antitrypsin (A1AT-mCherry) (Fig. 1A). This cell line is herein referred to as the EPO-eGFP/A1AT-mCherry reporter cell line or just as the reporter cell line. Importantly, EPO-eGFP and A1AT-mCherry are equally expressed from the same cytomegalovirus (CMV) promoter, with their respective coding sequences separated by a sequence encoding a P2A peptide (Fig. 1A).
FIG 1.
A reporter HEK293T cell line stably expressing EPO-eGFP and A1AT-mCherry. (A) A construct that expresses EPO-eGFP and A1AT-mCherry from the same CMV promoter was assembled and used to generate the reporter cell line. A P2A sequence separates EPO-eGFP from A1AT-mCherry. (B) Intracellular and extracellular EPO-eGFP and A1AT-mCherry protein abundance was determined by Western blotting using anti-eGFP and anti-mCherry antibodies, respectively. E, ER form of EPO; F, fully glycosylated EPO. (C) Protein abundance was quantified using ImageJ. (D) Inhibiting ER-to-Golgi transport with brefeldin A (BFA) leads to intracellular accumulation of EPO-eGFP and A1AT-mCherry, as measured by fluorescence intensity. (E) LMAN1 deletion results in intracellular accumulation of A1AT with no effect on EPO. Unless stated otherwise, data are represented as mean values.
We found that both EPO and A1AT are efficiently secreted from the reporter cell line (Fig. 1B and C) and that disruption of ER-to-Golgi transport with brefeldin A results in intracellular accumulation of EPO and A1AT (Fig. 1D). Deletion of the gene for the ER cargo receptor for A1AT, LMAN1, resulted in intracellular accumulation of A1AT, as expected, with no effect on intracellular EPO (Fig. 1E), ruling out a role for LMAN1 in EPO secretion. These studies demonstrate that the machinery required for the efficient secretion of EPO via the classical secretory pathway is intact in this cell line and establish the utility of this cell line to identify modifiers of intracellular EPO levels.
A CRISPR/Cas9 loss-of-function screen identified SURF4 as a putative regulator of intracellular EPO level.
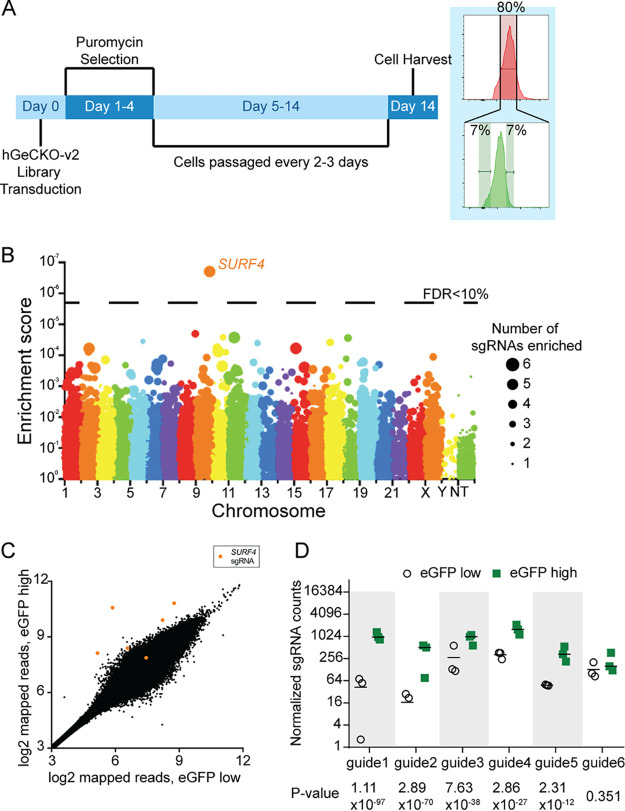
To identify proteins that affect EPO secretion, we mutagenized the EPO-eGFP/A1AT-mCherry reporter cell line with a CRISPR/Cas9 knockout library (hGeCKO-v2), which delivers SpCas9, a puromycin resistance cassette, and a pooled collection of 123,411 single guide RNAs (sgRNAs) that include 6 sgRNAs targeting nearly every gene in the human genome. Transduction of the library was performed at a low multiplicity of infection (MOI) (∼0.3), such that most infected cells receive one sgRNA to mutate one gene in the genome. Puromycin selection was applied from days 1 to 4 posttransduction. After an additional 9 days, cells with normal mCherry but increased (top ∼7%) or decreased (bottom ∼7%) eGFP fluorescence were isolated (Fig. 2A). This cell sorting strategy allows the identification of genes that affect EPO but not A1AT levels, therefore reducing the likelihood of identifying genes that affect global protein secretion. Integrated sgRNA sequences were quantified by deep sequencing and analyzed for their enrichment in the eGFP high compared to the eGFP low population.
FIG 2.
CRISPR/Cas9 loss-of-function screen to identify genes that affect intracellular EPO levels. (A) Screen strategy. Twenty-four hours following transduction of the CRISPR library, puromycin selection was applied for 3 days. At day 14, cells with unchanged mCherry but with top or bottom 7% eGFP fluorescence were isolated. sgRNA abundance was then determined in each cell population. (B) Gene level enrichment score was calculated for every gene using MAGeCK (see Materials and Methods). Each gene is represented by a bubble, the size of which is proportional to the number of sgRNAs with significant enrichment in the eGFP high population. SURF4 has the highest MAGeCK enrichment score and is the only gene for which the false discovery rate (FDR) is <10%. NT, nontargeting. (C) Normalized abundance of SURF4-targeting sgRNAs in the eGFP high and eGFP low populations. The abundance score was calculated from three biological replicates, using DEseq (see Materials and Methods). SURF4 sgRNAs are highlighted in orange. (D) Normalized counts for the six SURF4-targeting sgRNAs included in the library, for all three biological replicates. P values were calculated using MAGeCK. Unless stated otherwise, data are represented as mean values and the error bars represent standard deviations.
This screen, performed in biological triplicates, identified that the sgRNA sequences targeting only one gene, the surfeit locus protein 4 gene (SURF4), are enriched in the eGFP high population compared to the eGFP low population at a false discovery rate (FDR) of <10% (Fig. 2B). Five out of six sgRNAs targeting SURF4 were significantly enriched in the eGFP high population (Fig. 2C and D; also see Table S1 in the supplemental material).
SURF4 deletion results in intracellular accumulation and reduced secretion of EPO.
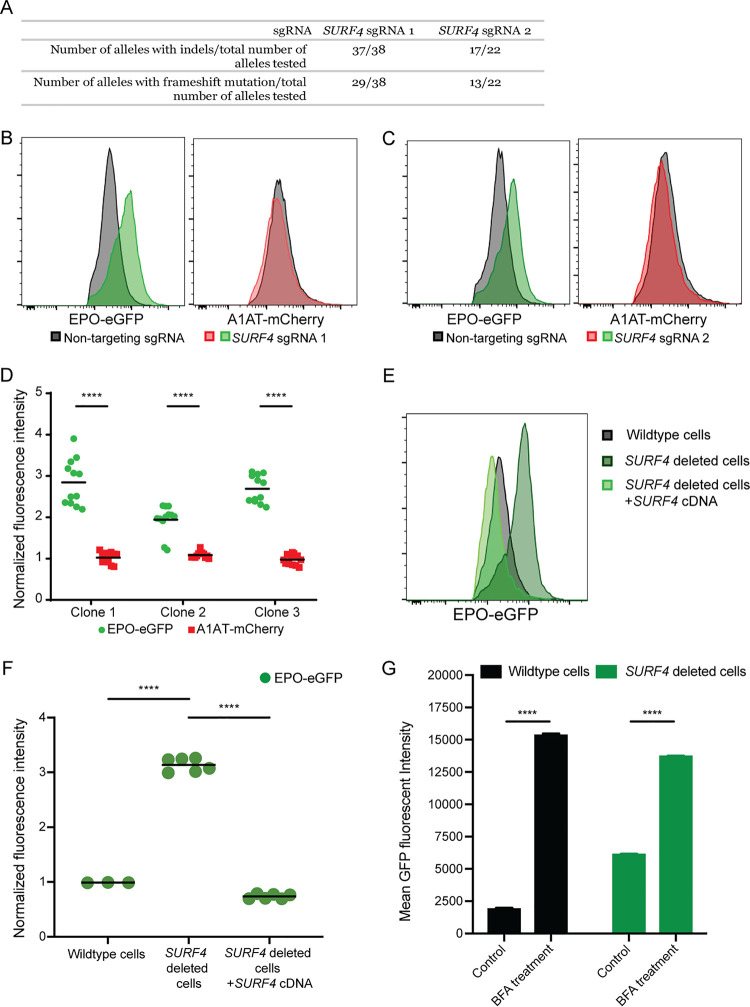
To validate the results of the screen, we targeted SURF4 with one sgRNA (sgRNA1) that showed significant enrichment in the whole-genome screen (Fig. 2D) and a second sgRNA (sgRNA2) not included in the hGeCKO-v2 library. SURF4 mutagenesis with sgRNA1 or sgRNA2 was highly efficient, resulting in insertions or deletions (indels) in ∼97% and 77% of alleles, respectively (Fig. 3A). Cells transduced with SURF4 sgRNA1 or sgRNA2 exhibited increased intracellular accumulation of EPO-eGFP, with no effect on A1AT-mCherry (Fig. 3B and C). This finding was confirmed in three independent EPO-eGFP/A1AT-mCherry reporter cell clones (Fig. 3D), ruling out an artifact unique to the clone used in the original screen.
FIG 3.
SURF4 deletion results in intracellular accumulation of EPO-eGFP. (A) SURF4-targeting sgRNA1 and sgRNA2 are highly efficient, causing indels in ∼97% and 77% of alleles, respectively. (B and C) Flow cytometry histograms showing intracellular accumulation of EPO, but not A1AT, following SURF4 deletion in HEK293T cells, using two independent sgRNAs, sgRNA1 (B) or sgRNA2 (C). (D) Quantification of intracellular mean fluorescence intensity in three independent clonal reporter cell lines transduced with SURF4-sgRNA1 (n = 12). Results were normalized to mean fluorescence intensity of cells transduced with nontargeting sgRNAs. (E and F) Flow cytometry histograms and normalized mean fluorescence intensity of EPO-eGFP in several clonal cell lines with sequence-confirmed SURF4 frameshift mutations (SURF4 deleted) with or without stable expression of wild-type SURF4 cDNA. Mean fluorescence intensity in panel F was normalized to that of wild-type cells. (G) Mean eGFP fluorescence intensity in wild-type and SURF4-deleted reporter cells with and without treatment with BFA (n = 3 per condition). ****, P < 0.0001.
To further confirm a direct effect of SURF4 deficiency on intracellular EPO accumulation, we next generated three clonal reporter cell lines with confirmed frameshift mutations of both SURF4 alleles by transient expression of SURF4 sgRNA1. The increased intracellular EPO protein levels observed in SURF4-deleted cells was completely rescued by a lentivirus expressing wild-type SURF4 cDNA (Fig. 3E and F), ruling out an off-target effect shared by sgRNA1 and sgRNA2. Taken together, these findings demonstrate that SURF4 disruption results in intracellular accumulation of EPO. Notably, disruption of ER-to-Golgi transport with brefeldin A further enhances the intracellular accumulation of EPO-eGFP in SURF4-deleted cells (Fig. 3G).
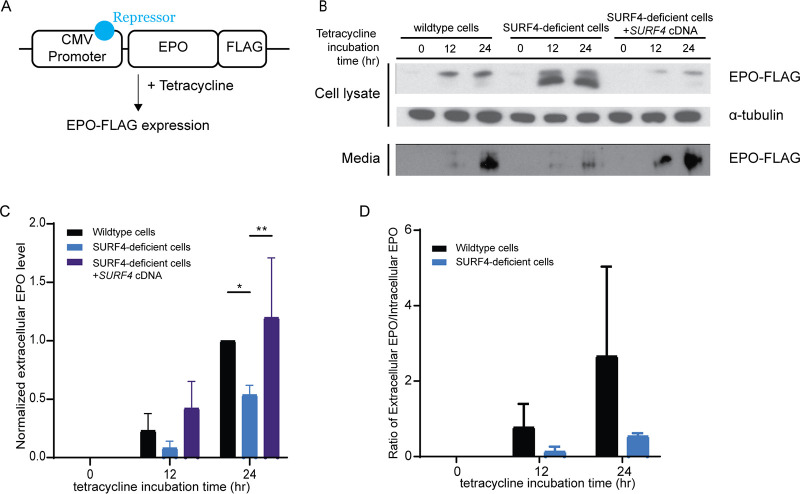
To rule out an indirect effect on EPO-eGFP secretion resulting from an interaction between eGFP and SURF4, we analyzed the dependence of FLAG-tagged EPO on SURF4 for secretion. We generated a wild-type and SURF4-deficient HEK293 cell line expressing FLAG-tagged EPO (EPO-FLAG) from a tetracycline-inducible promoter (Fig. 4A). Following tetracycline administration, the intracellular EPO accumulation was significantly more pronounced in SURF4-deficient cells than in wild-type cells (Fig. 4B), recapitulating the findings described above with EPO-eGFP and ruling out an indirect effect due to the eGFP tag.
FIG 4.
SURF4 mutagenesis causes reduced extracellular EPO-FLAG secretion. (A) We generated a Flp-In TREX HEK293 cell line with tetracycline-inducible EPO-FLAG expression. (B) Intracellular and extracellular EPO-FLAG abundance in wild-type, SURF4-deficient, and SURF4-rescued cells was measured by Western blotting (using anti-FLAG antibody) after 0, 12, and 24 h of incubation with tetracycline. α-Tubulin was used as a loading control. (C and D) Quantification of densitometry of extracellular EPO (C) and ratios of extracellular/intracellular EPO normalized to α-tubulin in 3 independent experiments (D). *, P < 0.05; **, P < 0.01 by two-way ANOVA.
SURF4 localizes to the ER membrane (31–33) and functions as an ER cargo receptor, suggesting that the increased accumulation of intracellular EPO in the setting of SURF4 deficiency is secondary to reduced EPO secretion. Consistent with this hypothesis, the extracellular EPO-FLAG protein level was considerably lower in the conditioned media of SURF4-deleted cells compared to the conditioned media of wild-type cells (Fig. 4B and C), as was the ratio of extracellular to intracellular EPO-FLAG levels (Fig. 4D). The latter findings observed in SURF4-deficient cells were rescued by stable expression of SURF4 cDNA (Fig. 4B and C). These results indicate that disruption of SURF4 results in a defect in EPO secretion.
SURF4 deletion results in accumulation of EPO in the ER.
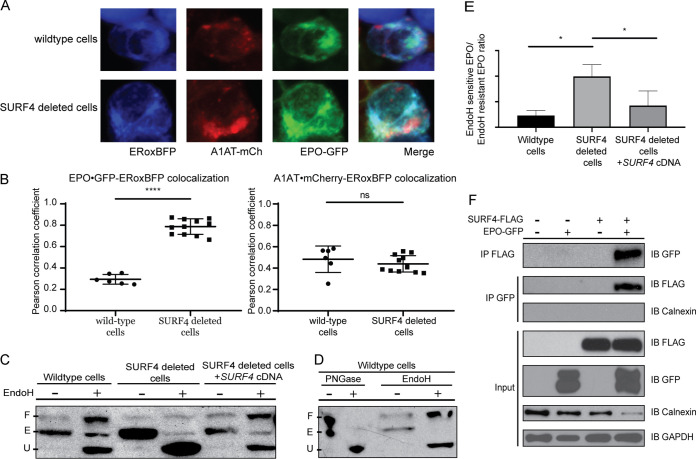
We next performed live-cell fluorescent confocal microscopy to determine the localization of accumulated EPO in the setting of SURF4 deletion. We cotransfected the EPO-eGFP/A1AT-mCherry reporter construct (Fig. 1A) with a vector expressing an ER blue fluorescent marker (ERoxBFP) into wild-type or SURF4-deficient HEK293 cells. We quantified the degree of colocalization between EPO and ERoxBFP (as well as A1AT and ERoxBFP, as a control) by Pearson correlation coefficient (PCC). SURF4-deficient cells exhibited an increased colocalization of EPO (but not A1AT) with ERoxBFP compared to wild-type cells (PCC of 0.7870 in SURF4-deleted cells versus 0.2934 in wild-type cells, P < 0.0001) (Fig. 5A and B).
FIG 5.
Disruption of SURF4 results in accumulation of EPO in the ER. (A) Live-cell fluorescent confocal microscopy of wild-type or SURF4-deleted reporter cells expressing the ER marker, ERoxBFP. (B) Quantification of the degree of colocalization between EPO and ERoxBFP, as well as A1AT and ERoxBFP as a control, by Pearson correlation coefficient. n = 6 for wild-type, n = 11 for SURF4-deficient cells. ****, P < 0.0001 by unpaired Student’s t test; ns, not significant. (C) Cell lysates were collected from wild-type, SURF4-deleted, or SURF4-rescued cells (SURF4-deleted cells with stable expression of wild-type SURF4 cDNA) expressing EPO-eGFP and were either treated with EndoH or left untreated. Immunoblotting was done with anti-eGFP antibody. The different forms of EPO (E, ER form of EPO [EndoH sensitive]; U, unglycosylated EPO; F, fully glycosylated EPO [post-Golgi form of EPO]) were confirmed by treating wild-type cells with either PNGase or EndoH (D). (E) Quantification of EndoH sensitivity from three independent experiments. *, P < 0.05. (F) FLAG antibody or eGFP antibody was used to immunoprecipitate EPO-eGFP or SURF4-FLAG, respectively, from lysates of cells expressing either EPO-eGFP, SURF4-FLAG, both, or neither. IP, immunoprecipitation; IB, immunoblotting.
To confirm the ER accumulation of EPO upon SURF4 disruption, we tested the glycosylation status of EPO in SURF4-deficient cells. EPO contains three N-glycosylation sites (34–36). In the ER, N-linked high-mannose oligosaccharides are added to EPO and further modifications are made in the Golgi apparatus. The ER form of EPO is cleavable by endoglycosidase H (EndoH) (37, 38), but the post-ER form is not. Therefore, the ratio of EndoH-cleaved to EndoH-uncleaved EPO will approximate the ratio of the amount of EPO in the ER versus the amount of EPO in the Golgi apparatus or beyond. In SURF4-deficient cells, the ratio of ER/post-ER form of EPO was significantly increased compared to that in wild-type cells (Fig. 5C to E), an effect that was decreased by stable expression of SURF4 cDNA (Fig. 5C and E). Taken together, these results demonstrate that SURF4 promotes the efficient ER exit and secretion of EPO.
SURF4 physically interacts with EPO.
To determine whether SURF4 binds to EPO, we tested for reciprocal coimmunoprecipitation of SURF4-FLAG and EPO-eGFP in HEK293T cells. An antibody against the FLAG epitope coimmunoprecipitated EPO-eGFP but not the ER luminal resident protein calnexin. Similarly, an antibody against eGFP coimmunoprecipitated FLAG-SURF4 (Fig. 5F). These results are consistent with a specific physical interaction between SURF4 and EPO.
Thrombopoietin (TPO) shares significant sequence homology with EPO. To test whether TPO, similarly to EPO, depends on SURF4 for efficient secretion, we generated two independent clonal HEK293 cells stably expressing and efficiently secreting TPO-eGFP and A1AT-mCherry (Fig. 6A and B). As expected, disruption of ER-to-Golgi transport with brefeldin A results in intracellular accumulation of TPO and A1AT (Fig. 6C). Notably, like A1AT, TPO did not accumulate intracellularly upon SURF4 deletion (Fig. 6D and E). These findings demonstrate the specificity of SURF4 for promoting EPO secretion and suggest that the SURF4-EPO interaction is mediated by one of the EPO domains not present in TPO.
FIG 6.
Thrombopoietin (TPO) secretion does not depend on SURF4. (A) A construct that expresses TPO-eGFP and A1AT-mCherry from the same CMV promoter was assembled and used to generate a reporter cell line stably expressing these two fusion proteins. (B) Intracellular and extracellular TPO-eGFP and A1AT-mCherry protein abundance was determined by Western blotting using anti-eGFP and anti-mCherry antibodies, respectively. (C) Inhibiting ER-to-Golgi transport with brefeldin A (BFA) leads to intracellular accumulation of TPO-eGFP and A1AT-mCherry, as measured by fluorescence intensity. (D) Flow cytometry histograms showing absence of intracellular accumulation of TPO following SURF4 deletion in HEK293T cells. (E) Quantification of cellular mean fluorescence intensity of TPO-eGFP and A1AT-mCherry in cells transduced with SURF4-targeting sgRNAs (n = 29). Results were normalized to mean fluorescence intensity of cells transduced with nontargeting sgRNAs. As a positive control, the same experiment was performed in parallel in reporter cell lines expressing EPO-eGFP and A1AT-mCherry (n = 48). ****, P < 0.0001; ns, not significant.
SURF4 promotes the secretion of endogenous EPO.
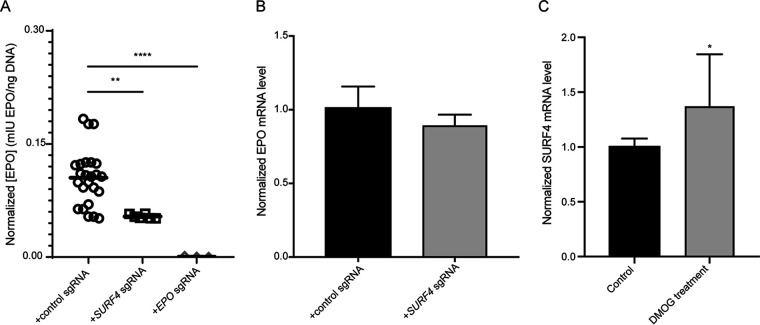
The experiments described above were performed in a heterologous cell line overexpressing EPO fused to either an eGFP or FLAG tag. To test the impact of SURF4 deletion on the secretion of endogenous EPO, we transduced human HEP3B cells with SURF4-targeting sgRNAs or control sgRNAs. As a positive control, a sgRNA targeting EPO resulted in profound reduction of extracellular EPO level to almost an undetectable (0.45% of control) level (Fig. 7A). Disruption of SURF4 in HEP3B cells using two independent sgRNAs resulted in a significant reduction (51.22% of control) of extracellular EPO levels compared to cells transduced with control sgRNAs (Fig. 7A), with no significant change in cellular EPO mRNA levels (Fig. 7B). Notably, SURF4 mRNA levels increased by ∼30% in cells treated with a prolyl hydroxylase inhibitor (Fig. 7C), suggesting that under conditions mimicking hypoxia, increased SURF4 expression may contribute to enhanced EPO secretion.
FIG 7.
SURF4 deletion in HEP3B cells results in reduced extracellular secretion of EPO expressed from its endogenous genomic locus. mIU, milli-international units. (A) HEP3B cells were transduced with lentivirus expressing SURF4-targeting sgRNAs, control sgRNAs, or EPO-targeting sgRNA as a positive control. EPO expression from its endogenous regulatory elements was subsequently induced with CoCl2 and measured in the conditioned media by ELISA and normalized to the amount of cellular DNA (a surrogate of the total number of cells). (B) EPO mRNA expression by qRT-PCR in wild-type (n = 8) and SURF4-deleted (n = 8) HEP3B cells following CoCl2 treatment. (C) SURF4 mRNA expression by qRT-PCR in wild-type HEP3B cells with and without dimethyloxalylglycine (DMOG) treatment (n = 8 per condition). *. P < 0.05; **, P < 0.01; ****, P < 0.0001.
SURF4 overexpression promotes more efficient EPO secretion.
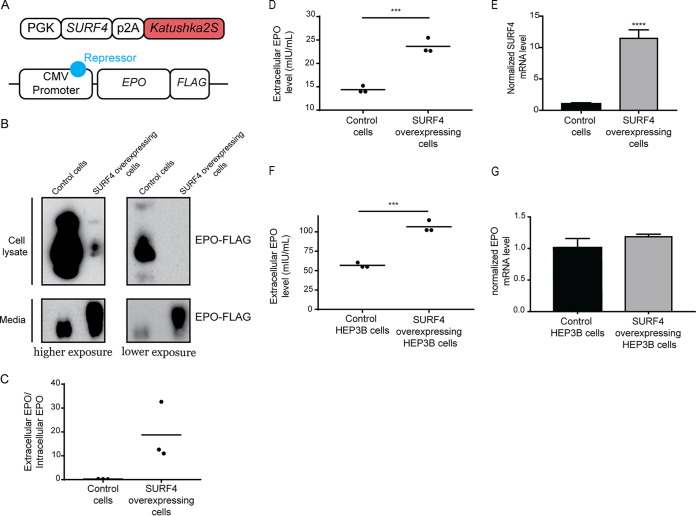
We next determined whether SURF4 overexpression promotes more efficient EPO secretion. We generated a lentivirus expressing equal amounts of SURF4 and Katushka2S (SURF4-p2A-Katushka2S) (Fig. 8A) and transduced it into HEK293 cells expressing EPO-FLAG from a tetracycline-inducible promoter. Cells with the highest (top 10%) and lowest (bottom 10%) SURF4 expression, as determined by Katushka2 fluorescence, were sorted by fluorescence-activated cell sorting (FACS). Following tetracycline administration, EPO level was found to be significantly increased in the conditioned media of cells overexpressing SURF4 compared to cells expressing low SURF4, with the reverse pattern observed intracellularly (Fig. 8B to E).
FIG 8.
SURF4 overexpression leads to enhanced EPO secretion. (A) A lentiviral construct that expresses equal amounts of SURF4 and Katushka2S from the same phosphoglycerate kinase (PGK) promoter was assembled and transduced into HEK293 cells expressing EPO-FLAG from a tetracycline-inducible promoter. Cells with top 10% and bottom 10% Katushka2S fluorescence were FACS sorted, corresponding to cells overexpressing SURF4 and control cells, respectively. (B and C) Intracellular and extracellular EPO abundance following a 12-h tetracycline incubation was analyzed by Western blotting (using anti-FLAG antibody) (B), and quantification of densitometry of the ratio of extracellular/intracellular EPO was determined in three independent experiments (C). (D) The extracellular EPO level was also measured by ELISA. (E) SURF4 mRNA expression by qRT-PCR in wild-type (n = 16) and top 10% SURF4-expressing reporter cells (n = 4). (F) HEP3B cells overexpressing SURF4 (and control cells) were generated as described above. Following incubation with CoCl2, the extracellular EPO level was measured by ELISA. (G) EPO mRNA expression by qRT-PCR in wild-type (n = 8) and SURF4-overexpressing (n = 3) HEP3B cells following CoCl2 treatment. ***, P < 0.001; ****, P < 0.0001 by unpaired t test.
To assess the impact of SURF4 overexpression on the secretion of EPO expressed from its endogenous genomic locus, we performed the same experiment described above in HEP3B cells. EPO level was increased in the conditioned media of cells expressing high compared to low SURF4 levels (Fig. 8F), with no significant change in cellular EPO mRNA levels (Fig. 8G). Taken together, these results demonstrate that SURF4 overexpression promotes more efficient EPO secretion.
DISCUSSION
In this report, we developed an unbiased genome-scale loss-of-function screen and identified SURF4 as a regulator of intracellular EPO levels. Deletion of SURF4 resulted in intracellular accumulation and extracellular depletion of EPO. Overexpression of SURF4 resulted in the reversed pattern. Consistent with the reported localization of SURF4 at the ER membrane (32, 33, 39), we found that disruption of SURF4 resulted in accumulation of EPO in the ER and that EPO and SURF4 physically interact. Taken together, these results strongly suggest that SURF4 is the ER cargo receptor that mediates the efficient secretion of EPO.
The screen described above was performed in a cell line with heterologous overexpression of EPO under the control of a CMV promoter. Therefore, it was important to examine whether SURF4 facilitates the secretion of EPO when expressed at a more physiological level. Accordingly, we deleted SURF4 in HEP3B cells that were induced to express EPO from its endogenous genomic locus and found that SURF4 also promotes EPO secretion under these conditions.
SURF4 is the mammalian ortholog of yeast Erv29p. Erv29p facilitates packaging of pro-alpha-factor in COPII vesicles promoting their ER-to-Golgi transport (31, 40, 41). Erv29p recycles back from the Golgi apparatus to ER via recognition of its well-conserved dilysine sorting signal by the COPI coat (42). In mammalian cells, only a handful of cargos (APOB, PCSK9, DSSP, AMLEX, and GH) have been shown to depend on SURF4 for efficient secretion (32, 33, 43). However, a recent report demonstrated that mice with germ line deletion of Surf4 exhibit early embryonic lethality (44) similar to Caenorhabditis elegans (33), suggesting the presence of one or more SURF4-dependent cargos with a critical function during embryogenesis. Future studies aimed at identifying the repertoire of cargos that depend on SURF4 for secretion are essential. Equally important are future studies that examine the transcriptional landscape of SURF4-deficient EPO-producing cells (such as HEP3B cells).
A recently published report suggested that the cargo proteins that depend on SURF4 for secretion contain an N-terminal tripeptide “ER-ESCAPE” motif (43). This motif is exposed following removal of the leader sequences and is recognized by SURF4 (43). However, an N-terminal “ER-ESCAPE” motif with high SURF4 binding affinity is not present in EPO. Additionally, we found that EPO, but not TPO, depends on SURF4 for efficient secretion. However, TPO has an N-terminal motif with a better predicted SURF4 binding affinity than EPO. These results suggest that the N-terminal “ER-ESCAPE” motif may not be a generalizable determinant of SURF4 interaction for all SURF4-dependent cargos.
Soluble cargos are exported from the ER via the passive “bulk flow” or the concentrative “cargo capture” processes, with several lines of evidence supporting one route versus the other (4). Though “bulk flow” and “cargo capture” are not mutually exclusive, this report provides support for the “cargo capture” model of EPO secretion. However, it is important to note that in our experimental conditions, ∼50 to 70% of extracellular EPO is reduced in the setting of SURF4 deficiency. Therefore, the secretion of the remaining EPO depends on either bulk flow or one or more separate and unidentified receptors.
Recent developments in genome engineering using CRISPR/Cas9 technology have dramatically enhanced the potential and efficacy of comprehensive, high-throughput genetic screens (45–59). Such strategies can be applied in vitro and in vivo to discover novel biologic insights. Our screen was designed to focus on posttranscriptional regulators of EPO by placing its expression under the control of a CMV promoter. Screening strategies similar to the one employed in this article and in a recently published report (32) might help identify additional ER cargo receptors for other soluble secreted proteins and shed more light into the extent of the contribution of “cargo capture” to recruitment of cargos into COPII vesicles.
Findings in this report may have important implications for erythropoiesis. EPO, the master regulator of erythropoiesis, is produced by specialized peritubular fibroblasts in the kidney. The transcriptional control of EPO via the hypoxia-inducible factor pathway has been well studied (28, 60–69), culminating in the development of prolyl hydroxylase inhibitors, a class of compounds that increase EPO production at the transcriptional level via activation of the hypoxia-inducible factor (70–77). These drugs are currently in clinical development, with several compounds in advanced phase 2 or 3 trials (78–82); however, there are numerous potential concerns and adverse effects of these drugs, including possible increased risks of malignancy and autoimmune disease (83–85). Similar to the transcriptional control of EPO, the intracellular signal transduction pathway downstream of the EPO receptor has also been well studied (86–88). In contrast, much less is known about the molecular basis of EPO trafficking. Our findings suggest that modulating SURF4 may be effective for the treatment of disorders of erythropoeisis that are driven by aberrant EPO levels (89–94). Though a handful of other cargos depend on SURF4 for their secretion (32, 33, 43), with additional cargos likely remaining to be identified, targeting SURF4 exclusively in the EPO-producing cells might alter plasma EPO levels and therefore regulate erythropoiesis without affecting other SURF4-dependent cargos that are expressed in other cells. Alternatively, an inhibitor that specifically disrupts the SURF4-EPO interaction would also be expected to have no effects on other cargos that bind SURF4.
Recombinant human EPO (rhEPO) is used clinically for the treatment of anemia due to chronic kidney disease, chemotherapy, or ziduvidine. rhEPO is also used to reduce the requirement of allogeneic red blood cell transfusion following certain elective surgeries. Though the use of rhEPO is indicated in only a subset of the above clinical scenarios, the rhEPO market size was valued at ∼7.4 billion U.S. dollars in 2016 (95). In this report, we demonstrate that SURF4 overexpression results in enhanced EPO secretion. This approach could be applied to increase the efficiency of rhEPO production, which might translate into reduced costs of this drug.
MATERIALS AND METHODS
Cell culture.
HEK293T and HEP3B cells were purchased from ATCC. Flp-In T-REx 293 cells were purchased from Invitrogen. HEK293T and Flp-In T-REx 293 cells were cultured in Dulbecco modified Eagle medium (DMEM) (Gibco) supplemented with 10% heat-inactivated fetal bovine serum (Peak Serum) and 1% penicillin-streptomycin (Gibco). HEP3B cells were cultured in alpha-MEM (Gibco) supplemented with 2 mM l-glutamine (Gibco), 10% heat-inactivated fetal bovine serum (Peak Serum), and 1% penicillin-streptomycin (Gibco). All cells were grown in a humidified 37°C incubator with 5% CO2.
Generation of the EPO-eGFP A1AT-mCherry reporter cell line.
The CMV-EPO-eGFP-p2A-A1AT-mCherry construct was assembled using the NEBuilder HiFi DNA assembly cloning kit (New England Biolabs [NEB]) using vector sequences derived from PCSK9-eGFP-p2A-A1AT-mCherry (32) and EPO cDNA obtained from Dharmacon. This construct expresses EPO linked to eGFP via a linker peptide formed of glycines, prolines, and alanines (GGAPAPAPAPAPAPAPAPG). HEK293T cells were transfected with CMV-EPO-eGFP-p2A-A1AT-mCherry using Fugene HD transfection reagent (Promega). Transfected cells were selected with 350 μg/ml hygromycin (Invitrogen). Five weeks following hygromycin selection, single cells were sorted into 96-well plates using a SY-3200 flow cytometer (Sony). Single cell clones were expanded and analyzed for stable expression of EPO-eGFP and A1AT-mCherry using an LSR Fortessa flow cytometer (BD Bioscience).
Generation of the TPO-eGFP A1AT-mCherry reporter cell line.
The TPO cDNA sequence was amplified from human liver RNA (deidentified tissue sample obtained from the tissue procurement core, University of Michigan, institutional review board [IRB] no. HUM00048303) and the CMV-TPO-eGFP-p2A-A1AT-mCherry construct was generated and transfected into HEK293T cells as described in the paragraph above. The same linker peptide linking TPO and eGFP (GGAPAPAPAPAPAPAPAPG) was used. Single cell clones were sorted, expanded, and analyzed for stable expression of TPO-eGFP and A1AT-mCherry as described above.
Expansion and lentiviral preparation of the pLentiCRISPRv2 library.
The pLentiCRISPRv2 whole-genome CRISPR library was obtained from Addgene (Addgene no. 1000000048, a gift from Feng Zhang [45]), expanded by 16 electroporations (8 for each half library) into Endura electrocompetent cells (Lucigen), and plated on 16 24.5-cm bioassay plates (ThermoFisher Scientific). Following a 12- to 14-h incubation at 37°C, colonies were harvested from agar plates, and pooled plasmids for each half library were isolated separately by Maxipreps using an EndoFree Plasmid Maxi kit (Qiagen). To prepare the pooled lentiviral library, 11.3 μg of each half library was cotransfected with 17 μg of psPAX2 (Addgene no. 12260, a gift from Didier Trono) and 11.3 μg of pCMV-VSV-G (Addgene no. 8454, a gift from Robert Weinberg [96]) using Lipofectamine LTX with PLUS reagent (ThermoFisher Scientific) into each of six T225 tissue culture flasks (ThermoFisher Scientific) containing HEK293T cells at ∼80 to 90% confluence. The medium was changed 24 h posttransfection, and viral supernatant was collected 12, 24, and 36 h afterwards. Media containing viral supernatant were centrifuged at 500 × g for 5 min, pooled, aliquoted, snap-frozen in liquid nitrogen, and stored at −80°C.
CRISPR/Cas9 loss-of-function genome-wide screen.
For each independent screen, more than 142 million reporter cells were plated in 15-cm tissue culture dishes (Corning) at 30% confluence. Cells were transduced with the lentiviral library (with 8 μg/ml Polybrene [Sigma]) at a multiplicity of infection (MOI) of ∼0.3. Twenty-four hours after viral transduction, puromycin selection (1 μg/ml; Sigma) was applied for 4 days. Subsequently, cells were kept at a logarithmic phase of growth and passaged every 2 or 3 days, maintaining more than 36 million cells in culture at all times in order to preserve library depth. Fourteen days posttransduction, ∼80 million cells were isolated from tissue culture dishes using 0.25% trypsin (Gibco), pelleted by centrifugation (350 × g, 4°C, 5 min), resuspended in cold phosphate buffered saline (PBS) plus 2% fetal bovine serum (FBS), and filtered through a 35-μm mesh into flow cytometry tubes (Corning). Cells were divided into 20 tubes and maintained on ice until sorting. Cells with normal mCherry fluorescence (mid 70 to 80% fluorescence) and top or bottom ∼7% eGFP fluorescence (∼4 million cells/population) were sorted using a BD FACSAria III (BD Biosciences) and collected into 15-ml polypropylene tubes (Cellstar) containing media. Genomic DNA was extracted using a DNeasy blood and tissue kit (Qiagen), and integrated lentiviral sgRNA sequences were amplified by a two-step PCR (20 cycles and 14 cycles, respectively) as previously described (32, 45) using a Herculase II fusion DNA polymerase kit (Agilent Biotechnologies). DNA was purified after each of the PCRs using a QIAquick PCR purification kit (Qiagen). Following the two-step PCR, DNA was analyzed with a bioanalyzer (Agilent), and the sgRNA amplicons were sequenced using a NextSeq 500 sequencing system (Illumina). On average, 23.5 million reads were generated for each sorted cell population of each screen. Overall, 98% of the reads had a per sequence quality score (phred-based base quality score) of greater than 30. A total of 104,331 sgRNA sequences were mapped and identified (along with the barcode corresponding to each cell population of each replicate) using a custom Perl script as previously described (32). Enrichment at the sgRNA and gene levels was analyzed using DESeq2 and MAGeCK, respectively (97, 98).
Disruption of candidate genes using CRISPR/Cas9.
sgRNAs targeting several genes and several nontargeting sgRNAs were cloned into the pLentiCRISPRv2 plasmid (Addgene no. 52961, a gift from Feng Zhang [45]) as previously described (46). pLentiCRISPR plasmids were packaged into lentivirus, using the same method described above. To disrupt genes in a population of cells, cells were transduced with lentivirus at an MOI of ∼0.3. Subsequently, transduced cells were selected with puromycin and passaged for 10 to 14 days prior to FACS analysis. For all validation experiments, a minimum of three biologic replicates were analyzed. The nontargeting sgRNA sequences (5′→3′) are sgRNA1 (GTTCATTTCCAAGTCCGCTG), sgRNA2 (CGTGTGTGGGTAAACGGAAA), sgRNA3 (GTATTACTGATATTGGTGGG), and sgRNA4 (TCATGCTTGCTTGGGCAAAA). SURF4-targeting sgRNA sequences (5′→3′) are sgRNA1 (TCAGACAGAGGCGCGCCACG) and sgRNA2 (CAGGTAGCCGCAGTTCCAGG).
Generation of SURF4-deficient clonal cell lines.
To generate clonal cell lines that are deficient for SURF4, a sgRNA targeting SURF4 exon 2 was cloned into the PX459 plasmid (Addgene no. 62988, a gift from Feng Zhang) as previously described (99), and the construct was transiently transfected into cells using Fugene HD transfection reagent (Promega). Twenty-four hours posttransfection, puromycin (1 μg/ml; Sigma) selection was applied for 3 days, and subsequently, single cells were sorted into each well of three 96-well plates using the SY-3200 flow cytometry instrument (Sony). Following expansion of the single cell clones, genomic DNA was extracted with QuickExtract (Epicentre) and indels were determined by amplification of the sgRNA target site by PCR using Herculase II Fusion DNA polymerase (Agilent Biotechnologies) and Sanger sequencing. The primers used for PCR and Sanger sequencing are SURF4 forward (TCTGTTCCTCACACACCCCGCCC) and SURF4 reverse (ACTCACTCAGCTGTCCCAGCAAG). Three independent single cell clones with homozygous frameshift indels in SURF4 (13-bp insertion, 1-bp insertion, and 10-bp deletion) were generated.
Flow cytometry analysis.
HEK293T cells were detached with 0.25% trypsin (Gibco), washed with PBS, collected by centrifugation (350 × g, 5 min, 4°C), resuspended in cold PBS with 0.1% bovine serum albumin (BSA) and 10 mM HEPES (Invitrogen), filtered with 70-μm cell strainers, and analyzed by BD LSR Fortessa (BD Bioscience). FlowJo (Tree Star) was used to calculate the mean fluorescence intensity and for further analysis.
Brefeldin A treatment.
HEK293 cells stably expressing EPO-eGFP (or TPO-eGFP) and A1At-mCherry were incubated with 1 μg/ml brefeldin A (Biolegend) for 12 h. Subsequently, cells were collected as described above and analyzed by flow cytometry for intracellular accumulation of EPO-eGFP (or TPO-eGFP) and A1AT-mCherry.
Western blots.
To prepare cell lysates, cells were washed in PBS, suspended in radioimmunoprecipitation assay (RIPA) buffer (Invitrogen) supplemented with cOmplete protease inhibitor cocktail (Sigma), briefly sonicated, and incubated for 30 min in the cold room with end-over-end rotation. Cell lysates were cleared by centrifugation to remove cell debris (20,000 × g, 30 min, 4°C) and were analyzed immediately or stored at −80°C until analysis. Protein quantification was performed using Pierce BCA protein assay kit (ThermoFisher Scientific) per the manufacturer’s instructions. Lysates were boiled for 5 min at 95°C with 4× Laemmli sample buffer (Bio-Rad) supplemented with β-mercaptoethanol. Equal amounts of proteins were loaded on either a 4 to 12% Bis-Tris gel or a 4 to 20% Tris-glycine gel (Invitrogen), and sodium dodecyl sulfate (SDS) gel electrophoresis was performed as previously described (100, 101). Proteins were then transferred onto a nitrocellulose membrane (Bio-Rad). Following blocking in 5% (wt/vol) milk−Tris-buffered saline with Tween (TBST), membranes were incubated with primary antibody at 4°C overnight, washed three times in TBST, probed with peroxidase-coupled secondary antibodies, washed again three times in TBST, and developed with SuperSignal West Pico Plus (ThermoFisher Scientific). For horseradish peroxidase (HRP)-conjugated primary antibodies, nitrocellulose membranes were incubated with these antibodies and immediately developed following three TBST washes. Densitometry was performed with ImageJ. To test for the secretion efficiency of various cargo proteins, cells were seeded at equal densities in six-well plates or 10-cm plates, and conditioned medium was collected at different time points, cleared by centrifugation (500 × g, 5 min, 4°C), and analyzed immediately (by Western blotting) as described above or stored at −80°C until analysis.
Antibodies.
The following antibodies were used for immunoblotting: anti-GFP (Abcam, ab290), anti-mCherry (Abcam, ab167453), anticalnexin (Cell Signaling, 2679S), anti-glyceraldehyde-3-phosphate dehydrogenase (anti-GAPDH) (Millipore, MAB374), HRP-conjugated anti-FLAG (Abcam, ab1238), anti-alpha-tubulin (Abcam, ab176560), HRP-conjugated anti-mouse IgG (Bio-Rad, catalog no. 1706516), and HRP-conjugated anti-rabbit IgG (Jackson ImmunoResearch Laboratories, catalog no. 111-035-003).
Tetracycline-induced EPO-FLAG expression.
The coding sequence of EPO with a C-terminal FLAG was cloned into pDEST-pcDNA5-BirA-FLAG (Invitrogen) using NEBuilder HiFi DNA assembly cloning kit (NEB). Wild-type, SURF4-deficient (with homozygous frameshift SURF4 indels), or SURF4-rescued (with homozygous frameshift SURF4 indels but with stable expression of SURF4 cDNA) Flp-In T-REx HEK293 cells with tetracycline-inducible expression of EPO-FLAG were generated as previously described (102). The canonical isoform of SURF4 encoded by NM_033161.3 was used. To induce the expression of EPO-FLAG, tetracycline (1 μg/ml) was added to the medium. Cells and medium were collected prior to the addition of tetracycline and 12 and 24 h following tetracycline. Intracellular and extracellular EPO levels were analyzed by Western blotting as described above.
EndoH assay.
HEK293T cells that are either wild type, SURF4 deficient, or SURF4 rescue (defined in the paragraph above) were transfected with a plasmid expressing EPO-eGFP. Thirty-six hours posttransfection, total cell lysates were prepared, and protein quantification was performed, both as described above. Lysates were incubated with denaturing buffer (NEB) 95°C for 10 min, and equal amounts of lysates (180 μg) were treated with either 1 μl of endoglycosidase H (EndoH) (NEB), peptide-N-glycosidase F (PNGase F) (NEB), or dimethyl sulfoxide (DMSO) as the control for 1 h (37°C). Subsequently, Laemmli buffer (Bio-Rad) was added, and the samples were boiled (95°C) for 5 min. Samples were loaded on a 4 to 12% Bis-Tris gel (Invitrogen), and Western blotting was performed as described above. This experiment was performed in biologic triplicates.
Live-cell confocal fluorescence microscopy.
Wild-type or SURF4-deficient HEK293T cells that stably express EPO-eGFP and A1AT-mCherry were transfected with a plasmid expressing ERoxBFP (Addgene no. 68126, a gift from Erik Snapp [103]). Twenty-four hours posttransfection, cells were seeded on Lab-Tek Chambered Coverglass (ThermoFisher). Fluorescent images were captured on a Nikon A2 confocal microscope. To quantify colocalization between two proteins, Pearson correlation coefficient was calculated using the Nikon Elements software. This experiment was performed and analyzed by an investigator in a blind manner (investigator unaware of the genotype of the cells).
Coimmunoprecipitation.
Flp-In T-Rex 293 cells that are either wild type, SURF4 deficient, or SURF4 rescued (defined above) were transfected with CMV-EPO-eGFP-p2A-A1AT-mCherry using Fugene HD transfection reagent (Promega). Twenty-four hours posttransfection, cells were washed with PBS and incubated in PBS containing 2 mM dithiobis (succinimidyl propionate) (Pierce) for 30 min at room temperature. Subsequently, 20 mM Tris-HCl (pH 7.5) was added to quench the reaction. Cells were then washed twice in PBS, and cell lysis was performed with the following lysis buffer (100 mM Tris, 10% glycerol, 1% NP-40, 130 mM NaCl, 5 mM MgCl2, 1 mM NaF, and 1 mM EDTA, supplemented with cOmplete protease inhibitor cocktail [pH 7.5]). Cell lysates were collected as described above and incubated overnight at 4°C with either anti-FLAG M2 magnetic beads (Sigma) or GFP-Trap beads (ChromoTek). Following five washes with lysis buffer, proteins were eluted from the beads via incubation with 2× Laemmli sample buffer containing β-mercaptoethanol for 15 min at room temperature.
Generation of cell lines expressing low or high SURF4 levels.
A construct expressing SURF4 and the Katushka2S fluorescent marker (PGK-SURF4-p2A-Katushka2S) was assembled with the NEBuilder HiFi DNA assembly cloning kit (NEB) using vector sequence derived from LV1-5 (Addgene no. 68411) and cDNAs of SURF4 and Katushka2S (a gift from Gary Luker [104]). The canonical isoform of SURF4 encoded by NM_033161.3 was used. The construct was packaged into lentivirus as described above and transduced at a MOI of ∼1 into Flp-In T-REx 293 or HEP3B cells. Transduced cells were selected with puromycin and passaged for 14 days prior to FACS sorting. Cells with top and bottom 10% Katushka2S fluorescence were sorted.
Generation of SURF4-deficient HEP3B cells.
Wild-type HEP3B cells were transduced with lentiviral sgRNA targeting SURF4, control sgRNA (combination of nontargeting sgRNAs and sgRNAs targeting genes that do not affect EPO such as BCL11A, MPL, and SERPINA1), or sgRNA targeting EPO as a positive control. Cells were selected with puromycin and passaged for at least 2 weeks prior to further analysis. EPO levels in the conditioned media were compared between SURF4-deleted cells and control cells, correcting for the total cell number at the time of EPO measurement. Genomic DNA was extracted from HEP3B cells using QuickExtract (Epicentre).
EPO ELISA.
Equal numbers of cells were seeded in 6-well or 24-well plates. For HEP3B cells, EPO production was stimulated with CoCl2 (75 μM, Sigma) for 24 h and conditioned medium was collected and cleared by centrifugation (500 × g, 5 min, 4°C). For Flp-In T-Rex HEK293 cells with tetracycline-inducible expression of EPO-FLAG, tetracycline (1 μg/ml) was added for 12 h and conditioned medium was collected, cleared by centrifugation (500 × g, 5 min, 4°C), and diluted 1:500. EPO level was measured in the conditioned medium using the Legend Max human erythropoietin enzyme-linked immunosorbent assay (ELISA) kit (Biolegend), according to the manufacturer’s instructions.
qRT-PCR.
RNA was isolated using the RNeasy kit (Qiagen), and cDNA was synthesized using the Superscript first-strand synthesis system for reverse transcription-PCR (RT-PCR) (Invitrogen) with random hexamers. Quantitative RT-PCR (qRT-PCR) was perfomed using Power SYBR green PCR Master Mix using the ViiA7 real-time qPCR System from ThermoFisher. Samples were analyzed in triplicates, and A1AT (forward primer [GGTCAACTGGGCATCACTAA] and reverse primer [GATGGTCAGCACAGCCTTAT]) was used as an internal control for EPO (forward primer [GGGAGCCCAGAAGGAAGCCAT] and reverse primer [CTGCAGGCCTCCCCTGTGTA]). TUBA1A (forward primer [CGATATTGAGCGTCCAACCTAT] and reverse primer [TTCAGGGCTCCATCAAATCTC]) was used as an internal control for SURF4 (forward primers CTCTTGTTGTGTGGCTCTTTG and GATGAGGAACCTGGCCCTGGG; respective reverse primers TGGTCTGGAAGAAGTCGTATTT and CCTGCCTCCGAGCTGCATGTA). The threshold cycle (2−ΔΔCT) method was used to determine relative gene expression.
Statistical analysis.
CRISPR screen data analysis was performed as described above. The statistical differences in mean fluorescence intensity between EPO-eGFP and A1AT-mCherry were compared by two-way analysis of variance (ANOVA). The difference in extracellular EPO-FLAG level among wild-type, SURF4-deficient, and SURF4-rescued Flp-In T-REx 293 cells were compared by two-way ANOVA. The Pearson correlation coefficient differences between wild-type and SURF4-deficient HEK293T cells were compared by unpaired t test. The statistical difference in extracellular EPO detected by EPO ELISA was assessed using an unpaired t test. The differences in the relative amounts of EndoH-sensitive EPO among wild-type, SURF4-deficient, and SURF4-rescued HEK 293T cells were assessed by one-way ANOVA.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grant R01 HL148333 (R. Khoriaty). This work was also supported by MCubed, a research seed-funding program for faculty at the University of Michigan (R. Khoriaty and K. Desch), and by the University of Michigan Rogel Cancer Center (R. Khoriaty). R. King was supported by NIH T32-CA009357. G. Balbin-Cuesta was supported by NIH T32-GM007315.
We declare that we have no competing interests.
Z. Lin and R. Khoriaty conceived the study and designed the experiments. Z. Lin performed the majority of the experiments. R. King, V. Tang, G. Myers, G. Balbin-Cuesta, A. Friedman, B. McGee, and B. Emmer performed additional experiments. Z. Lin, R. King, and R. Khoriaty analyzed most of the experimental data. K. Desch, P. Reddy, and B. Emmer helped analyze the results. V. Tang, A. B. Ozel, and D. Siemieniak analyzed the sequencing data. Z. Lin and R. Khoriaty wrote the manuscript with help from all authors. All the authors contributed to the integration and discussion of the results.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Braakman I, Bulleid NJ. 2011. Protein folding and modification in the mammalian endoplasmic reticulum. Annu Rev Biochem 80:71–99. doi: 10.1146/annurev-biochem-062209-093836. [DOI] [PubMed] [Google Scholar]
- 2.Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A, Olsson I, Edlund K, Lundberg E, Navani S, Szigyarto CA-K, Odeberg J, Djureinovic D, Takanen JO, Hober S, Alm T, Edqvist P-H, Berling H, Tegel H, Mulder J, Rockberg J, Nilsson P, Schwenk JM, Hamsten M, von Feilitzen K, Forsberg M, Persson L, Johansson F, Zwahlen M, von Heijne G, Nielsen J, Pontén F. 2015. Tissue-based map of the human proteome. Science 347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 3.Jensen D, Schekman R. 2011. COPII-mediated vesicle formation at a glance. J Cell Sci 124:1–4. doi: 10.1242/jcs.069773. [DOI] [PubMed] [Google Scholar]
- 4.Barlowe C, Helenius A. 2016. Cargo capture and bulk flow in the early secretory pathway. Annu Rev Cell Dev Biol 32:197–222. doi: 10.1146/annurev-cellbio-111315-125016. [DOI] [PubMed] [Google Scholar]
- 5.Bendayan M, Roth J, Perrelet A, Orci L. 1980. Quantitative immunocytochemical localization of pancreatic secretory proteins in subcellular compartments of the rat acinar cell. J Histochem Cytochem 28:149–160. doi: 10.1177/28.2.7354212. [DOI] [PubMed] [Google Scholar]
- 6.Kuehn MJ, Herrmann JM, Schekman R. 1998. COPII-cargo interactions direct protein sorting into ER-derived transport vesicles. Nature 391:187–190. doi: 10.1038/34438. [DOI] [PubMed] [Google Scholar]
- 7.Salama NR, Yeung T, Schekman RW. 1993. The Sec13p complex and reconstitution of vesicle budding from the ER with purified cytosolic proteins. EMBO J 12:4073–4082. doi: 10.1002/j.1460-2075.1993.tb06091.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Balch WE, McCaffery JM, Plutner H, Farquhar MG. 1994. Vesicular stomatitis virus glycoprotein is sorted and concentrated during export from the endoplasmic reticulum. Cell 76:841–852. doi: 10.1016/0092-8674(94)90359-x. [DOI] [PubMed] [Google Scholar]
- 9.Barlowe C, Orci L, Yeung T, Hosobuchi M, Hamamoto S, Salama N, Rexach MF, Ravazzola M, Mylene A, Schekman R. 1994. COPII: a membrane coat formed by Sec proteins that drive vesicle budding from the endoplasmic reticulum. Cell 77:895–907. doi: 10.1016/0092-8674(94)90138-4. [DOI] [PubMed] [Google Scholar]
- 10.Kappeler F, Klopfenstein DRC, Foguet M, Paccaud J-P, Hauri H-P. 1997. The recycling of ERGIC-53 in the early secretory pathway. J Biol Chem 272:31801–31808. doi: 10.1074/jbc.272.50.31801. [DOI] [PubMed] [Google Scholar]
- 11.Itin C, Roche A-C, Monsigny M, Hauri H-P. 1996. ERGIC-53 is a functional mannose-selective and calcium-dependent human homologue of leguminous lectins. Mol Biol Cell 7:483–493. doi: 10.1091/mbc.7.3.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nichols WC, Seligsohn U, Zivelin A, Terry VH, Hertel CE, Wheatley MA, Moussalli MJ, Hauri H-P, Ciavarella N, Kaufman RJ, Ginsburg D. 1998. Mutations in the ER-Golgi intermediate compartment protein ERGIC-53 cause combined deficiency of coagulation factors V and VIII. Cell 93:61–70. doi: 10.1016/S0092-8674(00)81146-0. [DOI] [PubMed] [Google Scholar]
- 13.Zhang B. 2009. Recent developments in the understanding of the combined deficiency of FV and FVIII. Br J Haematol 145:15–23. doi: 10.1111/j.1365-2141.2008.07559.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang B, Zheng C, Zhu M, Tao J, Vasievich MP, Baines A, Kim J, Schekman R, Kaufman RJ, Ginsburg D. 2011. Mice deficient in LMAN1 exhibit FV and FVIII deficiencies and liver accumulation of alpha1-antitrypsin. Blood 118:3384–3391. doi: 10.1182/blood-2011-05-352815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Appenzeller C, Andersson H, Kappeler F, Hauri H-P. 1999. The lectin ERGIC-53 is a cargo transport receptor for glycoproteins. Nat Cell Biol 1:330–334. doi: 10.1038/14020. [DOI] [PubMed] [Google Scholar]
- 16.Nyfeler B, Reiterer V, Wendeler MW, Stefan E, Zhang B, Michnick SW, Hauri HP. 2008. Identification of ERGIC-53 as an intracellular transport receptor of alpha1-antitrypsin. J Cell Biol 180:705–712. doi: 10.1083/jcb.200709100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gomez-Navarro N, Miller E. 2016. Protein sorting at the ER–Golgi interface. J Cell Biol 215:769–778. doi: 10.1083/jcb.201610031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bachmann S, Le Hir M, Eckardt KU. 1993. Co-localization of erythropoietin mRNA and ecto-5’-nucleotidase immunoreactivity in peritubular cells of rat renal cortex indicates that fibroblasts produce erythropoietin. J Histochem Cytochem 41:335–341. doi: 10.1177/41.3.8429197. [DOI] [PubMed] [Google Scholar]
- 19.Maxwell PH, Ferguson DJ, Nicholls LG, Iredale JP, Pugh CW, Johnson MH, Ratcliffe PJ. 1997. Sites of erythropoietin production. Kidney Int 51:393–401. doi: 10.1038/ki.1997.52. [DOI] [PubMed] [Google Scholar]
- 20.Kobayashi H, Liu Q, Binns TC, Urrutia AA, Davidoff O, Kapitsinou PP, Pfaff AS, Olauson H, Wernerson A, Fogo AB, Fong GH, Gross KW, Haase VH. 2016. Distinct subpopulations of FOXD1 stroma-derived cells regulate renal erythropoietin. J Clin Invest 126:1926–1938. doi: 10.1172/JCI83551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pan X, Suzuki N, Hirano I, Yamazaki S, Minegishi N, Yamamoto M. 2011. Isolation and characterization of renal erythropoietin-producing cells from genetically produced anemia mice. PLoS One 6:e25839. doi: 10.1371/journal.pone.0025839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gregory CJ. 1976. Erythropoietin sensitivity as a differentiation marker in the hemopoietic system: studies of three erythropoietic colony responses in culture. J Cell Physiol 89:289–301. doi: 10.1002/jcp.1040890212. [DOI] [PubMed] [Google Scholar]
- 23.Suzuki N, Ohneda O, Takahashi S, Higuchi M, Mukai HY, Nakahata T, Imagawa S, Yamamoto M. 2002. Erythroid-specific expression of the erythropoietin receptor rescued its null mutant mice from lethality. Blood 100:2279–2288. doi: 10.1182/blood-2002-01-0124. [DOI] [PubMed] [Google Scholar]
- 24.Wu H, Liu X, Jaenisch R, Lodish HF. 1995. Generation of committed erythroid BFU-E and CFU-E progenitors does not require erythropoietin or the erythropoietin receptor. Cell 83:59–67. doi: 10.1016/0092-8674(95)90234-1. [DOI] [PubMed] [Google Scholar]
- 25.Kim WY, Safran M, Buckley MRM, Ebert BL, Glickman J, Bosenberg M, Regan M, Kaelin WG. 2006. Failure to prolyl hydroxylate hypoxia-inducible factor alpha phenocopies VHL inactivation in vivo. EMBO J 25:4650–4662. doi: 10.1038/sj.emboj.7601300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kochling J, Curtin PT, Madan A. 1998. Regulation of human erythropoietin gene induction by upstream flanking sequences in transgenic mice. Br J Haematol 103:960–968. doi: 10.1046/j.1365-2141.1998.01081.x. [DOI] [PubMed] [Google Scholar]
- 27.Pugh CW, Tan CC, Jones RW, Ratcliffe PJ. 1991. Functional analysis of an oxygen-regulated transcriptional enhancer lying 3’ to the mouse erythrpoietin gene. Proc Natl Acad Sci U S A 88:10553–10557. doi: 10.1073/pnas.88.23.10553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang GL, Jiang BH, Rue EA, Semenza GL. 1995. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci U S A 92:5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suzuki N, Obara N, Pan X, Watanabe M, Jishage KI, Minegishi N, Yamamoto M. 2011. Specific contribution of the erythropoietin gene 3 enhancer to hepatic erythropoiesis after late embryonic stages. Mol Cell Biol 31:3896–3905. doi: 10.1128/MCB.05463-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jelkmann W. 2011. Regulation of erythropoietin production. J Physiol 589:1251–1258. doi: 10.1113/jphysiol.2010.195057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Belden WJ, Barlowe C. 2001. Role of Erv29p in collecting soluble secretory proteins into ER-derived transport vesicles. Science 294:1528–1531. doi: 10.1126/science.1065224. [DOI] [PubMed] [Google Scholar]
- 32.Emmer BT, Hesketh GG, Kotnik E, Tang VT, Lascuna PJ, Xiang J, Gingras AC, Chen XW, Ginsburg D. 2018. The cargo receptor SURF4 promotes the efficient cellular secretion of PCSK9. Elife 7:e38839. doi: 10.7554/eLife.38839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saegusa K, Sato M, Morooka N, Hara T, Sato K. 2018. SFT-4/Surf4 control ER export of soluble cargo proteins and participate in ER exit site organization. J Cell Biol 217:2073–2085. doi: 10.1083/jcb.201708115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Falck D, Haberger M, Plomp R, Hook M, Bulau P, Wuhrer M, Reusch D. 2017. Affinity purification of erythropoietin from cell culture supernatant combined with MALDI-TOF-MS analysis of erythropoietin N-glycosylation. Sci Rep 7:5324. doi: 10.1038/s41598-017-05641-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goldwasser E, Kung CK, Eliason J. 1974. On the mechanism of erythropoietin-induced differentiation. 13. The role of sialic acid in erythropoietin action. J Biol Chem 249:4202–4206. [PubMed] [Google Scholar]
- 36.Lai PH, Everett R, Wang FF, Arakawa T, Goldwasser E. 1986. Structural characterization of human erythropoietin. J Biol Chem 261:3116–3121. [PubMed] [Google Scholar]
- 37.Maley F, Trimble RB, Tarentino AL, Plummer TH Jr. 1989. Characterization of glycoproteins and their associated oligosaccharides through the use of endoglycosidases. Anal Biochem 180:195–204. doi: 10.1016/0003-2697(89)90115-2. [DOI] [PubMed] [Google Scholar]
- 38.Trumbly RJ, Robbins PW, Belfort M, Ziegler FD, Maley F, Trimble RB. 1985. Amplified expression of streptomyces endo-beta-N-acetylglucosaminidase H in Escherichia coli and characterization of the enzyme product. J Biol Chem 260:5683–5690. [PubMed] [Google Scholar]
- 39.Mitrovic S, Ben-Tekaya H, Koegler E, Gruenberg J, Hauri HP. 2008. The cargo receptors Surf4, endoplasmic reticulum-Golgi intermediate compartment (ERGIC)-53, and p25 are required to maintain the architecture of ERGIC and Golgi. Mol Biol Cell 19:1976–1990. doi: 10.1091/mbc.e07-10-0989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Malkus P, Jiang F, Schekman R. 2002. Concentrative sorting of secretory cargo proteins into COPII-coated vesicles. J Cell Biol 159:915–921. doi: 10.1083/jcb.200208074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Otte S, Barlowe C. 2004. Sorting signals can direct receptor-mediated export of soluble proteins into COPII vesicles. Nat Cell Biol 6:1189–1194. doi: 10.1038/ncb1195. [DOI] [PubMed] [Google Scholar]
- 42.Foley DA, Sharpe HJ, Otte S. 2007. Membrane topology of the endoplasmic reticulum to Golgi transport factor Erv29p. Mol Membr Biol 24:259–268. doi: 10.1080/09687860601178518. [DOI] [PubMed] [Google Scholar]
- 43.Yin Y, Garcia MR, Novak AJ, Saunders AM, Ank RS, Nam AS, Fisher LW. 2018. Surf4 (Erv29p) binds amino-terminal tripeptide motifs of soluble cargo proteins with different affinities, enabling prioritization of their exit from the endoplasmic reticulum. PLoS Biol 16:e2005140. doi: 10.1371/journal.pbio.2005140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Emmer BT, Lascuna PJ, Kotnik E, Saunders T, Khoriaty R, Ginsburg D. 2019. Murine Surf4 is essential for early embryonic development. bioRxiv 10.1101/541995. [DOI] [PMC free article] [PubMed]
- 45.Sanjana NE, Shalem O, Zhang F. 2014. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Joung J, Konermann S, Gootenberg JS, Abudayyeh OO, Platt RJ, Brigham MD, Sanjana NE, Zhang F. 2017. Genome-scale CRISPR-Cas9 knockout and transcriptional activation screening. Nat Protoc 12:828–863. doi: 10.1038/nprot.2017.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ihry RJ, Salick MR, Ho DJ, Sondey M, Kommineni S, Paula S, Raymond J, Henry B, Frias E, Wang Q, Worringer KA, Ye C, Russ C, Reece-Hoyes JS, Altshuler RC, Randhawa R, Yang Z, McAllister G, Hoffman GR, Dolmetsch R, Kaykas A. 2019. Genome-scale CRISPR screens identify human pluripotency-specific genes. Cell Rep 27:616–630.e6. doi: 10.1016/j.celrep.2019.03.043. [DOI] [PubMed] [Google Scholar]
- 48.Canver MC, Smith EC, Sher F, Pinello L, Sanjana NE, Shalem O, Chen DD, Schupp PG, Vinjamur DS, Garcia SP, Luc S, Kurita R, Nakamura Y, Fujiwara Y, Maeda T, Yuan GC, Zhang F, Orkin SH, Bauer DE. 2015. BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis. Nature 527:192–197. doi: 10.1038/nature15521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yamauchi T, Masuda T, Canver MC, Seiler M, Semba Y, Shboul M, Al-Raqad M, Maeda M, Schoonenberg VAC, Cole MA, Macias-Trevino C, Ishikawa Y, Yao Q, Nakano M, Arai F, Orkin SH, Reversade B, Buonamici S, Pinello L, Akashi K, Bauer DE, Maeda T. 2018. Genome-wide CRISPR-Cas9 screen identifies leukemia-specific dependence on a pre-mRNA metabolic pathway regulated by DCPS. Cancer Cell 33:386–400.e5. doi: 10.1016/j.ccell.2018.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grevet JD, Lan X, Hamagami N, Edwards CR, Sankaranarayanan L, Ji X, Bhardwaj SK, Face CJ, Posocco DF, Abdulmalik O, Keller CA, Giardine B, Sidoli S, Garcia BA, Chou ST, Liebhaber SA, Hardison RC, Shi J, Blobel GA. 2018. Domain-focused CRISPR screen identifies HRI as a fetal hemoglobin regulator in human erythroid cells. Science 361:285–290. doi: 10.1126/science.aao0932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu X, Zhang Y, Chen Y, Li M, Zhou F, Li K, Cao H, Ni M, Liu Y, Gu Z, Dickerson KE, Xie S, Hon GC, Xuan Z, Zhang MQ, Shao Z, Xu J. 2017. In situ capture of chromatin interactions by biotinylated dCas9. Cell 170:1028–1043.e19. doi: 10.1016/j.cell.2017.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang T, Wei JJ, Sabatini DM, Lander ES. 2014. Genetic screens in human cells using the CRISPR-Cas9 system. Science 343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Koike-Yusa H, Li Y, Tan E-P, Velasco-Herrera MDC, Yusa K. 2014. Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nat Biotechnol 32:267–273. doi: 10.1038/nbt.2800. [DOI] [PubMed] [Google Scholar]
- 54.Wang T, Yu H, Hughes NW, Liu B, Kendirli A, Klein K, Chen WW, Lander ES, Sabatini DM. 2017. Gene essentiality profiling reveals gene networks and synthetic lethal interactions with oncogenic Ras. Cell 168:890–903.e15. doi: 10.1016/j.cell.2017.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hart T, Chandrashekhar M, Aregger M, Steinhart Z, Brown KR, MacLeod G, Mis M, Zimmermann M, Fradet-Turcotte A, Sun S, Mero P, Dirks P, Sidhu S, Roth FP, Rissland OS, Durocher D, Angers S, Moffat J. 2015. High-resolution CRISPR screens reveal fitness genes and genotype-specific cancer liabilities. Cell 163:1515–1526. doi: 10.1016/j.cell.2015.11.015. [DOI] [PubMed] [Google Scholar]
- 56.Zhang R, Miner JJ, Gorman MJ, Rausch K, Ramage H, White JP, Zuiani A, Zhang P, Fernandez E, Zhang Q, Dowd KA, Pierson TC, Cherry S, Diamond MS. 2016. A CRISPR screen defines a signal peptide processing pathway required by flaviviruses. Nature 535:164–168. doi: 10.1038/nature18625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang E, Lu SX, Pastore A, Chen X, Imig J, Chun-Wei Lee S, Hockemeyer K, Ghebrechristos YE, Yoshimi A, Inoue D, Ki M, Cho H, Bitner L, Kloetgen A, Lin KT, Uehara T, Owa T, Tibes R, Krainer AR, Abdel-Wahab O, Aifantis I. 2019. Targeting an RNA-binding protein network in acute myeloid leukemia. Cancer Cell 35:369–384.e7. doi: 10.1016/j.ccell.2019.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.LaFleur MW, Nguyen TH, Coxe MA, Yates KB, Trombley JD, Weiss SA, Brown FD, Gillis JE, Coxe DJ, Doench JG, Haining WN, Sharpe AH. 2019. A CRISPR-Cas9 delivery system for in vivo screening of genes in the immune system. Nat Commun 10:1668. doi: 10.1038/s41467-019-09656-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sanson KR, Hanna RE, Hegde M, Donovan KF, Strand C, Sullender ME, Vaimberg EW, Goodale A, Root DE, Piccioni F, Doench JG. 2018. Optimized libraries for CRISPR-Cas9 genetic screens with multiple modalities. Nat Commun 9:5416. doi: 10.1038/s41467-018-07901-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Firth JD, Ebert BL, Pugh CW, Ratcliffe PJ. 1994. Oxygen-regulated control elements in the phosphoglycerate kinase 1 and lactate dehydrogenase A genes: similarities with the erythropoietin 3’ enhancer. Proc Natl Acad Sci U S A 91:6496–6500. doi: 10.1073/pnas.91.14.6496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Obara N, Suzuki N, Kim K, Nagasawa T, Imagawa S, Yamamoto M. 2008. Repression via the GATA box is essential for tissue-specific erythropoietin gene expression. Blood 111:5223–5232. doi: 10.1182/blood-2007-10-115857. [DOI] [PubMed] [Google Scholar]
- 62.Franke K, Gassmann M, Wielockx B. 2013. Erythrocytosis: the HIF pathway in control. Blood 122:1122–1128. doi: 10.1182/blood-2013-01-478065. [DOI] [PubMed] [Google Scholar]
- 63.Kapitsinou PP, Liu Q, Unger TL, Rha J, Davidoff O, Keith B, Epstein JA, Moores SL, Erickson-Miller CL, Haase VH. 2010. Hepatic HIF-2 regulates erythropoietic responses to hypoxia in renal anemia. Blood 116:3039–3048. doi: 10.1182/blood-2010-02-270322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zmajkovic J, Lundberg P, Nienhold R, Torgersen ML, Sundan A, Waage A, Skoda RC. 2018. A gain-of-function mutation in EPO in familial erythrocytosis. N Engl J Med 378:924–930. doi: 10.1056/NEJMoa1709064. [DOI] [PubMed] [Google Scholar]
- 65.Ang SO, Chen H, Hirota K, Gordeuk VR, Jelinek J, Guan Y, Liu E, Sergueeva AI, Miasnikova GY, Mole D, Maxwell PH, Stockton DW, Semenza GL, Prchal JT. 2002. Disruption of oxygen homeostasis underlies congenital Chuvash polycythemia. Nat Genet 32:614–621. doi: 10.1038/ng1019. [DOI] [PubMed] [Google Scholar]
- 66.Costa-Giomi P, Caro J, Weinmann R. 1990. Enhancement by hypoxia of human erythropoietin gene transcription in vitro. J Biol Chem 265:10185–10188. [PubMed] [Google Scholar]
- 67.Semenza GL, Nejfelt MK, Chi SM, Antonarakis SE. 1991. Hypoxia-inducible nuclear factors bind to an enhancer element located 3' to the human erythropoietin gene. Proc Natl Acad Sci U S A 88:5680–5684. doi: 10.1073/pnas.88.13.5680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Semenza GL, Koury ST, Nejfelt MK, Gearhart JD, Antonarakis SE. 1991. Cell-type-specific and hypoxia-inducible expression of the human erythropoietin gene in transgenic mice. Proc Natl Acad Sci U S A 88:8725–8729. doi: 10.1073/pnas.88.19.8725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Semenza GL. 2009. Involvement of oxygen-sensing pathways in physiologic and pathologic erythropoiesis. Blood 114:2015–2019. doi: 10.1182/blood-2009-05-189985. [DOI] [PubMed] [Google Scholar]
- 70.Ladroue C, Hoogewijs D, Gad S, Carcenac R, Storti F, Barrois M, Gimenez-Roqueplo AP, Leporrier M, Casadevall N, Hermine O, Kiladjian JJ, Baruchel A, Fakhoury F, Bressac-de Paillerets B, Feunteun J, Mazure N, Pouyssegur J, Wenger RH, Richard S, Gardie B. 2012. Distinct deregulation of the hypoxia inducible factor by PHD2 mutants identified in germline DNA of patients with polycythemia. Haematologica 97:9–14. doi: 10.3324/haematol.2011.044644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Percy MJ, Chung YJ, Harrison C, Mercieca J, Hoffbrand AV, Dinardo CL, Santos PC, Fonseca GH, Gualandro SF, Pereira AC, Lappin TR, McMullin MF, Lee FS. 2012. Two new mutations in the HIF2A gene associated with erythrocytosis. Am J Hematol 87:439–442. doi: 10.1002/ajh.23123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhuang Z, Yang C, Lorenzo F, Merino M, Fojo T, Kebebew E, Popovic V, Stratakis CA, Prchal JT, Pacak K. 2012. Somatic HIF2A gain-of-function mutations in paraganglioma with polycythemia. N Engl J Med 367:922–930. doi: 10.1056/NEJMoa1205119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gruber M, Hu CJ, Johnson RS, Brown EJ, Keith B, Simon MC. 2007. Acute postnatal ablation of Hif-2alpha results in anemia. Proc Natl Acad Sci U S A 104:2301–2306. doi: 10.1073/pnas.0608382104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rankin EB, Higgins DF, Walisser JA, Johnson RS, Bradfield CA, Haase VH. 2005. Inactivation of the arylhydrocarbon receptor nuclear translocator (Arnt) suppresses von Hippel-Lindau disease-associated vascular tumors in mice. Mol Cell Biol 25:3163–3172. doi: 10.1128/MCB.25.8.3163-3172.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Minamishima YA, Moslehi J, Bardeesy N, Cullen D, Bronson RT, Kaelin WG. 2008. Somatic inactivation of the PHD2 prolyl hydroxylase causes polycythemia and congestive heart failure. Blood 111:3236–3244. doi: 10.1182/blood-2007-10-117812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Querbes W, Bogorad RL, Moslehi J, Wong J, Chan AY, Bulgakova E, Kuchimanchi S, Akinc A, Fitzgerald K, Koteliansky V, Kaelin WG. 2012. Treatment of erythropoietin deficiency in mice with systemically administered siRNA. Blood 120:1916–1922. doi: 10.1182/blood-2012-04-423715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bernhardt WM, Wiesener MS, Scigalla P, Chou J, Schmieder RE, Gunzler V, Eckardt KU. 2010. Inhibition of prolyl hydroxylases increases erythropoietin production in ESRD. J Am Soc Nephrol 21:2151–2156. doi: 10.1681/ASN.2010010116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sankaran VG, Weiss MJ. 2015. Anemia: progress in molecular mechanisms and therapies. Nat Med 21:221–230. doi: 10.1038/nm.3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Joharapurkar AA, Pandya VB, Patel VJ, Desai RC, Jain MR. 2018. Prolyl hydroxylase inhibitors: a breakthrough in the therapy of anemia associated with chronic diseases. J Med Chem 61:6964–6982. doi: 10.1021/acs.jmedchem.7b01686. [DOI] [PubMed] [Google Scholar]
- 80.Maxwell PH, Eckardt KU. 2016. HIF prolyl hydroxylase inhibitors for the treatment of renal anaemia and beyond. Nat Rev Nephrol 12:157–168. doi: 10.1038/nrneph.2015.193. [DOI] [PubMed] [Google Scholar]
- 81.Kaplan J. 2019. Roxadustat and anemia of chronic kidney disease. N Engl J Med 381:1070–1072. doi: 10.1056/NEJMe1908978. [DOI] [PubMed] [Google Scholar]
- 82.Gupta N, Wish JB. 2017. Hypoxia-inducible factor prolyl hydroxylase inhibitors: a potential new treatment for anemia in patients with CKD. Am J Kidney Dis 69:815–826. doi: 10.1053/j.ajkd.2016.12.011. [DOI] [PubMed] [Google Scholar]
- 83.Yamamoto A, Hester J, Macklin PS, Kawai K, Uchiyama M, Biggs D, Bishop T, Bull K, Cheng X, Cawthorne E, Coleman ML, Crockford TL, Davies B, Dow LE, Goldin R, Kranc K, Kudo H, Lawson H, McAuliffe J, Milward K, Scudamore CL, Soilleux E, Issa F, Ratcliffe PJ, Pugh CW. 2019. Systemic silencing of PHD2 causes reversible immune regulatory dysfunction. J Clin Invest 129:3640–3656. doi: 10.1172/JCI124099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Semenza GL. 2003. Targeting HIF-1 for cancer therapy. Nat Rev Cancer 3:721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 85.Masoud GN, Li W. 2015. HIF-1alpha pathway: role, regulation and intervention for cancer therapy. Acta Pharm Sin B 5:378–389. doi: 10.1016/j.apsb.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kim AR, Ulirsch JC, Wilmes S, Unal E, Moraga I, Karakukcu M, Yuan D, Kazerounian S, Abdulhay NJ, King DS, Gupta N, Gabriel SB, Lander ES, Patiroglu T, Ozcan A, Ozdemir MA, Garcia KC, Piehler J, Gazda HT, Klein DE, Sankaran VG. 2017. Functional selectivity in cytokine signaling revealed through a pathogenic EPO mutation. Cell 168:1053–1064.e15. doi: 10.1016/j.cell.2017.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jun-Shen Huang L, Constantinescu SN, Lodish HF. 2001. The N-terminal domain of Janus kinase 2 is required for Golgi processing and cell surface expression of erythropoietin receptor. Mol Cell 8:1327–1338. doi: 10.1016/S1097-2765(01)00401-4. [DOI] [PubMed] [Google Scholar]
- 88.Halupa A, Bailey ML, Huang K, Iscove NN, Levy DE, Barber DL. 2005. A novel role for STAT1 in regulating murine erythropoiesis: deletion of STAT1 results in overall reduction of erythroid progenitors and alters their distribution. Blood 105:552–561. doi: 10.1182/blood-2003-09-3237. [DOI] [PubMed] [Google Scholar]
- 89.Victor RG, Stockton DW, Prchal JT. 2005. Congenital polycythemias/erythrocytoses. Haematologica 90:109–116. [PubMed] [Google Scholar]
- 90.Patnaik MM, Tefferi A. 2009. The complete evaluation of erythrocytosis: congenital and acquired. Leukemia 23:834–844. doi: 10.1038/leu.2009.54. [DOI] [PubMed] [Google Scholar]
- 91.Ng T, Marx G, Littlewood T, Macdougall I. 2003. Recombinant erythropoietin in clinical practice. Postgrad Med J 79:367–376. doi: 10.1136/pmj.79.933.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rizzo JD, Seidenfeld J, Piper M, Aronson N, Lichtin A, Littlewood TJ. 2001. Erythropoietin: a paradigm for the development of practice guidelines. Hematology Am Soc Hematol Educ Program 2001:10–30. doi: 10.1182/asheducation-2001.1.10. [DOI] [PubMed] [Google Scholar]
- 93.Rizzo JD, Lichtin AE, Woolf SH, Seidenfeld J, Bennett CL, Cella D, Djulbegovic B, Goode MJ, Jakubowski AA, Lee SJ, Miller CB, Rarick MU, Regan DH, Browman GP, Gordon MS, American Society of Clinical Oncology, American Society of Hematology. 2002. Use of epoetin in patients with cancer: evidence-based clinical practice guidelines of the American Society of Clinical Oncology and the American Society of Hematology. Blood 100:2303–2320. doi: 10.1182/blood-2002-06-1767. [DOI] [PubMed] [Google Scholar]
- 94.Jadersten M, Montgomery SM, Dybedal I, Porwit-MacDonald A, Hellstrom-Lindberg E. 2005. Long-term outcome of treatment of anemia in MDS with erythropoietin and G-CSF. Blood 106:803–811. doi: 10.1182/blood-2004-10-3872. [DOI] [PubMed] [Google Scholar]
- 95.Grand View Research. 2017. Erythropoietin (EPO) drugs market analysis by drug class (biologics, biosimilars), by product (epoetin-alfa, epoetin-beta, darbepoetin-alfa, others), by application, and segment forecasts, 2018–2025. https://www.grandviewresearch.com/industry-analysis/erythropoietin-epo-drugs-market.
- 96.Stewart SA, Dykxhoorn DM, Palliser D, Mizuno H, Yu EY, An DS, Sabatini DM, Chen IS, Hahn WC, Sharp PA, Weinberg RA, Novina CD. 2003. Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA 9:493–501. doi: 10.1261/rna.2192803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Li W, Xu H, Xiao T, Cong L, Love MI, Zhang F, Irizarry RA, Liu JS, Brown M, Liu S. 2014. MAGeCK enables robust identification of essential genes from genome-scale CRISPR:Cas9 knockout screens. Genome Biol 15:554. doi: 10.1186/s13059-014-0554-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. 2013. Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Khoriaty R, Everett L, Chase J, Zhu G, Hoenerhoff M, McKnight B, Vasievich MP, Zhang B, Tomberg K, Williams J, Maillard I, Ginsburg D. 2016. Pancreatic SEC23B deficiency is sufficient to explain the perinatal lethality of germline SEC23B deficiency in mice. Sci Rep 6:27802. doi: 10.1038/srep27802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Khoriaty R, Vasievich MP, Jones M, Everett L, Chase J, Tao J, Siemieniak D, Zhang B, Maillard I, Ginsburg D. 2014. Absence of a red blood cell phenotype in mice with hematopoietic deficiency of SEC23B. Mol Cell Biol 34:3721–3734. doi: 10.1128/MCB.00287-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Khoriaty R, Hesketh GG, Bernard A, Weyand AC, Mellacheruvu D, Zhu G, Hoenerhoff MJ, McGee B, Everett L, Adams EJ, Zhang B, Saunders TL, Nesvizhskii AI, Klionsky DJ, Shavit JA, Gingras AC, Ginsburg D. 2018. Functions of the COPII gene paralogs SEC23A and SEC23B are interchangeable in vivo. Proc Natl Acad Sci U S A 115:E7748–E7757. doi: 10.1073/pnas.1805784115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Costantini LM, Baloban M, Markwardt ML, Rizzo M, Guo F, Verkhusha VV, Snapp EL. 2015. A palette of fluorescent proteins optimized for diverse cellular environments. Nat Commun 6:7670. doi: 10.1038/ncomms8670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Luker KE, Pata P, Shemiakina II, Pereverzeva A, Stacer AC, Shcherbo DS, Pletnev VZ, Skolnaja M, Lukyanov KA, Luker GD, Pata I, Chudakov DM. 2015. Comparative study reveals better far-red fluorescent protein for whole body imaging. Sci Rep 5:10332. doi: 10.1038/srep10332. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.