Abstract
Background/Aim: Coronavirus disease 2019 (COVID-19) is an infectious disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). One drug that has attracted interest is the antiparasitic compound ivermectin, a macrocyclic lactone derived from the bacterium Streptomyces avermitilis. We carried out a docking study to determine if ivermectin might be able to attach to the SARS-CoV-2 spike receptor-binding domain bound with ACE2. Materials and Methods: We used the program AutoDock Vina Extended to perform the docking study. Results: Ivermectin docked in the region of leucine 91 of the spike and histidine 378 of the ACE2 receptor. The binding energy of ivermectin to the spike-ACE2 complex was -18 kcal/mol and binding constant was 5.8 e-08. Conclusion: The ivermectin docking we identified may interfere with the attachment of the spike to the human cell membrane. Clinical trials now underway should determine whether ivermectin is an effective treatment for SARS-Cov2 infection.
Keywords: COVID-19, ivermectin, ACE2, spike protein
Coronavirus disease 2019 (COVID-19) is an infectious disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). According to the World Health Organization, there are no available vaccines nor specific antiviral treatments for COVID-19. Management involves the treatment of symptoms, supportive care, isolation, and experimental measures, among them, the repurposing of old drugs. On 1 May 2020, the United States gave emergency use authorization to the antiviral remdesivir for people hospitalized with severe COVID-19.
One drug that has attracted interest is the antiparasitic compound ivermectin, a macrocyclic lactone derived from the bacterium Streptomyces avermitilis. Ivermectin kills parasites by interfering with nervous system and muscle function and enhancing inhibitory neurotransmission. Ivermectin binds to glutamate-gated chloride channels in the membranes of invertebrate nerve and muscle cells, causing increased permeability to chloride ions, resulting in cellular hyper-polarization, followed by paralysis and death (1).
Ivermectin shows broad-spectrum anti-viral activity in vitro. Caly et al. demonstrated that ivermectin inhibits SARS-CoV-2, with a single addition to Vero-hSLAM cells 2 h post infection. Caly et al. found ivermectin was able to achieve ~5,000-fold reduction in viral RNA at 48 h (2). But Caly et al. used high ivermectin concentrations. The relevance of their studies to human infection has been questioned (3) and FDA has warned that ivermectin in veterinary products should not be used for human therapy (4).
Patel et al. found that the administration of ivermectin during COVID-19 illness in hospitalized patients is associated with a lower mortality and hospital length of stay, although these findings have been called into question (5).
SARS-CoV-2 viral spike protein binds to the membrane-bound form of angiotensin-converting enzyme 2 (ACE2) to infect the host cell (6). For this reason, we carried out a docking study to determine if ivermectin might be able to attach to the SARS-CoV-2 spike receptor-binding domain bound with ACE2.
Materials and Methods
We used the program AutoDock Vina Extended to perform the docking study (7). The ivermectin molecule is from PubChem CID: 6321424 (Figure 1). Crystal structure of SARS-CoV-2 spike receptor-binding domain bound with ACE2 was deposited in the Protein Data Bank 2020-02-21, released: 2020-03-18 (8). We analyzed the human ACE2 receptor because this enzyme apparently differs among species, and the affinity of the virus for the human form may explain its particular infectivity for humans (9).
Figure 1. The ivermectin molecule. PubChem CID: 6321424.
Results
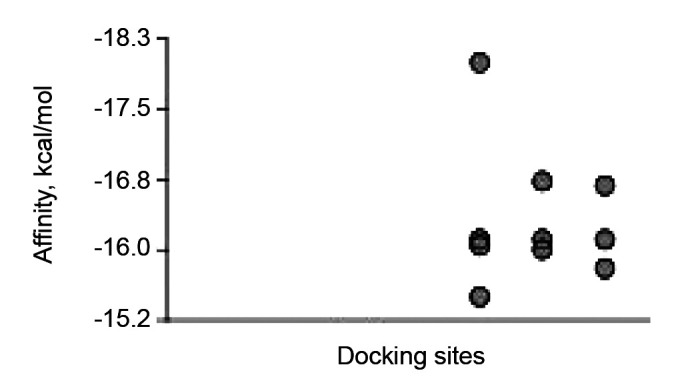
The root-mean-square deviations of atomic positions (RMSD in Angstroms) are tabulated in Table I. Lower values of RMSD indicate that docking is validated with higher accuracy. RMSD values of 3 or more indicate that no docking has occurred. Only one docking position with RMSD=0 is highly valid, and this docking position is shown in Figure 2 and Figure 3. The next 9 docking calculations were greater than 22 Angstroms RMSD, indicating that no docking was detected (Figure 4 and Table I).
Table I. Docking parameters calculated by AutoDock Vina Extended.
Lower values of root-mean-square deviations of atomic positions (RMSD) indicate that docking is validated with higher accuracy. RMSD values of 3 or more indicate no docking has occurred. Only one docking position, site 1, with RMSD=0 is highly valid.
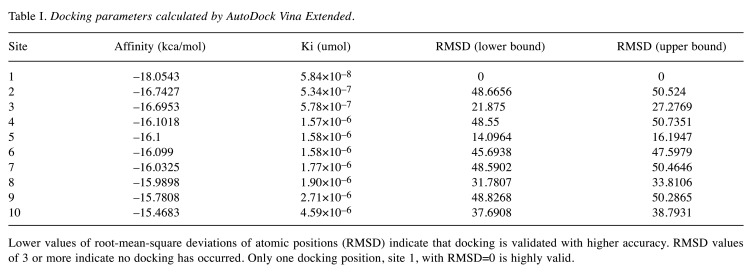
Figure 2. Ivermectin molecule docked with SARS-CoV-2 spike receptor-binding domain bound with ACE2. Triangle points to viral spike, arrow to ivermectin, star to ACE2 receptor.
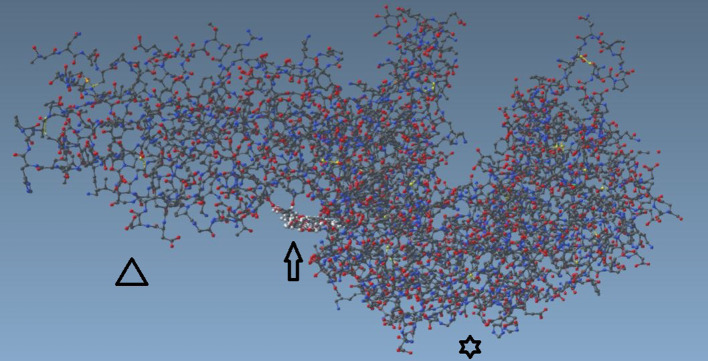
Figure 3. Ivermectin molecule docked with SARS-CoV-2 spike receptor-binding domain (left) bound with ACE2 (right) close-up view.
Figure 4. Binding affinity (kcal/mol) calculated for 10 docking sites. Only the one site with the highest affinity was a valid position. At the ther 9 no docking occurred.
Ivermectin docked in the region of leucine 91 of the spike and histidine 378 of the SARS Cov2-ACE2 receptor complex, between the SARS-Cov2 protein and the ACE2 protein. The binding energy of ivermectin to the spike-ACE2 complex was –18 kcal/mol and binding constant was 5.8×10–8.
Computational biochemical methods and docking software have been used to screen potential drugs in the structural protein and non-structural protein sites of SARS-Cov2. Ribavirin (a common antiviral drug), remdesivir, chloroquine and luteolin have been analyzed (10). SARS-CoV-2 main protease inhibitors have been predicted using Autodock (11), among them Hispidin and Lepidine E, two natural compounds, and folic acid (12).
The Vero-hSLAM cell assays performed by Caly et al. might not be entirely relevant to human SARS-Cov2 infection. Vero-hSLAM is an African green monkey kidney epithelial cell line that does not express human ACE2 (13). Therefore, Caly et al. had to use high ivermectin concentrations.
Conclusion
The ivermectin docking site we identified, between the viral spike and the ACE2 receptor, may interfere with the attachment of the spike to the human cell membrane. Our observation is consistent with the findings of Caly et al. and Patel et al. Clinical trials now underway should determine whether ivermectin is an effective treatment for SARS-Cov2 infection.
Conflicts of Interest
The Authors declare no conflicts of interest.
Authors’ Contributions
Drs. Lehrer and Rheinstein contributed equally to this work
References
- 1.Goa KL, McTavish D, Clissold SP. Ivermectin. A review of its antifilarial activity, pharmacokinetic properties and clinical efficacy in onchocerciasis. Drugs. 1991;42(4):640–658. doi: 10.2165/00003495-199142040-00007. [DOI] [PubMed] [Google Scholar]
- 2.Caly L, Druce JD, Catton MG, Jans DA, Wagstaff KM. The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antiviral Res. 2020;178:104787. doi: 10.1016/j.antiviral.2020.104787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bray M, Rayner C, Noel F, Jans D, Wagstaff K. Ivermectin and Covid-19: A report in antiviral research, widespread interest, an FDA warning, two letters to the editor and the authors’ responses. Antiviral Res. 2020;178:104805. doi: 10.1016/j.antiviral.2020.104805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.FDA: F.D.A. Letter to stakeholders: Do not use ivermectin intended for animals as treatment for covid-19 in humans. Available at: https://wwwfdagov/animal-veterinary/productsafety-information/fda-letter-stakeholders-do-not-use-ivermectinintended-animals-treatment-covid-19-humans [Last accessed on 18 Jun 2020]
- 5.Servick K, Enserink M. The pandemic’s first major research scandal erupts. Science. 2020;368(6495):1041–1042. doi: 10.1126/science.368.6495.1041. [DOI] [PubMed] [Google Scholar]
- 6.South AM, Diz DI, Chappell MC. Covid-19, ace2, and the cardiovascular consequences. Am J Physiol Heart Circ Physiol. 2020;318(5):H1084–H1090. doi: 10.1152/ajpheart.00217.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Trott O, Olson AJ. Autodock vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, Zhang Q, Shi X, Wang Q, Zhang L, Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ace2 receptor. Nature. 2020;581(7807):215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 9.Shang J, Ye G, Shi K, Wan Y, Luo C, Aihara H, Geng Q, Auerbach A, Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581(7807):221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu R, Chen L, Lan R, Shen R, Li P. Computational screening of antagonists against the SARS-CoV-2 (covid-19) coronavirus by molecular docking. Int J Antimicrob. 2020;Agents:106012. doi: 10.1016/j.ijantimicag.2020.106012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mothay D, Ramesh KV. Binding site analysis of potential protease inhibitors of Covid-19 using autodock. Virusdisease. 2020:1–6. doi: 10.1007/s13337-020-00585-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Serseg T, Benarous K, Yousfi M. Hispidin and lepidine e: Two natural compounds and folic acid as potential inhibitors of 2019-novel coronavirus main protease (2019-ncovmpro), molecular docking and sar study. Curr Comput Aided Drug Des. 2020 doi: 10.2174/1573409916666200422075440. [DOI] [PubMed] [Google Scholar]
- 13.Mossel EC, Huang C, Narayanan K, Makino S, Tesh RB, Peters CJ. Exogenous ace2 expression allows refractory cell lines to support severe acute respiratory syndrome coronavirus replication. J Virol. 2005;79(6):3846–3850. doi: 10.1128/JVI.79.6.3846-3850.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]