Abstract
Background: Antibody testing is necessary to identify immune individuals in the post-initial wave of the COVID-19 pandemic. Patients and Methods: We prospectively evaluated the performance of a quantitative point-of-care test (POCT) for SARS-CoV-2 antibodies. The patient group (PG) comprised of hospitalized confirmed COVID-19 cases. Asymptomatic healthcare volunteers with negative rRT-PCR were included in the control group (CG). Measurement of IgM and IgG was obtained by dry fluorescence immunoassay. Results: Twenty-six PG (65.9±15.4 years old, male 57.7%) and 18 CG (45.6±10.1 years old, male 33.3%) were included. By manufacturer’s cut-off (≥0.04 mIU/ml), sensitivity and specificity were 73.08% and 88.89% for IgM and 88.46% and 33.33% for IgG, respectively. Estimated areas under the ROC curve were 0.907 and 0.848 for IgM and IgG, respectively. Results were improved using a cut-off of IgM ≥0.05 mIU/ml and IgG ≥0.10 mIU/ml. Conclusion: Using stringent cut-off values, SARS-CoV-2 antibody POCT detects immune people and can be used during socioeconomic normalization of communities.
Keywords: SARS-CoV-2, novel coronavirus, COVID-19, antibodies, point-of-care testing, IgG, IgM, diagnostics
Since the onset of COVID-19 pandemic, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has affected more than 6,000,000 people, leading to more than 300,000 casualties worldwide (1). The ability of COVID-19 to cause asymptomatic or paucisynptomatic infection and explosively spread has overwhelmed even the most resilient and sustainable health systems (2). The containment of a pandemic of such magnitude imposes the implementation of tight precaution measures, such as widespread physical distancing and population-level movement restrictions, as well as vigilant screening for possible cases (3,4).
Nucleic acid detection-based approaches (real-time reverse transcriptase polymerase chain reaction – rRT-PCR) are used as reference diagnostic standards; however, challenges include the lack of capacity for laboratory-based testing in resource-limited areas, the global shortage of molecular reagents as well as the performance variability of rRT-PCR influenced by multiple factors (5-8). Thus, the need for widely available, rapid diagnostic methods is apparent (9-12).
Importantly, the employment of highly accurate, quick and widely available diagnostic tools for large-scale population screening is paramount and will be important for a safe transition back to a socioeconomic normalization of communities (2). Point-of-care tests (POCTs) detecting either viral antigens or human antibodies are in pipeline, but there is still inadequate evidence supporting their accuracy and clinical utility (9). In this pilot study we aimed to evaluate the performance of a new anti-SARS-CoV-2 antibody detection POCT in a European centre.
Patients and Methods
Participants. We conducted a single-centre prospective observational study in a tertiary teaching hospital between 30th March 2020 and 6th April 2020. Data collection was planned before the index test conduction. Subjects were prospectively selected based on the results of the real-time Reverse Transcription Polymerase Chain reaction (rRT-PCR) for SARS-CoV-2 and clinical symptoms.
Two groups of eligible participants were enrolled. The patient group (PG) consisted of hospitalized symptomatic patients with rRT-PCR confirmed COVID-19 infection. The control group (CG) consisted of hospital asymptomatic volunteers, with no clinical symptoms for the past month, with negative SARS-CoV-2 rRT-PCR at the day of sampling and no reported “close contact” history (based on the ECDC definitions for confirmed cases and close contacts) (13). All participants enrolled in this study were adults (≥18 years old). No additional exclusion criteria were applied. PG was further divided into three subgroups based on the days between onset of symptoms and testing with the IgG/IgM POCT: the early (<7 days), middle (7-14 days) and late COVID-19 group (>14 days), respectively. Definitions for disease severity were: 1) mild: mild symptoms, no imaging findings of pneumonia; 2) moderate: fever or respiratory symptoms; 3) severe: respiratory distress and respiratory rate >30/min or saturation <93% at rest or arterial partial pressure of oxygen to inspired fraction of oxygen ≤300 mmHg; 4) critical: respiratory failure requiring mechanical ventilation (including acute respiratory distress syndrome - ARDS) or shock or other organ failure requiring ICU.
Respiratory samples. Nasopharyngeal and/or oropharyngeal swabs were collected and transferred to the Clinical Microbiology laboratory, immersed in an appropriate virus transport medium (e.g. UTM Viral Transport, Copan Diagnostics Inc., Brescia, Italy). Flocked swabs made from synthetic material were preferred for sample collection in order to maximize viral recovery. Lower respiratory tract samples (e.g. bronchoalveolar lavage or aspirates, sputum, etc.) were also accepted.
RNA extraction and real time RT-PCR. Automated purification of viral RNA from either the viral transport medium or lower respiratory tract samples was performed using the QIAsymphony DSP virus/pathogen mini kit on the QIAsymphony SP platform (QIAGEN, Hilden, Germany). A real time, one – step reverse transcription – PCR, specific for ORF1ab gene of SARS-CoV-2 and for N gene of all, or other coronaviruses was performed on the Rotor-Gene Q MDx thermocycler (QIAGEN), using the VIASURE SARS-CoV-2 Real Time PCR Detection Kit (CerTest Biotec SL, Zaragoza, Spain).
Antibody assay and measurement. The (COVID-19) IgG/IgM Test Kit by Lansion Biotechnology Co., Ltd. (Nanjing, PR China) (14) was utilized in this study, based on the availability of quantitative antibody POCT kits for SARS-CoV-2 during the study period in Greece. The test obtains a quantitative in vitro measurement of both IgG and IgM anti-SARS-CoV-2 antibodies levels simultaneously, using dry fluorescence immunoassay via a portable analyzer. By a finger-prick, 5μl of whole blood were collected in a micropipette and delivered on a test strip. Following the immediate addition of 100μl-140μlof a sample diluent, the test strip was left in room temperature for 15 min and then inserted into the analyzer, as per the manufacturer’s instructions. Results were immediately displayed on the analyzer’s screen in mIU/ml for each antibody. The pre-defined manufacturer’s cut-off of antibody levels for positive results was ≥0.04 mIU/ml for both IgG and IgM antibodies.
Statistical analysis. Age comparisons between groups were based on independent samples t-test or one-way ANOVA, accordingly followed by Bonferroni post-hoc tests, where necessary. Comparison of sex distribution between patients and controls was based on Fisher’s exact test. ROC graphs and estimation of the corresponding area under the curve was performed using the non-parametric ROC method. Confidence intervals (CI) for sensitivity and specificity were based on exact results from the binomial distribution. Comparisons of IgM, IgG and IgM/IgG ratio between patients and controls were performed using the Man-Whitney U-test whereas trends according to time and severity of disease were assessed using the non-parametric test for trend.
All analyses were performed using the Stata Statistical Software (StataCorp. Stata Statistical Software: Release 15.1. College Station, TX: StataCorp LP). p-Values less than 0.05 were considered as indicating statistical significance.
Ethical statement. This study was approved by the Research Ethics Committee of the participating institution (protocol number: 151/30-3-2020) and was conducted according to the STARD 2015 reporting guidelines and in line with the Declaration of Helsinki, as revised in 2013 (15). Informed consent was obtained from the participants.
Results
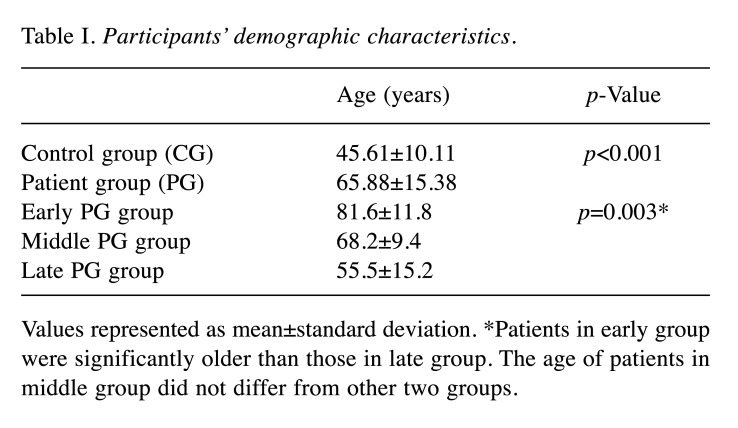
Overall, 44 participants (57.6±16.7 years old, male 47.7%), 26 PG (male 57.7%) and 18 CG (male 33.3%) were examined. Within PG, 5, 11 and 10 patients fulfilled the definition of early, middle and late COVID-19 infection respectively. Among patients 8, 8 and 10 were characterized with mild, moderate and severe and/or critical COVID-19 disease, respectively. Demographics are presented in Table I. Differences in sex distribution between patients and controls was not statistically significant (p=0.136).
Table I. Participants’ demographic characteristics.
Values represented as mean±standard deviation. *Patients in early group were significantly older than those in late group. The age of patients in middle group did not differ from other two groups.
Non-parametric ROC curves for IgM and IgG are shown in Figure 1. The respective areas under the ROC curve (95% CI) were 0.907 (0.824-0.990) and 0.848 (0.734-0.963). The calculated IgM sensitivity and specificity, using the manufacturer’s diagnostic cut-off of ≥0.04 mIU/ml and the corresponding results for two additional sets of cutoff values are shown in Table II Moreover, sensitivities of both tests were calculated separately in the three subgroups of patients (Table II). More specifically, increasing the cutoff values to 0.05 mIU/ml for IgM and 0.10 mIU/ml for IgG resulted in an increased specificity of IgM (94.4%) without any loss of sensitivity (73.08%) and increased specificity of IgG (94.4%) with a mild reduction of sensitivity (73.08%). Further increase of cutoff values to 0.08 mIU/ml for IgM and 0.19 mIU/ml for IgG gave a perfect sensitivity for both (100%) but significantly reduced their sensitivities to 65.38% and 69.23%, respectively.
Figure 1. Non-parametric ROC curves for IgM and IgG antibodies.
Table II. Sensitivity and Specificity percentages of the quantitative antibody POC test using different cut-off values.
Since this is a quantitative test, we further analyzed the IgM and IgG levels (Figure 2). Both IgM and IgG levels differed significantly (p<0.001) between patients and controls. Among patients, there was a significant trend for higher levels of IgG with increasing time of disease (p=0.022) whereas the corresponding trend for IgM was marginally not significant (p=0.083).
Figure 2. Quantitative analysis of anti-SARS-CoV-2 IgM and IgG antibodies by participant group [upper left, upper right] and by time of symptom onset [lower left, lower right]. Between participant groups differences [upper left, upper right]: p<0.001 for both IgM and IgG test. For time of symptom onset lower left, lower right]: p=0.083 for IgM, p=0.022 for IgG.
Considering the severity, neither IgM (p=0.686) nor IgG (p=0.448) showed significant trends. However, IgM/IgG ratio tended to be positively correlated with disease’s severity with the median (IQR) levels being 0.20 (0.17-0.32), 0.32 (0.14-0.52) and 0.59 (0.53-0.75) for patients with mild, moderate and severe/critical disease, respectively (p for trend=0.031).
Discussion
The performance of a dry fluorescence immunoassay POCT for dual IgG and IgM antibody quantitative detection as a diagnostic method for SAR-CoV-2 immunity levels was evaluated. To our knowledge, this is the first study appraising a quantitative POCT measuring SARS-CoV-2 antibodies in an actual clinical setting. Such immunoassays will play an important role in the future for epidemiological surveillance, evaluation of immunity and the outcome of vaccination studies (16).
Currently, the World Health Organization (WHO) recommends the utilization of nucleic acid-based molecular diagnostics in respiratory samples as the mainstay of COVID-19 diagnosis (9). The FDA has recently announced the authorization of rapid molecular tests that are capable of delivering results within minutes, but which are not globally available yet (17,18). Most importantly, although detection of SARS-CoV-2 RNA in nasopharyngeal swabs is useful for the detection of acutely infected subjects as we are moving towards the post-flattening curve era, it becomes evident that easily accessible and accurate detection of immune people is vital in order to move the economy forward without jeopardizing public health.
Antibody testing for monitoring the development of immunity in response to infection, coupled with rRT-PCR for the detection of acute infections, will be important for surveillance and may provide a tool for developing an exit strategy with selective restrictions as a reaction to the pandemic (16). In the short term, it could also contribute to informing whether people with a demonstrated immunity could be exempt from confinement measures. However, the variability of the results between the newly developed antibody kits, due to their different detection methods and/or their individual characteristics (i.e. sensitivity, specificity, accuracy, technical issues), will definitely shape their applicability in clinical practice. Therefore, large population based comparative trials among different tests are fundamental in order to define their role, their performance and eventually their utility in daily practice. Until more data are available, the use of different tests may be prudent.
Even though a total of 101 antibody tests have been currently CE-marked, limited data about their accuracy and utility are available in a clinical setting (19). Li et al. reported 88.66% sensitivity and 90.63% specificity of a qualitative combined IgM/IgG lateral flow immunoassay POCT (20). Most recently, a commercial qualitative test using whole blood obtained by fingerstick was evaluated by testing PCR-confirmed COVID patients as well as stored serum bank samples from 2018 used as controls. The test had high specificity (>99%) for both IgM and IgG antibodies indicating potential use for detection of past immunity (21). Both studies, however, evaluated qualitative tests performed in laboratory settings, whereas the present POC was quantitative and benchmarked at bedside settings, only with fresh samples.
In this study, highest sensitivities of both IgG and IgM antibodies were observed among the late PG subgroup. This is in concordance with previously published data, supporting that most patients develop anti-SARS-CoV-2 antibodies during the second week of symptoms (10,22-26). This could suggest that antibody POCTs may be used for the confirmation of asymptomatic or mildly symptomatic cases or close-contacts in home-quarantine that were not tested due the lack of available molecular reagents. However, it remains to be seen whether asymptomatic patients are able to mount a satisfactory antibody response. Additionally, numerical values can be important during the initial medical assessment to help limit the indeterminate cases, as well as in follow up.
Since SARS-CoV-2 antibody tests will be mainly used for the determination of the immune status against SARS-CoV-2, the diagnostic specificity is the most crucial parameter. The ROC analysis provides the potential of increasing the specificity of this test in the expense of sensitivity. Although the specificity of the test examined here was not ideal, the advantage of using quantitative immunoassays facilitates the differentiation of false positive results based on the measured antibody levels. Hence, despite the test’s observed low IgG specificity, false positive CG participants had only marginally higher levels above cut-off (0.04-0.10 mIU/ml), whereas true positive cases developed 5- to 180-fold higher titers of IgG (Figure 1). However, more data on antibody responses among asymptomatic patients is required. It is possible that false positive IgG, are possibly explained by a cross-reaction with other coronaviruses or by the exposure to a continuous ambient low virus load, since our volunteers were hospital staff (23,27,28).
The most important advantage of the test evaluated here was its quantitative aspect and the ease of use in a busy clinical setting or in remote areas. The system provided numerical data which can prove useful for the clinical evaluation, the confirmation and the follow up of antibody reaction during serial testing.
POC antibody testing represents a readily available portable kit and would facilitate the broad implementation of population-based testing even in areas without relevant infrastructures since such methods do not require complex laboratory equipment and expertise. Similarly, POC antibody tests, particularly serial quantitative measurements, may also be valuable for diagnostic purposes of patients with febrile respiratory illnesses in remote areas, such as small islands.
Limitations
The diagnostic specificity of antibody tests would require the availability of specimens from individuals that had never been in contact with the SARS-CoV-2 virus. We however, used hospital-based employees as negative controls; although this may have affected the specificity of the test, one may postulate that its quantitative nature allows the differentiation between false and true positive cases based on the titers detected. Considering the worldwide spread of the virus it is recommended to use specimens which were collected before November 2019 as negative controls.
Among the limitations of our study is the small number of included participants. Moreover, we did not include asymptomatic subjects and all patients were hospitalized while the number of patients with mild and early disease was small. Although in our study design we intended to include an equal number of participants from the early, middle and late groups, in order to produce more robust and balanced results during the different phases of the disease and of antibodies’ kinetic, this was not possible. In our small cohort patient with early infection were under-represented (19.2%). This is mainly due to the natural course of this disease since most patients who require hospitalization are usually admitted after the first week of symptoms, when respiratory failure develops. Evidently, larger trials are needed to establish the placement of POC testing in the diagnostic armamentarium, as well as the utility of numerical values.
Conclusion
The development of low-cost, accurate and widely available SARS-CoV-2 tests, including antibody POCTs, may represent an essential tool in the development of de-escalation strategies in which mobility and contact restrictions could be removed for people with proven immunity. There is a theoretical advantage of using quantitative immunoassays as they facilitate the numerical differentiation based on the measured antibody levels and allow for follow up evaluation. Large clinical studies are imperative in order to better understand and contextualize each test’s intended use in real-life clinical settings.
Conflicts of Interest
The Authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. Consumables, test strips and the reader were provided for free by Lansion Biotech.
Authors’ Contributions
PCF, VP, AA, SAP, ST and AK contributed in the conception and the design of the study. PCF, DK, TS, CP and AK contributed in data acquisition. PCF, VP, NP and AK drafted the article. All authors contributed in the analysis and interpretation of data, critically revised the manuscript for important intellectual content and approved the final submitted version of the manuscript.
Acknowledgements
Authors would like to express their sincere gratitude to Dr Emmanouil Karofylakis, Dr. Konstantinos Thomas, Dr. George Tsioulos, Dr. Christina Damoulari, Dr. Maria Paneta, Dr Sotiria Grigoropoulou and Mrs Aggeliki Perdikouli for providing us the patients’ clinical samples and for their daily input in the care of COVID-19 patients. Also, authors would like to thank Dr Gkikas Magiorkinis for his input in the study design.
Funding
For PCF: Supported by Doctorate scholarship by the State Scholarships Foundation (IKY), Partnership Agreement (PA) 2014-2020, co-financed by Greece and the European Union (European Social Fund - ESF) through the Operational Program “Human Resources Development, Education and Lifelong Learning 2014-2020”.
Data Availability Statement
The datasets for this study are available upon request by the corresponding author.
References
- 1.COVID-19 situation reports. The World Health Organization. Available at: https://www.who.int/emergencies/diseases/novelcoronavirus-2019/situation-reports/ [Last accessed on 01 June 2020]
- 2.Strategic preparedness and response plan for the novel coronavirus. The World Health Organization. Available at: https:// www.who.int/publications-detail/strategic-preparedness-andresponse-plan-for-the-new-coronavirus [Last accessed on 01 June 2020]
- 3.Infection prevention and control. The World Health Organization. Available at: https://www.who.int/emergencies/diseases/novelcoronavirus-2019/technical-guidance/infection-prevention-andcontrol [Last accessed on 01 June 2020]
- 4.COVID-19. European Centre for Disease Prevention and Control. Available at: https://www.ecdc.europa.eu/en/covid-19-pandemic [Last accessed on 01 June 2020] [DOI] [PubMed]
- 5.Li D, Wang D, Dong J, Wang N, Huang H, Xu H, Xia C. False-negative results of real-time reverse-transcriptase polymerase chain reaction for severe acute respiratory syndrome coronavirus 2: Role of deep-learning-based CT diagnosis and insights from two cases. Korean J Radiol. 2020;21(4):505–508. doi: 10.3348/kjr.2020.0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lippi G, Simundic A-M, Plebani M. Potential preanalytical and analytical vulnerabilities in the laboratory diagnosis of coronavirus disease 2019 (COVID-19). Clin Chem Lab Med. 2020 doi: 10.1515/cclm-2020-0285. [DOI] [PubMed] [Google Scholar]
- 7.Ai T, Yang Z, Hou H, Zhan C, Chen C, Lv W, Tao Q, Sun Z, Xia L. Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: A Report of 1014 Cases. Radiology. 2020;200642 doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.LabCorp Covid-19 RT-PCR Test EUA Summary: Accelerated Emergency Use Authorization (Eua) Summary Covid-19 RT-PCR Test (Laboratory Corporation Of America). The Food and Drug Administration (FDA). Available at: https://www.fda.gov/media/136151/download [Last accessed on 25 April 2020]
- 9.Advice on the use of point-ofcare immunodiagnostic tests for COVID-19. The World Health Organization. Available at: https://www.who.int/news-room/commentaries/detail/advice-onthe-use-of-point-of-care-immunodiagnostic-tests-for-covid-19[Last accessed on 25 April 2020]
- 10.Zhao J, Yuan Q, Wang H, Liu W, Liao X, Su Y, Wang X, Yuan J, Li T, Li J, Qian S, Hong C, Wang F, Liu Y, Wang Z, He Q, Li Z, He B, Zhang T, Fu Y, Ge S, Liu L, Zhang J, Xia N, Zhang Z. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. Clin Infect Dis. 2020;ciaa344 doi: 10.1093/cid/ciaa344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chan JF, Yuan S, Kok KH, To KK, Chu H, Yang J, Xing F, Liu J, Yip CC, Poon RW, Tsoi HW, Lo SK, Chan KH, Poon VK, Chan WM, Ip JD, Cai JP, Cheng VC, Chen H, Hui CK, Yuen KY. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395(10223):514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zou L, Ruan F, Huang M, Liang L, Huang H, Hong Z, Yu J, Kang M, Song Y, Xia J, Guo Q, Song T, He J, Yen HL, Peiris M, Wu J. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020;382(12):1177–1179. doi: 10.1056/NEJMc2001737.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Case definition and European surveillance for COVID-19, as of 2 March 2020. The European Centre for Disease Prevention and Control. Available at: https://www.ecdc.europa.eu/en/casedefinition-and-european-surveillance-human-infection-novelcoronavirus-2019-ncov [Last accessed on 25 April 2020]
- 14.(COVID-19) IgM/IgG Test Kit _Test Kits_Lansion Biotechnology Co., Ltd. Lansion Biotechnology Co. L. Available at: http://en.lansionbio.com/proArticle.asp?id=5234 [Last accessed on 25 April 2020]
- 15.Bossuyt PM, Reitsma JB, Bruns DE, Gatsonis CA, Glasziou PP, Irwig L, Lijmer JG, Moher D, Rennie D, de Vet HC, Kressel HY, Rifai N, Golub RM, Altman DG, Hooft L, Korevaar DA, Cohen JF; STARD Group. STARD 2015: an updated list of essential items for reporting diagnostic accuracy studies. Version 2. BMJ. 2015;351:h5527. doi: 10.1136/bmj.h5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rapid risk assessment: Coronavirus disease 2019 (COVID-19) pandemic: increased transmission in the EU/EEA and the UK – eighth update. The European Centre for Disease Prevention and Control. Available at: https://www.ecdc.europa.eu/en/publications-data/rapidrisk-assessment-coronavirus-disease-2019-covid-19-pandemiceighth-update [Last accessed on 25 April 2020]
- 17.Cepheid|Xpert® Xpress SARS-CoV-2 has received FDA Emergency Use Authorization. Available at: https://www.cepheid.com/coronavirus [Last accessed on 25 April 2020]
- 18.Detect COVID-19 in as Little as 5 Minutes. Abbott Newsroom. Available at: https://www.abbott.com/corpnewsroom/productand-innovation/detect-covid-19-in-as-little-as-5-minutes.html [Last accessed on 25 April 2020]
- 19.Communication from the Commission: Guidelines on COVID-19 in vitro diagnostic tests and their performance. The European Commission. Available at: https://ec.europa.eu/info/sites/info/files/testing_kits_communication.pdf [Last accessed on 25 April 2020]
- 20.Li Z, Yi Y, Luo X, Xiong N, Liu Y, Li S, Sun R, Wang Y, Hu B, Chen W, Zhang Y, Wang J, Huang B, Lin Y, Yang J, Cai W, Wang X, Cheng J, Chen Z, Sun K, Pan W, Zhan Z, Chen L, Ye F. Development and clinical application of a rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. J Med Virol. 2020 doi: 10.1002/jmv.25727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoffman T, Nissen K, Krambrich J, Rönnberg B, Akaberi D, Esmaeilzadeh M, Salaneck E, Lindahl J, Lundkvist Å. Evaluation of a COVID-19 IgM and IgG rapid test; an efficient tool for assessment of past exposure to SARS-CoV-2. Infect Ecol Epidemiol. 2020;10(1):1754538. doi: 10.1080/20008686.2020.1754538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Y, Liu Y, Diao B, Ren F, Wang Y, Ding J, Huang Q. Diagnostic Indexes of a rapid IgG/IgM combined antibody test for SARS-CoV-2. medRxiv. 2020 doi: 10.1101/2020.03.26.20044883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okba NMA, Müller MA, Li W, Wang C, GeurtsvanKessel CH, Corman VM, Lamers MM, Sikkema RS, de Bruin E, Chandler FD, Yazdanpanah Y, Le Hingrat Q, Descamps D, Houhou-Fidouh N, Reusken CBEM, Bosch BJ, Drosten C, Koopmans MPG, Haagmans BL. Severe acute respiratory syndrome coronavirus 2-specific antibody responses in coronavirus disease 2019 patients. Emerg Infect Dis. 2020;26(7) doi: 10.3201/eid2607.200841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wölfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, Müller MA, Niemeyer D, Jones TC, Vollmar P, Rothe C, Hoelscher M, Bleicker T, Brünink S, Schneider J, Ehmann R, Zwirglmaier K, Drosten C, Wendtner C. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581(7809):465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 25.Zhang W, Du RH, Li B, Zheng XS, Yang XL, Hu B, Wang YY, Xiao GF, Yan B, Shi ZL, Zhou P. Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerg Microbes Infect. 2020;9(1):386–389. doi: 10.1080/22221751.2020.1729071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang N, Li SY, Yang XL, Huang HM, Zhang YJ, Guo H, Luo CM, Miller M, Zhu G, Chmura AA, Hagan E, Zhou JH, Zhang YZ, Wang LF, Daszak P, Shi ZL. Serological evidence of bat SARS-related coronavirus infection in humans, China. Virol Sin. 2018;33(1):104–107. doi: 10.1007/s12250-018-0012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Che XY, Qiu LW, Liao ZY, Wang YD, Wen K, Pan YX, Hao W, Mei YB, Cheng VC, Yuen KY. Antigenic cross-reactivity between severe acute respiratory syndrome-associated coronavirus and human coronaviruses 229E and OC43. J Infect Dis. 2005;191(12):2033–2037. doi: 10.1086/430355. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets for this study are available upon request by the corresponding author.