Abstract
Background/Aim: Translation plays an important role in the carcinogenesis of various human tumors. Paip1 and eIF4A1 are translation-associated proteins that mediate the function of eukaryotic initiation factor 4F complex. This study aimed to analyse the relationship between the expression status of Paip1 and eIF4A1 and clinicopathologic features in hepatocellular carcinoma (HCC). Materials and Methods: Immunohistochemical analysis was used to evaluate the expression status of Paip1 and eIF4A1. Two pathologists independently interpreted the immunostained slides. The prognostic value of Paip1 and eIF4A1 was evaluated by the Kaplan–Meier plotter. Results: Among 173 HCC patients, 28 (16.1%) and 46 (26.6%) belonged in the Paip1 and eIF4A1 high-expression groups. High expression of Paip1 and eIF4A1 was associated with advanced TNM stage and more frequent vascular tumor invasion. Univariate analysis indicated that high Paip1 expression was associated with worse five-year overall survival (OS). Public dataset analysis by Kaplan–Meier plotter revealed that high mRNA expression of Paip1, and not of eIF4A1, was significantly associated with worse five-year OS and disease-free survival. Conclusion: Paip1 expression has a potential prognostic value in human HCC.
Keywords: Translation, hepatocellular carcinoma, prognosis, survival
Translation is one of the core processes in post-transcriptional gene expression regulation and is an important factor in tumorigenesis (1,2). The eIF4F complex is involved in eukaryote translation initiation and consists of eIF4E, eIF4G, and eIF4A. Poly (A)-binding protein (PABP) binds to eIF4G to induce mRNA circularization and stimulate mRNA translation. Poly (A)-binding protein-interacting protein 1 (Paip1) is encoded by the PAIP1 gene and binds to PABP to regulate its activity (3). High expression of Paip1 has been reported to be associated with poor overall survival in various tumors, including gastric, breast, pancreatic, uterine, and cervical cancers (4-7).
eIF4A1, which is one of three isoforms of eIF4A, is a constituent of the eIF4F complex and an ATP-dependent RNA helicase that plays a critical role in translation initiation by unwinding 5’-UTR structures of mRNA. Several reports suggest that eIF4A1 might be involved in the process of carcinogenesis (8-10).
This study aimed to examine the levels of Paip1 and eIF4A1 proteins that regulate human translation initiation in hepatocellular carcinoma (HCC). We investigated their association with various clinicopathological parameters. In addition, we divided patients into low- and high-expression groups and analyzed whether there was a significant difference in survival.
Materials and Methods
Patient selection and clinicopathologic data. We reviewed the electronic medical records of patients who underwent surgical resection of the liver as primary therapy for HCC in Korea University Guro Hospital from January 2000 to December 2013. Patients who underwent liver transplantation, had a history of any other local or systemic therapy before surgery, had other primary neoplasms, had positive surgical resection margins, or without available formalin-fixed paraffin-embedded tumor tissues were excluded. A total of 173 patients were finally enrolled in this study.
Demographic and clinical data including sex, age, clinical stage, underlying liver disease, presence of cirrhosis, presence of metastatic disease at diagnosis, and dates of death and recurrence were retrieved from the electronic medical records. Pathologic features of the tumor such as tumor size, vascular invasion, tumor multiplicity, and histologic grade were assessed by reviewing the histopathology slides. Tumor staging was performed according to the Staging Manual of the American Joint Committee on Cancer, 8th edition (11). Histologic grading of tumors was based on the Edmondson-Steiner histologic grading system (12). This study was approved by the Institutional Review Board of Korea University Guro Hospital (approval number: 2018GR0411).
Tissue microarray construction and immunohistochemical analysis. Tissue microarray (TMA) blocks were constructed by selecting representative tumor areas and transferring 2-mm-diameter tissue cores into recipient blocks. For each block, 4-um-thick tissue sections were serially cut and transferred onto charged glass slides. Sections were routinely processed with xylene and ethanol before immunostaining. Routine immunohistochemical (IHC) staining was performed using commercially available antibodies and a BOND-MAX automated staining system (Leica, Wetzlar, Germany). The antibodies used were: anti-Paip1 antibody [1:100, rabbit monoclonal (EPR13259); Abcam, Cambridge, MA, USA] and anti-eIF4A1 antibody (1:100, rabbit polyclonal; Abcam).
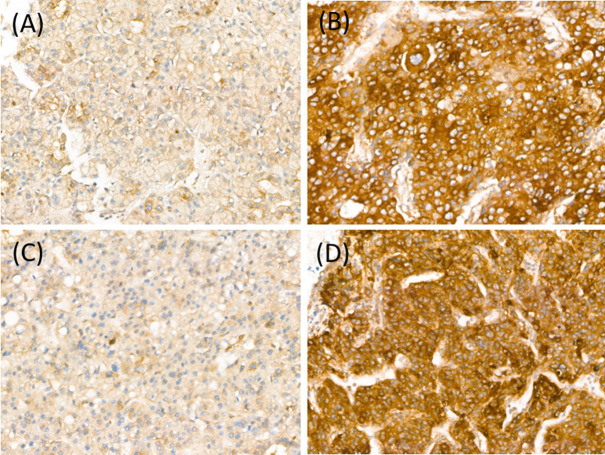
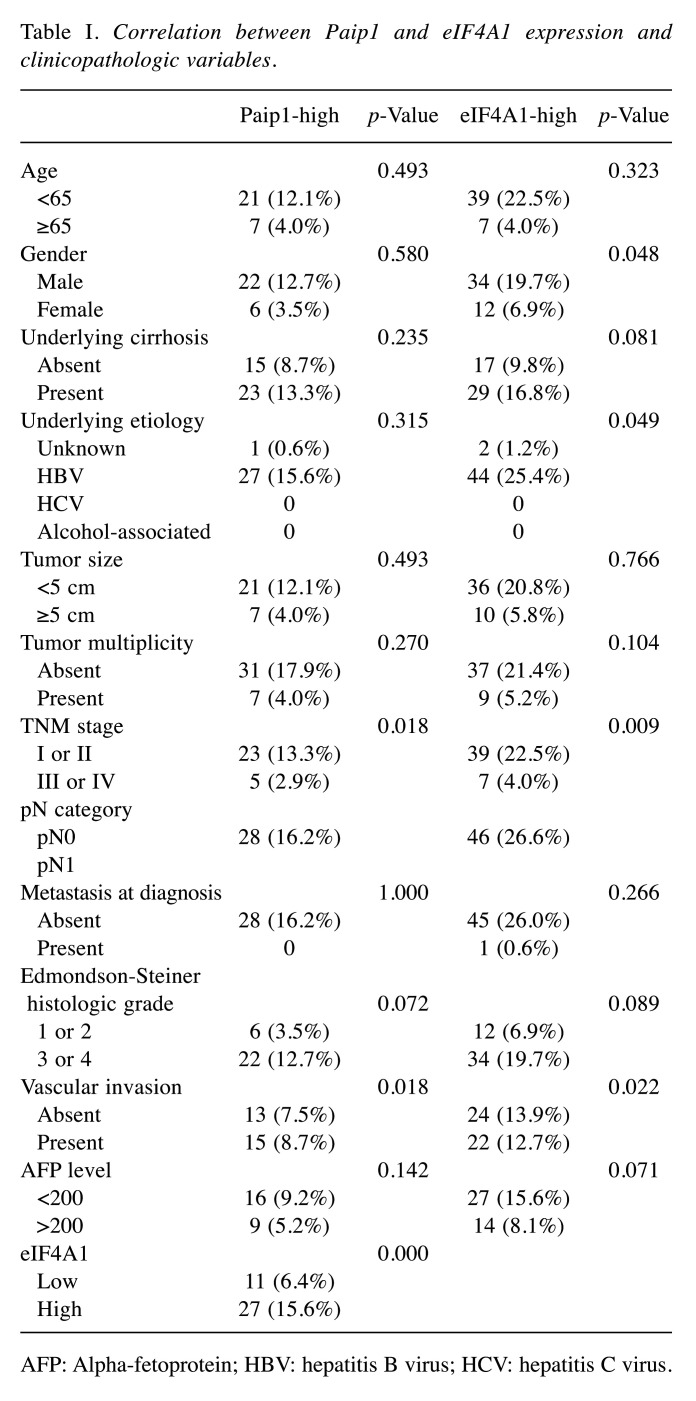
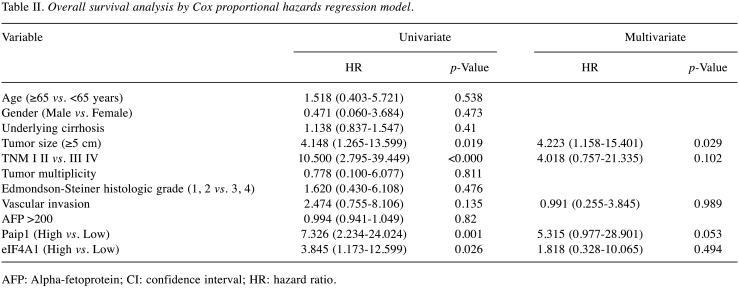
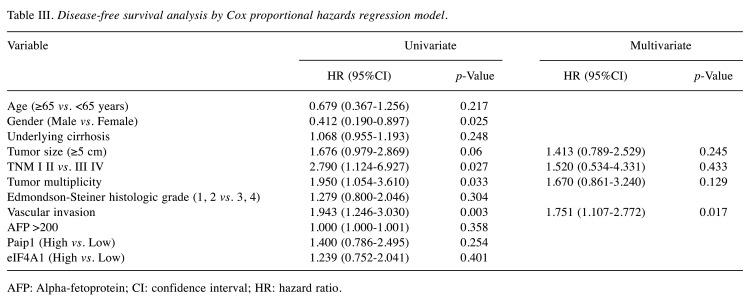
Stained sections were analyzed by two pathologists (HK and BHK) blinded to patients’ clinicopathologic characteristics and survival status and to each other’s interpretation results. The staining intensity of each antibody was graded as negative, weak, moderate, or strong (Figure 1). For Paip1 and eIF4A1, the percentage of tumor area showing each staining intensity was recorded in increments of 10%. Tumor cores with greater than 30% strong-intensity staining were assigned to the high-expression group of Paip1 and eIF4A1. All other cases were included in the low-expression group. When there was a discrepancy on expression group categorization, the pathologists discussed the case until a consensus was made.
Figure 1. Paip1 and eIF4A1 expression in hepatocellular carcinoma. Paip1, low (A), and high (B); eIF4A1, low (C), and high (D).
Statistical analysis. All statistical analyses were performed using IBM SPSS Statistics for Windows version 25.0 (IBM Corp., New York, NY, USA). Associations between clinicopathologic characteristics and protein expression status of Paip1 and eIF4A1 were analyzed using the chi-square test or Fisher’s exact test. For survival analysis, patients who died within one month after surgery were excluded. Kaplan–Meier survival curves were plotted, and the log-rank test was performed to test statistical significance between the curves. Univariate and multivariate statistical analyses were completed using the Cox proportional hazards regression model. Overall survival (OS) was defined as the time from the day of surgery to the day of death or the last follow-up date for censored cases. Disease-free survival (DFS) was defined as the duration from the day of surgery to the day of recurrence or, for censored cases, the last follow-up date. A p-value <0.05 was considered statistically significant.
Survival analysis by KM-plotter. The prognostic value of Paip1 and eIF4A1 was further evaluated by the Kaplan–Meier (KM) plotter (13). The liver cancer database in the KM-plotter contains survival data of 364 patients from The Cancer Genome Atlas repository. We submitted Paip1 and eIF4A1 as gene names into the KM-plotter and retrieved the results of OS and DFS analyses. The follow-up threshold was set to 60 months (5 years). The hazard ratio (HR) with a 95% confidence interval (CI) and the p-value of the log-rank test were obtained.
Results
Patient characteristics. The total number of patients included in the study was 173. Median age at the time of diagnosis was 56 years (range=36-80 years). Male and female patients accounted for 83.2% [144] and 16.8% [29] of the study population, respectively. One patient experienced metastasis, detected at the time of surgery. No patients showed lymphatic tumor invasion. Vascular tumor invasion was observed in 114 (65.9%) patients. Survival analysis was conducted in 168 patients. Median follow-up duration was 1,940 days (range=69-5,081 days).
Relationship of clinicopathologic characteristics and Paip1 and eIF4A1 expression. Both, Paip1 and eIF4A1 showed a predominantly cytoplasmic staining pattern. Lone cytoplasmic staining was considered positive in IHC interpretation. There were 28 (16.1%) and 46 (26.6%) cases in the Paip1 and eIF4A1 high-expression groups, respectively. The IHC expression levels of each protein were not significantly associated with older age (≥65 years), larger tumor size (≥5 cm), or tumor multiplicity. Advanced TNM stage (I, II vs. III, IV) was associated with high expression of Paip1 (p=0.018) and eIF4A1 (p=0.009). Vascular tumor invasion was more frequently associated with the high-expression group of Paip1 (p=0.018) and eIF4A1 (p=0.022) compared to the low-expression groups. The results are summarized in Table I.
Table I. Correlation between Paip1 and eIF4A1 expression and clinicopathologic variables.
AFP: Alpha-fetoprotein; HBV: hepatitis B virus; HCV: hepatitis C virus.
Correlation between the expression levels of Paip1 and eIF4A1. A significant association was found between the high-expression groups of Paip1 and eIF4A1 (p<0.000).
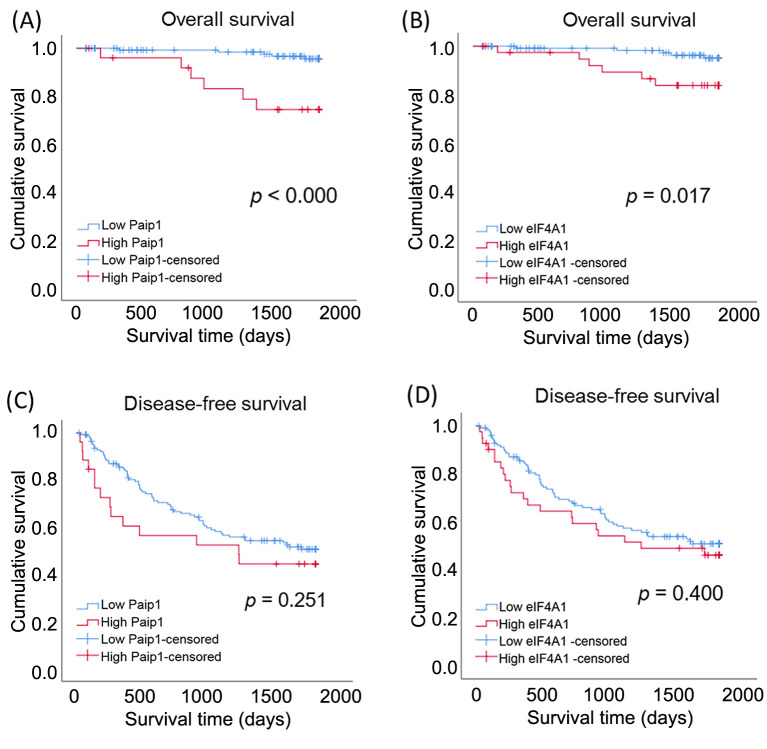
Survival analysis. In univariate survival analysis, large tumor size (≥5 cm) (HR=4.148; 95%CI=1.265-13.599; p=0.019), advanced TNM stage (I, II vs. III, IV) (HR=10.500; 95%CI=2.795-39.449; p<0.000), high Paip1 expression (HR=7.326; 95%CI=2.234-24.024; p=0.001), and high eIF4A1 expression (HR=3.845; 95%CI=1.173-12.599; p=0.026) were significantly associated with worse five-year OS. Male sex (HR=0.412; 95%CI=0.190-0.897, p=0.025), tumor multiplicity (HR=1.943; 95%CI=1.054-3.610; p=0.033), vascular invasion (HR=1.943; 95%CI=1.246-3.030; p=0.003), and advanced TNM stage (I, II vs. III, IV) (HR=2.790; 95%CI=1.124-6.927; p=0.027) were associated with worse five-year DFS. To assess the prognostic value of Paip1 and eIF4A1 overexpression in five-year OS, multivariate Cox proportional hazards regression analyses were performed. Large tumor size (≥5 cm) was an independent prognostic factor (HR=4.223; 95%CI=1.158-15.401; p=0.029). Although high Paip1 expression was not significantly associated with worse prognosis, it showed a trend toward worse OS (Table II and Table III, and Figure 2).
Table II. Overall survival analysis by Cox proportional hazards regression model.
AFP: Alpha-fetoprotein; CI: confidence interval; HR: hazard ratio.
Table III. Disease-free survival analysis by Cox proportional hazards regression model.
AFP: Alpha-fetoprotein; CI: confidence interval; HR: hazard ratio.
Figure 2. Kaplan–Meier curves and the results of log-rank tests of hepatocellular carcinoma patients stratified according to Paip1 and eIF4A1 expression. (A) 5-year overall survival (OS) curves stratified by Paip1 expression. (B) 5-year OS curves stratified by eIF4A1 expression. (C) 5-year disease-free survival (DFS) curves stratified by Paip1 expression. (D) 5-year DFS curves stratified by eIF4A1 expression.
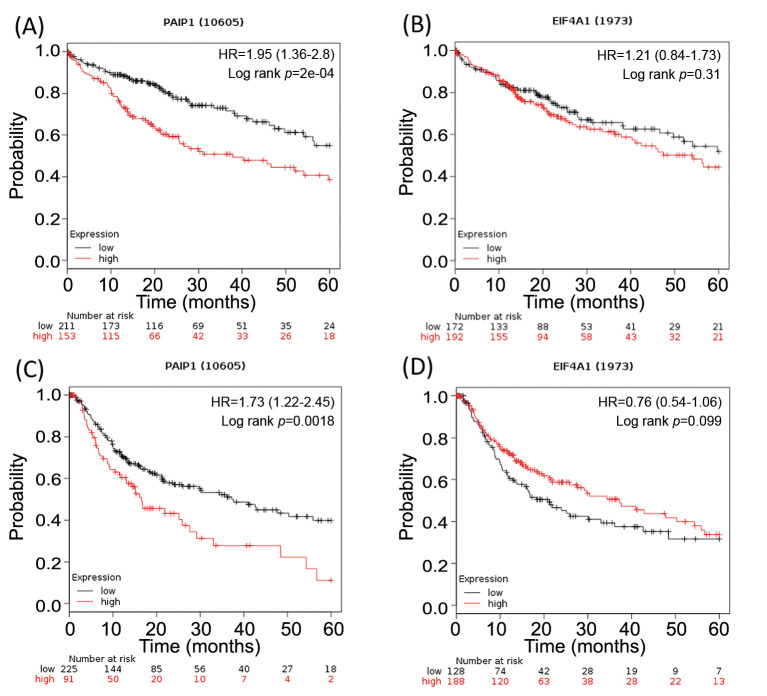
Survival analysis by KM-plotter. In KM plot analysis, high mRNA expression of Paip1 was significantly associated with worse five-year OS and DFS (p<0.000 and p=0.0018, respectively). High mRNA expression of eIF4A1 was not significantly associated with OS and DFS (p=0.31 and p=0.099, respectively) (Figure 3).
Figure 3. Kaplan–Meier curves and the results of log-rank tests in public datasets using Kaplan–Meier plotter. (A) 5-year overall survival (OS) curves stratified by Paip1 expression. (B) 5-year OS curves stratified by eIF4A1 expression. (C) 5-year disease-free survival (DFS) curves stratified by Paip1 expression. (D) 5-year DFS curves stratified by eIF4A1 expression.
Discussion
Gene expression regulation occurs at several stages, including genetic aberration, transcription regulation, and translation regulation. Translation is the last stage, and the point of convergence of the various changes in the upstream processes. Thus, translation dysregulation is important in carcinogenesis and is both a triggering factor and an outcome of abnormal gene expression (1,14). Among the four stages of translation (initiation, elongation, termination, and recycling), most of the regulation occurs at the initiation stage, which is the rate-limiting step (15). Paip1 and eIF4A1 are proteins that are engaged in human translation initiation. This study is the first to analyze IHC expression of Paip1 and eIF4A1 and their prognostic impact in human HCC.
Paip1 binds to PABP and contributes to eukaryotic translation initiation by modulating the function of the eIF4F complex. eIF4A1 is a member of the eIF4F complex and acts as an RNA helicase. Paip1 and eIF4A1 stimulate translation initiation by binding to each other and inducing circularization of mRNA (16). Thus, it can be inferred that the protein levels of Paip1 and eIF4A1 regulate protein translation. In this study, the groups with high expression of Paip1 and eIF4A1 had a statistically significant association, which agrees with their biochemical relationship.
In our study, high expression of Paip1 and eIF4A1 was associated with vascular tumor invasion, which occurs via a complex interaction of tumor cells and tumor microenvironment (17). Knockdown of Paip1 attenuated tumor invasion and migration in lung, gastric, and pancreatic carcinomas, while overexpression of Paip1 had the opposite effect (4,5,7). These findings support the hypothesis that Paip1 expression is an important factor in vascular tumor invasion and migration.
Recent studies have shown that Paip1 is overexpressed in uterine, cervical, gastric, and breast cancers compared with normal tissues. In addition, it has been reported that high expression of Paip1 was associated with poorer OS in these human cancers (4-7,18). The present study showed that the high Paip1 expression group trended to have worse OS in multivariate analysis. Though the prognostic association of Paip1 was not statistically significant in our study, high mRNA expression of Paip1 was associated with worse prognosis in the public data analysis using the KM-plotter. The small cohort size might have contributed to our equivocal results; further studies with larger cohorts are required to clarify the prognostic value of Paip1 expression assessed by IHC.
Our study showed that high expression of eIF4A1 was associated with worse OS in univariate analysis and with other adverse prognostic factors such as higher pT category and vascular tumor invasion. Accumulating studies suggest that translation is associated with tumor cell proliferation and cancer progression. Recent studies have shown that translation control via inhibition of eIF4A can be a novel anticancer therapy. The microRNAs miR-133a and miR-1248 have been shown to inhibit tumor cell growth in colorectal cancer and gastric adenocarcinoma cell lines, respectively, and nude mice transplantation models (8-10). eIF4A-inhibiting rocaglates such as Silvestrol suppressed tumor cell proliferation in pancreatic ductal adenocarcinoma (19). The therapeutic impact of eIF4A inhibitors and the prognostic value of eIF4A expression require further validation.
In conclusion, our results suggest that high immunohisto-chemical expression of Paip1 can be utilized as a risk stratification criterion in HCCs. Public cancer data on Paip1 mRNA expression also support the possibility of Paip1 as a prognostic marker of HCC. To validate the utility of Paip1 expression as a prognostic marker, further studies involving larger series of HCC cases are required.
Conflicts of Interest
The Authors have no conflicts of interest to declare regarding this study.
Authors’ Contributions
Conceptualization: HK, AR, HKK, BK; Data curation: HK, WJ; Formal analysis: HK, BK; Methodology: HK, WJ, AR, HKK, BK; Project administration: BK; Visualization: HK; Writing – original draft: HK, BK; Writing – review & editing: HK, WJ, AR, HKK, BK; Approval of final manuscript: all Authors.
Acknowledgements
The biospecimens and data used in this study were provided by the Biobank of Korea University Guro Hospital, a member of the National Biobank of Korea.
References
- 1.Vaklavas C, Blume SW, Grizzle WE. Translational dysregulation in cancer: Molecular insights and potential clinical applications in biomarker development. Front Oncol. 2017;7:158. doi: 10.3389/fonc.2017.00158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136(4):731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roy G, De Crescenzo G, Khaleghpour K, Kahvejian A, O’Connor-McCourt M, Sonenberg N. Paip1 interacts with poly(A) binding protein through two independent binding motifs. Mole Cell Biol. 2002;22(11):3769–3782. doi: 10.1128/mcb.22.11.3769-3782.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Y, Piao J, Wang Q, Cui X, Meng Z, Jin T, Lin Z. Paip1 predicts poor prognosis and promotes tumor progression through AKT/GSK-3beta pathway in lung adenocarcinoma. Hum Pathol. 2019;86:233–242. doi: 10.1016/j.humpath.2018.11.017. [DOI] [PubMed] [Google Scholar]
- 5.Wang Q, Han A, Chen L, Sun J, Lin Z, Zhang X, Ren X. Paip1 overexpression is involved in the progression of gastric cancer and predicts shorter survival of diagnosed patients. Onco Targets Ther. 2019;12:6565–6576. doi: 10.2147/OTT.S202698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lino-Silva LS, Gamboa-Dominguez A, Zuniga-Tamayo D, Lopez-Correa P. Interobserver variability in colorectal cancer and the 2016 ITBCC concensus. Mod Pathol. 2019;32(1):159–160. doi: 10.1038/s41379-018-0027-5. [DOI] [PubMed] [Google Scholar]
- 7.Guan H, Li N, Wang X, Shan X, Li Z, Lin Z. Role of Paip1 on angiogenesis and invasion in pancreatic cancer. Exp Cell Res. 2019;376(2):198–209. doi: 10.1016/j.yexcr.2019.01.017. [DOI] [PubMed] [Google Scholar]
- 8.Li W, Chen A, Xiong L, Chen T, Tao F, Lu Y, He Q, Zhao L, Ou R, Xu Y. miR-133a acts as a tumor suppressor in colorectal cancer by targeting eIF4A1. Tumour Biol. 2017;39(5):1010428317698389. doi: 10.1177/1010428317698389. [DOI] [PubMed] [Google Scholar]
- 9.Wei W, Cao W, Zhan Z, Yan L, Xie Y, Xiao Q. MiR-1284 suppresses gastric cancer progression by targeting EIF4A1. Onco Targets Ther. 2019;12:3965–3976. doi: 10.2147/OTT.S191015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cao W, Wei W, Zhan Z, Xie Y, Xiao Q. MiR-1284 modulates multidrug resistance of gastric cancer cells by targeting EIF4A1. Oncol Rep. 2016;35(5):2583–2591. doi: 10.3892/or.2016.4643. [DOI] [PubMed] [Google Scholar]
- 11.Amin MB, Edge S, Greene F, Byrd DR, Brookland RK, Washington MK, Gershenwald JE, Compton CC, Hess KR, Sullivan DC, Jessup JM, Brierley JD, Gaspar LE, Schilsky RL, Balch CM, Winchester DP, Asare EA, Madera M, Gress DM, Meyer LR. New York: Springer. 2017. AJCC Cancer Staging Manual. 8th ed. p. XVII, 1024 p. [Google Scholar]
- 12.Edmondson HA, Steiner PE. Primary carcinoma of the liver: a study of 100 cases among 48,900 necropsies. Cancer. 1954;7(3):462–503. doi: 10.1002/1097-0142(195405)7:3<462::aid-cncr2820070308>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 13.Kaplan-Meier Plotter. Available at: https://kmplot.com/analysis/index.php?p=service&cancer=liver_rnaseq [Last accessed July 13, 2020]
- 14.Spilka R, Ernst C, Mehta AK, Haybaeck J. Eukaryotic translation initiation factors in cancer development and progression. Cancer Lett. 2013;340(1):9–21. doi: 10.1016/j.canlet.2013.06.019. [DOI] [PubMed] [Google Scholar]
- 15.Pelletier J, Graff J, Ruggero D, Sonenberg N. Targeting the eIF4F translation initiation complex: a critical nexus for cancer development. Cancer Res. 2015;75(2):250–263. doi: 10.1158/0008-5472.CAN-14-2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Craig AW, Haghighat A, Yu AT, Sonenberg N. Interaction of polyadenylate-binding protein with the eIF4G homologue PAIP enhances translation. Nature. 1998;392(6675):520–523. doi: 10.1038/33198. [DOI] [PubMed] [Google Scholar]
- 17.Aleskandarany MA, Sonbul SN, Mukherjee A, Rakha EA. Molecular mechanisms underlying lymphovascular invasion in invasive breast cancer. Pathobiology. 2015;82(3-4):113–123. doi: 10.1159/000433583. [DOI] [PubMed] [Google Scholar]
- 18.Piao J, Chen L, Jin T, Xu M, Quan C, Lin Z. Paip1 affects breast cancer cell growth and represents a novel prognostic biomarker. Hum Pathol. 2018;73:33–40. doi: 10.1016/j.humpath.2017.10.037. [DOI] [PubMed] [Google Scholar]
- 19.Chan K, Robert F, Oertlin C, Kapeller-Libermann D, Avizonis D, Gutierrez J, Handly-Santana A, Doubrovin M, Park J, Schoepfer C, Da Silva B, Yao M, Gorton F, Shi J, Thomas CJ, Brown LE, Porco JA Jr., Pollak M, Larsson O, Pelletier J, Chio IIC. eIF4A supports an oncogenic translation program in pancreatic ductal adenocarcinoma. Nat Commun. 2019;10(1):5151. doi: 10.1038/s41467-019-13086-5. [DOI] [PMC free article] [PubMed] [Google Scholar]