Abstract
Animals counter specific environmental challenges with a combination of broad and tailored host responses. One protein family enlisted in the innate immune response includes the saposin-like antimicrobial proteins. We investigated the expression of a Caenorhabditis elegans saposin-like gene, spp-9, in response to different stresses. spp-9 expression was detected in the intestine and six amphid neurons, including AWB and AWC. spp-9 expression is increased in response to starvation stress. In addition, we discovered pathogen-specific regulation of spp-9 that was not clearly demarcated by Gram nature of the bacterial challenge. Multiple molecular innate immune response pathways, including DBL-1/TGF-β-like, insulin-like, and p38/MAPK, regulate expression of spp-9. Our results suggest spp-9 is involved in targeted responses to a variety of abiotic and bacterial challenges that are coordinated by multiple signaling pathways.
Subject terms: Gene regulation, Innate immunity
Introduction
All living organisms possess mechanisms to protect themselves from potentially harmful environments [1, 2]. Roundworms are a diverse group of organisms (comprising both free-living and parasitic members) that have adapted to various habitats ranging from terrestrial to marine environments [3]. In these environments, roundworms encounter a variety of challenges, including low food availability and pathogenic bacteria [4]. Caenorhabditis elegans, a free-living roundworm, is an established model organism to understand defense mechanisms nematodes use, including innate immune responses to bacteria [5]. It is important that these responses are well coordinated and appropriately regulated by the host [1, 2]. Aberrant activation of host immune responses could be a potential cellular stress for the host, making tight regulation of host immune responses important for host health in low-stress environments, as well [6, 7]. How animals respond to the various challenges in their natural environment by mounting specific defenses is not well understood.
Several pathogens infect C. elegans, including bacterial strains Pseudomonas aeruginosa, Staphylococcus aureus, Salmonella enterica serovar Typhimurium, Serratia marcescens, Enterococcus faecalis, Mycobacterium fortuitum, Bacillus megaterium, Microbacterium nematophilum, and Staphylococcus epidermidis. C. elegans is also infected by fungi, including Candida albicans and Drechmeria coniospora [8–15]. MAPK signaling, insulin-like signaling, and DBL-1/TGF-β-like signaling pathways are induced upon infection by these pathogens [8–13, 16–20]. These pathways regulate an overlapping set of antimicrobial genes, which suggests crosstalk between the pathways upon infection [18, 21, 22]. These antimicrobial genes encode lipases, lysozymes, defensin-like proteins, and saposin-like proteins (SAPLIPs or caenopores).
C. elegans caenopores have structural similarities with the protozoan amoebapores and the mammalian peptides NK-lysin and granulysin [23, 24]. 28 genes are predicted to encode 33 SAPLIP-domain containing proteins, but antimicrobial activity of only a few saposins (SPPs) has been characterized. Functional analyses of SPP-3, SPP-5, and SPP-12 show that they display pore-forming activity, permeabilize the cytoplasmic membrane of bacteria, and kill bacteria [24–26]. SPP-1 is required for protection of C. elegans against S. enterica serovar Typhimurium and P. aeruginosa through the DAF-2/DAF-16 signaling pathway [18, 27]. Some spp genes are highly up- or down-regulated in response to bacterial challenges [18, 22, 24–27]. In addition, one caenopore gene, spp-3, has been shown to be induced in starved animals [24, 25]. Although some caenopores are strongly expressed in the intestine, some are also expressed in specific nerves [18, 24–28]. One caenopore, spp-12, is expressed only in NSML/R and I6 pharyngeal neurons [26].
To expand our understanding of the function of caenopores within the context of innate immunity and stress, we chose spp-9, one member of the caenopore family of antimicrobial proteins. Phylogenetic analyses show that spp-9 is closely related to spp-3, whose role as an antimicrobial protein has been functionally characterized [24, 25]. spp-9 is expressed in intestine, an organ on the front line of innate immunity [21]. spp-9 expression is upregulated in response to loss of DKF-2/protein kinase D [29]. spp-9 is also negatively regulated by the DBL-1 signaling pathway and is used as DBL-1 pathway reporter [21, 30]. While spp-9 has been shown to be a DBL-1-responsive gene, here we characterized the role of spp-9 in the larger context of starvation stress and innate immunity. We also determined additional molecular innate immune/stress pathways that regulate spp-9 expression in C. elegans.
Materials and methods
Strains and maintenance
All C. elegans strains were maintained on EZ media plates at 20 °C except daf-2 strains, which were maintained at 17 °C, unless specified otherwise [31]. Strains used were: wild-type N2, NU3 dbl-1(nk3) V, TLG182 texIs100[dbl-1::dbl-1:gfp; ttx-3p::rfp] IV, NL2099 rrf-3(pk1426) II, TLG697 texIs127[spp-9p::gfp] X, LT998 wkIs40[spp-9p::gfp], TLG707 dbl-1(nk3) V; texIs127 X, TLG755 oyIs44 [odr-1::RFP; lin-15(+)] V; texIs127 X, TLG756 daf-16(mu86) I ; texIs127 X, TLG757 pmk-1(km25) IV; texIs127 X, TLG758 wkIs40; mut-2(r459) I; mek-1(pk97) X, TLG759 daf-2(e1370) III; texIs127 X, TLG760 wkIs40; sek-1(km4) X, TLG761 wkIs40; tir-1(tm3036) III. These strains were generated by standard genetic crosses and confirmed by PCR.
The bacteria used in this study include Bacillus megaterium (Carolina Biological Supply Company), Escherichia coli (OP50), Enterobacter cloacae (49141TM), Klebsiella oxytoca (49131TM), Serratia marcescens (Carolina Biological Supply Company), and Staphylococcus epidermidis (49134TM). All bacteria were grown in tryptic soy broth for 9 h at 37 °C. Bacterial cells were pelleted at 5000 rpm for 15 min and concentrated 20-fold. EZ media plates were freshly seeded with concentrated bacteria in full lawns. The plates were incubated at 37 °C overnight before they were used in experiments.
Dauer assay
daf-2(e1370) III; texIs127 X and texIs127 X populations were synchronized as embryos by bleaching [32]. Animals were transferred to 25 °C at the L2 stage. Dauered daf-2(e1370) III; texIs127 X animals and L4 texIs127 X controls were picked. Two days later, animals were imaged as dauer (daf-2(e1370) III; texIs127 X) or adults (texIs127 X). The experiment was performed in three independent trials.
Imaging
Fluorescence of the spp-9 reporter strain at different developmental stages was captured by a Nikon A1 confocal system (Nikon Instruments, Melville, NY). Colocalization studies of oyIs44 V; texIs127 X animals were performed on a Nikon swept-field confocal system (Nikon Instruments, Melville, NY). Hermaphrodites were synchronized as embryos by bleaching and imaged 48 h after the L4 stage at 20 °C, unless otherwise noted [32]. daf-16(mu86) I; texIs127 X animals and respective control animals (texIs127 X) were grown at 25 °C and imaged 48 h after the L4 stage. Fluorescence of the spp-9 reporter strains was captured by a Nikon DS-Ri2 camera mounted on a Nikon SMZ18 dissecting microscope (Nikon Instruments, Melville, NY). Animals were mounted on 2% agarose pads and anesthetized by using 1 mM levamisole. At least 15 animals were imaged per condition as determined by power analysis with a moderate effect size. The microscope conditions were optimized with respect to the control and test conditions and kept consistent within each trial. However, imaging exposure times were different between some trials to prevent saturation of signal in experimental conditions. All imaging experiments were performed in three independent trials. Mean fluorescence intensities were measured using the Nikon NIS Elements AR v5.02 software and were analyzed using the unpaired t test.
RNA isolation and qRT-PCR
Total RNA was extracted from animals at 48 h after the L4 stage. Animals were synchronized by bleaching [32]. Total RNA was extracted by the freeze cracking method as previously described [33]. After RNA isolation, 2 µg of total RNA was primed with oligo(dT) and reverse transcribed to yield cDNA using the SuperScript III reverse transcriptase kit as per manufacturer’s protocol (Invitrogen). Real-time PCR was performed on QuantStudio3 (Applied Biosystems by Thermo Fisher Scientific) instrument using the PowerUP SYBR Green master mix (Applied Biosystems) according to manufacturer’s instructions. The experiment was performed in three technical replicates for each condition. Primer sequences for dbl-1 are forward primer GCCATTCTCCACCTCTTCCT and reverse primer GGAACATCAATGCTCGGACC [34]. Primer sequences for spp-9 are forward primer GTTCTCTTTCTGGTTGCGGT and reverse primer GCTCTACAAACATCTTCTGGTGCA. Primer sequences for act-1 are forward primer CCATCATGAAGTGCGACATTG and reverse primer CATGGTTGATGGGGCAAGAG [13]. QuantStudio Design and Analysis Software v1.5.1 was used to calculate raw Ct values and to normalize the values for dbl-1 and spp-9 to the housekeeping gene act-1 (actin) (Applied Biosystems by Thermo Fisher Scientific). Fold change in gene expression between experimental sample and the wild-type control was determined by this software using the formula: 2(−ΔΔCt). Experimental ΔCt values were compared with wild-type ΔCt values using the unpaired t test.
Results
spp-9 localizes in the intestine and head neurons at all developmental stages
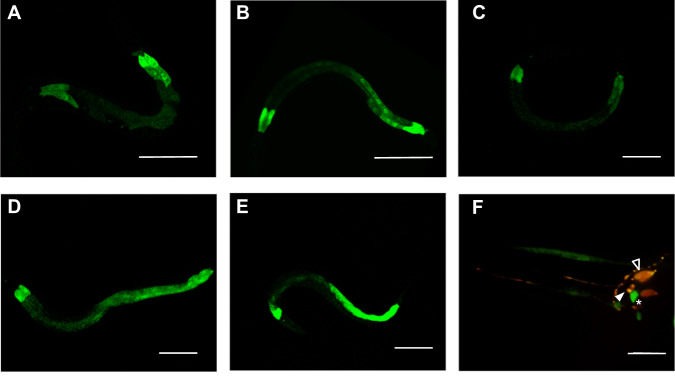
To determine where and when spp-9 is expressed, we used a strain expressing an integrated transgene with the spp-9 promoter driving expression of green fluorescent protein (spp-9p::GFP). Promoter activity of spp-9 was observed in the entire intestine of animals, strongly in the anterior and posterior ends (Fig. 1a–e). spp-9 transgene expression in the intestine was visible at all developmental stages starting from the first larval stage up to adulthood. We also observed spp-9 expression in six head neurons (Fig. 1f). We confirmed the expression of spp-9 in the AWB and AWC neurons by showing colocalization with a fluorescent marker, odr-1::RFP (oyIs44), that is specifically expressed in the two AWB and two AWC sensory neurons [35, 36]. The spp-9 reporter was also visible in a third amphid pair. VisCello for Visualization of Single Cell Data expression data suggests spp-9 is expressed in the ASH neurons, which is consistent with the observed position of this third neuron pair [37]. Because robust intestinal expression of spp-9 was observed only after L3, we quantitated spp-9 reporter activity at 24 h, 36 h, and 48 h after staging animals at L4. We observed an increase in reporter activity over this time frame, with the highest level of expression detected in this time frame in two-day old adults (Fig. S1). These results indicate that spp-9 is expressed majorly in the intestinal tissue and also in six head neurons throughout all the developmental stages of animals, with spp-9 intestinal expression increasing from late larval to adult stages.
Fig. 1. The spp-9 reporter is expressed throughout larval and adult stages in intestine and head neurons.
Confocal images show expression of spp-9p::GFP in (a) L1, (b) L2, (c) L3, (d) L4, and (e) 1-day adult in the intestine, with strong expression in the anterior and posterior ends of intestine. f Rotated swept-field confocal projection reveals spp-9p::GFP is expressed in six head neurons, colocalizing with ODR-1::RFP in AWB and AWC head neurons of an adult animal (marked by filled and unfilled arrowheads, respectively). An asterisks mark a third neuron that expresses the spp-9 reporter. Scale bar indicates a, b 50 µm, c, d, e 100 µm, and (f) 10 µm.
spp-9 expression is increased in starved animals
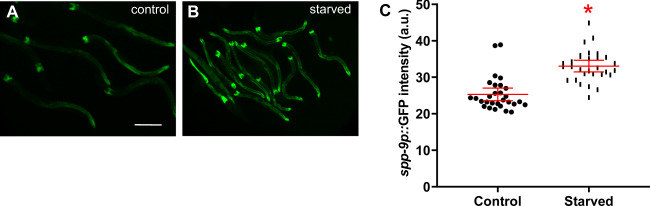
Animals can mount responses to a variety of environmental stresses. Loss of some dpy genes that encode cuticle collagens induce glycerol, osmotic, and detoxification responses, but do not change spp-9 reporter expression [30, 38–41]. Starvation is another stress, and spp-3 expression is induced in starved animals [24, 25]. However, spp-5 expression, which is constitutively expressed and remains unchanged in pathogenic conditions, is also high in starved animals [24]. We asked if starvation affects expression of spp-9. By 8 h without food starting in the L4 stage, C. elegans show dramatic organismal responses, including a global reduction in protein synthesis and changes in stress response pathway gene expression [42]. L4 animals that were starved were imaged 8 h after starvation and compared with identically staged animals that remained fed during that time. spp-9 expression was significantly increased when animals were deprived of food (Fig. 2). This suggests that expression of spp-9 is upregulated in starved animals.
Fig. 2. Starvation increases spp-9 reporter activity.
Fluorescence images show spp-9p::GFP intensities of (a) fed L4 animals and (b) age-matched animals starved for 8 h post L4 stage. Imaging conditions including exposure times were consistent. Scale bar indicates 100 µm. c Comparison of spp-9p::GFP intensities of fed animals (control) with starved animals indicates that spp-9 reporter activity was significantly increased in starved animals in comparison to fed animals (p = 0.0003). One representative trial of three is presented. Error bars represent 95% confidence intervals. n = 30 per condition. *p < 0.05 compared with fed control by unpaired t test.
spp-9 expression is altered in response to select Gram-positive and Gram-negative pathogens
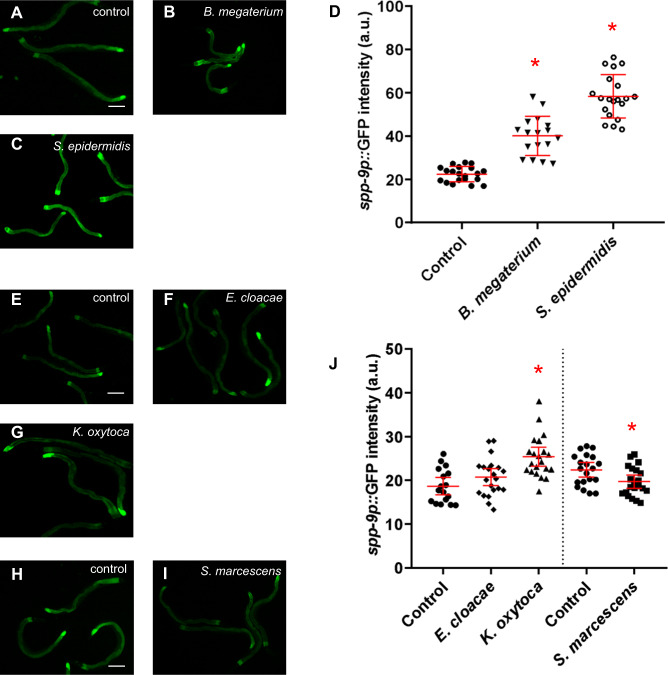
Because caenopores are predicted antimicrobial effector genes, a subset of this 23-member family has been tested for induction upon pathogen challenge. Expression of spp-1 is induced upon infection with S. enterica serovar Typhimurium [43]. spp-3 expression is induced upon confrontation with two Gram-positive bacterial species, B. megaterium and M. luteus, compared with normal lab food E. coli OP50, but was not induced on a different Gram-positive bacterium (Lactobacillus lactis) or any of the four Gram-negative species tested. Expression of spp-5, though, was consistent in animals exposed to different Gram-positive and Gram-negative bacteria, including E. coli [24]. To determine if expression of spp-9 is responsive to exposure to pathogenic bacteria, we detected activity of the spp-9p::gfp reporter in response to a panel of Gram-positive and Gram-negative bacterial exposures. This panel included two Gram-positive (B. megaterium and S. epidermidis) and three Gram-negative strains (E. cloacae, K. oxytoca, and S. marcescens), plus the standard lab food, Gram-negative E. coli OP50. Animals expressing spp-9p::gfp were synchronized as L4s and fed on Gram-positive or Gram-negative bacteria for 48 h. Animals were tested at 48 h because by that time, there was a robust response of the spp-9 reporter on E. coli (Fig. S1). Basal expression was measured for spp-9p::GFP fluorescence in animals fed E. coli OP50 (Fig. 3a, e, h). We observed a significant increase in the spp-9 reporter activity when animals were fed on select Gram-positive (B. megaterium and S. epidermidis), and Gram-negative (K. oxytoca) bacteria (Fig. 3b–d, g, j). On the other hand, exposure to S. marcescens caused a significant decrease in spp-9 expression in two of the three independent trials (Fig. 3i, j). This reduced spp-9 expression is consistent with previous studies showing exposure to S. marcescens induces the DBL-1 signaling pathway (which negatively regulates spp-9) [44]. Our results suggest that activity of spp-9 is differentially regulated in response to specific Gram-positive and Gram-negative bacterial challenges.
Fig. 3. spp-9 reporter activity is altered upon exposure to specific Gram-positive and Gram-negative bacteria.
Fluorescence images show spp-9p::GFP intensities in adult animals after a two-day exposure to the following bacteria; (a, e, h) control E. coli OP50 (n = 17, 20, 20), (b) B. megaterium (n = 17), (c) S. epidermidis (n = 20), (f) E. cloacae (n = 21), (g) K. oxytoca (n = 21), or (i) S. marcescens (n = 21). Imaging conditions including exposure times were consistent with respective controls. Scale bar indicates 100 µm. d Comparison of spp-9p::GFP intensities of wild-type animals fed on E. coli OP50 (control) with animals fed on B. megaterium or S. epidermidis. spp-9p::GFP intensity was significantly increased when animals were exposed to B. megaterium and S. epidermidis in comparison to the control (p < 0.0001). j Comparison of spp-9p::GFP intensities of wild-type animals fed on E. coli OP50 (control) with animals fed on E. cloacae, K. oxytoca, or S. marcescens. No significant change in spp-9p::GFP intensity was seen in animals fed on E. cloacae (p = 0.1353). Exposure to K. oxytoca caused a significant increase in the GFP intensity as compared with the control (p < 0.0001). On the other hand, exposure to S. marcescens caused a significant decrease in the GFP intensity in comparison to the control (p = 0.0153). One representative trial of three is presented. Error bars represent 95% confidence intervals. *p < 0.05 compared with E. coli OP50 by unpaired t test.
dbl-1 regulates endogenous spp-9 expression levels
Studies by our lab and others have shown that the activity of spp-9 is highly regulated by DBL-1 in an inverse fashion. Animals overexpressing DBL-1 show reduced spp-9p::GFP activity and animals lacking DBL-1 display high reporter activity [21, 30]. We tested if DBL-1 regulates endogenous levels of spp-9 expression by quantitative real-time PCR, comparing spp-9 expression levels from dbl-1 mutants to the wild-type control. Animals with the dbl-1(nk3) null allele, a deletion that deletes about 5 kb of the 3′ end of the 7 kb open reading frame, have almost no dbl-1 mRNA detected by qRT-PCR. Animals overexpressing dbl-1 have about a 10-fold increase in dbl-1 mRNA levels (Fig. S2a). We see an increase (about 2.6-fold) in the expression of spp-9 in animals lacking dbl-1, whereas we see a decrease (about 0.5-fold), however not significant, in spp-9 mRNA levels of animals overexpressing dbl-1 (Fig. S2b). These qRT-PCR results support previously reported microarray results and the use of spp-9p::GFP as a reporter for DBL-1 pathway signaling [21, 30].
spp-9 expression depends on DBL-1 and other innate immune response signaling pathways, depending on pathogen
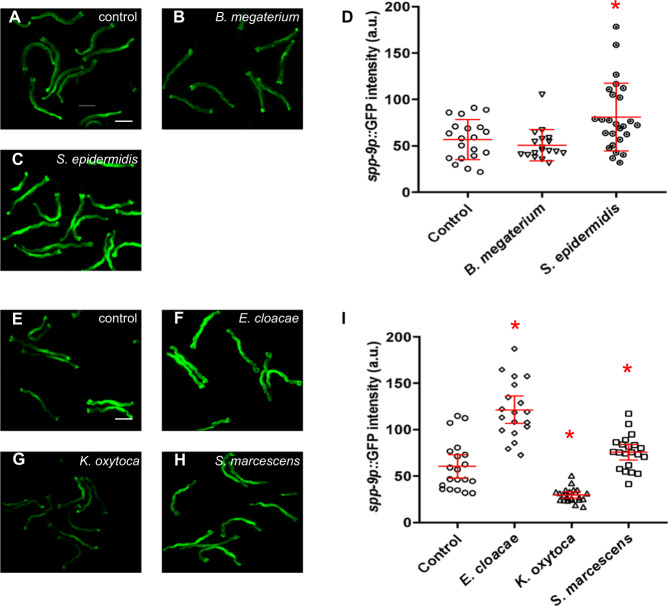
DBL-1 signaling pathway regulates expression of many innate immunity genes and is also involved in mounting protective immune responses against pathogens [13, 18, 44–48]. We showed that the DBL-1 pathway target gene spp-9 is differentially regulated upon exposure to different types of Gram-positive and Gram-negative bacteria. Therefore, we asked if the differential regulation of the spp-9 reporter activity in response to the panel of Gram-positive and Gram-negative bacteria is DBL-1 mediated. To test this, we exposed spp-9p::gfp and dbl-1(-); spp-9p::gfp animals to our panel of different bacteria at the L4 stage. These animals were imaged 48 h after L4 stage and we measured and compared GFP intensities of the two genotypes fed on different bacteria. In three independent trials, the dbl-1(-); spp-9p::gfp animals when fed on standard lab food, E. coli strain OP50, showed a significantly increased fluorescence, consistent with previous reports (Fig. 4a, e) [21, 30]. We observed that animals lacking dbl-1 when fed on S. epidermidis, E. cloacae, and S. marcescens showed a further increase in fluorescence in comparison to the control (dbl-1(-); spp-9p::gfp animals fed on E. coli OP50) (Fig. 4c, d, f, h, i). These findings indicate that loss of dbl-1 increases the spp-9 reporter activity and exposure to select Gram-positive (S. epidermidis) and Gram-negative (E. cloacae and S. marcescens) bacteria further increases the reporter activity when dbl-1 is absent, suggestive of an additive effect on spp-9 reporter activity, perhaps by other innate immune response pathways.
Fig. 4. spp-9 reporter activity is altered in response to specific pathogens by DBL-1 and other innate immunity signaling pathways.
Fluorescence images show spp-9p::GFP intensities in adult dbl-1(-) animals after a two-day exposure to the following bacteria; (a, e) control E. coli OP50 (n = 20), (b) B. megaterium (n = 18), (c) S. epidermidis (n = 26), (f) E. cloacae (n = 19), (g) K. oxytoca (n = 22), or (h) S. marcescens (n = 21). Imaging conditions including exposure times were consistent with respective controls. Scale bar indicates 100 µm. d Comparison of spp-9p::GFP intensities in dbl-1(-) animals fed on E. coli OP50 (control) with animals fed on B. megaterium or S. epidermidis. No significant change in spp-9p::GFP intensity was seen in animals fed on B. megaterium (p = 0.3534). The GFP intensity was significantly increased when animals were exposed to S. epidermidis in comparison to the control (p = 0.0117). i Comparison of spp-9p::GFP intensities in dbl-1(-) animals fed on E. coli OP50 (control) with animals fed on either E. cloacae, K. oxytoca, or S. marcescens. Exposure to E. cloacae (p = 0.0001) and S. marcescens (p = 0.0385) caused an increase in the GFP intensity as compared with the control. Exposure to K. oxytoca caused a decrease in the GFP intensity in comparison to the control (p = 0.0001). One representative trial of three is presented. Error bars represent 95% confidence intervals. *p < 0.05 compared with E. coli OP50 by unpaired t test.
On the other hand, dbl-1(-); spp-9p::gfp animals showed similar mean fluorescence intensities when fed on B. megaterium in comparison to dbl-1(-); spp-9p::gfp animals exposed to the E. coli OP50 control (Fig. 4b, d). This response is different from spp-9 reporter activity in the wild type, which showed significantly increased fluorescence with B. megaterium exposure as compared with the control (Fig. 3b, d). This lack of further spp-9 reporter induction in dbl-1 mutants on B. megaterium, like was observed on E. cloacae, S. marcescens, and S. epidermidis, indicates that spp-9 reporter activity induced on B. megaterium is DBL-1 mediated (Figs. 3 and 4).
Interestingly, while animals lacking dbl-1 fed on K. oxytoca displayed increased fluorescence than spp-9p::gfp animals, that increase was significantly less in comparison to the same genotype fed on E. coli in two out of three trials (Figs. 3 and 4, data from both genotypes was collected in each trial). This suggests that some other signaling pathway that negatively regulates spp-9 activity is induced in dbl-1(-) animals upon exposure to K. oxytoca, resulting in decrease of spp-9 activity. This suggests that the reporter activity response to K. oxytoca exposure depends on an innate immune response that is at least partially independent of dbl-1.
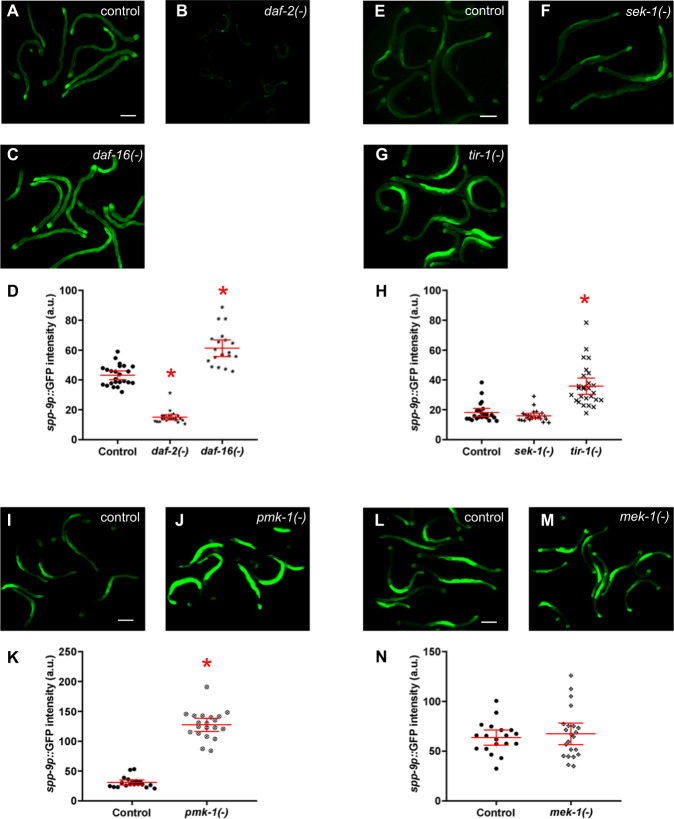
spp-9 reporter activity is regulated by innate immunity/stress signaling pathways
Because we see an additive effect of DBL-1 and infection on the activity of spp-9, and also DBL-1-independent but pathogen-dependent regulation of spp-9, we asked whether spp-9 is also regulated by other signaling pathways in the context of immunity. Besides the DBL-1 signaling pathway, two major pathways that are required for animals to respond to pathogens are the insulin-like and p38/MAPK signaling pathways [19, 49, 50]. The insulin-like pathway is defined by the insulin receptor DAF-2 and the downstream master transcriptional regulator DAF-16. Loss of daf-2 function not only causes animals to constitutively enter dauer but also increases resistance to pathogens. Loss of daf-16 prevents animals from entering dauer and increases sensitivity to infection [12, 49, 51–54]. We measured reporter fluorescence intensities in daf-2(-) and daf-16(-) backgrounds and compared it to the reporter in the wild-type background. Interestingly, we observed a significant reduction in daf-2(-) mutants (Fig. 5b, d). As expected, animals lacking daf-16 showed increased spp-9 reporter fluorescence (Fig. 5c, d). These findings suggest that spp-9 is responsive to changes in insulin-like signaling: DAF-2 promotes, while DAF-16 normally represses, spp-9 expression.
Fig. 5. spp-9 reporter activity is regulated by innate immunity signaling pathways.
Fluorescence images show spp-9p::GFP intensities in two-day adults with (a, e, i, l) wild-type (n = 24, 23, 19, 19), (b) daf-2(-) (n = 24), (c) daf-16(-) (n = 20), (f) sek-1(-) (n = 23), (g) tir-1(-) (n = 27), (j) pmk-1(-) (n = 20), and (m) mek-1(-) (n = 22) backgrounds. Imaging conditions including exposure times were consistent with respective controls. Scale bar indicates 100 µm. d Comparison of spp-9p::GFP intensities in wild-type (control), daf-2(-), and daf-16(-) backgrounds. Loss of daf-2 caused a decrease in the GFP intensity (p < 0.0001), whereas loss of daf-16 caused an increase in the GFP intensity (p < 0.0001) as compared with the control. h Comparison of spp-9p::GFP intensities in wild-type (control), sek-1(-), and tir-1(-) backgrounds. Loss of tir-1 caused an increase in the reporter activity in comparison to the control (p < 0.0001). k Comparison of spp-9p::GFP intensities in wild-type (control) and pmk-1(-) backgrounds. The GFP intensity increased in animals lacking pmk-1 (p < 0.0001). n Comparison of spp-9p::GFP intensities in wild-type (control) and mek-1(-) backgrounds. No significant change in spp-9p::GFP intensity was seen in mek-1(-) animals (p = 0.5674). One representative trial of three is presented. Error bars represent 95% confidence intervals. *p < 0.05 compared with spp-9p::GFP in wild-type background by unpaired t test.
The p38/MAPK signaling pathway includes TIR-1, TIR (Toll and Interleukin 1 Receptor) domain protein, which activates MAP3K, which activates MAP2K SEK-1, which activates MAPK PMK-1 [55–57]. We captured and measured GFP intensities of the reporter in pmk-1(-), sek-1(-), or tir-1(-) mutant backgrounds and compared them to the wild-type reporter control. We observed a significant increase in the mean GFP intensities in tir-1(-) and pmk-1(-) mutants (Fig. 5g, h, j, k). However, loss of MAP2K gene sek-1(-) had no effect on the fluorescent intensities in comparison to the control (Fig. 5f, h). We then tested if MEK-1, a stress-responsive MAP2K that can also activate PMK-1 independent of TIR-1, affected spp-9 reporter activity [56]. Loss of mek-1, like loss of sek-1, also had no effect on spp-9 reporter fluorescence (Fig. 5m, n). These findings suggest that the MAPK pathway defined by tir-1 and pmk-1 normally suppresses spp-9 activity, but does so using a MAP2K other than SEK-1 or MEK-1. Alternatively, these MAP2Ks act redundantly in the pathway that leads to expression of spp-9. Together, these results indicate spp-9 is regulated by multiple signaling pathways.
Discussion
Here, we show that SPP-9 has a unique role within its large family of saposin-like, pore-forming proteins in C. elegans, and its expression is modulated to respond to specific pathogens and other stressors.
spp-9 is visibly expressed from the first larval stage, when animals are initially exposed to environmental conditions, including pathogens, and this expression is maintained throughout the life of the animal. This observation is consistent with previous RNAseq and microarray analyses that do not detect spp-9 expression in embryos (WormBase.org). Basal expression of spp-9 is observed in the intestine of animals fed normal lab food, E. coli. Other saposin family members play roles in the digestion of the roundworm’s bacterial food. SPP-9 may also help digest bacteria and is upregulated in some environments with pathogenic bacteria. However, our work supports a role for spp-9 in stress response, not just digestion. For one, spp-9 expression is increased in starvation conditions (Fig. 2). spp-3 and spp-5 have also been shown to be expressed in starved adults [24]. Second, spp-9 is expressed in the AWB and AWC sensory neuron pairs, which play roles in sensing numerous stimuli, including bacterial food sources [58–61]. C. elegans uses AWB to sense and avoid serrawettin W2, a chemical secreted by the pathogenic S. marcescens [62]. spp-9 expression in amphid neurons might contribute to sensing different pathogens or other stresses. Some other C. elegans saposins are expressed not only in the intestine, but also or instead in a specific neuron or a few neurons. spp-3 is expressed in the SDQR interneuron, spp-12 is expressed in NSM/L and I6, which can sense bacteria, including pathogens, and spp-7 is expressed in head neurons [25, 26]. Neurons in C. elegans and other species express saposins and other antimicrobial proteins, possibly to protect the environmentally exposed neurons or surrounding tissues. Alternatively, the expression of different saposins in different environmentally exposed neurons could help specify distinct antimicrobial responses depending on the stimulus.
Third, a strong induction of spp-9 was observed in animals exposed to some bacterial challenges, but not all, suggesting specificity of recruiting SPP-9 in a response that is not demarcated by Gram nature (Figs. 3 and 4). Furthermore, we discovered that spp-9 is specifically upregulated by loss of specific innate immune/stress response pathways: the DBL-1/TGF-β-like pathway, the DAF-2/DAF-16 insulin-like pathway, and the p38/MAPK defined by TIR-1 and PMK-1, but independent of MAPKKs SEK-1 and MEK-1 (Figs. 4 and 5). This may explain why we see an additive induction of spp-9 in animals lacking DBL-1 signaling and challenged with E. cloacae, S. marcescens, or S. epidermidis: other innate immune response pathways may be further inducing spp-9 expression in this context. However, the response to B. megaterium, in which the spp-9 induction depends on functional DBL-1 signaling, suggests that the animals use DBL-1 but not other innate immune pathways to induce a spp-9 response to B. megaterium. However, the reduced spp-9 induction upon exposure to K. oxytoca in dbl-1 mutant animals suggests that other signaling pathways are dampening this response. These findings also support that DBL-1, insulin-like, and MAPK pathways act independently but converge to regulate spp-9 expression. These results suggest that spp-9 contributes to the “antimicrobial fingerprint” proposed by Alper et al., in which distinct molecular responses are generated against specific pathogen exposures [18].
spp-9 was identified as a highly regulated gene by the DBL-1 signaling pathway and a spp-9 promoter-GFP transgene has been used as a reporter for this pathway [21, 30]. The changes observed in the spp-9 transcriptional reporter are consistent with the changes in endogenous spp-9 expression levels in different dbl-1 backgrounds, indicating that the reporter activity is representative of endogenous spp-9 expression levels. Our findings support that spp-9 is a valid reporter of DBL-1 signaling, with the caveat that the experimental environment must be closely controlled.
In summary, we discovered that spp-9 expression is induced by starvation, specific bacteria, and by not only DBL-1, but other innate immune/stress pathways. It will be of interest to determine the molecular role of SPP-9 in replete and stress conditions, and to identify how multiple signaling pathways coordinate spp-9 expression. This work revealed further insights into the environmental responsiveness of the saposin family and how organisms generate complex, targeted molecular responses to physiological challenges.
Supplementary information
Acknowledgements
We thank Paul Yeatts and TWU CRDA for assistance with statistical analyses. The bacterial strains were provided by Amy Jo Hammett. We thank Robert Waterston for ASH neuron information. We thank Mehul Vora and Richard Padgett for wkIs40 and Maxwell Heiman for oyIs44. Some strains were obtained from the Caenorhabditis Genetics Center (CGC), which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). We thank WormBase. We thank Laura Hanson, Yasar Arfat Kasu, and members of the Gumienny lab for constructive feedback. This work was supported by NIH R01GM097591 to TLG, a TWU Research Enhancement Program grant to TLG, internal funding by Texas Woman’s University, a TWU Experiential Learning Scholar Award to BM, and a TWU Teaching and Research Grant for Equipment and Technology. This work is dedicated to Chimanlal Madhu and Darshan Madhu.
Data availability
Strains are available upon request. Supplementary figures are available in Figshare.
Compliance with ethical standards
Conflict of interest
TLG work was funded by the NIH. The other authors declare no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41435-020-0108-6) contains supplementary material, which is available to authorized users.
References
- 1.Medzhitov R, Janeway CA., Jr An ancient system of host defense. Curr Opin Immunol. 1998;10:12–5. doi: 10.1016/s0952-7915(98)80024-1. [DOI] [PubMed] [Google Scholar]
- 2.Cui J, Chen Y, Wang HY, Wang RF. Mechanisms and pathways of innate immune activation and regulation in health and cancer. Hum Vaccin Immunother. 2014;10:3270–85. doi: 10.4161/21645515.2014.979640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McSorley R. Adaptations of nematodes to environmental extremes. Fla Entomologist. 2003;86:138–42. [Google Scholar]
- 4.Murfin KE, Dillman AR, Foster JM, Bulgheresi S, Slatko BE, Sternberg PW, et al. Nematode-bacterium symbioses-cooperation and conflict revealed in the “omics” age. Biol Bull. 2012;223:85–102. doi: 10.1086/BBLv223n1p85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Engelmann I, Pujol N. Innate immunity in C. elegans. Adv Exp Med Biol. 2010;708:105–21. doi: 10.1007/978-1-4419-8059-5_6. [DOI] [PubMed] [Google Scholar]
- 6.Cheesman HK, Feinbaum RL, Thekkiniath J, Dowen RH, Conery AL, Pukkila-Worley R. Aberrant activation of p38 MAP kinase-dependent innate immune responses is toxic to Caenorhabditis elegans. G3. 2016;G3:541–9. doi: 10.1534/g3.115.025650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pukkila-Worley R. Surveillance immunity: an emerging paradigm of innate defense activation in Caenorhabditis elegans. PLoS Pathog. 2016;12:e1005795. doi: 10.1371/journal.ppat.1005795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Couillault C, Ewbank JJ. Diverse bacteria are pathogens of Caenorhabditis elegans. Infect Immun. 2002;70:4705–7. doi: 10.1128/IAI.70.8.4705-4707.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gravato-Nobre MJ, Nicholas HR, Nijland R, O’Rourke D, Whittington DE, Yook KJ, et al. Multiple genes affect sensitivity of Caenorhabditis elegans to the bacterial pathogen Microbacterium nematophilum. Genetics. 2005;171:1033–45. doi: 10.1534/genetics.105.045716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Begun J, Gaiani JM, Rohde H, Mack D, Calderwood SB, Ausubel FM, et al. Staphylococcal biofilm exopolysaccharide protects against Caenorhabditis elegans immune defenses. PLoS Pathog. 2007;3:e57. doi: 10.1371/journal.ppat.0030057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wong D, Bazopoulou D, Pujol N, Tavernarakis N, Ewbank JJ. Genome-wide investigation reveals pathogen-specific and shared signatures in the response of Caenorhabditis elegans to infection. Genome Biol. 2007;8:R194. doi: 10.1186/gb-2007-8-9-r194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Singh V, Aballay A. Regulation of DAF-16-mediated innate immunity in Caenorhabditis elegans. J Biol. Chem. 2009;284:35580–7. doi: 10.1074/jbc.M109.060905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zugasti O, Ewbank JJ. Neuroimmune regulation of antimicrobial peptide expression by a noncanonical TGF-β signaling pathway in Caenorhabditis elegans epidermis. Nat Immunol. 2009;10:249–56. doi: 10.1038/ni.1700. [DOI] [PubMed] [Google Scholar]
- 14.Pukkila-Worley R, Ausubel FM, Mylonakis E. Candida albicans infection of Caenorhabditis elegans induces antifungal immune defenses. PLoS Pathog. 2011;7:e1002074. doi: 10.1371/journal.ppat.1002074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dzakah EE, Waqas A, Wei S, Yu B, Wang X, Fu T, et al. Loss of miR-83 extends lifespan and affects target gene expression in an age-dependent manner in Caenorhabditis elegans. J Genet Genomics. 2018;45:651–62. doi: 10.1016/j.jgg.2018.11.003. [DOI] [PubMed] [Google Scholar]
- 16.Aballay A, Yorgey P, Ausubel FM. Salmonella typhimurium proliferates and establishes a persistent infection in the intestine of Caenorhabditis elegans. Curr Biol. 2000;10:1539–42. doi: 10.1016/s0960-9822(00)00830-7. [DOI] [PubMed] [Google Scholar]
- 17.Garsin DA, Sifri CD, Mylonakis E, Qin X, Singh KV, Murray BE, et al. A simple model host for identifying Gram-positive virulence factors. Proc Natl Acad Sci USA. 2001;98:10892–7. doi: 10.1073/pnas.191378698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alper S, McBride SJ, Lackford B, Freedman JH, Schwartz DA. Specificity and complexity of the Caenorhabditis elegans innate immune response. Mol Cell Biol. 2007;27:5544–53. doi: 10.1128/MCB.02070-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim DH, Ewbank JJ. Signaling in the innate immune response. WormBook. Pasadena, CA, 2018,1–35. [DOI] [PMC free article] [PubMed]
- 20.Sun L, Zhi L, Shakoor S, Liao K, Wang D. microRNAs involved in the control of innate immunity in Candida infected Caenorhabditis elegans. Sci Rep. 2016;6:36036. doi: 10.1038/srep36036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roberts AF, Gumienny TL, Gleason RJ, Wang H, Padgett RW. Regulation of genes affecting body size and innate immunity by the DBL-1/BMP-like pathway in Caenorhabditis elegans. BMC Dev Biol. 2010;10:61. doi: 10.1186/1471-213X-10-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dierking K, Yang W, Schulenburg H. Antimicrobial effectors in the nematode Caenorhabditis elegans: an outgroup to the Arthropoda. Philos Trans R Soc Lond B Biol Sci. 2016;371:20150299.. doi: 10.1098/rstb.2015.0299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Banyai L, Patthy L. Amoebapore homologs of Caenorhabditis elegans. Biochim Biophys Acta. 1998;1429:259–64. doi: 10.1016/s0167-4838(98)00237-4. [DOI] [PubMed] [Google Scholar]
- 24.Roeder T, Stanisak M, Gelhaus C, Bruchhaus I, Grotzinger J, Leippe M. Caenopores are antimicrobial peptides in the nematode Caenorhabditis elegans instrumental in nutrition and immunity. Dev Comp Immunol. 2010;34:203–9. doi: 10.1016/j.dci.2009.09.010. [DOI] [PubMed] [Google Scholar]
- 25.Hoeckendorf A, Leippe M. SPP-3, a saposin-like protein of Caenorhabditis elegans, displays antimicrobial and pore-forming activity and is located in the intestine and in one head neuron. Dev Comp Immunol. 2012;38:181–6. doi: 10.1016/j.dci.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 26.Hoeckendorf A, Stanisak M, Leippe M. The saposin-like protein SPP-12 is an antimicrobial polypeptide in the pharyngeal neurons of Caenorhabditis elegans and participates in defence against a natural bacterial pathogen. Biochem J. 2012;445:205–12. doi: 10.1042/BJ20112102. [DOI] [PubMed] [Google Scholar]
- 27.Evans EA, Kawli T, Tan MW. Pseudomonas aeruginosa suppresses host immunity by activating the DAF-2 insulin-like signaling pathway in Caenorhabditis elegans. PLoS Pathog. 2008;4:e1000175. doi: 10.1371/journal.ppat.1000175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murphy CT, McCarroll SA, Bargmann CI, Fraser A, Kamath RS, Ahringer J, et al. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature. 2003;424:277–83. doi: 10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- 29.Ren M, Feng H, Fu Y, Land M, Rubin CS. Protein kinase D is an essential regulator of C. elegans innate immunity. Immunity. 2009;30:521–32. doi: 10.1016/j.immuni.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lakdawala MF, Madhu B, Faure L, Vora M, Padgett RW, Gumienny TL. Genetic interactions between the DBL-1/BMP-like pathway and dpy body size-associated genes in Caenorhabditis elegans. Mol Biol Cell. 2019;30:3151–60. doi: 10.1091/mbc.E19-09-0500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Madhu BJ, Salazar AE, Gumienny TL. Caenorhabditis elegans egg-laying and brood-size changes upon exposure to Serratia marcescens and Staphylococcus epidermidis are independent of DBL-1 signaling. microPublication Biology. 2019. 10.17912/2r51-b476. [DOI] [PMC free article] [PubMed]
- 32.Stiernagle T. Maintenance of C. elegans. WormBook. Pasadena, CA, 2006;1–11. [DOI] [PMC free article] [PubMed]
- 33.Portman DS. Profiling C. elegans gene expression with DNA microarrays. WormBook. Pasadena, CA, 2006;1–11. [DOI] [PMC free article] [PubMed]
- 34.Kwon G, Lee J, Lim YH. Dairy Propionibacterium extends the mean lifespan of Caenorhabditis elegans via activation of the innate immune system. Sci Rep. 2016;6:31713. doi: 10.1038/srep31713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim K, Kim R, Sengupta P. The HMX/NKX homeodomain protein MLS-2 specifies the identity of the AWC sensory neuron type via regulation of the ceh-36 Otx gene in C. elegans. Development. 2010;137:963–74. doi: 10.1242/dev.044719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Low IIC, Williams CR, Chong MK, McLachlan IG, Wierbowski BM, Kolotuev I, et al. Morphogenesis of neurons and glia within an epithelium. Development. 2019;146:dev171124.. doi: 10.1242/dev.171124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Packer JS, Zhu Q, Huynh C, Sivaramakrishnan P, Preston E, Dueck H, et al. A lineage-resolved molecular atlas of C. elegans embryogenesis at single-cell resolution. Science. 2019;365:eaax1971.. doi: 10.1126/science.aax1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wheeler JM, Thomas JH. Identification of a novel gene family involved in osmotic stress response in Caenorhabditis elegans. Genetics. 2006;174:1327–36. doi: 10.1534/genetics.106.059089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lamitina T, Huang CG, Strange K. Genome-wide RNAi screening identifies protein damage as a regulator of osmoprotective gene expression. Proc Natl Acad Sci USA. 2006;103:12173–8. doi: 10.1073/pnas.0602987103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zugasti O, Thakur N, Belougne J, Squiban B, Kurz CL, Soule J, et al. A quantitative genome-wide RNAi screen in C. elegans for antifungal innate immunity genes. BMC Biol. 2016;14:35.. doi: 10.1186/s12915-016-0256-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dodd W, Tang L, Lone JC, Wimberly K, Wu CW, Consalvo C, et al. A damage sensor associated with the cuticle coordinates three core environmental stress responses in Caenorhabditis elegans. Genetics. 2018;208:1467–82. doi: 10.1534/genetics.118.300827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Larance M, Pourkarimi E, Wang B, Brenes Murillo A, Kent R, Lamond AI, et al. Global proteomics analysis of the response to starvation in C. elegans. Mol Cell Proteom. 2015;14:1989–2001. doi: 10.1074/mcp.M114.044289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alegado RA, Tan MW. Resistance to antimicrobial peptides contributes to persistence of Salmonella typhimurium in the C. elegans intestine. Cell Microbiol. 2008;10:1259–73. doi: 10.1111/j.1462-5822.2008.01124.x. [DOI] [PubMed] [Google Scholar]
- 44.Mallo GV, Kurz CL, Couillault C, Pujol N, Granjeaud S, Kohara Y, et al. Inducible antibacterial defense system in C. elegans. Curr Biol. 2002;12:1209–14. doi: 10.1016/s0960-9822(02)00928-4. [DOI] [PubMed] [Google Scholar]
- 45.Mochii M, Yoshida S, Morita K, Kohara Y, Ueno N. Identification of transforming growth factor-β-regulated genes in Caenorhabditis elegans by differential hybridization of arrayed cDNAs. Proc Natl Acad Sci. USA. 1999;96:15020–5. doi: 10.1073/pnas.96.26.15020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liang J, Yu L, Yin J, Savage-Dunn C. Transcriptional repressor and activator activities of SMA-9 contribute differentially to BMP-related signaling outputs. Dev Biol. 2007;305:714–25. doi: 10.1016/j.ydbio.2007.02.038. [DOI] [PubMed] [Google Scholar]
- 47.Zhang X, Zhang Y. DBL-1, a TGF-β, is essential for Caenorhabditis elegans aversive olfactory learning. Proc Natl Acad Sci USA. 2012;109:17081–6. doi: 10.1073/pnas.1205982109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Berg M, Monnin D, Cho J, Nelson L, Crits-Christoph A, Shapira M. TGF-β/BMP immune signaling affects abundance and function of C. elegans gut commensals. Nat Commun. 2019;10:604. doi: 10.1038/s41467-019-08379-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Garsin DA, Villanueva JM, Begun J, Kim DH, Sifri CD, Calderwood SB, et al. Long-lived C. elegans daf-2 mutants are resistant to bacterial pathogens. Science. 2003;300:1921. doi: 10.1126/science.1080147. [DOI] [PubMed] [Google Scholar]
- 50.Troemel ER, Chu SW, Reinke V, Lee SS, Ausubel FM, Kim DH. p38 MAPK regulates expression of immune response genes and contributes to longevity in C. elegans. PLoS Genet. 2006;2:e183. doi: 10.1371/journal.pgen.0020183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang RA. C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–4. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- 52.Kimura KD, Tissenbaum HA, Liu Y, Ruvkun G. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science. 1997;277:942–6. doi: 10.1126/science.277.5328.942. [DOI] [PubMed] [Google Scholar]
- 53.Zhao L, Zhao Y, Liu R, Zheng X, Zhang M, Guo H, et al. The transcription factor DAF-16 is essential for increased longevity in C. elegans exposed to Bifidobacterium longum BB68. Sci Rep. 2017;7:7408. doi: 10.1038/s41598-017-07974-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou M, Liu X, Yu H, Yin X, Nie SP, Xie MY, et al. Cell signaling of Caenorhabditis elegans in response to enterotoxigenic Escherichia coli Infection and Lactobacillus zeae protection. Front Immunol. 2018;9:1745. doi: 10.3389/fimmu.2018.01745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim DH, Feinbaum R, Alloing G, Emerson FE, Garsin DA, Inoue H, et al. A conserved p38 MAP kinase pathway in Caenorhabditis elegans innate immunity. Science. 2002;297:623–6. doi: 10.1126/science.1073759. [DOI] [PubMed] [Google Scholar]
- 56.Kim DH, Liberati NT, Mizuno T, Inoue H, Hisamoto N. Matsumoto K, et al. Integration of Caenorhabditis elegans MAPK pathways mediating immunity and stress resistance by MEK-1 MAPK kinase and VHP-1 MAPK phosphatase. Proc Natl Acad Sci USA. 2004;101:10990–4. doi: 10.1073/pnas.0403546101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liberati NT, Fitzgerald KA, Kim DH, Feinbaum R, Golenbock DT, Ausubel FM. Requirement for a conserved Toll/interleukin-1 resistance domain protein in the Caenorhabditis elegans immune response. Proc Natl Acad Sci USA. 2004;101:6593–8. doi: 10.1073/pnas.0308625101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bargmann CI, Hartwieg E, Horvitz HR. Odorant-selective genes and neurons mediate olfaction in C. elegans. Cell. 1993;74:515–27. doi: 10.1016/0092-8674(93)80053-h. [DOI] [PubMed] [Google Scholar]
- 59.Troemel ER, Kimmel BE, Bargmann CI. Reprogramming chemotaxis responses: sensory neurons define olfactory preferences in C. elegans. Cell. 1997;91:161–9. doi: 10.1016/s0092-8674(00)80399-2. [DOI] [PubMed] [Google Scholar]
- 60.Sagasti A, Hobert O, Troemel ER, Ruvkun G, Bargmann CI. Alternative olfactory neuron fates are specified by the LIM homeobox gene lim-4. Genes Dev. 1999;13:1794–806. doi: 10.1101/gad.13.14.1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Olofsson B. The olfactory neuron AWC promotes avoidance of normally palatable food following chronic dietary restriction. J Exp Biol. 2014;217:1790–8. doi: 10.1242/jeb.099929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pradel E, Zhang Y, Pujol N, Matsuyama T, Bargmann CI, Ewbank JJ. Detection and avoidance of a natural product from the pathogenic bacterium Serratia marcescens by Caenorhabditis elegans. Proc Natl Acad Sci USA. 2007;104:2295–300. doi: 10.1073/pnas.0610281104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Strains are available upon request. Supplementary figures are available in Figshare.