Abstract
Background
Metagenomic next-generation sequencing (mNGS) of bronchoalveolar lavage fluid (BALF) has the potential to improve the pathogen identification in severe community-acquired pneumonia (SCAP).
Methods
In this 1.5-year, multicenter, prospective study, we investigated the usefulness of mNGS of BALF for identifying pathogens of SCAP in hospitalized adults, comparing it with other laboratory methods.
Results
Of 329 SCAP adults, a microbial etiology was established in 304 cases (92.4%). The overall microbial yield was 90.3% for mNGS versus 39.5% for other methods (P < 0.05). The most frequently detected pathogens in immunocompetent patients were Streptococcus pneumoniae (14.8%), rhinovirus (9.8%), Haemophilus influenzae (9.1%), Staphylococcus aureus (8.7%), and Chlamydia psittaci (8.0%), while in immunocompromised patients they were Pneumocystis jirovecii (44.6%), Klebsiella pneumoniae (18.5%), Streptococcus pneumoniae (15.4%), Haemophilus influenzae (13.8%), and Pseudomonas aeruginosa (13.8%). Notably, novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was identified from two patients solely by mNGS in January 2020; uncommon pathogens including Orientia tsutsugamushi and Nocardia otitidiscaviarum were identified from one patient, respectively. Furthermore, mixed infections were detected in 56.8% of the patients.
Conclusions
A high microbial detection rate was achieved in SCAP adults using mNGS testing of BALF. The most frequently detected pathogens of SCAP differed between immunocompetent and immunocompromised patients. mNGS testing may be an powerful tool for early identification of potential pathogens for SCAP to initiate a precise antimicrobial therapy.
Keywords: Etiology, Metagenomic next-generation sequencing, Severe community-acquired pneumonia
Key Summary Points
| mNGS of BALF achieved a microbial etiology in 92.4% of SCAP adults. |
| Pathogen spectra differed between immunocompetent and immunocompromised cases. |
| mNGS played a unique role in identifying novel SARS-CoV-2 and uncommon pathogens. |
Digital Features
This article is published with digital features, including a summary slide, to facilitate understanding of the article. To view digital features for this article go to https://doi.org/10.6084/m9.figshare.13014236.
Introduction
Severe community-acquired pneumonia (CAP) is one of the most common infectious diseases in intensive care units (ICUs) and has a mortality rate of 30–50% [1]. Early identification of the causative pathogens is the basis for implementing precision antimicrobial therapy, which has a significant impact on patient prognosis. However, conventional microbial methods identify a pathogen in only 30–40% of CAP cases [2, 3]. Although molecular diagnostic techniques based on targeted polymerase chain reaction (PCR) assays have improved the detection rate of many common viral and bacterial pathogens [4, 5], the diagnostic value is confined to the targeted spectrum of pathogens and remains unsatisfying, particularly in immunocompromised patients with CAP who are challenged by a complex pathogen spectrum [6].
Recently, metagenomic next-generation sequencing (mNGS) has become a powerful technique in medical microbiology because of its high-throughput capacity and fast turnaround time [7–9]. Studies have examined microbial detection from cerebrospinal fluid in patients with central nervous system infections, from serum in septic patients, and from lower respiratory tract specimens in patients with pneumonia [10–15]. However, the use of mNGS for pathogen detection in CAP remains to be further investigated.
An understanding of the pathogens responsible for SCAP plays a crucial role in the selection of the initial antimicrobial treatment. Recently, the etiology of CAP in adults has been explored using conventional and new PCR-based methods; however, the etiology of SCAP remains poorly characterized.
This study examined the etiology of SCAP in adults and the diagnostic value of mNGS of BALF specimens compared to conventional, serology, and targeted PCR-based methods.
Methods
Patient Selection and Study Design
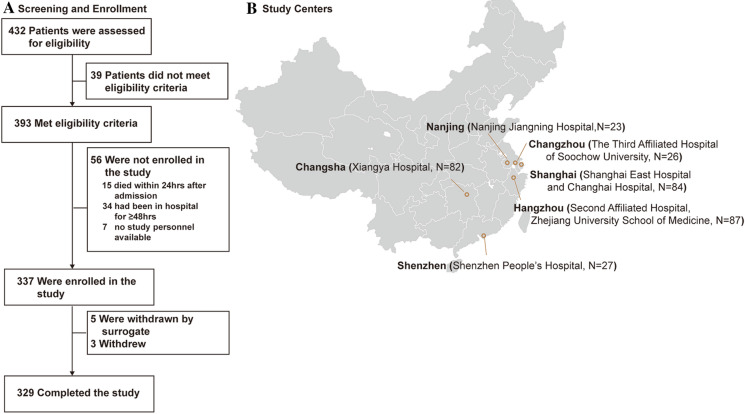
For this prospective study, we consecutively enrolled adult patients (aged ≥ 18 years) with SCAP admitted to ICUs in seven general hospitals in Shanghai, Changsha, Hangzhou, Changzhou, Nanjing, and Shenzhen from June 2018 to February 2020. SCAP was defined in patients with either one major criterion or at least three minor criteria of the Infectious Diseases Society of America (IDSA)/American Thoracic Society (ATS) criteria [16]. Patients were excluded if they had a life expectancy < 24 h according to the disease severity or had been in the hospital for ≥ 48 h prior to ICU admission. This study was performed in accordance with the Helsinki Declaration of 1964 and its later amendments. Ethical approval for this study was obtained from the institutional ethics committee at each study site (The Ethics Committee of Shanghai East Hospital Affiliated with Shanghai Tongji University, Xiangya Hospital Affiliated with Central South University, the Second Affiliated Hospital of Zhejiang University School of Medicine, the Third Affiliated Hospital of Soochow University, Fourth Affiliated Hospital of Zhejiang University School of Medicine, Changhai Hospital, The Affiliated Jiangning Hospital of Nanjing Medical University, and the Second Clinical Medical College Affiliated with Jinan University). Written informed consent was provided from each patient or family members at enrolment.
The definition of immunocompromised status was met when ≥ 1 of the following risk factors were found: (1) hematologic cancer; (2) chemotherapy during the last 3 months; (3) neutropenia; (4) chronic steroid or biologic drug use for autoimmune diseases; (5) immunosuppressive therapy due to hematologic cancer or solid organ transplantation; (6) solid tumor with either neutropenia or chemotherapy; (7) other immunocompromised state (any immunocompromised state, including congenital/genetic immunocompromise and asplenia). Individuals immunocompromised because of AIDS were not enrolled in this study.
All patients underwent bronchoscopy to obtain BALF. Both mNGS and conventional methods were used to detect pathogens.
Fiberoptic Bronchoscopy and BAL Sample Collection
Bronchoscopy was performed by experienced bronchoscopists in each respiratory ICU. Patients were moderately sedated with intravenous midazolam or dexmedetomidine before bronchoscopy. Local anesthesia with 2% lidocaine was applied sequentially during the procedures. Bedside bronchoalveolar lavage sampling was performed according to standard procedures using a flexible fiberoptic bronchoscope. Briefly, several aliquots of 20 ml sterile isotonic saline were instilled into the target subsegmental bronchi. The first 20 ml was usually discharged to avoid contamination, whereas the remaining samples were collected for detection.
Microbiologic Methods
The BALF specimens were divided into aliquots. Using conventional microbiologic methods, one aliquot was examined by microscopy with routine laboratory staining and cultures of bacteria, fungi, and mycobacteria. One aliquot was sent to detect influenza A/B, parainfluenza 1/2/3, human respiratory syncytial virus (hRSV), and adenovirus using commercial direct fluorescent-antibody assays (Diagnostic Hybrids, Athens, OH, USA). A multiplex real-time polymerase chain reaction (RT-PCR) was performed for the simultaneous detection of Chlamydia pneumoniae (C. pneumoniae), Mycoplasma pneumoniae (M. pneumoniae), Legionella pneumophila (L. pneumophila), human rhinovirus, hRSV, adenovirus, coronavirus, influenza A/B, and parainfluenza 1/2/3. For the other aliquots, a mNGS platform was used for both RNA-Seq and DNA-Seq of the nucleic acid samples extracted from the BALF.
Regarding the serology methods, peripheral blood samples were submitted for the detection of immunoglobulin G and M antibodies to influenza A/B, hRSV, adenovirus, C. pneumoniae, M. pneumoniae, and L. pneumophila using commercial enzyme-linked immunosorbent assays (Ani Labsystems) according to the manufacturer’s instructions.
An abundance score of the microbes detected by mNGS from each patient was calculated as the sum of the log(DNA-Seq) and log(RNA-Seq) reads per million (rpM) alignments at the genus level. The group of high-scoring microbes from each patient was characterized when the greatest score difference was determined between sequentially ranked microbes using the abundance score. The high-scoring microbes were considered significant microbes.
Subsequently, we classified the significant microbes as putative pathogens if two of the following criteria were met: both conventional testing and mNGS identified the microbe, there was robust evidence of pathogenicity in the lungs based on the clinical literature, and the score was as least double that of any other microbe of the same type (virus, bacterium, or fungus) identified in the patient. Microbes solely identified by mNGS were considered pathogens with a degree of clinical suspicion if the third criterion above was met along with discrete evidence of low or opportunistic pathogenicity in the lower respiratory tract. All other microbes were considered unlikely or uncertain pathogens.
Statistical Analyses
We tested for differences in continuous variables using the Wilcoxon rank-sum test and categorical variables with the Pearson chi-square test or Fisher’s exact test as appropriate. All analyses were done using SPSS 22.0 (IBM, Armonk, NY, USA). P values < 0.05 were considered significant and all tests were two-tailed.
Results
Patient Characteristics
Between June 1, 2018, and February 1, 2020, patients were screened across seven participating centers for prospective enrollment in this study (Fig. 1). A total of 393 patients met the eligibility criteria, 337 were enrolled, and 329 completed the study. The median age of the 329 patients (37.1% female) was 63.5 years (Table 1). At ICU admission, 280 patients (85.1%) had already received empirical antibiotic therapy. All patients underwent bronchoscopy to collect BALF within 48 h of ICU admission. Tracheal intubation and mechanical ventilation were required for 67.8% of the patients. The overall 30-day mortality was 44.1%.
Fig. 1.
Flow diagram of the study
Table 1.
Demographics and clinical characteristics of 329 patients with severe community-acquired pneumonia
| Patient characteristics | All patients (n = 329) |
|---|---|
| Age, years (median, IQR) | 64 (49–71) |
| Female | 122 (37.1%) |
| Comorbidity | |
| Immunocompromised statusa | 65 (19.8%) |
| CVD | 59 (17.9%) |
| Chronic obstructive pulmonary disease | 55 (16.7%) |
| Neoplastic disease | 48 (14.6%) |
| Diabetes mellitus | 41 (12.5%) |
| Renal disease | 37 (11.2%) |
| Bronchiectasis | 24 (7.3%) |
| Cerebrovascular disease | 19 (5.8%) |
| Prior antibiotic use | |
| Received antimicrobials in the 72 h prior to bronchoscopy | 280 (85.1%) |
| Clinical metrics | |
| HFNC or noninvasive ventilation | 106 (32.2%) |
| Intubation and invasive ventilation | 223 (67.8%) |
| ECMO | 19 (5.8%) |
| Vasopressors | 156 (47.4%) |
| APACHE III score | 20.5 ± 4.1 |
| Pneumonia severity index | 145.2 ± 17.8 |
| PaO2/FiO2 ratio | 108, 71–129.5 |
| Total 30-day mortality | 145 (44.1%) |
Data are number (%) of patients, median (interquartile range), or mean ± standard deviation
CVD cardiovascular disease, HFNC high-flow nasal cannula, ECMO extracorporeal membrane oxygenation
aImmunosuppressive drugs, hematologic malignancy, solid tumor with either neutropenia or chemotherapy, inherited immunodeficiency syndromes. AIDS patients were not included in this study
Pathogen Detection by mNGS Relative to Other Methods
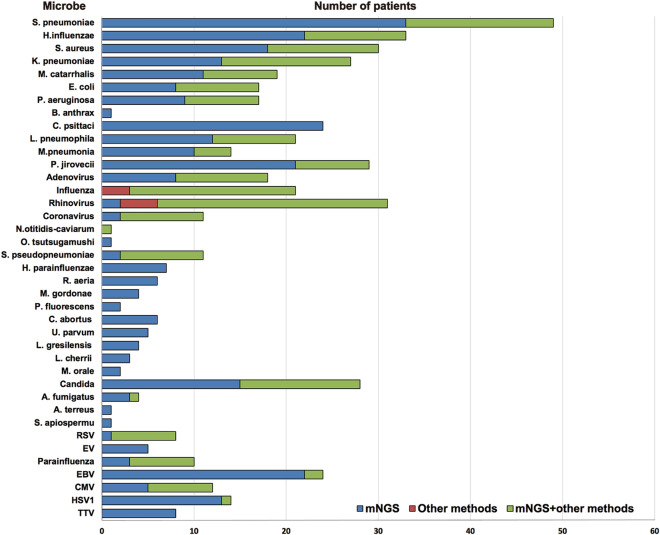
A definite or probable microbial etiology of SCAP was established for 92.4% of the 329 patients when mNGS was combined with the other methods for pathogen detection from BALF samples (Fig. 2). The overall microbial yield was 90.3% (297 of 329 patients) for mNGS versus 39.5% (130 of 329 patients) with other laboratory methods (P < 0.05).
Fig. 2.
Microbial yield in the study cohort and the contribution of mNGS testing and conventional methods to the determination of etiology. Of the 18 patients infected with human adenovirus, 8 had human adenovirus type 55 (HAdV-55), and 10 had human adenovirus type 7 (HAdV-7). Of the 11 patients infected with coronavirus, 2 had severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). A. fumigatus, Aspergillus fumigatus; A. terreus, Aspergillus terreus; B. anthrax, Bacillus anthrax; C. abortus, Chlamydophila abortus; CMV, Human cytomegalovirus; C. psittaci, Chlamydia psittaci; EBV, Epstein-Barr virus; E. coli, Escherichia coli; H. influenzae, Haemophilus influenzae; H. parainfluenzae, Heamophilus parainfluenzae; HSV 1, Herpes simplex virus 1; K. pneumoniae, Klebsiella pneumoniae; L. cherrii, Legionella cherrii; L. gresilensis, Legionella gresilensis; L. pneumophila, Legionella pneumophila; M. catarrhalis, Moraxella catarrhalis; M. gordonae, Mycobacterium gordonae; M. orale, Mycoplasma orale; M.pneumoniae, Mycoplasma pneumoniae; O. tsutsugamushi, Orientia tsutsugamushi; P. aeruginosa, Pseudomonas aeruginosa; P. fluorescens, Pseudomonas fluorescens; P. jirovecii, Pneumocystis jirovecii; R. aeria, Rothia aeria; S. aureus, Staphylococcus aureus; S. apiospermu, Scedosporium apiospermu; S. pneumoniae, Streptococcus pneumoniae; S. pseudopneumoniae, Streptococcus pseudopneumoniae; U. parvum, Ureaplasma parvum
A bacterial etiology was identified in 252 (76.6%) patients by mNGS, but in only 95 (28.9%) patients by conventional culture-based methods. The most frequently detected bacteria were Streptococcus pneumoniae (S. pneumoniae; 14.9%), Haemophilus influenzae (H. influenzae; 10.0%), Staphylococcus aureus (S. aureus; 9.1%), Klebsiella pneumoniae (K. pneumoniae; 8.2%), C. psittaci (7.3%), and L. pneumophila (6.4%).
A fungal pathogen was found in 41 (12.5%) patients by mNGS, including 29 (8.8%) with Pneumocystis jirovecii (P. jirovecii), 4 (1.2%) with Aspergillus fumigatus (A. fumigatus), and 1 (0.3%) with Aspergillus terreus (A. terreus). By contrast, only 8 of the 29 patients who had P. jirovecii were diagnosed by conventional methenamine-silver staining.
A viral pathogen was identified in 148 (45.0%) patients combining mNGS with conventional PCR methods. The most common viruses found were rhinovirus (9.4%), influenza A virus (6.4%), cytomegalovirus (CMV; 5.8%), and adenovirus (5.5%). Significantly, SARS-CoV-2 was identified from two patients solely by using mNGS methods in January, 2020.
Of note, rare pathogens including Orientia tsutsugamushi (O. tsutsugamushi) and Nocardia otitidiscaviarum (N.otitidis-caviarum) were detected solely by mNGS in one patient, respectively. Meanwhile, mNGS also identified pathogens with a degree of clinical suspicion, while conventional tests were negative, including Streptococcus pseudopneumoniae (S. pseudopneumoniae), Heamophilus parainfluenzae (H. parainfluenzae), Rothia aeria (R. aeria), Mycobacterium gordonae (M. gordonae), Legionella gresilensis (L. gresilensis), Legionella cherrii (L. cherrii), Chlamydophila abortus (C. abortus), Ureaplasma parvum (U. parvum), Candida, Epstein–Barr virus (EBV), and Torque teno virus (TTV), et al.
Differences of the Pathogen Spectrum Between Immunocompetent and Immunocompromised Individuals
As shown in Fig. 3, the spectrum of detected pathogens varied between immunocompetent and immunocompromised individuals. In immunocompetent patients, the most common pathogens were S. pneumoniae (14.8%), rhinovirus (9.8%), H. influenzae (9.1%), S. aureus (8.7%), C. psittaci (8.0%), and L. pneumophila (7.2%), while in immunocompromised patients, P. jirovecii (44.6%), K. pneumoniae (18.5%), S. pneumoniae (15.4%), H. influenzae (13.8%), and P. aeruginosa (13.8%) were the most prevalent pathogens. Multiple pathogens with a degree of clinical suspicion including Candida (43.1%), EBV (36.9%), and HSV1 (21.5%) were also frequently detected in immunocompromised patients.
Fig. 3.
Pathogen spectrum among immunocompetent and immunocompromised patients with SCAP
Mixed Infections
Metagenomic NGS combined with other diagnostic methods identified only one potential pathogen in 117 (35.9%) of the 329 patients, two potential pathogens in 60 patients (18.2%), three pathogens in 73 patients (22.2%), and at least four pathogens in 54 patients (16.4%). In immunocompetent patients, the most common combinations were bacterial-viral coinfection and bacterial-bacterial coinfection, while in immunocompromised patients, bacterial-fungal-viral coinfection and bacterial-fungal coinfection were the most frequent combinations (Fig. 4).
Fig. 4.
Common combinations of mixed infections (a) and findings of copathogens for various microbes (b). TTV Torque teno virus
Of the 304 bacterial microbes, 67 fungal microbes, and 162 viruses, 222 (73.0%), 38 (56.8%), and 138 (85.2%) were present as copathogens, respectively. The percentage of common microbes as a copathogen are shown in Fig. 4. Of note, all findings of S. pseudopneumoniae, C. abortus, U. parvum, L. gresilensis, L. cherrii, Aspergillus, Candida, parainfluenza virus, RSV, enterovirus, CMV, EBV, HSV1, and TTV were identified as copathogens.
Significant statistical differences were found between the patients with and without mixed infections in terms of immunocompromised status, chronic obstructive pulmonary disease (COPD) or bronchiectasis, and pneumonia severity index scores (13.7% vs. 27.3%, 6.8% vs. 30.5%, and 143.5 ± 14.6 vs. 159.2 ± 18.9, respectively; Table 2). However, there were no differences between the two groups in terms of diabetes, APACHE II scores, or total 30-day mortality (15.4% vs. 11.8%, 20.9 ± 5.5 vs. 18.7 ± 5.2, and 43.6% vs. 49.2%, respectively).
Table 2.
Association of various clinical variables with mixed infections in patients with SCAP
| No findings of copathogens (n = 117) | Findings of copathogens (n = 187) | P value | |
|---|---|---|---|
| COPD or bronchiectasis | 16 (13.7%) | 51 (27.3%) | 0.005 |
| Immunocompromised status | 8 (6.8%) | 57 (30.5%) | < 0.001 |
| Diabetes mellitus | 18 (15.4%) | 22 (11.8%) | 0.36 |
| APACHE III score, mean ± SD | 20.9 ± 5.5 | 18.7 ± 5.2 | 0.89 |
| Pneumonia severity index, mean ± SD | 143.5 ± 14.6 | 159.2 ± 18.9 | 0.04 |
| Total 30-day mortality | 51 (43.6%) | 92 (49.2%) | 0.34 |
COPD chronic obstructive pulmonary disease; SD standard deviation
Impact on Antibiotic Treatment
Records of antibiotic treatment during hospitalization were retrieved from all 329 patients. Based on the microbiologic results of mNGS testing, the number or spectrum of the initial empiric antimicrobial agents was de-escalated in 174 (52.9%) patients, escalated in 86 (26.1%), and not changed in 69 (21.0%) (Table 3). The majority of the escalation events were related to adding antimicrobial agents against specific pathogens, such as ganciclovir in cases where adenovirus was detected by mNGS and sulfamethoxazole and trimethoprim (TMP-SMZ) in combination with caspofungin when P. jirovecii was identified by mNGS.
Table 3.
Estimated potential impact of mNGS testing on application of antimicrobial agents in patients with severe community-acquired pneumonia
| Modifications | Antimicrobial agents | N (%) |
|---|---|---|
| De-escalation | 174 (52.9) | |
| Remove 1 agent | 109 (33.1) | |
| MXF | 30 | |
| MEM | 19 | |
| VAN | 12 | |
| TZP | 13 | |
| TMP-SMZ | 14 | |
| CAS | 21 | |
| Reduce spectrum of agent | 65 (19.8) | |
| MXF to LZD | 9 | |
| MXF + TZP to MXF + GCV | 42 | |
| MXF + TZP to MXF + Oseltamivir | 14 | |
| Escalation | 86 (26.1) | |
| Add 1 agent | 69 (20.9) | |
| VAN | 8 | |
| MEM | 10 | |
| DOX | 12 | |
| TMP-SMZ | 6 | |
| FLC | 14 | |
| VOR | 9 | |
| Oseltamivir | 10 | |
| Add 2 agent | 17 (5.2) | |
| TMP-SMZ + CAS | 17 | |
| No change | 69 (21.0) |
CAS caspofungin, DOX doxycycline, FLC fluconazole, LZD linezolid, MEM meropenem, MXF moxifloxacin, TMP-SMZ sulfamethoxazole and trimethoprim, TZP piperacillin-tazobactam, VAN vancomycin, VOR voriconazole
Discussion
A major strength of this study was that it was the largest prospective study of the use of mNGS to determine the etiology of SCAP in adults. The overall pathogen-detection yield with mNGS was significantly improved compared to that with other methods. The spectrum of pathogens for severe pneumonia differed between immunocompetent and immunocompromised patients. In addition, mixed infection was frequent and was associated with immunocompromised status, the presence of COPD or bronchiectasis, and higher PSI scores.
Obtaining optimal lower respiratory tract specimens is crucial for the microbial diagnosis of SCAP. BALF, which is easily acquired via bedside bronchoscopy from patients with severe pneumonia, has the following advantages over sputum or induced sputum: representing the component at the level of the alveoli; avoiding contamination by the oropharyngeal flora through delicate bronchoscopy performance; being easily obtained in severely ill patients with “no cough” or dry cough [17]. However, the indications for BAL for CAP remain poorly defined because of several factors, primarily because it is difficult for patients to consent to invasive procedures and their fear of unpleasant feelings. The recent wide use of conscious sedation in bronchoscopic practice has resulted in better procedural tolerance and patient satisfaction. Furthermore, several studies have shown the diagnostic value of BAL for pathogen identification in immunocompromised patients with pneumonia and in patients with non-responding or recurrent CAP [18–20]. In addition, BAL combined with the use of novel PCR-based diagnostic methods significantly improved pathogen detection in both adults and children with CAP [21]. Bedside BAL in the ICU is a safe, effective procedure for optimal specimen acquisition for pathogen detection from patients with SCAP. Therefore, the routine use of BAL for early pathogen diagnosis in SCAP should be considered.
There is mounting evidence that the lung is not a sterile environment and that potentially pathogenic organisms are commonly present in the lungs of healthy asymptomatic individuals [22, 23]. If there is a change in the physical or defense features of the lung environment, these microbes can dominate the airway microbial community and contribute to lower respiratory tract infection. Hence, distinguishing causative microbes from the background microbiota is a crucial challenge when interpreting mNGS data. Langelier et al. [24] used a custom bioinformatics pipeline to discriminate pathogens from respiratory commensals. First, they calculated a ranking score that consisted of the sum of the nucleotide and non-redundant protein Z scores for each microbial genus relative to no-template water controls multiplied by the nucleotide reads per million. Then, they characterized confirmed pathogens based on the ranking score and literature evidence of pathogenicity in the lungs. Langelier et al. [9] further developed two models based on mNGS data from respiratory samples to differentiate pathogens from the background microbiota in a derivation cohort of 20 patients with lower respiratory tract infections or noninfectious acute respiratory disease, and achieved accuracies of 95.5% for both models in an validation cohort of 24 patients. In this study, we classified the microbes identified by mNGS into three categories (putative, clinically suspicious, and unlikely pathogens) according to the abundance score of microbes and the degree of microbial pathogenicity in the lungs.
The superiority of mNGS in the detection of pathogens unidentifiable by routine diagnostic methods has been proven in nervous system infections, sepsis, and periprosthetic joint infections [13, 25, 26]. Besides, the significance of mNGS in the rapid identification and characterization of a potential pathogen for an outbreak of unknown pneumonia has also been highlighted [27, 28]. In this study, we used the mNGS approach to identify the novel coronavirus SARS-CoV-2 from two patients in January 2020 during the early stage of the COVID-19 epidemic. What is more, uncommon pathogens leading to severe pneumonia including O. tsutsugamushi and N.otitidis-caviarum were also detected soley by mNGS approach.
Although C. psittaci has been implicated as a causative pathogen of CAP, the epidemiology of C. psittaci infection in adults with CAP remains unclear. Serologic studies have indicated that the prevalence of C. psittaci in all CAP cases ranged from 0 to 6.7% [29, 30]. Dumke et al. [31] tested swab specimens from 783 adults with CAP using PCR methods and found that 2.1% of the specimens were positive for C. psittaci. Fulminant outbreaks of C. psittaci leading to SCAP in adults have been reported [32, 33]; however, the proportion of C. psittaci in SCAP has not been investigated. We found that 7.3% (24 of 329) of the adults with SCAP had this microorganism as the causative pathogen.
For detecting P. jirovecii, mNGS also showed remarkable advantages compared to conventional methods [34]. Of the 29 cases with P. jirovecii identified by mNGS in this study, only eight were detected by Wright-Giemsa stained smear. One reason for the lower detection rate with microscopic examination was that Wright-Giemsa staining was not performed routinely. The prevalence rates of Pneumocystis pneumonia have been reported ranging from 0 to 11% in non-HIV patients [35], while the incidence of severe pneumonia caused by P. jirovecii remained undetermined. In this study, P. jirovecii was responsible for 44.6% of the immunocompromised patients with SCAP, raising concerns of its contribution to severe pneumonia in immunocompromised individuals. Considering the variations in diagnostic yield of microscopic examination, more studies are needed to characterize the epidemiology of severe pneumonia caused by P. jirovecii by implementing nucleic acid amplification tests including mNGS methods.
This study has limitations. First, not all of the eligible patients were enrolled in this study; patients who had been in hospital for > 2 days were excluded, and eligible patients died during hospitalization before clinical samples could be obtained. Second, it is difficult to distinguish microbial colonization from infection because there are no widely accepted quantitative cutoffs for mNGS in diagnosing causative pathogens. Third, the relatively short time period of our study increased the potential for data bias for microbial pathogens in adults with SCAP. Finally, one main disadvantage of mNGS was that it cannot determine the antibiotic resistance of causative pathogens compared to conventional culture methods. A database of antibiotic-resistance genes for mNGS methods could resolve this issue in the foreseeable future [36, 37].
Conclusion
This study highlights the feasibility of using mNGS on BALF for the microbiologic diagnosis of adults with SCAP. However, further investigations are required because of the limited study time span.
Acknowledgements
The authors thank Chunfeng He and Rongzhang Chen for their support with data collection.
Funding
This work was supported by the Top-level Clinical Discipline Project of Shanghai Pudong (grant no. PWYgy2018-06), the National Natural Science Foundation of China (grant no. 81700006), and the National Key R&D Program of China (2018YFC1311900). The Rapid Service Fee is funded by the authors.
Authorship
All named authors meet the International Committee of Medical Journal Editors (ICMJE) criteria for authorship for this article, take responsibility for the integrity of the work as a whole, and have given their approval for this version to be published.
Authorship Contributions
LQ and XY are responsible for the study concept and designing. WXD, LYY, ZM, ZR, LX and LMM collected data. WXD and XY are responsible for the statistical analysis and interpretation of data. WXD and XY is responsible for the drafting of the manuscript. LQ and PPH are responsible for the critical revision of the manuscript. All authors read and approved the final manuscript.
Prior Presentation
This manuscript is based on work that has been previously presented (https://papers.ssrn.com/sol3/papers.cfm?abstract_id=3514590) in January, 2020.
Disclosures
Xiaodong Wu, Yuanyuan Li, Ming Zhang, Miaomiao Li, Rong Zhang, Xin Lu, Wei Gao, Qin Li, Yang Xia, Pinhua Pan, and Qiang Li declare that there are no conflicts of interest.
Compliance with Ethics Guidelines
This study was performed in accordance with the Helsinki Declaration of 1964 and its later amendments. Ethical approval for this study was obtained from the institutional ethics committee at each study site (The Ethics Committee of Shanghai East Hospital Affiliated with Shanghai Tongji University, Xiangya Hospital Affiliated with Central South University, the Second Affiliated Hospital of Zhejiang University School of Medicine, the Third Affiliated Hospital of Soochow University, Fourth Affiliated Hospital of Zhejiang University School of Medicine, Changhai Hospital, The Affiliated Jiangning Hospital of Nanjing Medical University, and the Second Clinical Medical College Affiliated with Jinan University). Written informed consent was provided from each patient or family members at enrolment.
Data Availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Footnotes
Xiaodong Wu, Yuanyuan Li and Ming Zhang contributed equally to this work.
Contributor Information
Yang Xia, Email: yxia@zju.edu.cn.
Pinhua Pan, Email: pinhuapan668@hotmail.com.
Qiang Li, Email: liqressh@hotmail.com.
References
- 1.Torres A, Chalmers JD, Dela Cruz CS, et al. Challenges in severe community-acquired pneumonia: a point-of-view review. Intensive Care Med. 2019;45(2):159–171. doi: 10.1007/s00134-019-05519-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jain S, Self WH, Wunderink RG et al. Community-acquired pneumonia requiring hospitalization among US adults. N Engl J Med. 2015;373(5):415–427. [DOI] [PMC free article] [PubMed]
- 3.Musher DM, Roig IL, Cazares G, Stager CE, Logan N, Safar H. Can an etiologic agent be identified in adults who are hospitalized for community-acquired pneumonia: results of a one-year study. J Infect. 2013;67(1):11–18. doi: 10.1016/j.jinf.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gadsby NJ, Russell CD, McHugh MP, et al. Comprehensive molecular testing for respiratory pathogens in community-acquired pneumonia. Clin Infect Dis. 2016;62(7):817–823. doi: 10.1093/cid/civ1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maartens G, Griesel R, Dube F, Nicol M, Mendelson M. Etiology of pulmonary infections in human immunodeficiency virus-infected inpatients using sputum multiplex real-time polymerase chain reaction. Clin Infect Dis. 2019;70(6):1147–1152. doi: 10.1093/cid/ciz332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Di Pasquale MF, Sotgiu G, Gramegna A, et al. Prevalence and etiology of community-acquired pneumonia in immunocompromised patients. Clin Infect Dis. 2019;68(9):1482–1493. doi: 10.1093/cid/ciy723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Simner PJ, Miller S, Carroll KC. Understanding the promises and hurdles of metagenomic next-generation sequencing as a diagnostic tool for infectious diseases. Clin Infect Dis. 2018;66(5):778–788. doi: 10.1093/cid/cix881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miao Q, Ma Y, Wang Q, et al. Microbiological diagnostic performance of metagenomic next-generation sequencing when applied to clinical practice. Clin Infect Dis. 2018;67(suppl_2):S231–s240. doi: 10.1093/cid/ciy693. [DOI] [PubMed] [Google Scholar]
- 9.Langelier C, Kalantar KL, Moazed F, et al. Integrating host response and unbiased microbe detection for lower respiratory tract infection diagnosis in critically ill adults. Proc Natl Acad Sci USA. 2018;115(52):E12353–e12362. doi: 10.1073/pnas.1809700115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chiu CY, Coffey LL, Murkey J, et al. Diagnosis of fatal human case of St. Louis encephalitis virus infection by metagenomic sequencing, California, 2016. Emerg Infect Dis. 2017;23(10):1964–1968. doi: 10.3201/eid2310.161986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guan H, Shen A, Lv X, et al. Detection of virus in CSF from the cases with meningoencephalitis by next-generation sequencing. J Neurovirol. 2016;22(2):240–245. doi: 10.1007/s13365-015-0390-7. [DOI] [PubMed] [Google Scholar]
- 12.Blauwkamp TA, Thair S, Rosen MJ, et al. Analytical and clinical validation of a microbial cell-free DNA sequencing test for infectious disease. Nat Microbiol. 2019;4(4):663–674. doi: 10.1038/s41564-018-0349-6. [DOI] [PubMed] [Google Scholar]
- 13.Wilson MR, Sample HA, Zorn KC, et al. Clinical metagenomic sequencing for diagnosis of meningitis and encephalitis. N Engl J Med. 2019;380(24):2327–2340. doi: 10.1056/NEJMoa1803396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xie Y, Du J, Jin W, et al. Next generation sequencing for diagnosis of severe pneumonia: China, 2010–2018. J Infect. 2019;78(2):158–169. doi: 10.1016/j.jinf.2018.09.004. [DOI] [PubMed] [Google Scholar]
- 15.Pan T, Tan R, Qu H, et al. Next-generation sequencing of the BALF in the diagnosis of community-acquired pneumonia in immunocompromised patients. J Infect. 2019;79(1):61–74. doi: 10.1016/j.jinf.2018.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Metlay JP, Waterer GW, Long AC, et al. Diagnosis and treatment of adults with community-acquired pneumonia. An official clinical practice guideline of the American Thoracic Society and Infectious Diseases Society of America. Am J Respir Crit Care Med. 2019;200(7):e45–e67. doi: 10.1164/rccm.201908-1581ST. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dubourg G, Abat C, Rolain J-M, Raoult D. Correlation between Sputum and Bronchoalveolar lavage fluid cultures. J Clin Microbiol. 2015;53(3):994–996. doi: 10.1128/JCM.02918-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hohenadel IA, Kiworr M, Genitsariotis R, Zeidler D, Lorenz J. Role of bronchoalveolar lavage in immunocompromised patients with pneumonia treated with a broad spectrum antibiotic and antifungal regimen. Thorax. 2001;56(2):115–120. doi: 10.1136/thorax.56.2.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsai CM, Wong KS, Lee WJ, et al. Diagnostic value of bronchoalveolar lavage in children with nonresponding community-acquired pneumonia. Pediatr Neonatol. 2017;58(5):430–436. doi: 10.1016/j.pedneo.2016.09.004. [DOI] [PubMed] [Google Scholar]
- 20.Magazine R, Chogtu B, Rao S, Chawla K. Bacterial isolates from the bronchoalveolar lavage fluid of patients with pneumonia not responding to initial antimicrobial therapy. Sahel Med J. 2013;16(3):102–106. [Google Scholar]
- 21.Baudel J-L, Tankovic J, Dahoumane R, et al. Multiplex PCR performed of bronchoalveolar lavage fluid increases pathogen identification rate in critically ill patients with pneumonia: a pilot study. Ann Intensive Care. 2014;4(1):35. doi: 10.1186/s13613-014-0035-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sundell N, Andersson L-M, Brittain-Long R, et al. PCR detection of respiratory pathogens in asymptomatic and symptomatic adults. J Clin Microbiol. 2019;57(1):e00716–00718. doi: 10.1128/JCM.00716-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dickson RP, Huffnagle GB. The lung microbiome: new principles for respiratory bacteriology in health and disease. PLoS Pathog. 2015;11(7):e1004923–e1004923. doi: 10.1371/journal.ppat.1004923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Langelier C, Zinter MS, Kalantar K, et al. Metagenomic sequencing detects respiratory pathogens in hematopoietic cellular transplant patients. Am J Respir Crit Care Med. 2018;197(4):524–528. doi: 10.1164/rccm.201706-1097LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang Z, Zhang C, Fang X, et al. Identification of musculoskeletal infection with non-tuberculous mycobacterium using metagenomic sequencing. J Infect. 2019;78(2):158–169. doi: 10.1016/j.jinf.2018.10.002. [DOI] [PubMed] [Google Scholar]
- 26.Grumaz S, Stevens P, Grumaz C, et al. Next-generation sequencing diagnostics of bacteremia in septic patients. Genome Med. 2016;8(1):73. doi: 10.1186/s13073-016-0326-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ai J-W, Zhang Y, Zhang H-C, Xu T, Zhang W-H. Era of molecular diagnosis for pathogen identification of unexplained pneumonia, lessons to be learned. Emerg Microbes Infect. 2020;9(1):597–600. doi: 10.1080/22221751.2020.1738905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen L, Liu W, Zhang Q, et al. RNA based mNGS approach identifies a novel human coronavirus from two individual pneumonia cases in 2019 Wuhan outbreak. Emerg Microbes Infect. 2020;9(1):313–319. doi: 10.1080/22221751.2020.1725399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hogerwerf L, Gier BDE, Baan B, Hoek WVD. Chlamydia psittaci (psittacosis) as a cause of community-acquired pneumonia: a systematic review and meta-analysis. Epidemiol Infect. 2017;145(15):3096–3105. [DOI] [PMC free article] [PubMed]
- 30.Marrie TJ, Peeling RW, Reid T, De Carolis E. Chlamydia species as a cause of community-acquired pneumonia in Canada. Eur Respir J. 2003;21(5):779–784. doi: 10.1183/09031936.03.00095403. [DOI] [PubMed] [Google Scholar]
- 31.Dumke R, Schnee C, Pletz MW, et al. Mycoplasma pneumoniae and Chlamydia spp. infection in community-acquired pneumonia, Germany, 2011–2012. Emerg Infect Dis. 2015;21(3):426–34. [DOI] [PMC free article] [PubMed]
- 32.Fischer N, Rohde H, Indenbirken D, et al. Rapid metagenomic diagnostics for suspected outbreak of severe pneumonia. Emerg Infect Dis. 2014;20(6):1072–1075. doi: 10.3201/eid2006.131526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shaw KA, Szablewski CM, Kellner S, et al. Psittacosis Outbreak among workers at Chicken Slaughter Plants, Virginia and Georgia, USA, 2018. Emerg Infect Dis. 2019;25(11):2143–2145. doi: 10.3201/eid2511.190703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, Ai JW, Cui P, Zhang WH, Wu HL, Ye MZ. A cluster of cases of pneumocystis pneumonia identified by shotgun metagenomics approach. J Infect. 2019;78(2):158–169. doi: 10.1016/j.jinf.2018.08.013. [DOI] [PubMed] [Google Scholar]
- 35.Cillóniz C, Dominedò C, Álvarez-Martínez MJ, et al. Pneumocystis pneumonia in the twenty-first century: HIV-infected versus HIV-uninfected patients. Expert Rev Anti Infect Ther. 2019;17(10):787–801. doi: 10.1080/14787210.2019.1671823. [DOI] [PubMed] [Google Scholar]
- 36.Lal Gupta C, Kumar Tiwari R, Cytryn E. Platforms for elucidating antibiotic resistance in single genomes and complex metagenomes. Environ Int. 2020;138:105667. doi: 10.1016/j.envint.2020.105667. [DOI] [PubMed] [Google Scholar]
- 37.Zinn MK, Schages L, Bockmühl D. The toothbrush microbiome: impact of user age, period of use and bristle material on the microbial communities of toothbrushes. Microorganisms. 2020;8:9. doi: 10.3390/microorganisms8091379. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.