Abstract
MADS-box genes take part in diverse biological functions especially in development of reproductive structures and control of flowering time. Recently, Cardamine hirsuta has emerged as an exclusively powerful genetic system in comparative studies of development. Although the C. hirsuta genome sequence is available but a comprehensive analysis of its MADS-box family genes is still lacking. Here, we determined 50 Cardamine MADS-box genes through bioinformatics tools and classified them into 2 Mβ, 6 Mα and 2 Mγ and 40 MIKC-type (35 MIKCc and 5MIKC*) genes based on a phylogenetic analysis. The C. hirsuta MIKC subfamily could be further classified into 14 subgroups as Arabidopsis. However the number of MADS-box proteins was not equal among these subgroups. Based on the structural diversity among 50 MADS-box genes, 2 lineages were obtained, type I and type II. The lowest number of introns (0 or 1) was found in the Mα, Mβ, and Mγ groups of the type I genes. The most Cardamine MADS-box genes were randomly distributed on only three chromosomes. C. hirsuta had a relatively lower number of flowering MADS-box genes than A. thaliana and probably tandem duplication event resulted in the expansion of FLC, SQUA and TM3 family members in Arabidopsis. Moreover among the conserved motifs, ChMADS5 of SQUA, ChMADS34 of TM3 and ChMADS51 of AGL15 families had no K-domain. This study provides a basis for further functional investigation of MADS-box genes in C. hirsuta.
Keywords: Arabidopsis, Brassicaceae, K-domain, MIKC, Phylogenetic analysis
Introduction
MADS-box genes play fundamental roles in diverse biological functions especially in the control of flowering time, vegetative development, flower architecture, pollen and embryo sac formation, seed and fruit development (Theißen and Gramzow 2016). The name of MADS-box is derived from the MINICHROMOSOME MAINTENANCE 1 (MCM1) genes in yeast, AGAMOUS (AG) in Arabidopsis, DEFICIENS (DEF) in Antirrhinum and Serum Response Factor (SRF) in humans (Medard and Yanofsky 2001). Members of this family have a highly conserved MADS domain containing 56–60 amino acids, which bind to specific DNA sequences acting as cis-regulatory elements in the promoters or enhancers of the genes (Riechmann et al. 1996; Smaczniak et al. 2012).
In phylogenetic analysis, MADS-box genes are divided into two groups of type I (SRF-like) and type II (MEF2-like) which are different in the amino acid consensus sequences in their MADS-box domains but both are found in animals, fungi, and plants. Type I, the M-type, contains the conserved M domain with a large variable region at the C-terminus and classifies into three subclasses (Mα, Mβ, and Mγ). MADS type II proteins known as MIKC domain are composed of MADS domain, I domain with approximately 30 amino acids, the K-domain and the C region from N- to C-termini (Theissen et al. 1996; De Folter et al. 2005; Xu et al. 2014). The K-domain with about 70 amino acids is also highly conserved, whereas I domain and C region are quite variable. The MIKC type has been further subdivided into MIKCC and MIKC* types based on the variable intervening domain (I) resulted from an ancestral gene duplication (Henschel et al. 2002; Parenicova et al. 2003; Zhang et al. 2020).
In plants, the functions of MADS-box genes are best understood during reproductive development. Unlike animals, plants are unable to change their location to escape adverse environmental conditions. Not surprisingly, complex mechanisms have evolved to determine when the environment is most favorable for plants reproductive stage (Castelán-muñoz et al. 2019). To date, MADS-box genes that regulate transition from vegetative to reproductive development have been identified.
Model plants provide biological insights in areas such as plant development, signaling, hormone biology, pathogen defense, disease resistance, and abiotic stress response (Chang et al. 2016). Many MADS-box gene functions were uncovered in the model species A. thaliana (Medard and Yanofsky 2001) but other model plant species including snapdragon (Antirrhinum majus) (Schwarz-sommer et al. 2003), petunia (Petunia hybrida) (Gerats and Vandenbussche 2005), gerbera (Gerbera hybrida) (Aelaitinen et al. 2006), rice (Oryza sativa) (Yoshida and Nagato 2011) and tomato (Solanum lycopersicum) have also been studied for MADS-box gene functions (Wang et al. 2019). Such comparative studies are particularly attractive in the Brassicaceae family. The complete genome sequence of Arabidopsis provided a clear picture of the complexity and diversity of MADS-box genes. In this plant, 107 MADS-box genes have been identified and functionally characterized (De Folter et al. 2005). The genetic studies have shown that these MADS-box transcription factor family play essential roles in almost every developmental processes such as meristem specification, flowering transition, seed, root and flower development, and fruit ripening (Smaczniak et al. 2012).
Recently, Cardamine hirsuta has emerged as an exclusively powerful genetic system for comparative studies of development. A. thaliana and C. hirsuta both belong to the family of Brassicaceae and have a close relationship in the phylogenetic tree. Cardamine L. with about 200 species is one of the largest genera in Brassicaceae. Historical and more recent reticulation events strongly affected on morphological and karyological diversity which provides an opportunity to study mechanisms of plant diversification. Although C. hirsuta occurs as a weed throughout the world but it is a nearly cosmopolitan diploid species (Zozomova-Lihova and Marhold 2006).
Cardamine hirsuta is a diploid and self-compatible annual plant with an abundant seed set, an 8-week seed-to-seed generation time and a small rosette growth habit that is amenable to large-scale cultivation. The C. hirsuta genome is estimated to be 1.5 times that of A. thaliana, with eight chromosomes. In addition, the constructed high-quality reference genome of the C. hirsuta strain ‘Oxford’ provided a powerful platform for molecular studies (Johnston et al. 2005; Hay et al. 2014; Hay and Tsiantis 2016; Gan et al. 2016). Its genome is largely syntenic to the genome of A. thaliana. According to the complete set of protein-coding genes of both, the divergence date was determined to around 32 Myr ago (Gan et al. 2016).
In this study, the flowering-related MADS-box genes of C. hirsuta were identified and then classified based on their phylogenetic relationships. Multiple bioinformatics methods were applied to perform a comprehensive survey of MADS-box genes. Gene structures, phylogenetic relationships, and conserved motifs of C. hirsuta MADS-box genes were analyzed, and mapped on the chromosome locations. The results would be useful for understanding the developmental processes in C. hirsuta and the Brassicaceae.
Methods
Database search of MADS-box sequences
The databases including Genomics Network (https://genomevolution.org/coge/CoGeBlast.pl), the Arabidopsis MADS Transcription Factor Family Network (https://www.arabidopsis.org/browse/genefamily/mads_tffamily.jsp), and the National Center for Biotechnology Information (NCBI, https://www.ncbi.nlm.nih.gov/) were used to comprehensively identify the whole MADS-box protein sequences of C. hirsuta. The first 50 MADS-box involved in flowering were selected and based on 52 Arabidopsis MADS-box protein sequences as queries, DNA and protein sequences were obtained from the CoGeBlast. The remaining two had no MADS-box in Cardamine. In addition, the molecular weight and isoelectric points of C. hirsuta MADS-box proteins were detected by the ExPASy proteomics server. To verify the MADS-box conserved domains, the protein sequences were inspected by the NCBI Batch CD Search Program (Marchler-Bauer and Bryant 2004) and sequences without MADS-box domains were discarded (Table 1).
Table 1.
The detailed information of the flowering-related MADS-box gene family in C. hirsuta
| Gene | Chromosome number | Length (aa) | MW (KDa) | IP | Genomic location | Arabidopsis MADS-box |
|---|---|---|---|---|---|---|
| ChMADS1 | Chr6 | 209 | 24.192 | 9.06 | 17,155,415–17,157,426 | IP(AT5G20240) |
| ChMADS2 | Chr5 | 232 | 27.283 | 8.41 | 18,167,809–18,169,621 | AP3(AT3G54340) |
| ChMADS3 | Chr1 | 334 | 38.162 | 4.9 | 7,918,920–7,920,923 | AGL104(AT1G22130) |
| ChMADS4 | Chr8 | 1020 | 116.195 | 6.11 | 11,828,548–11,831,971 | AGL81(AT5G39750) |
| ChMADS5 | Chr5 | 73 | 8.377 | 10.43 | 10,569,703–10,569,924 | AGL79(AT3G30260) |
| ChMADS6 | Chr8 | 198 | 23.132 | 8.41 | 13,763,972–13,766,666 | AGL72(AT5G51860) |
| ChMADS7 | Chr8 | 204 | 23.755 | 8.74 | 13,763,972–13,766,666 | AGL71(AT5G51870) |
| ChMADS8 | Chr6 | 191 | 21.188 | 8.87 | 21,028,257–21,033,375 | AGL70(AT5G65060) |
| ChMADS9 | Chr8 | 199 | 22.345 | 9.13 | 19,610,194–19,613,836 | AGL69(AT5G65070) |
| ChMADS10 | Chr8 | 200 | 22.628 | 7.66 | 19,604,875–19,608,953 | AGL68(AT5G65080) |
| ChMADS11 | Chr2 | 252 | 29.165 | 7.01 | 18,993,236–18,995,732 | AGL67(AT1G77950) |
| ChMADS12 | Chr2 | 331 | 37.971 | 4.85 | 19,000,496–19,002,464 | AGL66(AT1G77980) |
| ChMADS13 | Chr1 | 373 | 42.976 | 6.05 | 6,715,534–6,717,812 | AGL65(AT1G18750) |
| ChMADS14 | Chr1 | 234 | 26.939 | 8.88 | 11,676,303–11,678,113 | AGL63(AT1G31140) |
| ChMADS15 | Chr8 | 277 | 31.891 | 9.16 | 17,835,330–17,836,459 | AGL62(AT5G60440) |
| ChMADS16 | Chr1 | 213 | 24.310 | 9.50 | 10,551,430–10,552,071 | AGL61(AT2G24840) |
| ChMADS17 | Chr: NSCAFA.1966 | 191 | 21.301 | 5.65 | 8–580 | AGL56(AT1G60880) |
| ChMADS18 | Chr: NSCAFA.1966 | 191 | 21.301 | 5.65 | 8–580 | AGL55(AT1G60920) |
| ChMADS19 | Chr2 | 225 | 25.259 | 8.79 | 3,862,226–3,862,913 | AGL50(AT1G59810) |
| ChMADS20 | Chr2 | 225 | 25.259 | 8.79 | 3,862,226–3,862,913 | AGL49(AT1G60040) |
| ChMADS21 | Chr3 | 349 | 40.210 | 6.03 | 3,754,854–3,755,903 | AGL48(AT2G40210) |
| ChMADS22 | Chr8 | 144 | 17.245 | 9.34 | 18,536,660–18,538,665 | AGL42(AT5G62165) |
| ChMADS23 | Chr2 | 187 | 21.875 | 8.99 | 15,858,452–15,859,012 | AGL37(AT1G65330) |
| ChMADS24 | Chr6 | 236 | 27.873 | 6.11 | 16,135,563–16,137,470 | AGL32(AT5G23260) |
| ChMADS25 | Chr6 | 191 | 21.188 | 8.87 | 21,028,257–21,033,375 | AGL31(AT5G65050) |
| ChMADS26 | Chr5 | 389 | 44.034 | 6.40 | 952,455–954,313 | AGL30(AT2G03060) |
| ChMADS27 | Chr3 | 353 | 39.930 | 8.76 | 6,933,979–6,936,050 | AGL28(AT1G01530) |
| ChMADS28 | Chr6 | 191 | 21.188 | 8.87 | 21,028,257–21,033,375 | AGL27(AT1G77080) |
| ChMADS29 | Chr6 | 191 | 21.188 | 8.87 | 21,028,257–21,033,375 | AGL25(AT5G10140) |
| ChMADS30 | Chr7 | 221 | 25.371 | 6.87 | 17,573,656–17,576,270 | AGL24(AT4G24540) |
| ChMADS31 | Chr3 | 353 | 39.930 | 8.76 | 6,933,979–6,936,050 | AGL23(AT1G65360) |
| ChMADS32 | Chr4 | 238 | 26.891 | 5.57 | 1,247,182–1,250,310 | AGL22(AT2G22540) |
| ChMADS33 | Chr4 | 237 | 27.367 | 9.41 | 21,893,219–21,896,177 | AGL20(AT2G45660) |
| ChMADS34 | Chr8 | 144 | 17.245 | 9.34 | 18,536,660–18,538,665 | AGL19(AT4G22950) |
| ChMADS35 | Chr7 | 228 | 26.438 | 9.10 | 22,854,453–22,857,260 | AGL17(AT2G22630) |
| ChMADS36 | Chr5 | 132 | 15.336 | 5.44 | 19,308,194–19,308,882 | AGL16(AT3G57230) |
| ChMADS37 | Chr6 | 255 | 28.882 | 8.46 | 19,706,091–19,707,658 | AGL15(AT5G13790) |
| ChMADS38 | Chr8 | 221 | 25.594 | 8.91 | 2,843,832–2,848,366 | AGL14(AT4G11880) |
| ChMADS39 | Chr4 | 253 | 28.956 | 8.66 | 21,890,010–21,892,109 | AGL13(AT3G61120) |
| ChMADS40 | Chr: NSCAFB.269 | 211 | 23.953 | 8.66 | 97,801–99,942 | AGL12(AT1G71692) |
| ChMADS41 | Chr8 | 232 | 26.365 | 9.43 | 11,071,251–11,073,987 | AGL11(AT4G09960) |
| ChMADS42 | Chr1 | 256 | 30.306 | 7.16 | 9,238,176–9,241,438 | AGL10(AT1G26310) |
| ChMADS43 | Chr1 | 249 | 28.859 | 8.27 | 9,782,655–9,784,760 | AGL9(AT1G24260) |
| ChMADS44 | Chr1 | 256 | 30.306 | 7.16 | 9,238,176–9,241,438 | AGL7(AT1G69120) |
| ChMADS45 | Chr4 | 253 | 28.955 | 8.66 | 21,890,010–21,892,109 | AGL6(AT2G45650) |
| ChMADS46 | Chr4 | 249 | 28.484 | 9.33 | 20,822,078–20,824,951 | AGL5(AT2G42830) |
| ChMADS47 | Chr3 | 247 | 28.375 | 8.70 | 468,931–470,787 | AGL4(AT3G02310) |
| ChMADS48 | Chr5 | 259 | 29.472 | 9.07 | 1,210,632–1,212,666 | AGL3(AT2G03710) |
| ChMADS49 | Chr6 | 256 | 29.061 | 8.55 | 18,954,495–18,956,724 | AGL2(AT5G15800) |
| ChMADS50 | Chr5 | 248 | 28.329 | 9.07 | 19,954,253–19,957,550 | AGL1(AT3G58780) |
| ChMADS51 | Chr5 | 61 | 6.908 | 10.01 | 19,954,253–19,957,550 | AGL18(AT3G57390) |
| ChMADS52 | Chr7 | 256 | 30.306 | 7.16 | 15,096,060–15,100,461 | AG(AT4G18960) |
List of predicted genes and related information including gene name, chromosome number, molecular weight (MW), isoelectric point (IP), genomic location and Arabidopsis MADS-box are mentioned
Phylogenetic analysis
The C. hirsuta protein sequences, containing the MADS-box protein family, were selected for amino acid sequence multiple alignment and phylogenetic tree analysis. Using ClustalX 1.81, multiple sequence alignment for the two groups of all 50 C. hirsuta MADS-box genes was generated. The outputs were used to conduct a phylogenetic tree by the MEGA7 program and the evolutionary history was inferred using the neighbor-joining method with Poisson distances and the pair-wise deletion options. For the reliability of the tree, 1000 bootstrap replications were performed. The phylogenetic tree was further annotated by iTOL program (https://itol.embl.de/).
The analysis of conserved motif and gene structure
The C. hirsuta MADS-box coding domain sequences (CDS) and the corresponding genomic DNA sequences were collected from CoGeBlast and TAIR to predict gene structure. The online tool Gene Structure Display Server 2.0 (GSDS 2.0, Available online: https://gsds.cbi.pku.edu.cn/index.php) was used to construct an exon/intron map (Hu et al. 2015). To recognize the presence and distribution of conserved motifs in full length MADS-box protein sequences MEME 5.1. 1 database (https://meme-suit.org) was used as described by Bailey et al. (2015) (Available online: https://meme-suite.org/tools/meme). It was performed using the following parameters: 10 different motifs, a motif width of 6–200 amino acids, and any number of repetitions. The SMART database was used to annotate the MEME motifs. Tandem duplication events (TDs) on a single chromosome and segmental duplication (SDs) between different chromosomes (Qu et al. 2019; Liu and Ekramoddoullah, 2009) were investigated by BLASTp alignments (E value cutoff = 1e−20) to obtain the similarities between MADS-box genes and also to estimate gene duplication frequency in the MADS-box genes. Two criteria were considered to estimate gene duplication events (Bi et al. 2016) as (1) the coverage of the aligned sequence 80% of the longer gene; and (2) the similarity of the aligned regions 65%.
Results
Identification of MADS-box genes in C. hirsuta
To identify C. hirsuta MADS-box genes involvement in flowering transition and development, a set of 52 Arabidopsis MADS-box flowering-related genes were selected. The highly homologous ones to the MADS-box proteins reported in Arabidopsis were recovered using BLAST searches against the CoGe and NCBI databases. There were 50 C. hirsuta MADS-box genes which were identified and designated as CH1–CH51 (Table 1). The remaining two had no MADS-box in Cardamine (ChMADS4, ChMADS36 in Table 1). The molecular characteristics of these genes including length of amino acid sequences, the molecular weight, and the isoelectric points were analyzed (Table 1). The results showed that the amino acid sequence length of the 50 predicted C. hirsuta MADS-box proteins varied from 61 to 389 amino acids, with the relative molecular mass ranged from 6.908 (ChMADS51) to 44.034 KDa (ChMADS26), and the isoelectric point (IP) of 4.85 (ChMADS12) to 10.01(ChMADS51) as shown in Table 1.
Gene structure analysis of C. hirsuta MADS-box genes
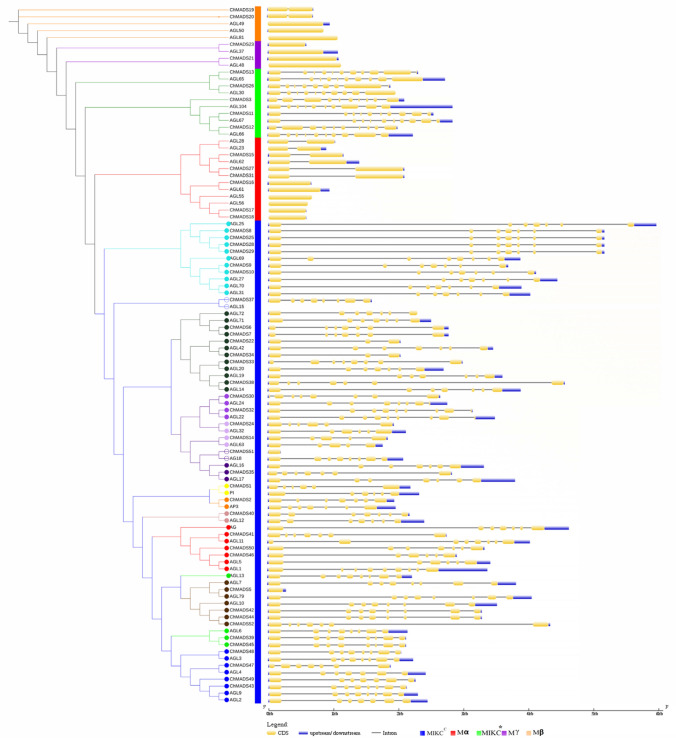
To analyze the organized structure of exon/intron, the complete sequence of 50 C. hirsuta MADS-box genes obtained by IGV program were compared with their CDs by Gene Structure Display Server (GSDS) program and two lineages were obtained, type I and type II. As shown in Fig. 1, the number of introns in C. hirsuta MADS-box genes was between one and eight. The lowest number of introns was found in the Mα, Mβ, and Mγ groups of the type I genes with no introns and ChMADS27, ChMADS31 and ChMADS15 in Mα group, and ChMADS19 and ChMADS20 in Mβ group with just one intron. The distribution of introns in type I and II was different, and also MIKCC and MIKC* types genes contained multiple introns, except ChMADS51 that lacked introns and ChMADS15 which only had two introns (Fig. 1).
Fig. 1.
Phylogenetic relationships and gene structure of 51 ChMADS from C. hirsuta and 52 from A. thaliana. The phylogenetic tree was constructed using MEGA 6.06 by the neighbor-joining (NJ) method with 1000 bootstrap replicates. The Gene Structure Display Server (GSDS) database was used to perform the exon–intron structure analyses. The blue boxes represent upstream/downstream, the yellow boxes represent exons, and the black lines represent introns. The lengths of exons and introns for each MADS-box genes are shown proportionally
Identification of conserved motifs
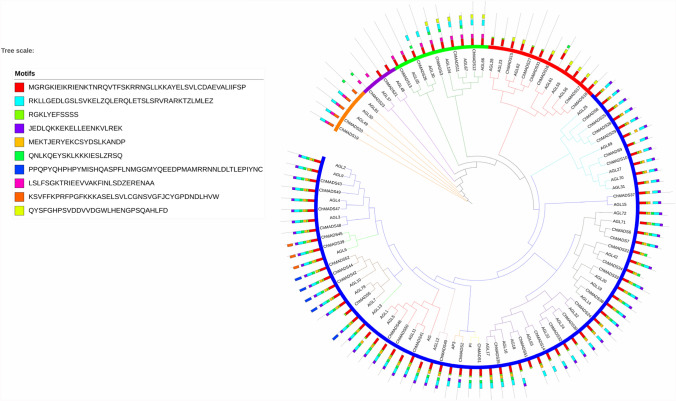
To gain insights into conserved motifs of C. hirsuta, MADS-box proteins selected from CoGeBlast the MEME program (Multiple Expectation Minimization for Motif Elicitation, (https://meme.sdsc.edu/meme/meme.html) version 5.1.1 (Bailey et al. 2015) were used. A total of ten conserved motifs, named 1–10, were identified (Fig. 2).
Fig. 2.
Phylogenetic relationships of flowering-related MADS-box genes family. The phylogenetic tree was constructed using MEGA 6.06 by the neighbor-joining (NJ) method with 1000 bootstrap replicates. It is representing relationships between 51 ChMADS proteins translated from C. hirsuta and 52 from A. thaliana. The MADS-box proteins are clustered into five subfamilies (marked as Mβ, Mα, MƳ MIKC*, and MIKCC). Colored solid circles are used to graphically distinguish the subfamilies in MIKCc. Distribution of conserved motifs in the MADS proteins were predicted by MEME. Different motifs were indicated by different colored number 1–10 and were added by ITOL program
As expected, MADS-box proteins type II genes contained the same motifs. Motif one was the most typical MADS-box domains. Motif two was a highly conserved K-domain motif which had an important role in protein–protein interactions among MADS-box proteins and was present in all MIKC-type genes except ChMADS34, ChMADS5 and ChMADS51.
Phylogenetic analysis of MADS-box genes
A phylogenetic tree was constructed using a multiple sequence alignment including C. hirsuta (ChMADS) and Arabidopsis (AtMADS) MADS-box proteins by the neighbor-joining (NJ) method in MEGA 6 (Fig. 2). Similar to Arabidopsis the 50 C. hirsuta MADS-box genes were classified into two types: type I and type II. The MADS-box family of C. hirsuta was consisted of five subfamilies of more closely related sequences, named Mα (6 genes), Mγ (2 genes), Mβ (2 genes), MICK*(5 genes), and MIKCC (35 genes).
The MIKCC group was classified into 14 clades including FLC family (6 genes), AGL17 (1 gene), AGL15 (2 genes), AG (3 genes), SVP (2 genes), TT16 (2 genes), AGL12 (1 gene), GLO (1 gene), DEF (1 gene), TM3 (6 genes), SQUA (4 genes), AGL6 (2 genes), SEP (4 genes). These results indicated that C. hirsuta and Arabidopsis MADS-box proteins were not equal within this clade and commonly two or more putative orthologs of AtMADSs from a single C. hirsuta gene was observed.
The orthologs of MIKCC in C. hirsuta MADS-box genes were screened with BLASTp e-value less than 10e-10 and more than 80% coverage in length and the ones with best-matching homology were selected (Fig. 2).
Chromosome distribution of the C. hirsuta MADS-box genes
For mapping chromosomal locations of C. hirsuta and Arabidopsis MADS-box genes, physical genome annotation files obtained from CoGe and TAIR and all MADS-box genes were detected on related chromosomes. The Map Chart software was used to map the physical position of MADS-box genes. In C. hirsuta, the most MADS-box genes mapped on chromosome eight while chromosomes three and seven contained the least. Moreover, chromosome five showed an enrichment region proving that the distribution of the MADS-box genes was not random in C. hirsuta (Figs. 3, 4).
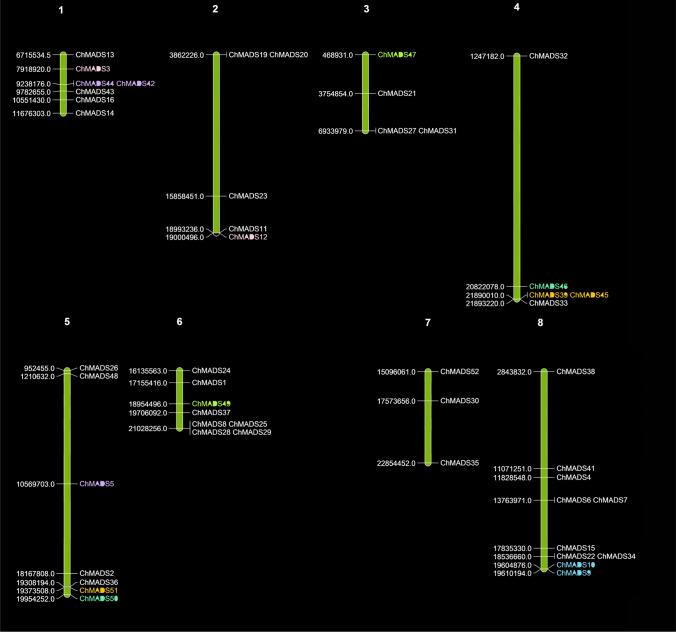
Fig. 3.
Chromosomal locations of C. hirsuta MADS-box genes. The eight chromosomes of C. hirsuta were labeled with their names. The position of C.hirsuta MADS-box genes on the chromosome was based on the CoGe database and Mapchart was used to draw the physical map of the C. hirsuta MADS-box genes. The approximate physical location of each MADS-box gene was shown on the left side of each chromosome. One tandemly duplicated gene (chMADS9-10) is shown in blue and five segmental duplication genes are shown in different colors
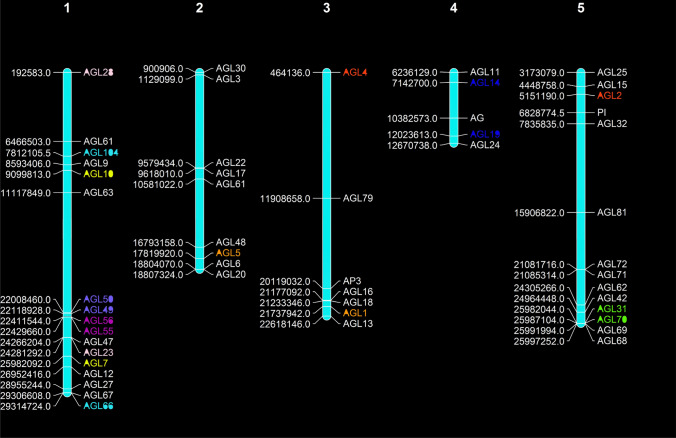
Fig. 4.
Chromosomal locations of A. thaliana MADS-box genes. The five chromosomes of A. thaliana were labeled with their names. The position of A. thaliana MADS-box genes on the chromosome was based on the TAIR database and Mapchart was used to draw the physical map of the A. thaliana MADS-box genes. The approximate physical location of each MADS-box gene was shown on the left side of each chromosome. Three tandemly duplications (AGL59-49, AGL56-55 and AGL31-70) and five segmental duplication genes are shown in different colors
The study of gene duplication events in the C. hirsuta MADS-box gene family revealed that 88.33% MADS-box genes (including 10 ChMADS genes) derived from segment duplications and 16.67% from tandem duplications (including 2 ChMADS genes) whereas in Arabidopsis segmental and tandem duplications accounted for 66.33% (including 12 AtMADS genes) and 33.33% (including 6 AtMADS genes) of homologous gene pairs, respectively (Figs. 3, 4).
Discussion
Floral development sequences are conserved among divergent species and MADS-box genes involved in a wide range of functions including the formation of flowers, development of reproductive structures and control of flowering time (Medard and Yanofsky 2001). A comparative study between model plant systems increases our understanding of the evolutionary event of MADS-box genes in plants, especially during reproductive development. Like A. thaliana, the C. hirsuta has been used to understand the genetic basis of morphological evolution (Hay and Tsiantis 2016). Genetic experiments in Arabidopsis have shown that several MADS-box genes are required to regulate the transition from vegetative to reproductive development. Analysis of the complete Arabidopsis genome sequence revealed 107 genes encoding MADS-box proteins which 52 were involved in flowering development such as FLC-like genes, MADS AFFECTING FLOWERING2(MAF2, also known as AGL31), MAF3 (AGL70), MAF4 (AGL69), MAF5, and FLOWERING LOCUSM (FLM, also known as MAF1 and AGL27) (Parenicova et al. 2003; Becker and Theißen 2003; Grimplet et al. 2016). There is no comparative report on the C. hirsuta MADS-box genes although a high-quality reference genome of C. hirsuta has been assembled (Gan et al. 2016). This high-quality reference genome of the C. hirsuta strain ‘Oxford’ allows the comparison between C. hirsuta and A. thaliana MADS-box genes family.
To do this a phylogenetic tree was produced using amino acid sequence of 50 MADS-box genes of C. hirsuta and A. thaliana. According to the phylogenetic analysis, 5 subfamilies (MIKCc, Mα, Mγ, Mβ, and MIKC*) were determined, 40 C. hirsuta MADS-box genes were classified as type II genes, including 35 MADS-box MIKCc type and 5 MADS-box MIKC* type, which is comparable to that in Arabidopsis and 11 C. hirsuta MADS-box genes were determined as type I genes, including the Mα, Mβ, and Mγ groups, which is similar to Arabidopsis (Fig. 2). Most MADS-box genes had their orthologs in Arabidopsis.
However the structural diversity of C. hirsuta MADS-box genes and chromosomal localization of the 50 C. hirsuta MADS-box genes showed that a number of MADS-box genes were similar; therefore, a MADS-box gene in C. hirsuta had several orthologs in A. thaliana. The total number of 43 flowering-related MADS-box genes was identified in C. hirsuta which was a relatively lower number of MADS-box genes than A. thaliana. A variation in the number of MADS-box genes among different species has already been reported. For example 153,107, 90 and 75 MADS-box genes were identified in potato, Arabidopsis, grapevine and rice, respectively (Gao et al. 2018a, b, Grimplet et al. 2016, Arora et al. 2007, Parenicova et al. 2003).
At least two rounds of duplications have probably occurred in the A. thaliana genome and MADS-box family genes expanded by tandem or local duplication as the most commonly evaluated mechanism for gene family expansion (Cannon et al. 2004). Whole-genome duplication and gene duplication events (segmental and tandem duplication) have always been considered to be the primary source of biological evolution. It is proposed that gene duplications have an important role in genomic rearrangement, expansion, and the diversification of gene function. Unequal crossing-over and transposable elements may also have played an important role in gene duplications and genome rearrangements in plants (Su et al. 2013; Zhang et al. 2013). In this study, six homologous gene pair duplication events were determined in C. hirsuta, including 12 MIKC-type (10 MIKCc and 2 MIKC*) which is lower than nine homologous gene pair duplication events in Arabidopsis. In C. hirsuta, about 83% of homologous gene pairs participated in SDs while only about 17% participated in TDs compared to Arabidopsis which is about 66% and 33%, respectively. These results suggest that both tandem and segmental duplication may play crucial roles in MADS-box gene expansion in C. hirsuta and A. thaliana genomes. Moreover, it seems the function of the MIKC type, particularly the MIKCc type, is more important in the evolution of both plants. The C. hirsuta MADS-box genes involved in flowering may have a lower duplication rate and/or a higher gene loss rate after duplication. The divergence of C. hirsuta and A. thaliana has been estimated in around 32 Myr ago (Gan et al. 2016).
MIKCC group of MADS-box genes are involved in control of many developmental processes in flowering plants such as the floral organ identity genes that provide the class A, B, C, D, and E homeotic functions, stamen development, pollen growth, flowering time, floral meristem specification, and ovule and fruit development (Yu-Ting et al. 2019). Based on phylogenetic analysis, type II MADS-box genes in C. hirsuta like Arabidopsis, contain 14 subfamilies including TM3, AGL6, and AGL17 which promote flowering and FLOWERING LOCUS C (FLC Family), SHORT VEGETATIVE PHASE (SVP), as MADS-box regulators and AGL15, AG, TT16, AGL12, GLO, DEF, SEP, and SQUA (Scortecci et al. 2001; Ratcliffe et al. 2001; Medard and Yanofsky 2001; Becker and Theißen 2003; Caicedo et al. 2009; Zhi et al. 2019) (Fig. 1).
FLC-like genes are mainly required for prolonged cold exposure to establish floral competency, known as vernalization (Smaczniak et al. 2012; Whittaker and Dean 2017). The comparison of the FLC family genes of A. thaliana and C. hirsuta, surprisingly showed that Arabidopsis had about two times more FLC family genes. C. hirsuta had three FLC genes (ChMADS9/25/28/29, ChMADS10 and ChMADS25) while A. thaliana had five FLC orthologs (AGL69, AGL68, AGL27, AGL31 and AGL70). However, there was a high similarity between FLC motifs sequences of two plants, which means FLC genes are highly conserved.
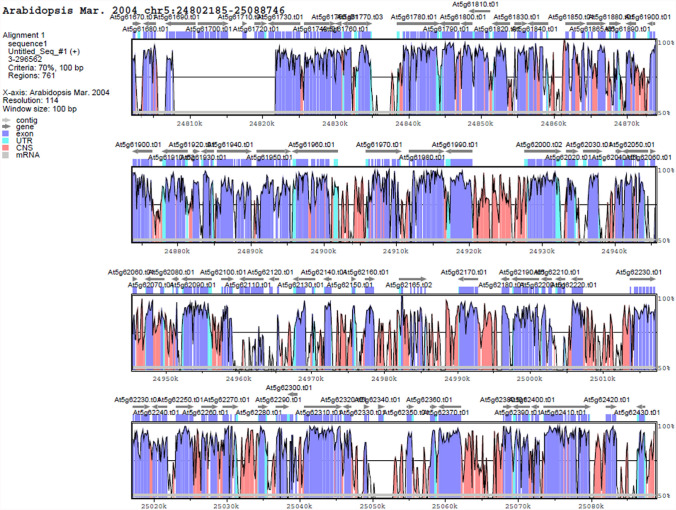
The chromosomal location analysis of the C. hirsuta FLC MADS-box genes showed that ChMADs8 and ChMADS9 FLC genes are distributed on chromosome eight terminal arm while in the A. thaliana AGL69, AGL68, AGL31, AGL70 and AGL25 FLC MADS-box genes located on the beginning of the chromosome five arm (Figs. 3, 4). Although the C. hirsuta genome was largely syntenic to the genome of A. thaliana, but C. hirsuta genome retained more ancestral features, including karyotype and genome size (Gan et al. 2016). The synteny between these two genomes indicated that the big part at the end of chromosome eight was similar to the beginning part of chromosome five. Therefore, tandem duplication event may result in the expansion of FLC family members in Arabidopsis (Fig. 5).
Fig. 5.
Synteny region between a part of the chromosomes of A. thaliana (ath) and C. hirsuta (chi). Identity plot compares the region from chromosome eight in the genome of C. hirsuta with chromosome five in A. thaliana as a reference sequence. The vertical scale indicates the percentage of identity 50–100%. The horizontal axis indicates the coordinates in the genome. Genome regions are color-coded as Contig, gene, exon, UTR, mRNA and conserved non-coding sequences (CNS)
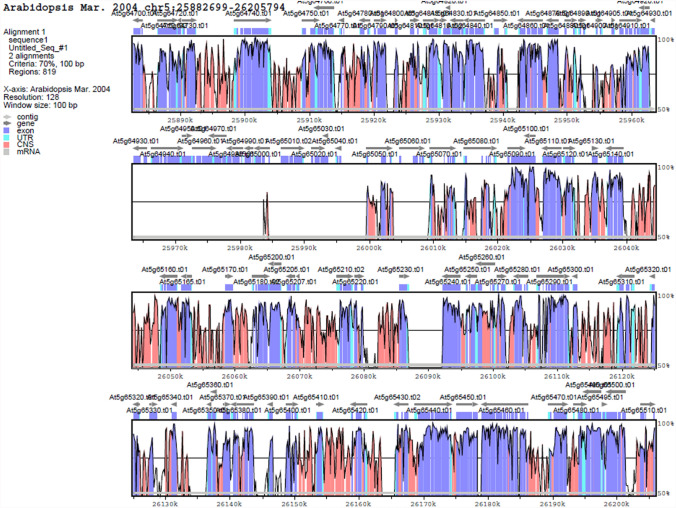
TM3 family is another MADS‐box genes involved in the autonomous pathway. AGAMOUS-LIKE 20(AGL20/SOC1) is the most well-known member of this family that negatively regulated by FLC and positively regulated by genes involved in autonomous pathway (Dorca-Fornell et al. 2011). The phylogenetic analysis showed three MADS-box genes (ChMADS6/ChMADS7, ChMADS22/ChMADS34 and ChMADS33) existed in C. hirsuta. The comparison of AGAMOUS-LIKE 20(AGL20/SOC1) family genes of Arabidopsis and C. hirsuta astonishingly showed that A. thaliana genome contained six AGAMOUS-LIKE20 family genes while C. hirsuta had four MADS-box. The results indicated that members of the same clade generally shared one or more motifs, nevertheless comparison between ChMADS34/ChMADS22 and AGL42 indicated that k-box motif in ChMADS34 was absent (Fig. 6). Furthermore, AGL42, AGL71 and AGL72 showed interactions with SOC1 which was involved in the floral transition (Dorca-Fornell et al. 2011).The AGL42 and ChMADS34/ChMADS22 were identified, respectively, on chromosome five in A. thaliana and chromosome eight in C. hirsuta with a high degree of synteny (Fig. 7). These results may suggest that a number of AGAMOUS-LIKE 20 family genes in C. hirsuta were altered or lost during the evolutionary process.
Fig. 6.
Protein classification and graphical summary of conserved domains of ChMADS34 with AGL42 of TM3 family gene in MIKCC group. AGL42 MADS-box gene of Arabidopsis has a k-box domain while ChMADS34 MADS-box gene in C. hirsuta lost the k-box domain
Fig. 7.
Synteny region between a part of A. thaliana (ath) and C. hirsuta (chi) chromosomes. Identity plot compares the region from chromosome eight in the genome of C. hirsuta with chromosome five in A. thaliana as a reference sequence. The vertical scale indicates the percentage of identity 50–100%. The horizontal axis indicates the coordinates in the genome. Genome regions are color-coded as Contig, gene, exon, UTR, mRNA and conserved non-coding sequences (CNS)
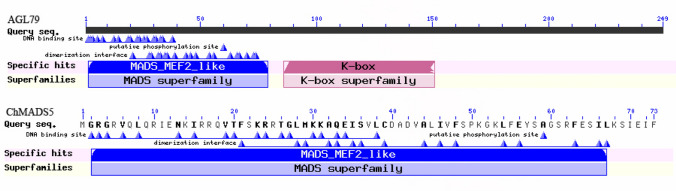
The SQUA family (AP1/FUL family) of MADS-box genes plays an important role in the initial activation of floral development of angiosperms (Chen et al. 2007). The genes belonging to this clade are APETALA1 (AP1 or AGL7), FRUITFULL (FUL or AGL8), CAULIFLOWER (CAL or AGL10), and AGL79. The chromosomal distribution of the SQUA family showed that three MADS-box genes AGL10, AGL7 and AGL79 were located on chromosomes one and three of A. thaliana whereas in C. hirsuta, ChMADS5, ChMADS52 and ChMADS44/42 were distributed on chromosome one, seven and five, respectively. The classification and evolution analysis by comparing conserved motifs indicated that ChMADS5 did not contain K-domain (Fig. 8) which is involved in protein dimer formation or higher order (multimeric) protein complexes formation (Kaufmann et al. 2005). Gao et al. (2018a, b) showed a linear relationship between SPL10 and AGL79 in regulating Arabidopsis plant development. The SPL (SQUAMOSA PROMOTER BINDING PROTEIN-LIKE) protein family members contain a conserved squamosal promoter binding protein (SBP) domain of 76 amino acids (Yamasaki et al. 2004; Preston). A genetic function study determined that SPL10 played crucial roles in vegetative-to-reproductive transition (Xu et al. 2016). On the other hand, enhanced or silenced expression of AGL79 in Arabidopsis plants showed fewer and smaller rosette leaves and earlier flowering time compared to WT plants (Gao et al. 2018a, b). Although data shows the absence of the K-domain in ChMADS5 however its role may be compensated by the K-domain of ChMADS44/42, in the same family (SQUA). Probably the K-domain of ChMADS5 was lost after event segmentation. Our data showed that ChMADS5 has also lost several exons and introns. This is already reported in potato (Gao et al. 2018a, b). Gene duplication events, gene mutation, and loss of certain domains, might also play important role in generating M-type MADS-box in the process of plant evolution (Gao et al. 2018a, b). Based on intron–exon structures of Cardamine MADS-box genes, it was determined that the MIKC* had less intron than MIKCC, and most of the M-type MADS-box genes were intronless.
Fig. 8.
Protein classification and graphical summary of conserved domains of ChMADS5 with AGL79 of SQUA family gene in MIKCC group. AGL79 MADS-box gene of Arabidopsis has a k-box domain while ChMADS5 MADS-box gene in C. hirsuta lost the k-box domain
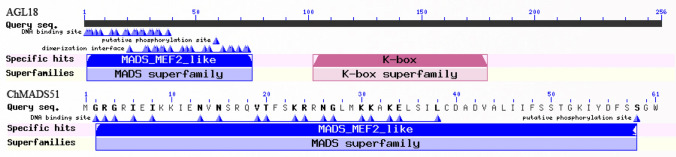
The MADS-box transcription factor AGAMOUS-LIKE15 (AGL15) and AGAMOUS-LIKE18 (AGL18) in Arabidopsis are able to promote somatic embryogenesis (SE). The phylogenetic analysis revealed two MADS-box genes of C. hirsuta (ChMADS51 and ChMADS37) are orthologs of AGL15 and AGL18. Compared to AGL15, the ChMADS51 lacks k-box and introns (Fig. 9). As there is an association between the level of evolutionary conservation, the size of intronic region and level of gene expression (Gorlova et al. 2014), the gene expression study can help more to understand the function of k-box and intron in ChMADS51.
Fig. 9.
Protein classification and graphical summary of conserved domains of ChMADS51 with AGL18 of AGL15 family gene in MIKCC group. AGL18 MADS-box gene in Arabidopsis has a k-box domain while ChMADS51 MADS-box gene in C. hirsuta lost the k-box domain
In conclusion, phylogenetic analyses provided a useful reference to identify 43 flowering-related MADS-box genes in the C. hirsuta genome. Several novel characteristics were found in the C. hirsuta MADS-box gene family including duplication and segmentation events, loss of introns and k-box in three ChMADS gene and lower number of MADS-box in C. hirsuta than A. thaliana. Further analysis is required to understand the biological functions of ChMADS genes.
Acknowledgements
This work was financially supported by a research Grant (no. 9424417001) to HB from Bu-Ali Sina University, Hamedan, Iran.
Author contributions
HB conceived and designed the analysis; MGM performed the analysis; HB and MG were involved in discussions and analysis; MGM wrote the manuscript and HB and MG revised the manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
References
- Aelaitinen R, Broholm S, Albert VA, Teeri TH, Elomaa P. Patterns of MADS-box gene expression mark flower-type development in gerbera hybrid (asteraceae) Open Access BMC Plant Biol. 2006;6:11. doi: 10.1186/1471-2229-6-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora R, Agarwal P, Ray S, Kumar AS, Singh VP, Tyagi AK, Kapoor S. MADS-box gene family in rice: genome-wide identification, organization and expression profiling during reproductive development and stress. BMC Genom. 2007;8:242. doi: 10.1186/1471-2164-8-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Johnson J, Grant CE, Noble WS. The MEME suite. Nucleic Acids Res. 2015 doi: 10.1093/nar/gkv416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker A, Theißen G. The major clades of MADS- box genes and their role in the development and evolution of flowering plants. Mol Phylogenet Evol. 2003;29:464–489. doi: 10.1016/S1055-7903(03)00207-0. [DOI] [PubMed] [Google Scholar]
- Bi C, Xu Y, Ye Q, Yin T, Ye N. Genome-wide identification and characterization of WRKY gene family in Salix suchowensis. Peer J. 2016;4:e2437. doi: 10.7717/peerj.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caicedo AL, Richards C, Ehrenreich IM, Purugganan MD. Complex rearrangements lead to novel chimeric gene fusion polymorphisms at the Arabidopsis thaliana MAF2-5 flowering time gene cluster. Mol Biol Evol. 2009;26:699–711. doi: 10.1093/molbev/msn300. [DOI] [PubMed] [Google Scholar]
- Cannon SB, Mitra A, Baumgarten A, Young ND, May G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004;4:10. doi: 10.1186/1471-2229-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelán-muñoz N, Herrera J, Cajero-sánchez W, Arrizubieta M, Trejo C, García-ponce B, De la Pazsánchez M, Alvarez-buylla ER, Garay-arroyo A. MADS-box genes are key components of genetic regulatory networks involved in abiotic stress and plastic developmental responses in plant sreview published: Front. Plant Sci. 2019 doi: 10.3389/fpls.2019.00853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Bowman JL, Meyerowitz EM. Field guide to plant model systems. Cell. 2016;167(2):325–339. doi: 10.1016/j.cell.2016.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Guo B, Hexige S, Zhang T, Shen D, Ming F. SQUA-like genes in the orchid Phalaenopsis are expressed in both vegetative and reproductive tissues. Planta. 2007;226:369–380. doi: 10.1007/s00425-007-0488-0. [DOI] [PubMed] [Google Scholar]
- De Folter S, Immink RGH, Kieffer M, Parenicova L, Henz SR, Weigel D, Busscher M, Kooiker M, Colombo L, Kater MM, Davies B, Angenenta GC. Comprehensive interaction map of the Arabidopsis MADS box transcription factors. Plant Cell. 2005;17:1424–1433. doi: 10.1105/tpc.105.031831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorca-Fornell C, Gregis V, Grandi V, Coupland G, Colombo L, Kater MM. The Arabidopsis SOC1-like genes AGL42, AGL71 and AGL72 promote flowering in the shoot apical and axillary meristems. Plant. 2011;J67:1006–1017. doi: 10.1111/j.1365-313X.2011.04653.x. [DOI] [PubMed] [Google Scholar]
- Gan X, Hay A, Kwantes M, Haberer G, Hallab A, DelloIoio R, Hofhuis H, Pieper B, Cartolano M, Neumann U, Nikolov LA, Song B, Hajheidari M, Briskine R, Kougioumoutzi E, Vlad D, Broholm S, Hein J, Meksem K, Lightfoot D, Shimizu KK, Shimizu-Inatsugi R, Imprialou M, Kudrna D, Wing R, Sato S, Huijser P, Filatov D, Mayer KFX, Mott R, Tsiantis M. The Cardamine hirsuta genome offers insight into the evolution of morphological diversity. Nat Plants. 2016;2(11):16167. doi: 10.1038/nplants.2016.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H, Wang Z, Li S, Hou M, Zhou Y, Zhao Y, LiZhaoMa GHH. Genome-wide survey of potato MADS-box genes reveals that StMADS1 and StMADS13 are putative downstream targets of tuberigen StSP6A. BMC Genom. 2018;19:726. doi: 10.1186/s12864-018-5113-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao R, Wang Y, Gruber MY, Hannoufa A. MiR156/SPL10 modulates lateral root development, branching and leaf morphology in Arabidopsis by silencing AGAMOUS-LIKE 79. Front Plant Sci. 2018;8:2226. doi: 10.3389/fpls.2017.02226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerats T, Vandenbussche MA. Model system for comparative research: petunia. Trends Plant Sci. 2005;10(5):251–256. doi: 10.1016/j.tplants.2005.03.005. [DOI] [PubMed] [Google Scholar]
- Gorlova O, Fedorov A, Logothetis C, Amos C, Gorlov I. Genes with a large intronic burden show greater evolutionary conservation on the protein level. BMC Evol Biol. 2014;14:50. doi: 10.1186/1471-2148-14-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimplet J, Miguelmartínez-zapater J, Josécarmona M. Structural and functional annotation of the MADS-box transcription factor family in grapevine. BMC Genom. 2016;17:80. doi: 10.1186/s12864-016-2398-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay A, Tsiantis M. Cardamine hirsuta: a comparative view current opinion in genetics & development. Curr Opin Genet Dev. 2016;39:1–7. doi: 10.1016/j.gde.2016.05.005. [DOI] [PubMed] [Google Scholar]
- Hay AS, Pieper B, Cooke E, Mandáková T, Cartolano M, Tattersall AD, Ioio RD, McGowan SJ, Barkoulas M, Galinha C, Rast MI, Hofhuis H, Then C, Plieske J, Ganal M, Mott R, Martinez-Garcia JF, Carine MA, Scotland RW, Gan X, Filatov DA, Lysak MA, Tsiantis M. Cardamine hirsuta: a versatile genetic system for comparative studies. Plant J. 2014;78:1–15. doi: 10.1111/tpj.12447. [DOI] [PubMed] [Google Scholar]
- Henschel K, Kofuji R, Hasebe M, Saedler H, Munster T, Ttheissen G. Two ancient classes of MIKC-type MADS-box genes are present in the moss Physcomitrella patens. Mol Biol Evol. 2002;19:801–814. doi: 10.1093/oxfordjournals.molbev.a004137. [DOI] [PubMed] [Google Scholar]
- Hu B, Jin J, Guo A, Zhang H, Luo J, Gao G. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics. 2015;31(8):1296–1297. doi: 10.1093/bioinformatics/btu817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston JS, Pepper AE, Hall AE, Chen ZJ, Hodnett G, Drabek J, LopezPrice JHJ. Evolution of genome size in Brassicaceae. Ann Bot. 2005;95(1):229–235. doi: 10.1093/aob/mci016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann K, Melzer R, Theissen G. MIKC-type MADS-domain proteins: structural modularity, protein interactions and network evolution in land plants. Gene. 2005;347(2):183–198. doi: 10.1016/j.gene.2004.12.014. [DOI] [PubMed] [Google Scholar]
- Liu JJ, Ekramoddoullah AKM. Identification and characterization of the WRKY transcription factor family in Pinus monticola. Genome. 2009;52:77–88. doi: 10.1139/G08-106. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Bryant SH. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 2004;32:327–331. doi: 10.1093/nar/gkh454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medard NG, Yanofsky MF. Function and evolution of the plant MADS-box gene family. Nat Rev Genet. 2001;2(3):186–195. doi: 10.1038/35056041. [DOI] [PubMed] [Google Scholar]
- Parenicova L, De Folter S, Kieffer M, Horner DS, Favalli C, Busscher J, Cook HE, Ingramrm RM, Kater MM, Davies B, Angenentg C, Colombo L. Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: new openings to the MADS world. Plant Cell. 2003;15:1538–1551. doi: 10.1105/tpc.011544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Y, Bi C, He B, Ye N, Yin T, Xu L. Genome-wide identification and characterization of the MADS-box gene family in Salix suchowensis. Peer J. 2019;7:e8019. doi: 10.7717/peerj.8019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliffe OJ, Nadzan GC, Reuber TL, Riechmann JL. Regulation of flowering in Arabidopsis by an FLC homologue. Plant Physiol. 2001;126:122–132. doi: 10.1104/pp.126.1.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann JL, Wang M, Meyerowitz EM. DNA-binding properties of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA and AGAMOUS. Nucleic Acids Res. 1996;24(16):3134–3141. doi: 10.1093/nar/24.16.3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz-sommerZ DB, Hudson A. An everlasting pioneer: the story of antirrhinum research. Nat Rev Genet. 2003;4(8):657–666. doi: 10.1038/nrg1127. [DOI] [PubMed] [Google Scholar]
- Scortecci KC, Michaels SD, Amasino RM. Identification of a MADS-box gene, FLOWERING LOCUSM, that represses flowering. Plant J. 2001;26:229–236. doi: 10.1046/j.1365-313x.2001.01024.x. [DOI] [PubMed] [Google Scholar]
- Smaczniak C, Immink RGH, Angenent GC, Kaufmann K. Developmental and evolutionary diversity of plant MADS domain factors: Insights from recent studies. Development. 2012;139:3081–3098. doi: 10.1242/dev.074674. [DOI] [PubMed] [Google Scholar]
- Su H, Zhang S, Yuan X, Chen C, Wang XF, Hao YJ. Genome-wide analysis and identification of stress-responsive genes of the NAM-ATAF1, 2-CUC2 transcription factor family in apple. Plant Physiol Biochem. 2013;71:11–21. doi: 10.1016/j.plaphy.2013.06.022. [DOI] [PubMed] [Google Scholar]
- Theißen G, Gramzow L. Structure and evolution of plant MADS domain transcription factors. In: Gonzalez DH, editor. Plant transcription factors: evolutionary, structural and functional aspects. Philadelphia: Elsevier; 2016. pp. 127–138. [Google Scholar]
- Theissen G, Kim JT, Saedler H. Classification and phylogeny of the MADS-box multigene family suggest defined roles of MADS-box gene subfamilies in the morphological evolution of eukaryotes. J Mol Evol. 1996;43(5):484–516. doi: 10.1007/BF02337521. [DOI] [PubMed] [Google Scholar]
- Wang Y, Zhang J, Hu Z, Guo X, Tian S, Chen G. Genome-wide analysis of the MADS-box transcription factor family in Solanum lycopersicum. Int J Mol Sci. 2019;20:2961. doi: 10.3390/ijms20122961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker C, Dean C. The FLC locus: a platform for discoveries in epigenetics and adaptation. Annu Rev Cell Dev Biol. 2017;33:555–575. doi: 10.1146/annurev-cellbio-100616-060546. [DOI] [PubMed] [Google Scholar]
- Xu M, Hu T, Zhao J, Park MY, Earley KW, Wu G, et al. Developmental functions of miR156-regulated SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) genes in Arabidopsis thaliana. PLoS Genet. 2016;12:e1006263. doi: 10.1371/journal.pgen.1006263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Zhang Q, Sun L, Du D, Cheng T, Pan H, Yang W, Wang J. Genome wide identification, characterisation and expression analysis of the MADS-box gene family in Prunus mume. Mol Gen Genom. 2014;289(5):903–920. doi: 10.1007/s00438-014-0863-z. [DOI] [PubMed] [Google Scholar]
- Yamasaki K, Kigawa T, Inoue M, Tateno M, Yamasaki T, Yabuki T, et al. A novel zinc-binding motif revealed by solution structures of DNA-binding domains of Arabidopsis SBP-family transcription factors. J Mol Biol. 2004;337:49–63. doi: 10.1016/j.jmb.2004.01.015. [DOI] [PubMed] [Google Scholar]
- Yoshida H, NagatoY, Flower development in rice. J Exp Bot. 2011;62(14):4719–4730. doi: 10.1093/jxb/err272. [DOI] [PubMed] [Google Scholar]
- Yu-Ting C, Chi-Chang C, Chi-Wei C, Kuan-Chun C, Yen-Wei C. MADS-box gene classification in angiosperms by clustering and machine learning. Approaches Front Genet. 2019;9:707. doi: 10.3389/fgene.2018.00707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Xu R, Luo X, Jiang Z, Shu H. Genome-wide identification and expression analysis of MAPK and MAPKK gene family in Malus domestica. Gene. 2013;531:377–387. doi: 10.1016/j.gene.2013.07.107. [DOI] [PubMed] [Google Scholar]
- Zhang X, Fatima M, ZhouMa PQ, Ming R. Analysis of MADS-box genes revealed modified flowering gene network and diurnal expression in pineapple. BMC Genom. 2020;21:8. doi: 10.1186/s12864-019-6421-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhi Wei NT, Wei Z, Lisha S. Dissecting the function of MADS-Box transcription factors in orchid reproductive development. Front Plant Sci. 2019;10:1474. doi: 10.3389/fpls.2019.01474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zozomova-Lihova J, Marhold K. Plant genome: biodiversity and evolution. Enfield: Science Publishers; 2006. Phylogenetic and diversity patterns in Cardamine (Brassicaceae)—a genus with conspicuous polyploid and reticulate evolution; pp. 149–186. [Google Scholar]