Abstract
Background
Hypertrophy of the nucleolus is a distinctive cytological feature of malignant cells and corresponds to aggressive behaviour. This study aimed to identify the key gene associated with nucleolar prominence (NP) in breast cancer (BC) and determine its prognostic significance.
Methods
From The Cancer Genome Atlas (TCGA) cohort, digital whole slide images identified cancers having NP served as label and an information theory algorithm was applied to find which mRNA gene best explained NP. Dyskerin Pseudouridine Synthase 1 (DKC1) was identified. DKC1 expression was assessed using mRNA data of Molecular Taxonomy of Breast Cancer International Consortium (METABRIC, n = 1980) and TCGA (n = 855). DKC1 protein expression was assessed using immunohistochemistry in Nottingham BC cohort (n = 943).
Results
Nuclear and nucleolar expressions of DKC1 protein were significantly associated with higher tumour grade (p < 0.0001), high nucleolar score (p < 0.001) and poor Nottingham Prognostic Index (p < 0.0001). High DKC1 expression was associated with shorter BC-specific survival (BCSS). In multivariate analysis, DKC1 mRNA and protein expressions were independent risk factors for BCSS (p < 0.01).
Conclusion
DKC1 expression is strongly correlated with NP and its overexpression in BC is associated with unfavourable clinicopathological characteristics and poor outcome. This has been a detailed example in the correlation of phenotype with genotype.
Subject terms: Breast cancer, Oncology
Background
Breast cancer (BC) is the most common cancer diagnosed in women worldwide, accounting for ~1 in 10 new cancer diagnoses each year and is the second most common cause of death1,2 due to cancer. BC is a heterogeneous disease with variable morphologies and response to therapy. Some morphological features, especially histological grade, have been well validated to have a strong prognostic value and their assessment helps in prognostic stratification of BC patients for treatment decisions.3
In the Nottingham cohort, nucleolar prominence (NP) has recently been shown to be a significant predictor for patient outcome as well as of being highly correlated with tumour grade. Since the NP is a distinctive morphological attribute, it is hypothesised to possibly serve as a substitute for the highly subjective pleomorphism component score of the Nottingham BC grading.4 Consequently, it is deemed imperative to explore the correlations between the nucleolar phenotype and genotype.
The major function of the nucleolus is synthesis and assembly of ribosomes5 where both are associated with malignant transformation and cancer progression.6,7 Indeed, ribosome biogenesis depends on the cancer growth rate, which is directly related to nucleolar size of malignant cells. Nucleolar size and cell kinetics’ parameters are interrelated because of the increasing rate of ribosome biogenesis in proliferating cells.8 In some solid cancer and haematological malignancies, the ribosome biogenesis rate increases as a consequence of overexpression of the oncogene c-Myc, which controls all the steps of ribosome biogenesis.9 Despite the biological and clinical significance of NP in BC, the key gene associated with NP and its prognostic significance remains to be defined.
Dyskerin Pseudouridine Synthase 1 (DKC1) is a predominantly nucleolar protein encoded by DKC1 gene and mapped at Xq28.10 DKC1 is a crucial component of the telomerase complex and is required for normal telomere maintenance and post-transcriptional processing of precursor rRNA. Therefore, DKC1 is necessary for tumour cell progression through mechanisms related to its function in the processing of rRNA precursor.11 Usually, clinically indolent and slow-growing tumours express lower levels of DKC1 and its inhibition slows or hinders the proliferation in most cell types.11,12 Through various deprivation of function approaches, emerging evidence suggests that DCK1 may regulate other cellular processes, including IRES-mediated translation, telomere maintenance independent of telomere length regulation, mitosis, transcription and possibly microRNA processing.13,14 Upregulation of DKC1 expression has been reported in several human cancers including hepatocellular carcinoma,15 neuroblastoma,16 lymphoma,17 melanoma,18 prostate cancer,19 colorectal cancer20 and ovarian carcinoma.21
Methods
Principle of DKC1 selection
We have applied an information theory (IT) approach to The Cancer Genome Atlas (TCGA) breast cancer dataset. The IT approach was used for feature selection to identify the key gene associated with NP, which was assessed morphologically in full face invasive BC sections stained with haematoxylin and eosin (H&E) using digital whole slide images (WSI) as explained in our previous study.4 The TCGA BC cohort was employed since it contains satisfactory whole slide images and mRNA-seq2 data present in 743 cancers. In the IT approach utilised, the nucleolar score served as a label and 20,339 mRNA transcripts served as predictor variables. The IT algorithm is a ‘greedy’ algorithm and reduces the number of features selected. “Greedy” is the term used in the machine learning community to describe an algorithm, which selects the optimal feature at each step and does not alter any choices already made based on findings from future choices.22 The analysis showed that the attribute, which exhibited the highest mutual information (information gain) with NP, was DKC1. Moreover, the detection of DKC1 was supported by LASSO regression feature selection. The required functions for LASSO were obtained from R library Glmnet.23 LASSO regression is also capable of reducing the number of predictors and thereby allowing for a focused study of a few attributes.24 Therefore, by applying the IT approach and LASSO regression, the selection was limited to a single gene (DKC1). This was followed by evaluating DKC1 mRNA and DKC1 protein expression in large clinically annotated cohorts of BC to evaluate its clinicopathological and prognostic value in invasive BC as described below.
Study cohorts for transcriptomic analysis
The discovery of DKC1 was by study of TCGA cohort. The TCGA was also used to assess the possible correlation between DKC1 mRNA expression and the variables recorded in this cohort.25 The Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) cohort (n = 1980) was used to evaluate DKC1 gene copy number (CN) aberrations and gene expression.26 Genomic and transcriptomic data for the METABRIC cohort had been obtained using the Affymetrix SNP 6.0 and Illumina HT-12v3 platforms, respectively.26 The association between DKC1 mRNA expression, copy number aberrations and clinicopathological parameters, molecular subtypes and patient outcome was investigated. Breast Cancer Gene Expression Miner online dataset v4.3 (http://bcgenex.centregauducheau.fr/BC-GEM/GEM-requete.php) was also used as external validation of DKC1 mRNA expression.
Study cohort for protein expression
The Nottingham BC patient cohort was used to evaluate the immunohistochemical (IHC) expression of DKC1. This cohort is a well-characterised large series (n = 943) of invasive BC patients aged ≤70 years and presented at Nottingham City Hospital between 1999 and 2006. The cohort has long-term clinical follow-up and clinicopathological data included patient’s age at diagnosis, histological tumour type, tumour grade, tumour size, lymph node status, Nottingham Prognostic Index (NPI) and lymphovascular invasion (LVI). Patient outcome data were obtained including BC-specific survival (BCSS), defined as the time (in months) from the date of primary surgical treatment to the time of death from BC and distant metastasis free survival (DMFS) defined as time (in months) from primary surgical treatment until the first event of distant metastasis. Patients were treated based on tumour features, NPI and hormone receptor status. Endocrine therapy was given to patients who had oestrogen receptor positivity (ER+) tumours with high NPI scores (>3.4), whereas no adjuvant therapy was given to patients with ‘good’ NPI scores (≤3.4). Premenopausal patients with moderate and poor NPI scores were candidates for chemotherapy, while postmenopausal patients with ‘moderate’ or ‘poor’ NPI scores were given hormonal therapy only. Classical treatment of cyclophosphamide, methotrexate and fluorouracil (CMF) was used as a therapy for patients presented with absence of ER expression and clinically fit to receive chemotherapy. None of the patients in the current study cohort received neoadjuvant therapy. Data related to the expression of ER, progesterone receptor (PR) and human epidermal growth factor receptor 2 (HER2) as well as Ki67 were available.27–30 Molecular subtypes were based on tumour immunohistochemical (IHC) profile and the Elston–Ellis31 mitotic score as ER+/HER2−; low proliferation (mitotic score 1), ER+/HER2− high proliferation (mitotic scores 2 and 3), HER2-positive class: HER2+ regardless of ER status, TN: ER−, PR− and HER2−.32 There was no significant differences in the distribution of the clinicopathological parameters between the Nottingham and the METABRIC cohorts (all correlation coefficients ≥0.948, all p < 0.0001)33 (Supplementary Table 1).
DKC1 validation by western blotting
The antibody specificity of anti-DKC1 antibody (EPR10399, Abcam, UK) was validated using western blotting (WB) performed on cell lysates of a wild and transfected MDA-MB-231 human breast cancer cell line (American Type Culture Collection; Rockville, MD, USA). The forward transfection of siRNA procedure was followed according to DKC1 siRNAs manufacturer’s instructions. In brief, the cells were seeded in 6-well plate at a cell density of 3 × 105 cells per well and incubated overnight in 37 °C 5% CO2 incubator. The following day, the cells reached about 40% confluence and were transfected with 10 nM of three different IDs of DKC1 siRNA (Cat#:4392420, ThermoFisher Scientific, UK). A transfection with 10 nM scrambled siRNA sequence (Cat#:4390843, ThermoFisher Scientific, UK) was carried out in the experiment and considered as a negative control. DKC1 protein expression of untransfected & transfected cells was then determined by the Western blotting. Briefly, after collecting the cell lysates, a dilution of 1:1000 of the primary antibody and 1:15000 IRDye 800CW Donkey anti-rabbit secondary antibody (LI-COR Biosciences) were applied, and 5% milk /PBS-Tween (0.1%) (Marvel Original Dried Skimmed Milk, Premier Food Groups Ltd., UK) was used for blocking and antibodies incubation. Mouse monoclonal anti-β-actin primary antibody (1:5000) (Sigma–Aldrich, UK) with IRDye 800CW Donkey anti-mouse fluorescent secondary antibody (LI-COR Biosciences) were used to visualise a marker of endogenous control. Visualisation of DKC1 band was done by using the Odyssey Fc with Image Studio 4.0 (LI-COR Biosciences).
Tissue microarrays and immunohistochemical analysis
Invasive BC tissues were previously arrayed as tissue microarrays (TMA) using the Grand Master® (3D HISTECH®, Budapest, Hungary).34 IHC staining was performed on 4 μm TMA thick sections using the Novocastra Novolink™ Polymer Detection Systems kit (Code: RE7280-K, Leica, Biosystems, Newcastle, UK). Antigen retrieval was performed in citrate buffer pH 6.0 using a microwave (Whirlpool JT359 Jet Chef 1000 W) for 20 min. Rabbit monoclonal DKC1 was diluted at 1:50 in Leica antibody diluent (RE AR9352, Leica, Biosystems, UK) and incubated with the sections for 60 min at room temperature. A negative control was obtained by omitting the incubation with primary antibody while formalin fixed placenta tissue was used as a positive control according to manufacturer’s datasheet.
Assessment of DKC1 protein expression
Scanning of TMA stained sections into high-resolution digital images was performed by using a NanoZoomer scanner (NanoZoomer; Hamamatsu Photonics, Welwyn Garden City, UK) at ×20 magnification. Scoring of DKC1 nuclear and nucleolar4 expression was evaluated based on a semi-quantitative scoring using modified histochemical score (H-score), where the intensity of staining was multiplied by the percentage of positive cells in the tissue for each intensity, producing a score ranging between 0 and 300.35 A score index of 0, 1, 2 and 3 corresponding to negative, weak, moderate and strong respectively were used for intensity. The percentage (0–100) of positive cells for each intensity was evaluated subjectively. All non-representative cores including folded tissue during processing and staining, cores with only normal breast tissue and cores with invasive tumour <15% of core surface area were excluded from scoring. All the cores were scored by a trained observer (K. Elsharawy) blinded of histopathological and patient outcome data. Further, to test the interobserver’s reproducibility of the scoring, a subset of TMA cores (10%) was randomly selected and double scored by a second trained observer (M. Aleskandarany). Moreover, for further evaluation of scoring reproducibility, 20% of the cases were double scored by the main observer (K. Elsharawy) after 5 months washout period blind from the first scores.
Statistical analysis
IBM-SPSS statistical software 24.0 (SPSS, Chicago, IL, USA) was used in statistical analysis. Interobserver agreement in DKC1 IHC scoring was assessed using intraclass correlation coefficient (ICC). Dichotomisation of DKC1 proteomic and transcriptomic levels expression was determined based on the prediction of BCSS using X-tile bioinformatics software version 3.6.1 (School of Medicine, Yale University, New Haven, USA).36 The H-scores of 110 and 10 were the optimal cut-off values of DKC1 nuclear and nucleolar protein expression. Continuous data of DKC1 mRNA and DKC1 protein expression were used to assess the association with clinicopathological parameters. Differences between three or more groups were investigated using one-way analysis of variance (ANOVA) with the post-hoc Tukey multiple comparison test (for parametric data) or Kruskal–Wallis test (for non-parametric distribution). Student t-test (parametric data) or Mann–Whitney test (non-parametric distribution) were used to evaluate the differences between two groups. Spearman’s correlation coefficient was calculated to examine the association between continuous variables. Univariate analysis was visualised using Kaplan–Meier curves and significance was assessed by log-rank test. Cox’s proportional hazard regression models were built for the multivariate survival analysis to adjust for confounding factors. P values were adjusted by using Bonferroni correction for multiple testing. For all tests, p value < 0.05 was considered as statistically significant. This study followed the reporting recommendations for tumour markers prognostic studies (REMARK) criteria.37
Results
In this study, we have applied the bespoke bioinformatics tools to identify the key genes associated with NP and this identified DKC1 as the target gene. Then, DKC1 was investigated at the transcriptomic, genetic and protein levels.
DKC1 mRNA expression and CN aberrations
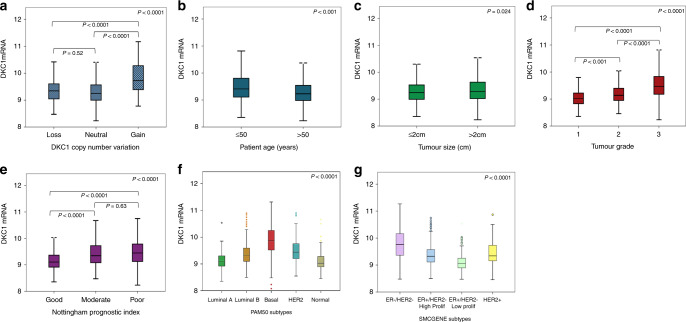
High DKC1 mRNA expression (log2 intensity >9.4) was observed in 709/1970 (36%) of the METABRIC cases. In all, 77/1980 (4%) of cases showed DKC1 CN gain, whereas 115/1980 (6%) showed a CN loss. A significant association was observed between DKC1 CN variation and DKC1 mRNA expression (p < 0.0001) (Fig. 1a).
Fig. 1.
DKC1 mRNA expression and its association with copy number variations, clinicopathological parameters and molecular subtypes a DKC1 and gene copy number variations b DKC1 and patient age. c DKC1 and tumour size. d DKC1 and tumour grade e DKC1 and Nottingham prognostic index. f DKC1 and PAM50 BC subtypes g DKC1 and SMCGENE subtypes in the METABRIC cohort using one-way analysis of variance with the post-hoc Tukey test.
DKC1 protein expression in breast cancer
Prior to IHC staining, the specificity of the antibody used was validated using WB performed on a DKC1 siRNA transfected BC cell line. A specific band at the predicted DKC1 molecular weight (58 kDa) was detected for proteins extracted from untransfected cells and those transfected with scrambled siRNA sequences. In addition, the DKC1 band intensity was significantly reduced with proteins extracted from DKC1 siRNAs transfected cells, confirming the specificity of the antibody utilised. A single band was observed at β-actin molecular weight (42 kDa) demonstrating the uniformity of loaded protein quantities (Fig. 2a).
Fig. 2.
This figure shows DKC1 immunohistochemistry in breast cancer. a A representative western blotting for DKC1 expression in cell lysate of MDA-MB-231 breast cancer cell line with the lanes (from left to right) of untransfected, transfected with scrambled siRNA and transfected with three different IDs ([ID1 represents ID s4110], [ID2 represents ID s4111] and [ID3 represents ID s4112]) of DKC1 siRNA b Negative DKC1 IHC expression c Positive DKC1 IHC nuclear expression in invasive breast cancer TMA cores and d Positive DKC1 IHC nucleolar expression.
DKC1 protein expression was observed in the nucleus and nucleoli of invasive BC cells, with expression levels varying from absent to strong (Fig. 2b–d). Strong concordance was observed between the two observers in DKC1 immuno-scoring in 10% of the cases (ICC = 0.864, p < 0.0001 for nuclear expression and ICC = 0.781, p < 0.001 for nucleolar expression). Moreover, second scoring of 20% of cases after 5 months washout period confirmed concordance ((ICC = 0.822, p < 0.0001 for nuclear expression and ICC = 0.804, p < 0.0001 for nucleolar expression). At the optimal DKC1 cut-off values (H-score 110 and 10, respectively), High DKC1 nuclear and nucleolar expression were observed in 574/942 (61%) and 153/942 (16%) of the informative tumours, respectively. There was a significant positive correlation between DKC1 nuclear and nucleolar expression (n = 429) (correlation coefficient = 0.143, p < 0.0001).
Correlation of DKC1 mRNA and protein expression with clinicopathological parameters
High DKC1 mRNA expression was significantly associated with younger patient age, larger tumour size, higher tumour grade and poorer NPI (p < 0.001, p = 0.024, p < 0.0001 and p < 0.0001) as shown in Fig. 1b-e, respectively. These associations were confirmed using the Breast Cancer Gene-Expression Miner v4.3 (Supplementary Fig. 1A-C)
In the TCGA BC dataset, similar associations, as described above, were observed with clinicopathological parameters. In particular, high DKC1 mRNA expression was significantly associated with high nucleolar score 34 (p < 0.0001, Supplementary Table 2).
High expression of DKC1 protein whether in the nucleus and/or nucleoli was associated with aggressive features of BC including higher tumour grade (p < 0.0001), larger tumour size (p = 0.04 only with nucleolar expression), higher mitotic scores (p < 0.0001), increased nuclear pleomorphism (p < 0.0001), higher scores of nucleolar prominence (p < 0.001), poor NPI (p < 0.0001) and the invasive ductal no special histological type (NST) (p < 0.0001), Table 1.
Table 1.
Clinicopathological associations of DKC1 protein nuclear and nucleolar expression in Nottingham cohort.
| Parameters | DKC1 nuclear protein expression | Adjusted p value | DKC1 nucleolar protein expression | Adjusted p value | ||||
|---|---|---|---|---|---|---|---|---|
| Number (%) | Mean rank | p value | Number (%) | Mean rank | p value | |||
| Patient age | ||||||||
| ≤50 | 348 (38) | 497.8 | 0.003 | 0.027 | 348 (38) | 477.3 | 0.09 | 0.27 |
| >50 | 580 (62) | 444.5 | 580 (62) | 456.8 | ||||
| Tumour size | ||||||||
| ≤2 cm | 575 (62) | 446.7 | 0.009 | 0.7 | 575 (62) | 452.7 | 0.01 | 0.04 |
| >2 cm | 353 (38) | 493.4 | 353 (38) | 483.7 | ||||
| Lymphovascular invasion | ||||||||
| Negative | 635 (68) | 447.9 | 0.005 | 0.012 | 635 (68) | 463.1 | 0.71 | 0.71 |
| Positive | 293 (32) | 500.4 | 293 (32) | 467.7 | ||||
| Axillary nodal stage | ||||||||
| Stage 1 | 572 (62) | 461.6 | 0.35 | 0.35 | 572 (62) | 464.7 | 0.1 | 0.2 |
| Stage 2 | 261 (28) | 457.4 | 261 (28) | 451.7 | ||||
| Stage 3 | 95 (10) | 501.1 | 95 (10) | 497.8 | ||||
| Nottingham prognostic index | ||||||||
| Good | 281 (31) | 379.3 | 3.5 × 10−9 | <0.0001 | 281 (31) | 416.8 | 0.000004 | <0.0001 |
| Moderate | 489 (53) | 492.1 | 489 (53) | 476.5 | ||||
| Poor | 150 (16) | 509.4 | 150 (16) | 490.2 | ||||
| IHC subtypes | ||||||||
| ER+/HER2− low proliferation | 390 (42) | 389.4 | 1.5 × 10−10 | <0.0001 | 390 (42) | 406.3 | 9.2 × 10−21 | <0.0001 |
| ER+/HER2− high proliferation | 272 (30) | 521.1 | 272 (30) | 453.1 | ||||
| HER2+ | 115 (13) | 504.8 | 115 (13) | 531.7 | ||||
| Triple negative | 140 (15) | 494.8 | 140 (15) | 557.6 | ||||
| Histological subtypes | ||||||||
| Lobular | 63 (7) | 383.9 | 4.9 × 10−7 | <0.0001 | 63 (7) | 393.7 | 7.5 × 10−7 | <0.0001 |
| Tubular | 128 (14) | 357.1 | 128 (14) | 409.1 | ||||
| No special type (NST) | 623 (67) | 497.4 | 623 (67) | 486.3 | ||||
| Mixed NST and lobular | 55 (6) | 440.9 | 55 (6) | 416.9 | ||||
| Other special type | 59 (6) | 458.4 | 59 (6) | 474.1 | ||||
| Tumour grade | ||||||||
| Grade 1 | 105 (11) | 269.1 | 9.8 × 10−20 | <0.0001 | 105 (11) | 402.6 | 7.8 × 10−17 | <0.0001 |
| Grade 2 | 392 (42) | 442.9 | 392 (42) | 421.8 | ||||
| Grade 3 | 432 (47) | 532.6 | 432 (47) | 519.4 | ||||
| Nuclear pleomorphism | ||||||||
| Score 1 | 10 (1) | 224.5 | 6.9 × 10−9 | <0.0001 | 10 (1) | 375.5 | 2.8 × 10−10 | <0.0001 |
| Score 2 | 239 (26) | 382.8 | 239 (26) | 396.3 | ||||
| Score 3 | 669 (73) | 490.4 | 669 (73) | 483.3 | ||||
| Mitosis | ||||||||
| Score 1 | 421 (46) | 385.2 | 4.9 × 10−16 | <0.0001 | 421 (46) | 411.1 | 6.6 × 10−15 | <0.0001 |
| Score 2 | 188 (20) | 476.1 | 188 (21) | 468.5 | ||||
| Score 3 | 308 (34) | 549.3 | 308 (33) | 518.7 | ||||
| Tubule formation | ||||||||
| Score 1 | 46 (5) | 296.7 | 0.000007 | 0.0006 | 46 (5) | 395.8 | 0.000012 | 0.0001 |
| Score 2 | 259 (28) | 436.9 | 259 (28) | 425.1 | ||||
| Score 3 | 613 (67) | 481.2 | 613 (67) | 478.9 | ||||
| Nucleoli | ||||||||
| Score 1 | 300 (39) | 346.2 | 0.0001 | 0.025 | 300 (39) | 333.7 | 7.8 × 10−33 | <0.0001 |
| Score 2 | 334 (43) | 411.6 | 334 (43) | 383.5 | ||||
| Score 3 | 143 (18) | 425.7 | 143 (18) | 517.9 | ||||
P values in bold means statistically significant.
DKC1 expression and other markers
The correlation of DKC1 mRNA with other relevant genes was investigated using the METABRIC and TCGA datasets. The genes were chosen based on published information, being either regulatory genes or those that share or support DKC1 biological function especially those primarily involved in the ribosomal biogenesis. DKC1 was positively associated with GAR1 (p < 0.0001), NOP10 (p < 0.001) and NHP2 (p < 0.0001). Moreover, there was a significant association between DKC1 MKI67 and the regulatory genes c-Myc (all p < 0.001) (Supplementary Table 3). High DKC1 mRNA expression was associated with those tumours, which showed TP53 mutations (p < 0.0001, Supplementary Table 4). Also, the statistical analysis showed a significant positive association of high DKC1 protein expression with high Ki67 (χ2 = 8.815, p = 0.003).
DKC1 mRNA and protein expression in BC molecular subtypes
At the transcriptomic level in METABRIC cohort, high DKC1 expression was significantly associated with hormone receptor negative (ER− and PR−), HER2+ tumours and TNBC (all p < 0.0001) as shown in Supplementary Table 4. Similar results were observed upon analysing the publicly available gene-expression data available on the Breast Cancer Gene-Expression Miner v4.3 online platform (Supplementary Fig. 1D–G) and TCGA datasets (Supplementary Table 4).
Regarding the association with the intrinsic PAM50 subtypes,38 high expression of DKC1 mRNA was observed in basal-like, HER2+ and Luminal B tumours (Fig. 1f, p < 0.0001). These findings were confirmed using the Breast Cancer Gene-Expression Miner v4.3 (Supplementary Fig. 1H). In the SCMGENE subtypes, high expression of DKC1 mRNA was observed in the ER−/HER2− cases followed by ER+/HER2− high proliferation class (p < 0.0001, Fig. 1g).
DKC1 nuclear and nucleolar protein expression was associated with negative ER status (p = 0.04 and p < 0.0001 respectively). Moreover, DKC1 nucleolar protein showed a significant correlation within HER2+ and triple negative (TN) tumours (both p < 0.0001), Table 2.
Table 2.
Association of DKC1 protein expression and the expression of other molecular biomarkers in the Nottingham cohort.
| Parameters | DKC1 nuclear protein expression | Adjusted p value | DKC1 nucleolar protein expression | Adjusted p value | ||||
|---|---|---|---|---|---|---|---|---|
| Number (%) | Mean rank | p value | Number (%) | Mean rank | p value | |||
| Oestrogen receptor | ||||||||
| Negative | 182 (20) | 511.1 | 0.01 | 0.04 | 182 (20) | 569.9 | 3.3 × 10−18 | <0.0001 |
| Positive | 748 (80) | 454.4 | 748 (80) | 440.1 | ||||
| Progesterone receptor | ||||||||
| Negative | 362 (39) | 462.9 | 0.92 | 0.92 | 362 (39) | 509.7 | 4.7 × 10−10 | <0.0001 |
| Positive | 565 (61) | 464.6 | 565 (61) | 434.7 | ||||
| HER2 status | ||||||||
| Negative | 815 (88) | 458.5 | 0.034 | 0.1 | 815 (88) | 455.1 | 0.000003 | <0.0001 |
| Positive | 115 (12) | 514.7 | 115 (12) | 539.4 | ||||
| Triple negative status | ||||||||
| Non-triple negative | 789 (85) | 458.5 | 0.058 | 0.12 | 789 (85) | 446.7 | 5.4 × 10−14 | <0.0001 |
| Triple negative | 141 (15) | 504.6 | 141 (15) | 570.9 | ||||
P values in bold means statistically significant.
There was a higher protein expression of DKC1 (nuclear & nucleolar) in the ER + high proliferative tumours than in the other molecular subtypes (p < 0.0001) as shown in Table 1.
Correlation of DKC1 mRNA and protein expression with patient outcome
In METABRIC cohort, high DKC1 mRNA expression was associated with poor BCSS in all cases (HR = 1.5, 95% CI = 1.3–1.8; p < 0.0001). Moreover, DKC1 mRNA expression was predictive of BCSS only in luminal B cases (HR = 1.5, 95%CI = 1.1–2.1; p = 0.015) as shown in Supplementary Fig. 2A–E. The relationship between high DKC1 mRNA expression and poor patient outcome in ER+ disease, but not ER− disease, was shown using the TCGA cohort (Supplementary Fig. 3).
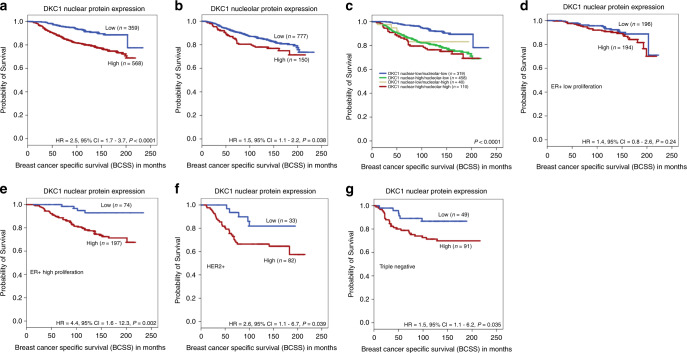
Both high DKC1 nuclear and nucleolar protein expressions, when assessed individually, were associated with poor outcome (HR = 2.5, 95%CI = 1.7–3.7; p < 0.0001 and HR = 1.5, 95%CI = 1.1–2.2; p = 0.038, respectively) Fig. 3a, b. When the analysis was limited to molecular subtypes, high expression of DKC1 nuclear protein was significantly associated with poor outcome in ER+ high proliferation tumours (HR = 4.4, 95% CI = 1.6–12.3; p = 0.002), HER2+ tumours (HR = 2.6, 95% CI = 1.1–6.7; p = 0.039) and TNBC (HR = 1.5, 95% CI = 1.1–6.2; p = 0.035) Fig. 3d–g. However, no significant association of DKC1 nucleolar protein expression was identified with outcome in BC subtypes (p > 0.05).
Fig. 3.
This figure shows the association between DKC1 protein expression and breast-cancer-specific survival (BCSS) as follows: a DKC1 nuclear expression and BCSS, b DKC1 nucleolar expression and BCSS c combinatorial DKC1 protein expression and BCSS d DKC1 and BCSS in oestrogen receptor (ER)+ low proliferation tumours e DKC1 and BCSS in (ER)+ high proliferation tumours f DKC1 and BCSS human epidermal growth factor receptor 2 positive (HER2+) tumours g DKC1 and BCSS of triple negative tumours in the studied cohort.
For further analysis, combinatorial DKC1 protein expression groups were created [i.e. low nuclear/low nucleolar, high nuclear/low nucleolar expression, low nuclear/high nucleolar and high nuclear/high nucleolar]. A significant difference in patient survival was observed between these four groups, where tumours with low nuclear and low nucleolar DKC1 expression showed the best outcome, whereas the tumours with high nuclear and high nucleolar expression showed the worst outcome (p < 0.0001) Fig. 3c.
The multivariate Cox-proportional models, including other prognostic covariates such as tumour size, grade and nodal stage, showed that DKC1 nuclear, combinatorial protein expression and DKC1 mRNA in the METABRIC dataset were independent predictors for poor prognosis in whole cases (p = 0.001, HR 2.037, 95% CI = 1.373–3.023, p = 0.003, HR 2.746, 95% CI = 1.484–5.083 and p = 0.006, HR 1.316, 95% CI = 1.097–1.579) as shown in Table 3 and Supplementary Table 5.
Table 3.
Multivariate Cox regression analysis results for predictors of breast-cancer-specific survival in Nottingham cohort.
| Models | Parameters | Hazard ratio (HR) | 95% confident interval (CI) | Significance P value | Adjusted P value | |
|---|---|---|---|---|---|---|
| Lower | Upper | |||||
| (A) | DKC1 nuclear protein expression | 2.037 | 1.373 | 3.023 | 0.000411 | 0.001 |
| Tumour size | 1.445 | 1.047 | 1.995 | 0.025 | 0.025 | |
| Nodal stage | 2.036 | 1.664 | 2.492 | 5.1 × 10−12 | <0.0001 | |
| Grade | 1.426 | 1.079 | 1.885 | 0.013 | 0.026 | |
| (B) | DKC1 nucleolar protein expression | 1.218 | 0.832 | 1.802 | 0.325 | 0.325 |
| Tumour size | 1.481 | 1.072 | 2.045 | 0.017 | 0.034 | |
| Nodal stage | 2.068 | 1.69 | 2.532 | 1.9 × 10−12 | <0.0001 | |
| Grade | 1.52 | 1.148 | 2.012 | 0.003 | 0.009 | |
| (C) | DKC1 combinatorial protein expression | 2.746 | 1.484 | 5.083 | 0.001 | 0.003 |
| Tumour size | 1.642 | 0.922 | 2.922 | 0.092 | 0.184 | |
| Nodal stage | 2.018 | 1.411 | 2.888 | 0.000123 | 0.0005 | |
| Grade | 1.037 | 0.637 | 1.69 | 0.883 | 0.883 | |
P values in bold means statistically significant.
In addition, high DKC1 nuclear protein expression was significantly associated with shorter distant metastases-free survival (DMFS) (HR = 2.1, 95% CI = 1.5–2.9; p < 0.0001). Likewise, combinatorial protein expression was associated with shorter DMFS (HR = 1.3, 95% CI = 1.1–1.4; p = 0.001) (Supplementary Fig. 4).
Discussion
In malignant tumours, the number and size of nucleoli are usually an indication of the rate of ribosome production, which is regarded as a major metabolic requisite for cell growth and proliferation.39,40 Nucleolar function and size are directly related to cell doubling time in cancer cells and quantitative morphometric evaluation of nucleolar size was considered as a cytological parameter of the tumour cells proliferation rate.4,41 In this study, we assessed NP in the TCGA breast cancer dataset as previously described,4 and used two greedy algorithms information theory and validated it using LASSO regression test22–24 to identify genes driving NP. This demonstrated that out of the 20,339 genes investigated, DKC1 was the top differentially expressed gene. Then DKC1 expression was evaluated at the proteomic, transcriptomic and genomic levels in large cohorts of invasive BC.
There were significant associations between high DKC1 mRNA in TCGA breast cancer dataset and DKC1 protein expression in Nottingham cohort with high nucleolar scoring.4 These findings supported our hypothesis that DKC1 plays a role in the nucleoli appearance and size likely through its mechanism in ribosomal biogenesis.
Our results also showed positive correlations between nuclear and nucleolar DKC1 protein expression in the breast tumour cells. It was reported that newly synthesised DKC1 initially localises to the nucleoplasm, followed by consecutive translocation to the nucleoli and the nuclear Cajal bodies. Usually, colocalisation of DKC1 on the nuclear Cajal bodies occurred only when it had already accumulated in the nucleoli.42 DKC1 is involved in the pseudouridination and processing of small spliceosomal RNAs through its binding to H/ACA small Cajal body RNAs.43
The current study confirms the significant association between the high expression of DKC1, at both protein and mRNA levels, and clinicopathological parameters characteristics of poor prognosis and with shorter survival; findings which are in-line with other studies.15,44,45 Some studies have also confirmed that DKC1 overexpression is involved in tumorigenic processes and has prognostic value in numerous types of cancer.19,21,46 The association between DKC1 mRNA and shorter survival was identified in both METABRIC and TCGA cohorts. Moreover, our analysis revealed that the prognostic significance of DKC1 protein and mRNA in BC was independent of other variables, demonstrating its potential clinical relevance in improving survival rate prediction.
When BC molecular subtypes were considered, the significant association between DKC1 protein and poor patient outcome was observed in the ER+ high proliferation (i.e. luminal B), HER2+ and TNBC classes whereas the high mRNA expression was only limited to the luminal B subtype. The most common type of BC constituting nearly 55–70% is the ER+/luminal tumour, and those tumours are variable in terms of recurrence, mortality rates and disease prognosis.47,48 These observations further endorse DKC1 functions in playing crucial roles in tumour growth and progression
DKC1 performs two fundamental functions for cell proliferation. First, DKC1 is a component of the H/ACA small nucleolar ribonucleoprotein particles (snoRNPs) involved the pseudouridylation of ribosomal RNA (rRNA) molecules and necessary for their processing. Second, it is required for telomerase activity by stabilising the telomerase RNA component.11 The faster the rate of cell proliferation, the higher the demand for protein production, which is compatible with increased rRNA synthesis.49,50 It has been reported that any dysregulation of DKC1 levels results in defects of ribosome biogenesis and a reduction of rRNA pseudouridylation, which in turn hinders the normal ribosome rRNA processing rate.51 For instance, Montanaro et al. have demonstrated that reduced DKC1 gene expression by specific RNA interference in BC cell lines resulted in a reduction of rRNA pseudouridylation, which subsequently effected the survival of proliferating cells.52 The role of DKC1 in mitosis was also confirmed,11 where dyskerin was identified as one of seventy genes which correlated with the development of aneuploidy. Alawi et al. have demonstrated that dyskerin expression peaks during G2/M and loss of dyskerin function has a widely disruptive effect on mitosis and triggers the spindle-assembly checkpoint.13 Our findings showed that high DKC1 expression was significantly associated with proliferation as assessed by Ki67 labelling index, which was also observed in other studies in BC52 hepatocellular carcinoma15 and prostate cancer,19 confirming that DKC1 is critical for mitotic progression and proliferation in these cancers.
DKC1 is the direct and conserved transcriptional target of c-Myc,53 which explains the strong correlation between its upregulation and active cell proliferation.54 In our study, we observed a significant positive association between DKC1 and c-Myc in mRNA expression. Previous studies have demonstrated that tumour oncogene c-Myc controls the transcription of DKC1 gene in addition to other proteins, which are required for rRNA processing.9,55 TP53 mutations were also highly prevalent in breast tumours with high DKC1 mRNA expression in METABRIC. There is mounting evidence that the usual increase of ribosome biogenesis (one of the main functions of DKC1) in cancer cells is the consequence of frequent alterations of two major tumour suppressors, TP53 and retinoblastoma (RB) genes.55 In addition, tumours with altered p53 and/or retinoblastoma protein pRb functions are characterised by significantly larger/more conspicuous nucleoli than tumours with normally functioning p53 and pRb.56
We further investigated the association of DKC1 expression with other H/ACA ribonucleoproteins, NHP2, NOP10 and GAR1, which play important roles in disease progression. DKC1, NHP2 and NOP10 form a core trimer that directly binds to H/ACA RNAs. The three proteins are interdependent with each other for stability and also regulate constancy of the bound RNAs.57 GAR1 binds only to DKC1 and is needed for a proper functioning of the H/ACA RNPs, but its absence does not reduce the stability of the rRNA.58 These findings confirmed the significant positive correlation between H/ACA ribonucleoproteins and DKC1 in BC.43 Alterations in DKC1 expression will potentially disrupt the biogenesis of H/ACA pathway and consequently affect ribosome synthesis and impair cell proliferation.
In the last decade, there have been a few attempts to construct DKC1 inhibitors. However, one in silico study successfully determined a small molecule inhibitor (Pyrazofurin) that exerted an ability to weaken the pharmacological and physiological activities of DKC1 through inhibiting its function in pseudouridylation of rRNA. Although Pyrazofurin failed to progress Phase 2 clinical trials; however, its chemical structure should continue to be exploited as a pharmacokinetic model to develop a potent, effective and safe DKC1 inhibitor that may eventually be used for BC highly expressing DKC1.59
A few limitations of this study findings are worth mentioning. On one hand, the semi-quantitative H-score method used to evaluate the immunohistochemical protein expression in the Nottingham cohort, might be regarded as having substantial subjectivity. This was addressed by double scoring a subset of the cancers to ensure the reproducibility and liability of the procedure. On the other hand, only one representative TMA core from each tumour tissue was arrayed and scored instead of considering replicates to express the tumour heterogeneity. This was due to the limited tissue resources in our biobank. However, to overcome the issue, 20 full face sections of randomly selected breast cancer cases were stained, prior to TMA application, with DKC1 antibody to assess the staining homogeneity and to evaluate the pertinence of using tissue microarrays (TMAs). These showed homogeneously distributed DKC1 expression, deeming the use of TMA to assess DKC1 an appropriate tissue platform, to mitigate the limited resources as well as testing the hypothesis a large BC cohort. Finally, the ‘weak’ but significant correlation between nuclear and nucleolar expression of DKC1 might also be regarded as a weakness point in the study. This might be due to the subjectivity of the method, which has been used in determining NP in our previous study.4
Conclusion
This study reveals a significant correlation between the morphological features of NP and an underlying molecular and protein description (DKC1). The importance of DKC1 was demonstrated in three independent datasets where each dataset contributed to the description of DKC1 from different perspectives. DKC1 is significantly associated with high nucleolar score and with poor prognostic characteristics and poor patients’ outcome. Overexpression of DKC1 appears to play a role in the proliferation and progression of the aggressive BC subtypes including the luminal B, TNBC and HER2 molecular subtypes. Findings here encourage further investigation of DKC1 as it might relate to guiding targeted therapies and to evaluate its role in response to chemotherapy.
Supplementary information
Acknowledgements
We thank Breast Cancer Now and PathLAKE project for supporting this study. These new Centres are supported by the Data to Early Diagnosis and Precision Medicine strand of the government’s Industrial Strategy Challenge, managed and delivered by UK Research and Innovation (UKRI).
Author contributions
K.E.: scored all the cases, took the lead in writing the manuscript, data analysis and interpretation. O.M. and M.A.: helped in double scoring, data interpretation and reviewing the article. A.H., H.G. and M.I.A.: contributed in data analysis, interpretation, writing and reviewing the article. A.G. and L.D.: contributed in data analysis, study design and reviewing the article. E.R.: conceived and planned the presented idea, data interpretation and reviewing the article. All authors reviewed and approved the final version of the manuscript. All authors declared their contribution(s) to the study
Ethics approval and consent to participate
This work obtained ethics approval by the North West—Greater Manchester Central Research Ethics Committee under the title; Nottingham Health Science Biobank (NHSB), reference number 15/NW/0685. All patients included were consented to participate in the study and to use their materials in research. All samples from Nottingham used in this study were pseudo-anonymised and stored in compliance with the UK Human Tissue Act. The study was performed in accordance with the Declaration of Helsinki.
Data availability
The authors confirm the data that have been used in this work are available on reasonable request.
Competing interests
The authors declare no competing interests.
Funding information
This research was supported and funded by Damietta University and Egyptian Ministry of Higher education and Scientific Research.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information is available for this paper at 10.1038/s41416-020-01045-7.
References
- 1.Parada H, Jr, Sun X, Tse CK, Olshan AF, Troester MA. Lifestyle patterns and survival following breast cancer in the Carolina Breast Cancer Study. Epidemiol. (Camb., Mass) 2019;30:83–92. doi: 10.1097/EDE.0000000000000933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andisha, N. M., McMillan, D. C., Gujam, F. J. A., Roseweir, A. & Edwards, J. The relationship between phosphorylation status of focal adhesion kinases, molecular subtypes, tumour microenvironment and survival in patients with primary operable ductal breast cancer. Cell. Signal.10.1016/j.cellsig.2019.04.006 (2019). [DOI] [PubMed]
- 3.Rakha EA, El-Sayed ME, Lee AH, Elston CW, Grainge MJ, Hodi Z, et al. Prognostic significance of Nottingham histologic grade in invasive breast carcinoma. J. Clin. Oncol. 2008;26:3153–3158. doi: 10.1200/JCO.2007.15.5986. [DOI] [PubMed] [Google Scholar]
- 4.Elsharawy, K. A., Toss, M. S., Abuelmaaty, S. R., Ball, G., Green, A. R., Aleskandarany, M. A. et al. Prognostic significance of nucleolar assessment in invasive breast cancer. Histopathology10.1111/his.14036 (2019). [DOI] [PubMed]
- 5.Lam YW, Trinkle-Mulcahy L, Lamond AI. The nucleolus. J. Cell Sci. 2005;118:1335. doi: 10.1242/jcs.01736. [DOI] [PubMed] [Google Scholar]
- 6.Montanaro L, Trere D, Derenzini M. Nucleolus, ribosomes, and cancer. Am. J. Pathol. 2008;173:301–310. doi: 10.2353/ajpath.2008.070752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bywater MJ, Poortinga G, Sanij E, Hein N, Peck A, Cullinane C, et al. Inhibition of RNA polymerase I as a therapeutic strategy to promote cancer-specific activation of p53. Cancer cell. 2012;22:51–65. doi: 10.1016/j.ccr.2012.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Derenzini M, Trerè D, Pession A, Govoni M, Sirri V, Chieco P. Nucleolar size indicates the rapidity of cell proliferation in cancer tissues. J. Pathol. 2000;191:181–186. doi: 10.1002/(SICI)1096-9896(200006)191:2<181::AID-PATH607>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 9.van Riggelen J, Yetil A, Felsher DW. MYC as a regulator of ribosome biogenesis and protein synthesis. Nat. Rev. Cancer. 2010;10:301–309. doi: 10.1038/nrc2819. [DOI] [PubMed] [Google Scholar]
- 10.Heiss NS, Knight SW, Vulliamy TJ, Klauck SM, Wiemann S, Mason PJ, et al. X-linked dyskeratosis congenita is caused by mutations in a highly conserved gene with putative nucleolar functions. Nat. Genet. 1998;19:32–38. doi: 10.1038/ng0598-32. [DOI] [PubMed] [Google Scholar]
- 11.Alawi F, Lin P. Dyskerin is required for tumor cell growth through mechanisms that are independent of its role in telomerase and only partially related to its function in precursor rRNA processing. Mol. Carcinog. 2011;50:334–345. doi: 10.1002/mc.20715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin P, Mobasher ME, Alawi F. Acute dyskerin depletion triggers cellular senescence and renders osteosarcoma cells resistant to genotoxic stress-induced apoptosis. Biochem. Biophys. Res. Commun. 2014;446:1268–1275. doi: 10.1016/j.bbrc.2014.03.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alawi F, Lin P. Dyskerin localizes to the mitotic apparatus and is required for orderly mitosis in human cells. PLoS ONE. 2013;8:e80805. doi: 10.1371/journal.pone.0080805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fong, Y. W., Ho, J. J., Inouye, C. & Tjian, R. The dyskerin ribonucleoprotein complex as an OCT4/SOX2 coactivator in embryonic stem cells. eLife 2014 10.7554/eLife.03573 (2014). [DOI] [PMC free article] [PubMed]
- 15.Liu B, Zhang J, Huang C, Liu H. Dyskerin overexpression in human hepatocellular carcinoma is associated with advanced clinical stage and poor patient prognosis. PLoS ONE. 2012;7:e43147. doi: 10.1371/journal.pone.0043147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Westermann F, Henrich K-O, Wei JS, Lutz W, Fischer M, König R, et al. High Skp2 expression characterizes high-risk neuroblastomas independent of MYCN status. Clin. Cancer Res. 2007;13:4695–4703. doi: 10.1158/1078-0432.CCR-06-2818. [DOI] [PubMed] [Google Scholar]
- 17.Piva R, Pellegrino E, Mattioli M, Agnelli L, Lombardi L, Boccalatte F, et al. Functional validation of the anaplastic lymphoma kinase signature identifies CEBPB and Bcl2A1 as critical target genes. J. Clin. Invest. 2006;116:3171–3182. doi: 10.1172/JCI29401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McDonald SL, Edington HD, Kirkwood JM, Becker D. Expression analysis of genes identified by molecular profiling of VGP melanomas and MGP melanoma-positive lymph nodes. Cancer Biol. Ther. 2004;3:110–120. doi: 10.4161/cbt.3.1.662. [DOI] [PubMed] [Google Scholar]
- 19.Sieron P, Hader C, Hatina J, Engers R, Wlazlinski A, Müller M, et al. DKC1 overexpression associated with prostate cancer progression. Br. J. Cancer (Mol. Diagnostics) 2009;101:1410. doi: 10.1038/sj.bjc.6605299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Witkowska A, Gumprecht J, Glogowska-Ligus J, Wystrychowski G, Owczarek A, Stachowicz M, et al. Expression profile of significant immortalization genes in colon cancer. Int. J. Mol. Med. 2010;25:321–329. doi: 10.3892/ijmm_00000348. [DOI] [PubMed] [Google Scholar]
- 21.Schaner ME, Ross DT, Ciaravino G, Sorlie T, Troyanskaya O, Diehn M, et al. Gene expression patterns in ovarian carcinomas. Mol. Biol. Cell. 2003;14:4376–4386. doi: 10.1091/mbc.E03-05-0279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brown G, Pocock A, Zhao M-J, Luján M. Conditional likelihood maximisation: a unifying framework for information theoretic feature selection. J. Mach. Learn Res. 2012;13:27–66. [Google Scholar]
- 23.Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J. Stat. Softw. 2010;33:1–22. [PMC free article] [PubMed] [Google Scholar]
- 24.Tibshirani R, Bien J, Friedman J, Hastie T, Simon N, Taylor J, et al. Strong rules for discarding predictors in lasso-type problems. J. R. Stat. Soc. 2012;74:245–266. doi: 10.1111/j.1467-9868.2011.01004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ciriello G, Gatza ML, Beck AH, Wilkerson MD, Rhie SK, Pastore A, et al. Comprehensive molecular portraits of invasive lobular breast cancer. Cell. 2015;163:506–519. doi: 10.1016/j.cell.2015.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, et al. The genomic and transcriptomic architecture of 2000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aleskandarany MA, Abduljabbar R, Ashankyty I, Elmouna A, Jerjees D, Ali S, et al. Prognostic significance of androgen receptor expression in invasive breast cancer: transcriptomic and protein expression analysis. Breast Cancer Res. Treat. 2016;159:215–227. doi: 10.1007/s10549-016-3934-5. [DOI] [PubMed] [Google Scholar]
- 28.Rakha EA, Agarwal D, Green AR, Ashankyty I, Ellis IO, Ball G, et al. Prognostic stratification of oestrogen receptor-positive HER2-negative lymph node-negative class of breast cancer. Histopathology. 2017;70:622–631. doi: 10.1111/his.13108. [DOI] [PubMed] [Google Scholar]
- 29.Rakha EA, Elsheikh SE, Aleskandarany MA, Habashi HO, Green AR, Powe DG, et al. Triple-negative breast cancer: distinguishing between basal and nonbasal subtypes. Clin. Cancer Res. 2009;15:2302–2310. doi: 10.1158/1078-0432.CCR-08-2132. [DOI] [PubMed] [Google Scholar]
- 30.Muftah AA, Aleskandarany MA, Al-Kaabi MM, Sonbul SN, Diez-Rodriguez M, Nolan CC, et al. Ki67 expression in invasive breast cancer: the use of tissue microarrays compared with whole tissue sections. Breast Cancer Res. Treat. 2017;164:341–348. doi: 10.1007/s10549-017-4270-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Elston CW, Ellis IO. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology. 1991;19:403–410. doi: 10.1111/j.1365-2559.1991.tb00229.x. [DOI] [PubMed] [Google Scholar]
- 32.Senkus E, Kyriakides S, Ohno S, Penault-Llorca F, Poortmans P, Rutgers E, et al. Primary breast cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2015;26:v8–v30. doi: 10.1093/annonc/mdt284. [DOI] [PubMed] [Google Scholar]
- 33.El Ansari R, Craze ML, Miligy I, Diez-Rodriguez M, Nolan CC, Ellis IO, et al. The amino acid transporter SLC7A5 confers a poor prognosis in the highly proliferative breast cancer subtypes and is a key therapeutic target in luminal B tumours. Breast Cancer Res. 2018;20:21. doi: 10.1186/s13058-018-0946-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abd El-Rehim DM, Ball G, Pinder SE, Rakha E, Paish C, Robertson JF, et al. High-throughput protein expression analysis using tissue microarray technology of a large well-characterised series identifies biologically distinct classes of breast cancer confirming recent cDNA expression analyses. Int. J. Cancer. 2005;116:340–350. doi: 10.1002/ijc.21004. [DOI] [PubMed] [Google Scholar]
- 35.McCarty KS, Jr, KS McCarty., Sr Histochemical approaches to steroid receptor analyses. Semin. Diagn. Pathol. 1984;1:297–308. [PubMed] [Google Scholar]
- 36.Camp, R. L., Dolled-Filhart, M. & Rimm, D. L. X-tile. A new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin. Cancer Res. 10, 7252–7259 (2004). [DOI] [PubMed]
- 37.McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM, et al. REporting recommendations for tumour MARKer prognostic studies (REMARK) Br. J. Cancer. 2005;93:387–391. doi: 10.1038/sj.bjc.6602678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J. Clin. Oncol. 2009;27:1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fischer AH, Bardarov S, Jr, Jiang Z. Molecular aspects of diagnostic nucleolar and nuclear envelope changes in prostate cancer. J. Cell. Biochem. 2004;91:170–184. doi: 10.1002/jcb.10735. [DOI] [PubMed] [Google Scholar]
- 40.Hernandez-Verdun D. The nucleolus: a model for the organization of nuclear functions. Histochem. Cell Biol. 2006;126:135–148. doi: 10.1007/s00418-006-0212-3. [DOI] [PubMed] [Google Scholar]
- 41.Derenzini M, Trere D, Pession A, Montanaro L, Sirri V, Ochs RL. Nucleolar function and size in cancer cells. Am. J. Pathol. 1998;152:1291–1297. [PMC free article] [PubMed] [Google Scholar]
- 42.Heiss NS, Girod A, Salowsky R, Wiemann S, Pepperkok R, Poustka A. Dyskerin localizes to the nucleolus and its mislocalization is unlikely to play a role in the pathogenesis of Dyskeratosis congenita. Hum. Mol. Genet. 1999;8:2515–2524. doi: 10.1093/hmg/8.13.2515. [DOI] [PubMed] [Google Scholar]
- 43.Ye K. H/ACA guide RNAs, proteins and complexes. Curr. Opin. Struct. Biol. 2007;17:287–292. doi: 10.1016/j.sbi.2007.05.012. [DOI] [PubMed] [Google Scholar]
- 44.Zhang M, Pan Y, Jiang R, Hou P, Shan H, Chen F, et al. DKC1 serves as a potential prognostic biomarker for human clear cell renal cell carcinoma and promotes its proliferation, migration and invasion via the NFkappaB pathway. Oncol. Rep. 2018;40:968–978. doi: 10.3892/or.2018.6484. [DOI] [PubMed] [Google Scholar]
- 45.von Stedingk K, Koster J, Piqueras M, Noguera R, Navarro S, Påhlman S, et al. snoRNPs regulate telomerase activity in neuroblastoma and are associated with poor prognosis. Transl. Oncol. 2013;6:447–457. doi: 10.1593/tlo.13112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Poncet D, Belleville A, t’kint de Roodenbeke C, Roborel de Climens A, Ben Simon E, Merle-Beral H, et al. Changes in the expression of telomere maintenance genes suggest global telomere dysfunction in B-chronic lymphocytic leukemia. Blood. 2008;111:2388–2391. doi: 10.1182/blood-2007-09-111245. [DOI] [PubMed] [Google Scholar]
- 47.Dawson SJ, Rueda OM, Aparicio S, Caldas C. A new genome-driven integrated classification of breast cancer and its implications. EMBO J. 2013;32:617–628. doi: 10.1038/emboj.2013.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rakha EA, El-Sayed ME, Green AR, Paish EC, Powe DG, Gee J, et al. Biologic and clinical characteristics of breast cancer with single hormone receptor positive phenotype. J. Clin. Oncol. 2007;25:4772–4778. doi: 10.1200/JCO.2007.12.2747. [DOI] [PubMed] [Google Scholar]
- 49.Mochizuki Y, He J, Kulkarni S, Bessler M, Mason PJ. Mouse dyskerin mutations affect accumulation of telomerase RNA and small nucleolar RNA, telomerase activity, and ribosomal RNA processing. Proc. Natl Acad. Sci. USA. 2004;101:10756–10761. doi: 10.1073/pnas.0402560101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Filipowicz W, Pogacic V. Biogenesis of small nucleolar ribonucleoproteins. Curr. Opin. Cell Biol. 2002;14:319–327. doi: 10.1016/s0955-0674(02)00334-4. [DOI] [PubMed] [Google Scholar]
- 51.Ruggero D, Grisendi S, Piazza F, Rego E, Mari F, Rao PH, et al. Dyskeratosis congenita and cancer in mice deficient in ribosomal RNA modification. Science. 2003;299:259–262. doi: 10.1126/science.1079447. [DOI] [PubMed] [Google Scholar]
- 52.Montanaro L, Brigotti M, Clohessy J, Barbieri S, Ceccarelli C, Santini D, et al. Dyskerin expression influences the level of ribosomal RNA pseudo-uridylation and telomerase RNA component in human breast cancer. J. Pathol. 2006;210:10–18. doi: 10.1002/path.2023. [DOI] [PubMed] [Google Scholar]
- 53.Alawi F, Lee MN. DKC1 is a direct and conserved transcriptional target of c-MYC. Biochem. Biophys. Res. Commun. 2007;362:893–898. doi: 10.1016/j.bbrc.2007.08.071. [DOI] [PubMed] [Google Scholar]
- 54.Alawi F, Lin P, Ziober B, Patel R. Correlation of dyskerin expression with active proliferation independent of telomerase. Head Neck. 2011;33:1041–1051. doi: 10.1002/hed.21579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Penzo M, Montanaro L, Treré D, Derenzini M. The ribosome biogenesis-cancer connection. Cells. 2019;8:55. doi: 10.3390/cells8010055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Trere D, Ceccarelli C, Montanaro L, Tosti E, Derenzini M. Nucleolar size and activity are related to pRb and p53 status in human breast cancer. J. Histochem. Cytochem. 2004;52:1601–1607. doi: 10.1369/jhc.4A6454.2004. [DOI] [PubMed] [Google Scholar]
- 57.Grozdanov PN, Roy S, Kittur N, Meier UT. SHQ1 is required prior to NAF1 for assembly of H/ACA small nucleolar and telomerase RNPs. RNA. 2009;15:1188–1197. doi: 10.1261/rna.1532109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Darzacq X, Kittur N, Roy S, Shav-Tal Y, Singer RH, Meier UT. Stepwise RNP assembly at the site of H/ACA RNA transcription in human cells. J. Cell Biol. 2006;173:207–218. doi: 10.1083/jcb.200601105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rocchi L, Barbosa AJ, Onofrillo C, Del Rio A, Montanaro L. Inhibition of human dyskerin as a new approach to target ribosome biogenesis. PLoS ONE. 2014;9:e101971. doi: 10.1371/journal.pone.0101971. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors confirm the data that have been used in this work are available on reasonable request.