Highlights
-
•
A protease from a psychrotolerant yeast was characterized.
-
•
Protease production was dependent on temperature and medium composition.
-
•
Mass spectrometry analysis indicated that the protein belongs to the pepsin family.
-
•
We propose that the enzyme reported here could be Rodothorulapepsin.
Keywords: Antarctic yeast, Aspartic protease, Rodothorulapepsin
Abstract
Enzymes from cold-adapted microorganisms are of high interest to industries due to their high activity at low and mild temperatures, which makes them suitable for their use in several processes that either require a supply of exogenous energy or involve the use of heat labile products. In this work, the protease production by the strain Rhodotorula mucilaginosa CBMAI 1528, previously isolated from the Antarctic continent, was optimized, and the purified enzyme analyzed. It was found that protease production was dependent on culture medium composition and growth temperature, being 20 °C and a culture medium containing both glucose and casein peptone (20 and 10 g/L, respectively) the optimal growing conditions in batch as well as in bioreactor. Moreover, mass spectrometry analysis revealed that the enzyme under study has a 100 % sequence identity with the deduced amino acid sequence of a putative aspartic protease from Rhodotorula sp. JG-1b (protein ID: KWU42276.1). This result was confirmed by the decrease of 95 % proteolytic activity by pepstatin A, a specific inhibitor of aspartic proteases. We propose that the enzyme reported here could be Rodothorulapepsin, a protein characterized in 1972 that did not have an associated sequence to date and has been classified as an orphan enzyme.
1. Introduction
In spite of severe environmental conditions, the Antarctic continent is inhabited by a highly diverse microbial population [1,2]. One of the most important challenges faced by the biota present there is to adapt to low temperatures by maintaining the rates of chemical reactions involved in the main cellular processes [3]. Some adaptations to these conditions are achieved by synthesizing cold active enzymes with more mobile or flexible catalytic centers than their mesophilic homologues, which allows them to have a higher specific activity at low temperatures and lower thermal stability [4]. Due to the possibility of reducing energy consumption, these enzymes are attractive for their use in industrial processes that involve the use of heat labile products and for those that require a supply of exogenous energy [5].
Antarctica is inhabited by a diverse group of fungi, of which the most abundant members are those of the genera Cryptococcus, Penicillium and Rhodotorula. These cold adapted cosmopolitan species were found mainly in association with seaweeds, due to the availability of organic substrates [6] and were found to be adapted to unfavorable conditions such as low temperatures, osmotic stress and high UV radiation [7,8].
Among the different species of the genus Rhodotorula, R. mucilaginosa has been receiving increasing attention due to its capability to grow in extreme ecosystems, and to produce valuable biotechnological products such as biosurfactants [9], proteases [10], lipases [11], carotenoids [12] and high levels of unsaturated fatty acids [13]. Moreover, it was found that R. mucilaginosa is able to remediate some specific contaminants [14], and that it represents an emergent opportunistic pathogen for humans and animals [15]. Altogether, this information indicates the importance of studying the biochemical properties of this yeast in order to be implemented in biotechnological processes.
Previously, Duarte et al. [16] recovered 97 yeast strains from marine and terrestrial Antarctic environments and analyzed their ability to produce lipases, xylanases and proteases. From these screenings, R. mucilaginosa CBMAI 1528 was selected as the main producer of extracellular proteases. Moreover, in previous assays, the protease secreted proved to be active at temperatures between 15 °C and 60 °C, with an optimal catalytic activity at pH 5.0 and 50 °C, and highly stable in the presence of NaCl [10]. Here, we report the optimization of protease production both in batch and bioreactor. In order to get a deeper characterization of the enzyme, we purified and then analyzed its sequence by mass spectrometry, which allowed us to identify it as an aspartic protease belonging to the peptidase A1 subfamily. We propose that this extracellular protease could be the Rodothorulapepsin characterized in 1972, which, until now, it was classified as one orphan enzyme, as it had an EC number without a related amino acid sequence. [17,18]. This is the first report that identifies the family and amino acid sequence of an extracellular protease from the genus Rhodotorula. To date, a large part of well- defined enzyme activities are still lacking an associated protein sequence, despite the immense progress of genomics and bioinformatics methods for function prediction [19]. The identification of the protease described in this work contributes to narrow the gap between knowledge of biochemical function and sequence information.
2. Materials and methods
2.1. Microorganism and growth conditions
The yeast under study (CBMAI 1528) was obtained during an expedition to Antarctica in the austral summer (2009 and 2010) by the Brazilian Antarctic Program team. The strain was selected among 97 isolated yeasts based on the highest extracellular proteolytic activity in milk agar Sabouraud plates (5 g/L tryptone from casein, 5 g/L peptone from animal tissue, 20 g/L dextrose) and based on the similarity of partial 26S rDNA gene, it was identified as R. mucilaginosa [16]. The yeast strain was stored in Sabouraud dextrose medium with 20 % (v/v) glycerol at −70 °C.
2.2. Effect of growth temperature on biomass and protease production
To evaluate the effect of growth temperature on biomass and extracellular protease production, 250 mL flasks containing 50 mL of fresh Sabouraud dextrose medium were inoculated with a 10−2 dilution of an overnight culture of R. mucilaginosa CBMAI 1528 grown at 20 °C. Each broth was incubated at 250 rpm under different temperatures (10, 15, 20, 25, 30 and 35 °C), and samples were taken every 24 h.
Biomass concentration was determined gravimetrically. Culture samples (1–10 mL) were filtered through membranes with 0.45 μm pore size (Sartorius, Goettingen, Germany) and then cells were dried at 200 mm Hg and 60 °C for 48 h until constant weight. Biomass concentration was expressed in grams of dry yeast per L of cultivation medium (g/L). Protease activity was determined on the extracellular medium of each sample by the azocasein method, as detailed below, and expressed as U/mL. Extreme care was taken to prevent volume losses. Data represent the mean values ± standard deviation of three independent measurements.
2.3. Protein quantification and protease assay
Total protein concentration was determined by Bradford method [20] using the Bio-Rad protein assay (Bio-Rad Laboratories, Munich, Germany) with Bovine Serum Albumin (BSA) as a standard. The crude extract and the purified enzyme were assayed for proteolytic activity using azocasein (Sigma, St. Louis, MO, USA) as a substrate [21], under previously selected optimal conditions [10].
2.4. Optimization of protease production
To assess the impact of the nitrogen source on the extracellular protease production by R. mucilaginosa CBMAI 1528, several combinations of different organic nitrogen sources (casein tryptone, peptone from animal tissue, yeast extract, casamino acids and peptone from gelatin) were tested at two different concentrations (0 or 2.5 g/L) according to the 25−1 fractional factorial design depicted in Table 1. The above organic nitrogen sources were selected because they are common components of culture media. Moreover, 0–2.5 g/L of each component in the different culture media was used in order to obtain a final concentration of organic nitrogen sources comparable to that of the standard medium (Saboraud).
Table 1.
Factor levels of the 25−1 fractional experimental design used for the optimization of protease production by R. mucilaginosa CBMAI 1528 using different organic nitrogen sources.
| Variable | Symbol parameter | Level |
|
|---|---|---|---|
| Low (-1) | High (+1) | ||
| Casein tryptone (g/L) | X1 | 0 | 2.5 |
| Peptone from animal tissue (g/L) | X2 | 0 | 2.5 |
| Yeast extract (g/L) | X3 | 0 | 2.5 |
| Casamino acids (g/L) | X4 | 0 | 2.5 |
| Peptone from gelatin (g/L) | X5 | 0 | 2.5 |
For all the assays, the growth temperature was 20 °C, and glucose (20 g/L) was used as the carbon source. Total protease activity (U/mL) and biomass concentration (g/L) were determined after 48 h of growth. The results were expressed as mean values ± standard deviation of three independent measurements and statistically analyzed using the software Statistica version 10 (StatSoft, Tulsa, OK, USA). Protease activity and biomass concentration at the end of each experiment were compared with the results obtained with the Sabouraud dextrose medium under the same conditions.
2.5. Elemental analysis
Elemental analyses of R. mucilaginosa biomass, Sabouraud-Dextrose medium, casein tryptone and yeast extract were performed using a CHNS analyzer, model 2400 series ii (Perkin Elmer, Waltham, MA, USA). Each sample (about 0.1 g) was packed in a silver boat and placed in the analyzer where it was turned to elements by heating at 1,150 °C, further oxidized and analyzed.
2.6. Growth experiments and bioreactor operation
Batch cultures were carried out in a lab scale 3 L bioreactor (Bioflo115 New Brunswick Scientific, Edison, NJ, USA) with 1.5 L working volume. From previous experiments, a positive correlation between airflow rate, agitation and protease production was observed, with a maximum protease production at 2.0 vvm and 500 rpm (data not shown). A kLa of 92 h−1 was determined under these conditions according to Potumarthi and Jetty [22]. After inoculation with 1.0 g/L biomass coming from a 18 h pre-inoculum culture, the bioreactor was operated at 20 °C in medium containing 20 g/L glucose and 10 g/L casein tryptone. Samples were collected periodically to monitor biomass and glucose concentrations as well as protease activity. Glucose concentration in the medium was determined colourimetrically by measuring the absorbance at 500 nm according to the glucose oxidase method (Laborclin, Pinhais, PR, Brazil).
2.7. Determination of maximum growth rate, yield coefficient and volumetric productivity
The maximum growth rate (μmax) and generation time were determined after growth curves preparation. Biomass concentration and glucose consumption were used to calculate the biomass/glucose yield coefficient (YX/S) expressed as g/g. The volumetric productivities of biomass (QX) and protease (QP) were calculated by dividing their concentrations by the corresponding culture time.
2.8. Protease purification, in-gel digestion, mass spectrometry, and database search
To purify the protease, 1.5 L of culture were centrifuged at 8,000 xg for 10 min, and the supernatant was filtered through a membrane with 0.47 mm-pore size to completely remove cells. Ammonium sulphate was added to the supernatant to reach 25 % (v/w) saturation. After centrifugation at 10,000 xg for 15 min at 4 °C, the precipitate was removed, and ammonium sulphate was added to the supernatant to reach 70 % (v/w) saturation. After centrifugation under the same conditions, the precipitate was dissolved in 50 mL of 50 mM sodium acetate buffer (pH 5.0) and dialyzed thoroughly against this buffer overnight. After that, the solution was applied to a CM-Sepharose Fast Flow column (GE Healthcare, Uppsala, Sweden), and purified as previously described [10].
The purified enzyme suspended in 50 mM sodium acetate buffer containing 2 % (w/v) sodium dodecyl sulfate (SDS), 5 % (v/v) β-mercaptoethanol, and 0.002 % (w/v) bromophenol blue was boiled at 100 °C for 5 min and then subjected to SDS-PAGE according to Laemmly [23]. The band was excised from the gel, transferred to 1.5 mL microcentrifuge tube and sent to CEBIQUIEM facilities (Facultad de Ciencias Exactas y Naturales, Universidad de Buenos Aires, Argentina) for analyses. The sample was reduced (20 mM DTT, 45 min, 56 °C), alkylated (50 mM iodoacetamide, 45 min, in the dark at room temperature) and digested overnight with trypsin (12 h, 37 °C). Peptides were extracted with acetonitrile and the digested mixture was lyophilized, resuspended in 10 μL formic acid 0.1 % (w/v), desalted and analyzed by nano-high performance liquid chromatography coupled with electrospray ionization mass spectrometry Q Exactive Plus Orbitrap LC-MS/MS System, Thermo Scientific technology (Waltham, MA, USA). The collected data were analyzed with Proteome Discoverer™ software (version 1.4).
2.9. Inhibition assay
Purified protease was preincubated for 15 min at 20 °C with different protease inhibitors, namely 1.0 mM 4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride (AEBSF), 5.0 mg/mL 6-aminohexanoic acid, 0.1 mM Antipain, 0.1 mM Aprotinin, 4.0 mM benzamidine HCl, 0.04 mM Bestatin, 0.1 mM Chymostatin, 10 μM E-64, 1.0 mM EDTA, 1.0 mM N-ethylmaleimide, 0.1 mM Leupeptin, 1.0 μg/mL Pepstatin A, 10 μM Phosphoramidon, and Trypsin Inhibitor in 1:1 stoichiometric binding. Purified protease without any inhibitor was included as control. After that, the proteolytic activity of the reaction mixture was measured using the azocasein assay as described above.
3. Results and discussion
Rhodotorula mucilaginosa CBMAI 1528 was previously reported to produce the highest amounts of extracellular proteases among 97 isolated yeasts from Antarctica [16]. The possibility of reducing the cost of the culture medium by using by-products of the food industry had previously been investigated [24]. In order to scale-up the microbial mass and protease production we optimized the temperature and media conditions. Then to obtain further characterization of the enzyme the sequence was analyzed by means of mass spectrometry.
3.1. Cell growth and protease production at different temperatures
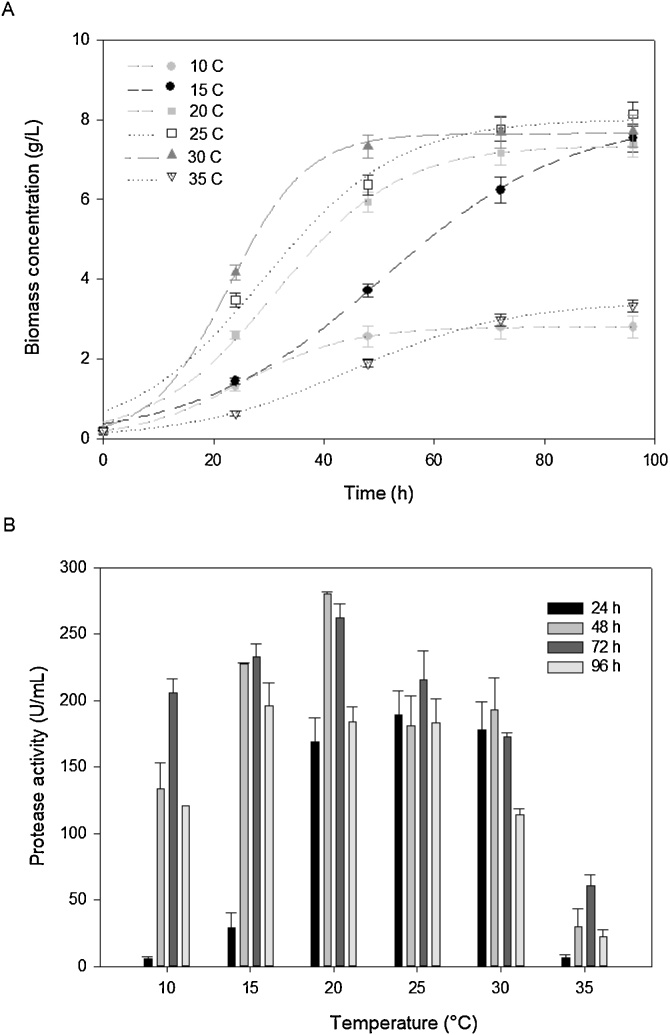
To assess the effect of temperature (T) on cell growth and extracellular protease production by R. mucilaginosa CBMAI 1528, cells were batch-wise cultured at temperatures from 10 to 35 °C in 250 mL flaks with Sabouraud dextrose medium. Samples were collected every 24 h up to 96 h. As shown in Fig. 1A, this cold adapted strain, which had previously been isolated from an Antarctic marine algae, was able to grow slowly even at temperatures as low as 10 °C or as high as 35 °C.However, it reached a mean maximum biomass concentration of 7.5 g/L at between 20 and 30 °C after 72 h, which corresponds to a volumetric biomass productivity (QX) of about 0.1 g/L.h.Furthermore, protease production was also dependent on growth temperature. In fact, the extracellular protease activity reached a maximum value of 280 ± 1.7 U/mL at 20 °C after 48 h of growth (Fig. 1B). Based on these results, 20 °C was selected as the optimal temperature for yeast growth in subsequent experiments.
Fig. 1.
Effect of temperature on A) R. mucilaginosa CBMAI 1528 dry biomass concentration (g/L) and B) extracellular protease activity (U/mL). The strain was cultured at 10, 15, 20, 25, 30 and 35 °C in 250 mL Erlenmeyer flaks with Sabouraud dextrose medium, and samples were collected every 24 h to perform both determinations. The data are expressed as mean values ± standard deviations of three independent assays.
According to Morita [25], psychrophilic taxa show an optimum growth at 15 °C or lower and cannot grow above 20 °C, while psychrotolerant taxa typically have maximum growth temperature higher than 20 °C, and still keep the ability to grow at low temperatures. Therefore, R. mucilaginosa CBMAI can be classified as a psychrotolerant organism.
3.2. Optimization of the culture media for protease production
Fungi have the ability to use many compounds as sole nitrogen sources. In the absence of the preferred nitrogen sources (ammonium and glutamine) they can also use less assimilated compounds such as nitrate, urea, purines, pyrimidines, uric acid and proteins [26], through the transcriptional activation of genes encoding enzymes and permeases required for the generation of ammonia performed by their metabolism. These responses to the quality of nitrogen source in the growth medium are termed nitrogen catabolite repression [27]. So, to evaluate the influence of the nitrogen source on the extracellular protease production by R. mucilaginosa CBMAI 1528, batch cultures were grown in 250 mL flasks for 48 h at 20 °C and 250 rpm. Glucose was used as the sole carbon source with 16 different combinations of 5 selected organic nitrogen sources, through a 25−1 fractional factorial design depicted in the Materials and Methods section 2.4 (Table 1).
Results in terms of protease activity and biomass concentrations are summarized in Table 2. The highest values of extracellular protease activity occurred in runs 4 (441.5 ± 35.5 U/mL), 6 (420.5 ± 47.6 U/mL) and 8 (422.8 ± 47.8 U/mL). As stressed in legend of Table 2, protease activity of R. mucilaginosa CBMAI 1528 on Sabouraud dextrose medium was 279.4 ± 12.4 U/mL; approximately 1.5 fold lower than in runs 4, 6 and 8, which contain casein tryptone as a component of the culture media. These results demonstrate the importance of the nitrogen source on the culture medium to stimulate protease production and/or enzyme release by the yeast.
Table 2.
Experimental values of extracellular protease activity (U/mL) and biomass concentration (g/L) at the end of batch runs carried out in flasks using different organic nitrogen sources, according to the 25−1 fractional factorial design depicted in Table 1.
| Run | X1 (g/L) | X2 (g/L) | X3 (g/L) | X4 (g/L) | X5 (g/L) | Protease activity (U/mL) | Biomass concentration (g/L) |
|---|---|---|---|---|---|---|---|
| 1 | 0 | 0 | 0 | 0 | 2.5 | 56.3 ± 10.2 | 0.78 ± 0.03 |
| 2 | 2.5 | 0 | 0 | 0 | 0 | 61.1 ± 6.8 | 0.53 ± 0.02 |
| 3 | 0 | 2.5 | 0 | 0 | 0 | 233.5 ± 2.9 | 0.63 ± 0.10 |
| 4 | 2.5 | 2.5 | 0 | 0 | 2.5 | 441.5 ± 35.5 | 8.05 ± 0.2 |
| 5 | 0 | 0 | 2.5 | 0 | 0 | 179.3 ± 32.0 | 0.60 ± 0.10 |
| 6 | 2.5 | 0 | 2.5 | 0 | 2.5 | 420.5 ± 47.6 | 11.73 ± 0.38 |
| 7 | 0 | 2.5 | 2.5 | 0 | 2.5 | 304.7 ± 21.3 | 12.29 ± 0.47 |
| 8 | 2.5 | 2.5 | 2.5 | 0 | 0 | 422.8 ± 47.8 | 12.79 ± 0.50 |
| 9 | 0 | 0 | 0 | 2.5 | 0 | 41.7 ± 15.7 | 0.60 ± 0.03 |
| 10 | 2.5 | 0 | 0 | 2.5 | 2.5 | 165.9 ± 0.2 | 5.54 ± 0.09 |
| 11 | 0 | 2.5 | 0 | 2.5 | 2.5 | 163.8 ± 5.0 | 6.05 ± 0.16 |
| 12 | 2.5 | 2.5 | 0 | 2.5 | 0 | 244.9 ± 1.4 | 11.61 ± 0.34 |
| 13 | 0 | 0 | 2.5 | 2.5 | 2.5 | 142.8 ± 3.4 | 6.21 ± 0.23 |
| 14 | 2.5 | 0 | 2.5 | 2.5 | 0 | 324.2 ± 36.9 | 12.76 ± 0.52 |
| 15 | 0 | 2.5 | 2.5 | 2.5 | 0 | 160.7 ± 30.2 | 12.22 ± 0.51 |
| 16 | 2.5 | 2.5 | 2.5 | 2.5 | 2.5 | 175.1 ± 3.6 | 15.30 ± 0.52 |
Under the same conditions (48 h of growth at 250 rpm, 20 °C and 20 g/L of dextrose), the protease activity of R. mucilaginosa CBMAI 1528 on Sabouraud dextrose medium was 279.4 ± 12.4 U/mL, and the biomass concentration 6.8 ± 0.3 g/L. X1: casein tryptone (g/L), X2: peptone from animal tissue (g/L), X3: yeast extract (g/L), X4: casamino acids (g/L), X5: peptone from gelatin (g/L).
On the other hand, the highest biomass production occurred in run 16 (15.30 ± 0.52 g/L) carried out in the simultaneous presence of all the nitrogen sources. These results suggest that a higher biomass production may be not associated to a higher protease production.
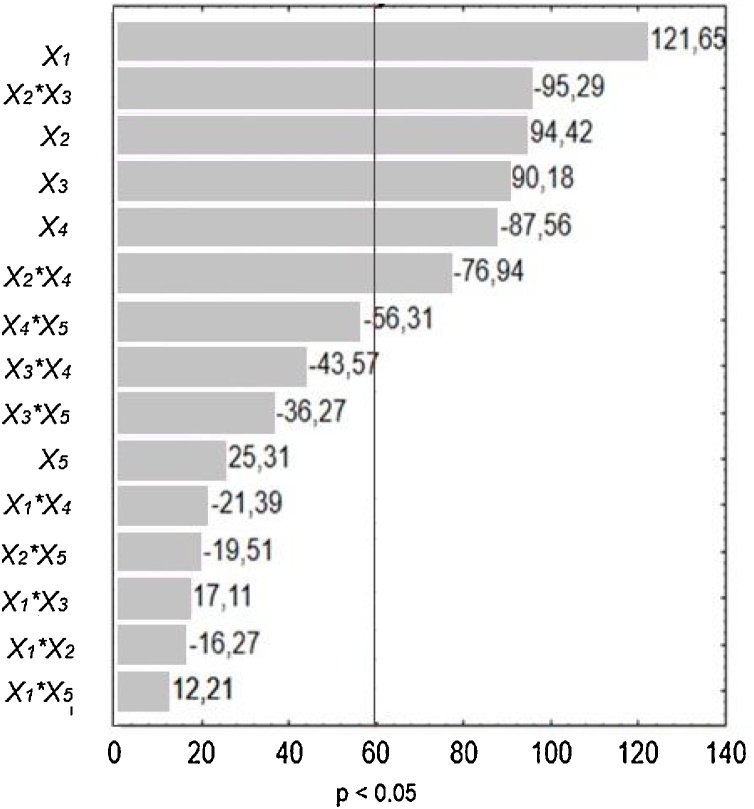
To identify the preferred nitrogen source for protease production, all these results were subjected to a Statistical analysis using a Pareto chart (Fig. 2). In this graph, the length of each bar is proportional to the standardized effect of the related variable or interaction, and the bars extending beyond the vertical line correspond to the statistically significant effects at a confidence level of 95 %. Thus, according to Fig. 2, protease production was stimulated in media containing casein tryptone (X1, positive sign), peptone from animal tissue (X2, positive sign), and yeast extract (X3, positive sign). However, it was negatively affected in media containing casamino acids (X4, negative sign). Moreover, consistent with the above simplified evaluation, casein tryptone (X1) was the culture component that most promoted protease production. The negative interaction between peptone from animal tissue (X2) and yeast extract (X3) indicates that these two medium components were mutually exclusive for protease production. Based on these results, we infer that the optimized medium for protease production should be composed of casein tryptone with yeast extract or peptone from animal tissue as organic nitrogen sources.
Fig. 2.
Pareto diagram showing the effects of five different organic nitrogen sources on extracellular protease production by R. mucilaginosa CBMAI 1528. Glucose at concentration of 20 g/L was used as a carbon source in runs carried out at 20 °C in flasks shaken at 250 rpm for 48 h. X1: Casein tryptone (g/L), X2: Peptone from animal tissue (g/L), X3: Yeast extract (g/L), X4: Casamino acids (g/L), X5: Peptone from gelatin (g/L).
These data demonstrate that protease production by R. mucilaginosa CBMAI 1528 greatly depends on medium composition. Similar results were previously reported for other microorganisms. For example, protease secretion by Candida humicola and Candida albicans was promoted when the culture media was composed by proteins and repressed when composed by amino acids and ammonium salts or by nitrogen starvation [[28], [29], [30]]. Moreover, casein and BSA are commonly used to induce the production and secretion of proteases [[31], [32], [33], [34]]. A possible explanation for this behavior is that proteins can be used as nitrogen sources by yeasts only when preferred nitrogen sources, such as amino acids, ammonium and urea, are unavailable [31]. In addition, protease-encoding genes are usually repressed at high concentrations of nitrogen sources other than proteins [35].
Based on these results and considering the elemental composition of both the yeast and the culture medium, a final optimization was attempted to improve extracellular protease production by R. mucilaginosa CBMAI 1528. To optimally cultivate any microorganism, not only must the medium contain all the chemical elements required for its growth, but also the elemental composition of such medium must reflect the elemental composition of the organism [36]. For this reason, the C:N ratio of a cell extract of R. mucilaginosa CBMAI 1528 was determined and then considered to optimize protease production. The C:N ratio of R. mucilaginosa cell extract was 9.86 (Table S1), a value close to that of the Sabouraud dextrose medium, in which the yeast was grown (C:N 9.47). Considering the results of the fractional factorial design, and the C:N ratio of the yeast under study, protease production was then evaluated on different culture media with a similar C:N ratio (9.72–10.87) and composed of glucose and casein tryptone and/or yeast extract as nitrogen source (Table S1 and Table 3).
Table 3.
Composition of the different culture media tested to optimize protease production by R. mucilaginosa CBMAI 1528.
| Medium | Glucose (g/L) | Yeast extract (g/L) | Casein tryptone (g/L) | C:N ratio |
|---|---|---|---|---|
| 1 | 20 | 0 | 10 | 9.72 |
| 2 | 20 | 5 | 5 | 10.25 |
| 3 | 20 | 10 | 0 | 10.87 |
| 4 | 30 | 0 | 15 | 9.72 |
| 5 | 30 | 7.5 | 7.5 | 10.25 |
| 6 | 30 | 15 | 0 | 10.87 |
| 7 | 40 | 0 | 20 | 9.72 |
| 8 | 40 | 10 | 10 | 10.25 |
| 9 | 40 | 20 | 0 | 10.87 |
The strain was grown on different media at 20 °C, pH 5.6, in flasks shaken at 250 rpm for 48 h. Aliquots from these cultures were taken to determine protease activity and biomass concentration.
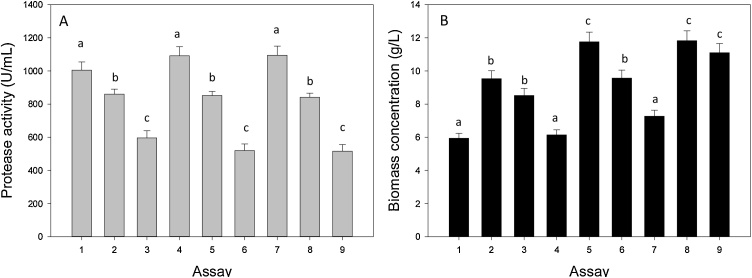
As observed in Fig. 3, protease production in all the media tested was higher than the one obtained in the previous assay (1.4–2.4 folds). This demonstrates the importance of considering the elemental composition of the organism to design the culture medium. The lowest biomass concentrations were obtained when casein tryptone was used as the only organic nitrogen source (media 1, 4 and 7), while the highest values were obtained when both components were present in the medium (media 2, 5 and 8). The highest extracellular protease activities were obtained in presence of casein tryptone (media 1, 4 and 7), and the lowest ones when yeast extract was used as nitrogen source (media 3, 6 and 9). Moreover, for each media (only casein tryptone, only yeast extract or both organic nitrogen sources), protease activity was very similar almost irrespectively of both glucose (20−40 g/L) and total nitrogen source (10−20 g/L) concentrations. It is a fact that to reduce the cultivation costs it is necessary to reduce to a minimum the number of ingredients of the culture medium and their concentrations [36]. Therefore, medium 1, which contains both glucose and casein tryptone at the lowest concentrations (20 and 10 g/L, respectively), was selected as the best one to perform batch R. mucilaginosa CBMAI 1528 culture in shaken flasks for protease production. It is interesting to note that between the three tested media, the selected one has the most similar C:N ratio to the yeast under study (C:N 9.72 and 9.86 respectively). However, more studies are required to assess the impact of the C:N ratio on protease production.
Fig. 3.
(A) Protease production (U/mL) and (B) biomass concentration (g/L) by R. mucilaginosa CBMAI 1528. The strain was grown on different culture media at 20 °C, pH 5.6, in flasks shaken at 250 rpm for 48 h. Compositions of the different culture media (1-9) are listed in Table 3. Data are means ± s.d. n = 3, where samples labelled with identical letters are not significant at P < 0.05.
3.3. Growth experiments and bioreactor operation
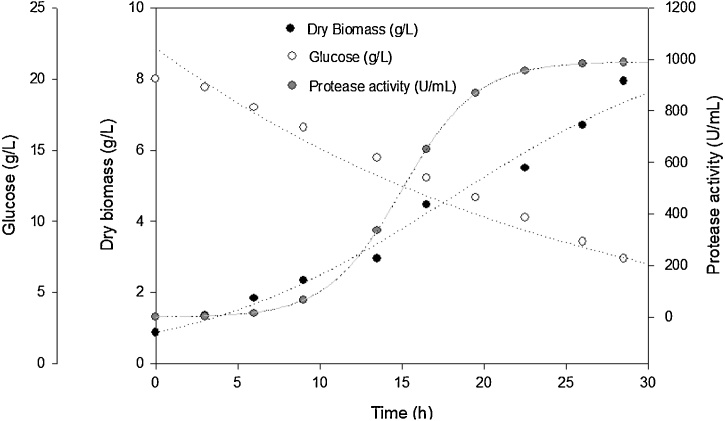
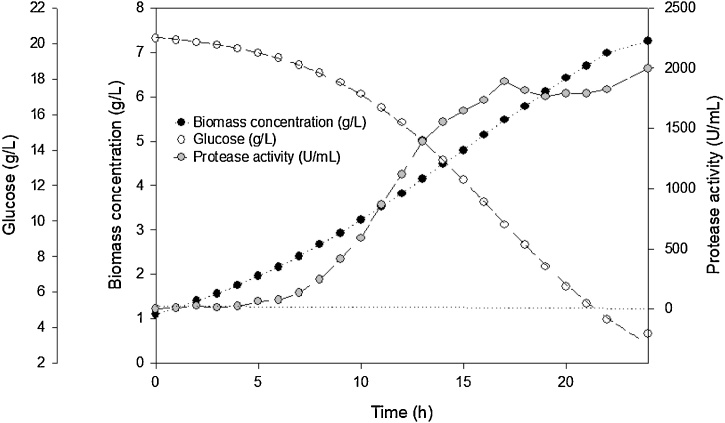
R. mucilaginosa CBMAI 1528 was cultured at 20 °C, pH 5.6 in shake flasks at 250 rpm for about 30 h, with an initial biomass concentration of 1 g/L in medium which contained 20 g/L glucose and 10 g/L of casein tryptone. Aliquots were taken every 3 h to determine protease activity and dry biomass concentration (Fig. 4). Under these conditions, the maximum specific growth rate (μmax) was 0.18 h−1, and the generation time (Tg) 3.85 h. After 28 h of culture, 56 % of the carbon source was consumed, with a biomass productivity (QX) of 0.28 g/L.h. On the other hand, this parameter achieved a maximum value (0.45 g/L.h) after only 3 h, whereas the maximum protease productivity (Qp) was 44.4 U/mL.h after 22.5 h.
Fig. 4.
Time course of a batch culture of R. mucilaginosa CBMAI 1528. The strain was cultured in medium containing 20 g/L glucose and 10 g/L of casein tryptone at 20 °C, pH 5.6, in shake flasks at 250 rpm for about 30 h, with an initial dry cell concentration of 1 g/L. Aliquots from this culture were taken every 3 h to determine protease activity, dry biomass and glucose concentration. (●) Dry biomass concentration (g/L); (○) glucose concentration (g/L); ( ) protease activity (U/mL).
) protease activity (U/mL).
Under similar conditions (20 °C, 20 g/L glucose, 10 g/L casein tryptone and 1 g/L of inoculum), the values obtained in bioreactor were very close to those obtained in shake flasks: μmax (0.20 h−1), Tg (3.46 h) and QX (0.35 g/L.h after 21 h). Nevertheless, the percentage of carbon source consumption (86 % after 24 h), maximum Qx (0.93 g/L.h after 2 h) and maximum Qp (111.2 U/mL.h after 17 h) were remarkably higher (Fig. 5), likely due to the positive effect of a more efficient aeration in bioreactor.
Fig. 5.
Time course of a batch culture of R. mucilaginosa CBMAI 1528 in bioreactor. The strain was cultured on medium containing 20 g/L glucose and 10 g/L of casein tryptone for 24 h at 20 °C, pH 5.6, 200 vvm, 500 rpm and KLa =92 h−1, with an initial dry biomass concentration of 1 g/L. Aliquots from this culture were taken every hour to determine protease activity, dry biomass and glucose concentration. (●) Dry biomass concentration (g/L); (○) glucose concentration (g/L); (●) protease activity (U/mL).
3.4. Protease characterization: mass spectrometry analysis and inhibition assay
In order to get a further characterization of the extracellular protease secreted by R. mucilaginosa CBMAI 1528, the enzyme was purified and then its sequence analyzed by mass spectrometry. To do that, 1.5 L of fermented broth obtained in bioreactor was centrifuged, and the enzymes present in the supernatant were precipitated with ammonium sulfate. The precipitate was then dissolved in sodium acetate buffer, dialyzed, and finally loaded on a CM-Sepharose Fast Flow column. Only one protein peak appeared in the elution profile, which coincided with the one displaying proteolytic activity (data not shown). A Coomassie stained SDS-PAGE showed a unique band of 34.5 kDa (Fig. S1). In order to ascertain the Rhodotorula extracellular protease identity, the protein band obtained from the SDS-PAGE was in-gel digested with trypsin, and peptides were analyzed by MS/MS analysis. The obtained data were used to search for matching peptides from proteins in the UNIPROT database using the Mascot search program.
Table 4 shows the sequences of the three tryptic peptides obtained. BLASTp search against UniprotKB reference proteomes plus Swiss-Prot database revealed 100 % identity with an acid protease from Rhodotorula sp. JG-1b (Gene RHOSPDRAFT_21543; protein ID: KWU42276.1) with an E- value of 2.6e−8. Fig. 6 shows the analysis of the nucleotide and deduced protein sequences of the enzyme under study. BLASTp search allowed us to classify the enzyme as an aspartic protease belonging to the peptidase A1 subfamily, also known as the pepsin family. All these peptidases are endopeptidases, mostly secreted from cells as inactive proenzymes that are activated autocatalytically under acidic conditions. This probably explains the lower molecular weight of the purified protease in contrast to the higher molecular mass of the predicted protein. Finally, to provide further proof that the protease characterized in this study is an aspartic protease, the effect of different inhibitors on its activity was investigated. As shown in Table 5, only pepstatin, a specific inhibitor of aspartic proteases, almost completely inhibited the enzyme activity.
Table 4.
Tryptic fragments from digestion of purified enzyme identified by LC–MS/MS against UNIPROT protein database.
| Sequence | Master Protein Accessions | Theoretical MH+ [Da] | XCorr Sequest HT | Confidence Sequest HT |
|---|---|---|---|---|
| LATQNSAASELYLGGLNSAK | A0A125PHT0 | 2008.03456 | 5.46 | High |
| VAANSFSFR | A0A125PHT0 | 998.50541 | 2.99 | High |
| ASTGSDSLTSYQNDNLWAGK | A0A125PHT0 | 2114.96252 | 2.62 | High |
Fig. 6.
R. mucilaginosa CBMAI 1528 protein with aspartic protease activity: partial nucleotide and amino acid sequences deducted from the homologous Rhodotorula sp. JG-01b gene RHOSPDRAFT_21543. Protease conserved Pfam domain PF00026 for members of the peptidase A1 family of proteases is highlighted (light grey) [39]. Tryptic peptides obtained by mass spectrometry are underlined.
Table 5.
Inhibitor reactivity.
| Inhibitor | Specificity | Solvent | Inhibitor concentration | Residual activity (%)a |
|---|---|---|---|---|
| None | 100.0 | |||
| AEBSFb | Serine proteases | Water | 1 mM | 87.4 |
| 6- Aminohexanoic acid | Chymotrypsin, Factor VIIa, lysine carboxypeptidase, plasmin, and plasminogen activator | Water | 40 mM | 94.8 |
| Antipain hydrochloride | Serine proteases | Water | 0.1 mM | 93.5 |
| Aprotinin | Serine proteases | Water | 0.1 mM | 74.8 |
| Benzamidine hydrochloride hydrate | Trypsin, trypsin-like enzymes, and serine proteases | Water | 4 mM | 91.6 |
| Bestatin | Metalloproteases | Water | 0.04 mM | 100.0 |
| Chymostatin | Chymotrypsin | DMSOc | 0.1 mM | 46.0 |
| E-64 | Cysteine proteases | Water | 10 μM | 94.2 |
| EDTA | Metalloproteases | Water | 1 mM | 86.6 |
| N-Ethylmaleimide | Cysteine proteases | Ethanol | 1 mM | 69.8 |
| Leupeptin | Serine and Cysteine proteases | Water | 0,1 mM | 68.7 |
| Pepstatin | Aspartic proteases | Ethanol | 1 μM | 4.9 |
| Phosphoaramidon | Metalloproteases | Water | 10 μM | 86.4 |
| Trypsin inhibitor | Trypsin | Water | 1:1 stoichiometric binding | 81.2 |
Enzyme samples were incubated with the inhibitor for 15 min at 20 °C and then the remaining activity in the sample was determined. Each value represents the mean of three experiments. The activity in the absence of inhibitor was considered 100 %.
AEBSF 4-(2-Aminoethyl) benzenesulfonyl fluoride hydrochloride.
Dimethyl Sulfoxide (DMSO).
4. Conclusions
In this work, we describe the production, purification and identification of an enzyme with protease activity secreted by R. mucilaginosa CBMAI 1528, previously isolated from the Antarctic continent. Analysis of the effect of the temperature on yeast cell growth indicates that the strain was able to grow at temperatures as low as 10 °C and as high as 35 °C with a maximum between 20 and 30 °C, allowing us to classify this strain as psychrotolerant. The temperature also influenced protease production, reaching a maximum activity at 20 °C.
Analysis of medium composition on protease production by R. mucilaginosa CBMAI 1528 demonstrated that it was promoted by casein tryptone, peptone from animal tissue and yeast extract but repressed by casamino acids. Further optimization of media composition was performed taking into account the C:N ratio. Medium containing both glucose and casein tryptone was selected as the best option to perform batch culture in shaken flasks. When the same culture medium was tested in the bioreactor, protease production and carbon source consumption were enhanced, thereby proving efficient scaling-up of the process.
The protein was purified by a two-step procedure, and the deduced amino acid sequence indicated that the protein is an aspartic protease belonging to the peptidase A1 subfamily, also known as the pepsin family. In accordance with this result, the activity of the enzyme was inhibited by pepstatin, a specific inhibitor of aspartic proteases. Interestingly, Murao et al. [37] reported during screening tests for yeast extracellular proteases, the isolation of a potent acid protease from Rhodotorula glutinis, which was subsequently purified and crystallized by Kamada et al. [18]. These authors found that the enzyme was stable in the pH range from 2.4 to 6.5 after 20 h of incubation at 37 °C. When this protease was compared with several other proteases obtained from various sources, it was found that the specific activity towards casein was potent and 1.7-fold higher than that of pepsin. Moreover, the enzyme was inactivated by a pepsin inhibitor produced from Streptomyces naniwaensis. Subsequently, Oda et al. [17] determined some physicochemical properties of the enzyme. The molecular weight was estimated to be approximately 30 kDa, and the isoelectric point 4.5. The enzyme was named as Rhodotorulapepsin (EC 3.4.23.26); however, until now, the enzyme has not had an associated sequence and has been classified as an orphan enzyme as it had a EC number without an associated sequence [38]. Based on the results collected here, we can reasonably propose that the aspartic protease described in the present study may be the Rodothorulapepsin described years ago.
Declaration of Competing Interest
The authors report no declarations of interest.
Acknowledgements
This work was supported by grants from the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP, PhD fellowship process Nº 2012/23726-4 awarded to LL), the Agencia Nacional de Promoción Científica y Tecnológica (PICT 2017-0356 and PICT 2016-2500 to LL and CPS, respectively). LDL, PC and CPS are members of the Researcher Career of CONICET and Faculty members of Biochemistry (LDL, PC and CPS), Facultad de Ciencias Bioquímicas y Farmacéuticas, Universidad Nacional de Rosario. We thank Dr. Matthew J. Smith (King's College London) and Prof. Sonia Ficarella for their kind review of this article. Authors declare that there are no conflicts of interest.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.btre.2020.e00546.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- 1.Niederberger T.D., McDonald I.R., Hacker A.L., Soo R.M., Barrett J.E., Wall D.H., Cary S.C. Microbial community composition in soils of Northern Victoria land, Antarctica. Environ. Microbiol. 2008;10:1713–1724. doi: 10.1111/j.1462-2920.2008.01593.x. [DOI] [PubMed] [Google Scholar]
- 2.Margesin R., Neuner G., Storey K.B. Cold-loving microbes, plants, and animals - Fundamental and applied aspects. Naturwissenschaften. 2007;94:77–99. doi: 10.1007/s00114-006-0162-6. [DOI] [PubMed] [Google Scholar]
- 3.Somero G.N. Proteins and temperature. Annu. Rev. Physiol. 1995;57:43–68. doi: 10.1146/annurev.ph.57.030195.000355. [DOI] [PubMed] [Google Scholar]
- 4.Feller G., Gerday C. Psychrophilic enzymes: hot topics in cold adaptation. Nat. Rev. Microbiol. 2003;1:200–208. doi: 10.1038/nrmicro773. [DOI] [PubMed] [Google Scholar]
- 5.Gerday C., Aittaleb M., Bentahir M., Chessa J.P., Claverie P., Collins T., D’Amico S., Dumont J., Garsoux G., Georlette D., Hoyoux A., Lonhienne T., Meuwis M.A., Feller G. Cold-adapted enzymes: from fundamentals to biotechnology. Trends Biotechnol. 2000:103–107. doi: 10.1016/S0167-7799(99)01413-4. [DOI] [PubMed] [Google Scholar]
- 6.Ogaki M.B., de Paula M.T., Ruas D., Pellizzari F.M., García-Laviña C.X., Rosa L.H. 2019. Marine Fungi Associated With Antarctic Macroalgae; pp. 239–255. [DOI] [Google Scholar]
- 7.Ruisi S., Barreca D., Selbmann L., Zucconi L., Onofri S. Fungi in Antarctica. Rev. Environ. Sci. Biotechnol. 2007:127–141. doi: 10.1007/s11157-006-9107-y. [DOI] [Google Scholar]
- 8.Durán P., Barra P.J., Jorquera M.A., Viscardi S., Fernandez C., Paz C., de la L. Mora M., Bol R. Occurrence of soil fungi in antarctic pristine environments. Front. Bioeng. Biotechnol. 2019;7 doi: 10.3389/fbioe.2019.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kawahara H., Hirai A., Minabe T., Obata H. Stabilization of astaxanthin by a novel biosurfactant produced by Rhodotorula mucilaginosa KUGPP-1. Biocontrol Sci. 2013;18:21–28. doi: 10.4265/bio.18.21. [DOI] [PubMed] [Google Scholar]
- 10.Lario L.D., Chaud L., das G. Almeida M., Converti A., Durães Sette L., Pessoa A. Production, purification, and characterization of an extracellular acid protease from the marine Antarctic yeast Rhodotorula mucilaginosa L7. Fungal Biol. 2015;119:1129–1136. doi: 10.1016/j.funbio.2015.08.012. [DOI] [PubMed] [Google Scholar]
- 11.Potumarthi R., Subhakar C., Vanajakshi J., Jetty A. Effect of aeration and agitation regimes on lipase production by newly isolated rhodotorula mucilaginosa - MTCC 8737 in stirred tank reactor using molasses as sole production medium. Appl. Biochem. Biotechnol. 2008;151:700–710. doi: 10.1007/s12010-008-8293-1. [DOI] [PubMed] [Google Scholar]
- 12.Aksu Z., Eren A.T. Carotenoids production by the yeast Rhodotorula mucilaginosa: use of agricultural wastes as a carbon source. Process Biochem. 2005;40:2985–2991. doi: 10.1016/j.procbio.2005.01.011. [DOI] [Google Scholar]
- 13.Singh P., Tsuji M., Singh S.M., Roy U., Hoshino T. Taxonomic characterization, adaptation strategies and biotechnological potential of cryophilic yeasts from ice cores of Midre Lovénbreen glacier, Svalbard, Arctic. Cryobiology. 2013;66:167–175. doi: 10.1016/j.cryobiol.2013.01.002. [DOI] [PubMed] [Google Scholar]
- 14.Liu B., Wang C., Liu D., He N., Deng X. Hg tolerance and biouptake of an isolated pigmentation yeast Rhodotorula mucilaginosa. PLoS One. 2017;12 doi: 10.1371/journal.pone.0172984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wirth F., Goldani L.Z. Epidemiology of Rhodotorula : An Emerging Pathogen. Interdiscip. Perspect. Infect. Dis. 2012;2012:1–7. doi: 10.1155/2012/465717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duarte A.W.F., Dayo-Owoyemi I., Nobre F.S., Pagnocca F.C., Chaud L.C.S., Pessoa A., Felipe M.G.A., Sette L.D. Taxonomic assessment and enzymes production by yeasts isolated from marine and terrestrial Antarctic samples. Extremophiles. 2013;17:1023–1035. doi: 10.1007/s00792-013-0584-y. [DOI] [PubMed] [Google Scholar]
- 17.Oda K., Kamada M., Murao S. Some physicochemical properties and substrate specificity of acid protease of Rhodotorula glutinis K-24. Agric. Biol. Chem. 1972;36:1103–1108. doi: 10.1271/bbb1961.36.1103. [DOI] [Google Scholar]
- 18.Kamada M., Oda K., Murao S. The purification of the extracellular acid protease of Rhodotorula glutinis K-24 and its general properties. Agric. Biol. Chem. 1972;36:1095–1101. doi: 10.1080/00021369.1972.10860377. [DOI] [Google Scholar]
- 19.Sorokina M., Stam M., Médigue C., Lespinet O., Vallenet D. Profiling the orphan enzymes. Biol. Direct. 2014:9–10. doi: 10.1186/1745-6150-9-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 21.Charney J., Tomarelli R.M. A colorimetric method for the determination of the proteolytic activity of duodenal juice. J. Biol. Chem. 1947;171:501–505. [PubMed] [Google Scholar]
- 22.Potumarthi R., Ch S., Jetty A. Alkaline protease production by submerged fermentation in stirred tank reactor using Bacillus licheniformis NCIM-2042: effect of aeration and agitation regimes. Biochem. Eng. J. 2007;34:185–192. doi: 10.1016/j.bej.2006.12.003. [DOI] [Google Scholar]
- 23.Laemmli U.K. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 24.Chaud L.C.S., Lario L.D., Bonugli-Santos R.C., Sette L.D., Pessoa Junior A., das G. de A. Felipe M. Improvement in extracellular protease production by the marine antarctic yeast Rhodotorula mucilaginosa L7. N. Biotechnol. 2016;33:807–814. doi: 10.1016/j.nbt.2016.07.016. [DOI] [PubMed] [Google Scholar]
- 25.Morita R.Y. Psychrophilic bacteria. Bacteriol. Rev. 1975;39:144–167. doi: 10.1128/MMBR.39.2.144-167.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koon H.W., Hynes M.J., Davis M.A. Recent advances in nitrogen regulation: a comparison between Saccharomyces cerevisiae and filamentous fungi. Eukaryot. Cell. 2008:917–925. doi: 10.1128/EC.00076-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Magasanik B., Kaiser C.A. Nitrogen regulation in Saccharomyces cerevisiae. Gene. 2002:1–18. doi: 10.1016/S0378-1119(02)00558-9. [DOI] [PubMed] [Google Scholar]
- 28.Ray M.K., Devi K.U., Kumar G.S., Shivaji S. Extracellular protease from the antarctic yeast Candida humicola. Appl. Environ. Microbiol. 1992;58:1918–1923. doi: 10.1128/AEM.58.6.1918-1923.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Remold H., Fasold H., Staib F. Purification and characterization of a proteolytic enzyme from Candida albicans. Biochim. Biophys. Acta - Enzymol. 1968;167:399–406. doi: 10.1016/0005-2744(68)90219-2. [DOI] [PubMed] [Google Scholar]
- 30.Ross I.K., De Bernardis F., Emerson G.W., Cassone A., Sullivan P.A. The secreted aspartate proteinase of Candida albicans: physiology of secretion and virulence of a proteinase-deficient mutant. J. Gen. Microbiol. 1990;136:687–694. doi: 10.1099/00221287-136-4-687. [DOI] [PubMed] [Google Scholar]
- 31.Banerjee A., Ganesan K., Datta A. Induction of secretory acid proteinase in Candida albicans. J. Gen. Microbiol. 1991;137:2455–2461. doi: 10.1099/00221287-137-10-2455. [DOI] [PubMed] [Google Scholar]
- 32.Gotoh T., Kikuchi K., Kodama K., Konno H., Kakuta T., Koizumi T., Nojiro K. Purification and properties of extracellular carboxyl proteinase secreted by Candida pulcherrima. Biosci. Biotechnol. Biochem. 1995;59:367–371. doi: 10.1271/bbb.59.367. [DOI] [PubMed] [Google Scholar]
- 33.Lagace L., Bisson L. Survey of yeast acid proteoses for effectiveness of wine haze reduction. Am. J. Enol. Vitic. 1990;41:147–155. [Google Scholar]
- 34.Togni G., Sanglard D., Quadroni M., Foundling S.I., Monod M. Acid proteinase secreted by Candida tropicalis: functional analysis of preproregion cleavages in C. Tropicalis and Saccharomyces cerevisiae. Microbiology. 1996;142:493–503. doi: 10.1099/13500872-142-3-493. [DOI] [PubMed] [Google Scholar]
- 35.Dabas N., Morschhäuser J. A transcription factor regulatory cascade controls secreted aspartic protease expression in Candida albicans. Mol. Microbiol. 2008;69:586–602. doi: 10.1111/j.1365-2958.2008.06297.x. [DOI] [PubMed] [Google Scholar]
- 36.Spaargaren D.H. The design of culture media based on the elemental composition of biological material. J. Biotechnol. 1996;45:97–102. doi: 10.1016/0168-1656(95)00152-2. [DOI] [Google Scholar]
- 37.Murao S., Kamada M., Nakase T., Ogura S., Oda K. Studies on the extracellular protease of yeasts part I. The identification of the yeast No. K-24 and the production of acid protease. Nippon NÅ□geikagaku Kaishi. 1972;46:167–170. doi: 10.1271/nogeikagaku1924.46.167. [DOI] [Google Scholar]
- 38.Lespinet O., Labedan B. Orphan enzymes could be an unexplored reservoir of new drug targets. Drug Discov. Today. 2006;11:300–305. doi: 10.1016/j.drudis.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 39.Bateman A., Coin L., Durbin R., Finn R.D., Hollich V., Griffiths-Jones S., Khanna A., Marshall M., Moxon S., Sonnhammer E.L.L., Studholme D.J., Yeats C., Eddy S.R. The Pfam protein families database. Nucleic Acids Res. 2004 doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.