Abstract
Background
The underlying mechanisms of incomplete immune reconstitution in treated HIV-positive patients are very complex and may be multifactorial, but perturbation of chemokine secretion could play a key role in CD4+T-cell turnover.
Methods
We evaluated the circulating baseline and 48-week follow-up concentrations of SDF-1/CXCL12, fractalkine/CX3CL1, MCP-1/CCL2, MIP-α/CCL3, MIP-β/CCL4 and RANTES/CCL5, and we estimated their association with CXCL12, CX3CR1, CCR2, CCL5 and CCR5 single nucleotide polymorphisms (SNPs) to investigate multiple chemokine-chemokine receptor signatures associated with immune dysregulation preceding poor immune recovery.
Findings
The circulating concentrations and gene expression patterns of SDF-1/CXCL12 (CXCL12 rs1801157) and MCP-1/CCL2 (CCR2 rs1799864_814) were associated with immune recovery status. CCR2 rs1799864_814 and CCR5 rs333_814 (Δ32) determine the baseline plasma RANTES and MIP-α concentrations, respectively, in participants with poor immune response.
Interpretation
SDF-1/CXCL12 and MCP-1/CCL2 could be considered prognostic markers of immune failure despite suppressive antiretroviral therapy. The strong linkage disequilibrium (LD) between CCR2 rs1799864_814 and CCR5 rs1800024 indicated that the alleles of each gene are inherited together more often than would be expected by chance.
Funding
This work was supported by Fondo de Investigacion Sanitaria and SPANISH AIDS Research Network (ISCIII-FEDER); AGAUR and Gilead Fellowship. FV and YMP are supported by grants from the Programa de Intensificación (ISCIII) and Servicio Andaluz de Salud, respectively. JVG,EY and LR are supported by the Instituto de Salud Carlos III (ISCIII). AR is supported by Departament de Salut, Generalitat de Catalunya and by the Instituto de Salud Carlos III (ISCIII).
Keywords: Chemokines, Chemokine receptors, HIV, Poor immune recovery, Polymorphisms variants
Research in context.
Evidence before this study
Chemokines are chemotactic cytokines that signal through cell surface G protein-coupled heptahelical chemokine receptors to control the migration and positioning of immune cells to the sites of infection and injury. They are critical for the function of the innate immune system. Accordingly, the chemokine-chemokine receptor system clearly plays an important role in the HIV life cycle, disease progression, and HIV reservoir establishment. Thus, imbalances of cytokine-chemokine levels were hypothesized to be responsible for a lack of appropriate CD4+ T-cell reconstitution after suppressive cART.
Added value of this study
The present study jointly evaluates a specific remarkable network of chemokines as prognostic and follow-up markers of immune recovery status by determining circulating concentrations and performing a genetic polymorphism study. This is the largest, longitudinal study assessing both chemokine genetics and circulating levels in immune restoration.
Implications of all the available evidence
The chemokine-chemokine receptor system is more complex than indicated by the study of a selected chemokine and its natural ligands due to the enormous network existing among the different chemokines and their receptors. However, we conclude that SDF-1/CXCL12, MCP-1/CCL-2 and CCR5 receptors are key effectors of susceptibility to HIV progression, especially in terms of immune reconstitution. Our data open a window for new proof-of-concept studies to explore the individual but also dual blockage of SDF-1 and MCP-1, as well as CCR2 and CCR5, as potential therapeutic options for advanced clinical trials with promising fortunate outcomes.
Alt-text: Unlabelled box
1. INTRODUCTION
Combination antiretroviral treatment (cART) in HIV-positive participants aims to suppress HIV replication below detectable levels and restore the CD4+ T-cell counts. [1] However, approximately 30% of people living with HIV (PLW) with optimal treatment and fully suppressed viral replication fail to recover their CD4+ T-cell counts. [2,3] These patients are referred to as “immunodiscordants” or “immunological nonresponders” (INRs), and are linked to an increased risk of disease progression and death compared with PLW who achieve complete immune reconstitution. [4,5]
The CD4+ T-cell count is the result of the production, destruction, and trafficking between secondary lymphoid organs and peripheral tissues of CD4+ T-cells, [2,6] and INRs may have both excessive destruction and alterations in the production of CD4+ T-cells, resulting in an increase in cycling and proliferation before cART. [7] Although the underlying mechanisms of incomplete immune reconstitution are very complex and may be multifactorial, perturbation of chemokine secretion could play a key role in CD4+ T-cell turnover, [8], [9], [10] and chemokine modulation has been proposed as a promising therapeutic strategy to improve immune reconstitution. [11,12]
Chemokines are chemotactic cytokines that control the migration and positioning of immune cells, most notably leukocytes, to the sites of infection and injury. [13] They are a large family of small (8–12 kDa) proteins that are characterized by the presence of three to four conserved cysteine residues and that can be classified into four families based on the positioning of the N-terminal cysteine residues. Chemokines signal through cell surface G protein-coupled heptahelical chemokine receptors to perform a variety of functions aside from chemotaxis, including T cell differentiation and function, as well as angiogenesis. Thus, investigations regarding multiple cytokine-chemokine signatures associated with immune dysregulation preceding poor immune recovery are needed to understand the molecular mechanism involved in this condition and therefore to develop a useful tool for early detection.
In fact, the connection between the expression of different pro-inflammatory cytokines and the discordant response encouraged us to recently explore the role of the IL-7/IL-7R axis in INRs. [10] Since our results confirmed that improved knowledge of this cytokine and its receptor in HIV participants could produce new insights regarding the immune response to ART, we hypothesized that the study of some selected chemokines related to IL-7/IL-7R axis could help to identify the molecular mechanisms associated with immune dysregulation preceding a discordant response to ART. Thus, in this study, we evaluated circulating baseline (pre-ART) and 48-week follow-up concentrations of stromal cell-derived factor 1 (SDF-1), also known as C-X-C motif chemokine 12 (CXCL12) (SDF-1/CXCL12), fractalkine/CX3CL1, monocyte chemoattractant protein-1 (MCP-1/CCL2), macrophage inflammatory protein-1 alpha (MIP-α/CCL3), macrophage inflammatory protein-1 beta (MIP-β/CCL4) and RANTES (regulated upon activation, normal T cell expressed and secreted), also known as CCL5, and we also determined whether some selected CXCL12, CX3CR1, CCR2, CCL5 and CCR5 single nucleotide polymorphisms (SNPs) are associated with CD4+ T-cell recovery in a previously characterized cohort of HIV participants. [10]
2. MATERIALS AND METHODS
2.1. Study design and participants
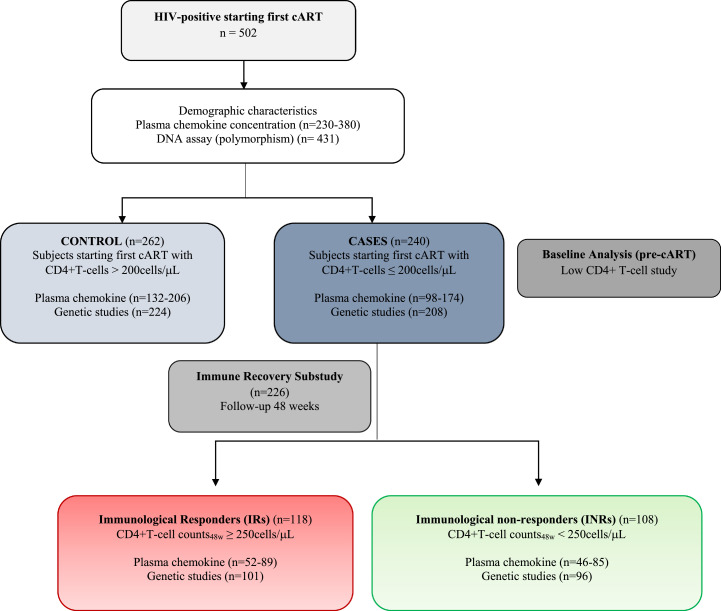
A multicentre, longitudinal case-controlled study of 502 adult HIV-positive participants who were consecutively recruited between 2011 and 2013 at the HIV outpatient clinics of the participating hospitals who started their first ART and achieved virological suppression after ART was performed. [10] Patients were selected from among those who were receiving a combination of two nucleoside reverse transcriptase inhibitors (NRTI) plus a nonnucleoside reverse transcriptase inhibitor (NNRTIs) or a protease inhibitor(s) (PI). A flowchart of patient selection and enrolment is provided in Fig. 1 with the previously defined inclusion/exclusion criteria. [10] Of the selected patients, 262 were defined as controls (baseline CD4+ T-cell counts >200 cells/μL), and 240 were cases (baseline CD4+ T-cell counts ≤ 200 cells/μL). Among the cases, 226 could be classified for the immune recovery study: 118 participants had more than 250 CD4+ T-cells/µL after 48 weeks of ART (“immunological responders”, IRs), and 108 participants did not reach the 250 cells/µL CD4+ T-cell threshold (“immunological nonresponders”, INRs). The threshold of 250 CD4+T-cells/µL to classify patients regarding immune recovery status was previously validated by the fact that patients receiving cART with CD4+ T-cells persistently below 250 cells/µL area associated with worse clinical outcomes [5] and thus, to be consistent with our previous related work, [10] in this study we maintain the same threshold criteria.
Fig. 1.
Flowchart illustrating subject cohort enrolment and analysis. HIV-infected subjects were included and categorized as controls and cases according to the pre-cART CD4+T-cell counts. For the immune recovery sub-study group, cases starting cART with T-cell counts below 200 cells/µL were categorized according to their immune status after 48 weeks of follow-up.
2.2. Ethics
The study and all research protocols were carried out in accordance with the recommendations of the Ethical and Scientific Committees from each participating institution and were approved by the Committee for Ethical Clinical Research by following the rules of Good Clinical Practice from the Institut d'Investigació Sanitària Pere Virgili (CEIm IISPV)(CEIC 61p/2013). The CEIm IISPV is an independent committee, made up of health and nonhealth professionals, which supervises the correct compliance of the ethical principles governing clinical trials and research projects that are carried out in our region, specifically in terms of methodology, ethics and laws. All participants gave written informed consent in accordance with the Declaration of Helsinki.
2.3. General laboratory measurements
Plasma was obtained by centrifugation and was stored at −80°C in the IISPV-Biobank until use. HIV-1 infection was diagnosed by a positive ELISA result and confirmed by Western blot analysis. The plasma HIV-1 viral load was determined by the Cobas Amplicor HIV-1 Monitoring Test v 1.5 (Roche Diagnostics, Barcelona, Spain). The limit of detectability was <20 copies/µL. CD4+ T-cell counts were analysed using a FACScan flow cytometer (Becton Dickinson, San Jose, CA, USA).
2.4. Genetic studies
We chose 13 single nucleotide polymorphisms (SNPs) in selected chemokine and chemokine receptors, with an allelic frequency greater than 20% in the Iberian Population in Spain (IBS) or the European Population (EU) in the NCBI SNP database. In summary, the following 13 SNPs in chemokines were analysed in this study: CXCL12 rs1801157; CCL5rs2280789, CCL5rs2280788 and CCL5rs2107538; in chemokine receptors: CX3CR1rs373278_814, CX3CR1rs3732379, CCR2rs1799864, CCR5rs2734648, CCR5rs1799987, CCR5rs1799988, CCR5rs1800023, CCR5rs1800024 and CCR5rs333_814 were analysed. Genomic DNA was extracted from peripheral blood with a Qiagen kit (Qiagen, Hilden, Germany) and then the extracted DNA samples (5 ng/μL) were sent to LGC Genomics Ltd. (formerly Kbioscience Ltd., Herts, UK) for genotyping. [10] The CCL3/4 copy number variation (CVN) was not considered in the present study due to the high level of discordance between different assays/techniques to measure it and also because CCL3L1–CCL4L1 CVN results will be bias by the study design as mentioned below in limitations of the study.
2.5. Plasma chemokine concentrations
Plasma concentrations of human SDF-1 (CXCL12), fractalkine (CX3CL1), MCP-1 (CCL2), MIP-α (CCL3), MIP-β (CCL4) and RANTES (CCL5) were measured by a double-antibody sandwich one-step process enzyme-linked immunosorbent assay (ELISA) with DuoSet DY350 (R&D Systems Inc), DuoSet DY365 (R&D Systems Inc), BMS281INST (eBioscience, The Affymetrix company), DuoSet DY270 (R&D Systems Inc) and DuoSet DY271 (R&D Systems Inc), respectively, according to the manufacturer's instructions.
2.6. Statistical analyses
Prior to the statistical analyses, the normal distribution and homogeneity of the variances were tested using a Kolmogorov-Smirnov test. Normally distributed data were expressed as the mean ± standard deviation (SD), whereas variables with a skewed distribution were represented as the median (25th percentile – 75th percentile) or transformed into a decimal logarithm. Categorical variables were reported as numbers (percentages). Qualitative variables were analysed using the χ2 test or Fisher's exact test when necessary. Comparisons between groups were performed with nonparametric Kruskal-Wallis (KW) and/or Mann-Whitney (MW) tests for unpaired samples and a Wilcoxon t-test for paired samples (W). When applicable, Bonferroni post-hoc approach was used in multiple comparison analyses. Associations between quantitative variables were evaluated using the Spearman correlation. Allele and genotype frequencies and the Hardy-Weinberg equilibrium (HWE) were evaluated using SNPstats software [14]. To estimate the association between chemokine and chemokine receptor genetic polymorphisms and immune recovery status, we used multiple inheritance models (codominant, dominant, recessive, overdominant and additive). For each SNP, the odds ratios (ORs) and 95% confidence intervals (CIs) were calculated using unconditional logistic regression analysis with adjustment of the baseline pre-ART CD4+ T-cell counts. In addition, the HaploReg v4.1 software (https://pubs.broadinstitute.org/mammals/haploreg/haploreg.php), a bioinformatic tool designed for exploring the annotations of the noncoding variants in haplotype blocks, was used to evaluate the mechanistic hypothesis of the queried SNP. Statistical analyses were performed using SPSS (version 21.0, SPSS Inc., Chicago, IL), and graphical representations were generated with GraphPad Prism software (version 5.0, GraphPad Inc., San Diego, CA). The results were considered significant at P < 0.05.
2.7. Role of funders
Funders had no role in the study design and participants’ recruitment, no role in general laboratory measurements, data analysis, and interpretation of data; no role in the writing of the manuscript, and no role in the decision to submit the final manuscript for publication.
3. RESULTS
3.1. Patient characteristics
The pre-cART clinical characteristics of the study cohort of HIV-positive participants (n = 502) were categorized according to the baseline CD4+ T-cell counts (Fig. 1) and are presented in Table 1. The patients associated with cases (baseline CD4+ T-cell ≤ 200 cells/µL) were older, presented significantly decreased CD4+ T-cell counts and increased plasma viral loads compared to controls (baseline CD4+ T-cell > 200 cells/µL), and they were categorized according to immune recovery criteria based on CD4+ T-cell counts after 48 weeks of suppressive cART (48w ART) as immunological recoverers (IRs, n = 118) or immunological nonrecoverers (INRs, n = 108). The INRs were older, presented significantly lower CD4+ T-cell counts at baseline and were associated with intravenous drug use at enrolment.
Table 1.
Study cohort (n = 502) characteristics of the according classification criteria .
| Control (n = 262) | Cases (n = 240) | P-value* | IR (n = 108) | INR (n = 118) | P-value** | |
|---|---|---|---|---|---|---|
| Pre-ART clinical characteristics | ||||||
| Age at cART initiation (years) | 37 [31–45] | 39 [34–48] | 0.001 | 37 [33–42] | 42 [36–50] | <0.001 |
| Male | 209 (80.38) | 192 (80.67) | 0.545 | 94 (79.67) | 83 (76.85) | 0.610 |
| Risk factor | 0.268 | 0.004 | ||||
| Heterosexual | 89 (34.23) | 92 (38.65) | 43 (37.07) | 47 (43.52) | ||
| Homo/Bisexual | 128 (49.23) | 94 (39.50) | 58 (50.00) | 35 (32.41) | ||
| Intravenous drug abuse | 37 (14.23) | 47 (19.75) | 13 (11.21) | 23 (21.3) | ||
| Other/Unknown | 6 (2.29) | 5 (2.10) | 2 (1.72) | 3 (2.78) | ||
| CD4+T-cell count (cells/µL) | 327 [265–439] | 92 [37–162] | <0.001 | 135 [59–182] | 59 [19–116] | <0.001 |
| Plasma HIV RNA load (log copies/mL) | 4.88 [4.30–5.26] | 5.15 [4.72–5.61] | <0.001 | 5.08 [4.74–5.61] | 5.27 [4.77–5.67] | 0.587 |
| HCV co-infection (Positive) | 38 (16.17) | 48 (22.53) | 0.257 | 22 (19.47) | 22 (22.68) | 0.116 |
| Pre-ART plasma chemokine concentrations | ||||||
| SDF-1 (pg/mL) (n = 377) | 0.25 [0.21–0.30] | 0.26 [0.22–0.30] | 0.074 | 0.25 [0.21–0.30] | 0.26 [0.24–0.31] | 0.036 |
| Log RANTES (pg/mL) (n = 380) | 1.45 [0.77–1.91] | 1.75 [1.04–1.97] | 0.006 | 1.61 [0.97–1.95] | 1.86 [1.11–1.98] | 0.104 |
| Log FK (pg/mL) (n = 345) | 0.66 [0.57–0.79] | 0.67 [0.55–0.76] | 0.583 | 0.66 [0.55–0.76] | 0.68 [0.55–0.78] | 0.304 |
| Log MCP-1 (pg/mL) (n = 345) | 2.40 [2.18–2.61] | 2.51 [2.31–2.73] | 0.002 | 2.47 [2.24–2.69] | 2.58 [2.39–2.78] | 0.037 |
| Log MIP-a (pg/mL) (n = 231) | 1.46 [1.24–1.70] | 1.34 [1.21–1.50] | 0.006 | 1.31 [1.18–1.49] | 1.39 [1.21–1.55] | 0.424 |
| Log MIP-b (pg/mL) (n = 223) | 1.69 [1.52–1.81] | 1.64 [1.49–1.80] | 0.471 | 1.63 [1.49–1.78] | 1.66 [1.45–1.81] | 0.571 |
Data are presented as n (%) or median (interquartile range). Categorical data were compared by means of a χ2 test, whereas continuous data were compared using non-parametric Mann-Whitney test (P* value for comparison between control and cases, P** value for comparison between IR and INR). P value 〈 0.05 was considered significant and is highlighted in bold. All P values 〉 0.05 but < 0.10 were considered relevant for results interpretation and are italicized. INR, incomplete immune recoverers; IR, immune recoverers.
MCP-1/CCL2, monocyte chemoattractant protein-1; MIP-α/CCL3, macrophage inflammatory protein-1 alpha; MIP-β/CCL4, macrophage inflammatory protein-1 beta; RANTES/CCL5, regulated upon activation, normal T cell expressed and secreted and SDF-1/CXCL12, stromal cell-derived factor 1.
3.2. SDF-1 and MCP-1 are prognostic markers of the immune response to cART
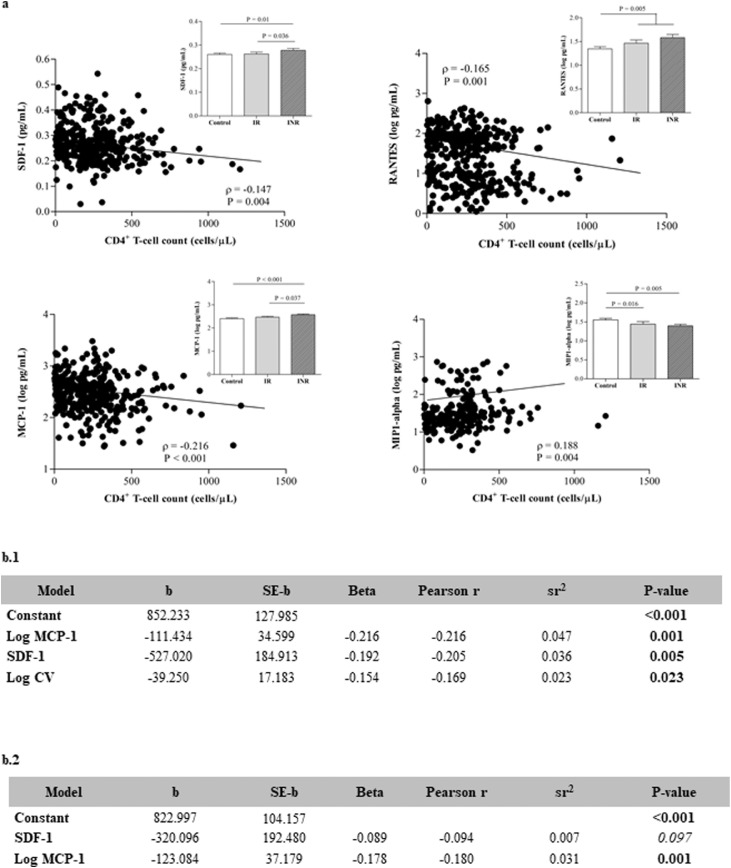
Higher SDF-1 (P = 0..074), RANTES (P = 0..006) and MCP-1 (P = 0..002) plasma concentrations (non-parametric Mann-Whitney was used to compare differences between groups, P values 〈0.05 considered statistically significant, and P values 〉 0.05 but < 0.10 were considered relevant for results interpretation) negatively correlated with these chemokines to baseline CD4+ T-cell counts (Fig. 2). Of interest, the baseline SDF-1 and MCP-1 plasma concentrations were higher in INRs than in IRs. Therefore, these two molecules are related not only to the pre-cART low CD4+ T-cell count but also to immune recovery prognosis. In contrast, MIP-alpha (MIP-α) showed a positive association with baseline CD4+ T-cell counts (P = 0.004, Spearman correlation test). In fact, plasma concentrations of MIP-α were significantly higher in controls compared to cases (Table 1, P = 0.006, non-parametric Mann-Whitney test), although no differences were observed between INRs and IRs.
Fig. 2.
a) Correlation analysis between the baseline (pre-cART) CD4+T-cell counts and selected chemokines (Spearman correlation test). Insert shows the baseline circulating concentrations of the selected chemokines in INRs (n = 85) and IRs (n = 89) compared with those in a group of control participants (n = 206) (non-parametric Mann-Whitney test). b) Association of plasma chemokine concentrations to pre-cART low CD4+T-cell counts (b.1) and immune recovery status (b.2) (Multiple regression analysis). b.1) Stepwise regression results for the prediction of the baseline pre-cART CD4+T-cell counts. The dependent variable was baseline pre-cART CD4+T-cell counts and the independent variables tested initially were log CV, SDF-1/CXCL12, log fractalkine/ CX3CL1, log MCP-1/CCL2, log MIP-α/CCL3, log MIP-β/CCL4 and log RANTES/CCL5. b.2) Standard regression results for the prediction of immune recovery status. The dependent variable was CD4+T-cell counts after 48 weeks of cART and the independent variables were SDF-1/CXCL12 and log MCP-1/CCL2. sr2 = squared semi-partial correlation used to calculated the percentage of the variance.
The independent association between baseline CD4+ T-cell counts and plasma chemokine concentrations was corroborated using stepwise multiple regression analysis with the baseline CD4+ T-cell count as the dependent variable. The model was statistically significant (F(3, 198) = 8.316, P < 0.001) and accounted for approximately 10% of the variance in the baseline CD4+ T-cell counts (R2 = 0.112, adjusted R2 = 0.098). These observations suggest that lower CD4+ T-cell counts are primarily predicted by higher plasma MCP-1 and SDF-1 concentrations and to a lesser extent by higher plasma viral loads, which accounted for approximately 5%, 4% and 2% of the variance in the baseline CD4+ T-cell counts, respectively (Fig. 2b.1). Thus, the potential for the circulating MCP-1 and SDF-1 baseline values to be prognostic markers of immune status due to poor CD4+ T-cell count recovery after 48 w cART was analysed using standard regression analysis with 48w cART CD4+ T-cell count as the dependent variable. In that case, the model was statistically significant (F(2, 333) = 7, P = 0.001, standard regression analysis) and accounted for approximately 4% of the variance in the 48w cART CD4+ T-cell counts (MCP-1 accounted for approximately 3% of the variance). Thus, we confirmed that these two molecules are related to low CD4+T-cell counts pre-cART and at 48w, playing an important role in immune recovery prognosis (Fig. 2b.2). It is important to highlight that the R-squared is a measure of exploratory power, not fit, and for that reason reporting the value of R-squared significantly different from 0, even less than 0.7 (generally considered a model with very good fit), indicated that the regression model had statistically significant exploratory power.
3.3. SDF-1 is a marker of poor progression, and the MCP-1& MIP-α are markers of ΔCD4+T-cells
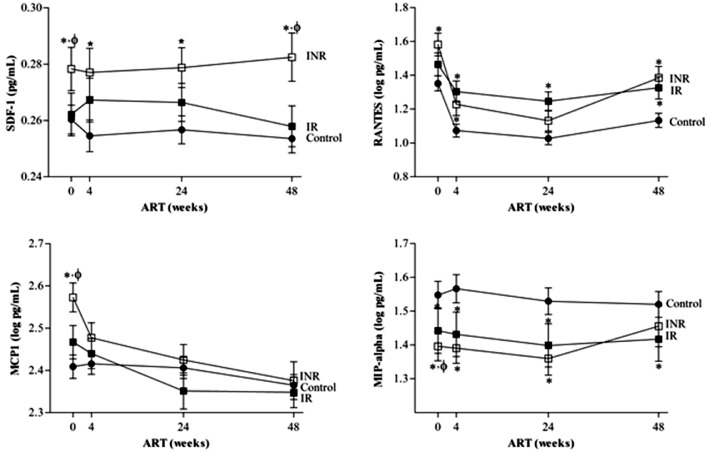
The circulating plasma SDF-1 and RANTES concentrations remained significantly elevated in cases compared to controls after cART for 48 weeks (Fig. 3). In fact, the level of SDF-1 was significantly different between INRs and IRs at both baseline (P = 0.036, non-parametric Mann-Whitney test) and after 48 w of cART (P = 0.036, non-parametric Mann-Whitney test), which indicates SDF-1 as not only a prognostic marker of the immune response but also disease progression. On the other hand, circulating plasma MIP-α levels were higher in controls than in cases after 48w of cART, although no differences were observed between INRs and IRs. Regarding plasma MCP-1 concentrations, the initiation of ART decreased the circulating chemokine concentration in all participants. In fact, the percentage decrease in MCP-1 (ΔMCP-1) during cART was significant between cases and controls (P = 0.001, non-parametric Mann-Whitney test) but not between INRs and IRs (P = 0.100, non-parametric Mann-Whitney test).
Fig. 3.
Longitudinal evaluation of SDF-1/CXCL12 (n = 208 controls, n = 85 INR and n = 88 IR), RANTES/CCL5 (n = 208 controls, n = 85 INR and n = 89 IR), MCP-1/CCL2 (n = 185 controls, n = 77 INR and n = 80 IR) and MIP-α/CCL-3 (n = 130 controls, n = 48 INR and n = 53 IR) during 48 weeks of cART in INRs and IRs compared to a control group of subjects.
Data are represented as the mean±SEM. Non-parametric Mann-Whitney test, * significant differences to control group and ϕ significant differences to IRs) .
Finally, the percentage increase in the CD4+ T-cell counts (ΔCD4+ T-cell) was calculated to evaluate its association with the percentage increase/decrease in each chemokine during the 48w of cART. The ΔCD4+ T-cell count was positively related to the ΔMIP-α concentrations (ρ= 0.137, P = 0.040, Spearman correlation test), and inversely correlated with the ΔMCP-1 plasma values (ρ= −0.124, P = 0.023, Spearman correlation test). These results corroborated MCP-1 and MIP-α as general markers of CD4+ T-cell responses to cART follow-up.
3.4. Genetic association study of chemokine (CXCL12 and CCL5) gene variants
Table 2 summarizes the distribution of CXCL12 rs1801157, located on chromosome 10, and CCL5 rs2280789, CCL5 rs2280788 and CCL5 rs2107538, located on chromosome 17, among the groups, which were in accordance with the data listed in the NCBI SNP database and consistent with the Hardy-Weinberg equilibrium (HWE).
Table 2.
General characteristics for the genetic study of CXCL12 (SDF-1) and CCL5 (RANTES) gene variants.
| Polymorphism | Chr Position | Genotype | Cases | Control | P-value | Cases |
P-value | HWE |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|
| INR | IR | Control | INR | IR | |||||||
| CXCL12 rs1801157 C>T | chr10:44,372,809 | CC | 144 (68%) | 151 (67%) | 0.439 | 64 (67%) | 73 (71%) | 0.065 | 0.054 | 0.33 | 0.12 |
| CT | 62 (29%) | 61 (27%) | 27 (28%) | 30 (29%) | |||||||
| TT | 7 (3%) | 13 (6%) | 5 (5%) | – | |||||||
| CCL5 rs2280789 T>C | chr17:35,879,999 | TT | 156 (75%) | 176 (80%) | 0.177 | 75 (77%) | 73 (74%) | 0.812 | 0.48 | 0.63 | 0.43 |
| TC | 47 (23%) | 45 (20%) | 20 (21%) | 23 (23%) | |||||||
| CC | 5 (2%) | 1 (0%) | 2 (2%) | 3 (3%) | |||||||
| CCL5 rs2280788 C>G | chr17:35,880,401 | CC | 205 (95%) | 219 (95%) | 0.948 | 98 (95%) | 96 (96%) | 0.767 | 1 | 1 | 1 |
| GC | 10 (5%) | 11 (5%) | 5 (5%) | 4 (4%) | |||||||
| CCL5 rs2107538 C>T | chr17:35,880,776 | CC | 135 (64%) | 144 (64%) | 0.548 | 64 (66%) | 64 (62%) | 0.348 | 0.41 | 0.16 | 1 |
| CT | 66 (31% | 73 (33%) | 30 (31%) | 31 (30%) | |||||||
| TT | 11 (5%) | 7 (3%) | 3 (3%) | 8 (8%) | |||||||
P-values were calculated by the Chi-square test. HWE, Hardy-Weinberg equilibrium.
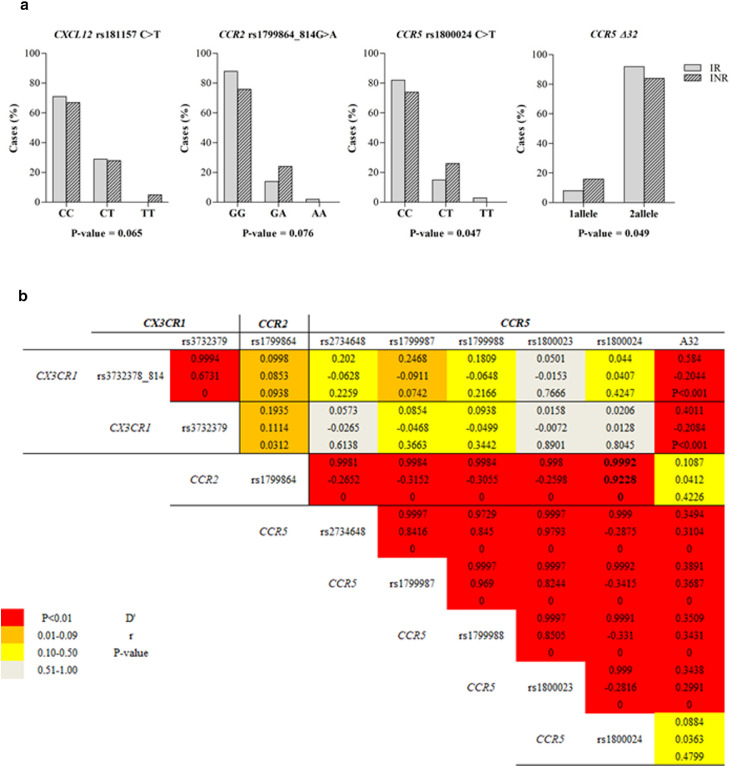
Regarding CXCL12 rs1801157, our results suggested a potential association between SDF-1 expression and the immune response to cART (P = 0.065, Fig. 4a). Five inheritance models were fitted, which correspond to different groupings of the genotypes, and the Akaike's Information Criterion (AIC) and Bayesian Information Criterion (BIC) calculated to select the best model for each specific SNP. In fact, considering the AIC and BIC scores adjusted for the baseline pre-cART CD4+ T-cell counts, an association with immune recovery status was detected with the recessive model (Table 3).
Fig. 4.
Genetic association study of selected chemokine and chemokine receptor gene variants. a) Distribution of CXCL12 rs1801157, CCR2 rs1799864_814, CCR5 rs1800024 and CCR5 rs333_814 (Δ32) among cases (n = 100 INR and n = 100 in IR) (P-values were calculated by the Chi-square test). b) Linkage disequilibrium analysis in cases (INRs versus IRs) for chemokine receptor located on chromosome 3 (n = 450) (Multiple SNP-analysis by SNPstats software).
Table 3.
Association between CXCL12 and CCL5 gene variants to immunological recovery status after 48 weeks of cART.
| Polymorphism | IHT model | Genotype | INR | IR | OR (95% CI) | P-value | AIC | BIC |
|---|---|---|---|---|---|---|---|---|
| A) CXCL12 rs1801177 was associated to the immune recovery status after 48 weeks of cART. Data analysis is summarized with n (%), odds ratio (OR) and 95% confidence interval (CI). The AIC and BIC values were used to choose the inheritance model (IHT) (adjusted by baseline CD4+T-cell counts) that best fits the data. AIC, Akaike Information Criteria; BIC, Bayesian Information Criteria. | ||||||||
| rs1801157 C>T |
Codominant | C/C | 64 (66.7%) | 73 (70.9%) | 1.00 | 0.038 | 249.6 | 262.8 |
| C/T | 27 (28.1%) | 30 (29.1%) | 0.86 (0.44–1.68) | |||||
| T/T | 5 (5.2%) | 0 (0%) | 0.00 (0.00-NA) | |||||
| Dominant | C/C | 64 (66.7%) | 73 (70.9%) | 1.00 | 0.34 | 253.3 | 263.2 | |
| C/T-T/T | 32 (33.3%) | 30 (29.1%) | 0.73 (0.38–1.39) | |||||
| Recessive | C/C—C/T | 91 (94.8%) | 103 (100%) | 1.00 | 0.012 | 247.8 | 257.7 | |
| T/T | 5 (5.2%) | 0 (0%) | 0.00 (0.00-NA) | |||||
| Overdominant | C/C-T/T | 69 (71.9%) | 73 (70.9%) | 1.00 | 0.82 | 254.1 | 264 | |
| C/T | 27 (28.1%) | 30 (29.1%) | 0.93 (0.48–1.80) | |||||
| Polymorphism | Cases-Controls |
INR-IR |
||||||
|---|---|---|---|---|---|---|---|---|
| D’ | r | P-value | D’ | r | P-value | |||
| B) Linkage disequilibrium analysis for CCL5 gene variants. | ||||||||
| rs2280789- rs2280788 | 0.9965 | 0.4176 | < 0.001 | 0.9964 | 0.3794 | < 0.001 | ||
| rs2280789- rs2107538 | 0.9995 | 0.7398 | < 0.001 | 0.9995 | 0.7723 | < 0.001 | ||
| rs2280789-rs2107538 | 0.9961 | 0.3090 | < 0.001 | 0.9961 | 0.2931 | < 0.001 | ||
No association was found between the selected CCL5 gene variants and a pre-cART low CD4+ T-cell count or immune response to cART. However, a pairwise linkage disequilibrium (LD) estimate was obtained for the CCL5 gene polymorphisms (Table 3b), revealing that all the SNP marker combinations exhibited perfect LD scores in each of the analysed groups. Any of the possible haplotypes were related to low pre-cART CD4+ T-cell counts or to cART-associated immune recovery (data not sown).
3.5. Genetic association study of selected chemokine receptor gene variants
Table 4 summarizes the characteristics of the CX3CR1 rs3732178_814 and rs3732379, CCR2 rs1799864_814, and CCR5 rs2734648, rs1799987, rs1799988, rs1800023, rs1800024 and rs333_814 polymorphisms located on chromosome 3 among the groups, which were in accordance with the data listed in the NCBI SNP database and the HWE. The genotype frequencies for all selected chemokine receptor gene variants explored in this study were consistent with the HWE. No association was found between the CX3CR1 gene variants, and pre-cART low CD4+ T-cell counts or immune recovery status. However, our results suggested a potential association between CCR2 rs1799864_814 and the immune response to cART (P = 0.076, Table 4 and Fig. 4). Considering the AIC and BIC scores (adjusted for the baseline pre-cART CD4+ T-cell counts), the association was detected for the codominant model (G/A, OR = 0.48, 95% CI = 0.22–1.06, P = 0.047). However, the confidence interval includes 1, and thus there is insufficient evidence to conclude that the groups are statistically significantly different.
Table 4.
General characteristics for the genetic study of CX3CR1, CCR2 and CCR5 gene variants.
| Polymorphism | Chr Position | Genotype | Cases | Control | P-value | Cases |
P-value | HWE |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|
| INR | IR | Control | INR | IR | |||||||
| CX3CR1rs3732378_814 G>A | chr3:39,265,671 | GG | 153 (73%) | 171 (76%) | 0.379 | 68 (72%) | 78 (76%) | 0.517 | 0.55 | 0.68 | 1 |
| GA | 54 (26%) | 49 (22%) | 26 (27%) | 23 (23%) | |||||||
| AA | 2 (1%) | 5 (2%) | 1 (1%) | 1 (1%) | |||||||
| CX3CR1 rs3732379 C>T | chr3:39,265,765 | CC | 111 (54%) | 118 (55%) | 0.733 | 47 (51%) | 58 (58%) | 0.347 | 0.49 | 0.28 | 0.59 |
| CT | 83 (4%) | 80 (37%) | 41 (45%) | 35 (35%) | |||||||
| TT | 11 (5%) | 17 (8%) | 4 (4%) | 7 (7%) | |||||||
| CCR2 rs1799864 G>A | chr3:46,357,717 | GG | 175 (82%) | 184 (83%) | 0.980 | 74 (76%) | 88 (85%) | 0.076 | 1 | 0.35 | 0.16 |
| GA | 37 (17%) | 37 (17%) | 23 (24%) | 14 (13%) | |||||||
| AA | 2 (1%) | 2 (1%) | – | 2 (2%) | |||||||
| CCR5 rs2734648 G>T | chr3:46,370,349 | GG | 78 (38%) | 76 (34%) | 0.583 | 34 (37%) | 40 (40%) | 0.919 | 0.58 | 1 | 1 |
| GT | 96 (47%) | 104 (47%) | 44 (47%) | 46 (46%) | |||||||
| TT | 31 (15%) | 42 (19%) | 15 (16%) | 14 (14%) | |||||||
| CCR5 rs1799987 A>G | chr3:46,370,444 | AA | 59 (28%) | 58 (26%) | 0.565 | 26 (27%) | 28 (27%) | 0.946 | 0.79 | 0.54 | 1 |
| AG | 107 (51%) | 109 (49%) | 52 (54%) | 52 (51%) | |||||||
| GG | 44 (21%) | 56 (25%) | 19 (20%) | 22 (22%) | |||||||
| CCR5 rs1799988 C>T | chr3:46,370,768 | CC | 59 (30%) | 62 (28%) | 0.842 | 28 (30%) | 28 (30%) | 0.851 | 0.34 | 1 | 1 |
| CT | 98 (49%) | 102 (47%) | 47 (51%) | 46 (49%) | |||||||
| TT | 41 (21%) | 55 (25%) | 18 (19%) | 20 (21%) | |||||||
| CCR5 rs1800023 A>G | chr3:46,370,817 | AA | 82 (40%) | 85 (38%) | 0.515 | 36 (38%) | 42 (42%) | 0.856 | 0.40 | 0.83 | 0.87 |
| AG | 92 (45%) | 107 (45%) | 43 (46%) | 44 (44%) | |||||||
| GG | 31 (15%) | 38 (17%) | 15 (16%) | 14 (14%) | |||||||
| CCR5 rs1800024 C>T | chr3:46,371,068 | CC | 169 (79%) | 181 (82%) | 0.749 | 70 (74%) | 86 (82%) | 0.047 | 1 | 0.36 | 0.08 |
| CT | 41 (19%) | 38 (17%) | 25 (26%) | 16 (15%) | |||||||
| TT | 3 (1%) | 2 (1%) | – | 3 (3%) | |||||||
| CCR5 rs333_814 (Δ32) |
chr3:46,373,453–46,373,487 |
1allele (AA) |
26 (13%) | 22 (10%) | 0.226 | 15 (16%) | 8 (8%) | 0.049 | – | – | – |
| 2 alleles (AB) |
108 (87%) | 201 (90%) | 76 (84%) | 95 (92%) | |||||||
Regarding CCR5 gene variants, only CCR5 rs1800024 and CCR5 rs333_814 (Δ32) were related to immune recovery status (Table 4 and Fig. 4). In the case of CCR5 rs1800024, considering the AIC and BIC scores (adjusted for baseline pre-ART CD4+ T-cell counts), an association was detected with the codominant model (C/T, OR = 0.48, 95% CI = 0.22–1.03, P = 0.024 from logistic regression analyses using SNPstats software). We also found a significant relationship between poor immune recovery status and the Δ32 homozygous genotype (1 allele, Table 5), although the risk was not found to be statistically significant in the SNP analysis (OR = 2.05, 95% CI = 0.79–5.34, P = 0.13).
Table 5.
Haplotype association analysis for the chemokine receptor variants explored in this study with immune recovery status (n = 207). The logistic regression model was adjusted by baseline pre-cART CD4+T-cell counts.
| Haplotype frequencies estimation (n = 207) |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CX3CR1 |
CCR2 | CCR5 |
Freq |
Haplotype association with response (n = 207, adjusted by CD4b) |
||||||||||
| rs3732378 | rs3732379 | rs1799864 | rs2734648 | rs1799987 | rs1799988 | rs1800023 | rs1800024 | Δ32 | INR | IR | Freq | OR (95% CI) | P-value | |
| 1 | G | C | G | T | G | T | G | C | B | 0.2237 | 0.1974 | 0.2165 | 1.00 | — |
| 2 | G | C | G | G | A | C | A | C | A | 0.1821 | 0.2123 | 0.1899 | 0.39 (0.10–1.44) | 0.16 |
| 3 | G | C | G | G | A | C | A | C | B | 0.104 | 0.0808 | 0.1019 | 0.54 (0.15–1.94) | 0.35 |
| 4 | G | C | G | T | G | T | G | C | A | 0.0930 | 0.1045 | 0.0886 | 0.12 (0.02–0.94) | 0.044 |
| 5 | A | T | G | G | A | C | A | C | A | 0.1002 | 0.0622 | 0.0688 | 0.11 (0.02–0.61) | 0.013 |
| 6 | G | T | G | G | A | C | A | C | A | 0.0343 | 0.0552 | 0.0578 | 1.06 (0.25–4.56) | 0.94 |
For the Δ32 analyses, the homozygous genotype (1 allele) was described as AA and the heterozygous genotype (2 alleles) was described as AB.
Only haplotype association with response that showed more than 0.05 frequencies was considered for the results.
Finally, multiple–SNP analysis was performed for all chemokine receptor variants located on chromosome 3 ordering them by the correlative chromosome position. LD was found between several genetic variants (Fig. 4b). Interestingly, among cases, a strong LD was found between the two chemokine receptor variants, CCR2 rs1799864_814 and CCR5 rs1800024, which were associated with the immune response in the single SNP analysis, (D’ = 0.9992, P < 0.001, LD analysis using SNPstats software), indicating that these alleles of each gene are inherited together more often than would be expected by chance (Fig. 4b). The haplotypes GCGTGTGCA (OR = 0.12, 95% CI = 0.02–0.94, P = 0.044, haplotype analyses using SNPstats software) and ATGGACACA (OR = 0.11, 95% CI = 0.02–0.61, P = 0.013, haplotype analyses using SNPstats software) were associated with the immune response according to the CD4+ T-cell counts after 48 weeks of cART (Table 5).
3.6. CXCL12 rs1801157 and CCL5 rs2280788 impact the baseline chemokine values
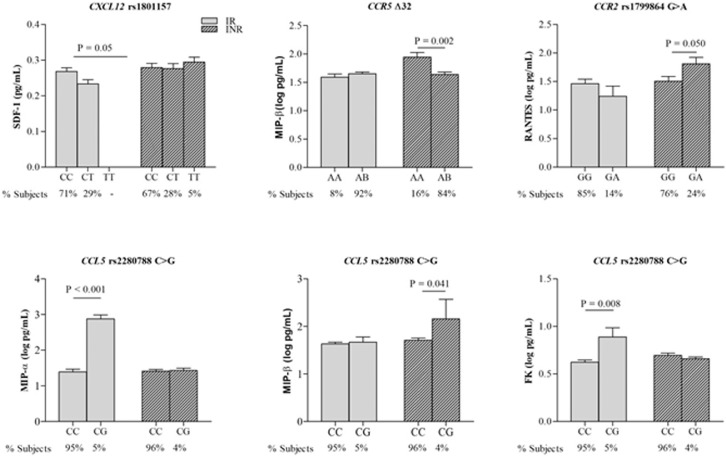
Then, the impact of the selected chemokine gene variants on circulating chemokine concentrations was explored in cases (INR and IR) to establish any possible association with immune recovery status in response to cART (Fig. 5). Interestingly, among the CCL5 gene variants explored in this study, only CCL5 rs2280788 was associated with circulating chemokine concentrations. Rather than being associated with plasma CCL5 values, CCL5 rs2280788 was strongly related to plasma fractalkine (P = 0.008, one-way ANOVA test) and MIP-α (P < 0.001, one-way ANOVA test) values in IRs and circulating MIP-β (P = 0.041, one-way ANOVA test) in INRs. Regarding CXCL12 rs1801157, an association was found between this SDF-1 gene variant and the circulating SDF-1 concentration in IRs (P = 0.05, one-way ANOVA test).
Fig. 5.
Influence of chemokine receptor genes in circulating chemokine concentrations in INRs compared to IRs. Data are represented as the mean±SD (One-Way ANOVA testing for association between listed parameters and SNP genotypes).
3.7. CCR2 rs1799864 and CCR5-Δ32 determine the chemokine concentration in INR
No association was found between the selected CX3CR1 variant and the vast majority of CCR5 gene variants and circulating chemokine values. Only CCR5-Δ32 was significantly related to the plasma MIP-β concentration in INRs (P = 0.002, one-way ANOVA test), although the expression of this chemokine in the circulation was not related to pre-cART CD4+ T-cell counts or to immune response in this study. On the other hand, the expression of CCR2 rs1799864_814 was significantly related to the plasma RANTES concentration in INRs, corroborating the binding of this chemokine to the CCR2 receptor (Fig. 5) and the potential role in the immune response.
4. DISCUSSION
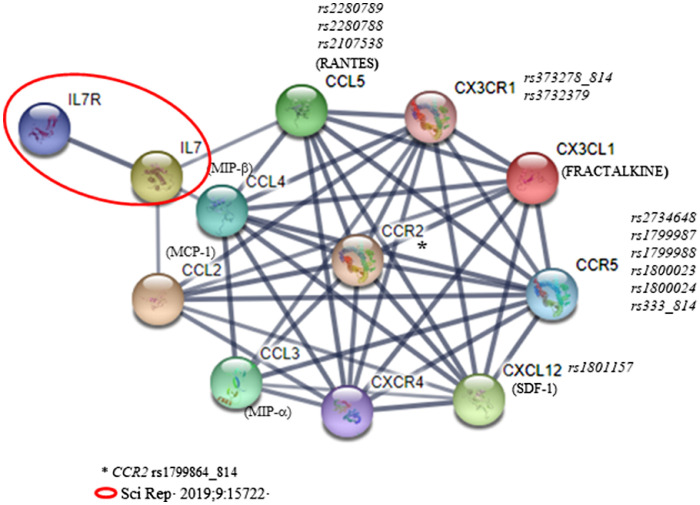
The cytokine-chemokine network is enormously complex with a large number of different involved molecules (ligands, receptors and regulatory proteins, among others) that play fundamental roles in the development and homeostasis of the immune system. [13] Accordingly, the chemokine-chemokine receptor system clearly plays an important role in the HIV life cycle, disease progression, and HIV reservoir establishment, [15], [16], [17] presenting new perspective for the development of effective therapeutic approaches in response to HIV/AIDS susceptibility during the last 10 years. [18] In this context, the present study aims to investigate the network associated with the selected CCL chemokines (Fig. 6) which are linked to our previous work focused on immune reconstitution. [9,10] Network modeling elucidated the relationship between glutamate metabolism to CCL2/CCR2, CCL5 and CX3CR1 axis in CD4+ T-recovery [9] and we anticipated that the study of IL-7 and its receptor could support new insights regarding T-cell reservoir in HIV-positive participants. [10] Based on these findings we firmly believed that the study of the previously identified chemokines related to IL-7/IL-7R axis may reveal promising results on immune reconstitution. For that reason, cytokine IL-7 and chemokines CCL2 and CCL5 have been selected as input in the STRING interface to expand the chemokine-chemokine receptor network and to establish which chemokine and chemokine receptor candidates will be included in the present work (Fig. 6). To our knowledge, this is the first work that jointly evaluates this network of chemokines as prognostic and follow-up markers of immune recovery status by determining circulating concentrations and performing a genetic polymorphism study. Concretely, our main findings were that the baseline plasma MCP-1/CCL2 and SDF-1/CXCL12 concentrations accounted for approximately 9% and 4% of the variance of the baseline and 48w cART CD4+ T-cell counts, respectively, and that, the CXCL12 rs1801157 was associated with the immune response. Additionally, regarding chemokine receptors, we demonstrated that while CCR2 rs1799864_814 and CCR5 rs1800024 revealed a strong LD, the combination of the different CX3CR1, CCR2 and CCR5 SNPs analyzed in chromosome 3 resulted in two haplotypes that were significantly associated with the immune response according to the CD4+ T-cell counts after 48 weeks of cART.
Fig. 6.
Chemokine-chemokine receptor interaction analysis based on previously available data [9,10] using the STRING database [38]. The cytokine IL-7 and chemokines CCL2 to CCL5 have been selected as input, expanded by an additional 5 proteins in the STRING interface and the confidence cut-off for showing interactions links has been set to high confidence (0.700).
Circulating concentrations of SDF-1/CXCL12 and CCL5/RANTES were higher in cases compared to those in controls at both baseline and after 48w cART, but only SDF-1/CXCL12 was crucial for the differentiation among INRs and IRs. Our results suggest that elevated plasma SDF-1/CXCL12 is a prognostic indicator of low CD4+ T-cell counts preceding immune failure in response to cART and a follow-up marker of poor immune reconstitution despite successful virological suppression during cART. Although increased plasma levels of SDF-1/CXCL12 were previously related to HIV infection, [19] data on its role as a prognostic marker of immune status are still scarce. SDF-1/CXCL12 is the most primitive chemokine that regulates development in multiple systems and diverse cell functions, including proliferation and survival in the immune system, through the two known receptors CXCR4 and ACKR3. [13,15,20] Interestingly, the affinity of CXCL12/SDF-1 for CXCR4 correlates well with its action as a suppressor of infection by T-tropic HIV-1 species and its ability to induce CXCR4 internalization. [21] In this context, the observed adverse effect of SDF-1 gene expression on the natural history of HIV-1 disease could be explained by the fact that the SDF-1–3′T allele was more likely to be associated with detectable X4- tropic viruses. [22] Consistent with these data, we observed that the expression of the CXCL12 gene influenced circulating SDF-1 concentrations in IRs, and according to Restrepo et al., [23] the expression of CXCL12 rs1801157 TT genotypes was associated with a higher probability of CD4 T-cell recovery failure in HIV patients despite complete viral suppression with cART. In fact, we observed that all cases included in the study carrying the CXCL12 rs1801157 TT genotypes were associated with INRs, and in contrast, the number of participants carrying the CXCL12 rs1801157 CC genotypes were higher in the group of IRs compared to the group of INRs (P = 0.065). By using the HaploReg software, we found that the CXCL12 rs1801157 is in high LD with SNPs involved in the promotion and enhancement of histone marks and motif changes. In the light of these results, protective role of the CXCL12 rs1801157 C-allele and CC genotype in chronic lymphocytic leukaemia development was observed, whereas the presence of the T variant was linked with the higher risk of the disease as well as impaired response to the therapy independently of the regimen used. Interestingly, the presence of the CXCL12 rs1801157 variant may be related with a decrease of CXCL12 and increase of its receptor (CXCR4) mRNA expression levels. [24]
The other chemokine that played a potential role in immune reconstitution was MCP-1/CCL2; increased baseline values were a good marker of poor immune prognosis, accounting for approximately 3% of the variance in CD4+ T cells after 48 weeks of treatment. MCP-1/CCL2 is an inflammatory chemokine produced by several types of cells and a chemoattractant for CD4+ T cells, monocytes/macrophages, and NK cells, recruiting them to the sites of infection and inflammation. [13,15] High CCL2/CCR2 levels were previously linked to several HIV-associated disorders through leukocyte recruitment and maintenance of the inflammatory status, [13,15,16,25] but their association with previous poor immune reconstitution in HIV participants on ART has been poorly studied, and the results remain unclear. In the present study, we found an association between the CCR2 rs1799864_814 AG genotype and the probability of presenting poor immune reconstitution after cART, which corroborates the recent work presented by Restrepo et al. [23] By using HaploReg software, we found that CCR2 rs1799864 is in very high LD with numerous SNPs implicated in the promotion and enhancement of histone marks, protein binding and motif changes. In fact, the stability or expression of CCR2 was previously related to Alzheimer's disease (AD) and mild cognitive impairment (MCI) as a consequence of higher MCP-1 levels associated with the A allele of CCR2 rs1799864. [26]
The CCR5 chemokine receptor receives great attention in the context of viral infection due to its role as an HIV-1 coreceptor [27] and because it is a natural ligand of multiple chemokines, including MIP-1α/CCL-3, MIP-1β/CCL-4, and RANTES/CCL-5. [27], [28], [29] In fact, chemokine receptors are firmly considered important therapeutic targets for the treatment of many human diseases, [30] and the dual CCR5/CCR2 targeting is emerging as a therapeutic strategy for complex human diseases, including HIV infection. [29] We found that the Δ32 homozygous genotype was related to poor immune recovery status and that the CCR5 rs1800024 CT polymorphism was significantly associated with INRs. CCR5Δ32 exerts a robust phenotypic effect on CCR5, [28] and CCR5Δ32 mutations were previously associated with differential gene expression which in most cases is critical for the immune response. [31] Moreover, the most interesting agreement with the literature is the synergy between CCR2 rs1799864_814 and CCR5 rs1800024, both located on chromosome 3. Mutations in CCR2 rs1799864 and CCR5 rs1800024 were found to predict HIV transmission, [32] but nothing has been described in relation to immune recovery status until now. The multiple SNP analysis showed a high LD between CCR2 rs1799864_814 and CCR5 rs1800024, and the expression of the CCR2 rs1799864_814 AG genotype was significantly related to increased plasma RANTES concentrations in INR participants. In this context, our data also revealed that the alleles of different genes in the same chromosome are inherited together more often than would be expected by chance. Notably, two haplotypes (GCGTGTGCA and ATGGACACA) resulting from combined CX3CR1, CCR2 and CCR5 chemokine receptor gene expression were associated with the immune response based on the CD4+ T-cell counts after 48 weeks of cART. Taken all together, our data supports the proof-of-concept studies of experimental chemokine receptors as promising candidates for advanced clinical trials. From the three chemokine antagonist that have been approved yet, one of them is a non-competitive allosteric that targets the chemokine recognition site 2 of CCR5 to stabilize the chemokine receptor in an inactive conformation. [30] It is Maraviroc, a small well-tolerated molecule approved by the US FDA to treat HIV-1 infection with special interest in participants with CCR5 tropism because their capability to prevent the binding of chemokine and HIV gp120. Additionally, other phase 3 trials are ongoing to evaluate this capability of CCR5 to be a potential therapeutic candidate (e.g. leronlimab or cenicriviroc), but of interest is cenicriviroc. Cenicriviroc is a well-tolerated dual CCR2 and CCR5 antagonist that has been evaluated for the treatment of liver fibrosis in HIV-negative adults with alcoholic steatohepatitis (AURORA study). In fact, promising data suggest that cenicriviroc could improve hepatic inflammation, insulin resistance and liver fibrosis by the inhibition of CCR2 monocyte recruitment which directly affects CCL2 signaling.
A limitation of our study is that the number of patients per group in genetic studies is not consistent with the number of samples available for the study of circulating chemokine-chemokine receptor concentrations, which would make our results more consistent in the search for predictive and diseases progression markers. Recently, Norris et al. [33] also evaluated serum levels of 30 different cytokines (including 17 chemokines) at baseline and at one and two years after viral control to determine whether cytokine levels correlated with INR status, and minimal differences were observed between IRs and INRs. The study was performed in a group of 50 INRs compared to 50 IRs from the Women's Interagency HIV Study and only circulating levels were tested. Thus, the present work is the largest study evaluating circulating chemokine levels and their association to genetic chemokine and chemokine receptor variants ever done. Another characteristic of the present study was that lower pre-cART CD4+ T cell counts median values in INR subjects can be observed compared to IR subjects, probably due to the relevance of the CD4+ T-cell count before cART onset as an intrinsic risk factor of immune failure to cART as previously described. [9,10] On the other hand, we did not include CCL3L1–CCL4L1 copy number variation (CVN) due to their complexity and controversial association to HIV infection. Ahuja et al. [34] postulated that variations in the CCL3L1-CCR5 axis influences the recovery of CD4+ T-cell counts and that the greatest impact of these genotypes appears to be on sustaining the initial CD4+T-cell gains after two years of cART in individuals initiating cART at more than 350 CD4+ T-cell counts. Thus, because our study design classified participants by their initial CD4+T-cell count, we considered that in this study CCL3L1–CCL4L1 CVN results would be biased by the study design.
In conclusion, taken together, our data corroborated that the chemokine-chemokine receptor system is more complex than indicated by the study of a selected chemokine and its natural ligands due to the network existing among the different chemokines and their receptors. We want to highlight that some therapeutic strategies involving the use of chemokine inhibitors (e.g. Il-2, IL-7) has already been tested but unfortunately the results obtained are still so far away to be effective. Thus, it is very likely that new individual or combinatorial approaches including cytokines and/or chemokines with immune checkpoint blockage are needed to achieve complete immune recovery status in poor immune recovery HIV-positive patients. In the present study, we suggest that the SDF-1/CXCL12, MCP-1/CCL-2 and CCR5 receptors are key effectors of susceptibility to HIV progression, especially in terms of immune reconstitution. Circulating concentrations and gene expression patterns of SDF-1/CXCL12 (CXCL12 rs1801157) and MCP-1/CCL2 (CCR2 rs1799864_814) are associated with immune recovery status and could be considered prognostic markers of immune failure despite cART. In fact, circulating concentrations of SDF-1 at 48 weeks of cART follow-up could be considered a marker of disease progression, whereas an increase in MCP-1 values could denote a decline in the CD4+ T-cell count. Thus, it is not surprising that targeting CXCL12/CXCR4 axis in tumor immunotherapy has emerged because of its critical role in the regulation of cancer stem cells, and in the activation of multiple signaling pathways (ERK1/2, ras, p38 MAPK, PLC/ MAPK, and SAPK/ JNK) for tumor initiation and progression. [35] Pharmacological inhibition of MCP-1/CCL2 with Spiegelmer mNOX-E36, a mirror-image oligonucleotide with good tolerability, has been investigated as adjunct immunosuppressive therapy in transplantation by their ability to bind to a relevant target molecule in a manner conceptually similar to the way antibodies recognize antigens. [36] Of interest, dual blockade of CCL2/MCP-1 and CXCL12/SDF-1 revealed additive therapeutic effects in murine proliferative lupus nephritis because the anti-inflammatory effects caused by CCL2/MCP-1 blockage and the glomerular filtration barrier protection due to CXCL12/SDF-1 inhibition. [37] Regarding CCR5 gene variants, only CCR5 rs1800024 and CCR5 rs333_814 (Δ32) were related to immune recovery, but interestingly, strong LD between CCR2 rs1799864_814 and CCR5 rs1800024 indicates that these alleles of each gene are inherited together more often than would be expected by chance. In summary, our results open a new window to proof-of-concept studies for chemokines (SDF-1/CXCL12 and MCP-1/CCL-2) and chemokine receptors (CCR2 and CCR5) blockage as potential therapeutic options for advanced clinical trials with promising fortunate outcomes.
Further investigation to clarify the synergy between the expression of different chemokines and chemokine receptors could provide new insights regarding the mechanisms implicated in the immune reconstitution, which could be exploited for emerging therapeutic strategies in INRs.
Data availability
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Funding
This work was supported by the Fondo de Investigacion Sanitaria [PI10/02635, PI13/0796, PI14/ 0063, PI14/0700, PI16/00503, PI17/0420, PI17/0498, PI18/01216, PI19/01337 and PI20/00326]-ISCIII-FEDER (co-funded by the European Regional Development Fund/European Social Fund; “A way to make Europe”/”Investing in your future”); Programa de Suport als Grups de Recerca AGAUR (2014SGR250, 2017SGR948); Gilead Fellowship Program GLD14/293and GLD19/00,008; and The SPANISH AIDS Research Network [RD12/0017/0005, RD16/0025/0002, RD16/0025/0006, RD16/0025/0019]-ISCIII-FEDER (Spain). FV is supported by grants from the Programa de Intensificación de Investigadores (INT20/00031)-ISCIII. PD is supported by grants from the Programa de Intensificación de Investigadores (INT19/00036)-ISCIII. YMP is supported by the Servicio Andaluz de Salud through Programa Nicolás Monardes (C-0013/17). JVG is supported by the Instituto de Salud Carlos III (ISCIII) under grant agreement “FI19/00330″ through the program “Contratos Predoctorales de Formación en Investigación en Salud”. EY is supported by the Instituto de Salud Carlos III (ISCIII) under grant agreement “FI20/00118″ through the program “Contratos Predoctorales de Formación en Investigación en Salud”. LR is supported by the Instituto de Salud Carlos III (ISCIII) under grant agreement “CD20/00105″ through the program “Sara Borrell”. AR is supported by a grant from the Acció Instrumental d'incorporació de científics i tecnòòlegs (PERIS SLT002/16/00,101), Departament de Salut, Generalitat de Catalunya, by IISPV through the project “2019/IISPV/05″ (Boosting Young Talent), by GeSIDA through the "III Premio para Jóvenes Investigadores 2019″ and by the Instituto de Salud Carlos III (ISCIII) under grant agreement “CP19/00,146″ through the Miguel Servet Program.
Author contributions
All authors have seen and approved the submitted version of the manuscript. The authors’ contributions are as follows: experimental design (EY, CV, PD, FV, JP, AR) and intellectual guidance (PD, YMP, SV, VF); recruitment of participants (EY, CV, PD, AC, SV, AI, VF, JM, JP) and sample procurement (EY, AC, VA, MV, AM); data collection (CV, JV-G, MP, CP, JM, LR); data analysis and interpretation (EY, VA, JM, LR, AR); manuscript preparation (EY, JM, LR, AR). EY, CV, PD, FV, JP and AR were responsible for the study design, data analysis, and article development. PD, YMP, VF, JM, FV, JP and AR reviewed and edited the manuscript.
Declaration of Competing interest
All authors declare they do not have anything to disclose regarding conflict of interest with respect to this manuscript.
Acknowledgments
This study would not have been possible without the collaboration of all the patients and medical and nursing staff who have taken part in the project. We also acknowledge the BioBanc IISPV (B.0000853 + B.0000854) integrated in the Spanish National Biobanks Platform (PT13/0010/0029 & PT13/0010/0062) for its collaboration. Authors greatly appreciate the comments and criticisms of the anonymous reviewers that greatly helped to improve the manuscript.
References
- 1.Battegay M., Nüesch R., Hirschel B., Kaufmann G.R. Immunological recovery and antiretroviral therapy in HIV-1 infection. Lancet Infect Dis. 2006;6:280–287. doi: 10.1016/S1473-3099(06)70463-7. [DOI] [PubMed] [Google Scholar]
- 2.Yang X., Su B., Zhang X., Liu Y., Wu H., Zhang T. Incomplete immune reconstitution in HIV/AIDS patients on antiretroviral therapy: challenges of immunological non-responders. J. Leukoc. Biol. 2020;107:597–612. doi: 10.1002/JLB.4MR1019-189R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gaardbo J.C., Hartling H.J., Gerstoft J., Nielsen S.D. Incomplete immune recovery in HIV infection: mechanisms, relevance for clinical care, and possible solutions. Clin Dev Immunol. 2012;2012 doi: 10.1155/2012/670957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aldrete S., Jang J.H., Easley K.A., Okulicz J., Dai T., Chen Y.N. CD4 rate of increase is preferred to CD4 threshold for predicting outcomes among virologically suppressed HIV-infected adults on antiretroviral therapy. PLoS ONE. 2020;15:1–16. doi: 10.1371/journal.pone.0227124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pacheco Y.M., Jarrin I., Rosado I., Campins A.A., Berenguer J., Iribarren J.A. Increased risk of non-AIDS-related events in HIV subjects with persistent low CD4 counts despite cART in the CoRIS cohort. Antiviral Res. 2015;117:69–74. doi: 10.1016/j.antiviral.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 6.Buggert M., Nguyen S., McLane L.M., Steblyanko M., Anikeeva N., Paquin-Proulx D. Limited immune surveillance in lymphoid tissue by cytolytic CD4+ T cells during health and HIV disease. PLoS Pathog. 2018;14:1–28. doi: 10.1371/journal.ppat.1006973. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 7.Rosado-Sánchez I., I H.-.F., AI Á.-.R., M G., MA A.-.C., E R.-.M. A Lower Baseline CD4/CD8 T-Cell Ratio Is Independently Associated with Immunodiscordant Response to Antiretroviral Therapy in HIV-Infected Subjects. Antimicrob Agents Chemother. 2017;61:8–13. doi: 10.1128/AAC.00605-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rosado-Sánchez I., Herrero-Fernández I., Tarancon-Diez L., Moreno S., Iribarren J.A., Dalmau D. Increased frequencies of Th17 cells and IL17a-producing regulatory T-cells preceding the immunodiscordant response to antiretroviral treatment. Journal of Infection. 2018;76:86–92. doi: 10.1016/j.jinf.2017.10.010. [DOI] [PubMed] [Google Scholar]
- 9.Rosado-Sánchez I., Rodríguez-Gallego E., Peraire J., Viladés C., Herrero P., Fanjul F. Glutaminolysis and lipoproteins are key factors in late immune recovery in successfully treated HIV-infected patients. Clin Sci. 2019;133:997–1010. doi: 10.1042/CS20190111. [DOI] [PubMed] [Google Scholar]
- 10.Ceausu A., Rodríguez-Gallego E., Peraire J., López-Dupla M., Domingo P., Viladés C. IL-7/IL-7R gene variants impact circulating IL-7/IL-7R homeostasis and ART-associated immune recovery status. Sci Rep. 2019;9:1–10. doi: 10.1038/s41598-019-52025-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arnold KB, Szeto GL, Alter G, Irvine DJ, Lauffenburger DA. CD4 + T cell–dependent and –independent cytokine-chemokine network changes in the immune responses of HIV-infected individuals HHS Public Access illustrate a broad approach for identifying key disease-associated nodes in a multicellular, multivariate sign. 2015;8. doi:10.1126/scisignal.aab0808. [DOI] [PMC free article] [PubMed]
- 12.Valdivia A., Ly J., Gonzalez L., Hussain P., Saing T., Islamoglu H. Restoring Cytokine Balance in HIV-Positive Individuals with Low CD4 T Cell Counts. AIDS Res. Hum. Retroviruses. 2017;33:905–918. doi: 10.1089/aid.2016.0303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hughes C.E., Nibbs R.J.B. A guide to chemokines and their receptors. FEBS Journal. 2018;285:2944–2971. doi: 10.1111/febs.14466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Solé X., Guinó E., Valls J., Iniesta R., Moreno V. SNPStats: a web tool for the analysis of association studies. Bioinformatics. 2006;22:1928–1929. doi: 10.1093/bioinformatics/btl268. [DOI] [PubMed] [Google Scholar]
- 15.Wang Z., Shang H., Jiang Y. Chemokines and chemokine receptors: accomplices for human immunodeficiency virus infection and latency. Front Immunol. 2017;8:1–12. doi: 10.3389/fimmu.2017.01274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chatterjee A., Rathore A., Vidyant S., Kakkar K., Dhole T.N. Chemokines and Chemokine Receptors in susceptibility to HIV-1 infection and progression to AIDS. Dis. Markers. 2012;32:143–151. doi: 10.3233/DMA-2011-0874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Evans V.A., Khoury G., Saleh S., Camerona P.U., Lewin S.R. HIV persistence: chemokines and their signalling pathways. Cytokine Growth Factor Rev. 2012;23:151–157. doi: 10.1016/j.cytogfr.2012.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lusso P. New EMBO Member's Review - HIV and the chemokine system: 10 years later. EMBO J. 2006;25:447–456. doi: 10.1038/sj.emboj.7600947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soriano A., Martínez C., García F., Plana M., Palou E., Lejeune M. Plasma Stromal Cell–Derived Factor (SDF)-1 Levels, SDF1-3′A Genotype, and Expression of CXCR4 on T Lymphocytes: their Impact on Resistance to Human Immunodeficiency Virus Type 1 Infection and Its Progression. J. Infect. Dis. 2002;186:922–931. doi: 10.1086/343741. [DOI] [PubMed] [Google Scholar]
- 20.Murphy Philip M., Heusinkveld L. Multisystem multitasking by CXCL12 and its receptors CXCR4 and ACKR3. Cytokine. 2018;109:2–10. doi: 10.1016/j.cyto.2017.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arenzana-Seisdedos F. SDF-1/CXCL12: a chemokine in the life cycle of HIV. Front Immunol. 2015;6:10–13. doi: 10.3389/fimmu.2015.00256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Daar E.S., Lynn H.S., Donfield S.M., Lail A., O'Brien S.J., Huang W. Stromal Cell–Derived Factor–1 Genotype, Coreceptor Tropism, and HIV Type 1 Disease Progression. J. Infect. Dis. 2005;192:1597–1605. doi: 10.1086/496893. [DOI] [PubMed] [Google Scholar]
- 23.Restrepo C., Gutierrez-Rivas M., Pacheco Y.M., García M., Blanco J., Medrano L.M. Genetic variation in CCR2 and CXCL12 genes impacts on CD4 restoration in patients initiating cART with advanced immunesupression. PLoS ONE. 2019;14:1–13. doi: 10.1371/journal.pone.0214421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Butrym A., Gebura K., Iwaszko M., Kuliczkowski K., Bogunia-Kubik K., Mazur G. Dual role of the CXCL12 polymorphism in patients with chronic lymphocytic leukemia. Hla. 2016;87:432–438. doi: 10.1111/tan.12810. [DOI] [PubMed] [Google Scholar]
- 25.Angela Covino D., Sabbatucci M., Fantuzzi L. The CCL2/CCR2 Axis in the Pathogenesis of HIV-1 Infection: a New Cellular Target for Therapy? Curr Drug Targets. 2015;17:76–110. doi: 10.2174/138945011701151217110917. [DOI] [PubMed] [Google Scholar]
- 26.Lee W.J., Liao Y.C., Wang Y.F., Lin I.F., Wang S.J., Fuh J.L. Plasma MCP-1 and cognitive decline in patients with Alzheimer's disease and mild cognitive impairment: a two-year follow-up study. Sci Rep. 2018;8:4–11. doi: 10.1038/s41598-018-19807-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barmania F., Pepper M.S. C-C chemokine receptor type five (CCR5): an emerging target for the control of HIV infection. Applied and Translational Genomics. 2013;2:3–16. doi: 10.1016/j.atg.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ellwanger J.H., Kulmann-Leala B., Kaminskia V de L, Rodrigues A.G., Bragatte MA de S, Chies J.A.B. Beyond HIV infection: neglected and varied impacts of CCR5 and CCR5Δ32 on viral diseases. Virus Res. 2020;286 doi: 10.1016/j.virusres.2020.198040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fantuzzi L., Tagliamonte M., Gauzzi M.C., Lopalco L. Dual CCR5/CCR2 targeting: opportunities for the cure of complex disorders. Cellular and Molecular Life Sciences. 2019;76:4869–4886. doi: 10.1007/s00018-019-03255-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miao M., De Clercq E., Li G. Clinical significance of chemokine receptor antagonists. Expert Opinion on Drug Metabolism and Toxicology. 2020;16:11–30. doi: 10.1080/17425255.2020.1711884. [DOI] [PubMed] [Google Scholar]
- 31.Hütter G., Neumann M., Nowak D., Klein S., Klüter H., Hofmann W.K. The effect of the CCR5-delta32 deletion on global gene expression considering immune response and inflammation. J. Inflamm. 2011;8:29. doi: 10.1186/1476-9255-8-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu Q., Zhu P., Zhang Y., Li J., Ma X., Li N. Analysis of social and genetic factors influencing heterosexual transmission of HIV within serodiscordant couples in the henan cohort. PLoS ONE. 2015;10:1–13. doi: 10.1371/journal.pone.0129979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Norris P.J., Zhang J., Worlock A., Nair S.V, Anastos K., Minkoff H.L. Systemic cytokine levels do not predict CD4+ T-cell recovery after suppressive combination antiretroviral therapy in chronic human immunodeficiency virus Infection. Open Forum Infect Dis. 2016;3:1–9. doi: 10.1093/ofid/ofw025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ahuja S.K., Kulkarni H., Catano G., Agan B.K., Camargo J.F., He W. CCL3L1-CCR5 genotype influences durability of immune recovery during antiretroviral therapy of HIV-1-infected individuals. Nat. Med. 2008;14:413–420. doi: 10.1038/nm1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou W, Guo S, Liu M, Burow ME, Wang G, District H. et al. HHS Public Access. 2019;26:3026–41.
- 36.Kalnins A., Thomas M.N., Andrassy M., Müller S., Wagner A., Pratschke S. Spiegelmer Inhibition of MCP-1/CCR2 - Potential as an Adjunct Immunosuppressive Therapy in Transplantation. Scand. J. Immunol. 2015;82:102–109. doi: 10.1111/sji.12310. [DOI] [PubMed] [Google Scholar]
- 37.Devarapu S.K., Kumar VR S., Rupanagudi K.V., Kulkarni O.P., Eulberg D., Klussmann S. Dual blockade of the pro-inflammatory chemokine CCL2 and the homeostatic chemokine CXCL12 is as effective as high dose cyclophosphamide in murine proliferative lupus nephritis. Clinical Immunology. 2016;169:139–147. doi: 10.1016/j.clim.2016.07.003. [DOI] [PubMed] [Google Scholar]
- 38.Szklarczyk D., Gable A.L., Lyon D., Junge A., Wyder S., Huerta-Cepas J. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.