Highlights
-
•
FOXD3-AS1 downregulation inhibits cell migration, invasion and EMT in osteosarcoma.
-
•
Overexpression of miR-296-5p hinders the malignant behaviors of osteosarcoma cells.
-
•
FOXD3-AS1 sponges miR-296-5p to elevate ZCCHC3 to facilitate malignancy in osteosarcoma.
-
•
ELF1 activates FOXD3-AS1 to mediate tumor development in osteosarcoma.
Keywords: Osteosarcoma, FOXD3-AS1, ELF1, miR-296-5p, ZCCHC3
Abstract
Osteosarcoma (OS) is a malignant carcinoma often occurring in adolescents. The critical function of long non-coding RNAs (lncRNAs) in cancer arouses increasing attention. Nevertheless, the specific function of FOXD3 Antisense RNA 1 (FOXD3-AS1) in OS has not been understood yet. In this research, FOXD3-AS1 showed strengthened level in OS specimens and cell lines, and its deficiency restrained cell migration, invasion and epithelial-to-mesenchymal transition (EMT) in OS. Then, we confirmed the interaction of FOXD3-AS1 with microRNA-296-5p (miR-296-5p) and that miR-296-5p overexpression blocked OS cell migration, invasion and EMT. Besides, miR-296-5p targeted zinc finger CCHC-type containing 3 (ZCCHC3), and FOXD3-AS1 released ZCCHC3 via sequestering miR-296-5p. Moreover, rescue assays delineated that ZCCHC3 upregulation neutralized the inhibitory effect of FOXD3-AS1 depletion on in vitro behaviors and in vivo tumorigenesis in OS. In addition, E74 like ETS transcription factor 1 (ELF1) stimulated FOXD3-AS1 transcription, and ELF1 silence-suppressed malignant phenotypes of OS cells were offset by FOXD3-AS1 upregulation. Overall, present work elucidated that ELF1-activated FOXD3-AS1 aggravated cell migration, invasion and EMT in OS via absorbing miR-296-5p to augment ZCCHC3 expression, which might provide potential guidance for researchers to find effective targets for OS treatment.
1. Introduction
Osteosarcoma (OS) is considered to be the most common primary malignant bone tumor occurring in skeletal system. OS accounts for about 55.8% of bone tumor cases and jumps to the third in the list of the most common malignancies in teenagers [1]. Its characteristics, such as frequent occurrence, high malignant degree, and fast metastasis, lead to unpleasing prognosis. Efficacy of OS treatment has been dramatically upgraded due to the employment of neoadjuvant chemotherapy and the progress in contemporary surgical techniques [1], and past 2 decades have witnessed the increase of survival rate in OS from 49.8% to 68.2% [2]. Nevertheless, multidrug resistance (MDR) seriously impedes the application of neoadjuvant chemotherapy. Despite the constantly updated molecular researches on OS, the underlying molecular mechanisms in OS remain to be further inquired due to the complexity of cellular behaviors during OS progression.
Increasing researches have implied the effective impact of non-coding RNAs (ncRNAs) on gene expression [3], [4], [5]. NcRNAs with exceeding 200 nucleotides are named as lncRNAs [6]. Researchers have recently focused on exploring and elucidating the underlying functions of lncRNAs in cancer development [3], [7]. Increasing studies have illustrated that lncRNAs are deeply implicated in multiple physiological processes like cell growth, migration and invasion in various cancers, including OS. For example, androgen-stimulated SOCS2-AS1 encourages cell proliferation and impedes cell apoptosis in prostate cancer [8]. Enhanced FEZF1-AS1 leads to a bad prognosis of gastric cancer patients and it functions as an oncogene in gastric cancer through activating Wnt pathway [9]. SNHG15 enhances the proliferation, migration and invasion of breast cancer cells through downregulating miR-211-3p [10]. TCF3-activated LINC00152 elicits the competing endogenous RNA (ceRNA) function to target miR-1182/CDK14 axis and facilitate OS progression [11]. Further, the functions that FOXD3-AS1 promotes tumor progression have been documented in some cancers, such as breast cancer [12] and glioma [13]. Interestingly, the fact that FOXD3-AS1 functions as a ceRNA by regulating miR-135a-5p/SIRT1 axis has been proved in colorectal cancer [14]. Nonetheless, the biological behavior of FOXD3-AS1 in OS has never been illustrated yet.
In this research, we aimed to explore the function and probable mechanism of FOXD3-AS1 in OS. As a result, we revealed that ELF1-activated FOXD3-AS1 drives the migration, invasion and EMT of OS cells via sponging miR-296-5p to upregulate ZCCHC3, which might be conducive to find out a promising target for treating OS.
2. Materials and methods
2.1. Tissue Collection
The paired OS tissue specimens and non-tumor tissue specimens were collected from 52 patients from 2013 to 2018. The ethical approval of tissue samples was acquired from the Ethics Committee of the First Affiliated Hospital of Chongqing Medical University. They were 31 males (59.62%) and 21 females (40.38%) in this study, with the age range of 10–64 years old (the mean age: 19 years old). All the participants never received any treatment of chemotherapy or radiotherapy before our research, and they all signed the informed consents. These tissue samples were instantly frozen in liquid nitrogen and then preserved at −80℃.
2.2. Cell culture
Human OS cell lines (U2OS, MG-63, HOS, SAOS2 and 143B) and human osteoblast cell line (OB3) were procured from the American Type Culture Collection (ATCC; Manassas, VA, USA). Cell culture was processed in DMEM (Corning, Tewksbury, MA, USA) with additional 10% fetal bovine serum (FBS; Gibco, Gaithersburg, MD, USA) under 5% CO2 at 37 °C. Medium was replaced every 3 days.
2.3. Cell transfection
Specific shRNAs targeting FOXD3-AS1 (sh-FOXD3-AS1#1/2) or ELF1 (sh-ELF1#1/2) and respective NC (sh-NC) were designed by Genechem (Shanghai, China). To obtain pcDNA3.1/FOXD3-AS1, pcDNA3.1/ZCCHC3 or pcDNA3.1/ELF1, corresponding cDNA sequence was inserted into pcDNA3.1 vector (Invitrogen, Carlsbad, CA, USA), with the empty vector as negative control. Also, miR-296-5p mimics, miR-296-5p inhibitor, NC mimics and NC inhibitor were constructed by GenePharma (Shanghai, China). Lipofectamine 3000 (Invitrogen) was utilized for the transfection of respective plasmids into MG-63 or HOS cells.
2.4. Real-time reverse-transcription polymerase chain reaction (RT-qPCR)
Total RNA was extracted by applying Trizol reagent (Invitrogen), and then cDNA was reverse-transcribed from total RNA. GAPDH or U6 served as the internal control. RT-qPCR was implemented on Light Cycler system (Roche, Basel, Switzerland) using SYBR Green I kit (TaKaRa, Dalian, China). Data were assayed by 2−ΔΔCt.
2.5. Wound healing assay
Transfected MG-63 or HOS cells were cultured in 6-well plates (5 × 105 cells/well) to 90% confluence. Later, scratches were made through using a 10 µl sterile pipette tip. Cellular debris was obliterated through washing the plates thrice with PBS (Sigma-Aldrich, St. Louis, MO, USA). The wound was detected at 0 and 24 h via a light microscope (Nikon, Tokyo, China).
2.6. Transwell invasion assay
MG-63 or HOS cells were added to the upper transwell chambers (Corning Costar, Cambridge, MA, USA) which were precoated with Matrigel (Sigma-Aldrich). Medium with 10% FBS was loaded in the bottom chambers. Non-invasive cells on the upper surface were wiped off, whereas invasive cells on the lower surface were immobilized for 30 min by 4% paraformaldehyde (PFA; Sigma-Aldrich) and stained for 10 min using crystal violet (Sigma-Aldrich). At length, the invasive cells were imaged and counted via a microscope (Nikon).
2.7. Western blot
MG-63 or HOS cells were lysed by lysis buffer (Beyotime, Shanghai, China). The protein concentration was examined via utilizing a BCA Protein Assay Kit (Beyotime, P0010). Equal quantities of protein were run on 10% SDS-PAGE (Bio-Rad, Hercules, CA, USA) prior to being electro-transferred to PVDF membranes (Millipore, Bedford, MA, USA). Membranes were blocked in 5% nonfat milk before incubation with primary antibodies as follows: E-cadherin (ab40772, Abcam, Cambridge, MA, USA), N-cadherin (ab76057, Abcam), ZCCHC3 (SAB1408147, Sigma-Aldrich) and GAPDH (ab9485, Abcam). After incubating with secondary antibodies, blots were examined by ECL Fuazon Fx (Vilber Lourmat, Marne La Vallee, France). Images were taken via FusionCapt Advance Fx5 software (Vilber Lourmat).
2.8. Immunofluorescence (IF)
Transfected MG-63 or HOS cells were placed in glass slides and washed by PBS, after which cells were fixed for 10 min by 4% PFA and permeabilized for 15 min with 0.5% Triton X-100 (Sigma-Aldrich). Next, cells were capped for 1 h using 5% bovine serum albumin (BSA; Sigma-Aldrich) in PBS. Upon this, at 4℃, cells were respectively subjected to primary antibodies against E-cadherin or N-cadherin overnight. Subsequently, cells were treated for 1 h with Alexa Fluor 488-conjugated goat anti-rabbit secondary antibody in the dark environment. Thereafter, cells were incubated for 5 min with DAPI (Sigma-Aldrich) before the examination with a fluorescence microscope (Nikon). In addition, parameters of fluorescent images were UV monochromatic filter block: excitation 340–380 nm; obstruction 400 nm; launch: 425 nm. B monochromatic filter block: excitation 450–490 nm; obstruction 510 nm; launch: 512–542 nm. G monochromatic filter block: excitation 515–560 nm; obstruction 580 nm; launch: 590 nm. The exposure time for DAPI was 13 ms and for the others were 350 ms.
2.9. Subcellular fractionation
Cytoplasmic and nuclear RNAs from MG-63 or HOS cells were centrifuged and purified through a Cytoplasmic and Nuclear RNA Purification Kit (Norgen, Thorold, ON, Canada). Expression levels of FOXD3-AS1, U6 (nuclear control) and GAPDH (cytoplasmic control) were verified by RT-qPCR.
2.10. Luciferase reporter assay
MG-63 or HOS cells with miR-296-5p mimics or NC mimics were co-transfected with dual-luciferase pmirGLO reporter vectors (Promega, Madison, WI, USA) covering full-length FOXD3-AS1 sequence with wild-type (WT) or mutant (Mut) miR-296-5p binding sites. Likewise, ZCCHC3 3′UTR fragments with wild-type and mutant miR-296-5p binding sites were also cloned into the pmirGL vectors to establish ZCCHC3 3′UTR-WT/Mut reporters. On the other hand, the sequence of FOXD3-AS1 promoter was amplified by PCR and then inserted into pGL3-basic vector (Promega). The pGL3 vectors covered FOXD3-AS1 promoter sequence with mutated ELF1 binding sites were synthesized similarly. Recombinant luciferase reporter plasmids were co-transfected with the pRL-TK plasmid expressing Renilla luciferase into MG-63 or HOS cells. Examination of relative luciferase activity was performed by dual-luciferase reporter assay systems (Promega).
2.11. RNA immunoprecipitation (RIP)
RIP experiment was achieved by a Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore). Anti-Ago2 antibody (Millipore) and anti-IgG antibody (Millipore) were mixed with magnetic beads (Invitrogen) and the extracts of MG-63 or HOS cells in RIP buffer. In the end, the levels of FOXD3-AS1, miR-296-5p and ZCCHC3 in the immunoprecipitate were exposed via RT-qPCR.
2.12. Chromatin immunoprecipitation (ChIP)
ChIP was conducted via the EZ-ChIP™ Chromatin immunoprecipitation kit (Millipore). MG-63 or HOS cells were incubated with anti-ELF1 antibody (Millipore) or anti-IgG antibody. After that, the complexes were precipitated by magnetic beads (Invitrogen), and the obtained DNA fragments were measured by RT-qPCR after purification.
2.13. In vivo assay
Three-week-old nude mice were acquired from the National Laboratory Animal Center (Beijing, China) and maintained under SPF (specific pathogen free) conditions. Then mice were subcutaneously injected with indicated MG-63 cells that were transfected with sh-NC, sh-FOXD3-AS1#1, or sh-FOXD3-AS1#1 + pcDNA3.1/ZCCHC3. Animal research was conducted upon the approval of the Institutional Animal Care and Use Committee of the First Affiliated Hospital of Chongqing Medical University. Tumor growth was analyzed by recording tumor volume every fourth day, and the mice were sacrificed 28 days later. Besides, according to IACUC criteria, if the nude mice lost more than 20% body weight and exhibited tumor metastasis, lethargy, or other painful symptoms, they wound be sacrificed via cervical dislocation. Tumor volume and weight were examined after tumors were excised from the sacrificed mice. Each mouse was burden with one tumor.
2.14. Immunohistochemistry (IHC) assay
Fresh tissues gotten from in vivo assay were fixed in 4% PFA. Then, they were dehydrated in ethanol solutions, embedded in paraffin and cut into slices with 4-μm thickness. Afterwards, the sections were processed with primary antibodies against Ki67, PCNA, E-cadherin or N-cadherin overnight at 4 °C, followed by cultivation with HRP-conjugated secondary antibodies. Finally, all these sections were observed and imaged under a light microscope (Olympus).
2.15. Statistical analysis
Data were reported as mean ± SD. Tests were conducted three copies. Statistical data analyses were carried out using GraphPad Prism 5.0 (GraphPad Software, San Diego, CA, USA). Student’s t test and one-way or two-way ANOVA were conducted for assessing significant differences between groups. P < 0.05 denoted the threshold of statistical significance.
3. Results
3.1. FOXD3-AS1 downregulation inhibits cell migration, invasion and EMT in OS
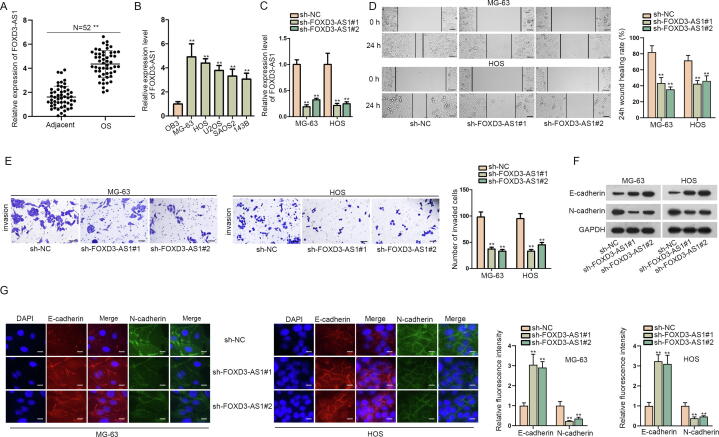
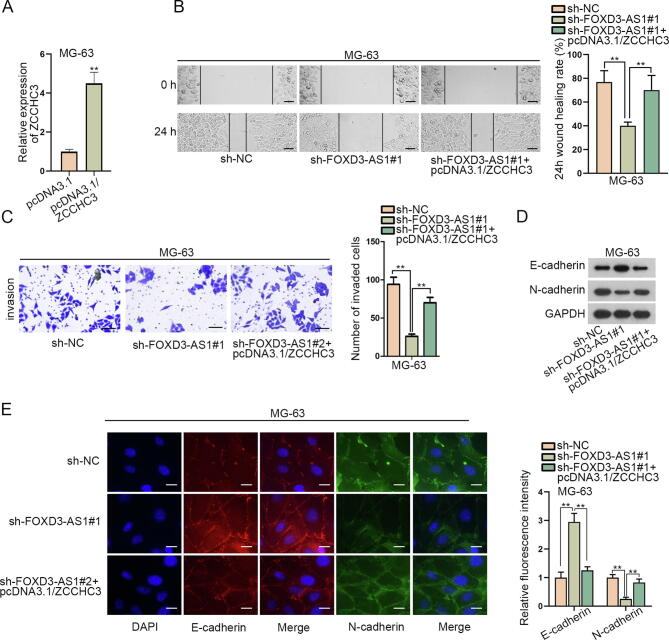
To figure out whether FOXD3-AS1 was related to OS development, we tested its expression under different conditions. RT-qPCR revealed relatively high FOXD3-AS1 expression in OS samples in comparison with adjacent non-tumor tissues (Fig. 1A). Additionally, FOXD3-AS1 expression was upregulated in OS cell lines, particularly in MG-63 and HOS cells, versus normal osteoblasts (OB3) (Fig. 1B). Later, data of RT-qPCR showed that FOXD3-AS1 expression was obviously inhibited by sh-FOXD3-AS1#1/#2 in MG-63 and HOS cells (Fig. 1C). To figure out the biological impact of FOXD3-AS1 on OS cells, loss-of-function assays were unfolded using MG-63 and HOS cells. As shown in Fig. 1D, wound healing assay results verified that FOXD3-AS1 knockdown suppressed the migration of MG-63 and HOS cells. Likewise, transwell assay data unveiled that the number of invasive OS cells was decreased under FOXD3-AS1 deficiency (Fig. 1E), implying that downregulating FOXD3-AS1 weakened the invasive ability of OS cells. Meanwhile, we also wanted to know the influence of FOXD3-AS1 on EMT process in OS cells. Interestingly, data from western blot analyses uncovered that the level of E-cadherin protein was promoted whereas that of N-cadherin protein was lessened when FOXD3-AS1 was silenced in these two OS cells (Fig. 1F). IF images consistently showed the enhanced fluorescence of E-cadherin and weakened fluorescence of N-cadherin under FOXD3-AS1 deficiency (Fig. 1G). In conclusion, FOXD3-AS1 was upregulated in OS cells, and its knockdown prevented the events of migration, invasion and EMT in OS cells.
Fig. 1.
FOXD3-AS1 downregulation restrained cell migration, invasion and EMT in OS. (A) The expression of FOXD3-AS1 was examined via RT-qPCR in 52 OS tissues and matched non-tumor tissues. Student’s t test. (B) RT-qPCR detected FOXD3-AS1 expression in OS cell lines versus normal OB3 cells. One-way ANOVA. (C) The efficiency of FOXD3-AS1 knockdown was analyzed by RT-qPCR. One-way ANOVA. (D) Wound healing assay (scale bar = 200 μm) revealed the migration ability of MG-63 and HOS cells under FOXD3-AS1 knockdown. One-way ANOVA. (E) Transwell assay (scale bar = 200 μm) tested the invasion of transfected MG-63 and HOS cells. One-way ANOVA. (F) Western blot measured the protein levels of E-cadherin and N-cadherin in transfected MG-63 and HOS cells. (G) IF assay (scale bar = 50 μm) assessed the fluorescence intensity of E-cadherin and N-cadherin in transfected MG-63 and HOS cells. One-way ANOVA. **P < 0.01.
3.2. FOXD3-AS1 sponges miR-296-5p to block the function of miR-296-5p in OS
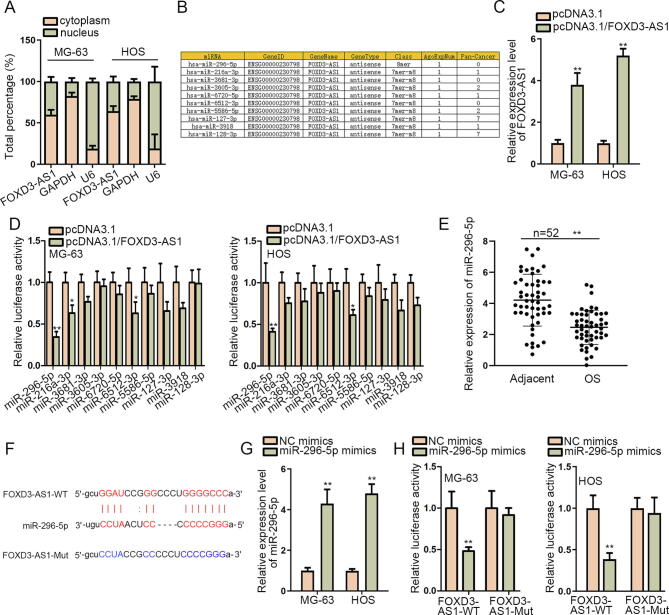
To better understand the molecular mechanism of FOXD3-AS1 in OS, we analyzed the subcellular distribution of FOXD3-AS1 in OS cells. As shown by the outcomes of subcellular fractionation assay, FOXD3-AS1 mainly located in the cytoplasm of both MG-63 and HOS cells (Fig. 2A), which indicated the possibility that FOXD3-AS1 participated in post-transcriptional regulation. Through starBase v2.0 (http://starbase.sysu.edu.cn/index.php), 10 miRNAs possessing the potential interaction with FOXD3-AS1 were obtained (Fig. 2B). Then, FOXD3-AS1 expression was remarkably elevated after the transfection of pcDNA3.1/FOXD3-AS1 (Fig. 2C). Luciferase reporter assay results demonstrated that the reporter containing miR-296-5p presented the most significant decrease in the luciferase activity under FOXD3-AS1 overexpression (Fig. 2D), highly suggesting the suppression of upregulated FOXD3-AS1 on the function of miR-296-5p. Further, we discovered the weakened expression of miR-296-5p in 52 OS tissues in contrast to matched non-cancerous ones (Fig. 2E). Afterwards, the miR-296-5p binding site in FOXD3-AS1 was predicted using starBase (Fig. 2F). Subsequently, RT-qPCR revealed that miR-296-5p level was significantly upregulated after miR-296-5p mimics was transfected into MG-63 and HOS cells (Fig. 2G). Consequently, it was proved that miR-296-5p upregulation reduced the luciferase activity of FOXD3-AS1-WT, while the luciferase activity of FOXD3-AS1-Mut showed no obvious difference (Fig. 2H), which confirmed the binding between FOXD3-AS1 and miR-296-5pat predicted sites. Thus, we recognized that miR-296-5p is the downstream molecule sponged by FOXD3-AS1 in OS.
Fig. 2.
FOXD3-AS1 sponged miR-296-5p in OS. (A) Subcellular fractionation assay measured the distribution of FOXD3-AS1 in OS cells. (B) Ten miRNAs that might bind with FOXD3-AS1 were predicted via starBase. (C) RT-qPCR detected FOXD3-AS1 expression after the transfection of pcDNA3.1/FOXD3-AS1 or pcDNA3.1. (D) Luciferase reporter assay measured the impact of FOXD3-AS1 overexpression on the luciferase activity of reporters covering above ten miRNAs. (E) RT-qPCR determined the expression of miR-296-5p in 52 OS samples and matched non-tumor controls. (F) The binding sequences between miR-296-5p and FOXD3-AS1 were predicted by starBase. (G) RT-qPCR detected miR-296-5p expression in OS cells with the transfection of NC mimics or miR-296-5p mimics. (H) The luciferase activity of FOXD3-AS1-WT or FOXD3-AS1-Mut in different groups was measured by luciferase reporter assay. Above data were all analyzed via Student’s t test. *P < 0.05, **P < 0.01.
3.3. Overexpression of miR-296-5p inhibits OS cell migration, invasion and EMT
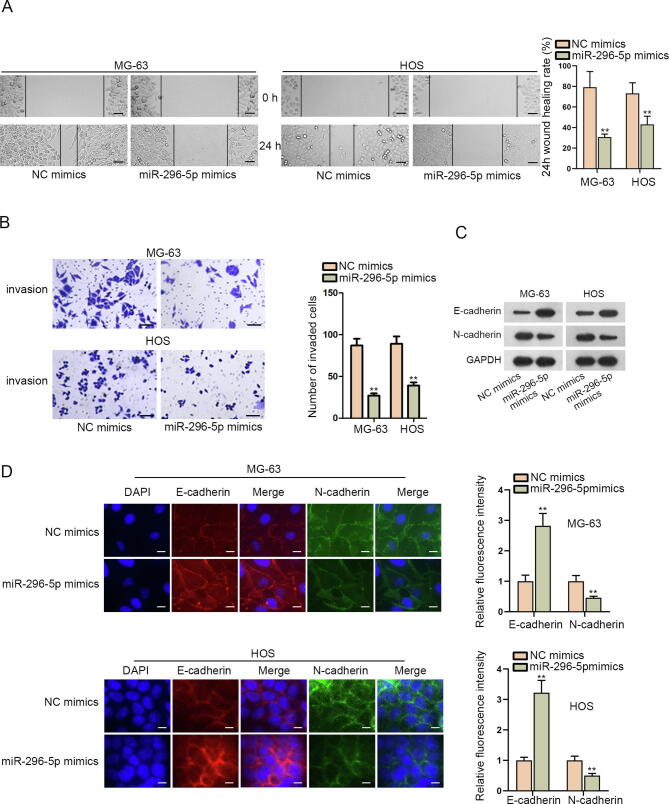
Then, we planned to probe into the impact of miR-296-5p on the behaviors of OS cells. Interestingly, the outcomes of wound healing and transwell assays determined that the migration and invasion of MG-63 and HOS cells were restrained by enhanced miR-296-5p (Fig. 3A–B). Data from western blot revealed that miR-296-5p upregulation hampered EMT process in MG-63 and HOS cells, evidenced by enhanced E-cadherin level and declined N-cadherin level under such circumstances (Fig. 3C). Likewise, results from IF assay further proved the restraining effect of augmented miR-296-5p on EMT process in these two OS cells (Fig. 3D). In general, ectopic miR-296-5p expression subdued OS cell migration, invasion and EMT process.
Fig. 3.
Overexpression of miR-296-5p hindered OS cell migration, invasion and EMT. (A) The migration of MG-63 and HOS cells with or without miR-296-5p elevation was monitored by wound healing assay (scale bar = 200 μm). (B) Transwell assay (scale bar = 200 μm) tested the invasion of indicated MG-63 and HOS cells. (C) Western blot measured E-cadherin and E-cadherin protein levels in MG-63 and HOS cells after different transfections. GAPDH was the loading control. (D) IF assay (scale bar = 50 μm) evaluated the fluorescence intensity of E-cadherin and N-cadherin in indicated MG-63 and HOS cells. These results were all analyzed via Student’s t test. *P < 0.05, **P < 0.01.
3.4. FOXD3-AS1 boosts the level of miR-296-5p-targted ZCCHC3 in OS
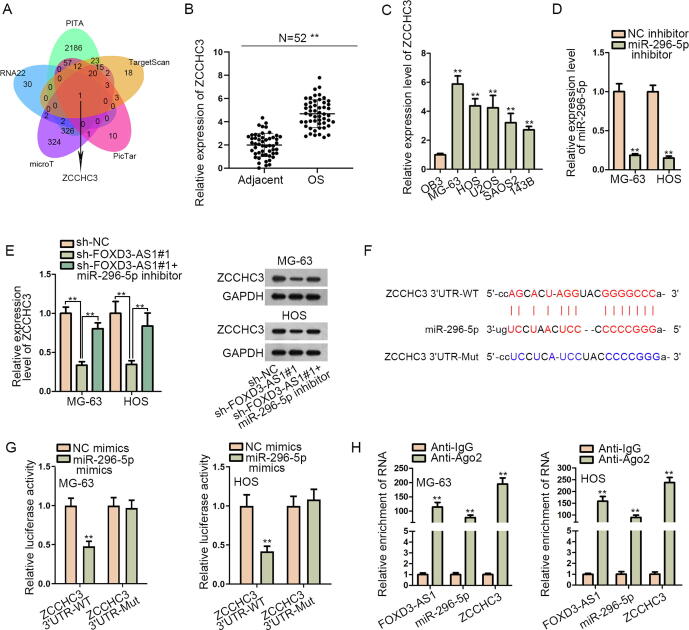
Thereafter, we searched the targets of miR-296-5p. Through combining the prediction data from PITA, TargetScan, PicTar, microT and RNA22 databases in starBase, ZCCHC3 was recognized as the only target of miR-296-5p that was shared by above five tools (Fig. 4A). Then, RT-qPCR analyzed that ZCCHC3 expression was remarkably strengthened in OS specimens and cells (Fig. 4B–C). Subsequently, whether FOXD3-AS1 could affect ZCCHC3 through miR-296-5p was determined. As expected, silencing FOXD3-AS1 significantly decreased ZCCHC3 expression at both mRNA and protein levels, and this effect was remedied after the co-inhibition of miR-296-5p (Fig. 4D–E). Afterwards, the miR-296-5p binding site in ZCCHC3 3′UTR was predicted via starBase (Fig. 4F). Importantly, it was uncovered that overexpressing miR-296-5p in MG-63 and HOS cells resulted in a decline in the luciferase activity of ZCCHC3 3′UTR-WT but not that of ZCCHC3 3′UTR-Mut (Fig. 4G). Further, we monitored the co-enrichment of FOXD3-AS1, miR-296-5p and ZCCHC3 in RIP products of anti-Ago2 (Fig. 4H), which suggested the interaction of miR-296-5p with FOXD3-AS1 and ZCCHC3 in RNA-induced silencing complexes (RISCs). Altogether, FOXD3-AS1 fortifies ZCCHC3 expression in OS cells through sequestering miR-296-5p.
Fig. 4.
FOXD3-AS1 enhanced ZCCHC3 expression in OS by absorbing miR-296-5p. (A) ZCCHC3 was screened out to be the target gene of miR-296-5p through PITA, TargetScan, PicTar, microT and RNA22 programs in starBase. (B) ZCCHC3 expression was detected via RT-qPCR in 52 OS tissues and paired non-tumor tissues. Student’s t test. (C) RT-qPCR detected ZCCHC3 expression in OB3 cells and OS cells. One-way ANOVA. (D) RT-qPCR detected miR-296-5p expression in MG-63 and HOS cells after transfection with NC inhibitor or miR-296-5p inhibitor. Student’s t test. (E) RT-qPCR and western blot analyses measured the mRNA and protein levels of ZCCHC3 in different groups. One-way ANOVA. (F) The binding site between miR-296-5p and ZCCHC3 was predicted by starBase. (G) The luciferase activity of ZCCHC3 3′UTR-WT and ZCCHC3 3′UTR-Mut in different groups was determined by luciferase reporter assay. Student’s t test. (H) RIP assay detected the relative enrichment of FOXD3-AS1, miR-296-5p and ZCCHC3 in anti-Ago2 and anti-IgG groups. Student’s t test. **P < 0.01.
3.5. FOXD3-AS1 depends on ZCCHC3 to facilitate malignancy in OS
Thereafter, we focused on whether FOXD3-AS1 mediated OS progression via targeting ZCCHC3. Seen from Fig. 5A, ZCCHC3 was overexpressed in MG-63 cells by transfection with pcDNA3.1/ZCCHC3. Next, the results of wound healing and transwell assays revealed that ZCCHC3 overexpression reversed the restraining effect of silenced FOXD3-AS1 on the migration and invasion of MG-63 cells (Fig. 5B–C). Besides, the data from western blot and IF assays testified that ZCCHC3 upregulation counteracted the suppression of FOXD3-AS1 depletion on EMT process in MG-63 cells (Fig. 5D–E). In short, FOXD3-AS1 contributes to OS cell migration, invasion and EMT through elevating ZCCHC3 expression.
Fig. 5.
FOXD3-AS1 functioned in OS by targeting ZCCHC3. (A) RT-qPCR detected ZCCHC3 expression in MG-63 cells after the transfection with pcDNA3.1 or pcDNA3.1/ZCCHC3. Student’s t test. (B) Wound healing assay (scale bar = 200 μm) measured the migration of MG-63 cells under diverse conditions. One-way ANOVA. (C) Transwell assay (scale bar = 200 μm) tested the invasion of MG-63 cells with different transfections. One-way ANOVA. (D) Western blot measured E-cadherin and N-cadherin protein levels in indicated MG-63 cells. (E) IF assay (scale bar = 50 μm) estimated the fluorescence intensity of E-cadherin and N-cadherin in indicated MG-63 cells. One-way ANOVA. **P < 0.01.
3.6. FOXD3-AS1 mediates tumorigenesis in OS via regulating ZCCHC3
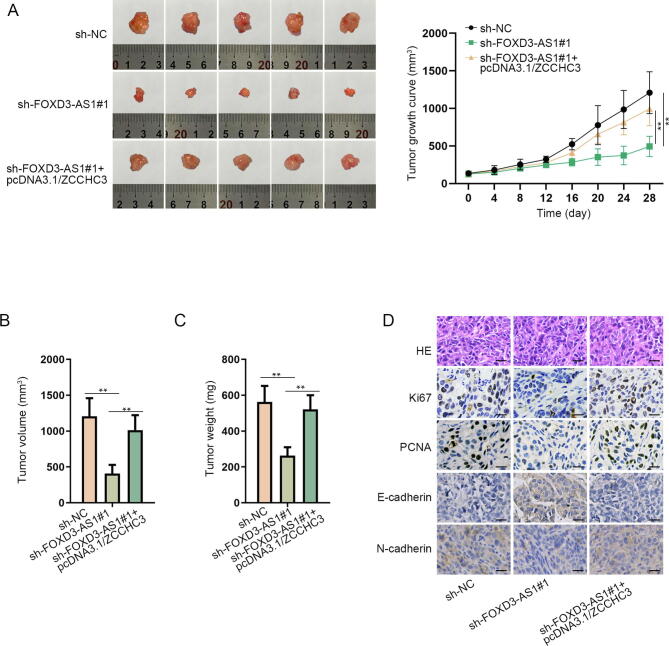
In order to testify the modulation of FOXD3-AS1 in OS tumorigenesis, in vivo assays were carried out. As shown in Fig. 6A, FOXD3-AS1 depletion hindered in vivo tumor growth, while such suppression could be offset under further overexpression of ZCCHC3. Eventually, the volume and weight of in vivo tumors were much lowered in response to reduced FOXD3-AS1 level but counteracted when further facing ZCCHC3 upregulation (Fig. 6B–C). Moreover, the suppressive influence of silenced FOXD3-AS1 on the expression level of proliferation-related proteins (Ki67 and PCNA) in these tumors was reversed by elevating ZCCHC3 expression (Fig. 6D). Besides, the increase of E-cadherin and decline of N-cadherin in xenografts caused by FOXD3-AS1 inhibition were also rescued by ZCCHC3 overexpression (Fig. 6D). Above data indicated that ZCCHC3 upregulation countervailed the inhibitive effect of FOXD3-AS1 knockdown on cell proliferation and EMT process in vivo. In sum, FOXD3-AS1 facilitates OS tumorigenesis through upregulating ZCCHC3 expression.
Fig. 6.
FOXD3-AS1 contributed to OS tumorigenesis through ZCCHC3-mediated manner. (A) Representative images and the growth curves of tumors originated from indicated MG-63 cells. Two-way ANOVA. (B-C) Tumor volume and weight were detected after mice were sacrificed. One-way ANOVA. (D) IHC analyses of Ki67, PCNA, E-cadherin and N-cadherin in tumors from different groups. Scale bar = 20 μm. **P < 0.01.
3.7. ELF1 activates FOXD3-AS1 transcription in OS
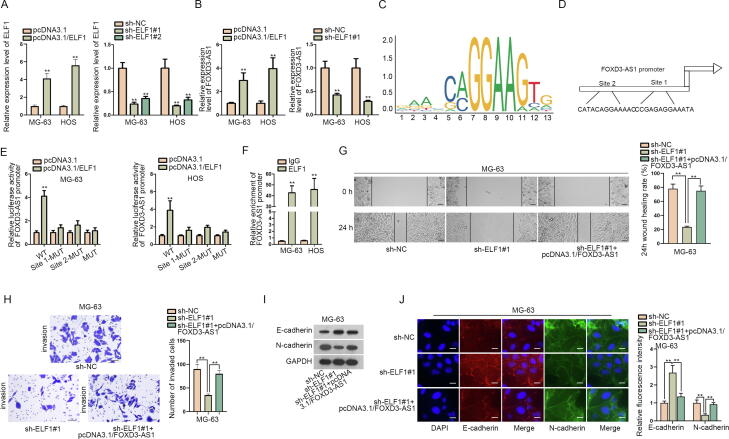
Further, we also wondered the upstream of FOXD3-AS1 in regulating OS development. The role of ELF1 as a transcription activator has been supported in a number of cancer-related researches [15], [16]. Thus, we tested whether ELF1 could activate FOXD3-AS1 in OS. Firstly, it was evidenced that the transfection of pcDNA3.1/ELF1 effectively upregulated ELF1 expression in MG-63 and HOS cells, while that of sh-ELF1#1/#2 obviously decreased ELF1 expression in both cells (Fig. 7A). Then, RT-qPCR revealed that FOXD3-AS1 expression was increased after overexpressing ELF1 but declined when silencing ELF1 (Fig. 7B). Additionally, by using JASPAR (http://jaspar.genereg.net/), the DNA motif of ELF1 was obtained (Fig. 7C). Furthermore, we found two ELF1 binding sites (“CATACAGGAAAAC” and “CCGAGAGGAAATA”) in FOXD3-AS1 promoter region by comparing the ELF1 motif with FOXD3 promoter sequence gained from NCBI (https://www.ncbi.nlm.nih.gov/gene/) (Fig. 7D). Expectedly, the luciferase activity of FOXD3-AS1 promoter was promoted by ELF1 overexpression, and mutating either site 1 or 2 could weaken such promotion, whereas the luciferase activity of FOXD3-AS1 promoter reporter with mutations of both site 1 and 2 presented no change under the same circumstance (Fig. 7E). Such results indicated that both site 1 and 2 were in charge of ELF1 binding to FOXD3-AS1 promoter. Also, data from ChIP assay ensured the binding of ELF1 with FOXD3-AS1 promoter in MG-63 and HOS cells (Fig. 7F). Thereafter, rescue assays proved that FOXD3-AS1 upregulation offset the inhibitory influence of ELF1 depletion on cell migration, invasion and EMT process in MG-63 cells (Fig. 7G–J). Collectively, ELF1 activates FOXD3-AS1 to aggravate OS progression.
Fig. 7.
ELF1 activated FOXD3-AS1 transcription in OS. (A) RT-qPCR analyzed the efficiency of ELF1 overexpression (Student’s t test) and knockdown (one-way ANOVA) in both MG-63 and HOS cells. (B) RT-qPCR detected FOXD3-AS1 expression in OS cells under ELF1 upregulation or downregulation. Student’s t test. (C) DNA motif of ELF1 from JASPAR. (D) Two binding sites for ELF1 in FOXD3-AS1 promoter were predicted by JASPAR. (E) Luciferase reporter assay measured the luciferase activity of indicated FOXD3-AS1 promoter in MG-63 and HOS cells after the transfection of pcDNA3.1/ELF1 or pcDNA3.1. Student’s t test. (F) ChIP assay verified the binding between ELF1 and FOXD3-AS1 promoter in these two OS cells. Student’s t test. (G-H) Wound healing (scale bar = 200 μm) and transwell assays (scale bar = 200 μm) measured the migration and invasion of indicated MG-63 cells. One-way ANOVA. (I) Western blot analyzed the protein levels of E-cadherin and N-cadherin in indicated MG-63 cells. (J) IF assay (scale bar = 50 μm) determined the fluorescence intensity of E-cadherin and N-cadherin in MG-63 cells under diverse transfections. One-way ANOVA. **P < 0.01.
4. Discussion
Usually, the youth account for a large proportion of OS cases. However, the molecular pathology of OS is still uncertain [17]. Patients without signs of tumor spread show a five-year survival rate of 59.2–78.6%, but those diagnosed with metastatic OS show only 19.7–29.3% of 5-year survival rate [18]. Consequently, it is badly in need of finding effective approaches for treating OS patients.
The researching value of lncRNAs in cancers has been increasingly proved. For example, lncRNA MEG3 modulates colorectal cancer development via regulating ADAR1 [19]. LncRNA ABHD11-AS1 serves as a ceRNA regulating miR-199a-5p/SLC1A5 axis to affect papillary thyroid cancer progression [20]. LncRNA MACC1-AS1 accelerates pancreatic carcinoma development via activating PAX8/NOTCH1 pathway [21]. Besides, existing evidence has emphasized the significant effect of lncRNAs on OS progression, and ceRNA network regulated by lncRNAs has been largely validated in OS. For instance, lncRNA SNHG16 facilitates OS cell proliferation, migration and invasion by its regulation on miR-1301/BCL9 axis [22]. LncRNA SNHG1 downregulates miR-101-3p to increase ROCK1 level and accelerate OS development [23]. FOXD3-AS1 has been previously validated as a progression-promoter in cancers and diseases [24], [25]. More importantly, FOXD3-AS1 has been proved to mediate ceRNA networks to facilitate the progression of cancers like cutaneous melanoma [26] and colorectal cancer [14]. Herein, we provided the first evidence that FOXD3-AS1 participated in OS development. We depicted the remarkably high FOXD3-AS1 expression in OS tissues and cells, and illustrated that FOXD3-AS1 knockdown prevented cell migration, invasion and EMT process in OS. All the findings supported that FOXD3-AS1 was a cancer-promoting gene in OS.
Meanwhile, the aberrantly expressed miRNAs, which are transcripts with around 22 nucleotides, are also recognized to be significant for cancer progression [27]. As is largely documented, miRNAs can be sponged by lncRNAs and therefore mRNAs are released to exert restraining or promoting functions in the progression of various cancers. For instance, lncRNA HULC motivates liver cancer development via upregulating HMGA2 expression by sequestering miR-186 [28]. LncRNA SNHG1 up-regulates MTDH by sponging miR-145-5p to promote the progression of non-small cell lung cancer [29]. According to existing researches, the tumor-suppressing role of miR-296-5p has been confirmed in multiple cancers, such as hepatocellular carcinoma [30], non-small cell lung cancer [31], and breast cancer [32]. In this study, we first discovered that FOXD3-AS1 could bind with miR-296-5p in OS cells, and miR-296-5p overexpression suppressed OS cell migration, invasion and EMT. Moreover, ZCCHC3 was verified to be a target gene of miR-296-5p by bioinformatics prediction and molecular mechanism assays. Significantly, here we first uncovered that ZCCHC3 was overexpressed in OS cells and FOXD3-AS1 could upregulate ZCCHC3 expression by sequestering miR-296-5p. Subsequently, rescue assays showed that ZCCHC3 overexpression offset the suppressive influence of FOXD3-AS1 deficiency on OS cell migration, invasion and EMT, indicating that ZCCHC3 mediated the contribution of FOXD3-AS1 to OS progression.
ELF1 has been researched and reported as a transcription factor in many diseases including diverse cancers, such as Hodgkin lymphoma [15], breast cancer [33] and non-small cell lung cancer [34]. The present study was the first to relate ELF1 to FOXD3-AS1 in OS. We demonstrated that ELF1 bound with FOXD3-AS1 promoter to activate FOXD3-AS1 transcription in OS cells. Then, rescue experiments proved that FOXD3-AS1 upregulation restored the repressive effect of ELF1 depletion on OS cell migration, invasion and EMT, which indicated that FOXD3-AS1 could be induced by ELF1 to promote OS progression. Further, in vivo assays verified that FOXD3-AS1 could promote tumorigenesis in OS through elevating ZCCHC3 expression.
5. Conclusion
In conclusion, ELF1-activated FOXD3-AS1 promotes the migration, invasion and EMT of OS cells via sponging miR-296-5p to upregulate ZCCHC3 expression. The findings may pave the way for improving molecular-targeted therapies for patients with OS.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
CRediT authorship contribution statement
Lei Wang: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Writing - original draft, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
Thanks a lot to all individuals or teams that were involved in our research.
References
- 1.Mirabello L., Troisi R.J., Savage S.A. Osteosarcoma incidence and survival rates from 1973 to 2004: data from the surveillance, epidemiology, and end results program. Cancer. 2009;115(7):1531–1543. doi: 10.1002/cncr.24121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bielack S.S., Kempf-Bielack B., Delling G., Exner G.U., Flege S., Helmke K., Kotz R., Salzer-Kuntschik M., Werner M., Winkelmann W., Zoubek A., Jurgens H., Winkler K. Prognostic factors in high-grade osteosarcoma of the extremities or trunk: an analysis of 1,702 patients treated on neoadjuvant cooperative osteosarcoma study group protocols. J. Clin. Oncology Official J. Am. Soc. Clin. Oncol. 2002;20(3):776–790. doi: 10.1200/JCO.2002.20.3.776. [DOI] [PubMed] [Google Scholar]
- 3.Wang K.C., Chang H.Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell. 2011;43(6):904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang X., Arai S., Song X., Reichart D., Du K., Pascual G., Tempst P., Rosenfeld M.G., Glass C.K., Kurokawa R. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature. 2008;454(7200):126–130. doi: 10.1038/nature06992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shao Y., Zhang D., Li X., Yang J., Chen L., Ning Z., Xu Y., Deng G., Tao M., Zhu Y., Jiang J. MicroRNA-203 increases cell radiosensitivity via directly targeting Bmi-1 in hepatocellular carcinoma. Mol. Pharm. 2018;15(8):3205–3215. doi: 10.1021/acs.molpharmaceut.8b00302. [DOI] [PubMed] [Google Scholar]
- 6.Prensner J.R., Chinnaiyan A.M. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1(5):391–407. doi: 10.1158/2159-8290.CD-11-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mercer T.R., Dinger M.E., Mattick J.S. Long non-coding RNAs: insights into functions. Nat. Rev. Genet. 2009;10(3):155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 8.Misawa A., Takayama K., Urano T., Inoue S. Androgen-induced long noncoding RNA (lncRNA) SOCS2-AS1 promotes cell growth and inhibits apoptosis in prostate cancer cells. J. Biol. Chem. 2016;291(34):17861–17880. doi: 10.1074/jbc.M116.718536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.X. Wu, P. Zhang, H. Zhu, S. Li, X. Chen, L. Shi, Long noncoding RNA FEZF1-AS1 indicates a poor prognosis of gastric cancer and promotes tumorigenesis via activation of Wnt signaling pathway, Biomed. Pharmacother. Biomed. Pharmacother. 96 (2017) 1103–1108. [DOI] [PubMed]
- 10.Kong Q., Qiu M. Long noncoding RNA SNHG15 promotes human breast cancer proliferation, migration and invasion by sponging miR-211-3p. Biochem. Biophys. Res. Commun. 2018;495(2):1594–1600. doi: 10.1016/j.bbrc.2017.12.013. [DOI] [PubMed] [Google Scholar]
- 11.Zheng L., Hu N., Zhou X. TCF3-activated LINC00152 exerts oncogenic role in osteosarcoma through regulating miR-1182/CDK14 axis. Pathol. Res. Pract. 2019;215(2):373–380. doi: 10.1016/j.prp.2018.12.031. [DOI] [PubMed] [Google Scholar]
- 12.Guan Y., Bhandari A., Xia E., Yang F., Xiang J., Wang O. lncRNA FOXD3-AS1 is associated with clinical progression and regulates cell migration and invasion in breast cancer. Cell Biochem. Funct. 2019;37(4):239–244. doi: 10.1002/cbf.3393. [DOI] [PubMed] [Google Scholar]
- 13.Chen Z.H., Hu H.K., Zhang C.R., Lu C.Y., Bao Y., Cai Z., Zou Y.X., Hu G.H., Jiang L. Down-regulation of long non-coding RNA FOXD3 antisense RNA 1 (FOXD3-AS1) inhibits cell proliferation, migration, and invasion in malignant glioma cells. Am. J. Transl. Res. 2016;8(10):4106–4119. [PMC free article] [PubMed] [Google Scholar]
- 14.Wu Q., Shi M., Meng W., Wang Y., Hui P., Ma J. Long noncoding RNA FOXD3-AS1 promotes colon adenocarcinoma progression and functions as a competing endogenous RNA to regulate SIRT1 by sponging miR-135a-5p. J. Cell. Physiol. 2019;234(12):21889–21902. doi: 10.1002/jcp.28752. [DOI] [PubMed] [Google Scholar]
- 15.Paczkowska J., Soloch N., Bodnar M., Kiwerska K., Janiszewska J., Vogt J., Domanowska E., Martin-Subero J.I., Ammerpohl O., Klapper W., Marszalek A., Siebert R., Giefing M. Expression of ELF1, a lymphoid ETS domain-containing transcription factor, is recurrently lost in classical Hodgkin lymphoma. Brit. J. Haematol. 2019;185(1):79–88. doi: 10.1111/bjh.15757. [DOI] [PubMed] [Google Scholar]
- 16.Ando M., Kawazu M., Ueno T., Koinuma D., Ando K., Koya J., Kataoka K., Yasuda T., Yamaguchi H., Fukumura K., Yamato A., Soda M., Sai E., Yamashita Y., Asakage T., Miyazaki Y., Kurokawa M., Miyazono K., Nimer S.D., Yamasoba T., Mano H. Mutational landscape and antiproliferative functions of ELF transcription factors in human cancer. Cancer Res. 2016;76(7):1814–1824. doi: 10.1158/0008-5472.CAN-14-3816. [DOI] [PubMed] [Google Scholar]
- 17.McManus M.M., Weiss K.R., Hughes D.P. Understanding the role of Notch in osteosarcoma. Adv. Exp. Med. Biol. 2014;804:67–92. doi: 10.1007/978-3-319-04843-7_4. [DOI] [PubMed] [Google Scholar]
- 18.Poos K., Smida J., Nathrath M., Maugg D., Baumhoer D., Neumann A., Korsching E. Structuring osteosarcoma knowledge: an osteosarcoma-gene association database based on literature mining and manual annotation. Database J. Biol. Databases Curation. 2014;2014 doi: 10.1093/database/bau042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang W., Xie Y., Chen F., Liu X., Zhong L.L., Wang H.Q., Li Q.C. LncRNA MEG3 acts a biomarker and regulates cell functions by targeting ADAR1 in colorectal cancer. World J. Gastroenterol. 2019;25(29):3972–3984. doi: 10.3748/wjg.v25.i29.3972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhuang X., Tong H., Ding Y., Wu L., Cai J., Si Y., Zhang H., Shen M. Long noncoding RNA ABHD11-AS1 functions as a competing endogenous RNA to regulate papillary thyroid cancer progression by miR-199a-5p/SLC1A5 axis. Cell Death Dis. 2019;10(8):620. doi: 10.1038/s41419-019-1850-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qi C., Xiaofeng C., Dongen L., Liang Y., Liping X., Yue H., Jianshuai J. Long non-coding RNA MACC1-AS1 promoted pancreatic carcinoma progression through activation of PAX8/NOTCH1 signaling pathway. J. Experiment. Clin. Cancer Res. CR. 2019;38(1):344. doi: 10.1186/s13046-019-1332-7. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Wang X., Hu K., Chao Y., Wang L. LncRNA SNHG16 promotes proliferation, migration and invasion of osteosarcoma cells by targeting miR-1301/BCL9 axis. Biomed. Pharmacother. Biomed. Pharmacother. 2019;114:108798. doi: 10.1016/j.biopha.2019.108798. [DOI] [PubMed] [Google Scholar]
- 23.Deng R., Zhang J., Chen J. lncRNA SNHG1 negatively regulates miRNA1013p to enhance the expression of ROCK1 and promote cell proliferation, migration and invasion in osteosarcoma. Int. J. Mol. Med. 2019;43(3):1157–1166. doi: 10.3892/ijmm.2018.4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu Q., Shi M., Meng W., Wang Y., Hui P., Ma J. Long noncoding RNA FOXD3-AS1 promotes colon adenocarcinoma progression and functions as a competing endogenous RNA to regulate SIRT1 by sponging miR-135a-5p. J. Cell. Physiol. 2019 doi: 10.1002/jcp.28752. [DOI] [PubMed] [Google Scholar]
- 25.Zhang D., Lee H., Haspel J.A., Jin Y. Long noncoding RNA FOXD3-AS1 regulates oxidative stress-induced apoptosis via sponging microRNA-150. FASEB J. Publ. Federation Am. Soc. Experiment. Biol. 2017;31(10):4472–4481. doi: 10.1096/fj.201700091R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.X. Chen, J. Gao, Y. Yu, Z. Zhao, Y. Pan, LncRNA FOXD3-AS1 promotes proliferation, invasion and migration of cutaneous malignant melanoma via regulating miR-325/MAP3K2, Biomed. Pharmacother. Biomed. Pharmacother. 120 (2019) 109438. [DOI] [PubMed]
- 27.Guo W., Yu Q., Zhang M., Li F., Liu Y., Jiang W., Jiang H., Li H. Long intergenic non-protein coding RNA 511 promotes the progression of osteosarcoma cells through sponging microRNA 618 to upregulate the expression of maelstrom. Aging. 2019;11(15):5351–5367. doi: 10.18632/aging.102109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y., Chen F., Zhao M., Yang Z., Li J., Zhang S., Zhang W., Ye L., Zhang X. The long noncoding RNA HULC promotes liver cancer by increasing the expression of the HMGA2 oncogene via sequestration of the microRNA-186. J. Biol. Chem. 2017;292(37):15395–15407. doi: 10.1074/jbc.M117.783738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu Q., Shan S., Li Y., Zhu D., Jin W., Ren T. Long noncoding RNA SNHG1 promotes non-small cell lung cancer progression by up-regulating MTDH via sponging miR-145-5p. FASEB J. Publ. Federation Am. Soc. Experiment. Biol. 2018;32(7):3957–3967. doi: 10.1096/fj.201701237RR. [DOI] [PubMed] [Google Scholar]
- 30.Shi D.M., Li L.X., Bian X.Y., Shi X.J., Lu L.L., Zhou H.X., Pan T.J., Zhou J., Fan J., Wu W.Z. miR-296-5p suppresses EMT of hepatocellular carcinoma via attenuating NRG1/ERBB2/ERBB3 signaling. J. Experiment. Clin. Cancer Res. CR. 2018;37(1):294. doi: 10.1186/s13046-018-0957-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu C., Li S., Chen T., Hu H., Ding C., Xu Z., Chen J., Liu Z., Lei Z., Zhang H.T., Li C., Zhao J. miR-296-5p suppresses cell viability by directly targeting PLK1 in non-small cell lung cancer. Oncol. Rep. 2016;35(1):497–503. doi: 10.3892/or.2015.4392. [DOI] [PubMed] [Google Scholar]
- 32.Maia D., de Carvalho A.C., Horst M.A., Carvalho A.L., Scapulatempo-Neto C., Vettore A.L. Expression of miR-296-5p as predictive marker for radiotherapy resistance in early-stage laryngeal carcinoma. J. Transl. Med. 2015;13:262. doi: 10.1186/s12967-015-0621-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gerloff A., Dittmer A., Oerlecke I., Holzhausen H.J., Dittmer J. Protein expression of the Ets transcription factor Elf-1 in breast cancer cells is negatively correlated with histological grading, but not with clinical outcome. Oncol. Rep. 2011;26(5):1121–1125. doi: 10.3892/or.2011.1409. [DOI] [PubMed] [Google Scholar]
- 34.Yang D.X., Li N.E., Ma Y., Han Y.C., Shi Y. Expression of Elf-1 and survivin in non-small cell lung cancer and their relationship to intratumoral microvessel density. Chin. J. Cancer. 2010;29(4):396–402. doi: 10.5732/cjc.009.10547. [DOI] [PubMed] [Google Scholar]