Abstract
Background
Malignant pleural mesothelioma (MPM) is an aggressive tumor but approximately 12% of patients survive more than 3 years. The biological differences underlying better outcomes are not known. Several targeted agents and immunotherapy have been ineffective. Hedgehog (Hh) is one emerging pathway. We compared the biological profiles of patients with different survival, investigating the most frequently altered genes, including the Hh pathway.
Methods
We analyzed 56 MPM. A 36-month overall survival (OS) cut-off divided patients into 32 normo (NS) and 24 long (LS) survivors. We used next generation sequencing to test 21 genes, immunohistochemistry to evaluate SMO expression. Mutation differences between NS and LS and their associations with clinical features were analysed by Fisher’s test, OS with the Kaplan-Meier method and its association with mutations by univariate and multivariate Cox proportional hazard models.
Results
Clinical features were similar in both groups. Eighteen out of 56 patients (32%) were wild-type for the genes analysed. At least five had mutations in BAP1, NF2, TP53, SMO and PTCH1 with no significant differences between the groups except for SMO. SMO, a member of the Hh pathway, was mutated only in NS (15.6%) and only SMO mutations were significantly associated with poor prognosis at univariate (HR =4.36, 95% CI: 2.32–8.18, P<0.0001) and multivariate (HR =9.2, 95% CI: 3.0–28.4, P=0.0001) analysis. All SMO mutated patients expressed high protein levels.
Conclusions
SMO mutations were clearly associated with worse prognosis. SMO may be a therapeutic target but this needs to be confirmed in a prospective trial.
Keywords: Gene mutations, hedgehog pathway, malignant pleural mesothelioma (MPM), prognosis, SMO
Introduction
Malignant pleural mesothelioma (MPM) is a rare cancer associated with asbestos exposure (1). Thus, incidence rates differ across countries. Although the use of asbestos has been banned in 55 countries for at least the last 20 years, about 125 million people worldwide are still exposed to it (2). On the basis of global asbestos consumption over the last decades, a further mesothelioma wave can be expected, involving large geographic areas (3), and may peak in developed countries by 2030 (4).
In the last few decades, the identification of specific molecular targets and genetic alterations has radically changed the treatment paradigms for different cancers, improving outcomes. Unfortunately, this is not the case for MPM patients, whose prognosis remains poor, with median survival of about 12 months (5). The roles of surgery and radiotherapy are debated (6). Since 2003, the only treatment that has slightly improved survival is platinum-based doublet with an antifolate agent (7,8). The association of the antiangiogenic drug bevacizumab to chemotherapy has been explored in several studies and was recently reported to give significant improvement of survival, although the clinical application of this association remains uncertain (9-11). Despite a consistent biological rationale and promising preclinical data, several targeted agents and immunotherapy with anti-CTLA4 have shown no efficacy in unselected patients. Other immune-checkpoint inhibitors and new-generation compounds are now under investigation (12,13).
In the single arm phase II MERIT trial, monotherapy with anti-PD-1 nivolumab was administered in 34 MPM patients as second- or third-line treatment; objective response rate (ORR), median progression-free survival (PFS) and overall survival (OS) were 29%, 6.1 and 17.3 months, respectively (14). Based on these results, nivolumab was approved in Japan for unresectable recurrent MPM. Other phase II studies demonstrated a potential activity of nivolumab in MPM (15). However, in a randomized phase III trial the anti-PD-1 pembrolizumab failed to show a PFS or OS benefit in advanced pretreated MPM patients in comparison with single agent chemotherapy (16). In MPM also the predictive role of PD-L1 expression for immunotherapy is still debatable (15).
Nevertheless, according to population studies, approximately 12% of MPM patients survive more than 3 years (17). Biological or molecular differences that might explain the better outcome of these longer-term survivors are still unknown. Histological subtype, age, and stage are recognized prognostic factors (18,19).
From the biological point of view, the main genetic alterations in MPM patients involve a handful of genes (i.e., TP53, NF2, BAP1 and CDKN2A) (20-25). However, their real predictive or prognostic value is still uncertain (22-25). Furthermore, these alterations are not easily druggable and first attempts to target them have already failed (26).
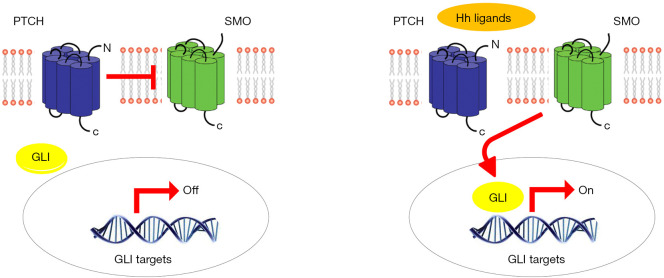
Among the new pathways reported in MPM, Hedgehog (Hh) is emerging. Hh is involved in cell proliferation, survival, epithelial-to-mesenchymal transition, stemness and differentiation during embryonic development. Hh ligands bind to PTCH1 and PTCH2 receptors. If the ligand is absent, PTCH1 or PTCH2 binds to the SMO co-receptor, repressing its activity. Consequently, the transcription factors (GLI family) are not activated and transcription is stopped. In contrast, when the ligand binds to the receptor, the SMO repression is released, leading to transcriptional activation (Figure 1) (27). Normally the Hh pathway is inactive in adult tissues (28). Inappropriate reactivation of Hh signaling, mainly due to mutations in key pathway regulators (e.g., PTCH, SUFU or SMO) or over-expression of pathway activators (Hh ligands, SMO, GLI1, GLI2), has been linked to different sporadic malignancies, including basal cell carcinoma (BCC), pancreatic adenocarcinoma and gastrointestinal stromal tumors (GIST) (29,30). In particular a strong correlation between higher SMO, SHH, GLI expression levels and poorer OS was observed in MPM patients. High levels of Hh involved genes were detected in MPM compared to normal pleura (31,32). Furthermore, data available in public database at the beginning of the study reported SMO and PTCH1 among the most frequent mutated genes in MPM (33). This study was designed to distinguish possible differences in the biological profiles of MPM patients with different survival periods, investigating the most frequently altered genes, including those in the Hh pathway.
Figure 1.
Hedgehog (Hh) ligands bind to PTCH receptors. If Hh ligands are absent, PTCH binds and represses SMO co-receptor. Thus, the transcription factors GLI are not activated and transcription is stopped. Conversely, when Hh ligands bind to PTCH, the SMO repression is released, leading to transcriptional activation. PTCH, patched homologue; SMO, smoothened protein; GLI, glioma-associated oncogene.
We present the following article in accordance with the MDAR Reporting Checklist (available at http://dx.doi.org/10.21037/tlcr-19-425).
Methods
Selection of patients and sample collection
Following the evidence from our Italian epidemiologic study on pleural mesothelioma, LUME (LUngo sopravviventi nel MEsotelioma pleurico) (17), we established an OS cut-off of 36 months to divide patients into normo survivors (NS) and long survivors (LS). This cut-off guarantees the identification of true LS, three times the median survival (5).
We designed a pilot study in which 60 patients (30 LS and 30 NS) had to be retrospectively enrolled. The recruitment period was 2002–2014 and three Italian institutions contributed: INT, Istituto Oncologico Veneto-Padua and Azienda Ospedaliera-Parma. We considered only patients with enough tissue samples to perform molecular tests and with almost all clinical and pathological data available. For each patient, clinical data (disease characteristics, surgical treatments and outcomes) and formalin-fixed paraffin-embedded (FFPE) tissue samples at diagnosis were collected. FFPE were centrally reviewed and analysed at the Fondazione IRCSS Istituto Nazionale dei Tumori-Milan (INT). We included in the study all the LS available and we randomly identified, from the remaining patients, the MPM NS to see whether they had a biological profile different from the MPM LS. No differences in clinical characteristics (i.e., stage, age, sex and treatment) were observed between MPM patients included and excluded from the study.
The present study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). All the experimental protocols were approved by INT Independent Ethics Committee, code INT 91/13. Because the study was retrospective, the patients were not in treatment or in active follow-up; therefore, their informed consent was not required in accordance with Italian law (Gazzetta Ufficiale n. 72, 26/03/2012; n. 303, 29/12/2016).
Genomic DNA extraction and next generation sequencing (NGS)
FFPE sample was sliced in 5-µm sections and manually microdissected to isolate the highest percentage of neoplastic cells as possible. Sample was treated with xylene and 100% ethanol to remove paraffin and then DNA was isolated using the GeneRead DNA FFPE kit (Qiagen). DNA amount was quantified with Qbit dsDNA BR kit (ThermoFisher). gDNA (40 ng) was profiled using a customized panel (Ampliseq Designer, ThermoFisher) that amplify 21 genes (CDKN2A, NF2, GSTM1, NAT2, BAP1, TERT, TP53, PTCH1, SMO, LATS2, KEAP1, PI3KCA, KRAS, NRAS, STK11, WT1, FBXW7, CTNNB1, KIT, KDR, and REV3). Our genes selection was based on those reported in the public MPM database (33). The library was prepared by IonAmpliSeq Library kit 2.0 (ThermoFisher) according to the manufacturer’s instructions. Emulsion PCR was performed on the Ion One Touch 2 instrument (ThermoFisher) using Ion PGM™ template OT2 200 kit, according to the manufacturer’s instructions. Sequencing was carried out on the Ion PGM System (ThermoFisher) using Ion 318 v2 Chip and Ion PGM™ sequencing 200 kit v2 according to the manufacturer’s instructions. Data was processed by using Torrent Suite SoftwareTM v4.4.2; the variant calling from sequencing data was generated by Variant Caller plugin. The resulting variants was annotated using Ensemble Variant Effect Predictor pipeline, Ion Reporter™ analysis software, ClinVar db and COSMIC database. The filtered variants were examined using the Integrative Genomic Viewer IGV tool (34). The coverage depth was always more than 500× and the reported mutations had a frequency of at least 5%. Matched normal DNA was used for six patients; where normal tissue is not available, we filtering germline variants by using publically-available or proprietary database of known polymorphisms (e.g., dbSNP, ExAC, 1000 Genomes), excluding variant with minor allele frequency (MAF) >10−6.
Immunohistochemistry (IHC)
IHC was done on FFPE whole-tissue sections. SMO mouse monoclonal antibody was purchased from Origene, TA318627, clone 3E5 and used at the dilution of 1:500. In accordance with the Ventana BenchMark ultra-automated system protocol, the antigen retrieval Ultra Cell Conditioning and standard reagents, provided by Ventana (OptiView DAB IHC Detection Kit; Ventana Medical Systems), were used. The staining intensity (I) and the percentage of positive cells (P) were evaluated by a trained pathologist and a semi-quantitative score S = I × P was calculated.
Statistical analysis
Gene mutation differences between NS and LS and the association between gene mutations and clinical features were assessed by the non-parametric Fisher’s exact test. The t-test was used for the association of gene mutations with age. OS was analyzed from the date of diagnosis to the date of death or last follow-up with the Kaplan-Meier method.
We intentionally included all LS available at the centres contributing to the data collection. This choice was taken to increase the number of LS in the biological analyses in order to better analyze the population of interest although it led to a sample with a LS proportion higher than the 12% observed at population-level. To test the association between the gene mutation and survival, we removed the bias of having a sample with a high proportion of LS readjusting their number. In other words, we decreased the number of LS in order to have a 12% of them. To this extend, we randomly selected LS simulating 100 different hypothetical samples. In each of this hypothetical sample, the NS were always all the patients of our sample whereas the LS were randomly selected from the LS in the database. To assess the association between mutated genes and OS, we applied the univariate Cox proportional hazard model to each sample and used Rubin’s rule to pool the 100 estimates from these models (35).
To examine the association between SMO mutations and OS we applied Cox proportional hazard model adjusted for the available prognostic factors (sex, age, stage, histology and treatment) to each sample and used Rubin’s rule to pool the 100 estimates from these models. P values (P) less than 0.05 were considered significant.
For the IHC analysis, the median semi-quantitative score was used as a good initial threshold to divide our sample into high and low SMO expression groups. To compare median survival across SMO expression groups in NS we used Laplace regression models for percentiles (36).
Results
The final analysis comprised 56 patients, 24 LS and 32 NS. No other LS with available tissue were found in our database.
Patients characteristics
Patients were mainly male, with epithelioid histotype, and diagnosed at stage III (Table 1). The clinical features did not differ significantly in the two groups. Out of the 29 patients who underwent surgery, the surgical approach was extra-pleural pneumonectomy in 20 cases, pleurectomy/decortication in 9 patients. The chemotherapy regimens, administered as neoadjuvant, adjuvant or first-line chemotherapy were platinum-based doublets in 39 out of 42 cases; the second drug was pemetrexed in 35 and gemcitabine in 4 patients. Three patients underwent monotherapy with pemetrexed. The systemic treatments administered as second or further lines were generally monotherapy with gemcitabine, vinorelbine, pemetrexed or treatments within clinical trials. Postoperative radiotherapy was performed in 23 patients; it was delivered to the ipsilateral hemithorax area in 18 patients and as prophylactic radiotherapy to chest wall tracts after surgery in 5 cases. Two patients, who did not undergo surgery, performed radiotherapy on the ipsilateral pleura. Palliative radiant treatments or palliative surgical approaches were not considered.
Table 1. Patients main characteristics overall and for NS and LS.
| Patients main characteristics | Subcategories | ALL (56 pts), N (%) | NS (32 pts), N (%) | LS (24 pts), N (%) | P (NS vs. LS) |
|---|---|---|---|---|---|
| Age, years | Mean | 61.4 | 60.9 | 62.1 | 0.71 |
| Sex | Female | 15 [27] | 8 [25] | 7 [29] | 0.77 |
| Male | 41 [73] | 24 [75] | 17 [71] | ||
| Histology | Epithelioid | 51 [91] | 29 [91] | 22 [92] | 0.36 |
| Mixed | 4 [7] | 3 [9] | 1 [4] | ||
| Sarcomatoid | 1 [2] | 0 [0] | 1 [4] | ||
| Stage | I | 5 [9] | 3 [9] | 2 [8] | 0.42 |
| II | 14 [25] | 8 [25] | 6 [25] | ||
| III | 26 [46] | 13 [41] | 13 [54] | ||
| IV | 10 [18] | 8 [25] | 2 [8] | ||
| Missing | 1 [2] | 0 [0] | 1 [4] | ||
| Radical surgery | Yes | 29 [52] | 16 [50] | 13 [54] | 0.76 |
| No | 27 [48] | 16 [50] | 11 [46] | ||
| Chemotherapy | Yes | 42 [75] | 23 [72] | 19 [79] | 0.76 |
| No | 12 [21] | 8 [25] | 4 [17] | ||
| Missing | 2 [4] | 1 [3] | 1 [4] | ||
| Radiotherapy | Yes | 25 [45] | 11 [34] | 14 [58] | 0.10 |
| No | 30 [54] | 20 [63] | 10 [42] | ||
| Missing | 1 [2] | 1 [3] | 0 [0] | ||
| Treatment | None | 5 [9] | 4 [13] | 1 [4] | 0.53 |
| One | 22† [39] | 13 [41] | 9 [38] | ||
| Two | 13‡ [23] | 8 [25] | 5 [21] | ||
| All three | 16 [29] | 7 [22] | 9 [38] |
†, 18 chemotherapy, 3 surgery, 1 radiotherapy; ‡, 5 chemotherapy and surgery, 3 chemotherapy and radiotherapy, 5 surgery and radiotherapy. NS, normo-survivors, OS ≤3 years; LS, long survivors, OS >3 years.
Gene variations in the 21 MPM associated genes
Table 2 reports the sequencing analysis, with the mutated gene and the amino-acid residue involved. Eighteen out of 56 patients (32%) were wild-type for the genes analysed: 12 were NS and 6 were LS. The overall mutated genes and their distribution in NS and LS groups are reported in Table 3.
Table 2. Mutations across 56 MPM patients.
| MPM case | Histo type | Mutated genes | Survival group |
|---|---|---|---|
| 1 | E | BAP1 (E600D) | LS |
| TP53 (G187S) | |||
| KEAP1 (R596Q, A321V) | |||
| 2 | E | LATS2 (E505 stop) | NS |
| NF2 (E421 stop) | |||
| 3 | E | No mutations | NS |
| 4 | E | PTCH1 (P725S) | LS |
| 5 | E | TP53 (R248W) | NS |
| KEAP1 (R554Q, E289K) | |||
| NF2 (R424C) | |||
| 6 | E | BAP1 (ins) | NS |
| 7 | E | No mutations | NS |
| 8 | E | CDKN2A (A148T) | NS |
| 9 | E | NF2 (L436fs) | LS |
| 10 | E | BAP1 (Q156 stop) | LS |
| 11 | E | No mutations | LS |
| 12 | E | KDR (T1038I) | NS |
| SMO (R257Q) | |||
| 13 | E | BAP1 (R713Q) | NS |
| SMO (T245M) | |||
| TP53 (R175H) | |||
| FBXW7 (R564C) | |||
| 14 | E | NF2 (I210T) | LS |
| 15 | E | No mutations | NS |
| 16 | E | BAP1 (ins) | LS |
| 17 | E | No mutations | LS |
| 18 | E | No mutations | LS |
| 19 | E | NF2 (R57STOP) | LS |
| 20 | E | REV3L (I691M) | LS |
| 21 | E | No mutations | LS |
| 22 | E | BAP1 (K51fs) | NS |
| 23 | E | PTCH1 (G1212S) | LS |
| 24 | E | PTCH1 (K842R) | NS |
| NF2 (R187fs) | |||
| 25 | E | No mutations | NS |
| 26 | E | NF2 (R341STOP) | NS |
| 27 | E | KDR (C482R) | LS |
| 28 | M | No mutations | NS |
| 29 | E | TERT (H815N) | LS |
| TP53 (Y327 stop) | |||
| 30 | E | No mutations | LS |
| 31 | E | BAP1 (del) | NS |
| 32 | M | No mutations | NS |
| 33 | E | No mutations | NS |
| 34 | E | TP53 (A119fs) | NS |
| 35 | E | BAP1 (R699Q) | LS |
| TERT (T292M) | |||
| 36 | E | BAP1 (R717W) | LS |
| 37 | E | LATS2 (V212M) | NS |
| 38 | E | BAP1 (E685 stop) | LS |
| 39 | E | BAP1 (H193R) | NS |
| 40 | M | NF2 (R196 stop) | NS |
| 41 | E | No mutations | LS |
| 42 | M | NF2 (fs) | LS |
| WT1 (513V) | |||
| 43 | E | No mutations | NS |
| 44 | E | TP53 (Y234C) | NS |
| 45 | E | No mutations | NS |
| 46 | S | BAP1 (R610 stop) | LS |
| TP53 (start loss M1K) | |||
| 47 | E | BAP1 (R60 stop) | LS |
| 48 | E | No mutations | NS |
| 49 | E | TERT (R1084 stop) | NS |
| TP53 (R181C) | |||
| 50 | E | SMO (R257W) | NS |
| PTCH1 (G1363S) | |||
| 51 | E | SMO (A601V) | NS |
| 52 | E | PTCH1 (T1052M) | NS |
| 53 | E | No mutations | NS |
| 54 | E | No mutations | NS |
| 55 | E | NF2 (E260 stop) | LS |
| 56 | E | BAP1 (V447I) | NS |
| FBXW7 (M269I) |
MPM, malignant pleural mesothelioma; E, epithelioid; M, mixed; S, sarcomatoid; NS, normo-survivors, OS ≤3 years; LS, long-survivors, OS >3 years.
Table 3. Distribution of gene mutations.
| Gene | All, N (%) | NS, N (%) | LS, N (%) | P (NS vs. LS) |
|---|---|---|---|---|
| Wild-type | 18 (32.1) | 12 (37.5) | 6 (25.0) | 0.243 |
| BAP1 | 14 (25.0) | 6 (18.8) | 8 (33.3) | 0.232 |
| NF2 | 10 (17.9) | 5 (15.6) | 5 (20.8) | 0.730 |
| P53 | 8 (14.3) | 5 (15.6) | 3 (12.5) | 0.686 |
| SMO | 5 (8.9) | 5 (15.6) | 0 (0.0) | 0.063 |
| PTCH1 | 5 (8.9) | 3 (9.4) | 2 (8.3) | 1.000 |
| TERT | 3 (5.4) | 1 (3.1) | 2 (8.3) | 0.571 |
| FBXW7 | 2 (3.6) | 2 (6.3) | 0 (0.0) | 0.501 |
| LATS2 | 2 (3.6) | 2 (6.3) | 0 (0.0) | 0.501 |
| KDR | 2 (3.6) | 1 (3.1) | 1 (4.2) | 1.000 |
| KEAP1 | 2 (3.6) | 1 (3.1) | 1 (4.2) | 1.000 |
| WT1 | 1 (1.8) | 0 (0.0) | 1 (4.2) | 0.429 |
| REV3L | 1 (1.8) | 0 (0.0) | 1 (4.2) | 0.429 |
| CDKN2A | 1 (1.8) | 1 (3.1) | 0 (0.0) | 1.000 |
NS, normo-survivors, OS ≤3 years; LS, long survivors, OS >3 years. P values refer to the comparison between NS and LS.
The only association between gene mutations and clinical features was LATS2 and age: LATS2 mutated patients were younger (43 vs. 62 years, P=0.01).
Mutations in at least five patients were observed only for BAP1, NF2, TP53, SMO and PTCH1 genes. There were no significant differences in mutation frequency between the two groups (Table 3). Of note, SMO was mutated only in NS (15.6%) with epithelioid histotype (Table 2).
In NS patients the median survival was 21.6 (min 0.6; max 32.4) months, while in LS was 53 (min 37; max 86) months. To investigate the prognostic role of the more frequent mutated genes, we calculated the OS hazard ratio (HR) as reported in Table 4.
Table 4. Association between mutated genes in at least five patients and OS.
| Gene | HR (95% CI) | P |
|---|---|---|
| Any mutation-wild type | 1.09 (0.71–1.67) | 0.70 |
| BAP1 | 1.14 (0.57–2.30) | 0.73 |
| NF2 | 1.16 (0.66–2.07) | 0.60 |
| TP53 | 0.91 (0.54–1.54) | 0.36 |
| SMO | 4.36 (2.32–8.18) | <0.0001 |
| PTCH1 | 1.16 (0.66–2.07) | 0.71 |
OS is expressed as hazard ratio (HR) with 95% confidence interval (CI). Wild-type patients for the specific gene were used as reference to calculate P values.
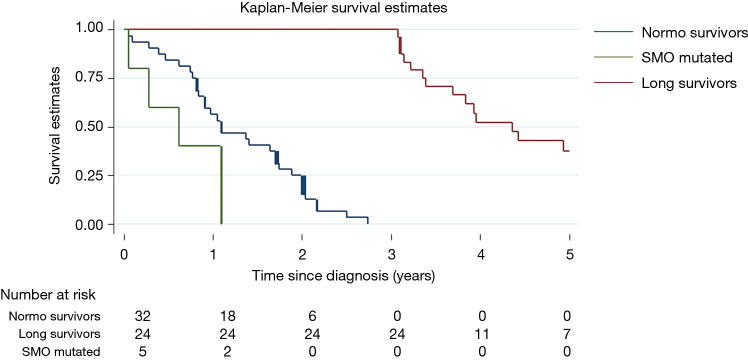
Only SMO mutational status was significantly associated with poor prognosis (P<0.0001) and remained a prognostic factor in multivariable analysis (HR =9.2, 95% CI: 3.0–28.4 P=0.0001). Kaplan-Meier curves show the prognostic role of SMO, considering that SMO mutated patients died within 13 months (Figure 2).
Figure 2.
Kaplan-Meier survival estimates. Normo-survivors patients, OS ≤3 years, in blue; long survivors patients, OS >3 years, in red; SMO mutated patients in green.
All the SMO mutations were missense and different. The same amino-acid residue was involved only in two patients, and the prediction of functional effects of all the variants was verified by PolyPhen-2 analysis (Table 5). Three mutations had never been described before, while mutation T245M was found in esophageal squamous cell carcinoma and mutation A601V in pancreatic cancer (37,38). SMO was mutated as a single gene or with mutations in other genes. We confirmed by Sanger the only mutation occurring with frequency higher than the limit of detection of this technique.
Table 5. SMO mutations.
| SMO mutations (frequency) | Function prediction (polyphen) | Topology protein domain | Co-mutated genes |
|---|---|---|---|
| R257Q (16%) | Benign | 1° cytoplasmic domain | KDR |
| T245M (11%) | Probably damaging | 1° transmembrane domain | BAP1, TP53, FBXW |
| F484L (47%) | Probably damaging | 4° extracellular domain | BAP1 |
| R257W (9%) | Probably damaging | 1° cytoplasmic domain | PTCH1 |
| A601V (5%) | Probably damaging | 4° cytoplasmic domain | – |
SMO IHC
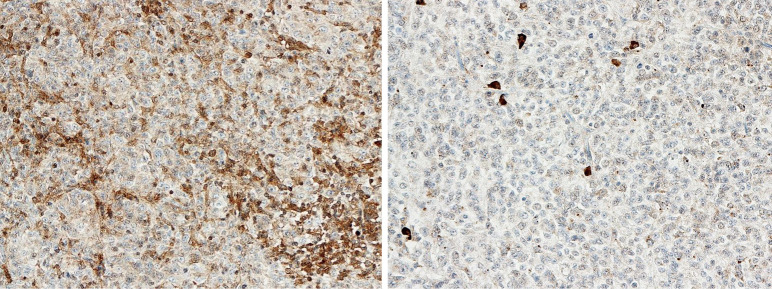
The expression of SMO was analyzed by IHC in 53 patients with leftover tissue to investigate the association between mutational status and the protein level (Figure 3). The median value of the semi quantitative score was 160 and divided the 53 patients into two groups: those with a high level of protein (≥160) and the others, with low expression (<160). The SMO mutated patients (4 of the 5 cases were available) all expressed a high level of protein. In addition, there were equal numbers of NS and LS (12 each) among the patients with low protein expression, while there were more NS with high expression (18 NS and 11 LS). The median survival of NS patients expressing high protein levels was about eight months less than those expressing low levels (11.6 vs. 19.6 months).
Figure 3.
An example of high (on the left) and low (on the right) SMO expression level. IHC, 20×.
Discussion
We sequenced 56 MPM samples using a customized 21-gene panel. We confirmed that MPM has a low mutational burden and that BAP1, NF2 and TP53 are the most frequent mutated genes, as reported by other authors (20-25). For the first time, to our knowledge, we observed that SMO mutations were associated with a worse prognosis, with a HR of 9.2 (95% CI: 3.0–28.4, P=0.0001), although only 5 patients with SMO mutation were identified. We found SMO mutations in about 9% of our samples, and only in NS patients. We also found five PTCH1 mutations, raising to 16% the overall frequency of alterations in the Hh pathway; however, these mutations did not show any clear correlation with survival. We evaluated patients managed in MPM expert centres in which interventional procedures, such as surgery, are more common, so that the rate of operated patients (52%) is higher than usual. However, we applied Cox proportional hazard model adjusted for the known prognostic factors (i.e., sex, age, stage, histology and treatment, including surgery) to assess the impact of SMO mutations on prognosis at the net of the available prognostic factors. In the past few years, some studies have shown that Hh signalling is active in MPM and regulates cell proliferation, in cell lines and clinical samples (32).
At preclinical level, Hh pathway was reported to be up-regulated in MPM cancer cell lines with increased expression level of GLI1, GLI2, SMO, SHH, PTCH1 and PTCH2, that are indicative of an active Hh signaling. Zhang et al., in 46 MPM sample tissues, identified a correlation between higher SMO expression and worse survival (31). Non-synonymous mutation affecting SMO, PTCH and SUFU were detected, in both MPM cell lines and patients: the biological significance was unclear; however, the patient with the SMO insertion survived 3.4 months (39). At least six other studies have analyzed MPM mutations using NGS technology (20-25); the results were similar as regards the low mutational load and the most frequent mutated genes. However, the frequencies and the prognostic roles of each mutation differed across the studies, on account of their heterogeneity. All of them were retrospective and used different NGS approaches. Two studies reported a negative prognostic role for TP53 mutations (22,23), which was not confirmed by De Rienzo or by this present work. No study showed a prognostic role of BAP1 mutations although De Rienzo et al. found a correlation between higher BAP1 expression and worse survival. In addition, deletions in CDKN2A were associated with poorer outcome (25). Our panel included this gene but it was not designed to detect large genetic losses (gene amplifications/losses and translocations). Consequently, we could describe only one CDKN2A missense mutation in our series.
Despite the methodological limits, all these studies have opened up some research areas in MPM. A proper prospective trial with adequate statistical power is warranted to define the true frequency of the reported mutations, their final prognostic role and the possibility of developing targeted treatments.
SMO and all the alterations of the Hh pathway might be potential therapeutic targets. In recent years, targeting Hh components has proved an interesting approach, at both preclinical and clinical levels (40). These compounds showed good activity in vitro, inhibiting the Hh downstream signaling and dramatically suppressing cell proliferation when used in range of nanomolar (41). Of note, they reduced tumor volume, although the majority of used models (mostly medulloblastoma, BCC and pancreas carcinoma) were not characterized for the mutational status of the Hh components. Vismodegib and sonidegib, two SMO inhibitors, have already been approved for the treatment of locally advanced BCC (42). In a rat MPM model, vismodegib gave good results in terms of tumor shrinkage and growth delay, with significant down-regulation of downstream transcriptional factor GLI1 in the stromal compartment (43). Other authors have reported suppression of cell growth in cell lines and animal models treated with different inhibitors directed against Hh components (44). Despite this preclinical evidence, however, in a phase I trial vismodegib failed to show any activity in the only three MPM patients included (45); however, these patients were not evaluated for alterations in the Hh pathway.
Although SMO protein expression was revealed by IHC in all our SMO mutated tumors, we still do not know whether these mutations are linked to a deregulated Hh pathway. We are therefore now working in vitro to clarify their final biological role.
Genomics has given us an important lesson on MPM including the discovery of a large number of wild-type patients which suggests that mechanisms different from activating mutations are implicated in this malignancy. Unfortunately, we are still far from finding effective strategies for this disease for which a wave of new cases is expected in many parts of the world. Thus, there is a pressing need for identification of new targets and therapeutic strategies for MPM.
Supplementary
The article’s supplementary files as
Acknowledgments
Thanks to Pietro Seghini (Cancer Registry of Piacenza) for the follow-up data.
Funding: The study was granted by AIRC (Investigator Grant - IG 2012 number 13534) and by Italian Ministry of Health (GR-2016-02361229).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The present study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). All the experimental protocols were approved by INT Independent Ethics Committee, code INT 91/13. Because the study was retrospective, the patients were not in treatment or in active follow-up; therefore, their informed consent was not required in accordance with Italian law (Gazzetta Ufficiale n. 72, 26/03/2012; n. 303, 29/12/2016).
Footnotes
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/tlcr-19-425
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tlcr-19-425
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr-19-425). DS reports personal fees from Astra Zeneca, BMS, Lilly, Boehringer Ingelheim, non-financial support from Astra Zeneca, Roche, MSD, Pfizer outside the submitted work. CP reports personal fees from BMS, MSD outside the submitted work. MT reports personal fees from Astra Zeneca, Pfizer, Eli-Lilly, BMS, Novartis, Roche, MSD, Boehringer Ingelheim, Otsuka, Takeda, Pierre Fabre, grants from Astra Zeneca, Boehringer Ingelheim outside the submitted work. MT serves as an unpaid editorial board member of Translational Lung Cancer Research from Dec 2019 to Nov 2021. GP reports ADVISOR/CONSULTANT: BOEHRINGER ING, AstraZeneca, Roche, BMS, MSD, LILLY ONC. GLR reports personal fees from BMS, MSD, Astra Zeneca outside the submitted work. AP reports personal fees from Roche, AstraZeneca, BMS outside the submitted work. FdB reports personal fees from TIZIANA LIFE SCIENCES, BMS, CELGENE, SERVIER, PHARMA RESEARCH, DAIICHI SANKYO, IGNYTA, NOVARTIS, AMGEN, PFIZER, OCTIMET ONCOLOGY, INCYTE, PIERRE FABRE, ELI LILLY, ROCHE, ASTRA ZENECA, GENTILI, DEPHAFORUM, NOVARTIS, MSD, BAYER, FONDAZIONE MENARINI, SANOFI outside the submitted work. Dr. G Gatta reports grants from AIRC during the conduct of the study. MCG reports grants and personal fees from Eli Lilly, Otsuka Pharma, Astra Zeneca, Novartis, BMS, Roche, Pfizer, Celgene, Incyte, Bayer, MSD, GlaxoSmithKline S.p.A., Spectrum Pharmaceutcials, Blueprint Medicine, personal fees from Boehringer Ingelheim, Inivata, Takeda, Sanofi-Aventis, Seattle Genetics, Daiichi Sankyo, Jannesen, grants from Tiziana Sciences, Clovis, Merck Serono, UNITED THERAPEUTICS CORPORATION, Merck KGaA, TURNING POINT, IPSEN, Medlmmune, EXELISIS, non-financial support from MSD, Pfizer, Eli-Lilly outside the submitted work. MG reports grants from MOH, during the conduct of the study. The other authors have no conflicts of interest to declare.
References
- 1.Tunesi S, Ferrante D, Mirabelli D, et al. Gene-asbestos interaction in malignant pleural mesothelioma susceptibility. Carcinogenesis 2015;36:1129-35. 10.1093/carcin/bgv097 [DOI] [PubMed] [Google Scholar]
- 2.Stayner L, Welch LS, Lemen R, et al. The worldwide pandemic of asbestos-related diseases. Annu Rev Public Health 2013;34:205-16. 10.1146/annurev-publhealth-031811-124704 [DOI] [PubMed] [Google Scholar]
- 3.Bianchi C, Bianchi T. Global mesothelioma epidemic: Trend and features. Indian J Occup Environ Med 2014;18:82-8. 10.4103/0019-5278.146897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robinson BM. Malignant pleural mesothelioma: an epidemiological perspective. Ann Cardiothorac Surg 2012;1:491-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Damhuis RA, Schroten C, Burgers JA, et al. Population-based survival for malignant mesothelioma after introduction of novel chemotherapy. Eur Respir J 2012;40:185-9. 10.1183/09031936.00153611 [DOI] [PubMed] [Google Scholar]
- 6.Takuwa T, Hasegawa S. Current surgical strategies for malignant pleural mesothelioma. Surg Today 2016;46:887-94. 10.1007/s00595-015-1275-3 [DOI] [PubMed] [Google Scholar]
- 7.Vogelzang NJ, Rusthoven JJ, Symanowski J, et al. Phase III study of pemetrexed in combination with cisplatin versus cisplatin alone in patients with malignant pleural mesothelioma. J Clin Oncol 2003;21:2636-44. 10.1200/JCO.2003.11.136 [DOI] [PubMed] [Google Scholar]
- 8.van Meerbeeck JP, Gaafar R, Manegold C, et al. Randomized phase III study of cisplatin with or without raltitrexed in patients with malignant pleural mesothelioma: an intergroup study of the European Organisation for Research and Treatment of Cancer Lung Cancer Group and the National Cancer Institute of Canada. J Clin Oncol 2005;23:6881-9. 10.1200/JCO.20005.14.589 [DOI] [PubMed] [Google Scholar]
- 9.Kindler HL, Karrison TG, Gandara DR, et al. Multicenter, double-blind, placebo-controlled, randomized phase II trial of gemcitabine/cisplatin plus bevacizumab or placebo in patients with malignant mesothelioma. J Clin Oncol 2012;30:2509-15. 10.1200/JCO.2011.41.5869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ceresoli GL, Zucali PA, Mencoboni M, et al. Phase II study of pemetrexed and carboplatin plus bevacizumab as first-line therapy in malignant pleural mesothelioma. Br J Cancer 2013;109:552-8. 10.1038/bjc.2013.368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zalcman G, Mazieres J, Margery J, et al. Bevacizumab for newly diagnosed pleural mesothelioma in the Mesothelioma Avastin Cisplatin Pemetrexed Study (MAPS): a randomised, controlled, open-label, phase 3 trial. Lancet 2016;387:1405-14. 10.1016/S0140-6736(15)01238-6 [DOI] [PubMed] [Google Scholar]
- 12.Signorelli D, Macerelli M, Proto C, et al. Systemic approach to malignant pleural mesothelioma: what news of chemotherapy, targeted agents and immunotherapy? Tumori 2016;102:18-30. 10.5301/tj.5000436 [DOI] [PubMed] [Google Scholar]
- 13.Maio M, Scherpereel A, Calabrò L, et al. Tremelimumab as second-line or third-line treatment in relapsed malignant mesothelioma (DETERMINE): a multicentre, international, randomised, double-blind, placebo-controlled phase 2b trial. Lancet Oncol 2017;18:1261-73. 10.1016/S1470-2045(17)30446-1 [DOI] [PubMed] [Google Scholar]
- 14.Okada M, Kijima T, Aoe K, et al. Clinical Efficacy and Safety of Nivolumab: Results of a Multicenter, Open-label, Single-arm, Japanese Phase II study in Malignant Pleural Mesothelioma (MERIT). Clin Cancer Res 2019;25:5485-92. 10.1158/1078-0432.CCR-19-0103 [DOI] [PubMed] [Google Scholar]
- 15.Hotta K, Fujimoto N. Current evidence and future perspectives of immune-checkpoint inhibitors in unresectable malignant pleural mesothelioma. J Immunother Cancer 2020;8:e000461. 10.1136/jitc-2019-000461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Popat S, Curioni-Fontecedro A, Polydoropoulou V, et al. A multicentre randomized phase III trial comparing pembrolizumab vs single agent chemotherapy for advanced pre-treated malignant pleural mesothelioma – results from the European Thoracic Oncology Platform (ETOP 9-15) PROMISE-meso trial. Ann Oncol 2019;30:v851-v934. 10.1093/annonc/mdz394.091 [DOI] [PubMed] [Google Scholar]
- 17.Trama A, Botta L, Garassino MC, et al. I pazienti con mesotelioma pleurico lungo sopravviventi in Italia ed il progetto LUME. Abstract 19, XVIII Riunione Scientifica Annuale. Associazione Italiana Registri Tumori, Taranto, 2014. [Google Scholar]
- 18.Montanaro F, Rosato R, Gangemi M, et al. Survival of pleural malignant mesothelioma in Italy: a population-based study. Int J Cancer 2009;124:201-7. 10.1002/ijc.23874 [DOI] [PubMed] [Google Scholar]
- 19.Taioli E, Wolf AS, Camacho-Rivera M, et al. Determinants of Survival in Malignant Pleural Mesothelioma: A Surveillance, Epidemiology, and End Results (SEER) Study of 14,228 Patients. PLoS One 2015;10:e0145039. 10.1371/journal.pone.0145039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo G, Chmielecki J, Goparaju C, et al. Whole-Exome Sequencing reveals Frequent Genetic Alterations in BAP, NF2, CDKN2A and CUL1 in Malignant Pleural Mesothelioma. Cancer Res 2015;75:264-9. 10.1158/0008-5472.CAN-14-1008 [DOI] [PubMed] [Google Scholar]
- 21.Kato S, Tomson BN, Buys TP, et al. Genomic Landscape of Malignant Mesotheliomas. Mol Cancer Ther 2016;15:2498-507. 10.1158/1535-7163.MCT-16-0229 [DOI] [PubMed] [Google Scholar]
- 22.Bueno R, Stawiski EW, Goldstein LD, et al. Comprehensive genomic analysis of malignant pleural mesothelioma identifies recurrent mutations, gene fusions and splicing alterations. Nat Genet 2016;48:407-16. 10.1038/ng.3520 [DOI] [PubMed] [Google Scholar]
- 23.Lo Iacono M, Monica V, Righi L, et al. Targeted Next-Generation Sequencing of Cancer Genes in Advanced Stage Malignant Pleural Mesothelioma. J Thorac Oncol 2015;10:492-9. 10.1097/JTO.0000000000000436 [DOI] [PubMed] [Google Scholar]
- 24.Ugurluer G, Chang K, Gamez ME, et al. Genome-based Mutational Analysis by Next Generation Sequencing in Patients with Malignant Pleural and Peritoneal Mesothelioma. Anticancer Res 2016;36:2331-8. [PubMed] [Google Scholar]
- 25.De Rienzo A, Archer MA, Yeap BY, et al. Gender-Specific Molecular and Clinical Features Underlie Malignant Pleural Mesothelioma. Cancer Res 2016;76:319-28. 10.1158/0008-5472.CAN-15-0751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bonelli MA, Fumarola C, La Monica S, et al. New therapeutic strategies for malignant pleural mesothelioma. Biochem Pharmacol 2017;123:8-18. 10.1016/j.bcp.2016.07.012 [DOI] [PubMed] [Google Scholar]
- 27.Hanna A, Shevde LA. Hedgehog signaling: modulation of cancer properties and tumor microenvironment. Mol Cancer 2016;15:35. 10.1186/s12943-016-0522-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Katoh Y, Katoh M. Hedgehog target genes: mechanisms of carcinogenesis induced by aberrant hedgehog signaling activation. Curr Mol Med 2009;9:873-86. 10.2174/156652409789105570 [DOI] [PubMed] [Google Scholar]
- 29.Barakat MT, Humke EW, Scott MP. Learning from Jekyll to control Hyde: Hedgehog signaling in development and cancer. Trends Mol Med 2010;16:337-48. 10.1016/j.molmed.2010.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tang CM, Lee TE, Syed SA, et al. Hedgehog pathway dysregulation contributes to the pathogenesis of human gastrointestinal stromal tumors via GLI-mediated activation of KIT expression. Oncotarget 2016;7:78226-41. 10.18632/oncotarget.12909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y, He J, Zhang F, et al. SMO expression level correlates with overall survival in patients with malignant pleural mesothelioma. J Exp Clin Cancer Res 2013;32:7. 10.1186/1756-9966-32-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shi Y, Moura U, Opitz I, et al. Role of Hedgehog Signaling in Malignant Pleural Mesothelioma. Clin Cancer Res 2012;18:4646-56. 10.1158/1078-0432.CCR-12-0599 [DOI] [PubMed] [Google Scholar]
- 33.Available online: http://cancer.sanger.ac.uk/cosmic
- 34.Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 2013;14:178-92. 10.1093/bib/bbs017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Little RJA., Rubin DB. Statistical analyses with missing data. Journal of Educational Statistics 1991;16:150-5. 10.2307/1165119 [DOI] [Google Scholar]
- 36.Bottai M, Zhang J. Laplace regression with censored data. Biom J 2010;52:487-503. 10.1002/bimj.200900310 [DOI] [PubMed] [Google Scholar]
- 37.Zhang L, Zhou Y, Cheng C, et al. Genomic analyses reveal mutational signatures and frequently altered genes in esophageal squamous cell carcinoma. Am J Hum Genet 2015;96:597-611. 10.1016/j.ajhg.2015.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Biankin AV, Waddell N, Kassahn KS, et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature 2012;491:399-405. 10.1038/nature11547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lim CB, Prele CM, Cheah HM, et al. Mutational Analysis of Hedgehog Signaling Pathway Genes in Human Malignant Mesothelioma. PLoS One 2013;8:e66685. 10.1371/journal.pone.0066685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Felley-Bosco E, Opits I, Meerang M. Hedgehog Signaling in Malignant Pleural Mesothelioma. Genes 2015;6:500-11. 10.3390/genes6030500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xin M, Ji X, De La Cruz LK, et al. Strategies to target the Hedgehog signaling pathway for cancer therapy. Med Res Rev 2018;38:870-913. 10.1002/med.21482 [DOI] [PubMed] [Google Scholar]
- 42.Yin VT, Esmaeli B. Targeting the Hedgehog Pathway for Locally Advanced and Metastatic Basal Cell Carcinoma. Curr Pharm Des 2017;23:655-9. 10.2174/1381612822666161208100325 [DOI] [PubMed] [Google Scholar]
- 43.Meerang M, Bérard K, Felley-Bosco E, et al. Antagonizing the Hedgehog Pathway with Vismodegib Impairs Malignant Pleural Mesothelioma Growth In Vivo by Affecting Stroma. Mol Cancer Ther 2016;15:1095-105. 10.1158/1535-7163.MCT-15-0583 [DOI] [PubMed] [Google Scholar]
- 44.You M, Varona-Santos J, Singh S, et al. Targeting of the Hedgehog signal transduction pathway suppresses survival of malignant pleural mesothelioma cells in vitro. J Thorac Cardiovasc Surg 2014;147:508-16. 10.1016/j.jtcvs.2013.08.035 [DOI] [PubMed] [Google Scholar]
- 45.LoRusso PM, Rudin CM, Reddy JC, et al. Phase I trial of hedgehog pathway inhibitor vismodegib (gdc-0449) in patients with refractory, locally advanced or metastatic solid tumors. Clin Cancer Res 2011;17:2502-11. 10.1158/1078-0432.CCR-10-2745 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as