Abstract
Crocins are a group of highly valuable apocarotenoid-derived pigments mainly produced in Crocus sativus stigmas and Gardenia jasminoides fruits, which display great pharmacological activities for human health, such as anticancer, reducing the risk of atherosclerosis, and preventing Alzheimer’s disease. However, traditional sources of crocins are no longer sufficient to meet current demands. The recent clarification of the crocin biosynthetic pathway opens up the possibility of large-scale production of crocins by synthetic metabolic engineering methods. In this review, we mainly introduce the crocin biosynthetic pathway, subcellular route, related key enzymes, and its synthetic metabolic engineering, as well as its challenges and prospects, with a view to providing useful references for further studies on the synthetic metabolic engineering of crocins.
Keywords: Synthetic biology, Metabolic engineering, Biosynthesic pathway, Apocarotenoid, Crocins, Crocetin, UDP-glycosyltransferase
1. Introduction
Crocus sativus L., a perennial herb belonging to the Iridaceae family, is well known for its dried red stigmas, called saffron [1]. Due to its complicated harvesting process, low yield, and expensive labor costs, saffron, known as “red gold”, is one of the most expensive spices on Earth [2], [3]. Saffron not only has powerful pharmacological activities but is also the source of some unique apocarotenoids, including crocins (crocetin glycosyl esters), picrocrocin, and safranal, responsible for its red color, bitter taste, and pungent aroma, respectively [4], [5]. Gardenia jasminoides has been cultivated in China for nearly 1000 years with great ornamental and medicinal value. The dried ripe fruit of G. jasminoides, a natural yellow dye, is also a popular traditional Chinese medicine because of its various biological activities [6], [7].
Crocins, belonging to apocarotenoid glycosides, are the main bioactive ingredients and colorants in C. sativus stigmas and G. jasminoides fruits [8], [9]. Glycosylation reactions are catalyzed by glycosyltransferases acting on the carboxyl- and glycosyl- groups of crocetins and crocins, and form five kinds of crocins (crocin-Ⅰ, crocin-Ⅱ, crocin-III, crocin-Ⅳ, and crocin-Ⅴ) [10]. Among these, crocin-Ⅰ is the main constituent [4], [11]. Different from most of the lipid-soluble carotenoids (including lycopene, α-carotene, β-carotene, zeaxanthin, and lutein), crocins are water-soluble, which can be attributed to these glycosylations/ their glycosylation [12]. Modern pharmacological studies show that crocins have anti-oxidation [12], [13], anti-inflammatory [14], [15], anti-hyperlipidemic [16], anticancer [17], [18], [19], and antilithiatic [20] properties, as well as potential properties for treatment of Alzheimer’s disease [12]. In recent years, crocins have drawn increasing attention in the medical, food, and cosmetics fields for their excellent pharmacological and coloring functions. Traditionally, crocins have been obtained by extraction and purification from plants (mainly saffron), but the scarce sources and low yield limit their commercial utilization. In addition, C. sativus (2n = 3× = 24) is an autotriploid evolved from diploid C. cartwrightianus [21]. The vegetative propagation of triploid C. sativus from corms is the main factor limiting its genetic improvement. Therefore, it is necessary to develop economical and efficient methods for producing crocins to complement traditional sources.
Crocins are difficult to be produced through chemosynthesis because of their complex structure and abundant chiral centers, and it is easy to form inactive or toxic optical isomers in the chemosynthetic process [22]. Therefore, crocin biosynthesis is a subject of considerable interest. In the last few years, significant progress has been made in elucidating the crocin biosynthetic pathway based on the development of next-generation sequencing (NGS) technologies. With the in-depth study of the synthetic pathway and regulatory mechanism of crocins, genetic and metabolic engineering technology will become one of the effective means to increase crocins production. Here, we review recent advances in the crocin biosynthetic pathway, which will lay the foundation for the industrial production of crocins and the development of crocin-rich functional food using metabolic engineering.
2. Crocin biosynthesis
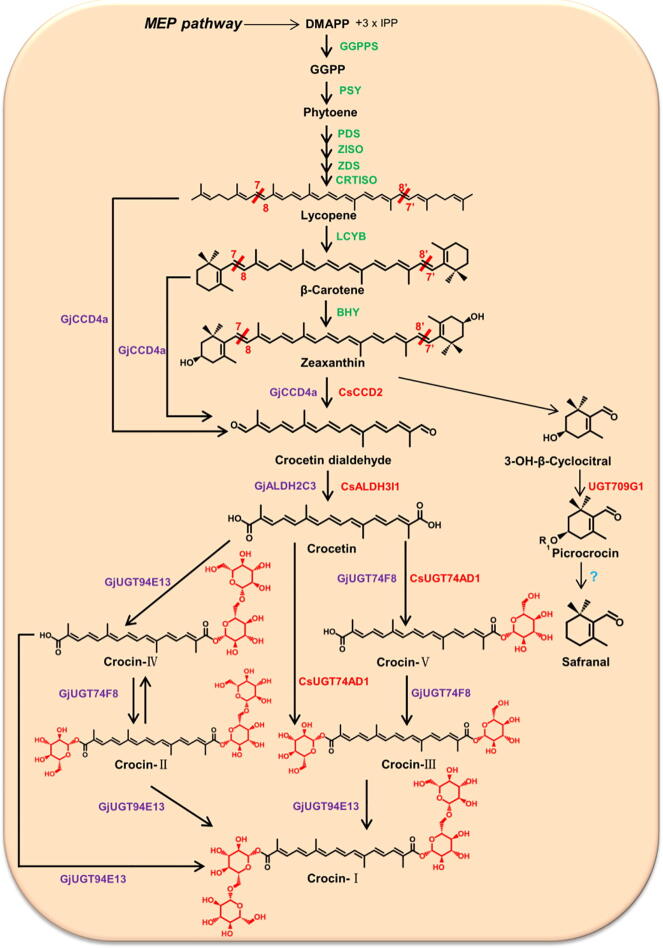
Carotenoids are a group of important pigments found in plants, alage, bacteria and fungi. Carotenoids contribute to the bright colors of vegetables, fruits, and flowers, and play a crucial role in human health [23]. Their cleavage products, called apocarotenoids, are formed after oxidative cleavage catalyzed by a family of double bond-specific carotenoid cleavage dioxygenases (CCDs) [24]. There are some unique apocerotenoids, including crocetins, crocins, picrocrocins, and safranals, which accumulate at high levels only in saffron and gardenia. Among these, crocins, the glycosylated forms of apocerotenoids, are the most valuable and stable constituents in C. sativus stigmas [25], [26]. Crocins have two glycosyls, including β-D-glucosyl and β-D-gentiobiosyl, and according to the position and numbers of these two glycosyls, crocins could be divided into five forms, crocin-I, crocin-II, crocin-III, crocin-IV, and crocin-Ⅴ. To date, the crocin biosynthetic pathway in plants has been elucidated by transcriptome and genome sequencing of C. sativus and G. jasminoides (Fig. 1) [3], [24], [27], [28], [29], [30].
Fig. 1.
The crocin biosynthetic pathways in C. sativus and G. jasminoides. The precursors of carotenoids are generated via the 2-C-methyl-D-erythritol 4-phosphate (MEP) pathway. The enzymes marked in green are known enzymes from the upstream carotenoid biosynthetic pathway, and those in red and purple are identified from C. sativus and G. jasminoides, respectively. GGPPS, geranylgeranyl diphosphate synthase; PSY, phytoene synthase; PDS, phytoene desaturase; Z-ISO, ζ-carotene isomerase; ZDS, ζ-carotene desaturase; CRTISO, carotenoid isomerase; LCYB, lycopene β-cyclase; BHY, β-carotene hydrolase; CCD, carotenoid cleavage dioxygenase; ALDH, aldehyde dehydrogenase; UGT, UDP-glucosyltransferase. Information on these enzymes can be found in Table 1. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
2.1. Crocin biosynthetic pathway in plants
Crocins are accumulated in stigmas of C. sativus and fruits of G. jasminoides in a tissue-specific manner, and their biosynthesis requires a high degree of coordination of several pathways, including the upstream methylerythritol phosphate (MEP) pathway, the midstream carotenoid biosynthetic pathway, and the downstream crocin biosynthetic pathway [3], [31].
In the MEP pathway, deoxyxylulose-5-phosphate (DXP) synthase utilizes pyruvate and glyceraldehyde-3-phosphate (GAP) as initial substrates to form isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP). Carotenoid biosynthesis occurs in plastid and begins with geranylgeranyl diphosphate (GGPP), which is produced by the condensation of IPP and DMAPP [27]. Although IPP and GGPP can be synthesized by the cytoplasm-localized mevalonate pathway (MEV) pathway and the plastid-localized MEP pathway, only the latter provides precursors for plant carotenoid biosynthesis in plastid. Firstly, phytoene synthase (PSY), the first rate-limiting enzyme of the carotenoid biosynthetic pathway, catalyzes the condensation of two molecules of GGPP to form the first carotenoid, 15-cis-phytoene [32]. Then, 15-cis-phytoene is converted into all-trans-lycopene by desaturation and isomerization, which is a multi-step process requiring two desaturases, phytoene desaturase (PDS) and ζ-carotene desaturase (ZDS) [33], as well as two isomerases, ζ-carotene isomerase (Z-ISO) [34] and carotenoid isomerase (CRTISO) [35], [36]. Lycopene, the branching point in carotenogenesis, acts as the substrate for lycopene ε-cyclase (ε-LCY) and lycopene β-cyclase (β-LCY) leading to formation of α-carotene, which is further hydroxylated to lutein, or for β-LCY alone leading to formation of β-carotene, which is further hydroxylated to zeaxanthin by β-carotene hydroxylase (BCH) [37]. The key enzymes involved in general carotenoid biosynthesis, including GPPS, GGPPS, PSY, PDS, BCH, ZDS, Z-ISO, CRTISO, β-LCY, and ε-LCY, have so far been identified from C. sativus [24], [27], [30], [38], and further functional characterization of enzyme-coding genes indicates that CsBCH1 [38], CstLcyB2a [39], and CsPSY2 [40] are responsible for carotenoid accumulation in stigmas of C. sativus. Recently, Ji et al. [3] performed the first transcriptome sequencing of fruits in G. jasminoides and identified candidate genes encoding the key enzymes involved in the MEP and carotenoid biosynthesis pathways.
Both β-carotene and zeaxanthin are important precursors for apocarotenoid biosynthesis and are converted by different CCDs leading to different apocarotenoid compounds [38], [41]. For example, CsCCD1 and CsCCD4 cleave β-carotene at the 9, 10 and 9′,10′ double bonds into β-ionone and β-cyclocitral [24], [41], while Arabidopsis thaliana CCD7 (AtCCD7) and Arabidopsis thaliana CCD8 (AtCCD8) are responsible for strigolactone production by cleavage of β-carotene [42]. In the crocin biosynthetic pathway, CCD2, a recently identified CCD family member from C. sativus, cleaves the 7, 8 and 7′, 8′ double bonds of zeaxanthin to produce crocetin dialdehyde and 3-OH-β-cyclocitral, which is the first step of saffron crocin biosynthesis [2]. Aldehyde dehydrogenase (ALDH) catalyzes the conversion of crocetin dialdehyde to crocetin. UDP-glucosyl transferases (UGT) transform crocetin and β-cyclocitral into crocins (crocin-Ⅰ to crocin-Ⅴ) and picrocrocin, respectively. Finally, picrocrocin is converted into a safranal under the combined action of heating and β-glucosidase (β-GS) [27]. The reported transcriptome data and functional assays show that CsCCD2 [2], CsALDH3I1 [43], and CsUGT74AD1 [43], as well as GjCCD4a [3], [29], GjALDH2C3 [29], GjUGT74F8 [29], [44], and GjUGT94E13 [29], [44], have been identified to be involved in the complete crocin biosynthetic pathway in stigmas of C. sativus and fruits of G. jasminoides, respectively.
2.2. Subcellular routes for crocin biosynthesis
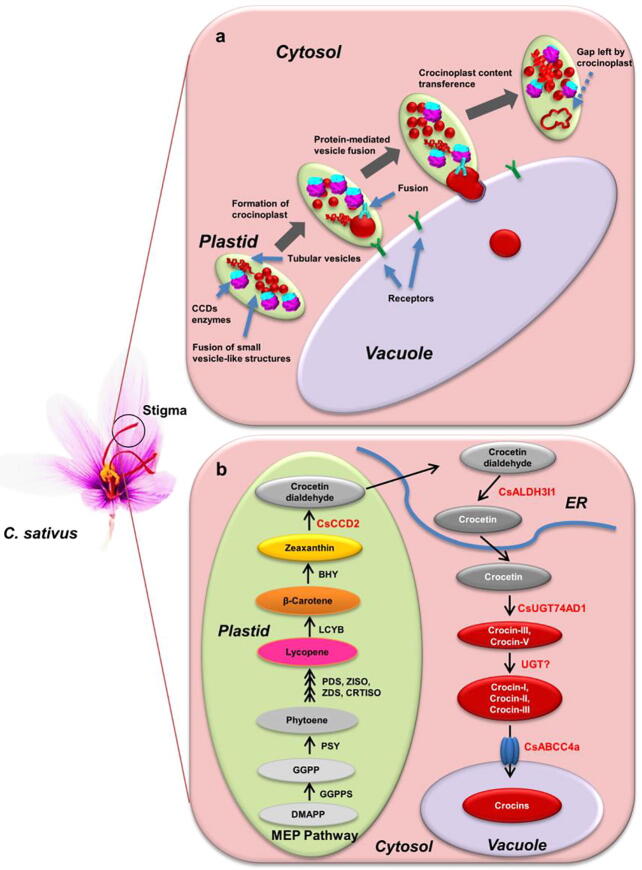
It has been reported that zeaxanthin and crocin are localized in plastids and vacuoles, respectively, suggesting that the crocin biosynthetic pathway might involve multiple subcellular compartments [43], [45], [46]. Initially, CsCCD2 that cleaves zeaxanthin to produce apocarotenoid was reported to be localized in the cytoplasm [2]. However, Ahrazem et al. [47] identified a longer CsCCD2 version, CsCCD2L, and the transient expression experiment in tobacco leaves indicated that CCD2 is a plastid-localized enzyme rather than a cytoplasm-localized enzyme, which was further confirmed by immunogold electron microscopy on C. sativus stigmas and confocal fluorescence microscopy in N. benthamiana leaves [43]. At present, there are two models regarding the subcellular route of crocin biosynthesis in saffron. In 2017, Gómez-Gómez et al. [46] proposed the first model through a proteome analysis and microscopy studies: crocins are synthesized in the chromoplast and then accumulated in chromoplast-localized vesicles to form crocinoplasts, which are further transported from the end of the chromoplast to the vacuole for storage. In 2018, Demurtas et al. [43], [48] analyzed the localization of CsCCD2, CsALDH3I, and CsUGT74AD1, which have been characterized as responsible for crocin biosynthesis, and proposed a different model: crocetin dialdehyde, crocetin, and crocins are synthesized in the chromoplast, the endoplasmic reticulum (ER), and the cytoplasm, respectively. Finally, crocins are transported from the cytosol to the vacuole for storage through unidentified tonoplast transporters. Recently, Demurtas et al. [49] developed a transportomic approach to identify saffron tonoplast transporters mediating crocin accumulation in the vacuole, and CsABCC4a, a tonoplast transporter belonging to the ATP-binding cassette C (ABCC) transporter family, was characterized to be involved in crocins transport in saffron. Since plastid-localized ALDH and UGT have not been identified and the subcellular route of stevioside biosynthesis in Stevia rebaudiana leaves is similar to the second model described above [50], we prefer to consider the second model as the true subcellular route of crocin biosynthesis in saffron (Fig. 2).
Fig 2.
Proposed models of subcellular routes for crocin compartmentation in stigmas of C. sativus. (a) Crocins are synthesized in the plastid, and then accumulated in plastid-localized small vesicle-like structures, which gradually fuse together to form crocinoplasts that could be directly transferred from the polarized end of the plastid to the vacuole. This is modified from Gomez-Gomez et al. 2017 [46]. (b) CsCCD2 cleaves zeaxanthin in the plastid to produce crocetin dialdehyde, which is further converted to crocetin by CsALDH3I1 in the endoplasmic reticulum (ER). CsUGT74AD1 catalyzes the formation of crocins in the cytoplasm, and then crocins are transported into the vacuole through CsABCC4a. This is modified from Demurtas et al. 2018 [43].
2.3. Key enzymes involved in crocin biosynthesis
In recent years, with the increasing number of reports on genomic and transcriptomic analyses of C. sativus and G. jasminoides [3], [24], [27], [28], [29], [30], the functional characterization and localization of three key enzymes (CCD, ALDH, and UGT) involved in crocin biosynthesis have been described [2], [23], [29], [43], [44], [47], [51]. From these studies, we have a better understanding of the crocin biosynthetic pathway and provide guidance for using metabolic engineering to enhance crocins production. The key enzymes for crocin biosynthesis that have been identified are summarized in Table1.
Table 1.
Key enzymes involved in crocin biosynthesis.
| Key enzymes | Name | Species | GenBank accession no. | Identification methods | Refs |
|---|---|---|---|---|---|
| PSY | CsPSY2 | Crocus sativus | MH124239 | bioinformatics analysis, subcellular localization, expression analysis, functional complementation | [40] |
| β-LCY | CstLcyB2a | Crocus sativus | GQ202141 | bioinformatics analysis, expression analysis, in vivo functional analysis | [39] |
| BCH | CsBCH1 | Crocus sativus | CAC95130 | various PCR-based methods, bioinformatics analysis, expression analysis | [38] |
| CCD | CsCCD2 | Crocus sativus | KJ541749 | transcriptome sequencing, in vivo and in vitro assays, subcellular localization | [2] |
| CsCCD2L | Crocus sativus | KP887110 | subcellular localization, bioinformatics analysis, in silico analysis | [47] | |
| CaCCD2 | Crocus ancyrensis | KP792756 | bioinformatics analysis, in silico analysis, subcellular localization, expression analysis, in vivo functional analysis | [47] | |
| BdCCD4.1 | Buddleja davidii | KX374547 | bioinformatics analysis, in silico analysis, subcellular localization, expression analysis, in vivo and in vitro assays | [23] | |
| BdCCD4.3 | Buddleja davidii | KX374549 | bioinformatics analysis, in silico analysis, subcellular localization, expression analysis, in vivo and in vitro assays | [23] | |
| GjCCD4a | Gardenia jasminoides | ARU08109 | Genome-wide analysis, bioinformatics analysis, expression analysis, in vivo and in vitro assays, LC/LC-MS analyses | [29] | |
| ALDH | CsALDH3I1 | Crocus sativus | MF596165 | subcellular localization, transcripts analysis, expression analysis, in vivo and in vitro assays | [43] |
| GjALDH2C3 | Gardenia jasminoides | KY631926 | Genome-wide analysis, bioinformatics analysis, expression analysis, in vivo and in vitro assays, LC/LC-MS analyses | [29] | |
| UGT | UGTCs2 | Crocus sativus | AY262037 | Genomic analysis, expression analysis, in vitro functional assays | [25] |
| CsUGT74AD1 | Crocus sativus | MF596166 | subcellular localization, transcripts analysis, expression analysis, in vitro functional assays | [43] | |
| GjUGT75L6 | Gardenia jasminoides | F8WKW8 | expression analysis, bioinformatics analysis, in vitro functional assays | [51] | |
| GjUGT94E5 | Gardenia jasminoides | F8WKW0 | expression analysis, bioinformatics analysis, in vitro functional assays | [51] | |
| GjUGT74F8 | Gardenia jasminoides | MN944054 | Genome-wide analysis, bioinformatics analysis, expression analysis, in vivo and in vitro assays, LC/LC-MS analyses | [29], [44] | |
| GjUGT94E13 | Gardenia jasminoides | MN944055 | Genome-wide analysis, bioinformatics analysis, expression analysis, in vivo and in vitro assays, LC/LC-MS analyses | [29], [44] | |
| Bs-GT | Bacillus subtilis | WP_003220110.1 | bioinformatics analysis, in vitro functional assays | [10] |
2.3.1. Enzyme catalyzing the cleavage reaction: carotenoid cleavage dioxygenase (CCD)
The first step in crocin biosynthesis is the cleavage of carotenoids by CCD to produce crocetin dialdehyde. CCDs, a class of double bond-specific enzymes involved in apocarotenoid biosynthesis, have been grouped into five clusters in plants according to their substrate preference and/or the cleavage position, named CCD1, CCD4, CCD7, CCD8, and nine-cis-epoxy-carotenoid cleavage dioxygenases (NCEDs) [24], [52], [53]. Structurally, all CCDs contain a conserved seven-bladed β-propeller structure, a less-conserved dome region formed by a-helix elements, strands and loops, and a Fe2+ ion essential for the catalytic activity [54], [55]. Zeaxanthin cleavage dioxygenase (ZCD) was first reported to mediate crocetin production by cleavage of zeaxanthin at the 7, 8 and 7′, 8′ double bonds [45]. However, later studies including sequence and structure analyses, in vivo and in vitro assays, showed that ZCD is an N-truncated CCD4 form with a lack of activity to convert zeaxanthin into crocetin dialdehyde [2], [41].
Through deep transcriptome sequencing, Frusciante et al. [2] identified a new dioxygenase named CCD2 from C. sativus, which belongs to a novel CCD clade closely related to the CCD1 subfamily, and found that the expression pattern of CCD2 is consistent with crocins accumulation during stigma development. Further in vivo and in vitro functional analyses revealed that CsCCD2 is able to catalyze the conversion of zeaxanthin into the crocins precursor crocetin dialdehyde [2]. The second member of the CCD2 subfamily, CaCCD2, was identified from Crocus ancyrensis by Ahrazem et al. [47], and like CsCCD2, it is functionally characterized as involved in crocetin formation. The identification and functional characterization of CsCCD2 and CaCCD2 confirm that the cleavage of zeaxanthin by the CCD2 enzymes can give rise to crocetin dialdehyde. Lately, according to full-length transcriptome data of C. sativus, Yue et al. [30] proposed that CCD2 might have evolved from the CCD1 subfamily via the whole-genome duplication event.
To date, the CCD2 enzymes have only been identified from Crocus species [2], [47], but crocetin has also been found in other plant organs, such as fruits of G. jasminoides [56] and flowers of Buddleja davidii [57]. Recently, BdCCD4.1 and BdCCD4.3 were identified from Buddleja davidii and exhibited cleavage activity over zeaxanthin in vivo and in vitro, leading to production of crocetin dialdehyde [23]. It is reported that BdCCD4.1 and BdCCD4.3 are located in plastids, which is consistent with CCD2 localization [23]. However, it is worth noting that GjCCD4a identified from G. jasminoides not only has the same catalytic activity as CsCCD2, CaCCD2, BdCCD4.1, and BdCCD4.3 to cleave zeaxanthin, but also catalyzes the production of crocetin dialdehyde from lycopene and β-carotene [29].
Therefore, the first dedicated step in crocin biosynthesis can be catalyzed by the CCD2 enzymes from Crocus [2], [47] and by the CCD4 enzymes from Buddleja [23] and Gardenia [29], indicating that these CCD enzymes acquire the ability to cleave zeaxanthin at the 7, 8 and 7′, 8′ double bonds through convergent evolution in different species [29].
2.3.2. Enzyme catalyzing the dehydrogenation reaction: ALDH
The second step in crocin biosynthesis is the dehydrogenation of crocetin dialdehyde to produce crocetin. The ALDH superfamily is generally characterized by the use of NAD(P)+ as a cofactor to catalyze the conversion of aldehyde groups to carboxyl groups [58]. The plant ALDHs have been grouped into 13 subfamilies [58]. In 2018, Demurtas et al. [43] identified six candidate genes encoding ALDHs from C. sativus stigmas, and only one of their corresponding proteins, CsALDH3I1, was characterized as an enzyme capable of converting crocetin dialdehyde into crocetin by using an Escherichia coli expression system and an in vitro enzyme activity assay. Like those ER-localized proteins containing a C-terminal KKXX signal, CsALDH3I1 harboring a C-terminal KKPK signal was also confirmed to be localized in the ER [43], [59]. Recently, GjALDH2C3 from G. jasminoides, whose expression accompanied the accumulation of crocins in fruits of G. jasminoides, was reported to be involved in the second step in crocin biosynthesis by Xu et al., and they also identified crocetin semialdehyde as an intermediate product in the catalytic process from crocetin dialdehyde to crocetin [29]. In addition, some candidate genes that might encode ALDHs responsible for crocetin production have also been identified from plants by transcriptome analysis, such as ADH2946, ADH11367, and ADH54788 derived from C. sativus [46], and ALDH12 and ALDH14 derived from G. jasminoides [3]. However, further in vivo and in vitro studies are needed to determine their functions.
2.3.3. Enzyme catalyzing the glycosylation reaction: uridine diphosphate glycosyltransferase (UGT)
The final step in crocin biosynthesis is the glycosylation of crocetin by UGTs to produce crocins. UGTs catalyze glucose molecules and specific receptors to be linked together by glycosidic bonds. In saffron, the glycosylation of crocetin results in the transformation of lipid-soluble carotenoid into water-soluble and stable apocarotenoid glycosides, crocins. Moraga et al. [25] cloned two UGTase-encoding genes (UGTCs2 and UGTCs3) containing the plant secondary product GTase (PSPG) box from C. sativus using degenerate primers, and found that UGTCs2 and UGTCs3 were expressed at high levels in stigmas and stamens, respectively. Expression of UGTCs2 in E. coli and an in vitro enzyme activity assay show that UGTCs2 exhibits strong glucosylation activity toward crocetin, β-D-glucosyl ester, and crocetin β-D-gentibiosyl ester [25]. To further verify the function of UGTCs2, Demurtas et al. [43] searched for the corresponding transcript in the transcriptome derived from their own and others’ studies [27], and found a new gene encoding a different version of UGTCs2, CsUGT74AD1, whose corresponding protein CsUGT74AD1 was identified to be responsible for the primary glycosylation of crocetin [43]. It is reported that CsUGT74AD1 displays a cytoplasmic localization [43]. Recently, UGT709G1, a novel UGT involved in picrocrocin biosynthesis, was identified and characterized from saffron, which contributes to the industrial production of safranal [60].
Besides saffron, crocins are also highly enriched in gardenia. In 2012, Nagatoshi et al. [51] identified and functionally characterized two glucosyltransferases, GjUGT75L6 and GjUGT94E5, which mediate crocin production in G. jasminoides. An in vitro enzyme activity assay demonstrate that the carboxyl group of crocetin is first glycosylated by UGT75L6 forming crocetin glucosyl esters, which is further glycosylated on the 6-position hydroxyl group of the glucose moiety by UGT94E5, yielding other forms of crocins [50]. However, the expression profiles of the genes encoding these two UGTs are not correlated with the distribution of crocins in gardenia, suggesting that there may be other UGTs involved in crocin synthesis in gardenia [51]. Recently, GjUGT74F8 and GjUGT94E13 from G. jasminoides identified by Xu et al. showed an expression pattern consistent with crocin accumulation, and in vitro functional analysis confirmed that their corresponding proteins were responsible for the primary and secondary glycosylation of crocetin [29]. Furthermore, using E. coli as an expression system, GjUGT74F8 and GjUGT94E13 can lead to efficient conversion of crocetin to five types of crocins [44], indicating that GjUGT74F8 and GjUGT94E13 are involved in crocin biosynthesis in gardenia.
Based on the substrate and position of glycosylation, the above UGTs can be divided into three types: UGT-I type (UGTCs2/CsUGT74AD1/GjUGT75L6/ GjUGT74F8/Bs-GT) for the primary glycosylation of crocetin to produce crocetin monoglucosyl and diglucosyl esters; UGT-II type (GjUGT94E5) for the secondary glycosylation of Glc groups to form one or two gentiobiose groups; UGT-III type (GjUGT94E13) for the primary and secondary glycosylations. Since the functions of most of the reported UGTs are determined by in vitro enzyme activity assays, it is necessary to further verify their functions in plants.
3. Crocin metabolic engineering
As a high-value carotenoid, crocins have great potential in pharmacology. Currently, the main production method of crocins is extraction and purification from C. sativus. However, complicated production methods and expensive raw materials have been the main factors limiting the mass production and broad application [44]. To improve the manufacturing technology, the research of crocin metabolic engineering has attracted increasing attention, and the development of engineered microorganisms and biofortified plants to produce crocin in vivo is the main research direction.
The biosynthesis of apocarotenoids in C. sativus was studied and analyzed by transcriptomics and dynamic metabolomics [28]. The synthetic pathway of crocins has been largely revealed, and many key enzymes have also been found and applied in metabolic engineering. The biosynthetic pathway of carotenoids in C. sativus includes: starting with geranylgeranyl diphosphate, zeaxanthin was gradually synthesized, and the specific synthesis of crocetin and crocins by cleavage of zeaxanthin. Enzymes of the upstream crocin synthesis pathway, such as PSY, PDS, and CRT, have been used to synthesize carotenoids such as lycopene, β-carotene, and zeaxanthin in various crops. The combined overexpression strategy of multiple biosynthetic genes co-transformation can effectively improve the accumulation of target carotenoids. Zeaxanthin is the key intermediate product in the synthetic pathway, an important direct precursor for the synthesis of crocins [8]. In the published studies, researchers either integrated the β-carotene hydroxylase gene to synthesize zeaxanthin, or directly optimized a microbial strain already producing zeaxanthin. According to the existing reports (Table 2), many microbes have been successfully transformed to synthesize crocetin or crocins, including E. coli [39], [44], Chlorella vulgaris [61], and yeast [62]. Recently, the transient transformation of Nicotiana benthamiana to synthesize crocins were reported [63].
Table 2.
Crocin biosynthesis by metabolic engineering.
| Species | Transgene | Operational methods | Products | Yield | Refs |
|---|---|---|---|---|---|
| Escherichia coli | GjUGT94E13, GjUGT74F8 | Co-transformation of expression vectors | Crocins | 60.81 ± 1.0 mg/L | [44] |
| Escherichia coli | CsCCD2, NcALD8, BsYjiC, BsYdhE, BsYojK | Integrated genes into the E. coli chromosome by multigene vector and electroporation- transformation | Crocins | [39] | |
| Saccharomyces cerevisiae | PsCrtZ, CCD2, SsALD | Transformation in steps via the lithium acetate method and integrated genes into chromosome | Crocetin | 6.28 mg /L | [62] |
| Chlorella vulgaris | HpCrtRB, CsZCD1 | Agrobacterium-mediated transformation and integrated genes into chromosome | Crocetin | [61] | |
| Nicotiana benthamiana | CsCCD2L, BdCCD4.1, BdCCD4.3, PaCrtB, CsBCH2, | Virus vectors transient expression in leaves by Agrobacterium-mediated co-trasformation | Crocins and Picrocrocin (derived from 3-OH-β-cyclocitral, the side product of zeaxanthin cleavage) | 2.2 ± 0.2 mg/g crocins and 8.2 ± 2.93 mg/g picrocrocin | [63] |
Note: NcALD8, Neurospora crassa aldehyde dehydrogenases gene; BsYjiC, BsYdhE and BsYojK, Bacillus subtilis glycosyltransferase genes; PsCrtZ Pantoea stewartii β-carotene hydroxylase gene; SsALD Synechocystis sp. aldehyde dehydrogenase gene; HpCrtRB, Haematococcus pluvialis β-carotene hydroxylases gene; CsZCD1, Crocus sativus 7, 8 (7′, 8′) -zeaxanthin cleavage dioxygenase gene; PaCrtB, Pantoea ananatis phytoene synthase gene.
E. coli has a simple and clear genetic background, and its related genetic engineering technologies are mature and widely used, which make it an excellent host bacterium for the production of crocin by metabolic engineering. Based on the terpenoid producing strains, Wang et al. used pyruvic acid and glyceraldehyde-3-phosphate as substrates, and introduced a series of genes, crtE, crtB, crtI, crtY, and crtZ, to gradually synthesize lycopene to β-carotene from upstream prenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP), then finally obtained the zeaxanthin-producing strains YL4 and YL5. Also, by integrating and optimizing the expression of heterologous genes, CsCCD2 from C. sativus and ALD8 from Neurospora crassa in YL4 and YL5, crocetin was produced. Finally, the glycosyltransferase genes YjiC, YdhE, and YojK from Bacillus subtilis were introduced, and five crocins were produced [26].
In the research of eukaryotes, Saccharomyces cerevisiae and Chlorella vulgaris have been successfully engineered to produce crocetin. Chai et al. introduced heterologous CrtZ, CCD, and ALD into a β-carotene producing strain (S. cerevisiae, SyBE_SC0014CY06), and successfully accomplished the biosynthesis of crocetin in S. cerevisiae [62]. The enzyme source combination PsCrtZ (from Pantoea stewartii) /CCD2 with the highest yield was found by introducing nine CrtZs and four CCDs from various sources. By overexpressing CCD2 and ALD (from Synechocystis sp.) and controlling the reaction temperature, the reported highest titer in shake flask was 1219 μg/L. The titer of crocetin in a 5-L bioreactor reached 6278 μg/L through fed-batch fermentation, which is the highest crocetin titer reported in eukaryotic cells. In C. vulgaris, Lou et al. transformed crtRB and ZCD1 into a wild type strain with a similar design as above, and produced crocetin without exogenous ALD by using the rich secondary metabolites and potential aldehyde oxidase activity in C. vulgaris [61]. Recently, Martí et al. [63] constructed transient transformation vectors for Nicotiana tabacum by using the Tobacco etch virus (TEV), and transformed the tobacco plants with a series of CCD genes and several carotenoid synthesis related genes. The accumulation of yellow pigments of crocins and picrocrocin can be seen with naked eyes in leaves infected by the virus. Crocins (0.2%) and picrocrocin (0.8%) were detected in the plants expressing only the CCD gene, which were in relatively low levels. Meanwhile, expression of the phytoene synthase gene (PaCrtB) could increase the crocin content, reaching as much as 0.35% of dry weight [63]. This is a successful case of crocin synthesis in higher plants. However, stably inherited plants cannot be obtained by the transient transformation. To develop valuable bioreactors, it will be necessary to create de novo synthetic genetic engineering plants using stable transformation methods.
The above cases provide solid theoretical support and practical basis for further synthesizing crocins in higher plants, especially in crop plants. There have been many reports on the successful synthesis of carotenoids, such as β-carotene, zeaxanthin, canthaxanthin and astaxanthin, in crops like rice, maize, and tomato [65], [66], which prove the feasibility of synthesizing crocins in plants. Many plants have robust carotenoid biosynthesis and accumulation providing potential precursors for crocins. For example, tomato contains a high content of lycopene, and even zeaxanthin, the direct precursor of crocetin, which is the main carotenoid in maize. When these plants are genetically engineered, the carotenoids present in them can be used as the synthetic raw materials. During the transformation, the existing endogenous synthetic pathways can be utilized, and the silenced or low-expression genes in them can be complemented, which can simplify the operation. For plants with little carotenoid accumulation and lack of suitable endogenous genes, such as rice endosperm, it is necessary to introduce a complete synthetic pathway to complete the de novo synthesis of crocins. Using multi-gene stacking technology, such as TransGene Stacking II (TGSII) [64], multiple gene expression cassettes can be transferred at the same time, and with high-expression promoters, so that high-level enzymes can be accumulated at specific tissues to synthesize target products [64], [65], [66].
4. Challenges and prospects
In recent years, with the mining of genomic and transcriptomic data of crocin-producing species (mainly saffron and gardenia) [3], [24], [29], [30], the complete crocin biosynthetic pathway in plants has been elucidated, and some crocin biosynthetic genes have been identified from several plants and microorganisms by in vivo and/or vitro assays. However, there are still some challenges to overcome, such as the lack of studies of the regulatory mechanism of crocin biosynthesis and genome-wide data of C. sativus, as well as the so far missing identification of the enzyme catalyzing secondary glycosylation of crocetin in saffron. In addition, the production of crocins through metabolic engineering to meet current demands remains a challenge, despite relevant studies on crocin or crocetin production in microbial cell factories such as E. coli [10], [26], [44] and yeast [28], [62]. How to improve the productivity and yield of industrial platform strains has become the first problem to be solved in the industrialization of crocin metabolic engineering. Some new technologies and methods may be used to improve metabolic engineering of microbial strain for production of glycosylated crocins, for examples: (1) development of specific CRISPR engineered E. coli strains or yeast to redirect metabolic fluxes for crocins production [67]; (2) using bacterial microcompartments (BMCs) technology to efficiently synthesize crocins without affecting normal metabolic activities of cells [68]; and (3) Using the synthetic microbial consortia method [69], which the synthesis process of zeaxanthin, crocetin and the final product crocin can be given to three different engineering microorganisms, which can maximize the synthesis efficiency.
On the other hand, there are many directions for future research on crocin biosynthesis and metabolic engineering in plants, for examples: (1) genome-wide sequencing of C. sativus to gain an in-depth understanding of the evolution of crocin biosynthesis; (2) identification of the enzyme(s) responsible for secondary glycosylation of crocetin in saffron to understand the complete crocin biosynthetic pathway in C. sativus; (3) identification of more genes encoding transcription factors (TFs) involved in the crocin biosynthetic pathway by comprehensive analysis of genomics, transcriptomics, proteomics, and metabolomics to reveal the regulatory mechanism of crocin biosynthesis; (4) determination of the subcellular localization of more key enzymes to provide novel insights into the subcellular route of crocin biosynthesis; (5) characterization of more key enzymes responsible for crocin biosynthesis to construct suitable cell factories in plants or microorganisms for crocin biosynthesis to address crocins shortages; and (6) introduction of genes involved in the crocin biosynthetic pathway into important agricultural or industrial crops by using a multi-gene stacking system to develop crocin-rich functional foods or raw materials.
In conclusion, further researches on crocin biosynthesis and metabolic engineering will contribute to the industrial production of crocins, which will not only bring huge economic benefits but also have beneficial effect on human health.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (31971915; 31771740), the Major Program of Guangdong Basic and Applied Research (2019B030302006), and the Guangdong special support program of young top-notch talent in science and technology innovation (2019TQ05N147).
Contributor Information
Yao-Guang Liu, Email: ygliu@scau.edu.cn.
Qinlong Zhu, Email: zhuql@scau.edu.cn.
References
- 1.Fernández J. Biology, biotechnology and biomedicine of saffron. Recent Res. Dev. Plant Sci. 2004;2:127–159. [Google Scholar]
- 2.Frusciante S., Diretto G., Bruno M., Ferrante P., Pietrella M. Novel carotenoid cleavage dioxygenase catalyzes the first dedicated step in saffron crocin biosynthesis. Proc Natl Acad Sci. 2014;111:12246–12251. doi: 10.1073/pnas.1404629111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ji A., Jia J., Xu Z., Li Y., Bi W. Transcriptome-Guided Mining of Genes Involved in Crocin Biosynthesis. Front Plant Sci. 2017;8:518. doi: 10.3389/fpls.2017.00518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xi L, Qian Z. Pharmacological properties of crocetin and crocin (digentiobiosyl ester of crocetin) from saffron. Natural Product Communications 2006;1:1934578X0600100112.
- 5.Caballero-Ortega H., Pereda-Miranda R., Abdullaev F.I. HPLC quantification of major active components from 11 different saffron (Crocus sativus L.) sources. Food Chem. 2007;100:1126–1131. [Google Scholar]
- 6.Chen Y., Zhang H., Tian X., Zhao C., Cai L. Antioxidant potential of crocins and ethanol extracts of Gardenia jasminoides ELLIS and Crocus sativus L.: A relationship investigation between antioxidant activity and crocin contents. Food Chem. 2008;109:484–492. [Google Scholar]
- 7.Xiao W., Li S., Wang S., Ho C.T. Chemistry and bioactivity of Gardenia jasminoides. J Food Drug Anal. 2017;25:43–61. doi: 10.1016/j.jfda.2016.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen X., Tan H., Zhang L. Research progress on biosynthetic pathway of apo-carotenoids in Crocus sativus. Chinese Traditional Herbal Drugs. 2018;49:4702–4709. (in chinese with english abstract) [Google Scholar]
- 9.Zhu A., Xia Q., Li X., Wang Z., Wang W. Research progress of purification, determination and structure-activity relationship of crocins from gardenia and saffron. Chin J Pharm Anal. 2018;38:735–747. (in chinese with english abstract) [Google Scholar]
- 10.Ding F., Liu F., Shao W., Chu J., Wu B. Efficient Synthesis of Crocins from Crocetin by a Microbial Glycosyltransferase from Bacillus subtilis 168. J Agric Food Chem. 2018;66:11701–11708. doi: 10.1021/acs.jafc.8b04274. [DOI] [PubMed] [Google Scholar]
- 11.Liang Z., Yang M., Xu X., Xie Z., Huang J. Isolation and purification of geniposide, crocin-1, and geniposidic acid from the fruit of Gardenia jasminoides Ellis by high-speed counter-current chromatography. Sep Sci Technol. 2014;49:1427–1433. [Google Scholar]
- 12.Finley J.W., Gao S. A Perspective on Crocus sativus L. (Saffron) Constituent Crocin: A Potent Water-Soluble Antioxidant and Potential Therapy for Alzheimer's Disease. J Agric Food Chem. 2017;65:1005–1020. doi: 10.1021/acs.jafc.6b04398. [DOI] [PubMed] [Google Scholar]
- 13.Amin B., Abnous K., Motamedshariaty V., Hosseinzadeh H. Attenuation of oxidative stress, inflammation and apoptosis by ethanolic and aqueous extracts of Crocus sativus L. stigma after chronic constriction injury of rats. An Acad Bras Cienc. 2014;86:1821–1832. doi: 10.1590/0001-3765201420140067. [DOI] [PubMed] [Google Scholar]
- 14.Yarijani Z.M., Pourmotabbed A., Pourmotabbed T., Najafi H. Crocin has anti-inflammatory and protective effects in ischemia-reperfusion induced renal injuries. Iran J Basic Med Sci. 2017;20:753–759. doi: 10.22038/IJBMS.2017.9005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu W., Sun Y., Cheng Z., Guo Y., Liu P. Crocin exerts anti-inflammatory and anti-arthritic effects on type II collagen-induced arthritis in rats. Pharm Biol. 2018;56:209–216. doi: 10.1080/13880209.2018.1448874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sheng L., Qian Z., Zheng S., Xi L. Mechanism of hypolipidemic effect of crocin in rats: crocin inhibits pancreatic lipase. Eur J Pharmacol. 2006;543:116–122. doi: 10.1016/j.ejphar.2006.05.038. [DOI] [PubMed] [Google Scholar]
- 17.Hoshyar R., Mollaei H. A comprehensive review on anticancer mechanisms of the main carotenoid of saffron, crocin. J Pharm Pharmacol. 2017;69:1419–1427. doi: 10.1111/jphp.12776. [DOI] [PubMed] [Google Scholar]
- 18.Rahaiee S., Hashemi M., Shojaosadati S.A., Moini S., Razavi S.H. Nanoparticles based on crocin loaded chitosan-alginate biopolymers: Antioxidant activities, bioavailability and anticancer properties. Int J Biol Macromol. 2017;99:401–408. doi: 10.1016/j.ijbiomac.2017.02.095. [DOI] [PubMed] [Google Scholar]
- 19.Jiang Z., Gu M., Liu J., Li H., Peng J. Anticancer activity of crocin against cervical carcinoma (HeLa cells): Bioassessment and toxicity evaluation of crocin in male albino rats. J Photochem Photobiol B. 2018;180:118–124. doi: 10.1016/j.jphotobiol.2018.01.013. [DOI] [PubMed] [Google Scholar]
- 20.Ghaeni F.A., Amin B., Hariri A.T., Meybodi N.T., Hosseinzadeh H. Antilithiatic effects of crocin on ethylene glycol-induced lithiasis in rats. Urolithiasis. 2014;42:549–558. doi: 10.1007/s00240-014-0711-y. [DOI] [PubMed] [Google Scholar]
- 21.Nemati Z., Harpke D., Gemicioglu A., Kerndorff H., Blattner F.R. Saffron (Crocus sativus) is an autotriploid that evolved in Attica (Greece) from wild Crocus cartwrightianus. Mol Phylogenet Evol. 2019;136:14–20. doi: 10.1016/j.ympev.2019.03.022. [DOI] [PubMed] [Google Scholar]
- 22.Dai Z., Wang Y., Zhou Z., Li S., Zhang X. Synthetic biology for production of plant-derived natural products. Bull Chinese Acad Sci. 2018;33:1228–1238. (in chinese with english abstract) [Google Scholar]
- 23.Ahrazem O., Diretto G., Argandoña J., Rubio-Moraga Á., Julve J.M. Evolutionarily distinct carotenoid cleavage dioxygenases are responsible for crocetin production in Buddleja davidii. J Exp Bot. 2017;68:4663–4677. doi: 10.1093/jxb/erx277. [DOI] [PubMed] [Google Scholar]
- 24.Baba S.A., Mohiuddin T., Basu S., Swarnkar M.K., Malik A.H. Comprehensive transcriptome analysis of Crocus sativus for discovery and expression of genes involved in apocarotenoid biosynthesis. BMC Genomics. 2015;16:698. doi: 10.1186/s12864-015-1894-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moraga A.R., Nohales P.F., Perez J.A., Gomez-Gomez L. Glucosylation of the saffron apocarotenoid crocetin by a glucosyltransferase isolated from Crocus sativus stigmas. Planta. 2004;219:955–966. doi: 10.1007/s00425-004-1299-1. [DOI] [PubMed] [Google Scholar]
- 26.Wang W., He P., Zhao D., Ye L., Dai L. Construction of Escherichia coli cell factories for crocin biosynthesis. Microb Cell Fact. 2019;18 doi: 10.1186/s12934-019-1166-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jain M., Srivastava P.L., Verma M., Ghangal R., Garg R. De novo transcriptome assembly and comprehensive expression profiling in Crocus sativus to gain insights into apocarotenoid biosynthesis. Sci Rep. 2016;6:22456. doi: 10.1038/srep22456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tan H., Chen X., Liang N., Chen R., Chen J. Transcriptome analysis reveals novel enzymes for apo-carotenoid biosynthesis in saffron and allows construction of a pathway for crocetin synthesis in yeast. J Exp Bot. 2019;70:4819–4834. doi: 10.1093/jxb/erz211. [DOI] [PubMed] [Google Scholar]
- 29.Xu Z., Pu X., Gao R., Demurtas O.C., Fleck S.J. Tandem gene duplications drive divergent evolution of caffeine and crocin biosynthetic pathways in plants. BMC Biol. 2020;18:63. doi: 10.1186/s12915-020-00795-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yue J., Wang R., Ma X., Liu J., Lu X. Full-length transcriptome sequencing provides insights into the evolution of apocarotenoid biosynthesis in Crocus sativus. Comput Struct Biotechnol J. 2020;18:774–783. doi: 10.1016/j.csbj.2020.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ashraf DN, Baba SA. Apocarotenoids Of Crocus Sativus L: From biosynthesis to pharmacology. 2016.
- 32.Cazzonelli C.I., Pogson B.J. Source to sink: regulation of carotenoid biosynthesis in plants. Trends Plant Sci. 2010;15:266–274. doi: 10.1016/j.tplants.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 33.Matthews P.D., Luo R., Wurtzel E.T. Maize phytoene desaturase and zeta-carotene desaturase catalyse a poly-Z desaturation pathway: implications for genetic engineering of carotenoid content among cereal crops. J Exp Bot. 2003;54:2215–2230. doi: 10.1093/jxb/erg235. [DOI] [PubMed] [Google Scholar]
- 34.Li F., Murillo C., Wurtzel E.T. Maize Y9 encodes a product essential for 15-cis-zeta-carotene isomerization. Plant Physiol. 2007;144:1181–1189. doi: 10.1104/pp.107.098996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Isaacson T., Ronen G., Zamir D., Hirschberg J. Cloning of tangerine from tomato reveals a carotenoid isomerase essential for the production of beta-carotene and xanthophylls in plants. Plant Cell. 2002;14:333–342. doi: 10.1105/tpc.010303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park H., Kreunen S.S., Cuttriss A.J., DellaPenna D., Pogson B.J. Identification of the carotenoid isomerase provides insight into carotenoid biosynthesis, prolamellar body formation, and photomorphogenesis. Plant Cell. 2002;14:321–332. doi: 10.1105/tpc.010302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cunningham F.X., Jr., Pogson B., Sun Z., McDonald K.A., DellaPenna D. Functional analysis of the beta and epsilon lycopene cyclase enzymes of Arabidopsis reveals a mechanism for control of cyclic carotenoid formation. Plant Cell. 1996;8:1613–1626. doi: 10.1105/tpc.8.9.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Castillo R., Fernández J.A., Gómez-Gómez L. Implications of carotenoid biosynthetic genes in apocarotenoid formation during the stigma development of Crocus sativus and its closer relatives. Plant Physiol. 2005;139:674–689. doi: 10.1104/pp.105.067827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ahrazem O., Rubio-Moraga A., Lopez R.C., Gomez-Gomez L. The expression of a chromoplast-specific lycopene beta cyclase gene is involved in the high production of saffron's apocarotenoid precursors. J Exp Bot. 2010;61:105–119. doi: 10.1093/jxb/erp283. [DOI] [PubMed] [Google Scholar]
- 40.Ahrazem O., Diretto G., Argandona Picazo J., Fiore A., Rubio-Moraga A. The Specialized Roles in Carotenogenesis and Apocarotenogenesis of the Phytoene Synthase Gene Family in Saffron. Front Plant Sci. 2019;10:249. doi: 10.3389/fpls.2019.00249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rubio A., Rambla J.L., Santaella M., Gomez M.D., Orzaez D. Cytosolic and plastoglobule-targeted carotenoid dioxygenases from Crocus sativus are both involved in beta-ionone release. J Biol Chem. 2008;283:24816–24825. doi: 10.1074/jbc.M804000200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alder A., Jamil M., Marzorati M., Bruno M., Vermathen M. The path from beta-carotene to carlactone, a strigolactone-like plant hormone. Science. 2012;335:1348–1351. doi: 10.1126/science.1218094. [DOI] [PubMed] [Google Scholar]
- 43.Demurtas O.C., Frusciante S., Ferrante P., Diretto G., Azad N.H. Candidate Enzymes for Saffron Crocin Biosynthesis Are Localized in Multiple Cellular Compartments. Plant Physiol. 2018;177:990–1006. doi: 10.1104/pp.17.01815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pu X., He C., Yang Y., Wang W., Hu K. In Vivo Production of Five Crocins in the Engineered Escherichia coli. ACS Synth Biol. 2020;9:1160–1168. doi: 10.1021/acssynbio.0c00039. [DOI] [PubMed] [Google Scholar]
- 45.Bouvier F., Suire C., Mutterer J., Camara B. Oxidative remodeling of chromoplast carotenoids: identification of the carotenoid dioxygenase CsCCD and CsZCD genes involved in Crocus secondary metabolite biogenesis. Plant Cell. 2003;15:47–62. doi: 10.1105/tpc.006536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gomez-Gomez L., Parra-Vega V., Rivas-Sendra A., Segui-Simarro J.M., Molina R.V. Unraveling Massive Crocins Transport and Accumulation through Proteome and Microscopy Tools during the Development of Saffron Stigma. Int J Mol Sci. 2017;18 doi: 10.3390/ijms18010076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ahrazem O., Rubio-Moraga A., Berman J., Capell T., Christou P. The carotenoid cleavage dioxygenase CCD2 catalysing the synthesis of crocetin in spring crocuses and saffron is a plastidial enzyme. New Phytol. 2016;209:650–663. doi: 10.1111/nph.13609. [DOI] [PubMed] [Google Scholar]
- 48.Yeats T.H., Nagel R. Subcellular Spice Trade Routes: Crocin Biosynthesis in the Saffron Crocus (Crocus sativus) Plant Physiol. 2018;177:869–870. doi: 10.1104/pp.18.00662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Demurtas O.C., de Brito Francisco R., Diretto G., Ferrante P., Frusciante S. ABCC Transporters Mediate the Vacuolar Accumulation of Crocins in Saffron Stigmas. Plant Cell. 2019;31:2789–2804. doi: 10.1105/tpc.19.00193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brandle J., Telmer P. Steviol glycoside biosynthesis. Phytochemistry. 2007;68:1855–1863. doi: 10.1016/j.phytochem.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 51.Nagatoshi M., Terasaka K., Owaki M., Sota M., Inukai T. UGT75L6 and UGT94E5 mediate sequential glucosylation of crocetin to crocin in Gardenia jasminoides. FEBS Lett. 2012;586:1055–1061. doi: 10.1016/j.febslet.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 52.Auldridge M.E., McCarty D.R., Klee H.J. Plant carotenoid cleavage oxygenases and their apocarotenoid products. Curr Opin Plant Biol. 2006;9:315–321. doi: 10.1016/j.pbi.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 53.Walter M.H., Strack D. Carotenoids and their cleavage products: biosynthesis and functions. Nat Prod Rep. 2011;28:663–692. doi: 10.1039/c0np00036a. [DOI] [PubMed] [Google Scholar]
- 54.Kloer D.P., Ruch S., Al-Babili S., Beyer P., Schulz G.E. The structure of a retinal-forming carotenoid oxygenase. Science. 2005;308:267–269. doi: 10.1126/science.1108965. [DOI] [PubMed] [Google Scholar]
- 55.Sui X., Kiser P.D., Lintig J., Palczewski K. Structural basis of carotenoid cleavage: from bacteria to mammals. Arch Biochem Biophys. 2013;539:203–213. doi: 10.1016/j.abb.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Carmona M., Zalacain A., Sánchez A.M., Novella J.L., Alonso G.L. Crocetin esters, picrocrocin and its related compounds present in Crocus sativus stigmas and Gardenia jasminoides fruits. Tentative identification of seven new compounds by LC-ESI-MS. J Agric Food Chem. 2006;54:973–979. doi: 10.1021/jf052297w. [DOI] [PubMed] [Google Scholar]
- 57.Liao Y.H., Houghton P.J., Hoult J.R. Novel and known constituents from Buddleja species and their activity against leukocyte eicosanoid generation. J Nat Prod. 1999;62:1241–1245. doi: 10.1021/np990092+. [DOI] [PubMed] [Google Scholar]
- 58.Brocker C., Vasiliou M., Carpenter S., Carpenter C., Zhang Y. Aldehyde dehydrogenase (ALDH) superfamily in plants: gene nomenclature and comparative genomics. Planta. 2013;237:189–210. doi: 10.1007/s00425-012-1749-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mohammed Benghezal, Geoffrey O. Wasteneys, Jones DA. The C-terminal dilysine motif confers endoplasmic reticulum localization to type I membrane proteins in plants. 2000. [DOI] [PMC free article] [PubMed]
- 60.Diretto G., Ahrazem O., Rubio-Moraga A., Fiore A., Sevi F. UGT709G1: a novel uridine diphosphate glycosyltransferase involved in the biosynthesis of picrocrocin, the precursor of safranal in saffron (Crocus sativus) New Phytol. 2019;224:725–740. doi: 10.1111/nph.16079. [DOI] [PubMed] [Google Scholar]
- 61.Lou S., Wang L., He L., Wang Z., Wang G. Production of crocetin in transgenic Chlorella vulgaris expressing genes crtRB and ZCD1. J Appl Phycol. 2015;28:1657–1665. [Google Scholar]
- 62.Chai F., Wang Y., Mei X., Yao M., Chen Y. Heterologous biosynthesis and manipulation of crocetin in Saccharomyces cerevisiae. Microb Cell Fact. 2017;16 doi: 10.1186/s12934-017-0665-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Marti M., Diretto G., Aragones V., Frusciante S., Ahrazem O. Efficient production of saffron crocins and picrocrocin in Nicotiana benthamiana using a virus-driven system. Metab Eng. 2020 doi: 10.1016/j.ymben.2020.06.009. [DOI] [PubMed] [Google Scholar]
- 64.Zhu Q., Yu S., Zeng D., Liu H., Wang H., Yang Z., Xie X., Shen R., Tan J., Li H., Zhao X., Zhang Q., Chen Y., Guo J., Chen L., Liu Y.-G. Development of ‘‘purple endosperm rice’’ by engineering anthocyanin biosynthesis in the endosperm with a high-efficiency transgene stacking system. Mol Plant. 2017;10:918–929. doi: 10.1016/j.molp.2017.05.008. [DOI] [PubMed] [Google Scholar]
- 65.Zhu Q., Zeng D., Yu S., Cui C., Li J. From Golden Rice to aSTARice: bioengineering astaxanthin biosynthesis in rice endosperm. Mol Plant. 2018;11:1440–1448. doi: 10.1016/j.molp.2018.09.007. [DOI] [PubMed] [Google Scholar]
- 66.Zhu Q., Wang B., Tan J., Liu T., Li L. Plant synthetic metabolic engineering for enhancing crop nutritional quality. Plant Commun. 2020;1 doi: 10.1016/j.xplc.2019.100017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schultenkämper K, Brito LF, Wendisch VF. Impact of CRISPR interference on strain development in biotechnology. 2020;67(1):7–21. [DOI] [PubMed]
- 68.Kirst H, Kerfeld CA. Bacterial microcompartments: catalysis-enhancing metabolic modules for next generation metabolic and biomedical engineering. BMC Biology. 2019;17(1):79 [DOI] [PMC free article] [PubMed]
- 69.Sgobba E., Wendisch V.F. Synthetic microbial consortia for small molecule production. Curr Opin Biotechnol. 2020;62:72–79. doi: 10.1016/j.copbio.2019.09.011. [DOI] [PubMed] [Google Scholar]