Abstract
The COVID-19 disease is caused by SARS-CoV-2 and spreading rapidly worldwide with extremely high infection rate. Since effective and specific vaccine is not available to combat the deadly COVID-19, the objective of our study was to design a multi-epitope vaccine using immunoinformatics approach with translational implications. Nucleocapsid (N) protein of SARS-CoV-2 is stable, conserved and highly immunogenic along with being less prone to mutations during infection, which makes it a suitable candidate for designing vaccine. In our study, B- and T-cells epitopes were identified from N protein and screened based on crucial parameters to design the multi-epitope vaccine construct. Additionally, human beta-defensin-2 was incorporated into the vaccine construct as an adjuvant along with suitable linkers followed by its further evaluation based on crucial parameters including allergenicity, antigenicity, stability etc. Combined major histocompatibility complexes (MHC-I and MHC-II) binding epitopes presented broader population coverage of the vaccine throughout the world. The three-dimensional structure of vaccine candidate implied strong interaction with toll-like receptor 3 (TLR3) using molecular docking. The vaccine-TLR3 complex was observed to be highly stable during simulation and electrostatic free energy was foremost contributor for stabilization of the structure. Subsequently, in silico cloning of vaccine candidate was carried out to generate the construct into pET-28a(+) expression vector succeeded by its virtual confirmation. Altogether, our results advocate that the designed vaccine candidate could be an effective and promising weapon to fight with COVID-19 infection worldwide.
Keywords: SARS-CoV-2, Nucleocapsid protein, Multi-epitope vaccine, Immunoinformatics approach, TLR3 receptor, Molecular dynamics simulation
Introduction
A new betacoronavirus, SARS-CoV-2 (Severe Acute Respiratory Syndrome Corona Virus 2), which belongs to Coronaviridae family is the causative agent of the recent world health hazard COVID-19 (coronavirus disease 2019). SARS-CoV-2 represents 79.6% sequence identity to SARS-CoV (Zhou et al. 2020), which was the causative agent of an epidemic in the year 2003. The recent outbreak of COVID-19 has been declared as a public health crisis of global concern by World Health Organization (WHO), which has reached to over 216 countries in a short span with affecting approximately thirty million people worldwide (https://covid19.who.int/). The case fatality rate (CFR) is approximately 3% and looks to be dependent on age, with an elevated percentage in the elderly, in particular men. The number of cases with asymptomatic and mild infection might be to a great extent than the official number of cases, which would go ahead to a lesser infection fatality rate (IRF). Some antiviral drugs such as remdesivir have been tested against the deadly COVID-19 (Wang et al. 2020); however, no drug has been specifically approved for the fatal disease till date. The lack of medical counter measures and very high infection rate suggests vaccination as the efficient way to control and prevent COVID-19.
SARS-CoV-2 has a single-stranded, positive sense, RNA genome of approximately 30 kb that contains majorly five ORFs for nonstructural polyproteins and structural proteins. Envelope (E), spike (S), nucleocapsid (N) and membrane (M) proteins are the structural proteins (Phan 2020) that are also present in SARS-CoV with similar molecular weight. The S protein of SARS-CoV-2 is considered as the foremost antigen target in development of vaccines (Chen et al. 2020). It shares 76% similarity with its homolog in SARS-CoV (Grifoni et al. 2020), but nonsynonymous type of mutations were observed in the S protein with the progression of coronavirus infection (Ruan et al. 2003). Conversely, the SARS-CoV-2 N protein is stable with very less mutations and also reveals very high homology with its SARS-CoV counterpart (Grifoni et al. 2020). Various reports suggest that N protein of SARS coronavirus plays critical role in numerous steps of life cycle of the virus including RNA replication, mRNA transcription, formation of nucleocapsid and virus budding (McBride et al. 2014). It is also known to be highly antigenic and shows upregulated expression during viral infection (Cong et al. 2020). Elevated levels of IgG antibodies were detected against N protein in sera from SARS-CoV patients (Leung et al. 2004) and considered as a promising antigen for the response of T-cells during a vaccine development along with helping in inducing cytotoxic activity and T-cell proliferation specific to SARS-CoV (Gao et al. 2003). Significantly, it has been observed that the middle and C-terminal portion of the N protein is crucial to generate antibodies during the immune response against SARS-CoV (Dutta et al. 2008).
The SARS-CoV-2 N protein contains non-specific RNA-binding activity and has suppressed RNAi by restraining dsRNA in host cells (Mu et al. 2020). Additionally, N gene of SARS-CoV-2 was found to be safe and strongly antigenic as a DNA vaccine (Ahlén et al. 2020). The crystal structure of N protein of SARS-CoV-2 is highly similar to that of the earlier reported N protein of coronavirus except different characteristics of surface electrostatic potential (Kang et al. 2020). The significant and well-timed insights related to the SARS-CoV-2 N protein presents diverse advantages as compared with other potential antigenic targets of this virus (Dutta et al. 2020). Moreover, distinct features of N protein including conserved protein sequence, improved knowledge of its biochemistry and genetics along with its exceptionally higher immunogenicity envisioned this protein as a strong vaccine candidate for the dreaded COVID-19 disease.
As current advancements in computational tools work as a valuable substitute for antigenic epitopes prediction with translational implications, the immunoinformatics approach was employed here to design immunogenic multi-epitope vaccine from N protein of SARS-CoV-2. The vaccine candidate consists of helper T-lymphocyte (HTL), cytotoxic T-lymphocyte (CTL) and B-cell lymphocyte (BCL) epitopes from the middle and C-terminal portion of N protein sequence as these regions are crucial for antibodies generation. Subsequently, molecular docking and MD simulation were preformed to authenticate the interaction, binding mode and stability of vaccine candidate with TLR3 immune receptor of human host to design novel and potential multi-epitope vaccine that can pave the way to fight against life threatening SARS-CoV-2.
Materials and Methods
Sequence Analysis of SARS-CoV-2 N Protein
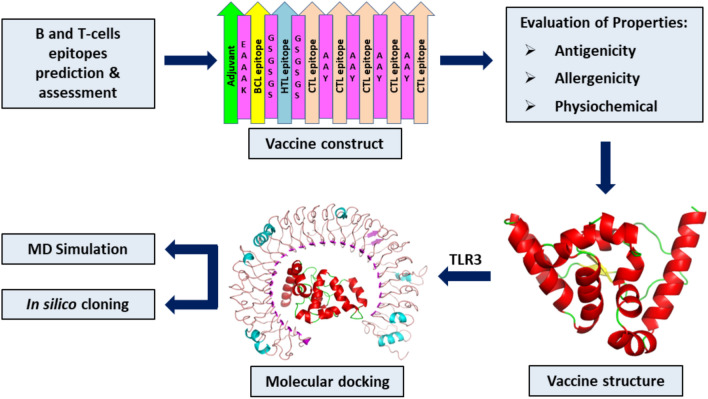
The amino acid sequence of SARS-CoV and SARS-CoV-2 N protein was retrieved from the Uniprot database with the respective ID of P59595 and P0DTC9 and then pairwise sequence alignment was performed using software Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/). Subsequently, antigenic peptides prediction tool (http://imed.med.ucm.es/Tools/antigenic.pl) and AllerTop v2.0 servers (http://ddg-pharmfac.net/AllergenFP/) were employed to screen the N protein of SARS-CoV-2 for the average antigenic propensity and allergenicity, respectively. Furthermore, N protein amino acid sequence was utilized for multi-epitope vaccine designing with the procedure presented in Fig. 1.
Fig. 1.
Diagrammatic representation of the steps involved for in silico designing of the multi-epitope vaccine candidate from N protein of SARS-CoV-2
Prediction of CTL, HTL and BCL Epitopes
The cytotoxic T-lymphocyte (CTL) epitopes for N protein was predicted by NetCTL1.2 (http://www.cbs.dtu.dk/services/NetCTL/) for all the available serotypes (A1, A2, A3, A24, A26, B7, B8, B27, B39, B44, B58, B62) taking 0.75 as threshold value with 0.97 specificity and 0.80 sensitivity, while weight on C-terminal cleavage and TAP transport efficiency were kept as default parameters. Successively, immunogenicity and antigenicity were analyzed through Class I Immunogenicity of IEDB server (http://tools.iedb.org/immunogenicity/) and VaxiJen v2.0 (http://www.ddg-pharmfac.net/vaxijen/VaxiJen/VaxiJen.html), respectively. The MHC-I binding alleles of selected CTL epitopes were predicted by MHC-I binding predictions of IEDB server (http://tools.iedb.org/mhci/) using consensus method with percentile rank of less than 2. The helper T-lymphocyte (HTL) epitopes of 15 amino acids long were identified by the IEDB MHC-II epitope prediction tool (http://tools.iedb.org/mhcii/) utilizing NN_Align method to calculate percentile rank and IC50 value for the interaction of peptide with MHC-II. Human was taken as source species, while loci HLA-DR, HLA-DP and HLA-DQ were considered for further analysis. The IC50 values and percentile rank of less than 10 nM and 1.5 respectively, were considered for prediction as these values represent higher binding affinity. Predicted HTL epitopes were assessed for their antigenic properties.
Simultaneously, B-cell lymphocyte (BCL) epitopes for the N protein of SARS-CoV-2 were identified through the server ABCPred (http://crdd.osdd.net/raghava/abcpred/) followed by evaluation using BepiPred (http://tools.iedb.org/bcell/help/#Bepipred) of IEDB. VaxiJen v2.0 was used to evaluate the antigenicity of B-cell epitopes. Furthermore, B-cell epitopes were analyzed for many crucial parameters like hydrophilicity, surface accessibility, flexibility and beta-turn prediction using respective tools of IEDB (http://tools.iedb.org/bcell/) including Parker hydrophilicity, Emini surface accessibility, Karplus & Schultz flexibility and Chou & Fasman beta-turn predictions. Finally, AllerTop v2.0 and ToxinPred (http://crdd.osdd.net/raghava/toxinpred/) servers were employed to estimate the respective allergenic and toxic nature of HTL, CTL and BCL epitopes.
Analysis of Cross-Reactivity and Population Coverage
The selected epitopes were analyzed for cross-reactivity with human proteome. Concurrently, population coverage tool (http://tools.iedb.org/population/) of IEDB was utilized to analyze the CTL and HTL as expression and distribution of HLA alleles deviate within the regions, ethnicities and countries globally that can influence the advancement of multi-epitope based vaccines.
Construction of Multi-epitope Vaccine
The vaccine was constructed by combining adjuvant, CTL, HTL and BCL epitopes with the suitable linkers to give enough space to epitopes in vivo. To increase immunogenicity of the vaccine candidate, human β-defensin-2 (PDB ID: 1FD3) was utilized as an adjuvant and connected to B-cell epitope using EAAAK linker. Similarly, BCL to HTL, HTL to CTL and intra CTL epitopes were joined through GSGSGS, GSGSGS and AAY linkers, respectively.
Evaluation of Multi-epitope Vaccine
The allergenicity of vaccine candidate was determined by servers AllerTop v2.0 and Allergen FP v1.0 (http://ddg-pharmfac.net/AllergenFP/). The antigenicity of the constructed vaccine was analyzed through VaxiJen v2.0 and ANTIGENpro (http://scratch.proteomics.ics.uci.edu/). Afterwards, ProtParam tool (http://web.expasy.org/protparam/) of ExPASy server was employed to determine the physiochemical properties of the vaccine construct. In addition, SOLpro (http://scratch.proteomics.ics.uci.edu/) and Protein-Sol (https://protein-sol.manchester.ac.uk/) servers were employed to check the solubility of the vaccine construct after expression in Escherichia coli (E. coli). Simultaneously, solvent accessibility of vaccine construct was assessed by RaptorX server (http://raptorx.uchicago.edu/StructurePropertyPred/predict/).
Structure Prediction and Validation
The secondary structure content of the designed vaccine was analyzed using software PSIPRED v4.0 (http://bioinf.cs.ucl.ac.uk/psipred/). The three-dimensional structure of vaccine construct was predicted by iterative threading modelling method through I-TASSER server (https://zhanglab.ccmb.med.umich.edu/I-TASSER/) followed by refinement with two-step procedure. Firstly, the GalaxyRefine tool (http://galaxy.seoklab.org/cgi-bin/submit.cgi?type=REFINE) was applied for the modelled structure refinement succeeded by loop refinement using GalaxyLoop server (http://galaxy.seoklab.org/cgi-bin/submit.cgi?type=LOOP). Subsequently, Procheck (https://servicesn.mbi.ucla.edu/PROCHECK/) and ProSA-web (https://prosa.services.came.sbg.ac.at/prosa.php) were used for the validation of vaccine structure.
Conformational B-cell Epitopes Screening
IEDB Ellipro tool (http://tools.iedb.org/ellipro/) was employed with default parameters for prediction of B-cell conformational epitopes. It is a structure based approach and predicts the antibody epitopes based on certain parameters like neighboring residues, estimation of shape, and protrusion index (PI).
Molecular Docking Analysis
To evaluate the interaction between vaccine candidate and host immune receptor, molecular docking was executed through ClusPro 2.0 (https://cluspro.bu.edu/publications.php) server using TLR3 and vaccine construct as receptor and ligand, respectively. Consequently, complexes were generated based on three consecutive steps that include rigid body docking, clustering of lowest energy structure, and structural refinement. PyMol (http://www.pymol.org) was used for analysis of docked structure and the best complex was chosen for further study on the basis of the lowest energy score.
Molecular Dynamics Simulation
The molecular dynamics (MD) simulation is a crucial and effective technique to study the induced structural stability and molecular interactions in the protein complex during the dynamics. Therefore, MD simulation was performed for the vaccine-TLR3 complex by GROMACS 5.1.4 version (Abraham et al., 2005) with charm36 all atom force field and TIP3P water model. Firstly, Na+ ions were added to the system for neutralization and then steepest descent algorithm was utilized for minimization of system energy in 50,000 steps. After energy minimization, 1 ns each of canonical assemble NVT (at 300 K) and isothermal-isobaric assemble NPT (at 1 bar) equilibrations were done for the minimized system followed by 50 ns production simulation. Further, simulation data was used for calculating root mean square fluctuations (RMSF), root mean square deviation (RMSD) and radius of gyration (Rg) of the system. The average binding energy of vaccine-TLR3 complex was calculated for the last 10 ns by g_MMPBSA package (Kumari et al. 2014) of GROMACS, for each 0.1 ns frame of the simulation trajectories. The types of interactions like hydrophobic, cation-pi, aromatic, and ionic were calculated by server Protein Interactions Calculator (PIC, http://pic.mbu.iisc.ernet.in/) and hydrogen bonds by the GROMACS for the vaccine-TLR3 complex from initial and stable trajectory. In addition, iMODS server (http://imods.chaconlab.org/) was utilized to analyse the deformability and rigidity in each residue of the vaccine-TLR3 complex by the normal mode analysis (NMA).
Codon Optimization and In Silico Cloning
Java Codon Adaptation Tool (JCat) (http://www.jcat.de/) was employed for codon optimization to obtain maximal expression of vaccine construct in E. coli strain K12. Further, NEBcutter (http://nc2.neb.com/NEBcutter2/) was utilized to analyze restriction enzyme cleavage sites, while pET-28a(+) plasmid vector was used for cloning. The vaccine construct was incorporated into the multiple cloning sites (MCS) of vector with 6xHis-tag at N-terminus for in silico cloning followed by virtual confirmation using the SnapGene tool (http://www.snapgene.com/) to obtain the expression of designed vaccine construct.
Results
Evaluation of SARS CoV-2 N Protein Sequence
The amino acid sequence of N protein from SARS-CoV-2 was aligned with its SARS-CoV homolog, and the identity and similarity between these proteins were found to be 91.5% and 94%, respectively. The N protein of SARS-CoV-2 is comprised of 419 amino acids with approximately 46 kDa molecular weight. Protein with an antigenic predicted score higher than 0.8 is generally considered for vaccine designing using epitopes prediction. The average antigenic propensity of N protein was found to be 0.9871, while subsequent analysis depicted it to be non-allergenic. Hence, SARS-CoV-2 N protein was selected for designing of multi-epitope vaccine.
Prediction and Analysis of T-Lymphocyte Epitopes
As the cytotoxic T-lymphocyte (CTL) epitopes play vital role in activating MHC-I cellular immune response, CTL epitopes of N protein were predicted by server NetCTL 1.2. In total, 74 epitopes (9-mer) with higher than 0.75 combined score were predicted from all MHC-I serotypes. Subsequently, 14 epitopes were selected after assessing the various parameters like allergenicity, immunogenicity, antigenicity and toxicity (Table 1). Out of these, five epitopes (GDAALALLL, EVTPSGTWL, TWLTYTGAI, KTFPPTEPK and SRIGMEVTP) were further selected for the study as the epitope GDAALALLL is the part of middle region of SARS-CoV-2 N protein, whereas SRIGMEVTP, TWLTYTGAI, EVTPSGTWL and KTFPPTEPK epitopes belong to C-terminal dimerization domain.
Table 1.
Cytotoxic T-lymphocyte (CTL) epitopes prediction from N protein of SARS-CoV-2
| S. no. | Peptide | Position | Antigenicity | Allergenicity | Toxicity | No. of alleles bind (Percentile rank < 2) |
|---|---|---|---|---|---|---|
| 1. | QRNAPRITF | 9–17 | Yes | No | No | HLA-C*07:02 |
| 2. | SSPDDQIGY | 78–86 | Yes | No | No | HLA-A*01:01 HLA-A*30:02 |
| 3. | SPDDQIGYY | 79–87 | Yes | No | No | HLA-B*35:01 HLA-A*01:01 |
| 4. | LSPRWYFYY | 104–112 | Yes | No | No | HLA-A*01:01 HLA-A*29:02 HLA-A*30:02 |
| 5. | DLSPRWYFY | 103–111 | Yes | No | No | HLA-A*29:02 HLA-A*30:02 HLA-A*26:01 HLA-A*01:01 |
| 6. | SPRWYFYYL | 105–113 | Yes | No | No | HLA-B*08:01 HLA-B*07:02 HLA-B*35:03 HLA-B*51:01 HLA-B*53:01 |
| 7. | NNAAIVLQL | 153–161 | Yes | No | No | HLA-B*14:02 |
| 8. | LPQGTTLPK | 161–169 | Yes | No | No | HLA-B*53:01 HLA-B*07:02 |
| 9. | GDAALALLL | 215–223 | Yes | No | No | HLA-B*40:02 HLA-B*48:01 HLA-E*01:01 |
| 10. | GMSRIGMEV | 316–324 | Yes | No | No | HLA-A*02:01 |
| 11. | EVTPSGTWL | 323–331 | Yes | No | No | HLA-A*68:02 HLA-A*26:01 |
| 12. | TWLTYTGAI | 329–337 | Yes | No | No | HLA-A*24:02 |
| 13. | SRIGMEVTP | 318–326 | Yes | No | No | NONE |
| 14. | KTFPPTEPK | 361–369 | Yes | No | No | HLA-A*11:01 HLA-A*30:01 HLA-A*03:01 HLA-A*31:01 HLA-A*68:01 HLA-A*32:01 |
Helper T-lymphocytes are known to participate in the B- and cytotoxic T-cells activation for the production of antibodies and killing of infected target cells, respectively. N protein was subjected to prediction of HTL epitopes from MHC-II epitope module of IEDB. Based on the IC50 values (less than 10 nM) and percentile rank (less than 1.5), HTL epitopes for N protein were predicted for HLA-DR, HLA-DQ and HLA-DP loci. The HTL epitopes of HLA-DR locus were found to fulfill the different parameters and eight HTLs were further analyzed (Table 2). Finally, one epitope (AQFAPSASAFFGMSR) was selected for construction of vaccine as it is present in C-terminal dimerization domain.
Table 2.
Evaluation of SARS-CoV-2 N protein helper T-lymphocyte (HTL) epitopes
| S. no. | Peptide | Position | Alleles | Antigenicity | Allergenicity | Toxicity | IFN-γ |
|---|---|---|---|---|---|---|---|
| 1. | QIAQFAPSASAFFGM | 303–317 | HLA-DRB1*09:01 | Yes | No | No | Positive |
| 2. | AQFAPSASAFFGMSR | 305–319 | HLA-DRB1*09:01 | Yes | No | No | Positive |
| 3. | QFAPSASAFFGMSRI | 306–320 | HLA-DRB1*09:01 | Yes | No | No | Positive |
| 4.. | QIGYYRRATRRIRGG | 83–97 | HLA-DRB1*11:01 HLA-DRB4*01:03 HLA-DRB1*13:01 HLA-DRB5*01:01 | Yes | No | No | Positive |
| 5. | IGYYRRATRRIRGGD | 84–98 | HLA-DRB1*11:01 HLA-DRB4*01:03 HLA-DRB1*13:01 HLA-DRB5*01:01 | Yes | No | No | Positive |
| 6. | ASAFFGMSRIGMEVT | 311–325 | HLA-DRB1*11:01 | Yes | No | No | Positive |
| 7. | SASAFFGMSRIGMEV | 310–324 | HLA-DRB1*11:01 | Yes | No | No | Positive |
| 8. | PSASAFFGMSRIGME | 309–323 | HLA-DRB1*11:01 | Yes | No | No | Positive |
Prediction and Evaluation of B-Lymphocyte Epitopes
B cells are the vital component of humoral immunity for the duration of the adaptive type immune response for antibodies production against pathogens. Hence, linear B-cell epitopes with 0.5 or above score of 16-mer length were selected using ABCPred followed by confirmation with BepiPred server. Subsequently, three BCL epitopes (TGSNQNGERSGARSKQ, RSGARSKQRRPQGLPN and NSSRNSTPGSSRGTSP) were found to fulfill the parameters of both of the servers and also observed to be antigenic, non-allergenic and non-toxic. These epitopes were further assessed for several parameters like surface accessibility, hydrophilicity, flexibility and beta-turn prediction (Table 3).
Table 3.
Assessment of linear B-cell lymphocyte epitopes from N protein of SARS-CoV-2
| S. no. | Peptide | Position | BepiPred | Antigenicity | Allergenicity | Toxicity | Surface accessibility | Hydrophilicity | Flexibility | Beta-turn |
|---|---|---|---|---|---|---|---|---|---|---|
| 1. | TGSNQNGERSGARSKQ | 24–39 | Yes | Yes | No | No | Yes | Yes | Yes | Yes |
| 2. | RSGARSKQRRPQGLPN | 32–47 | Yes | Yes | No | No | Yes | Yes | Yes | Yes |
| 3. | NSSRNSTPGSSRGTSP | 192–207 | Yes | Yes | No | No | Yes | Yes | Yes | Yes |
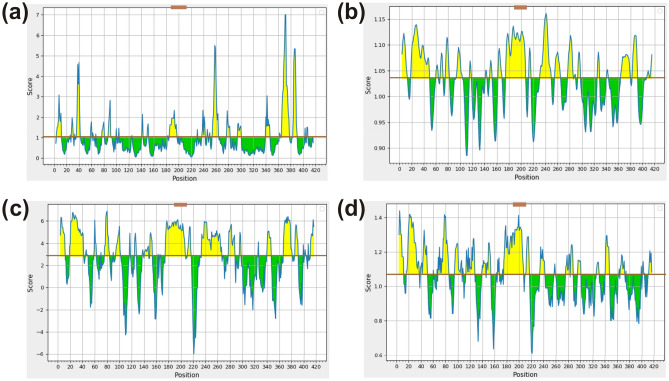
Remarkably, antigenic sites should be accessible or present on the protein surface (probably hydrophilic) as they are recognized by antibodies, while there is high correlation between the propensity for protein sequence to form beta-turn and reactivity of protein to anti-peptide antibodies (Petersen et al., 2010). All three BCL epitopes have shown significant values for Emini surface accessibility, Karplus & Schulz flexibility, Parker hydrophilicity and Chou & Fasman beta-turn algorithm (Fig. 2). However, one (NSSRNSTPGSSRGTSP) out of three epitopes was considered for vaccine construction as it is present in middle portion of N protein that is known as an immunogenic region.
Fig. 2.
Evaluation of BCL epitopes from SARS-CoV-2 N protein. a Emini surface accessibility, b Karplus and Schulz flexibility, c Parker hydrophilicity and d Chou and Fasman beta-turn prediction, where X- and Y-axis represent the amino acid position and respective scores. The brown line represents the average values of the respective calculated scores and the yellow regions above the average are considered as good, while the selected peptide region (NSSRNSTPGSSRGTSP) is marked as brown (Color figure online)
Cross-Reactivity and Population Coverage Analysis
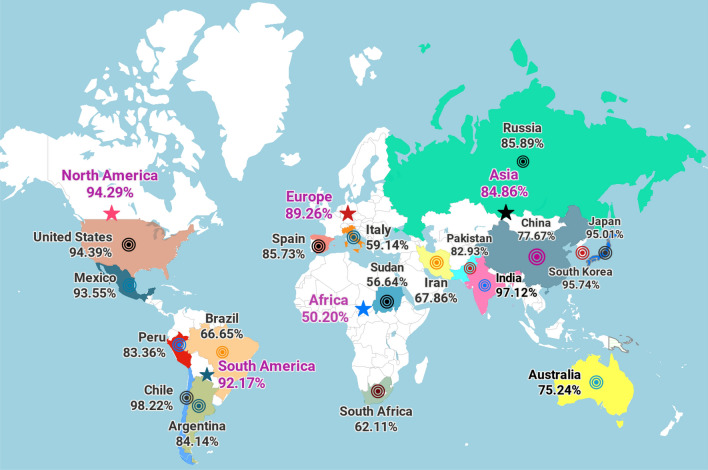
To avoid auto immunity, selected vaccine epitopes should not have homologs in human proteome. Cross-reactivity evaluation depicted no homolog to N protein of SARS-CoV-2 in human proteome. Simultaneously, population coverage was analyzed globally as WHO has declared COVID-19 as a pandemic. The selected T-cell epitopes (5 CTL and 1 HTL) with cognate HLA alleles were utilized and combined MHC-I and MHC-II epitopes highlighted 92.57% population coverage worldwide. Maximal population coverage was found to be 97.43% for South Asia, while it was observed to be 97.12% and 77.67% for India and China, respectively (Fig. 3). Notably, our vaccine candidate has shown broader population coverage all over the world to fight against deadly SARS-CoV-2 virus.
Fig. 3.
Population coverage by T-cell epitopes. Worldwide population coverage was mapped on the basis of the selected T-cell epitopes with their combined MHC-I and II scores. The continents and countries are represented as colored stars and circles, respectively (Color figure online)
Construction of Multi-epitope Vaccine
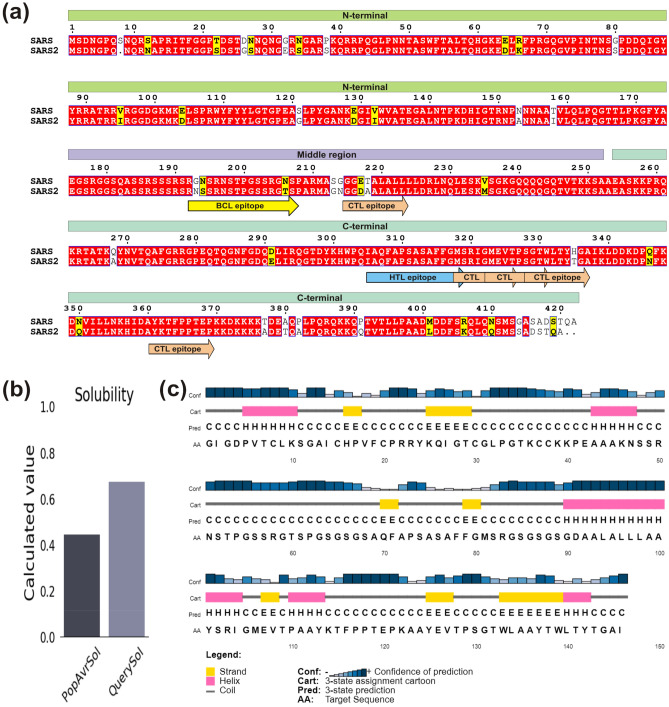
To design a multi-epitope vaccine, the selected epitopes (5 CTL, 1 HTL and 1 BCL) were stitched by suitable linkers (Fig. 4a). Human β-defensin-2 (hBD-2, 41 amino acids) sequence was also incorporated as an adjuvant to increase the immunogenicity of desired vaccine construct. To prevent interaction of adjuvant with vaccine construct, we employed EAAAK linker for joining the adjuvant with BCL epitope. CTL epitopes were combined together through AAY linker that maintains structural conformations of epitopes and increases the probability of antigen presentation. In addition, BCL to HTL and HTL to CTL epitopes were joined through linker GSGSGS, which is known to provide structural flexibility to protein without interfering with vaccine candidate function. Altogether, the final vaccine construct encompasses 146 amino acids containing an adjuvant and seven N-protein epitopes connected by the linkers.
Fig. 4.
Sequence analysis of N protein and its vaccine construct. a Sequence alignment between SARS-CoV and SARS-CoV-2 nucleocapsid protein. The domains are labelled and presented as box with different colors, while identical and similar residues are highlighted in red and yellow color, respectively. The selected epitopes are shown as arrowed box with different colors and labelled. b Solubility and c Secondary structure analysis of vaccine construct using Protein-Sol and PSIPRED server, respectively (Color figure online)
Assessment of Multi-epitope Vaccine
One of the most important aspect for vaccine candidate construction is immunogenicity i.e. stimulation of cell-mediated and humoral immune responses against pathogens. The constructed multi-epitope vaccine reveals good antigenic property as VaxiJen v2.0 and ANTIGENpro servers depicted the antigenicity scores of 0.646 and 0.926, respectively. Simultaneously, vaccine construct was prediced to be non-allergenic through Allergen FP v1.0 and AllerTOP v2.0. Subsequently, ProtParam server was used to evaluate the physiochemical properties of vaccine construct and it was found to possess molecular weight and theoretical PI as 14.81 kDa and 9.39, respectively. The predicted half-life of vaccine was 30 h in mammalian reticulocytes (in vivo), >20 h in yeast (in vivo), and >10 h in E. coli (in vivo), signifying the stability of vaccine construct. Further, the instability index was found to be 39.22 that advocates the vaccine candidate to be stable. The grand average of hydropathicity and aliphatic index were calculated to be − 0.133 and 56.37, respectively, suggesting the hydrophilic and thermostable nature of vaccine candidate. The vaccine candidate was found to be soluble upon over expression with respective scores of 0.829 and 0.675 using SOLpro and Protein-sol server (Fig. 4b). Simultaneously, solvent accessibility of vaccine construct was determined through RaptorX that indicated 59% residues to be exposed, 16% medium and 23% buried.
Structure Prediction and Validation
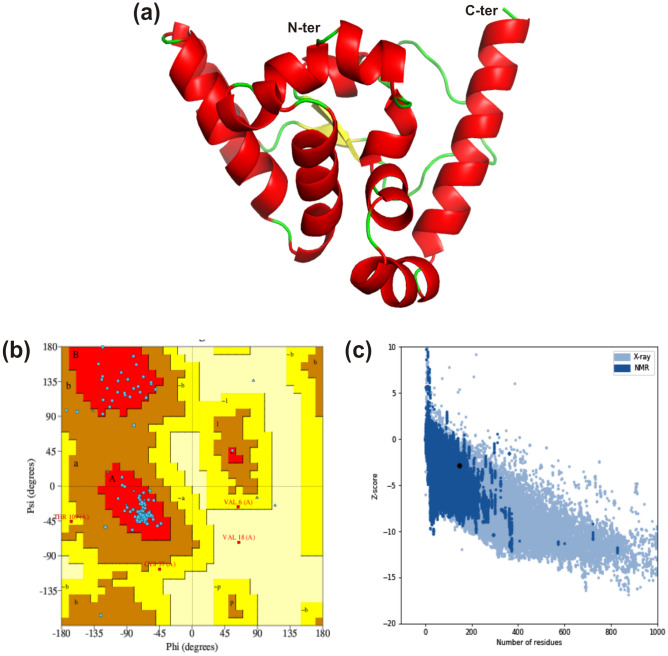
Secondary structure content of vaccine construct was determined through the server PSIPRED 4.0 that depicted presence of 22.60% α-helices, 15.75% β-sheets and 61.65% random coils (Fig. 4c). Furthermore, three-dimensional structure was predicted using I-TASSER server where five models were attained and the structure with the best C score value (− 4.05) was further refined. The refinement helps model to attain the structure close to native form by increasing the model quality, which was achieved through two-step refinement using Galaxy web server (Fig. 5a).
Fig. 5.
Prediction and validation of three-dimensional structure. a Modelled three-dimensional structure of the multi-epitope vaccine showing α-helices (red), β-sheets (yellow) and random coils (green). b Ramachandran plot indicating the presence of amino acid residues in favored, allowed and disallowed region. c ProSA-web validation presenting the Z-score of predicted 3D structure (Color figure online)
The refined structure was evaluated using Ramachandran plot with 91.2%, 5.3%, 2.7% and 0.9% residues in core, allowed, generously allowed and disallowed regions, respectively. Only one residue (Val18) was in the disallowed region, which belongs to adjuvant hBD-2 (Fig. 5b). There were two bad contacts in modelled structure, while planar groups were found to be 100% within the limits. Subsequently, modelled structure was analyzed through ProSA-web server and the Z-score of vaccine candidate was observed to be -2.81 that falls within range of NMR structure (Fig. 5c). Therefore, validation analysis suggested good quality of vaccine structure and could be used for downstream analysis.
Prediction of Discontinuous BCL Epitopes
Discontinuous (conformational) BCL epitopes are the segments present distantly in the protein sequence of pathogen and come closer during protein folding (Van Regenmortel 1996). Ellipro tool from IEDB was employed to identify the conformational B cell epitopes in vaccine construct. In total, four BCL conformational epitopes comprised of 84 residues were observed with predicted score from 0.58 to 0.84 and epitopic size ranging from 3 to 27 residues (Table 4). Predicted top scorer epitope was of 9 residues stretch consisting of residues K25, T138, L140, T141, Y142, T143, G144, A145 and I146 with the score of 0.84. Hence, the existence of conformational BCL epitopes in vaccine construct could initiate strong antibody responses against SARS-CoV-2 virus.
Table 4.
Predicted discontinuous epitopes from multi-epitope vaccine of SARS CoV-2 N protein
| S. no. | Residues | No. of residues | Score |
|---|---|---|---|
| 1. | K25, T138, L140, T141, Y142, T143, G144, A145, I146 | 9 | 0.84 |
| 2. | V18, F19, C20, P21, R22, R23, Y24, Y113, K114, T115, F116, P117, P118, T119, E120, P121, K122, A123, A124, Y125, E126, V127, S130, L134, Y137 | 25 | 0.69 |
| 3. | C38, K39, K40, P41, E42, A43, A44, A45, K46, S48, S49, R50, N51, S52, T53, P54, G55, S56, S57, R58, G59, T60, S82, G84, G86, S87, G88 | 27 | 0.66 |
| 4. | A95, L96, L97 | 3 | 0.58 |
Molecular Docking
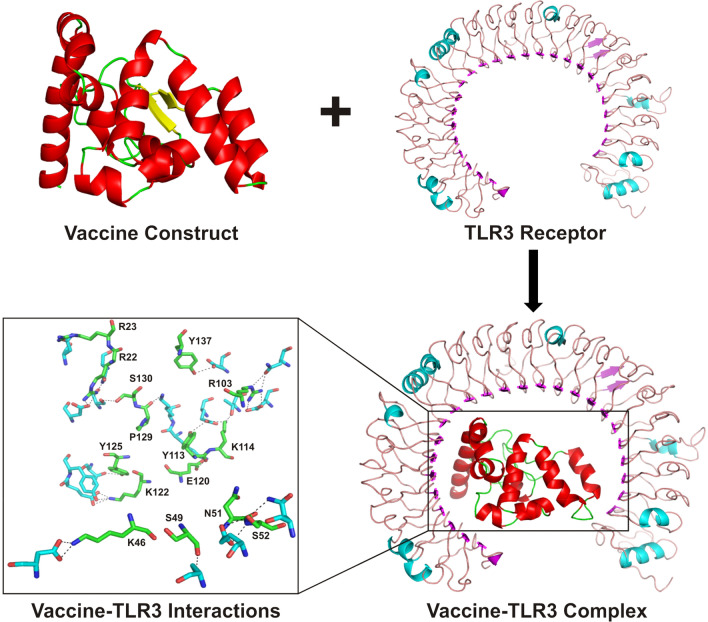
To assess the interaction of the vaccine with TLR3 receptor, molecular docking was carried out by ClusPro 2.0 server. Among the docked complexes, the best complex of vaccine-TLR3 was selected on the basis of the lowest energy score (− 1006.7 kJ mol−1) with the center energy (the energy between receptor and ligand) of − 852.3 kJ mol−1. The interactions between the TLR3 receptor and vaccine candidate were visualized using Pymol, where residues (K46, S49, N51, S52, R103, Y113, K114, E120, K122, Y125, P129, S130 and Y137) of vaccine candidate revealed polar contacts with the residues (N105, E127, T151, D153, E175, N229, T277, E301, Y302, D536, S614, T638 and E639) of TLR3 receptor (Fig. 6).
Fig. 6.
Molecular docking of vaccine candidate and TLR3. The docked complex of TLR3 and multi-epitope vaccine obtained from ClusPro is presented in cartoon mode. The residues depicting interactions between TLR3 and vaccine candidate are shown as stick model and colored in cyan and green, respectively, while vaccine construct residues are labelled and polar contacts are represented by black dashed lines (Color figure online)
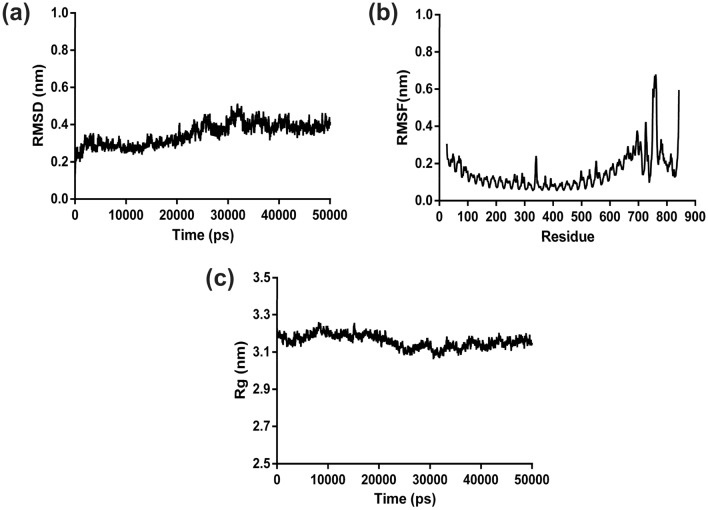
MD Simulation Analysis
The stability and compactness of docked vaccine-TLR3 complex was analysed by 50 ns molecular dynamics simulation. The enumerated root mean square deviation of complex revealed the average RMSD value of 0.40 nm during the simulation. It also depicted that initially RMSD remained between 0.20 to 0.51 nm up to 32 ns, but structural deviations for complex got stabilized from 33 ns with a RMSD value of ~ 0.42 nm throughout the remaining simulation time (Fig. 7a). The analysis of RMSD data delineates that the vaccine-TLR3 complex was stabilized during the dynamics. Simultaneously, the average RMSF was estimated to be of 0.31 nm for the backbone atoms of the residues in protein complex (Fig. 7b). The RMSF analysis of complex showed that the fluctuation in the residues was low during the simulation and infers the stable interaction between the vaccine and TLR3. In addition, the average radius of gyration was calculated to be 3.19 nm for the docked complex. The compactness analysis of the complex elucidates that the Rg of backbone atom of the vaccine-TLR3 structure remained approximately close to the average Rg value throughout simulation (Fig. 7c), which was lower than the Rg value of the initial conformation (~ 3.23 nm). The Rg analysis depicted that the complex structure gained a stable compactness during the dynamics.
Fig. 7.
MD simulation analysis of docked complex. a RMSD, b RMSF and c Rg have been represented as graphs for the backbone-atoms of the docked multi-epitope vaccine and TLR3 complex
Total number of different types of interactions between the vaccine and TLR3 in the initial and stable trajectory complexes depicts the interface stability during simulation. The hydrogen bonds and other interactions calculated by GROMACS and PIC highlighted that the initial complex structure has lesser interactions than the stable trajectory (Table 5).
Table 5.
Interactions of the vaccine-TLR3 complex during MD simulation
| Hydrophobic | Hydrogen bond | Ionic | Aromatic-aromatic | Cation-π | |
|---|---|---|---|---|---|
| Initial complex | 14 | 16 | 09 | 3 | 3 |
| Stable complex | 14 | 21 | 10 | 1 | 5 |
The average binding energy of the complex was − 1672.88 kJ mol−1, which was lower than that of the initial complex (− 1217.21 kJ mol−1) and major contribution was from electrostatic free energy (− 3112.04 kJ mol−1). Subsequent analysis showed that the interface between the vaccine and TLR3 was more stable, which could be due to enhanced binding affinity between the vaccine and TLR3 receptor (Table 6).
Table 6.
The Van der Wall, Electrostatic, Polar solvation and Binding energy of the vaccine-TLR3 complex calculated by g_MMPBSA method
| Electrostatic (kJ/mol) | Van der wall (kJ/mol) | Polar solvation (kJ/mol) | Binding (kJ/mol) | |
|---|---|---|---|---|
| Initial complex | − 2840.63 | − 551.85 | 2259.85 | − 1217.21 |
| Stable complex | − 3240.70 | − 479.77 | 2181.77 | − 1616.98 |
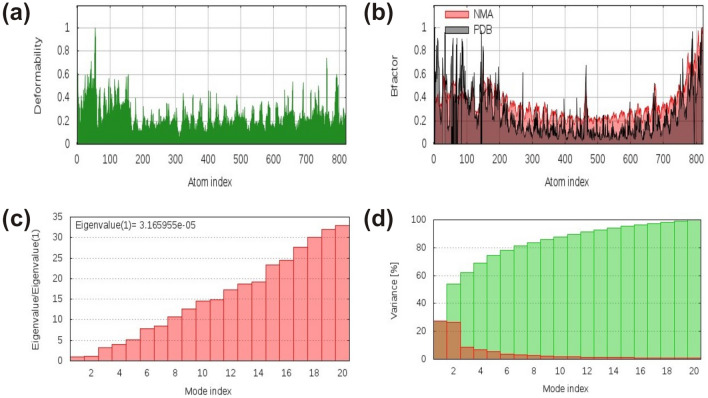
Concomitantly, lowering of the average polar solvation in comparison to that of initial structure showed increase in compactness of the complex that supports our findings for Rg of the complex. Additionally, the stiffness in the mobility and deformability in the residues of the complex were analysed by enumerating normal mode analysis (NMA) via server iMODS. The deformability plot of the complex depicted less mobility in the residues, while the analysed NMA was also less deviated from the B-factor of the complex (Fig. 8a–b). The computed eigenvalue of the complex structure was 3.165955e−05, which increased gradually in every mode (Fig. 8c). The variance plot analysis showed moderate decrease in the individual variance of every consecutive mode (Fig. 8d). Altogether, iMODS analysis concludes the decrease in mobility and deformability in the coordinates of the complex structure that signify the stability of vaccine-TLR3 complex interface.
Fig. 8.
iMODS analysis of the vaccine–TLR3 complex showing plots of a Deformability, b B-factor, c Eigenvalue and d NMA variance
In Silico Cloning
Vaccine construct was codon optimized by JCat server with the sequence length and GC content of 441 nucleotides and 50.73% respectively. As optimal range of GC content is 30-70%, our result shows higher probability of multi-epitope vaccine expression in bacterial system.
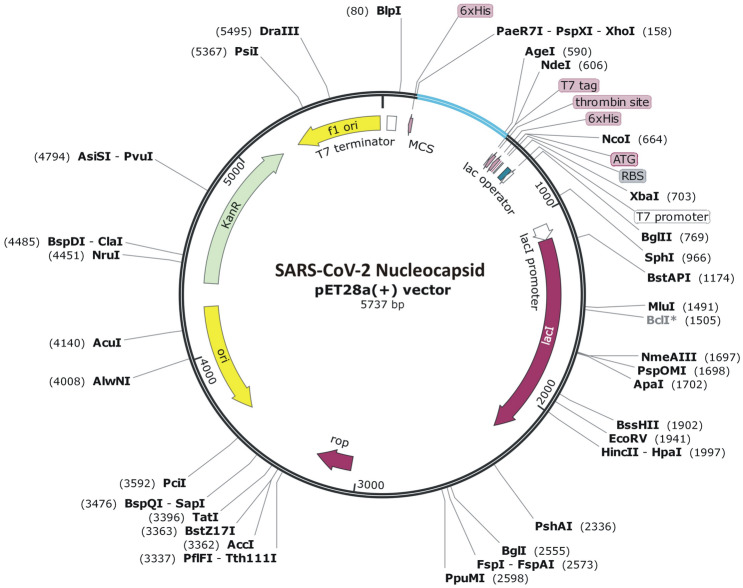
Concurrently, NdeI and XhoI were incorporated at respective N- and C-terminal of optimized DNA sequence and then these restriction sites were framed within multiple cloning sites (MCS) of pET-28a(+) expression vector (Fig. 9). Existence of poly-histidine affinity tag in pET-28a(+) plasmid presents it as universal vector for cloning and expression of recombinant proteins. Furthermore, virtual confirmation of clone using agarose gel simulation tool of SnapGene revealed presence of insert (~ 0.5 kb) along with vector (~ 5.5 kb) after digestion with NdeI and XhoI enzymes (Fig. 10). The size of insert was found to be in accordance of the calculated molecular weight of vaccine candidate.
Fig. 9.
In silico cloning of multi-epitope vaccine. Codon optimized gene sequence of the vaccine candidate (represented in cyan color) was cloned between NdeI and XhoI restriction sites of pET-28a(+) expression vector (shown as black circle) (Color figure online)
Fig. 10.
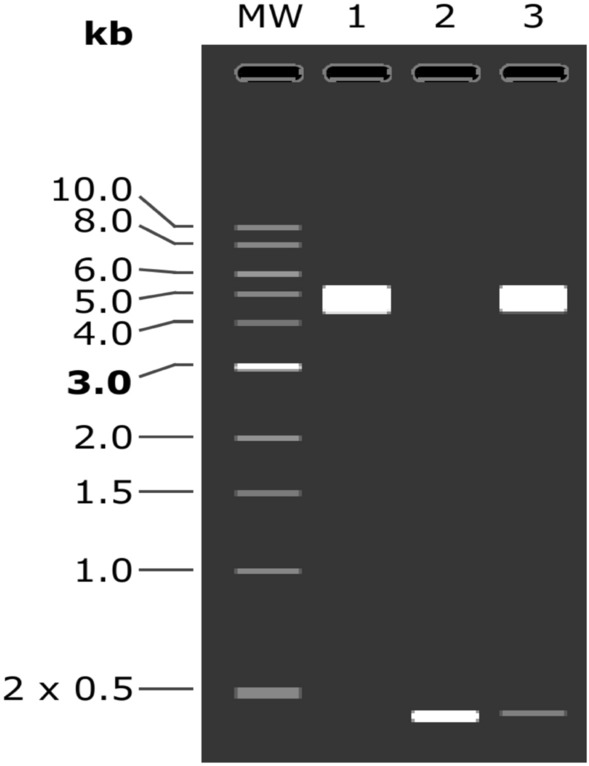
Virtual conformation of SARS-CoV-2 N protein vaccine clone by double digestion. Lane 1 represents native pET-28a(+) vector digested with NdeI and XhoI, Lane 2 indicates double digested insert (Vaccine) and Lane 3 shows double digestion of vaccine construct (Vaccine + Vector)
Discussion
Vaccine has been an effective and improved option as compared to drugs for the management of viral diseases. SARS-CoV-2 causes severe respiratory infections leading to millions of death worldwide. Due to negligible success through chemotherapy, the competition for vaccine production against COVID-19 is attaining a speedy pace globally. At present, many vaccines are in clinical trial that includes mRNA vaccine (LNP encapsulated) and vector based vaccine (Adenovirus Type-5), while many vaccines have failed during preclinical trials (WHO 2020). The immunoinformatics approach has been widely utilized to design novel and efficient epitope based vaccine against several virulent pathogens including Zika virus, Ebola virus etc. (Kumar Pandey et al. 2018; Ahmad et al. 2019). Consequently, it has also been employed to search for multi-epitopes from various proteins (including S, N, etc.) to design the vaccine candidate for COVID-19 (Ahmad et al. 2020; Kalita et al. 2020; Jyotisha et al. 2020; Enayatkhani et al. 2020). The N protein of the coronavirus is the most abundant, highly conserved and immunogenic protein. It contributes in packaging of viral genome into ribonucleopcapsid and plays very significant role for the period of self-assembly of virus (McBride et al. 2014). The middle and C-terminal portions of SARS-CoV N protein are known to play significant roles in antibody generation against SARS-CoV and interestingly SARS-CoV-2 nucleocapsid protein is analogous to that of SARS-CoV at level of protein sequence and three-dimensional structure. Notably, N protein of SARS-CoV-2 is observed to be antigenic and non-allergenic suggesting the generation of immune response without causing any side effect that makes it suitable for designing of effective vaccine.
Epitopes from viral protein are known to elicit the immune system to release various cytokines through helper T-lymphocytes, which leads to activation of cytotoxic T- lymphocytes and B-cell lymphocytes for the removal of pathogens (Channappanavar et al. 2014). Hence, owing to perform crucial tasks during immune response, CTL, HTL and BCL epitopes were employed in this study to acquire both humoral and cell-mediated immunity. On the basis of several parameters including antigenic, immunogenic, non-allergenic, non-toxic and number of MHC-I & II binding allele properties, five CTL and one HTL epitopes were selected from SARS-CoV-2 N protein. Importantly, selected HTL epitope was found to be immunogenic during in vitro T cell assays (Lee and Koohy 2020) and also one of the CTL epitopes (KTFPPTEPK) has been utilized in other recent reports to construct vaccine candidate for SARS-CoV-2 (Kalita et al. 2020; Mukherjee et al. 2020). Simultaneously, one linear B-cell epitope was selected on the basis of various criteria like antigenicity, allergenicity, surface accessibility and flexibility etc. The population coverage was high worldwide for combined MHC-I and MHC-II binding epitopes that advocates this vaccine candidate could be a potent tool against this fatal disease. COVID-19 has badly affected people throughout the world with India and United States being among the most affected countries. Significantly, our vaccine candidate has shown very high parentage of population coverage with 97.12% and 94.39% for India and United States, respectively. Simultaneously, no homolog of the selected epitopes was found in human to negate the cross-reactivity.
Consequently, the multi-epitope vaccine was constructed by amalgamating the selected BCL, HTL and CTL epitopes from SARS-CoV-2 N protein with appropriate adjuvant as well as linkers. As human β-defensin-2 (hBD-2) is known to enhance chemotactic activity for memory T cells, monocytes and immature dendritic cells (Duits et al. 2002), it was used as an adjuvant at N-terminus of vaccine to increase the antigenicity. In addition, it could protect the vaccines from degradation and also improve their delivery (Perrie et al. 2008). Simultaneously, suitable linkers were employed to connect the selected epitopes that improve the recognition of the vaccine candidate through increased stability and folding (Chen et al. 2013). Remarkably, the vaccine candidate was found to be non-allergenic, antigenic, stable after expression and highly soluble in the bacterial expression system. Higher solvent accessibility to the epitopes further reinforced the candidature of this vaccine construct due to the positive association between solvent accessible residues of vaccine protein and generation of profound humoral and cellular immune responses (Kar and Srivastava 2018). Thus, overall distinct outputs for various features confer this multi-epitope vaccine as an effective candidate against SARS-CoV-2.
The vaccine construct showed well defined secondary structure containing more α-helices and a tertiary structure highlighting proper conformation. Conformational B-cell epitopes are located proximally in the three-dimensional structure of the vaccine protein, but are discontinuous in the protein sequence with majority of the B-cell epitopes being conformational (Pellequer et al. 1991). We also found approximately 60% residues of constructed vaccine to be present in the conformational B-cell epitopes indicating towards a proficient antibody response against the virus. TLR3 is a vital immune receptor for host pathogenesis as it is involved in providing protective immunity. Moreover, generation of immune response majorly contributes to the pathophysiology of COVID-19 disease and TLR3 could be targeted to initiate anti-viral host immune response to eliminate SARS-CoV-2 related infection (Astuti and Ysrafil 2020). The molecular docking of multi-epitope vaccine and TLR3 immune receptor suggested high propensity of interaction, which can positively contribute in infection-inhibitory activity. Molecular dynamics simulation is an effective tool to evaluate the basis of the three-dimensional structure of any protein and its interaction with another molecule. The docked vaccine-TLR3 complex structure showed stability and compactness during MD simulation. Subsequent analysis revealed increment in the interactions and binding affinity at interface of the stable vaccine-TLR3 complex structure in comparison to initial complex. Furthermore, to increase the efficiency of transcription and translation, codon optimization of vaccine construct was performed to attain elevated vaccine expression in E. coli host (Makrides 1996). In addition, codon optimization was executed to provide the stability to vaccine candidate within the microbial host for maximum production followed by in silico cloning into bacterial expression vector.
Conclusion
Due to the lack of effective chemotherapy and unavailability of specific vaccine, COVID-19 has become the deadliest disease worldwide. Hence, a multi-epitope vaccine containing BCL, HTL and CTL epitopes was designed from nucleocapsid protein of SARS-CoV-2 using immunoinformatics approach. The selected epitopes belong to either middle region or C-terminal dimerization domain of SARS-CoV-2 N protein that are known to be the immunogenic regions, while most of the chosen T-cell epitopes have also been proved to be immunogenic through in vitro assays. Successively, designed vaccine candidate was found to be non-allergenic for host cells with high antigenicity, solubility and stability. Subsequent study highlights specific and stable interaction between vaccine and TLR3 immune receptor followed by its stable and efficient expression into the bacterial system. Therefore, our vaccine construct could elicit stronger memory response when administrated into the human body and will be capable to generate both cell-mediated and humoral immune responses against deadly SARS-CoV-2 worldwide.
Acknowledgements
The authors are thankful to Centre for Modelling Simulation & Design (CMSD), University of Hyderabad, Hyderabad for computational resources for MD simulation studies. We also acknowledge Jyotisha and Samayaditya Singh for discussion during this work.
Funding
No funding was received for this study.
Compliance with Ethical Standards
Conflict of interest
The authors declare that they have no conflict of interest.
Consent for Publication
All authors concur for the publication.
Ethics Approval
The article does not contain any studies on human/ animals.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Abraham MJ, Murtola T, Schulz R, Pall S, Smith JC, Hess B, Lindahl E. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2005;1(2):19–25. [Google Scholar]
- Ahlén G, Frelin L, Nikouyan N, Weber F, Höglund U, Larsson O, Westman M, Tuvesson O, Gidlund EK, Cadossi M, Appelberg S, Mirazimi A, Sällberg M, OPENCORONA Consortium The SARS-CoV-2 N protein is a good component in a vaccine. J Virol. 2020;94(18):e01279-20. doi: 10.1128/JVI.01279-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad B, Ashfaq UA, Rahman MU, Masoud MS, Yousaf MZ. Conserved B and T cell epitopes prediction of Ebola virus glycoprotein for vaccine development: an immuno-informatics approach. Microb Pathog. 2019;132:243–253. doi: 10.1016/j.micpath.2019.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad S, Navid A, Farid R, Abbas G, Ahmad F, Zaman N, Parvaiz N, Syed SS. Design of a novel multi epitope-based vaccine for pandemic coronavirus disease (COVID-19) by vaccinomics and probable prevention strategy against avenging zoonotics. Eur J Pharm Sci. 2020;151:105387. doi: 10.1016/j.ejps.2020.105387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astuti I, Ysrafil Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2): an overview of viral structure and host response. Diabetes Metab Syndr. 2020;14(4):407–412. doi: 10.1016/j.dsx.2020.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Channappanavar R, Zhao J, Perlman S. T cell-mediated immune response to respiratory coronaviruses. Immunol Res. 2014;59(1–3):118–128. doi: 10.1007/s12026-014-8534-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Zaro JL, Shen WC. Fusion protein linkers: property, design and functionality. Adv Drug Deliv Rev. 2013;65(10):1357–1369. doi: 10.1016/j.addr.2012.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WH, Strych U, Hotez PJ, Bottazzi ME. The SARS-CoV-2 vaccine pipeline: an overview. Curr Trop Med Rep. 2020;7:61–64. doi: 10.1007/s40475-020-00201-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong Y, Ulasli M, Schepers H, Mauthe M, V’kovski P, Kriegenburg F, Thiel V, de Haan C, Reggiori F. Nucleocapsid protein recruitment to replication-transcription complexes plays a crucial role in coronaviral life cycle. J Virol. 2020;94(4):e01925-19. doi: 10.1128/JVI.01925-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duits LA, Ravensbergen B, Rademaker M, Hiemstra PS, Nibbering PH. Expression of beta-defensin 1 and 2 mRNA by human monocytes, macrophages and dendritic cells. Immunology. 2002;106(4):517–525. doi: 10.1046/j.1365-2567.2002.01430.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta NK, Mazumdar K, Lee BH, Baek MW, Kim DJ, Na YR, Park SH, Lee HK, Kariwa H, Mai LQ, Park JH. Search for potential target site of nucleocapsid gene for the design of an epitope-based SARS DNA vaccine. Immunol Lett. 2008;118(1):65–71. doi: 10.1016/j.imlet.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta NK, Mazumdar K, Gordy JT. The nucleocapsid protein of SARS-CoV-2: a target for vaccine development. J Virol. 2020;94(13):e00647-20. doi: 10.1128/JVI.00647-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enayatkhani M, Hasaniazad M, Faezi S, Guklani H, Davoodian P, Ahmadi N, Einakian MA, Karmostaji A, Ahmadi K. Reverse vaccinology approach to design a novel multi-epitope vaccine candidate against COVID-19: an in silico study. J Biomol Struct Dyn. 2020 doi: 10.1080/07391102.2020.1756411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao W, Tamin A, Soloff A, D’Aiuto L, Nwanegbo E, Robbins PD, Bellini WJ, Barratt-Boyes S, Gambotto A. Effects of a SARS-associated coronavirus vaccine in monkeys. Lancet. 2003;362(9399):1895–1896. doi: 10.1016/S0140-6736(03)14962-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A, Sidney J, Zhang Y, Scheuermann RH, Peters B, Sette A. A Sequence homology and bioinformatic approach can predict candidate targets for immune responses to SARS-CoV-2. Cell Host Microbe. 2020;27(4):671–680. doi: 10.1016/j.chom.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jyotisha, Singh S, Qureshi IA. Multi-epitope vaccine against SARS-CoV-2 applying immunoinformatics and molecular dynamics simulation approaches. J Biomol Struct Dyn. 2020 doi: 10.1080/07391102.2020.1844060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalita P, Padhi AK, Zhang KYJ, Tripathi T. Design of a peptide-based subunit vaccine against novel coronavirus SARS-CoV-2. Microb Pathog. 2020;145:104236. doi: 10.1016/j.micpath.2020.104236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S, Yang M, Hong Z, Zhang L, Huang Z, Chen X, He S, Zhou Z, Zhou Z, Chen Q, Yan Y, Zhang C, Shan H, Chen S. Crystal structure of SARS-CoV-2 nucleocapsid protein RNA binding domain reveals potential unique drug targeting sites. Acta Pharm Sin. 2020;10(7):1228–1238. doi: 10.1016/j.apsb.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kar PP, Srivastava A. Immuno-informatics analysis to identify novel vaccine candidates and design of a multi-epitope based vaccine candidate against Theileria parasites. Front Immunol. 2018;9:2213. doi: 10.3389/fimmu.2018.02213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar Pandey R, Ojha R, Mishra A, Kumar Prajapati V. Designing B- and T-cell multi-epitope based subunit vaccine using immunoinformatics approach to control Zika virus infection. J Cell Biochem. 2018;119(9):7631–7642. doi: 10.1002/jcb.27110. [DOI] [PubMed] [Google Scholar]
- Kumari R, Kumar R, Open Source Drug Discovery Consortium. Lynn A. g_mmpbsa–a GROMACS tool for high-throughput MM-PBSA calculations. J Chem Inf Model. 2014;54:1951–1962. doi: 10.1021/ci500020m. [DOI] [PubMed] [Google Scholar]
- Lee CH, Koohy H. In silico identification of vaccine targets for 2019-nCoV. F1000Res. 2020;9:145. doi: 10.12688/f1000research.22507.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung DT, Tam FC, Ma CH, Chan PK, Cheung JL, Niu H, Tam JS, Lim PL. Antibody response of patients with severe acute respiratory syndrome (SARS) targets the viral nucleocapsid. J Infect Dis. 2004;190(2):379–386. doi: 10.1086/422040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makrides SC. Strategies for achieving high-level expression of genes in Escherichia coli. Microbiol Rev. 1996;60(3):512–538. doi: 10.1128/MR.60.3.512-538.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride R, van Zyl M, Fielding BC. The coronavirus nucleocapsid is a multifunctional protein. Viruses. 2014;6(8):2991–3018. doi: 10.3390/v6082991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu J, Xu J, Zhang L, Shu T, Wu D, Huang M, Ren Y, Li X, Geng Q, Xu Y, Qiu Y, Zhou X. SARS-CoV-2-encoded nucleocapsid protein acts as a viral suppressor of RNA interference in cells. Sci China Life Sci. 2020;63(9):1–4. doi: 10.3390/v6082991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S, Tworowski D, Detroja R, Mukherjee SB, Frenkel-Morgenstern M. Immunoinformatics and structural analysis for identification of immunodominant epitopes in SARS-CoV-2 as potential vaccine targets. Vaccines. 2020;8(2):290. doi: 10.3390/vaccines8020290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellequer JL, Westhof E, Van Regenmortel MH. Predicting location of continuous epitopes in proteins from their primary structures. Methods Enzymol. 1991;203:176–201. doi: 10.1016/0076-6879(91)03010-e. [DOI] [PubMed] [Google Scholar]
- Perrie Y, Mohammed AR, Kirby DJ, McNeil SE, Bramwell VW. Vaccine adjuvant systems: enhancing the efficacy of sub-unit protein antigens. Int J Pharm. 2008;364(2):272–280. doi: 10.1016/j.ijpharm.2008.04.036. [DOI] [PubMed] [Google Scholar]
- Petersen B, Lundegaard C, Petersen TN. NetTurnP–neural network prediction of beta-turns by use of evolutionary information and predicted protein sequence features. PLoS ONE. 2010;5(11):e15079. doi: 10.1371/journal.pone.0015079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan T. Novel coronavirus: from discovery to clinical diagnostics. Infect Genet Evol. 2020;79:104211. doi: 10.1016/j.meegid.2020.104211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan YJ, Wei CL, Ee AL, Vega VB, Thoreau H, Su ST, Chia JM, Ng P, Chiu KP, Lim L, Zhang T, Peng CK, Lin EO, Lee NM, Yee SL, Ng LF, Chee RE, Stanton LW, Long PM, Liu ET. Comparative full-length genome sequence analysis of 14 SARS coronavirus isolates and common mutations associated with putative origins of infection. Lancet. 2003;361(9371):1779–1785. doi: 10.1016/s0140-6736(03)13414-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Regenmortel MHV. Mapping epitope structure and activity: from one-dimensional prediction to four-dimensional description of antigenic specificity. Methods. 1996;9(3):465–472. doi: 10.1006/meth.1996.0054. [DOI] [PubMed] [Google Scholar]
- Wang M, Cao R, Zhang L, Yang X, Liu J, Xu M, Shi Z, Hu Z, Zhong W, Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30(3):269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO (2020) Draft landscape of COVID-19 candidate vaccines. https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines
- Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]