Abstract
In insect midgut, prostaglandins (PGs) play a crucial role in defending bacterial and malarial pathogens. However, little is known about the PG signalling pathway in the midgut. A dual oxidase (Se-Duox) with presumed function of catalysing reactive oxygen species (ROS) production in the midgut was identified in beet armyworm, Spodoptera exigua. Se-Duox was expressed in all developmental stages, exhibiting relatively high expression levels in the midgut of late larval instars. Se-Duox expression was upregulated upon bacterial challenge. RNA interference (RNAi) of Se-Duox expression significantly suppressed ROS levels in the midgut lumen. The suppression of ROS levels increased insecticidal activity of Serratia marcescens after oral infection. Interestingly, treatment with a PLA2 inhibitor prevented the induction of Se-Duox expression in response to bacterial challenge. On the other hand, addition of its catalytic product rescued the induction of Se-Duox expression. Especially, PG synthesis inhibitor significantly suppressed Se-Duox expression, while the addition of PGE2 or PGD2 rescued the inhibition. Subsequent PG signals involved cAMP and downstream components because specific inhibitors of cAMP signal components such as adenylate cyclase (AC) and protein kinase A (PKA) significantly inhibited Se-Duox expression. Indeed, addition of a cAMP analogue stimulated Se-Duox expression in the midgut. Furthermore, individual RNAi specific to PGE2 receptor (a trimeric G-protein subunit), AC, PKA or cAMP-responsive element-binding protein resulted in suppression of Se-Duox expression. These results suggest that PGs can activate midgut immunity via cAMP signalling pathway by inducing Se-Duox expression along with increased ROS levels.
Keywords: dual oxidase, prostaglandin, cAMP, reactive oxygen species, gut immunity, Spodoptera exigua
1. Introduction
The insect gut is usually exposed to various microbes. The insect foregut and hindgut are originated from embryonic ectoderm. They can protect insects from pathogen infection through their cuticle linings. The midgut lacking a cuticle barrier is known to be relatively susceptible to various microbial pathogens such as bacteria, viruses, nematodes and protozoa [1]. Instead of a cuticle layer, the midgut in many insects possesses a mucus layer and a peritrophic matrix that can act as physical barriers against microbial infections [2]. In addition, insect midguts possess commensal or mutualistic microbes required for assisting digestion, supplementing essential nutrients, detoxifying xenochemicals or defending against other pathogens [3].
In addition to these protective defence lines, insect midguts exhibit direct antimicrobial activities by producing reactive oxygen species (ROS) and antimicrobial peptides (AMPs) [4]. However, these chemical defences should be tightly regulated so that beneficial microbes could be protected in insect midguts, while pathogens could be selectively removed [5]. For example, peptidoglycans derived from invading bacterial pathogens can trigger AMP expression [6]. However, Caudal, a homeobox protein, can selectively repress the expression of IMD/NF-kB-dependent AMP genes to protect beneficial symbiotic bacteria in the midgut [7]. In addition, ROS are specifically produced by pathogens [8–10].
ROS are produced in insect midguts by two different kinds of enzymes: NADPH-dependent oxidase (Nox) and dual oxidase (Duox) [11]. Inducing activities of these enzymes is crucial for the regulation of toxic ROS production. Nox expression can be induced by lactic acid produced by anaerobic metabolism of pathogens [12], while Duox expression is regulated by uracil released from pathogens in Drosophila [10]. However, it remains unclear how pathogen-derived factors can activate oxidases to upregulate ROS levels.
Eicosanoids are a group of oxygenated C20 polyunsaturated fatty acids that can mediate various physiological processes including immune responses in metazoan animals [13]. In Spodoptera exigua, a lepidopteran insect, Nox gene expression and subsequent ROS production in haemocytes are regulated by eicosanoids [14]. In the midgut of Plutella xylostella, another lepidopteran insect, Duox gene expression and subsequent ROS production that are essential for defending against bacterial pathogens are inhibited by treatment with eicosanoid biosynthesis inhibitors [11]. These results suggest that eicosanoids can mediate ROS production by activating Nox or Duox in these lepidopteran insects. Among eicosanoids, PGs play crucial role in mediating various immune responses in midguts of lepidopteran and dipteran insects [11,15]. In particular, several PGs along with PGE2 receptor (PGR) and its downstream signal components have been found in S. exigua [16]. Thus, the hypothesis of this study was that PGE2 could mediate ROS production by inducing Duox expression in S. exigua through cAMP signalling pathway.
2. Results
2.1. Identification of Se-Duox and domain analysis
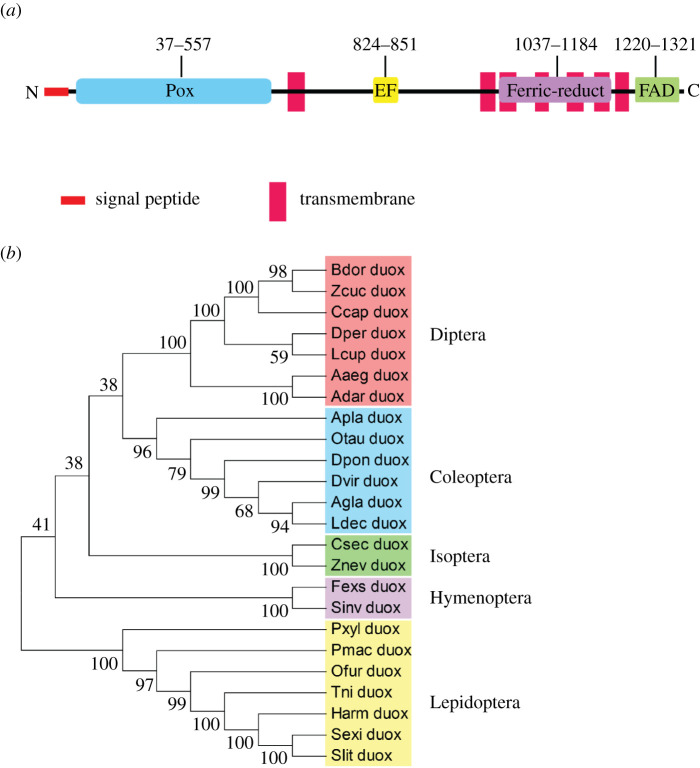
Transcriptome of S. exigua was interrogated with Duox gene of Drosophila melanogaster. A highly matched sequence (Se-Duox) encoding 1594 amino acid residues (figure 1) was found. It was predicted to be a transmembrane protein because of the presence of a signal peptide and transmembrane domains (figure 1a). It contains two oxidase domains (peroxidase at the N terminus and FAD-linked oxidase at C-terminus). When Se-Duox was phylogenetically analysed with several insect Duox genes, it formed a monophyletic cluster with other lepidopteran orthologues (figure 1b).
Figure 1.
Molecular characterization of Se-Duox. (a) Functional domain analysis of Se-Duox. Domains were predicted using HMMER (https://www.ebi.ac.uk) and Pfam (http://pfam.xfam.org). Predicted domains include ‘Pox’ for peroxidase, ‘EF’ for calcium-binding EF hand, ‘Ferric-reduct’ for ferric chelate reductase, ‘FAD’ for FAD-binding domain and ‘NAD’ for NAD-binding domain. (b) Phylogenetic analysis of Se-Duox with other insect dual oxidases based on their amino acid sequences. The tree was generated by the neighbour-joining method using MEGA 6.0. Bootstrapping values were obtained with 1000 repetitions to support branch and clustering. Amino acid sequences were retrieved from GenBank. Accession numbers of genes are shown in electronic supplementary material, table S2.
2.2. Inducible expression of Se-Duox upon bacterial challenge
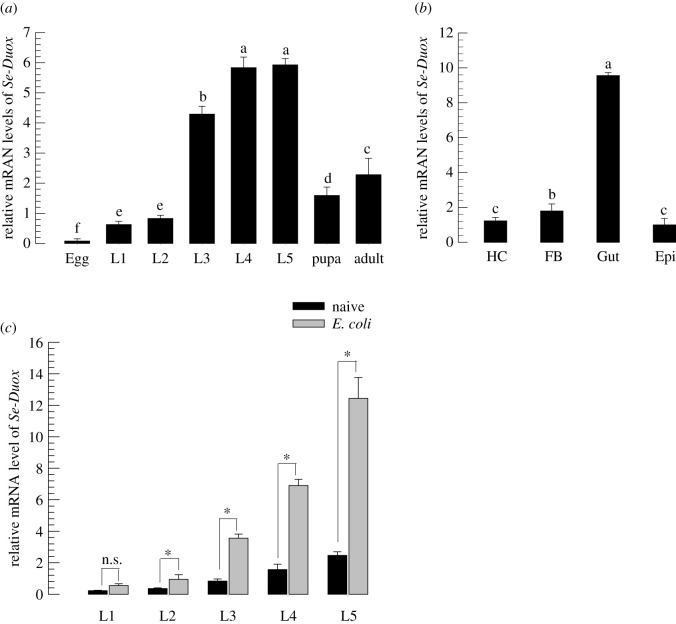
Se-Duox was expressed in all developmental stages of S. exigua (figure 2a). It was highly expressed in late larval instars. In the last larval instar (L5), Se-Duox was highly expressed in the midgut (figure 2b). Bacterial challenge significantly (p < 0.05) increased its gene expression levels in all larval instars (figure 2c).
Figure 2.
Expression profile of Se-Duox. (a) Expression patterns of Se-Duox in different developmental stages: egg, first to fifth instar larvae (‘L1–L5’), pupa and adult. (b) Expression patterns of Se-Duox in indicated tissues of L5D2 larvae, including haemocyte (HC), fat body (FB), midgut (Gut) and epidermis (Epi). (c) Induction of Se-Duox expression in response to bacterial challenge. L1–L5 larvae were fed with E. coli (2 × 104)-treated artificial diet for 8 h. Midguts were then dissected to study expression patterns of Se-Duox. A ribosomal gene, RL32, was used as a reference gene. Each treatment was replicated three times with independent tissue preparations. Different letters above standard deviation bars indicate significant differences among means at Type I error = 0.05 (LSD test).
2.3. RNA interference of Se-Duox suppresses ROS level and enhances bacterial pathogenicity
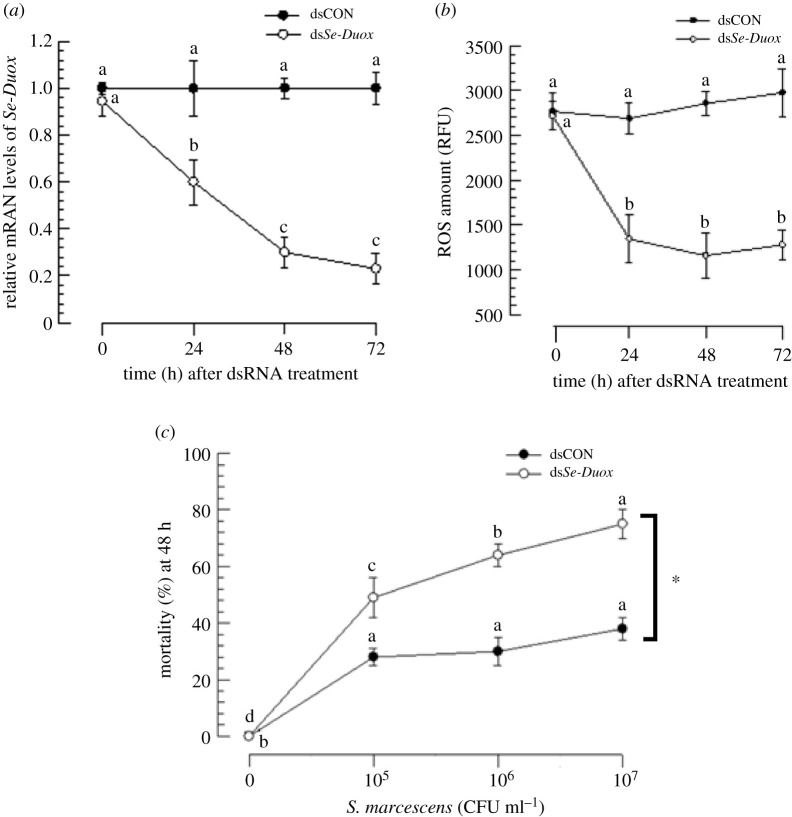
Injection of dsRNA specific to Se-Duox to L5 larvae significantly (p < 0.05) suppressed its expression. Such RNA interference (RNAi) efficiency was maintained for at least 72 h PI (figure 3a). Along with decrease in Se-Duox expression, ROS levels in the gut lumen were also significantly (p < 0.05) reduced (figure 3b). Such a decrease in ROS levels increased insecticidal activity of S. marcescens after oral administration (figure 3c).
Figure 3.
RNAi of Se-Duox expression and subsequent influence on ROS level and larval susceptibility to S. marcescens. (a) Effects of RNAi on Se-Duox expression at different time points in midguts of S. exigua larvae (L5). One microgram of gene-specific dsRNA (dsSe-Duox) was injected into each larva. CpBV302, a viral gene, was used to generate control dsRNA (dsCON). (b) Inhibitory effects of RNAi specific to Se-Duox expression on total ROS levels in the midgut. To induce Duox expression and ROS, after dsRNA treatments, larvae were fed with E. coli (2 × 104 cells) treated artificial diet for 12 h. (c) Effects of Se-Duox RNAi on pathogenicity of S. marcescens against L4 larvae of S. exigua. RNAi-treated larvae were exposed to different concentrations of S. marcescens. Mortality was recorded at 48 h post feeding. Each treatment was replicated three times. Each replication used 10 larvae. Different letters above standard deviation bars indicate significant difference among means in each treatment and control at Type I error = 0.05 (LSD test). Asterisk indicates the statistical difference between control and treatment at 107 CFU ml−1 dose.
2.4. PGs activate Se-Duox expression
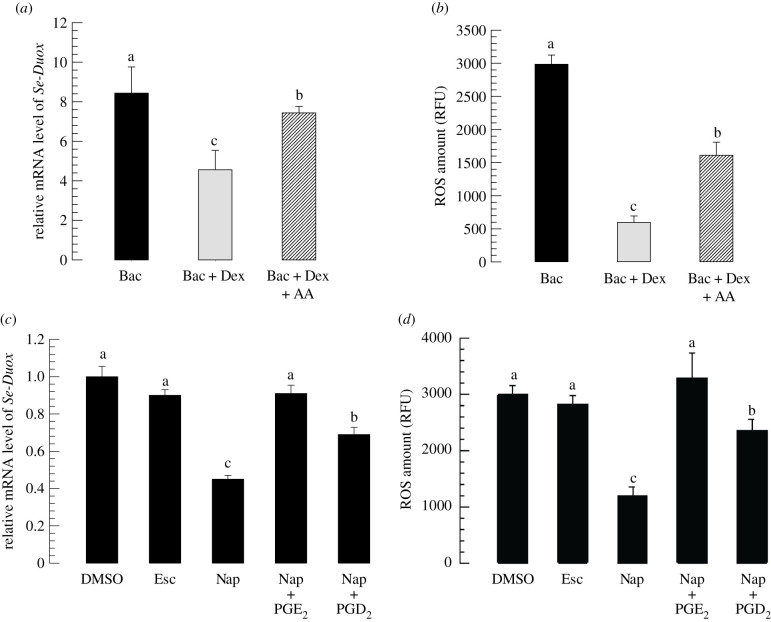
Next, the effect of eicosanoids on induction of Se-Duox expression upon bacterial challenge was analysed. PLA2 activity was significantly (p < 0.05) inhibited by its specific inhibitor dexamethasone (Dex). Bacterial induction of Se-Duox expression was reduced by such inhibitor treatment (figure 4a). However, an addition of arachidonic acid (‘AA’, a catalytic product of PLA2) to Dex-treated larvae significantly (p < 0.05) rescued such inhibition of Se-Duox expression. ROS levels in gut lumen were highly dependent on Se-Duox expression levels. Dex treatment suppressed ROS levels, whereas the addition of AA rescued ROS levels suppressed by Dex treatment (figure 4b).
Figure 4.
Effects of PLA2 inhibitor and COX and LOX inhibitors on expression levels of Se-Duox. (a) Inhibitory effect of a PLA2 inhibitor, dexamethasone (DEX), on Se-Duox expression in L5 larvae. For bacterial challenge, larvae were fed with E. coli (2 × 104 cells/larva) treated artificial diet for 12 h. (b) Effects of DEX on ROS levels in gut lumen. To induce ROS, E. coli (2 × 104 cells)-treated artificial diet was fed to larvae at 8 h post-injection of DEX. At 12 h after bacterial treatment, intestinal ROS levels were measured. (c) Influence of naproxene (‘Nap’, a COX inhibitor) and esculetin (‘Esc’, a LOX inhibitor) on expression of Se-Duox. (d) Effects of Nap and Esc on ROS levels in gut lumen. Each inhibitor was injected into larvae at a final concentration of 10 µg per larva. At 8 h post-injection of inhibitor, E. coli (2 × 104 cells)-treated artificial diet were fed to larvae to induce Se-Duox expression and ROS. To rescue Se-Duox expression and ROS production, PGD2 or PGE2 was injected at 1 µg/larva. Each treatment was replicated three times. Each replication used 15 larvae. Different letters indicate significant differences among means at Type I error = 0.05 (LSD test).
Modulation of Se-Duox expression by Dex or AA treatment suggested that Se-Duox expression could be controlled by eicosanoids. To clarify the types of eicosanoids that could modulate Se-Duox expression, esculetin (Esc) known to inhibit LOX or naproxen (Nap) known to inhibit COX was subjected to assessment for their abilities to modulate Se-Duox expression (figure 4c). Both inhibitors significantly (p < 0.05) suppressed Se-Duox expression, with Nap being much more potent than Esc. Inhibited expression of Se-Duox after Nap treatment was rescued by the addition of PGE2 or PGD2. ROS level in the gut lumen was downregulated by Nap treatment (figure 4d). However, the suppressed ROS level caused by Nap treatment was significantly (p < 0.05) rescued by the addition of PGE2 or PGD2.
2.5. cAMP signal mediates Se-Duox expression
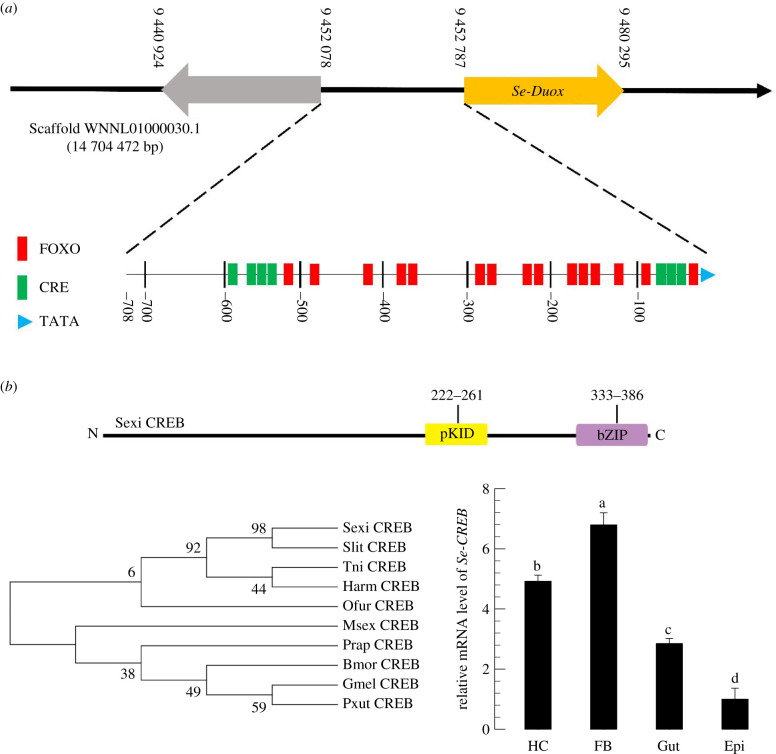
PGR of S. exigua can activate cAMP secondary messenger [16]. To predict the involvement of cAMP signal pathway in modulating Se-Duox expression, the upstream region (708 bp) of Se-Duox open reading frame (ORF) was analysed to determine the presence of any cAMP-related promoter element (figure 5a). In addition to the TATA box, the putative promoter region contained cAMP-responsive elements (CREs) at 0 to −100 bp and −500 to −600 bp. The presence of CRE in the promoter region suggested a functional interaction with CRE-binding protein (CREB). CREB of S. exigua (Se-CREB) was predicted from the transcriptome of S. exigua (GenBank accession number: WNNL01000001.1) after interrogating with CREB gene of S. litura (NCBI GenBank accession number: XM_022964340.1). Its sequence (390 amino acids) was highly homologous to other CREB genes (figure 5b). This predicted Se-CREB was found to be expressed in different larval tissues (figure 5c).
Figure 5.
Identification of CRE and CREB associated with Se-Duox. (a) Promoter analysis of dual oxidase gene of S. exigua using PROMO and GPMiner programs. The upstream region from ATG start codon of Se-Duox was analysed for promoter components. (b) Functional domain analysis and phylogenetic analysis of Se-CREB with other lepidopteran CREBs based on their amino acid sequences. Domains were predicted using HMMER (https://www.ebi.ac.uk) and Pfam (http://pfam.xfam.org). Predicted domains include ‘pKID’ for phosphorylated kinase-inducible-domain and ‘bZIP’ for basic leucine zipper. The tree was generated by the neighbour-joining method using MEGA 6.0. Bootstrapping values were obtained with 1000 repetitions to support branch and clustering. Amino acid sequences were retrieved from GenBank. Accession numbers of genes are shown in electronic supplementary material, table S3. (c) Expression patterns in indicated tissues of L5D2 larvae, including haemocyte (HC), fat body (FB), midgut (Gut) and epidermis (Epi). A ribosomal gene, RL32, was used as a reference gene. Each treatment was replicated three times with independent tissue preparations. Different letters above standard deviation bars indicate significant differences among means at Type I error = 0.05 (LSD test).
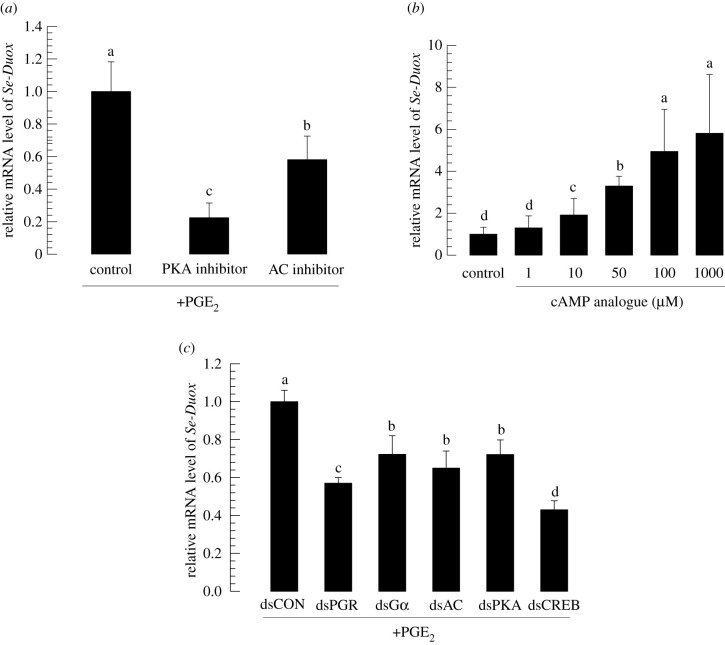
Specific inhibitors against adenylate cyclase (AC) producing cAMP and protein kinase A (PKA) phosphorylating proteins in response to cAMP were then used to treat larvae of S. exigua (figure 6a). Larval midguts from treated larvae expressed Se-Duox at significantly lower levels compared with those from control larvae without such treatment. By contrast, the administration of cAMP analogue (8-(4-chlorophenylthio)adenosine 3′,5′-cyclic monophosphate sodium salt) increased Se-Duox expression in a dose-dependent manner (figure 6b).
Figure 6.
Se-Duox expression is regulated by PKA signalling pathway. (a) Inhibitory effects of PKA and AC inhibitors on Se-Duox expression in fifth instar larvae (L5). (b) Dose-dependent effect of cAMP analogue on Se-Duox expression in L5. For bacterial challenge and induction of Duox expression, at 8 h post-injection of inhibitors or cAMP analogue, larvae were fed with E. coli (2 × 104 cells)-treated artificial diet for 12 h. (c) Downregulatory effects of RNAi specific to PKA signalling pathway components on expression levels of Se-Duox. At 48 h after dsRNA injection, larvae were fed with E. coli (2 × 104 cells)-treated artificial diet for 12 h to induce Duox expression. Each treatment was replicated three times. Each replication used five larvae. Different letters indicate significant differences among means at Type I error = 0.05 (LSD test).
The role of cAMP in modulating Se-Duox expression suggested a signalling pathway from PGE2 to Se-Duox expression via PGR, Gαs, AC, PKA and CREB. All these five genes were downregulated after injecting their gene-specific dsRNAs (electronic supplementary material, figure S1). When Se-Duox expression was assessed at 72 h after each dsRNA treatment, its expression levels were significantly reduced after RNAi treatment (all five gene-specific dsRNAs) compared with those in controls (figure 6c).
3. Discussion
This study focused on gut immunity associated with ROS production. To this end, we identified a Se-Duox gene through bioinformatics prediction. Its expression was confirmed in all developmental stages and different larval tissues. Functional domains of Se-Duox contain two different oxidases, like other Duox genes [8]. Such domain prediction results suggest a pathway of ROS production in midgut epithelium. It begins at the intracellular oxidase domain by extracting two electrons from NADPH + H+. These electrons are then delivered to haem structures in the transmembrane domain for forming superoxide and hydrogen peroxide in the extracellular (luminal) side of the midgut. Hydrogen peroxide is then subjected to the catalytic activity of peroxidase to generate microbicidal HOCl. Mutant Duox lacking the N-terminal peroxidase domain results in severely impaired host defence system [17]. Our current study showed that RNAi specific to Se-Duox suppressed ROS level. Suppressed ROS level after RNAi treatment caused larvae to become susceptible to bacterial infection. This suggests that Se-Duox activity is required for immune defence but should be tightly controlled because excessive ROS production by uncontrolled Duox activity would be detrimental to insect midgut tissues. In Drosophila, Duox activity is activated through increased gene expression or post-translational modification [9]. Uracil derived from pathogenic bacteria can bind to G-protein-coupled receptor and trigger Gαq-PLCβ-Ca2+ pathway to activate MEKK1–MKK3–p38 MAPK with subsequent upregulation of Duox gene expression or increased enzymatic activity through Ca2+-binding domain of Duox. By contrast, Duox in another lepidopteran host, P. xylostella, is controlled by eicosanoid biosynthesis inhibitor [11], suggesting that there might be a cross-talk between Gαq-PLCβ-Ca2+ pathway and eicosanoid signalling.
Se-Duox expression was dependent on PLA2 activity. The inhibition of PLA2 activity suppressed Se-Duox expression. Such suppression in the expression of Se-Duox was then rescued by the addition of AA, suggesting that AA and/or its oxygenated eicosanoids could mediate gene expression of Se-Duox. To clarify the types of eicosanoids involved in the regulation, esculetin (leukotriene biosynthesis inhibitor) or naproxen (prostaglandin biosynthesis inhibitor) was used for treatment. The higher inhibitory activity of naproxen against Se-Duox expression suggested the role of PGs in mediating the gene expression. This was further supported by the rescue effect of PGE2 or PGD2. A variety of PGs other than PGE2 and PGD2 along with epoxyeicosatrienoic acids (EETs) are not assessed in this current study, and thus it remains that other types of PGs and EETs may mediate Se-Duox induction.
Changed PLA2 activities by its specific inhibitor, dexamethasone (DEX), modulated ROS production, suggesting that PGE2 could activate Se-Duox to produce ROS in the midgut. To explain this pathway, we speculate that MAPK p38 could mediate ROS production via activation of Duox gene expression in Drosophila's midgut [18]. In mammalian intestine, MAPK p38 can also activate COX-2 to elevate PGE2 levels [19], suggesting that MAPK p38 plays a role in mediating the cross-talk between the Gαq-PLCβ-Ca2+ pathway and eicosanoid signal to produce ROS. Thus, we wondered how the upregulated level of PGE2 could activate Se-Duox expression.
Promoter analysis of Se-Duox gene indicated the presence of a CRE at the upstream region of Se-Duox ORF. PGR of S. exigua can transmit its signal using cAMP [16]. To determine the significance of CRE, this study identified a CRE-binding protein (Se-CREB) of S. exigua. Its sequence exhibited high homologies with other known CREB genes. The presence of a highly conserved serine residue was predicted to be activated after phosphorylation by PKA [20]. The leucine zipper motif in the α-helix was predicted to form a characteristic leucine zipper motif, which binds to CRE for transactivation [21]. Se-CREB was expressed in all developmental stages and different tissues of S. exigua. These results suggest a cAMP signal pathway of S. exigua from PGR to CREB via Gαs, AC and PKA signalling components.
Specific inhibitors against AC or PKA prevented the induction of Se-Duox expression. By contrast, the addition of cAMP analogue significantly induced the expression of Se-Duox in a dose-dependent manner. Thus, cAMP signalling pathway is likely to mediate Se-Duox expression in response to PGE2 in S. exigua. Indeed, RNAi of PGR (= PGE2 receptor) gene expression prevented the induction of Se-Duox gene. Furthermore, the cAMP signalling pathway to induce Se-Duox expression was supported by suppressed Se-Duox expression after individual RNAi treatments of its signalling components. Alternatively, calcium signalling pathway may play a crucial role in activating Duox enzyme activity [4]. Especially, Se-Duox contains EF domain suggesting its catalytic activity depending on Ca2+. A specific PG receptor in oenocytoi haemocytes of S. exigua activates calcium signalling pathway [13]. PG-Ca2+-Duox activity needs to be clarified in the subsequent study.
This study showed that Se-Duox expression in the gut was mediated by PGE2 via cAMP secondary messenger (figure 7). MAPK p38 induced by pathogenic bacterial infection might induce PG production via a COX-like enzyme in S. exigua. A specific peroxidase called peroxynectin (Pxt) is known to mediate PG production in S. exigua [22]. PGE2, a main PG member in S. exigua [16], can activate the cAMP signal via its specific receptor, leading to the expression of Se-Duox to produce ROS in the midgut to defend microbial pathogens. PGE2 is produced in midguts of different insects including Manduca sexta and Helicoverpa zea (two lepidopteran insects), Periplanata americana (cockroach) and Anopheles gambiae (mosquito) [15,23]. In addition to ROS production caused by induced Duox expression, PGE2 can also mediate other gut immune responses by recruiting haemocytes to infection foci [15] and activating gene expression of antimicrobial peptides [24] in mosquitoes. All these immune mediation by PGE2 should be preceded by pathogen recognition in the midgut. This study suggests that uracil-based Gαq-PLCβ-Ca2+ recognition signal can activate eicosanoid signal pathway via p38 MAPK in S. exigua. Further study is needed to identify a specific p38 kinase that can activate Pxt to produce PGE2.
Figure 7.
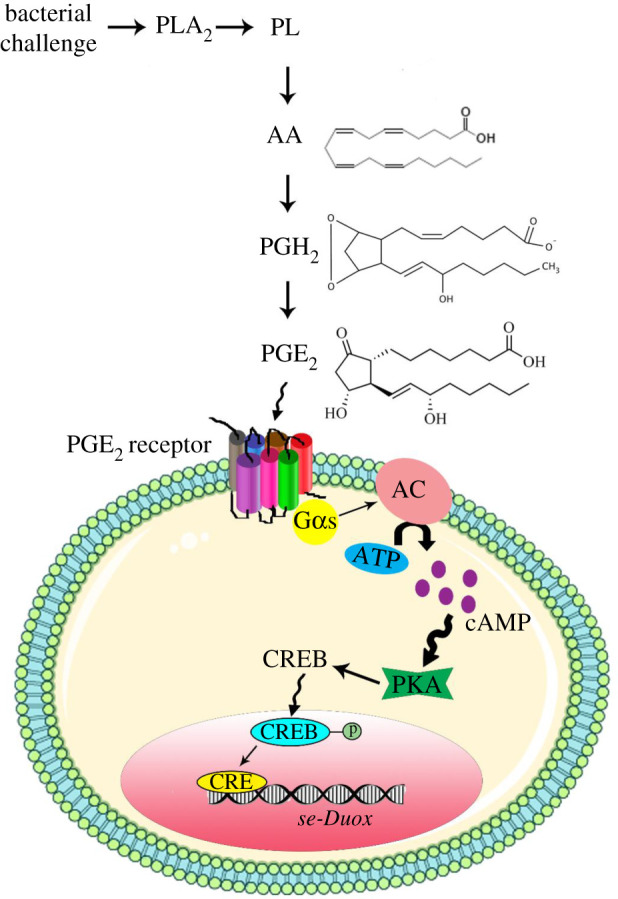
A schematic of Se-Duox expression under the control of PKA signalling pathway. In response to bacterial challenge, PGE2 receptor on the cell surface is activated. AC can increase the amount of cAMP in the cytosol which induces PKA. Induced PKA causes CREB phosphorylation and subsequent translocation to the nucleus. CREB in the nucleus acts as a transcription factor and upregulates Se-Duox expression.
4. Methods
4.1. Insect rearing and bacterial culture
All experiments were conducted with a lepidopteran model, S. exigua larvae. Rearing of S. exigua larvae followed the method described previously [25]. Under rearing conditions, S. exigua underwent five larval instars (L1–L5) before pupation. Escherichia coli Top10A, a Gram-negative bacterium, was obtained from Invitrogen (Carlsbad, CA, USA) and cultured overnight in Luria-Bertani (LB) medium at 37°C. Serratia marcescens, a Gram-negative bacterium, was cultured in nutrient broth medium for 20 h at 28°C in a shaking (200 rpm) incubator. After centrifuging the cultured broth at 8000g for 20 min, bacterial cells were collected with 100 mM phosphate-buffered saline (PBS, pH 7.4) and used for immune challenge. Escherichia coli (2 × 104 cells larva−1) and S. marcescens (105, 106 and 107 colony-forming unit (CFU) ml−1) were used for immune challenge through feeding assays after dipping food/diet in bacterial suspensions. For each bacterial concentration, 10 test larvae were used. Each treatment was replicated three times.
4.2. Chemicals
Arachidonic acid (AA: 5,8,11,14-eicosatetraenoic acid), dexamethasone (DEX: (11β,16α)-9-fluoro-11,17,21-trihydroxy-16-methylpregna-1,4-diene-3), naproxen (Nap: (S)-(+)-6-methoxy-α-methyl-2-naphthaleneacetic acid), esculetin (Esc: 6,7-dihydroxycoumarin) and cAMP analogue (8-(4-chlorophenylthio)adenosine 3′,5′-cyclic monophosphate sodium salt) were purchased from Sigma-Aldrich Korea (Seoul, Korea). Prostaglandin D2 (PGD2: 9α,15S-dihydroxy-11-oxo-prosta-5Z,13E-dien-1-oic acid) and prostaglandin E2 (PGE2: 9-oxo-11α,15S-dihydroxy-prosta-5Z,13E-dien-1-oic acid) were purchased from Cayman Chemical (Ann Arbor, MI, USA). PKA inhibitor H89 dihydrochloride (N-2-3-(4-bromophenyl)-2-propenylaminoethyl-5-isoquinolinesulfonamide dihydrochloride) was purchased from Tocris (Bristol, UK). Adenylyl cyclase type V inhibitor (2-amino-7-(furanyl)-7,8-dihydro-5(6H)-quinazolinone) was purchased from Calbiochem (San Diego, CA, USA). All these chemicals were dissolved in dimethyl sulfoxide (DMSO).
4.3. Midgut tissue preparation
Midguts were isolated from larval instars (L1–L5), pupae (2–3 days old) and adults (2–3 days old). For L1–L2 stages, 50 individuals were used to collect a midgut sample. For L3–L4 stages, 10 individuals were used. By contrast, only one individual was used to collect a midgut sample from L5 larva, pupa or adult.
4.4. Bioinformatics
Duox sequence (NCBI GenBank accession number: NM_134871.3) of D. melanogaster was used to obtain its orthologue (Se-Duox) from S. exigua using whole-genome shotgun database deposited at GenBank with BlastN search engine. To localize the promoter region of Se-Duox, a scaffold (accession number: WNNL01000030.1) was subjected to analysis with FGENESH (http://www.softberry.com) to predict gene loci. Putative promoter (708 bp upstream from ATG start codon) of Se-Duox was analysed using PROMO (http://alggen.lsi.upc.es). GPMiner (http://gpminer.mbc.nctu.edu.tw) was used to identify putative transcription factor-binding sites. Protein domains of Se-Duox were predicted using HMMER (https://www.ebi.ac.uk) and Pfam (http://pfam.xfam.org). Phylogenetic analyses and phylogenetic tree construction with the neighbour-joining method were performed using MEGA6 and ClustalW programs. Bootstrapping values were obtained with 1000 repetitions to support branching and clustering.
4.5. RNA extraction and cDNA construction
Total RNAs were extracted from all developmental stages of S. exigua (egg stage, more than 1000 eggs were used for each extracton; L1, 30 larvae; L2, 20 larvae; L3, 10 larvae; L4, three larvae; L5, one larva; one pupa; one adult) using Trizol reagent (Invitrogen) according to the manufacturer's instruction. Different tissue samples were isolated from L5 larvae, in which fat body, midgut and epidermis were collected from five larvae as an experimental unit. Haemocytes were collected from 30 larvae per replication. Extracted RNAs were dissolved in 50 µl of diethyl pyrocarbonate-treated deionized and distilled water. First-strand cDNAs were synthesized from 1 µg of RNAs using Maxime RT PreMix (Intron Biotechnology, Seoul, Korea) containing oligo dT primers according to the manufacturer's instruction.
4.6. RT-PCR and RT-qPCR
Synthesized cDNAs were used as templates for PCR amplification or for constructing dsRNAs. Real-time quantitative polymerase chain reaction (qPCR) was carried out in a total volume of 20 µl consisting of 2× SYBR Green Realtime PCR Master Mix (Toyobo, Osaka, Japan), 5 mM of gene-specific forward and reverse primers and 80 ng cDNA as template. PCR amplification was performed at 95°C for 10 min as an initial heat treatment step followed by 40 cycles of 98°C for 30 s, 52°C for 30 s and 72°C for 30 s. It was then finished with a final extension step at 72°C for 7 min. As an endogenous control for constitutive expression, a stably expressed ribosomal gene, RL32, was assessed along with test samples. Melting curves of products were obtained to confirm amplification specificity. A comparative CT method [26] was used to estimate relative gene expression. All experiments were replicated three times with independent samples of test stages or tissues. All primer sequences used in this study for RT-PCR and RT-qPCR are presented in electronic supplementary material, table S1.
4.7. dsRNA preparation and RNAi
Double-stranded RNAs (dsRNAs) specific to Se-Duox and other genes were prepared as described previously [11]. Briefly, a partial Se-Duox sequence was produced by PCR using gene-specific primers containing a T7 promoter sequence at the 5′ end (electronic supplementary material, table S1). The PCR product was used as a template to generate dsRNA using a MEGAscript RNAi kit (Ambion, Austin, TX, USA) according to the manufacturer's instruction. Sense and antisense RNA strands were synthesized using T7 RNA polymerase at 37°C for 4 h. A control dsRNA (dsCON) was also prepared by synthesizing 520 bp fragment dsRNA of CpBV302, a viral gene. The resulting dsRNA was purified and mixed with transfection reagent Metafectene PRO (Biontex, Plannegg, Germany) at a ratio of 1 : 1 (v/v) followed by incubation at 25°C for 30 min to form liposomes. One microgram of dsRNA was injected to larval haemocoel (3-day-old L4 larvae) using a microsyringe (Hamilton, Reno, NV, USA) equipped with a 26 gauge needle. At 24, 48 and 72 h post-injection (PI), RNAi efficacies were determined using RT-qPCR as described above. At 48 h PI, treated larvae were used for immune challenge and measurement of Se-Duox expression after an oral infection. Each treatment was replicated three times.
4.8. ROS measurement
ROS quantification was conducted using an OxiSelect Intracellular ROS Assay Kit (cat. no. STA-342, Cell Biolabs Inc., San Diego, CA, USA). After squeezing out contents of the dissected midguts, remaining gut tissues were then washed twice with 1 ml of PBS by repetitive resuspension followed by centrifugation at 800g for 5 min. Washed tissues were then suspended in TC-100 cell culture medium containing 0.1× DCFH-DA (dichlorofluorescein diacetate, 20× DCFH-DA stock, Part no. 234201) and incubated at 37°C for 30 min with gentle mix (inverting). After washing tissues three times with PBS, tissues were dissolved in 500 µl of Cell Lysis Buffer (Part no. 234203) diluted with TC100 cell culture medium and incubated at room temperature for 5 min. Cell lysate (150 µl) was then transferred to a 96-well plate and fluorescence was read at emission wavelength of 530 nm after excitation at 480 nm. For extracellular ROS measurement, DCFH-DA was mixed with midgut content and incubated at 37°C for 30 min. A calibration curve was drawn using serial dilutions of DCF (dichlorofluorescein) standard (part no. 234202) in TC-100 cell culture medium.
4.9. Applications of different inhibitors
All inhibitors used in this study were dissolved in DMSO at 10 µM and injected into L5 larvae (1 µl larva−1). At 8 h PI, treated larvae were used to estimate Se-Duox expression levels. For oral administration, gene expression analysis was performed at 12 h after feeding treatment along with the bacterial challenge.
4.10. Data analysis
All data are presented as mean ± s.e. and plotted with Sigma plot. To determine statistical differences, means were compared with Fisher's least squared difference (LSD) test and discriminated at Type I error = 0.05. Significant differences in RNAi efficiency were tested using the t-test of Sigma plot software v. 12.5. A p-value of less than 0.05 was considered statistically significant. All experiments were performed using three independent biological replicates.
Supplementary Material
Acknowledgements
The authors would like to thank Ahmed Shabbir for revision of figures.
Data accessibility
This article has no additional data.
Authors' contributions
Y.K. designed and supervised the research. S.M.S. performed the research and wrote the manuscript.
Competing interests
We declare we have no competing interests.
Funding
This work was supported by a grant no. (2017R1A2133009815) of the National Research Foundation (NRF) funded by the Ministry of Science, ICT and Future Planning (MSIP), Republic of Korea.
References
- 1.Wu K, Yang B, Huang W, Dobens L, Song H, Ling E. 2016. Gut immunity in lepidopteran insects. Dev. Comp. Immunol. 64, 65–74. ( 10.1016/j.dci.2016.02.010) [DOI] [PubMed] [Google Scholar]
- 2.Terra WR, Dias RO, Oliveira PL, Ferreira C, Venancio TM. 2018. Transcriptomic analyses uncover emerging roles of mucins, lysosome/secretory addressing and detoxification pathways in insect midguts . Curr. Opin. Insect Sci. 29, 34–40. ( 10.1016/j.cois.2018.05.015) [DOI] [PubMed] [Google Scholar]
- 3.Engel P, Moran NA. 2013. The gut microbiota of insects-diversity in structure and function. FEMS Microbiol. Rev. 37, 699–735. ( 10.1111/1574-6976.12025) [DOI] [PubMed] [Google Scholar]
- 4.Lee KA, et al. 2018. Inflammation-modulated metabolic reprogramming is required for DUOX-dependent gut immunity in Drosophila. Cell Host Microbe 23, 338–352. ( 10.1016/j.chom.2018.01.011) [DOI] [PubMed] [Google Scholar]
- 5.Lemaitre B, Miguel-Aliaga I. 2013. The digestive tract of Drosophila melanogaster. Annu. Rev. Genet. 47, 377–404. ( 10.1146/annurev-genet-111212-133343) [DOI] [PubMed] [Google Scholar]
- 6.Crava CM, Jakubowska AK, Escriche B, Herrero S, Be Y. 2015. Dissimilar regulation of antimicrobial proteins in the midgut of Spodoptera exigua larvae challenged with Bacillus thuringiensis toxins or baculovirus. PLoS ONE 10, e0125991 ( 10.1371/journal.pone.0125991) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ryu JH, et al. 2008. Innate immune homeostasis by the homeobox gene caudal and commensal-gut mutualism in Drosophila. Science 319, 777–782. ( 10.1126/science.1149357) [DOI] [PubMed] [Google Scholar]
- 8.Bae YS, Choi MK, Lee WJ. 2010. Dual oxidase in mucosal immunity and host-microbe homeostasis. Trends Immunol. 31, 278–287. ( 10.1016/j.it.2010.05.003) [DOI] [PubMed] [Google Scholar]
- 9.Ha EM, Lee KA, Park SH, Kim SH, Nam HJ, Lee HY, Kang D, Lee WJ. 2009. Regulation of DUOX by the Gαq-phospholipase Cβ-Ca2+ pathway in Drosophila gut immunity. Dev. Cell 16, 386–397. ( 10.1016/j.devcel.2008.12.015) [DOI] [PubMed] [Google Scholar]
- 10.Lee KA, et al. 2013. Bacterial-derived uracil as a modulator of mucosal immunity and gut-microbe homeostasis in Drosophila. Cell 153, 797–811. ( 10.1016/j.cell.2013.04.009) [DOI] [PubMed] [Google Scholar]
- 11.Sajjadian SM, Kim Y. 2020. Dual oxidase-derived reactive oxygen species against Bacillus thuringiensis and its suppression by eicosanoid biosynthesis inhibitors. Front. Microbiol. 11, 528 ( 10.3389/fmicb.2020.00528) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iatsenko I, Boquete JP, Lemaitre B. 2018. Microbiota-derived lactate activates production of reactive oxygen species by the intestinal NADPH oxidase Nox and shortens Drosophila lifespan. Immunity 49, 929–942. ( 10.1016/j.immuni.2018.09.017) [DOI] [PubMed] [Google Scholar]
- 13.Kim Y, Ahmed S, Stanley D, An C. 2018. Eicosanoid-mediated immunity in insects. Dev. Comp. Immunol. 83, 130–143. ( 10.1016/j.dci.2017.12.005) [DOI] [PubMed] [Google Scholar]
- 14.Park Y, Stanley D, Kim Y. 2015. Eicosanoids up-regulate production of reactive oxygen species by NADPH-dependent oxidase in Spodoptera exigua phagocytic haemocytes. J. Insect Physiol. 79, 63–72. ( 10.1016/j.jinsphys.2015.06.005) [DOI] [PubMed] [Google Scholar]
- 15.Barletta ABF, Trisnadi N, Ramirez JL, Barillas-Mury C. 2019. Mosquito midgut prostaglandin release establishes systemic immune priming. iScience 19, 54–62. ( 10.1016/j.isci.2019.07.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim Y, Ahmed S, Al Baki MA, Kumar S, Kim K, Park Y, Stanley D.. 2020. Deletion mutant of PGE2 receptor using CRISPR-Cas9 exhibits larval immunosuppression and adult infertility in a lepidopteran insect, Spodoptera exigua. Dev. Comp. Immunol. 111, 103743 ( 10.1016/j.dci.2020.103743) [DOI] [PubMed] [Google Scholar]
- 17.Ha EM, Oh CT, Bae YS, Lee WJ. 2005. A direct role for dual oxidase in Drosophila gut immunity. Science 310, 847–850. ( 10.1126/science.1117311) [DOI] [PubMed] [Google Scholar]
- 18.Chakrabarti S, Poidevin M, Lemaitre B. 2014. The Drosophila MAPK p38c regulates oxidative stress and lipid homeostasis in the intestine. PLoS Genet. 10, e1004659 ( 10.1371/journal.pgen.1004659) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim H, Rhee SH, Kokkotou E, Na X, Savidge T, Moyer MP, Pothoulakis C, LaMon JT. 2005. Clostridium difficile toxin A regulates inducible cyclooxygenase-2 and prostaglandin E2 synthesis in colonocytes via reactive oxygen species and activation of p38 MAPK. J. Biol. Chem. 280, 21 237–21 245. ( 10.1074/jbc.M413842200) [DOI] [PubMed] [Google Scholar]
- 20.Shaywitz AJ, Greenberg ME. 1999. CREB: a stimulus-induced transcription factor activated by a diverse array of extracellular signals. Annu. Rev. Biochem. 68, 821–861. ( 10.1146/annurev.biochem.68.1.821) [DOI] [PubMed] [Google Scholar]
- 21.Dwarki VJ, Montminy M, Verma IM. 1990. Both the basic region and the ‘leucine zipper’ domain of the cyclic AMP response element binding (CREB) protein are essential for transcriptional activation. EMBO J. 9, 225–232. ( 10.1002/j.1460-2075.1990.tb08099.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park J, Stanley D, Kim Y. 2014. Roles of peroxinectin in PGE2-mediated cellular immunity in Spodoptera exigua. PLoS ONE 9, e105717 ( 10.1371/journal.pone.0105717) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Büyükgüzel K, Tunaz H, Putnam SM, Stanley D. 2002. Prostaglandin biosynthesis by midgut tissue isolated from the tobacco hornworm, Manduca sexta. Insect Biochem. Mol. Biol. 32, 435–443. ( 10.1016/S0965-1748(01)00121-7) [DOI] [PubMed] [Google Scholar]
- 24.García Gil de Muñoz FL, Martínez-Barnetche J, Lanz-Mendoza H, Rodríguez MH, Hernández-Hernández FC. 2008. Prostaglandin E2 modulates the expression of antimicrobial peptides in the fat body and midgut of Anopheles albimanus. Arch. Insect Biochem. Physiol. 68, 14–25. ( 10.1002/arch.20232) [DOI] [PubMed] [Google Scholar]
- 25.Shrestha S, Stanley D, Kim Y. 2011. PGE2 induces oenocytoid cell lysis via a G protein-coupled receptor in the beet armyworm, Spodoptera exigua. J. Insect Physiol. 57, 1568–1576. ( 10.1016/j.jinsphys.2011.08.010) [DOI] [PubMed] [Google Scholar]
- 26.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. ( 10.1006/meth.2001.1262) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This article has no additional data.