Abstract
Background
COVID-19 has caused a global pandemic, and there is no wonder drug for epidemic control at present. However, many clinical practices have shown that traditional Chinese medicine has played an important role in treating the outbreak. Among them, ephedra-bitter almond is a common couplet medicine in anti-COVID-19 prescriptions. This study aims to conduct an exploration of key components and mechanisms of ephedra-bitter almond anti-COVID-19 based on network pharmacology.
Material and methods
We collected and screened potential active components of ephedra-bitter almond based on the TCMSP Database, and we predicted targets of the components. Meanwhile, we collected relevant targets of COVID-19 through the GeneCards and CTD databases. Then, the potential targets of ephedra-bitter almond against COVID-19 were screened out. The key components, targets, biological processes, and pathways of ephedra-bitter almond anti-COVID-19 were predicted by constructing the relationship network of herb-component-target (H-C-T), protein-protein interaction (PPI), and functional enrichment. Finally, the key components and targets were docked by AutoDock Vina to explore their binding mode.
Results
Ephedra-bitter almond played an overall regulatory role in anti-COVID-19 via the patterns of multi-component-target-pathway. In addition, some key components of ephedra-bitter almond, such as β-sitosterol, estrone, and stigmasterol, had high binding activity to 3CL and ACE2 by molecular docking simulation, which provided new molecular structures for new drug development of COVID-19.
Conclusion
Ephedra-bitter almonds were used to prevent and treat COVID-19 through directly inhibiting the virus, regulating immune responses, and promoting body repair. However, this work is a prospective study based on data mining, and the findings need to be interpreted with caution.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13040-020-00229-4.
Keywords: COVID-19, Ephedra-bitter almond, Molecular docking simulation, Traditional Chinese medicine, Molecular mechanisms of pharmacological action
Background
The recent emergence of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has resulted in a WHO-declared public health emergency of international concern [1]. Since December 2019, the sudden epidemic of Corona Virus Disease 2019 (COVID-19) has seriously threatened the healthy life of the people. As of April 3, 2020, there have been approximately 1,020,000 confirmed cases of COVID-19 worldwide, with 54,000 deaths. Among them, a total of 82,813 cases of COVID-19 (including overseas imported cases) were diagnosed in China, with 76,791 cured cases and 3331 deaths. From this data, it is not difficult to find that China’s prevention and control of COVID-19 epidemic are beginning to bear fruit, which is not only due to measures such as restricting the flow of people and vigorously publicizing but also to the important factor of medical assistance. In particular, the widespread use of Traditional Chinese Medicine (TCM) has played a huge role in the prevention and control of this epidemic [2].
During the epidemic, the administrations of TCM and health committees of various provinces and cities in China have formulated and issued Chinese medicine prevention and treatment programs for COVID-19 based on over-all symptoms of the patients. Some studies [3, 4] have explored the law of drug use through data mining on these prescriptions for the prevention and treatment of COVID-19, and found that ephedra (Ephedrae Herba) - bitter almond (Armeniacae Semen Amarum) is one of the most frequently used couplet medicines (called “yaodui” in Chinese, this is a relatively fixed combination of two herbs commonly used in TCM). For example, Qingfei Paidu Decoction, as a recommended prescription in “the Diagnosis and Treatment Protocol for Novel Coronavirus Pneumonia” formulated by the National Health Commission of the People’s Republic of China [5], contains the couplet medicines of ephedra-bitter almond, and the effective cure rate of this prescription for COVID-19 is more than 90% [6]. When it was used as an adjuvant treatment to western medicine, it could relieve symptoms, improve the regression of lung inflammation, and show a tendency to reduce the degree of multi-organ impairment [7]. However, due to the complexity of TCM components and the limitations of the current experimental conditions, we cannot know the pharmacological substance basis and mechanism of ephedra-bitter almond against COVID-19.
Traditionally, the hypothesis that “one drug for one target for one disease” has influenced various aspects of drug discovery strategies. However, advances in systems biology have shown that interventions with a single node may not be effective in the treatment of complex diseases [8]. Hence, network biology seems to clarify our understanding of drug action more clearly. Network pharmacology is a cross-discipline which is based on systems biology and combines polypharmacology, molecular network data, bioinformatics, and computer simulation [9–11]. This discipline has resulted in widespread public interest, and humanity has made extraordinary progress in further understanding diseases through this knowledge and technology over recent years. Some studies try to redefine diseases based on the methods and ideas of network pharmacology, and provide new ideas for the treatment of diseases and the reuse of drugs [12]. For example, Feixiong Cheng et al. identified new drug-disease associations for old drugs by quantifying the proximity of disease genes and drug targets in the protein-protein interaction (PPI) network [13]; they also proposed a network-based method to identify clinically effective drug combinations for specific diseases, with a view to discovering the synergistic and attenuating rules of drug compatibility [14]. Excitingly, during the COVID-19 pandemic people used methods such as system pharmacology and network medicine to quickly identify reusable drugs and potential drug combinations for SARS-CoV-2 from candidate drugs [15, 16]. Compared with the traditional model of developing new drugs from scratch, this method can minimize the conversion gap between preclinical test results and clinical results, and facilitate the rapid formulation of effective treatment strategies for the epidemic.
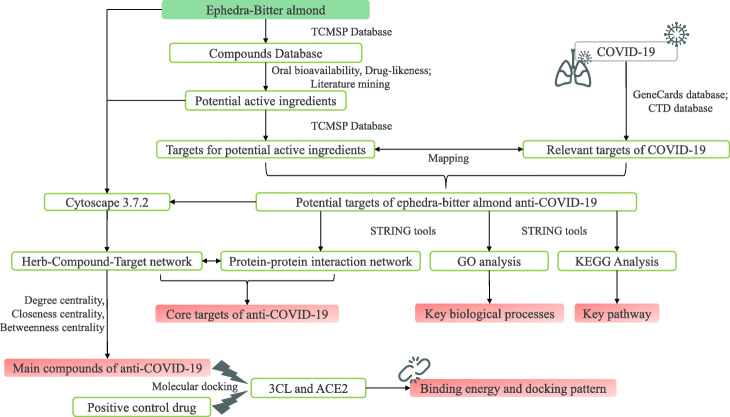
In recent years, network pharmacology has become more and more widely used in the research of TCM formulations, including targets discovery, bioactive compounds screening, toxicity evaluation, mechanism research, and quality control research, and has also derived new frontier disciplines such as network toxicology and TCM-Gut microbiota network pharmacology [17]. It is considered to be one of the most suitable techniques for TCM formulation research [18]. By constructing a complex network of herbs-components-targets-disease, the action mechanism of TCM prescriptions can be explained from a system level, which is consistent with the “holistic view” of TCM for treating diseases. Its ability to analyze and process huge data information provides a powerful tool for the analysis of the overall action mechanism of TCM prescription. In this study, we explored the potential pharmacodynamic material basis and molecular mechanism of ephedra-bitter almond against COVID-19 using network pharmacology and molecular docking technology, to provide some basis for its experimental research (Fig. 1).
Fig. 1.
Research process
Material and methods
Screening of potentially active components from ephedra-bitter almond
All candidate compounds of ephedra and bitter almond were collected from the TCMSP Database [19] (Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform, http://tcmspw.com/tcmsp.php), a unique systems pharmacology platform of Chinese herbal medicines that captures the relationships between drugs, targets, and diseases.
Oral bioavailability (OB) is a significant indicator of the efficiency of drug or active compound delivery to the body’s blood circulation [20]. Drug-likeness (DL) means “molecule which holds functional groups and/or has physical properties consistent with the majority of known drugs” [21]. Therefore, for new drug candidates, good OB and DL are one of their most important pharmacokinetic parameters. In the TCMSP database, the OB values of each compound were predicted by the OBioavail 1.1 model, which was built based on 805 drug-like molecules of known human OB [22]. The DL values of each compound were predicted based on a construction model of molecular descriptors and Tanimoto coefficients. These models have been shown to be effective in screening drugs or active ingredients [23, 24]. In this study, we selected compounds with OB ≥ 30% and DL ≥ 0.18 as candidate ingredients.
In addition, in order to avoid filtering out the effective ingredients with exact curative effects of ephedra and bitter almond in the screening process, we also supplemented the active ingredients by means of literature research.
Prediction and screening of targets
Target prediction of chemical composition is essential to elucidate its pharmacological effects. Therefore, this study predicted the potential targets of the components through the TCMSP database [19]. The Weighted Ensemble Similarity method and support vector machine method were applied to the target prediction in the TCMSP database. The model showed better prediction performance of drug-target interaction, with a consistency of 82.83%, a sensitivity of 81.33%, and a specificity of 93.62% [25, 26], which had been widely proven to be effective [27]. The predicted target proteins were imported into the UniProt database (The Universal Protein Resource, http://www.uniprot.org/), and the species was set to “Homo sapiens” to standardize its name. Meanwhile, we used “coronavirus disease 2019” OR “COVID-19” as keywords to search the GeneCards database [28] (https://www.genecards.org/) and the CTD database (Comparative Toxicogenomics Database, http://ctdbase.org/) to get the relevant targets of COVID-19. Then, based on the mapping analysis of the components targets and the disease targets, the potential action targets of ephedra-bitter almond against COVID-19 were screened out.
Construction of “herb-component-target” network
In order to investigate the multi-component and multi-target therapeutic feature of ephedra-bitter almond in combating COVID-19, we completed a relationship mapping of the herbs of ephedra-bitter almond, the candidate compounds, and potential targets using Cytoscape 3.7.2, a standard tool for biological network visualization and data integration [29, 30]. The herb-component-target (H-C-T) network was constructed in this work. In addition, we analyzed the quantitative properties of the network using plug-ins in the Cytoscape Network Analyzer [31]. Through the parameters such as degree centrality (DC), closeness centrality (CC), and betweenness centrality (BC), we screened out the potential pharmacodynamic material basis and main action targets of ephedra-bitter almond against COVID-19. Centrality is an important parameter to describe the characteristics of network topology. DC is the sum of the number of nodes directly connected to a node, which is the most direct indicator of the importance of nodes in network analysis. CC is the sum of distances from one node to all other nodes, reflecting the extent of the proximity of a node to other nodes. BC is the number of shortest paths through a node, which reflects the degree of cohesion of nodes in the network. In this study, if the three-parameter values of a node were greater than the corresponding median, then the node was considered to have an important position in the network, and the substance represented by the node was the key component or target.
Construction of protein-protein interaction (PPI) network
We imported the potential targets of ephedra-bitter almond anti-COVID-19 into the STRING tool (Search Tool for Retrieval of Interacting Genes/Proteins, https://string-db.org/) [32], and set the network edge in Basic Settings to confidence; the active interaction sources were set to Textmining, Experiments, Databases, Co-expression, Neighborhood, Gene Fusion, and Co-occurrence; minimum required interaction score was set to highest confidence 0.900 for PPI analysis. Then, the interaction information was further visually analyzed by Cytoscape, and the main targets were predicted by the parameters such as DC, CC, and BC. Combined with the main targets predicted by H-C-T network analysis in the previous step, the core targets of ephedra-bitter almond against COVID-19 were obtained.
Gene ontology (GO) and Kyoto encyclopedia of genes and genomes (KEGG) analysis
We imported the potential targets of ephedra-bitter almond anti-COVID-19 into the STRING database for enrichment analysis of GO and KEGG pathways [32], aiming to explore the relevant biological processes and signal pathways of potential anti-COVID-19 targets. After selecting the GO and KEGG terms with False Discovery Rate (FDR) ≤ 0.05, the top 10 terms and top 15 pathways with the highest number of observed genes were selected, and the bubble diagrams were drawn by R statistical modeling software to further analyze and predict the key pathways of ephedra-bitter almond anti-COVID-19.
Molecular docking prediction
Currently, it is generally believed that 3CL hydrolase (Mpro) from 2019-nCoV coronavirus and angiotensin-converting enzyme 2 (ACE2) from host cells are potential targets for anti-COVID-19 [33, 34]. Therefore, we performed molecular docking simulation between the key components based on H-C-T network analysis and the target proteins.
We searched and downloaded the protein structure of 3CL hydrolase (PDB ID: 6 LU7) and ACE2 (PDB ID: 1R42) from Protein Data Bank (PDB, http://www.rcsb.org), and the 3D structure of key components from ZINC database (http://zinc.docking.org/), respectively. The protein structure and active components were modified by Pymol and AutoDockTools [35, 36]. The processed protein and components were introduced into AutoDock Vina for molecular docking, and the binding energy was calculated using the Iterated Local Search global optimizer [37]. The cluster with the maximum absolute value of binding energy was selected from the docking results, and the lowest value in the cluster was the binding energy between the component and the protein molecule.
Meanwhile, we selected hydroxychloroquine, which is a potential antiviral drug for anti-COVID-19, as a positive control [38], and downloaded its structure. The molecular docking simulation was performed under the same conditions, and the obtained binding energy was compared with the active components of ephedra-bitter almond.
Results
Potential active components of ephedra-bitter almond
In the TCMSP Database, we collected a total of 363 candidate components from ephedra, and a total of 113 candidate components from bitter almond. Through observation and analysis, it was found that ephedra mainly contains alkaloids, flavonoids, volatile oils, and organic acids, while bitter almonds mainly contain glycosides, fatty oils, proteins, and amino acids.
Next, we screened potential active components based on their ADME properties. As a result, 40 potential active molecules with OB ≥ 30% and DL ≥ 0.18 were obtained, accounting for 9.37% of the total. In addition, although the OB or DL values of some compounds do not meet the screening criteria, the literature shows that they have related pharmacological activities and belong to higher content and more typical components in the herb. Therefore, in order to obtain more accurate results, these compounds were also included in the candidate compounds for further analysis. For instance, amygdalin with relatively small OB value was retained for further analysis since it was an active ingredient of bitter almond [39]. It has been reported that amygdalin may play a protective effect on acute lung injury by inhibiting NF-κB and NLRP3 signal pathways [40]. Furthermore, the sympathomimetic effect of alkaloids in ephedra was the material basis for the pharmacological effects of ephedra [41], so we also included ephedrine, pseudoephedrine, norephedrine, norpseudoephedrine, methylephedrine, and (+)-N-Methylpseudoephedrine in this study. Finally, we screened a total of 47 “potentially active compounds” (Table 1).
Table 1.
Basic information of 47 potentially active compounds obtained by ADME screening
| No. | Mol ID | Molecule Name | OB (%) | DL | Herb |
|---|---|---|---|---|---|
| 1 | MOL000006 | luteolin | 36.16 | 0.25 | Ephedra |
| 2 | MOL000098 | quercetin | 46.43 | 0.28 | Ephedra |
| 3 | MOL000358 | β-sitosterol | 36.91 | 0.75 | Ephedra |
| 4 | MOL000422 | kaempferol | 41.88 | 0.24 | Ephedra |
| 5 | MOL001494 | ethyl linoleate | 42 | 0.19 | Ephedra |
| 6 | MOL001506 | squalene | 33.55 | 0.42 | Ephedra |
| 7 | MOL001755 | Stigmast-4-en-3-one | 36.08 | 0.76 | Ephedra |
| 8 | MOL001771 | clionasterol | 36.91 | 0.75 | Ephedra |
| 9 | MOL002823 | herbacetin | 36.07 | 0.27 | Ephedra |
| 10 | MOL002881 | diosmetin | 31.14 | 0.27 | Ephedra |
| 11 | MOL004328 | naringetol | 59.29 | 0.21 | Ephedra |
| 12 | MOL004576 | taxifolin | 57.84 | 0.27 | Ephedra |
| 13 | MOL004798 | delphinidin | 40.63 | 0.28 | Ephedra |
| 14 | MOL005043 | campesterol | 37.58 | 0.71 | Ephedra |
| 15 | MOL005190 | eriodictyol | 71.79 | 0.24 | Ephedra |
| 16 | MOL005573 | genkwanin | 37.13 | 0.24 | Ephedra |
| 17 | MOL005842 | pectolinarigenin | 41.17 | 0.3 | Ephedra |
| 18 | MOL006594 | ephedrine | 43.35 | 0.03 | Ephedra |
| 19 | MOL006637 | pseudoephedrine | 52.25 | 0.03 | Ephedra |
| 20 | MOL007214 | (+)-leucocyanidin | 37.61 | 0.27 | Ephedra |
| 21 | MOL009189 | methylephedrine | 44.08 | 0.04 | Ephedra |
| 22 | MOL009190 | norephedrine | 66.05 | 0.03 | Ephedra |
| 23 | MOL009191 | norpseudoephedrine | 68.94 | 0.03 | Ephedra |
| 24 | MOL009194 | (+)-N-Methylpseudoephedrine | 37.12 | 0.04 | Ephedra |
| 25 | MOL010489 | leucocianidol | 30.84 | 0.27 | Ephedra |
| 26 | MOL010788 | leucopelargonidin | 57.97 | 0.24 | Ephedra |
| 27 | MOL011319 | Butyl octyl phthalate | 43.74 | 0.24 | Ephedra |
| 28 | MOL000449 | stigmasterol | 43.83 | 0.76 | Ephedra, Bitter almond |
| 29 | MOL000492 | cianidanol | 54.83 | 0.24 | Ephedra, Bitter almond |
| 30 | MOL000211 | betulinic acid | 55.38 | 0.78 | Bitter almond |
| 31 | MOL000359 | sitosterol | 36.91 | 0.75 | Bitter almond |
| 32 | MOL000953 | cholesterol | 37.87 | 0.68 | Bitter almond |
| 33 | MOL001320 | amygdalin | 4.42 | 0.61 | Bitter almond |
| 34 | MOL002211 | 11,14-eicosadienoic acid | 39.99 | 0.2 | Bitter almond |
| 35 | MOL002311 | glycyrol | 90.78 | 0.67 | Bitter almond |
| 36 | MOL002372 | (E,E,E,E)-Squalene | 33.55 | 0.42 | Bitter almond |
| 37 | MOL003410 | Ziziphin_qt | 66.95 | 0.62 | Bitter almond |
| 38 | MOL004355 | α-spinasterol | 42.98 | 0.76 | Bitter almond |
| 39 | MOL004841 | licochalcone B | 76.76 | 0.19 | Bitter almond |
| 40 | MOL004903 | liquiritin | 65.69 | 0.74 | Bitter almond |
| 41 | MOL004908 | glabridin | 53.25 | 0.47 | Bitter almond |
| 42 | MOL005017 | phaseol | 78.77 | 0.58 | Bitter almond |
| 43 | MOL005030 | 11-Eicosenoic acid | 30.7 | 0.2 | Bitter almond |
| 44 | MOL007207 | (R)-coclaurine | 79.64 | 0.24 | Bitter almond |
| 45 | MOL010921 | estrone | 53.56 | 0.32 | Bitter almond |
| 46 | MOL010922 | butanedioic acid | 31.62 | 0.23 | Bitter almond |
| 47 | MOL012922 | l-Stepholidine | 87.35 | 0.54 | Bitter almond |
Target prediction and disease mapping
In this study, a total of 243 targets of the compounds were predicted by the TCMSP database (after deduplication). Meanwhile, a total of 5329 targets related to COVID-19 were obtained in the GeneCards database and the CTD database. Then, the compound targets were mapped to COVID-19-related targets. After screening, 178 targets related to herbal ingredients were identified. These targets were potential targets of ephedra-bitter almond against COVID-19 (Table 2).
Table 2.
The information on potential targets of ephedra-bitter almond against COVID-19
| No. | UniProt ID | Gene Symbol | No. | UniProt ID | Gene Symbol | No. | UniProt ID | Gene Symbol |
|---|---|---|---|---|---|---|---|---|
| 1 | P80404 | ABAT | 61 | Q9NRD8 | DUOX2 | 121 | P01106 | MYC |
| 2 | P33527 | ABCC1 | 62 | Q01094 | E2F1 | 122 | P14598 | NCF1 |
| 3 | Q9UNQ0 | ABCG2 | 63 | Q14209 | E2F2 | 123 | Q15788 | NCOA1 |
| 4 | Q13085 | ACACA | 64 | P01133 | EGF | 124 | Q15596 | NCOA2 |
| 5 | P22303 | ACHE | 65 | P00533 | EGFR | 125 | Q16236 | NFE2L2 |
| 6 | Q08462 | ADCY2 | 66 | P19419 | ELK1 | 126 | P25963 | NFKBIA |
| 7 | Q15848 | ADIPOQ | 67 | P04626 | ERBB2 | 127 | P35228 | NOS2 |
| 8 | P08913 | ADRA2A | 68 | P21860 | ERBB3 | 128 | P29474 | NOS3 |
| 9 | P07550 | ADRB2 | 69 | P03372 | ESR1 | 129 | P15559 | NQO1 |
| 10 | P35869 | AHR | 70 | Q92731 | ESR2 | 130 | O75469 | NR1I2 |
| 11 | O95433 | AHSA1 | 71 | P13726 | F3 | 131 | P08235 | NR3C2 |
| 12 | P31749 | AKT1 | 72 | P49327 | FASN | 132 | Q9BZD4 | NUF2 |
| 13 | P09917 | ALOX5 | 73 | P01100 | FOS | 133 | P11926 | ODC1 |
| 14 | P04114 | APOB | 74 | P17302 | GJA1 | 134 | P09874 | PARP1 |
| 15 | P10275 | AR | 75 | P49841 | GSK3B | 135 | P12004 | PCNA |
| 16 | Q92934 | BAD | 76 | P00390 | GSR | 136 | P06401 | PGR |
| 17 | Q07812 | BAX | 77 | P09488 | GSTM1 | 137 | P11309 | PIM1 |
| 18 | P10415 | BCL2 | 78 | P09211 | GSTP1 | 138 | P00750 | PLAT |
| 19 | Q07817 | BCL2L1 | 79 | Q16665 | HIF1A | 139 | P00749 | PLAU |
| 20 | Q9BXY8 | BEX2 | 80 | P52789 | HK2 | 140 | P27169 | PON1 |
| 21 | O15392 | BIRC5 | 81 | P04035 | HMGCR | 141 | P16435 | POR |
| 22 | P00918 | CA2 | 82 | P09601 | HMOX1 | 142 | Q07869 | PPARA |
| 23 | P42574 | CASP3 | 83 | Q00613 | HSF1 | 143 | Q03181 | PPARD |
| 24 | P55210 | CASP7 | 84 | P07900 | HSP90AA1 | 144 | P37231 | PPARG |
| 25 | Q14790 | CASP8 | 85 | P11021 | HSPA5 | 145 | P17252 | PRKCA |
| 26 | P55211 | CASP9 | 86 | P04792 | HSPB1 | 146 | P05771 | PRKCB |
| 27 | P04040 | CAT | 87 | P05362 | ICAM1 | 147 | O43242 | PSMD3 |
| 28 | Q03135 | CAV1 | 88 | P01579 | IFNG | 148 | P43115 | PTGER3 |
| 29 | P13500 | CCL2 | 89 | P01344 | IGF2 | 149 | P35354 | PTGS2 |
| 30 | P20248 | CCNA2 | 90 | P17936 | IGFBP3 | 150 | P06400 | RB1 |
| 31 | P14635 | CCNB1 | 91 | O14920 | IKBKB | 151 | Q04206 | RELA |
| 32 | P24385 | CCND1 | 92 | P22301 | IL10 | 152 | Q06455 | RUNX1T1 |
| 33 | P29965 | CD40LG | 93 | P01583 | IL1A | 153 | Q13950 | RUNX2 |
| 34 | P06493 | CDK1 | 94 | P01584 | IL1B | 154 | P19793 | RXRA |
| 35 | P24941 | CDK2 | 95 | P60568 | IL2 | 155 | Q14524 | SCN5A |
| 36 | P11802 | CDK4 | 96 | P05112 | IL4 | 156 | P16581 | SELE |
| 37 | P38936 | CDKN1A | 97 | P05231 | IL6 | 157 | P05121 | SERPINE1 |
| 38 | O14757 | CHEK1 | 98 | P06213 | INSR | 158 | P14672 | SLC2A4 |
| 39 | O96017 | CHEK2 | 99 | P10914 | IRF1 | 159 | P23975 | SLC6A2 |
| 40 | Q15822 | CHRNA2 | 100 | P05412 | JUN | 160 | Q01959 | SLC6A3 |
| 41 | O15111 | CHUK | 101 | Q12809 | KCNH2 | 161 | P31645 | SLC6A4 |
| 42 | O14493 | CLDN4 | 102 | P35968 | KDR | 162 | P35610 | SOAT1 |
| 43 | P02452 | COL1A1 | 103 | P01130 | LDLR | 163 | P00441 | SOD1 |
| 44 | P02461 | COL3A1 | 104 | P21397 | MAOA | 164 | P10451 | SPP1 |
| 45 | P02741 | CRP | 105 | P27338 | MAOB | 165 | P36956 | SREBF1 |
| 46 | P17538 | CTRB1 | 106 | P11137 | MAP2 | 166 | P42224 | STAT1 |
| 47 | P07339 | CTSD | 107 | P28482 | MAPK1 | 167 | P01137 | TGFB1 |
| 48 | P02778 | CXCL10 | 108 | Q16539 | MAPK14 | 168 | P07204 | THBD |
| 49 | P19875 | CXCL2 | 109 | P27361 | MAPK3 | 169 | P01375 | TNF |
| 50 | P10145 | CXCL8 | 110 | P45983 | MAPK8 | 170 | P11387 | TOP1 |
| 51 | P11511 | CYP19A1 | 111 | Q07820 | MCL1 | 171 | P11388 | TOP2A |
| 52 | P04798 | CYP1A1 | 112 | Q00987 | MDM2 | 172 | P04637 | TP53 |
| 53 | P05177 | CYP1A2 | 113 | P08581 | MET | 173 | P14679 | TYR |
| 54 | Q16678 | CYP1B1 | 114 | O43451 | MGAM | 174 | Q9HAW9 | UGT1A8 |
| 55 | P08684 | CYP3A4 | 115 | P03956 | MMP1 | 175 | P19320 | VCAM1 |
| 56 | Q96PD7 | DGAT2 | 116 | P08253 | MMP2 | 176 | P15692 | VEGFA |
| 57 | P27487 | DPP4 | 117 | P08254 | MMP3 | 177 | P47989 | XDH |
| 58 | P21728 | DRD1 | 118 | P14780 | MMP9 | 178 | P98170 | XIAP |
| 59 | P14416 | DRD2 | 119 | P05164 | MPO | |||
| 60 | P35462 | DRD3 | 120 | P55157 | MTTP |
Network construction
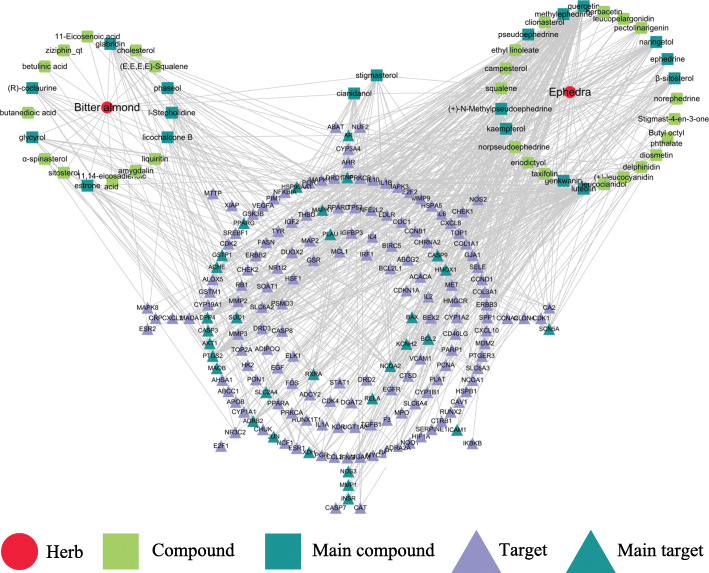
In this step, the H-C-T network was constructed including 227 nodes (2 herbs, 47 active compounds, and 178 potential targets) and 569 edges (Fig. 2). The circles represent herbs, the squares represent compounds and the triangles represent the potential targets of ephedra-bitter almond against COVID-19. The DC, CC, and BC values of compound nodes and target nodes were shown in Supplementary Material Table S1 and Table S2, respectively. The results show that the compound nodes had a median DC value of 7, a median CC of 0.3565, and a median BC of 0.0033. Among them, the three centralities of 19 compounds are higher than the median, including quercetin, luteolin, kaempferol, naringetol, β-sitosterol, glabridin, stigmasterol, licochalcone B, cianidanol, ephedrine, pseudoephedrine, phaseol, methylephedrine, (+)-N-Methylpseudoephedrine, l-Stepholidine, estrone, glycyrol, genkwanin and (R)-coclaurine, which indicates that these compounds play a relatively important role in the network. They may be the main active components of ephedra-bitter almond to exert anti-COVID-19.
Fig. 2.
Herb-compound-target (H-C-T) network of ephedra-bitter almond against COVID-19. The red circles represent the herbs, the green squares represent the compounds and the blue squares represent main compounds, the purple triangles represent the potential targets and the blue triangles represent main potential targets
Also, the target nodes have a median DC value of 2, a median CC of 0.3628, and a median BC of 0.0001. Among them, the three centralities of 31 targets are higher than the median, including PTGS2, HSP90AA1, AR, PPARG, DPP4, ACHE, NCOA2, NOS3, RELA, AKT1, BCL2, BAX, TNF, JUN, CASP3, XDH, MMP1, HMOX1, ICAM1, GSTP1, SLC2A4, INSR, KCNH2, SCN5A, ADRB2, RXRA, MAOB, CASP9, PLAU, MAPK1 and SOD1, which indicates that these targets also play a relatively important role in the network. They may be the main targets of ephedra-bitter almond to anti-COVID-19.
PPI analysis
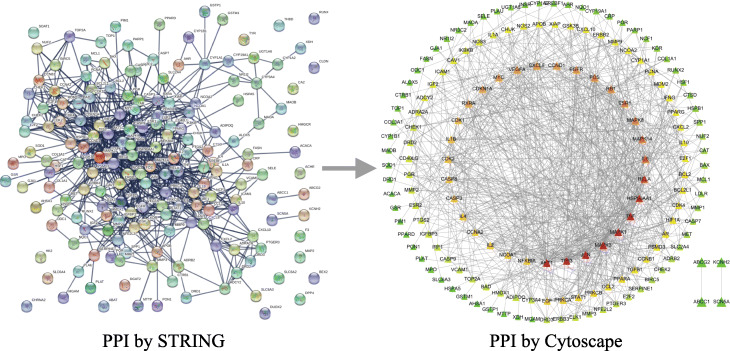
The PPI network was obtained by importing 178 potential targets into the STRING database and setting the species as “Homo sapiens”. Subsequently, the PPI network was further visualized and topologically analyzed using Cytoscape software (Fig. 3). The network consists of 159 nodes and 778 edges. It was analyzed by NetworkAnalyzer, the node color was set by the DC value of each node, the size of the node was set by the CC value, and the edge thickness was set according to the BC value of the node. The redder the node color, the larger the DC, and the larger the node size, the larger the CC, indicating that it is more critical in the network; the thicker the edge, the larger the combined score, indicating a closer relationship between the targets. The target nodes have a median DC value of 7, a median CC of 0.3624, and a median BC of 0.0036. Among them, the three centralities of 55 compounds are higher than the median, including AKT1, TP53, JUN, MAPK1, MAPK3, TNF, HSP90AA1, RELA, IL6, MAPK14, ESR1, MAPK8, RB1, FOS, EGFR, CXCL8, VEGFA, CCND1, MYC, RXRA, CDKN1A, CDK1, CASP3, CASP8, IL1B, CDK2, IL4, PRKCA, EGF, NCOA1, IL2, CCNA2, NFKBIA, STAT1, PRKCB, PPARA, CCL2, PSMD3, TGFB1, AR, HIF1A, BCL2, BCL2L1, PPARG, PCNA, IFNG, MMP9, NCOA2, APOB, NOS2, XIAP, IGF2, CAV1, IGFBP3 and PTGS2, which indicates that these targets also play a relatively important role in the PPI network.
Fig. 3.
Protein-protein interaction (PPI) network. PPI by Cytoscape: node color represents the size of the degree. The node color was from green to red, and the corresponding degree gradually larger. Node size represents closeness centrality. The node size is proportional to its closeness centrality. Line thickness represents combined score, indicating a closer relationship between the targets
The 55 main targets based on the PPI network analysis were compared with the 31 main targets based on the H-C-T network analysis. It is found that 13 targets play an important role in both H-C-T network and PPI network, including PTGS2, HSP90AA1, AR, PPARG, NCOA2, RELA, AKT1, BCL2, TNF, JUN, CASP3, RXRA and MAPK1. These targets not only play a significant role in the anti-COVID-19 process of ephedra-bitter almond but also play a key role in the gene regulatory network. Therefore, they are considered to be the core targets of active component anti-COVID-19.
Functional annotations
GO enrichment analysis
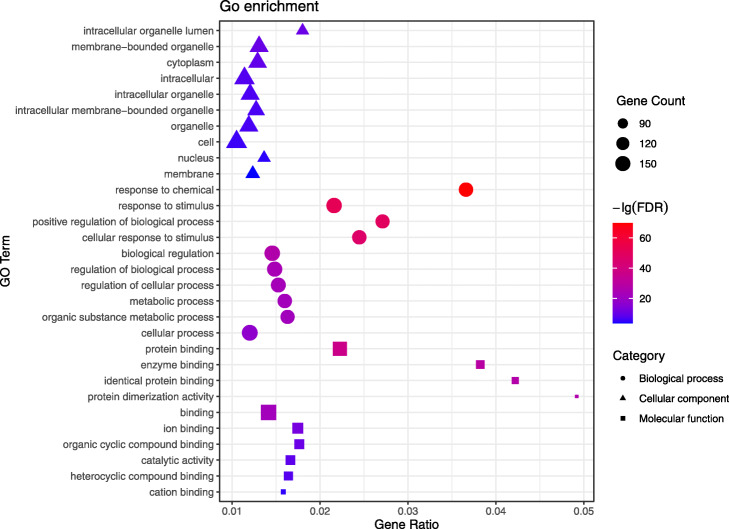
GO annotation analysis includes three parts: cellular component, biological process, and molecular function. We analyzed the top 10 GO terms that were the most significant, respectively (See Fig. 4 and Supplementary Material Table S3).
Fig. 4.
Cellular component, biological process, and molecular function analysis of 179 potential targets. The horizontal axis (Gene Radio) of the bubble diagram represents the ratio of the core targets involved in each term to the total number of targets in the term; the size of the bubble represents the number of core targets involved in the term; and the color from red to blue indicates the FDR value from small to large, that is, the redder it is, the higher the significance of the term
From cellular component ontology analysis, it was significantly correlated with the intracellular organelle lumen (GO: 0070013), membrane-bounded organelle (GO: 0043227), and cytoplasm (GO: 0005737) in cellular components, indicating that the active components of ephedra-bitter almond anti-COVID-19 interact primarily with related targets in the cytoplasm and organelles.
From biological process ontology analysis, the active ingredients of ephedra-bitter almond are mainly through the response to chemical (GO: 0042221), response to stimulus (GO: 0050896), positive regulation of biological process (GO: 0048518), and cellular response to stimulus (GO: 0051716) to against COVID-19. These processes are mainly related to the changes in the state or activity of cells or organisms caused by stimulation, which is consistent with the stimulation of the body after the virus infects the human.
From molecular function ontology analysis, ephedra-bitter almond anti-COVID-19 was mainly associated with functions such as protein binding (GO: 0005515), enzyme binding (GO: 0019899), identical protein binding (GO: 0042802), and protein dimerization activity (GO: 0046983). These molecular functions were primarily involved in the selective interaction with proteins and enzymes, thereby affecting the physiological and biochemical processes of the body.
KEGG pathway enrichment analysis
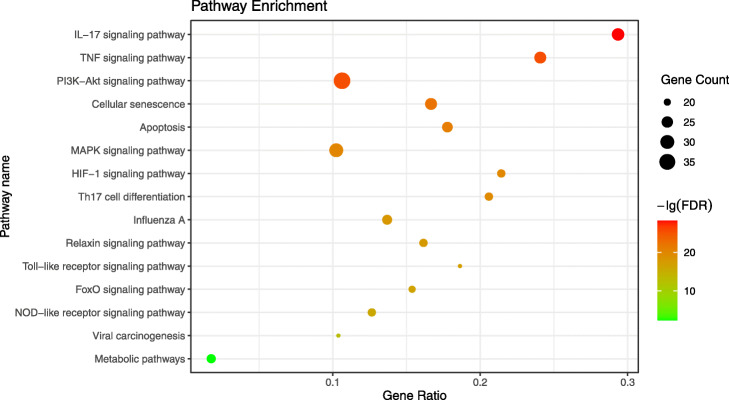
The result showed that the 178 potential targets were mapped to a total of 197 pathways. Next, we excluded pathways unrelated to COVID-19, such as “prostate cancer”, “pancreatic cancer”, “Chagas disease”, identified 58 pathways, and selected the top 15 pathways with the highest number of observed genes for analysis (See Fig. 5 and Supplementary Material Table S4). Through the bubble diagram, it was not difficult to find that the IL-17 signaling pathway (hsa04657), TNF signaling pathway (hsa04668), and PI3K-Akt signaling pathway (hsa04151) were more important pathways.
Fig. 5.
KEGG pathway enrichment analysis of 178 potential targets. The horizontal axis (Gene Radio) of the bubble diagram represents the ratio of the core targets involved in each pathway to the total number of targets in the pathway; the size of the bubble represents the number of core targets involved in the pathway; and the color from red to green indicates the FDR value was from small to large, that is, the redder it is, the higher the significance of the pathway
Docking results
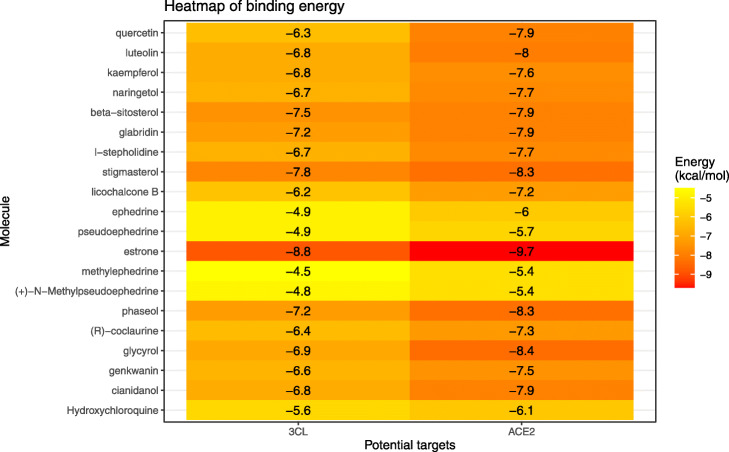
We simulated the docking of 19 main compounds of ephedra-bitter almond with two potential targets, 3CL and ACE2, respectively. The binding energies are shown in Table 3 and Fig. 6. Compared with the positive control drug hydroxychloroquine, the binding energies of these active components with 3CL and ACE2 are generally ideal. This further suggests that ephedra-bitter almond anti-COVID-19 may be performed through a multi-component-target-pathway mode. Meanwhile, we also drew the docking patterns of three compounds with higher binding energy and hydroxychloroquine with target proteins (Fig. 7). The prediction of docking patterns and binding residues could provide an important basis for further exploration of drug targets.
Table 3.
Binding energies of 19 main compounds and positive control drug to two potential targets
| No. | Mol ID | Molecule Name | 3CL (kcal/mol) | ACE2 (kcal/mol) |
|---|---|---|---|---|
| 1 | MOL000098 | quercetin | −6.3 | −7.9 |
| 2 | MOL000006 | luteolin | −6.8 | −8 |
| 3 | MOL000422 | kaempferol | −6.8 | −7.6 |
| 4 | MOL004328 | naringetol | −6.7 | −7.7 |
| 5 | MOL000358 | β-sitosterol | −7.5 | −7.9 |
| 6 | MOL004908 | glabridin | −7.2 | −7.9 |
| 7 | MOL012922 | l-stepholidine | −6.7 | −7.7 |
| 8 | MOL000449 | stigmasterol | −7.8 | −8.3 |
| 9 | MOL004841 | licochalcone B | −6.2 | −7.2 |
| 10 | MOL006594 | ephedrine | −4.9 | −6 |
| 11 | MOL006637 | pseudoephedrine | −4.9 | −5.7 |
| 12 | MOL010921 | estrone | −8.8 | −9.7 |
| 13 | MOL009189 | methylephedrine | −4.5 | −5.4 |
| 14 | MOL009194 | (+)-N-Methylpseudoephedrine | −4.8 | −5.4 |
| 15 | MOL005017 | phaseol | −7.2 | −8.3 |
| 16 | MOL007207 | (R)-coclaurine | −6.4 | −7.3 |
| 17 | MOL002311 | glycyrol | −6.9 | −8.4 |
| 18 | MOL005573 | genkwanin | −6.6 | −7.5 |
| 19 | MOL000492 | cianidanol | −6.8 | −7.9 |
| 20 | Positive control | Hydroxychloroquine | −5.6 | −6.1 |
Fig. 6.
Binding energies heatmap of 19 main compounds and positive control drug to two potential targets. The color from yellow to red indicates the binding energy was from small to large
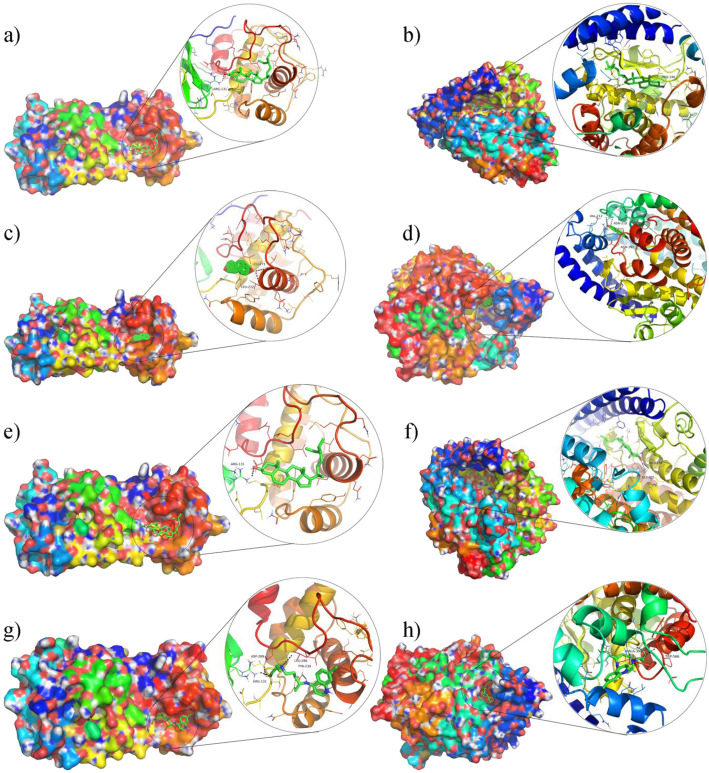
Fig. 7.
The docking complex of two targets and four components. Colored irregular clumps represent proteins, green chemical structures represent compounds, and each picture shows the details of the docking part. a β-sitosterol-3CL, (b) β-sitosterol-ACE2; (c) estrone-3CL, (d) estrone-ACE2, (e) stigmasterol-3CL, (f) stigmasterol-ACE2, (g) Hydroxychloroquine-3CL, (h) Hydroxychloroquine-ACE2
Discussion
The rapid spread of COVID-19 has alarmed many people [42]. The disease is characterized by fulminated onset and develops into respiratory failure [43]. With no wonder drugs for SARS-CoV-2, some people are turning to TCM, often on the advice of their doctors [44]. TCM drugs have proven to be effective in the treatment of COVID-19, especially for mild and general cases. They have effectively relieved symptoms, cut the rate of patients developing severe conditions, reduced the mortality rate, and boosted patients’ recovery [2]. Nonetheless, no single method is ever going to be universally applicable. Hence, the goal of management is to achieve optimal symptom control.
Ephedra-bitter almond is a common couplet medicine in classic TCM prescriptions for the treatment of upper respiratory tract infections. For instance, Mahuang decoction and Ma Xing Shi Gan decoction both contain ephedra-bitter almond. In this study, network pharmacology combined with molecular docking techniques was used to explore the active components, key targets, and related pathways of ephedra-bitter almond against COVID-19. Recently, some similar studies have also been carried on the beneficial exploration of TCM against COVID-19. For example, Yu-Liang Zhang et al. [45] studied the mechanism of action of Xuebijing injection in the treatment of COVID-19 based on network pharmacology, revealing that this Chinese medicine injection may alleviate the symptoms of COVID-19 by affecting angiotensin-converting enzyme 2 and some key pathways. Compared with the reference, the advantage of our study is that the collection of prescription ingredients is not limited to the TCMSP database, as we also combined with literature mining to supplement the potential active ingredients. Moreover, in order to collect disease targets more comprehensively, we have collected and integrated COVID-19-related targets in the GeneCards and CTD databases respectively, which seems to be more credible and rigorous compared with the method of expanding the collection of targets by STRING tool. Next, we used all the potential targets of the herbs anti-COVID-19 for functional annotation, which explained the potential biological process of herbal treatment of diseases more comprehensively than using key targets alone. We also simultaneously constructed the H-C-T network and PPI network to collect the key targets, taking into account not only the process of prescription treatment of diseases but also the interaction between target genes. Through the study of ephedra-bitter almond, we can initially understand its pharmacodynamic material basis and molecular mechanism of action, thus providing a certain theoretical basis for the development and clinical application of new drugs.
We found that some important components in ephedra-bitter almond, such as quercetin, luteolin, kaempferol, naringetol, β-sitosterol, and glabridin, may play a key role in the prevention and treatment of COVID-19. Here, quercetin, as one of the components with the highest degree value, had certain preventive or therapeutic effects on murine coronavirus, enterovirus 71, human immunodeficiency virus type 1, and dengue virus infection [46–49]. And luteolin was also resistant to dengue virus, influenza A virus, Japanese encephalitis virus, and so on [50–52]. In addition, ephedrine alkaloids, such as ephedrine, pseudoephedrine, and methylephedrine, had potential therapeutic effects on viral-induced respiratory infections [53]. This indicated that ephedra-bitter almond may resist COVID-19 through antiviral and sympathomimetic effects.
H-C-T network and PPI network analysis showed that the active components of ephedra-bitter almond were anti-COVID-19 mainly by regulating PTGS2, HSP90AA1, AR, PPARG, NCOA2, RELA, AKT1, BCL2, TNF, JUN, CASP3, RXRA and MAPK1. Here, PTGS2, also known as cyclooxygenase 2 (COX-2), is a key enzyme in prostaglandin biosynthesis. It was regulated by specific stimulating events and was responsible for the biosynthesis of prostaglandins in the process of inflammation [54]. COX-2 also was regarded as playing an important role in the pathogenesis of airway inflammation in respiratory diseases. Therefore, ephedra-bitter almond may regulate the expression of PTGS2 in the process of anti-COVID-19 and thus treat respiratory inflammation [55]. When pathogens invade cells, autophagy can be activated as an innate immune mechanism to control infection [56]. And there is a highly complex interplay between autophagy and invading viruses. As a highly conserved molecular chaperone, HSP90AA1 may initiate natural cellular defense against invading pathogens [57]. Therefore, ephedra-bitter almond may be helpful against COVID-19 by enhancing the destructive aspects of autophagy on the life cycle of the virus. In the latest network pharmacology research, the mechanism of Qingfei Paidu Decoction and Ma Xing Shi Gan Decoction in the treatment of COVID-19 was studied. It was found that these TCM prescriptions could be anti-COVID-19 through anti-viral, anti-inflammatory activity, and metabolic processes, of which the regulation of immune function may be the main channel [58–60]. These TCM prescriptions all contained ephedra-bitter almond, and the research results were basically consistent with this study.
Functional enrichment analysis showed that ephedra-bitter almond may play a role in anti-COVID-19 by regulating different biological processes and signaling pathways. Virus-infected host cells act as an important immune niche during viral infection and replication, and they stimulate the host’s immune response through molecular signaling [61]. However, as the virus continues to mutate, the body sometimes cannot respond as quickly as needed. At this time, the intervention of drugs or vaccines may increase the body’s sensitivity to viral stimulation, thereby restoring the balance of the immune ecosystem in the infected host tissue [62]. As shown by the results of the GO analysis in this study, ephedra-bitter almond may participate in biological processes such as immune regulation by enhancing the body’s response to pathogen stimulation. As one of the most significant signaling pathways, the PI3K-Akt signaling pathway was involved in the regulation of various cellular functions such as proliferation, differentiation, apoptosis, and glucose transport [63]. It had been reported that the nucleocapsid protein of SARS-CoV may promote the phosphorylation of Akt and JNK in host cells, and the PI3K-Akt pathway played a key role in avoiding apoptosis in SARS-CoV-infected cells [64, 65]. Therefore, the intervention of ephedra-bitter almond on SARS-CoV-2 infected cells may also be carried out through the PI3K-Akt pathway. The excessive inflammatory response in the process of pathogen infection is destructive to the host, and when the production of pro-inflammatory cytokines increases, it causes serious damage to the lungs [66]. A study showed that IL-17 produced during viral infection specially enhanced the pro-inflammatory response by directly cooperating with antiviral signaling [67]. Therefore, the IL-17 signaling pathway played a key role in regulating the immune pathophysiology of viral infection. In addition, COVID-19 can cause a strong immune response and inflammatory storm [68]. And, the inflammatory process extensively mediated by the TNF signaling pathway also had a certain regulatory effect on the occurrence and development of infectious diseases [69, 70]. Also, many studies have confirmed that the active ingredients in ephedra-bitter almond have a regulatory effect on these pathways. For example, ephedrine can reduce the secretion of proinflammatory cytokines through the PI3K-Akt pathway to inhibit the inflammation induced by peptidoglycan [71]. Amygdalin can relieve the symptoms of acute lung injury by inhibiting the production of TNF-α [72]. Luteolin could inhibit inflammatory response via inactivation of the PI3K-Akt pathway in LPS-stimulated RAW 264.7 cells [73]. In conclusion, ephedra-bitter almond may act against COVID-19 mainly through the PI3K-Akt signaling pathway, IL-17 signaling pathway, and TNF signaling pathway.
ACE2 is widely distributed, and is not only a necessary receptor for the invasion of coronaviruses such as SARS-CoV-2, but also a key substance leading to organ damage [74]. Therefore, the search for possible treatment strategies from ACE2 has broad application prospects and clinical value. 3CLpro, as a major protease encoded by the viral genome, is also one of the most attractive drug targets, because it plays a key role in the cleavage of viral polyproteins into functional proteins. Therefore, inhibition of this enzyme is also an effective strategy to block virus replication [75]. In this study, the results of molecular docking indicated that some key components of ephedra-bitter almond, such as β-sitosterol, estrone, and stigmasterol, had higher binding activities to the potential targets of anti-COVID-19. These natural small molecules may play the role of anti-inflammation or direct inhibition of virus replication by regulating 3CL and ACE2 [76, 77], so it is speculated that ephedra-bitter almond could play an anti-COVID-19 role by regulating 3CL and ACE2. This work provides the possibility to discover or design and synthesize effective protease inhibitors as antivirals for COVID-19.
Conclusion
In summary, ephedra-bitter almonds were used to prevent and treat COVID-19, not only by directly inhibiting the virus but also by regulating immune responses and promoting body repair. However, this work is a prospective study based on data mining, and the findings need to be interpreted with caution. This study can provide a certain theoretical basis for subsequent experiments.
Supplementary Information
Additional file 1: Table S1. Basic information and network topology parameter values of 47 potentially active compounds obtained by ADME screening. Table S2. The information and network topology parameter values of 178 potential targets of ephedra-bitter almond against COVID-19. Table S3. GO enrichment analysis. Table S4. KEGG enrichment analysis.
Abbreviations
- ACE2
Angiotensin-converting enzyme 2,
- BC
Betweenness centrality
- CC
Closeness centrality
- CTD database
Comparative Toxicogenomics Database
- COVID-19
Corona Virus Disease 2019
- DC
Degree centrality
- DL
Drug-likeness
- FDR
False Discovery Rate
- GO
Gene Ontology
- H-C-T
Herb-component-target
- KEGG
Kyoto encyclopedia of genes and genomes
- OB
Oral bioavailability
- PPI
Protein-protein interaction
- STRING
Search Tool for Retrieval of Interacting Genes/Proteins
- SARS-CoV-2
Severe acute respiratory syndrome coronavirus 2
- TCM
Traditional Chinese Medicine
- TCMSP Database
Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform
- UniProt database
The Universal Protein Resource
Authors’ contributions
Methodology: Kai Gao and Anna Song. Visualization: Kai Gao. Project administration: Kai Gao. Software: Kai Gao. Supervision: Yan-Ping Song. Writing – original draft: Kai Gao and Anna Song. Writing – review & editing: Yan-Ping Song. The authors read and approved the final manuscript.
Availability of data and materials
All data are available in the manuscript and they are shown in figures and tables.
Ethics approval and consent to participate
Our goal is to publish this research article in a peer-reviewed journal. Since there are no issues about participant privacy, the review will not require ethical approval.
Competing interests
All the authors declare that there is no conflict of interest regarding the publication of this paper.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Lai CC, Shih TP, Ko WC, Tang HJ, Hsueh PR. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): the epidemic and the challenges. Int J Antimicrob Agents. 2020;55:105924. doi: 10.1016/j.ijantimicag.2020.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang Y, Islam MS, Wang J, Li Y, Chen X. Traditional Chinese medicine in the treatment of patients infected with 2019-new coronavirus (SARS-CoV-2): a review and perspective. Int J Biol Sci. 2020;16:1708–1717. doi: 10.7150/ijbs.45538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheng YQ, Chen X, Wu YQ, Sun D, Yu X, Li SB , et al. Analysis on rules of TCM prescriptions in treating and preventing COVID-19 based on data mining. Shanghai Journal of Traditional Chinese Medicine 2020;54:32-39.
- 4.Ling XY, Jiang MC, Xu QY, Wu YW, Yuan B. Study on medication regularity of Traditional Chinese Medicine in prevention and treatment of novel coronavirus pneumonia based on data mining. J Chinese Med Mater. 2020;(07):1766–71.
- 5.Zhao JY, Yan JY, Qu JM. Interpretations of "Diagnosis and treatment protocol for novel coronavirus pneumonia (trial version 7)". Chinese Med J. 2020;133:1347–9. [DOI] [PMC free article] [PubMed]
- 6.Ren JL, Zhang AH, Wang XJ. Traditional Chinese medicine for COVID-19 treatment. Pharmacol Res. 2020;155:104743. doi: 10.1016/j.phrs.2020.104743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xin S, Cheng X, Zhu B, Liao X, Yang F, Song L, et al. Clinical retrospective study on the efficacy of Qingfei Paidu decoction combined with Western medicine for COVID-19 treatment. Biomed Pharmacother. 2020;129:110500. doi: 10.1016/j.biopha.2020.110500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hopkins AL. Network pharmacology. Nat Biotechnol. 2007;25:1110–1111. doi: 10.1038/nbt1007-1110. [DOI] [PubMed] [Google Scholar]
- 9.Berger SI, Iyengar R. Network analyses in systems pharmacology. Bioinformatics (Oxford, England) 2009;25:2466–2472. doi: 10.1093/bioinformatics/btp465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hao da C, Xiao PG. Network pharmacology: a Rosetta stone for traditional Chinese medicine. Drug Dev Res. 2014;75:299–312. doi: 10.1002/ddr.21214. [DOI] [PubMed] [Google Scholar]
- 11.Huang C, Zheng C, Li Y, Wang Y, Lu A, Yang L. Systems pharmacology in drug discovery and therapeutic insight for herbal medicines. Brief Bioinform. 2014;15:710–733. doi: 10.1093/bib/bbt035. [DOI] [PubMed] [Google Scholar]
- 12.Casas AI, Hassan AA, Larsen SJ, Gomez-Rangel V, Elbatreek M, Kleikers PWM, et al. From single drug targets to synergistic network pharmacology in ischemic stroke. Proc Natl Acad Sci U S A. 2019;116:7129–7136. doi: 10.1073/pnas.1820799116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng F, Desai RJ, Handy DE, Wang R, Schneeweiss S, Barabási AL, et al. Network-based approach to prediction and population-based validation of in silico drug repurposing. Nat Commun. 2018;9:2691. doi: 10.1038/s41467-018-05116-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng F, Kovács IA, Barabási AL. Network-based prediction of drug combinations. Nat Commun. 2019;10:1197. doi: 10.1038/s41467-019-09186-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gysi DM, Do Valle Í, Zitnik M, Ameli A, Gan X, Varol O, et al. Network medicine framework for identifying drug repurposing opportunities for COVID-19. ArXiv [Preprint]. 2020:arXiv:2004.07229v1. [DOI] [PMC free article] [PubMed]
- 16.Zhou Y, Hou Y, Shen J, Huang Y, Martin W, Cheng F. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov. 2020;6:14. doi: 10.1038/s41421-020-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang R, Zhu X, Bai H, Ning K. Network pharmacology databases for traditional Chinese medicine: review and assessment. Front Pharmacol. 2019;10:123. doi: 10.3389/fphar.2019.00123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo TT, Lu Y, Yan SK, Xiao X, Rong XL, Guo J. Network pharmacology in research of Chinese medicine formula: methodology, application and prospective. Chin J Integr Med. 2020;26:72–80. doi: 10.1007/s11655-019-3064-0. [DOI] [PubMed] [Google Scholar]
- 19.Ru J, Li P, Wang J, Zhou W, Li B, Huang C, et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. Aust J Chem. 2014;6:13. doi: 10.1186/1758-2946-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alam MA, Al-Jenoobi FI, Al-Mohizea AM, Ali R. Understanding and managing oral bioavailability: physiological concepts and patents. Recent Pat Anticancer Drug Discov. 2015;10:87–96. doi: 10.2174/1574892809666140917103834. [DOI] [PubMed] [Google Scholar]
- 21.Tian S, Wang J, Li Y, Li D, Xu L, Hou T. The application of in silico drug-likeness predictions in pharmaceutical research. Adv Drug Deliv Rev. 2015;86:2–10. doi: 10.1016/j.addr.2015.01.009. [DOI] [PubMed] [Google Scholar]
- 22.Xu X, Zhang W, Huang C, Li Y, Yu H, Wang Y, et al. A novel chemometric method for the prediction of human oral bioavailability. Int J Mol Sci. 2012;13:6964–6982. doi: 10.3390/ijms13066964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu H, Wang J, Zhou W, Wang Y, Yang L. Systems approaches and polypharmacology for drug discovery from herbal medicines: an example using licorice. J Ethnopharmacol. 2013;146:773–793. doi: 10.1016/j.jep.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 24.Liu J, Pei T, Mu J, Zheng C, Chen X, Huang C, et al. Systems pharmacology uncovers the multiple mechanisms of Xijiao Dihuang decoction for the treatment of viral hemorrhagic fever. Evid Based Complement Alternat Med. 2016;2016:9025036. doi: 10.1155/2016/9025036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng C, Guo Z, Huang C, Wu Z, Li Y, Chen X, et al. Large-scale direct targeting for drug repositioning and discovery. Sci Rep. 2015;5:11970. doi: 10.1038/srep11970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu H, Chen J, Xu X, Li Y, Zhao H, Fang Y, et al. A systematic prediction of multiple drug-target interactions from chemical, genomic, and pharmacological data. PLoS One. 2012;7:e37608. doi: 10.1371/journal.pone.0037608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee WY, Lee CY, Kim YS, Kim CE. The methodological trends of Traditional herbal medicine employing network pharmacology. Biomolecules. 2019;9(8):362. [DOI] [PMC free article] [PubMed]
- 28.Fishilevich S, Zimmerman S, Kohn A, Iny Stein T, Olender T, Kolker E, et al. Genic insights from integrated human proteomics in GeneCards. Database (Oxford). 2016;2016:baw030. [DOI] [PMC free article] [PubMed]
- 29.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics (Oxford, England) 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Assenov Y, Ramirez F, Schelhorn SE, Lengauer T, Albrecht M. Computing topological parameters of biological networks. Bioinformatics (Oxford, England) 2008;24:282–284. doi: 10.1093/bioinformatics/btm554. [DOI] [PubMed] [Google Scholar]
- 32.Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–d613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang L, Lin D, Sun X, Curth U, Drosten C, Sauerhering L, et al. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science (New York, N.Y.). 2020;368:409–12. [DOI] [PMC free article] [PubMed]
- 34.Batlle D, Wysocki J, Satchell K. Soluble angiotensin-converting enzyme 2: a potential approach for coronavirus infection therapy? Clin Sci (London, England : 1979) 2020;134:543–545. doi: 10.1042/CS20200163. [DOI] [PubMed] [Google Scholar]
- 35.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, et al. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seeliger D, de Groot BL. Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J Comput Aided Mol Des. 2010;24:417–422. doi: 10.1007/s10822-010-9352-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu J, Cao R, Xu M, Wang X, Zhang H, Hu H, et al. Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro. Cell Discov. 2020;6:16. doi: 10.1038/s41421-020-0156-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shi J, Chen Q, Xu M, Xia Q, Zheng T, Teng J, et al. Recent updates and future perspectives about amygdalin as a potential anticancer agent: a review. Cancer Med. 2019;8:3004–3011. doi: 10.1002/cam4.2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang A, Pan W, Lv J, Wu H. Protective effect of amygdalin on LPS-induced acute lung injury by inhibiting NF-kappaB and NLRP3 signaling pathways. Inflammation. 2017;40:745–751. doi: 10.1007/s10753-017-0518-4. [DOI] [PubMed] [Google Scholar]
- 41.Ibragic S, Sofic E. Chemical composition of various Ephedra species. Bosnian J Zbasic Med Sci. 2015;15:21–27. doi: 10.17305/bjbms.2015.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cao W, Fang Z, Hou G, Han M, Xu X, Dong J, et al. The psychological impact of the COVID-19 epidemic on college students in China. Psychiatry Res. 2020;287:112934. doi: 10.1016/j.psychres.2020.112934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ahn DG, Shin HJ, Kim MH, Lee S, Kim HS, Myoung J, et al. Current status of epidemiology, diagnosis, therapeutics, and vaccines for novel coronavirus disease 2019 (COVID-19) J Microbiol Biotechnol. 2020;30:313–324. doi: 10.4014/jmb.2003.03011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wan S, Xiang Y, Fang W, Zheng Y, Li B, Hu Y, et al. Clinical features and treatment of COVID-19 patients in Northeast Chongqing. J Med Virol. 2020;92:797–806. doi: 10.1002/jmv.25783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Y-L, Cui Q, Zhang D, Ma X, Zhang G-W. Efficacy of Xuebijing injection for the treatment of coronavirus disease 2019 via network pharmacology. Tradit Med Res. 2020;5:201–215. [Google Scholar]
- 46.Chiow KH, Phoon MC, Putti T, Tan BK, Chow VT. Evaluation of antiviral activities of Houttuynia cordata Thunb. Extract, quercetin, quercetrin and cinanserin on murine coronavirus and dengue virus infection. Asian Pac J Trop Med. 2016;9:1–7. doi: 10.1016/j.apjtm.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu W, Li R, Li X, He J, Jiang S, Liu S, et al. Quercetin as an antiviral agent inhibits influenza a virus (IAV) entry. Viruses. 2015;8(1):6. [DOI] [PMC free article] [PubMed]
- 48.Yang X, Zhu X, Ji H, Deng J, Lu P, Jiang Z, et al. Quercetin synergistically reactivates human immunodeficiency virus type 1 latency by activating nuclear factorkappaB. Mol Med Rep. 2018;17:2501–2508. doi: 10.3892/mmr.2017.8188. [DOI] [PubMed] [Google Scholar]
- 49.Yao C, Xi C, Hu K, Gao W, Cai X, Qin J, et al. Inhibition of enterovirus 71 replication and viral 3C protease by quercetin. Virol J. 2018;15:116. doi: 10.1186/s12985-018-1023-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fan W, Qian S, Qian P, Li X. Antiviral activity of luteolin against Japanese encephalitis virus. Virus Res. 2016;220:112–116. doi: 10.1016/j.virusres.2016.04.021. [DOI] [PubMed] [Google Scholar]
- 51.Peng M, Watanabe S, Chan KWK, He Q, Zhao Y, Zhang Z, et al. Luteolin restricts dengue virus replication through inhibition of the proprotein convertase furin. Antivir Res. 2017;143:176–185. doi: 10.1016/j.antiviral.2017.03.026. [DOI] [PubMed] [Google Scholar]
- 52.Yan H, Ma L, Wang H, Wu S, Huang H, Gu Z, et al. Luteolin decreases the yield of influenza a virus in vitro by interfering with the coat protein I complex expression. J Nat Med. 2019;73:487–496. doi: 10.1007/s11418-019-01287-7. [DOI] [PubMed] [Google Scholar]
- 53.Wei W, Du H, Shao C, Zhou H, Lu Y, Yu L, et al. Screening of antiviral components of Ma Huang Tang and investigation on the Ephedra alkaloids efficacy on influenza virus type a. Front Pharmacol. 2019;10:961. doi: 10.3389/fphar.2019.00961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ricciotti E, FitzGerald GA. Prostaglandins and inflammation. Arterioscler Thromb Vasc Biol. 2011;31:986–1000. doi: 10.1161/ATVBAHA.110.207449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rumzhum NN, Ammit AJ. Cyclooxygenase 2: its regulation, role and impact in airway inflammation. Clin Exp Allergy. 2016;46:397–410. doi: 10.1111/cea.12697. [DOI] [PubMed] [Google Scholar]
- 56.Kudchodkar SB, Levine B. Viruses and autophagy. Rev Med Virol. 2009;19:359–378. doi: 10.1002/rmv.630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hu B, Zhang Y, Jia L, Wu H, Fan C, Sun Y, et al. Binding of the pathogen receptor HSP90AA1 to avibirnavirus VP2 induces autophagy by inactivating the AKT-MTOR pathway. Autophagy. 2015;11:503–515. doi: 10.1080/15548627.2015.1017184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang R, Liu H, Bai C, Wang Y, Zhang X, Guo R, et al. Chemical composition and pharmacological mechanism of Qingfei Paidu decoction and Ma Xing Shi Gan decoction against coronavirus disease 2019 (COVID-19): in silico and experimental study. Pharmacol Res. 2020;157:104820. doi: 10.1016/j.phrs.2020.104820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen J, Wang YK, Gao Y, Hu LS, Yang JW, Wang JR, et al. Protection against COVID-19 injury by qingfei paidu decoction via anti-viral, anti-inflammatory activity and metabolic programming. Biomed Pharmacother. 2020;129:110281. doi: 10.1016/j.biopha.2020.110281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang YX, Ma JR, Wang SQ, Zeng YQ, Zhou CY, Ru YH, et al. Utilizing integrating network pharmacological approaches to investigate the potential mechanism of Ma Xing Shi Gan decoction in treating COVID-19. Eur Rev Med Pharmacol Sci. 2020;24:3360–3384. doi: 10.26355/eurrev_202003_20704. [DOI] [PubMed] [Google Scholar]
- 61.Maarouf M, Rai KR, Goraya MU, Chen JL. Immune ecosystem of virus-infected host tissues. Int J Mol Sci. 2018;19(5):1379. [DOI] [PMC free article] [PubMed]
- 62.Unterholzner L, Almine JF. Camouflage and interception: how pathogens evade detection by intracellular nucleic acid sensors. Immunology. 2019;156:217–227. doi: 10.1111/imm.13030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xie Y, Shi X, Sheng K, Han G, Li W, Zhao Q, et al. PI3K/Akt signaling transduction pathway, erythropoiesis and glycolysis in hypoxia (review) Mol Med Rep. 2019;19:783–791. doi: 10.3892/mmr.2018.9713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mizutani T, Fukushi S, Ishii K, Sasaki Y, Kenri T, Saijo M, et al. Mechanisms of establishment of persistent SARS-CoV-infected cells. Biochem Biophys Res Commun. 2006;347:261–265. doi: 10.1016/j.bbrc.2006.06.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mizutani T, Fukushi S, Saijo M, Kurane I, Morikawa S. Importance of Akt signaling pathway for apoptosis in SARS-CoV-infected Vero E6 cells. Virology. 2004;327:169–174. doi: 10.1016/j.virol.2004.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bohmwald K, Galvez NMS, Canedo-Marroquin G, Pizarro-Ortega MS, Andrade-Parra C, Gomez-Santander F, et al. Contribution of cytokines to tissue damage during human respiratory syncytial virus infection. Front Immunol. 2019;10:452. doi: 10.3389/fimmu.2019.00452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ryzhakov G, Lai CC, Blazek K, To KW. Hussell T, Udalova I. IL-17 boosts proinflammatory outcome of antiviral response in human cells. J Immunol (Baltimore, Md : 1950) 2011;187:5357–5362. doi: 10.4049/jimmunol.1100917. [DOI] [PubMed] [Google Scholar]
- 68.Xu Z, Shi L, Wang Y, Zhang J, Huang L, Zhang C, et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. The lancet. Respir Med. 2020;8:420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kalliolias GD, Ivashkiv LB. TNF biology, pathogenic mechanisms and emerging therapeutic strategies. Nat Rev Rheumatol. 2016;12:49–62. doi: 10.1038/nrrheum.2015.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang SF, Tseng SP, Yen CH, Yang JY, Tsao CH, Shen CW, et al. Antibody-dependent SARS coronavirus infection is mediated by antibodies against spike proteins. Biochem Biophys Res Commun. 2014;451:208–214. doi: 10.1016/j.bbrc.2014.07.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zheng Y, Yang Y, Li Y, Xu L, Wang Y, Guo Z, et al. Ephedrine hydrochloride inhibits PGN-induced inflammatory responses by promoting IL-10 production and decreasing proinflammatory cytokine secretion via the PI3K/Akt/GSK3β pathway. Cell Mol Immunol. 2013;10:330–337. doi: 10.1038/cmi.2013.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang A, Pan W, Lv J, Wu H. Protective effect of amygdalin on LPS-induced acute lung injury by inhibiting NF-κB and NLRP3 signaling pathways. Inflammation. 2017;40:745–751. doi: 10.1007/s10753-017-0518-4. [DOI] [PubMed] [Google Scholar]
- 73.Park CM, Jin KS, Lee YW, Song YS. Luteolin and chicoric acid synergistically inhibited inflammatory responses via inactivation of PI3K-Akt pathway and impairment of NF-κB translocation in LPS stimulated RAW 264.7 cells. Eur J Pharmacol. 2011;660:454–459. doi: 10.1016/j.ejphar.2011.04.007. [DOI] [PubMed] [Google Scholar]
- 74.Li SR, Tang ZJ, Li ZH, Liu X. Searching therapeutic strategy of new coronavirus pneumonia from angiotensin-converting enzyme 2: the target of COVID-19 and SARS-CoV. Eur J Clin Microbiol Infect Dis. 2020;39:1021–1026. doi: 10.1007/s10096-020-03883-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li Q, Kang C. Progress in developing inhibitors of SARS-CoV-2 3C-like protease. Microorganisms. 2020;8(8):1250. [DOI] [PMC free article] [PubMed]
- 76.Liu Y, Liang C, Xin L, Ren X, Tian L, Ju X, et al. The development of coronavirus 3C-like protease (3CL(pro)) inhibitors from 2010 to 2020. Eur J Med Chem. 2020;206:112711. doi: 10.1016/j.ejmech.2020.112711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Huang YF, Bai C, He F, Xie Y, Zhou H. Review on the potential action mechanisms of Chinese medicines in treating coronavirus disease 2019 (COVID-19) Pharmacol Res. 2020;158:104939. doi: 10.1016/j.phrs.2020.104939. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Basic information and network topology parameter values of 47 potentially active compounds obtained by ADME screening. Table S2. The information and network topology parameter values of 178 potential targets of ephedra-bitter almond against COVID-19. Table S3. GO enrichment analysis. Table S4. KEGG enrichment analysis.
Data Availability Statement
All data are available in the manuscript and they are shown in figures and tables.