Abstract
Hearing loss is the most common neurosensory deficit. It results from a variety of heritable and acquired causes and is linked to multiple deleterious effects on a child's development that can be ameliorated by prompt identification and individualized therapies. Diagnosing hearing loss in newborns is challenging, especially in mild or progressive cases, and its management requires a multidisciplinary team of healthcare providers comprising audiologists, pediatricians, otolaryngologists, and genetic counselors. While physiologic newborn hearing screening has resulted in earlier diagnosis of hearing loss than ever before, a growing body of knowledge supports the concurrent implementation of genetic and cytomegalovirus testing to offset the limitations inherent to a singular screening modality. In this review, we discuss the contemporary role of screening for hearing loss in newborns as well as future directions in its diagnosis and treatment.
Keywords: genetic hearing loss, deafness, cytomegalovirus, newborn screening, precision medicine
Introduction
Hearing loss is the most common neurosensory deficit. It affects about 1 in 500 newborns, and by the age of 80 approximately half the population has hearing loss significant enough to interfere with effective communication.1 While causality is multifactorial, on aggregate, at least 50% of cases are linked to genetic causes. For deafness arising pre-lingually (prior to the development of language), genetic causes comprise an even higher percentage.2 Non-genetic causes in neonates and young infants arise from a wide variety of prenatal, perinatal, and postnatal etiologies, including infections, developmental anomalies, hypoxia, trauma, and the use of certain medications.
Prior to the implementation of universal newborn hearing screening (NBHS) in the USA, even until the mid-1990s children born with even profound (>90 dB) hearing loss had significant delays in their diagnosis and treatment. History and physical examination in these patients may reveal no obvious abnormalities, and traditional means of evaluating auditory function in newborns such as behavioral tests are fraught with the potential for bias.3–5 In 1994, the Joint Committee on Infant Hearing (JCIH) endorsed the goal of universal detection of infants with hearing impairment and promoted continued research to improve detection of, and intervention for, deafness as early as possible.6 Laws mandating NBHS were subsequently passed by 43 states and territories, with the remainder of states implementing universal NBHS without legislation. Consequently, the most recent data show that 98.3% of newborns in the USA undergo NBHS,7 adhering to the current JCIH recommendations of NBHS by 1 month of age, diagnosis by 3 months of age, and early intervention by 6 months of age.6,8
In this review, we will discuss the strengths and weaknesses of the current NBHS in the context of permanent, bilateral sensorineural congenital hearing loss. We will also discuss the future direction of hearing screening, including adjunctive hearing screening tests for genetic and infectious causes. Finally, we will focus on the personalized current and future treatments for hearing loss in newborns that are afforded by rapidly advancing diagnostic technology for heritable deafness. Throughout this review, we will use the terms “deaf” with a lowercase “d”, “hearing loss”, or “hard of hearing” to refer to a diminished or lack of a sense of hearing. For many within the Deaf (written with a capital “D”) community, “hearing impairment” is an inappropriate and antiquated term, as they do not consider themselves impaired.9
Physiologic NBHS
Prior to the widespread adoption of screening for hearing loss in newborns, hearing screening was accomplished via distraction or other behavioral tests in infants.10 Only children with specific risk factors for hearing loss were screened at birth. A list of risk factors for hearing loss in childhood is given in Table 1.11,12 That this list was suboptimal is reflected by the fact that >50% of children with hearing loss have none of these risk factors.13 In addition, the testing procedures were inadequate, with validation studies of behavioral tests of hearing in children revealing unacceptably poor inter-observer reliability, high false positive rates, and high false negative rates.3–5 As a consequence, many children with clinically actionable hearing loss were missed during this critical period of auditory cortex neuroplasticity, language acquisition, and development.14 In response to these shortfalls, universal NBHS was endorsed by the American Academy of Pediatrics and the United States Preventative Services Task Force with the goal of screening all infants by 1 month of age, diagnosing hearing loss by 3 months of age, and enacting appropriate intervention by 6 months (1-3-6 paradigm).6,8
Table 1.
Risk factors associated with the development of deafness in newborns.
| 1. Family history of childhood deafness |
| 2. Neonatal intensive care unit stay of >5 days |
| 3. Use of ototoxic medications (e.g. aminoglycosides) |
| 4. Signs and symptoms of maternal infection during pregnancy |
| 5. Asphyxia at birth |
| 6. Prematurity (<32–34 gestational weeks) |
| 7. Low birth weight (<1200 g) |
| 8. Hyperbilirubinemia |
| 9. Craniofacial anomalies of the head and neck |
| 10. Parental consanguinity |
| 11. Parental concerns about their child's hearing |
| 12. Signs associated with syndromic hearing loss (e.g. a white forelock) |
Contemporary physiologic NBHS is a two-tiered approach, with an initial screen via portable automated auditory brainstem response (AABR) or detection of transient evoked or distortion product otoacoustic emissions (OAEs) followed by a formal auditory brainstem response (ABR) for an abnormal initial test.15 Some institutions choose to perform OAE testing followed by AABR prior to formal ABR. The International Pediatric Otolaryngology Group recommends performing both OAE and AABR in children with risk factors for deafness.16 These tests are simple to perform, inexpensive, require only 10–20 minutes to complete, and do not require sedation. Parental response has been overwhelmingly positive,17 and in 2017 over 98% of newborns in the USA were screened.7 The results are astounding—the median age of diagnosis of severe-to-profound hearing loss has improved from >2 years of age prior to universal NBHS to just 2 months of age.18
Despite the unquestionable success of universal NBHS, there are several limitations to the current screening methods. The two-tiered system was designed for the identification of children with moderate, severe, and profound hearing loss as the effect of mild hearing loss was not recognized in the mid-1990s. Both AABR and OAE testing have thresholds of detection of approximately 30–40 dB,8 which means that newborns with mild-to-moderate hearing loss have a longer median time-to-diagnosis and time-to-treatment than newborns with severe-to-profound hearing loss.18 There is now a growing body of literature that links even mild hearing loss to delays in linguistic development, diminished academic achievement, and social difficulties that can be obviated with early identification and individualized resources.19,20 Furthermore, some forms of genetic hearing loss are progressive and because they may begin as mild or even normal hearing, affected children will be under diagnosed or missed completely by the current screening methods.21 False positive test results secondary to environmental factors, debris in the ear canal, and fluid in the middle ear are also common with both OAE and AABR testing,8 and as a consequence, over five normal-hearing newborns will be erroneously referred for evaluation for every one newborn with true hearing loss.22 Poor inter-test reliability between OAE and AABR testing has been reported, with up to a quarter of children with abnormal OAE results having a normal AABR despite ultimately abnormal hearing findings on diagnostic ABR.23 Thus, despite the successes of universal physiologic NBHS, all infants with hearing loss are not identified with current screening techniques.
Current role of genetic diagnosis
Prior to the late 1990s when GJB2 was identified as the cause of hearing loss in three consanguineous Pakistani families, segregating autosomal recessive deafness, genetic testing for non-syndromic hearing loss (NSHL) was not possible.24 In the decade following this discovery, a large number of genes was implicated in non-syndromic hearing loss, although establishing a genetic diagnosis for hearing loss remained challenging. Candidate genes were typically screened for mutations using Sanger sequencing, a highly reliable but relatively expensive and slow technique if many genes were being considered. GJB2 is an exceptional gene—not only is it the leading cause of severe-to-profound autosomal recessive NSHL in many populations around the world, it is also simple to screen, comprising a single coding exon. As a result, in the late 1990s, a few clinical laboratories began to offer genetic testing for deaf and hard-of-hearing patients and their families focusing primarily on testing GJB2.
In 2003, the Human Genome Project (HGP) was completed ahead of schedule and under budget.25 This monumental achievement provided a foundation for the development of massively parallel sequencing (MPS) technologies, which could amplify millions upon millions of 150–300 base pair fragments of DNA and align them to the reference genome established by the HGP. This technology led to the development of OtoSCOPE™, a targeted "deafness" panel of genes for the clinical diagnosis of genetic hearing loss. Initially described in 201026 and made clinically available in 2012, OtoSCOPE™ was the first clinically validated comprehensive genetic panel. MPS also made novel gene discovery much easier, and, consequently, the number of genes associated with NSHL increased markedly.
A variety of MPS genetic panels targeting deafness are now available in the United States, including from 23 to 252 genes (Table 2). Obtaining a specific genetic diagnosis of deafness has significant implications for patients regarding counseling, prognosis, and individualized treatment. Furthermore, information regarding the genetic diagnosis of a child and associated risk of having another affected child may influence future reproductive decisions for parents.27 Obtaining this knowledge is empowering for the patient and their family members alike.
Table 2.
Comparison of the current MPS genetic panels in the United States that target deafness, which range from 23 to 252 genes implicated in autosomal dominant, autosomal recessive, and X-linked NSHL, mitochondrial deafness, a number of the more common syndromic forms of hearing loss, and non-syndromic mimics.
| Name | Institution | Number of genes |
|---|---|---|
| Blueprint Genetics Comprehensive Hearing Loss and Deafness Panel28 | Blueprint Genetics, Seattle, WA | 239 |
| GeneDx Hearing Loss Test29 | GeneDx, Gaithersburg, MD | 150 |
| Hereditary Hearing Loss and Deafness Panel30 | Prevention Genetics, Marshfield, WI | 202 |
| Comprehensive Hearing Loss Panel31 | sema4, Stamford, CT | 92 |
| Fulgent Comprehensive Hearing Loss NGS Panel32 | Fulgent Genetics, Temple City, CA | 167 |
| OtoGenome™33 | Partners Healthcare, Boston, MA | 110 |
| OtoSeq®34 | Cincinnati Children's Hospital Medical Center Laboratory of Genetics and Genomics, Cincinnati, OH | 23 |
| OtoSCOPE® (version 9)35 | Molecular Otolaryngology and Renal Research Laboratories at the University of Iowa, Iowa City, IA | 252 |
| Hearing Loss Panel36 | EGL Genetics, Tucker, GA | 131 |
| Hearing Loss Sequencing Panel37 | Greenwood Genetic Center, Greenwood, SC | 91 |
| Expanded Hearing Loss Panel38 | ARUP Laboratories, Salt Lake City, UT | 56 |
Current role of cytomegalovirus testing and treatment
Congenital cytomegalovirus (cCMV) is estimated to affect ∼0.3%–1.2% of newborns, making it the most common prenatal infection in developed countries.39,40 A primary maternal infection during pregnancy has a fetal transmission rate of ∼32%, but in seropositive mothers the rate of vertical transmission is only about 1.4%. Infection of the newborn is classified as either symptomatic or asymptomatic cCMV.41 Symptomatic cCMV, which affects up to 10% of infected fetuses, manifests as intrauterine growth restriction, preterm delivery, and end-organ dysfunction. Half of this overtly symptomatic group of newborns develop sensorineural hearing loss.42 Asymptomatic cCMV is more difficult to diagnose as there is typically no evidence that an in utero infection has occurred. However, approximately 10% of newborns with asymptomatic cCMV infection present with hearing loss at birth as their only symptom. This association has led to the progressive implementation of hearing-targeted cCMV screening in the USA, with several states having legislation to mandate CMV screening after failed NBHS to identify hearing loss associated with asymptomatic cCMV. Although the presentation of this type of hearing loss is extremely variable and can be unilateral or bilateral and stable, fluctuating or progressive, it is estimated that ∼15%–20% of children with bilateral moderate-to-profound sensorineural hearing loss have CMV-related hearing loss, making CMV the second most common etiology of hearing loss in this subgroup of children after genetic causes.43
Testing all newborns for cCMV may not necessarily be beneficial or cost-effective, as most infected newborns develop no symptoms and have no sequelae. If testing is pursued, the virus must be isolated either prenatally or within the first 2–3 weeks of life, as the presence of CMV after this point cannot be distinguished from a peri- or postnatal infection.44 Culture and polymerase chain reaction (PCR) testing of amniotic fluid via amniocentesis is the method of choice for diagnosing CMV in the fetus41, and this is recommended in the presence of verified primary CMV infection in an expectant mother.41 Because of an attributable risk of 0.49% for fetal demise following amniocentesis,45 this step is not recommended as a routine screening test. In newborns, PCR assays from saliva swabs or urine samples are the preferred method of testing and have sensitivities and specificities approaching 100%.42 Serological testing for CMV-specific IgM antibodies in newborns is not recommended, as only 70% of cCMV-positive newborns have detectable levels of antibody. PCR of dried blood spots also has a limited role in screening for cCMV, with a disappointing sensitivity of 28.3%.46
To optimize value and maximize the yield of cCMV testing, Park and colleagues developed a sequential diagnostic protocol for children with abnormal hearing screenings.47 Because of its relatively low cost in comparison to imaging studies or targeted genetic panels, saliva or urine-based cCMV PCR is performed as the initial test for idiopathic deafness. In a cohort of 83 newborns with confirmed hearing loss who were evaluated with this method over a 5-year period, 30% had confirmed or probable cCMV-associated deafness. This protocol, known as hearing-directed cCMV testing, was enacted into the Utah Health Code in 2013, requiring newborns with abnormal physiologic hearing screenings to undergo cytomegalovirus testing.48 A follow-up study 2 years after implementation revealed that the rate of definitive audiologic evaluation before the age of 3 months for all patients with an abnormal screening examination increased from 56% to 77%.49 This increase may be related to a secondary provision contained within the 2013 bill, which set aside funds for education and outreach regarding CMV and hearing loss for pregnant women and healthcare providers.48 At the time of writing this manuscript, legislation requiring hearing-directed cCMV testing has been enacted in Utah, Connecticut, Iowa, New York, and Virginia.50
Symptomatic cCMV has classically been treated with the guanosine analog ganciclovir parenterally, or its oral prodrug valganciclovir. This medication's triphosphate derivative has a particular affinity for the viral DNA polymerase of CMV.51 A randomized clinical trial in 2003 examining audiometric outcomes via ABR in neonates (≤1 month of age) with symptomatic cCMV who received ganciclovir versus a control showed statistically significant preservation of hearing in the treatment arm.52 The primary toxicity was neutropenia, which developed in over half of children on treatment. A continuation of this study was published in 2015 and investigated the effect of a 6-week versus a 6-month course of valganciclovir against placebo.53 Although no difference in hearing thresholds was noted 6 months after treatment, a modest improvement was noted 12–24 months after treatment in the longer treatment arm. Unfortunately, neither study enrolled significant numbers of patients with asymptomatic cCMV and sensorineural hearing loss and so conclusions regarding treatment for these babies could not be made.
Small, uncontrolled case series and case reports have been published investigating valganciclovir for hearing loss in asymptomatic cCMV, but the lack of a placebo control group in these studies has made it impossible to conclude whether there is an improvement in hearing as a result of a medication effect versus the naturally waxing and waning hearing loss associated with cCMV.54–57 Currently, there are three clinical trials investigating the effect of valganciclovir treatment on the progression and severity of cCMV-associated sensorineural hearing loss.58–60 Other lingering questions regarding long-term use of ganciclovir or valganciclovir revolve around as-yet-unknown sequelae. Animal models have demonstrated a risk for gonadotoxicity and carcinogenicity,61 and although these complications have not been reported in humans to date, parents or legal guardians should be counseled on the potential for these effects prior to initiating therapy.
Finally, it is important to recognize that hearing loss associated with asymptomatic cCMV is characterized by its variability. In contrast to the typically symmetric hearing loss seen with nonsyndromic genetic deafness, the hearing loss with asymptomatic cCMV can be asymmetric, fluctuating, and progressive.62 In up to half of cases, the hearing loss is late-onset and does not appear until the child is several years of age. Furthermore, a positive diagnosis of cCMV does not exclude genetic hearing loss. In one study, for example, of 12 children with hearing loss and a diagnosis of asymptomatic cCMV infection, two had a genetic cause for their hearing loss.63 Both of these children had bilateral, symmetric, severe-to-profound hearing loss. While some children with asymptomatic cCMV have audioprofiles indistinguishable from genetic deafness, a positive viral test with a symmetric severe-to-profound sensorineural hearing loss warrants suspicion for an underlying genetic cause and comprehensive genetic testing should be completed.
CMV testing cannot be used as a stand-alone diagnostic tool for deafness in newborns. However, as an adjunct to a thorough history and physical examination and comprehensive genetic testing, it provides valuable information that narrows the list of potential etiologies for hearing loss. In selected clinical scenarios, it also allows for initiation of antiviral treatment with the potential for amelioration of hearing loss in affected children.
The future of hearing screening and challenges to implementation
Physiologic hearing screening, genetic testing, and cCMV screening each have strengths and weaknesses, which are summarized in Table 3. A screening strategy that combines these methods will maximize the number of infants diagnosed with hearing loss and in doing so, society will benefit via a reduction of economic burden and lost wages.64 More importantly, individual patients will benefit from the personalized therapy, counseling, and appropriate treatment afforded by an early diagnosis of hearing loss.
Table 3.
Comparison of strengths and weaknesses of current and future newborn hearing screening techniques.
| Physiologic | Genetic | cCMV | |
|---|---|---|---|
| Strengths | Quick and easy to perform | Provides definitive diagnosis and etiology | Highly accurate |
| Inexpensive | Phenotype-genotype correlation | Inexpensive | |
| Minimal risk | Highly accurate | Minimal risk | |
| Weaknesses | High false positive rate | Most expensive option | High rate of asymptomatic carriers |
| No information on etiology of deafness | Potential for genetic discrimination | Must be collected within 2–3 weeks of birth |
cCMV—congenital cytomegalovirus
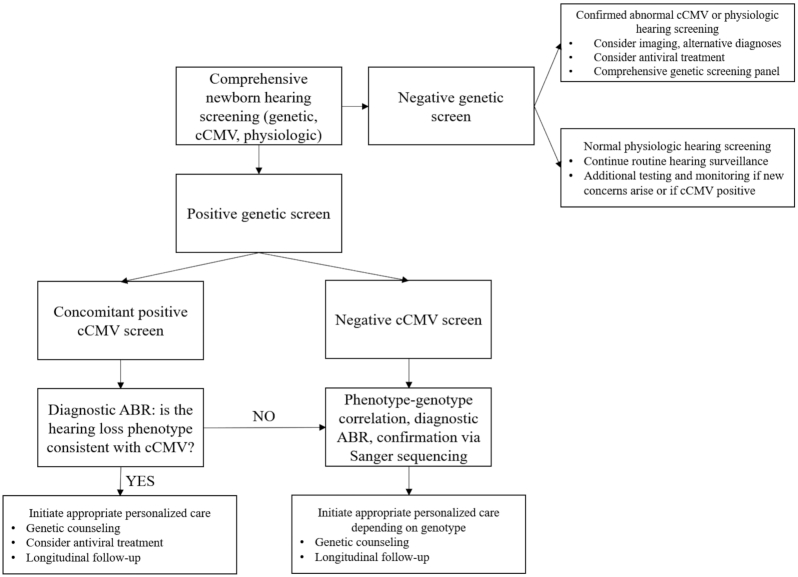
A proposal for comprehensive hearing screening involving physiologic, genetic, and cCMV testing was described in 2019,65 and a flowchart incorporating all three methods of NBHS for otherwise healthy-appearing newborns is shown in Fig. 1. While whole genome sequencing is likely to be the primary method by which newborns are screened within the next few decades,66 the method of delivery and timing of genetic screening for deafness in the newborn is currently debated. Clinicians in China recently implemented concurrent physiologic and genetic hearing screening using microarrays that tested between 7 and 18 variants associated with deafness in GJB2, SLC26A4, and GJB3.67,68 This panel also tested for two variants that predispose for aminoglycoside-related ototoxicity in MT-RNR1. Although the microarrays included 16 variants that are relatively common causes of genetic deafness, bi-allelic genotypes involving these variants alone would only diagnose 134 out of 2050 patients (6.5%) who received a definitive genetic diagnosis via a comprehensive MPS panel (Table 4). For SLC26A4, 464 variants are currently classified as likely pathogenic or pathogenic in the Deafness Variation Database (DVD), a comprehensive database of all known variants responsible for genetic hearing loss, highlighting the difficulty of obtaining an acceptable diagnostic yield via microarray.69 Nonetheless, these studies have provided promising results. In total, 420 105 babies received genetic screening during the study periods. 111 babies received a diagnosis of genetic deafness as a result of the presence of bi-allelic GJB2 or SLC26A4 variants. Thirty-two of these 111 patients (28.8%) passed their NBHS. Twenty of these 32 patients (71.9%) eventually developed hearing loss by 60 months of age. By receiving a diagnosis of genetic deafness at birth, these children were able to access personalized therapies immediately, including cochlear implantation in at least 12.
Figure 1.
Proposed molecular diagnostic algorithm for genetic hearing screening.
Table 4.
List of patients diagnosed by OtoSCOPE™ stratified by type of genetic deafness.
| OtoSCOPE patient groups, from its inception to October 2019 | Number of patients | Number of unique variants |
|---|---|---|
| All unique patients with positive diagnoses (% of total) | 2050 (100.0%) | 1692 |
| All patients with autosomal recessive deafness (% of total) | 1525 (74.4%) | 1250 |
| All patients with autosomal dominant, X-linked, or mitochondrial deafness (% of total) | 525 (25.6%) | 442 |
| All patients with GJB2-associated deafness (% of total) | 405 (19.8%) | 80 |
| All patients with SLC26A4-associated deafness (% of total) | 116 (5.7%) | 97 |
| Patients with bi-allelic genotypes for microarray variants described by Dai et al.67 and Guo et al.68 (% of total) | 134 (6.5%) | 16 |
This is further stratified by number of patients who were diagnosed with GJB2 or SLC26A4-associated deafness, and the number of patients who would be diagnosed using a microarray as proposed by Dai et al.67 and Guo et al.68 The GJB2 variants on the microarray are NM_004004.5: c.35delG, c.167delT, c.176_191del16, c.235delC, and c.299_300delAT. The SLC26A4 variants on the microarray are NM_000441.1: c.281C > T, c.589G > A, c.919–2A > G, c.1174A > T, c.1226G > A, c.1229C > T, c.1707 + 5G > A, c.1975G > C, c.2027T > A, c.2162C > T, and c.2168A > G.
Suitable genetic screening technology for the heterogenous population of the United States has yet to be incorporated on a large scale. With over 8000 deafness-associated variants classified as likely pathogenic or pathogenic, an optimal screening platform must be capable of testing for a large number of mutations.65,70 As the numbers of targeted genomic regions and patients being tested increase, MPS becomes the most cost-effective option.71,72 Furthermore, copy number variants (CNVs)—insertions or deletions in the genome that result in structural variations—are implicated in about ∼20% of diagnoses, making them a common contributor to inherited deafness and mandating their detection on screening platforms.73 Although microarrays are capable of detecting CNVs via comparative genomic hybridization or single nucleotide polymorphism arrays, the diagnostic yield is low and the false negative and false positive rates are high.74 MPS, by comparison, is a high yield diagnostic method for the detection of CNVs.75
In accordance with the updated Wilson and Jungner criteria for the implementation of genetic screening programs, the overall benefits of the screening must outweigh its harms.76 Sequencing portions of the genome must be performed judiciously; although MPS can generate incredible amounts of data, this comes at a cost of an ever-higher number of variants of uncertain significance (VUS). Per the American College of Medical Genetics and Genomics, a VUS is a variant with clinical relevance unknown, and thus should not be used in clinical decision making.77 The DVD reports >695 000 VUSs within genes that are associated with deafness.70 Some bioethicists argue that VUSs should be reported to patients under the principle of hypothetical utilitarianism (i.e. the potential that the VUS may someday be impactful), whereas others posit counterarguments to reporting on principles of avoiding harm.78
Childbirth is a highly stressful time for parents. For the purposes of a NBHS, every effort must be made to minimize false positive results.79 For this reason, we advocate a middle ground between a small microarray panel and a full diagnostic deafness panel with a targeted MPS panel that uses high throughput detection of thousands of confirmed pathogenic or likely pathogenic variants at costs comparable to other newborn screening examinations. These variants must be thoughtfully selected based on population-specific frequencies to prevent ethnic bias and should include both autosomal recessive and autosomal dominant causes of deafness. Furthermore, targeting genetic deafness associated with mild-to-moderate hearing loss will be particularly valuable as a complement to physiologic hearing screening, as AABR and OAE may miss these phenotypes. Variants included in this type of targeted detection panel are easily modifiable to populations of interest and as our knowledge of the genetics of deafness increases.
Other ethical concerns to genetic testing (and other hearing screening methods for newborns) include ethnic biases. Especially in the context of a targeted genetic screening panel, ethnic bias must be addressed. For Mendelian inheritance, genetic diseases common in European populations are over diagnosed compared with their relative frequency, whereas diseases common in African populations are under diagnosed.80 Significant differences exist between racial groups in their access to genetic research and genetic testing as well as their concerns regarding the misuse of genetic testing.81 This disparity is reflected in the Genome Aggregation Database, a collection of exome and genome sequencing data, which contains 110 588 alleles from non-Finnish Europeans compared with only 15 152 from African populations.82 Accordingly, the design of a targeted genetic deafness panel must use ethnicity-specific minor allele frequencies for deafness-associated variants to ensure equity and reduce disparities in healthcare.
Over 90% of deaf children are born to hearing parents and most, therefore, do not have frequent contact with other deaf persons.73,83 Among normal hearing parents with deaf children, there is overwhelmingly positive sentiment in regards to the benefits of genetic testing from both those who have and have not already sought out genetic tests.84,85 Of parents who receive a genetic diagnosis for their child, reported benefits include a better understanding of risk of recurrence in future children, a reduction in blame toward themselves for their child's deafness, and personalized guidance of treatment.86 However, despite having a generally positive opinion of genetic testing, parents who are members of the Deaf community are more cautious and skeptical than hearing parents.87,88 As a linguistic minority, the potential for “ethnocide” of Deaf communities must be considered: the advent of treatments for deafness including cochlear implantation could be viewed as a means of forced integration of these communities.
We contend that even if a parent within the Deaf community is not interested in cochlear implantation, hearing amplification, or other treatments for their child with hereditary deafness, a genetic diagnosis would provide value through counseling on potential comorbid conditions and the expected trajectory of deafness. Sign language is a sophisticated form of nonverbal communication that offers the full depth of language,89 and an earlier diagnosis of hearing loss would result in earlier access to individualized resources to acquire this skill. Ultimately, despite strong evidence for a high benefit-risk ratio, newborn screening requires informed consent from parents. Under the auspices of parental autonomy, parents would possess the moral and legal rights to refuse any part of the comprehensive NBHS for their child.
In regards to cCMV, with PCR already being a cost-effective and accurate method of detection,42 the most important lingering question for screening implementation is the timing of testing. Should all infants be screened at birth or should testing be limited to those with abnormal physiologic hearing screenings? Screening may not be beneficial for all newborns, as many with cCMV will be asymptomatic and never develop hearing loss or other sequelae.42 With the poor positive predictive value of physiologic hearing screening22 and high rate of asymptomatic cCMV infections, most children with both a positive cCMV test and an abnormal physiologic screen will ultimately have normal hearing. Furthermore, the requirement that a sample for PCR of cCMV be taken within 2–3 weeks of birth44 removes the possibility of waiting until a confirmatory ABR shows hearing loss to test for the virus. As the second leading cause of congenital deafness, it is reasonable to incorporate the simultaneous collection of saliva or urine for cCMV PCR along with collection of dried blood spot or saliva for targeted genetic screening and performance of physiologic hearing screening. Without concurrent data from genetic testing and a confirmatory ABR, however, current data do not justify antiviral treatment for a positive cCMV test in an otherwise healthy newborn. A flowchart incorporating these three methods is described in Fig. 1.
Hearing amplification and cochlear implantation
Early hearing detection and intervention (EHDI) is the process by which children navigate from detection of hearing loss via screening or clinician referral to an individualized intervention program. When an infant is confirmed to have hearing loss, a multidisciplinary team is required for audiologic, medical, and educational management.90 This team includes, but is not limited to, audiologists, pediatricians, otolaryngologists, and genetic counselors. The strongest influence of success in language development in deaf children is early identification and therapy.91 Six months of age is the critical intervention period before which adequate habilitation (including learning sign language) can allow for similar outcomes to normal-hearing peers.92
For the development of spoken language in a deaf child, current treatment options include hearing amplification and cochlear implantation. Hearing amplification is an excellent option for children with mild-to-moderate congenital sensorineural loss, as hearing aids have a maximum gain of approximately 41–58 dB.93 With severe-to-profound hearing loss, use of conventional hearing aids is limited by a small dynamic range and potential for uncomfortable overamplification.94 In children with diagnosed autosomal dominant forms of hearing loss, audioprofiles are a useful tool for prognostication. For example, a child born with a pathogenic variant in KCNQ4 would be expected to progress to a severe high frequency hearing loss between 20 and 40 years of age. Conventional hearing amplification may suffice prior to that point, but the family should be counseled on the expected temporal course of hearing loss and available treatment options. A child with bi-allelic pathogenic variants in STRC, however, would be expected to have a stable level of mild-to-moderate hearing loss and may obtain satisfactory results for their entire life with hearing aids alone.
Cochlear implants, the first effective treatment for severe-to-profound deafness, have revolutionized the field of otology over the past several decades.95 Genetic testing using a commercially available genetic deafness panel offers useful pre-operative information for cochlear implantation regarding the etiology and expected benefit of the surgery. Some authors suggest its implementation in the routine pre-operative workup of all potential cochlear implant recipients.96,97 Patients with pathogenic variants in genes involved in the structure and function of the spiral ganglion—OPA1, DIAPH3, DFNB59, AIFM1, TBC1D24, MT-RNR1,and TMPRSS3—perform significantly worse on post-operative speech measurements compared with those who had other deafness-associated pathogenic variants.98 Patients with pathogenic variants in GJB2 and SLC26A4 have been noted to have particularly successful results after cochlear implantation,97,99 although positive outcomes have also been reported with mutations in other organ of Corti-associated genes such as MYH9100 and MYO15A.96,101 These differences in outcome based on genetic etiology may explain the small subset of deaf children that do not gain significant benefit from cochlear implantation.101 Thus, another benefit of early genetic diagnosis of deafness is earlier eligibility and improved pre-operative counseling for cochlear implantation.
Gene therapy
Alongside advancements in genetic sequencing technology, significant progress has been made regarding our knowledge of the molecular and biochemical pathways interrupted in genetic deafness.102–104 After the first Food and Drug Administration (FDA)-approved clinical protocol for human gene therapy was successfully implemented in 1990,105 interest has risen in novel treatments for various genetic diseases, including deafness. Contemporary methods for gene therapy include gene replacement, gene suppression, targeted gene editing, and cell replacement.106 For deafness, the application of these methods is made more challenging by difficulty in accessing the inner ear and its encasing bony labyrinth, which places limitations on the volume of vector that can be delivered to the cochlea. Furthermore, the final common outcome of many types of deafness as audiometric thresholds fall into the severe range, is irreversible damage to the inner and outer hair cells. Nonetheless, several deafness-associated genes have emerged as targets potentially amenable to curative gene therapy.
Because of their favorable tolerance by the host immune system, lack of pathogenicity, and excellent transduction potential of many different cell types in the cochlea, adeno-associated viruses (AAV) are the most commonly studied and used vectors for gene therapy delivery in deafness.106 As recently reviewed by Askew and Chien,107 improvements in hearing threshold have been demonstrated in murine models via AAV-mediated inner ear gene delivery for Vglut3,108Kcnq1,109Tmc1,109,110Whrn,111Pjvk,112Msrb3,113Lhfpl5,114Ush1c,115Ush1g,116Ush3a,117–119Slc26a4,120 and Otof.121,122 Although these results are promising, differences between murine and human inner ear development limit the translational applicability to humans. For example, most of the gene therapies were administered to mice between P0 and P10 (0 and 10 days after birth) when the murine inner ear is still immature.123 Equivalent experiments on a human would need to be delivered in utero at ∼19 weeks' gestational age. Newer surgical techniques in mice in which a second hole is made in the bony labyrinth have been developed to allow gene therapy to be delivered to mice at any age. This advance is important as it will help to identify a series of candidate genes to move forward for consideration in human trials based on murine data that show benefit in preventing progression and even reversing progression of specific types of genetic hearing loss.
Potential candidates for human trials include OTOF and TMC1.121 The first clinical trials for gene therapy of non-syndromic hearing loss will soon begin with OTOF. Akouos, Inc. has issued patent US20190185864A1 in relation to its novel adeno-associated viral vector AK-OTOF, with plans to begin recruiting for a clinical trial in 2020.124 Bi-allelic pathogenic mutations in OTOF, which encodes for the otoferlin transmembrane protein, are associated either with prelingual severe-to-profound bilateral sensorineural hearing loss or a normal-to-mild hearing loss that progresses to severe or profound hearing loss with fevers (temperature-sensitive nonsyndromic auditory neuropathy).125 These phenotypes present in a variant-dependent fashion and affect different components of the auditory pathway, which may be important for both the effectiveness of gene therapy and cochlear implantation. This highlights the importance of obtaining a specific diagnosis via MPS for both characterizing phenotype and for guiding treatment.
Conclusions
Physiologic NBHS has improved the time to diagnosis and treatment of severe-to-profound hearing loss in newborns. Unfortunately, many newborns with mild or progressive forms of hearing loss remain undiagnosed during their critical period of language development and neuroplasticity. The time has come to expand NBHS to include genetic and cCMV testing to maximize our ability to diagnose these patients. Advancing the diagnosis and care of congenital permanent hearing loss will require development of a novel, economic method of universal genetic screening. Providing patients with a genetic diagnosis of hearing loss is useful for identifying comorbid conditions, providing reproductive counseling, and guiding treatments including cochlear implantation and soon, gene therapy. With the use of targeted high throughput MPS technology, genetic screening in newborns will soon become a reality. Its implementation will require a therapeutic alliance with clinicians and parents in both the Deaf and hearing communities.
Acknowledgements
This project was supported by NIH-NIDCD (Grants No. R01 DC002842, DC012049, and DC017955) (RJHS), and NIH-NIDCD (Grant No. 5T32DC000040) (RKT).
Contributor Information
Ryan K Thorpe, Molecular Otolaryngology and Renal Research Laboratories, Carver College of Medicine, University of Iowa, 375 Newton Rd, Iowa City, IA 52242, USA; Department of Otolaryngology – Head and Neck Surgery, University of Iowa, 200 Hawkins Dr, Iowa City, IA 52242, USA.
Richard J H Smith, Molecular Otolaryngology and Renal Research Laboratories, Carver College of Medicine, University of Iowa, 375 Newton Rd, Iowa City, IA 52242, USA; Department of Otolaryngology – Head and Neck Surgery, University of Iowa, 200 Hawkins Dr, Iowa City, IA 52242, USA; The Interdisciplinary Graduate Program in Genetics, University of Iowa, 375 Newton Rd, Iowa City, IA 52242, USA; Iowa Institute of Human Genetics, University of Iowa, 375 Newton Rd, Iowa City, IA 52242, USA.
Author contributions
RKT and RJHS conceived the presented idea. RKT was responsible for initial drafting of the manuscript. RJHS was responsible for critical revision and editing of the manuscript. RJHS supervised the project.
Conflict of interest
RKT has no conflicts of interest to report. RJHS directs the Molecular Otolaryngology and Renal Research Laboratories, which offers MPS as a clinical diagnostic test for hearing loss.
References
- 1. Morton NE. Genetic epidemiology of hearing impairment. Ann N Y Acad Sci. 1991;630:16–31. doi:10.1111/j.1749-6632.1991.tb19572.x [DOI] [PubMed] [Google Scholar]
- 2. Shearer AE, Hildebrand MS, Smith RJ. Hereditary Hearing Loss and Deafness Overview. 1993. http://www.ncbi.nlm.nih.gov/pubmed/20301607. [Google Scholar]
- 3. Ling D, Ling AH, Doehring DG. Stimulus, response, and observer variables in the auditory screening of newborn infants. J Speech Hear Res. 1970;13:9–18. doi:10.1044/jshr.1301.09. [DOI] [PubMed] [Google Scholar]
- 4. Gans DP, Flexer C. Observer bias in the hearing testing of profoundly involved multiply handicapped children. Ear Hear. 1982;3:309–13. doi:10.1097/00003446-198211000-00004. [DOI] [PubMed] [Google Scholar]
- 5. Russ SA, Poulakis Z, Wake M, et al. The distraction test: The last word?. J Paediatr Child Health. 2005;41:197–200. doi:10.1111/j.1440-1754.2005.00587.x. [DOI] [PubMed] [Google Scholar]
- 6. Joint Committee on Infant Hearing 1994 Position Statement. American Academy of Pediatrics Joint Committee on Infant Hearing. Pediatrics. 1995;95:152–6. http://www.ncbi.nlm.nih.gov/pubmed/7770297. [PubMed] [Google Scholar]
- 7. 2017 Annual Data Early Hearing Detection and Intervention (EHDI) Program. Centers for Disease Control and Prevention, National Center on Birth Defects and Developmental Disabilties. https://www.cdc.gov/ncbddd/hearingloss/ehdi-data2017.html. Published 2017.
- 8. Patel H, Feldman M. Universal newborn hearing screening. Paediatr Child Health. 2011;16:301–5. doi:10.1093/pch/16.5.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Community and Culture – Frequently Asked Questions. National Association of the Deaf. https://www.nad.org/resources/american-sign-language/community-and-culture-frequently-asked-questions/.
- 10. Stewart-Brown S, Haslum MN. Screening for hearing loss in childhood: a study of national practice. BMJ. 1987;294:1386–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Joint committee on infant hearing position statement 1982. Ear Hear. 1983;4:3–4. doi:10.1136/bmj.294.6584.1386. [DOI] [PubMed] [Google Scholar]
- 12. Kountakis SE, Psifidis A, Chang CJet al. Risk factors associated with hearing loss in neonates. Am J Otolaryngol. 1997;18:90–3. doi:10.1016/S0196-0709(97)90093-4. [DOI] [PubMed] [Google Scholar]
- 13. Mauk GW, White KR, Mortensen LBet al. The effectiveness of screening programs based on high-risk characteristics in early identification of hearing impairment. Ear Hear. 1991;12:312–19. doi:10.1097/00003446-199110000-00003. [DOI] [PubMed] [Google Scholar]
- 14. Cardon G, Campbell J, Sharma A. Plasticity in the developing auditory cortex: evidence from children with sensorineural hearing loss and auditory neuropathy spectrum disorder. J Am Acad Audiol. 2012;23:396–411. doi:10.3766/jaaa.23.6.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Choo D, Meinzen-Derr J. Universal newborn hearing screening in 2010. Curr Opin Otolaryngol Head Neck Surg. 2010;18:399–404. doi:10.1097%2FMOO.0b013e32833d475d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liming BJ, Carter J, Cheng A, et al. International Pediatric Otolaryngology Group (IPOG) consensus recommendations: Hearing loss in the pediatric patient. Int J Pediatr Otorhinolaryngol. 2016;90:251–8. doi: 10.1016/j.ijporl.2016.09.016. [DOI] [PubMed] [Google Scholar]
- 17. Magnuson M, Hergils L. The parents’ view on hearing screening in newborns: Feelings, thoughts and opinions on otoacoustic emissions screening. Scand Audiol. 1999;28:47–56.doi:10.1080/010503999424905. [DOI] [PubMed] [Google Scholar]
- 18. Harrison M, Roush J, Wallace J. Trends in age of identification and intervention in infants with hearing loss. Ear Hear. 2003;24:89–95. doi:10.1097/01.AUD.0000051749.40991.1F. [DOI] [PubMed] [Google Scholar]
- 19. Tomblin JB, Harrison M, Ambrose SE, et al. Language outcomes in young children with mild to severe hearing loss. Ear Hear. 2015;36:76S–91S. doi:10.1097%2FAUD.0000000000000219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Stika CJ, Eisenberg LS, Johnson KC, et al. Developmental outcomes of early-identified children who are hard of hearing at 12 to 18 months of age. Early Hum Dev. 2015;91:47–55. doi:10.1016/j.earlhumdev.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Korver AMH, Smith RJH, Van Camp G, et al. Congenital hearing loss. Nat Rev Dis Prim. 2017;3:16094. doi:10.1038/nrdp.2016.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Szyfter W, Wróbel M, Radziszewska-Konopka M, et al. Polish universal neonatal hearing screening program—4-year experience (2003–2006). Int J Pediatr Otorhinolaryngol. 2008;72:1783–7. doi:10.1016/j.ijporl.2008.08.015. [DOI] [PubMed] [Google Scholar]
- 23. Levit Y, Himmelfarb M, Dollberg S. Sensitivity of the automated auditory brainstem response in neonatal hearing screening. Pediatrics. 2015;136:e641–7. doi:10.1542/peds.2014-3784. [DOI] [PubMed] [Google Scholar]
- 24. Kelsell DP, Dunlop J, Stevens HPet al. Connexin 26 mutations in hereditary non-syndromic sensorineural deafness. Nature. 1997;387:80–3. doi:10.1038/387080a0. [DOI] [PubMed] [Google Scholar]
- 25. Collins FS. The human genome project: lessons from large-scale biology. Science. 2003;300:286–90. doi:10.1126/science.1084564. [DOI] [PubMed] [Google Scholar]
- 26. Shearer AE, DeLuca AP, Hildebrand MS, et al. Comprehensive genetic testing for hereditary hearing loss using massively parallel sequencing. Proc Natl Acad Sci. 2010;107:21104–9. doi:10.1073%2Fpnas.1012989107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Frets PG, Duivenvoorden HJ, Verhage F, et al. Factors influencing the reproductive decision after genetic counseling. Am J Med Genet. 1990;35:496–502. doi:10.1002/ajmg.1320350411. [DOI] [PubMed] [Google Scholar]
- 28. Blueprint Comprehensive Hearing Loss and Deafness Panel. Genetic Testing Registry. https://www.ncbi.nlm.nih.gov/gtr/tests/552698/. Published2018. Accessed July 1, 2020.
- 29. GeneDx Hearing Loss Test. Genetic Testing Registry. https://www.ncbi.nlm.nih.gov/gtr/tests/569708/. Published 2019. Accessed July 1, 2020.
- 30. Hereditary Hearing Loss and Deafness Panel. Genetic Testing Registry. https://www.ncbi.nlm.nih.gov/gtr/tests/567331/. Published2020. Accessed July 1, 2020.
- 31. Comprehensive Hearing Loss Panel. Genetic Testing Registry. https://www.ncbi.nlm.nih.gov/gtr/tests/570440/. Published2020. Accessed July 1, 2020.
- 32. Fulgent Comprehensive Hearing Loss NGS Panel. Genetic Testing Registry. https://www.ncbi.nlm.nih.gov/gtr/tests/566571/. Published2019. Accessed July 1, 2020.
- 33. OtoGenome Test for Hearing Loss. Genetic Testing Registry. https://www.ncbi.nlm.nih.gov/gtr/tests/509148/. Published 2020. Accessed July 1, 2020.
- 34. OtoSeq Hearing Loss Panel by next-generation sequencing. Genetic Testing Registry. https://www.ncbi.nlm.nih.gov/gtr/tests/500213/. Published2019. Accessed July 1, 2020.
- 35. OtoSCOPE. Genetic Testing Registry. https://www.ncbi.nlm.nih.gov/gtr/tests/323596/. Published2019. Accessed July 1, 2020.
- 36. Hearing Loss: Sequencing and Deletion/Duplication Panel. Genetic Testing Registry. https://www.ncbi.nlm.nih.gov/gtr/tests/512601/. Published 2020. Accessed July 1, 2020.
- 37. Hearing Loss Sequencing Panel. Genetic Testing Registry. https://www.ncbi.nlm.nih.gov/gtr/tests/548231/. Published 2020. Accessed July 1, 2020.
- 38. Expanded Hearing Loss Panel, Sequencing and Deletion/Duplication. Genetic Testing Registry. https://www.ncbi.nlm.nih.gov/gtr/tests/515701/. Published 2019. Accessed July 1, 2020.
- 39. Jones CA. Congenital cytomegalovirus infection. Curr Probl Pediatr Adolesc Health Care. 2003;33:70–93. doi:10.1067/mps.2003.3. [DOI] [PubMed] [Google Scholar]
- 40. Ross DS, Dollard SC, Victor M, et al. The epidemiology and prevention of congenital cytomegalovirus infection and disease: activities of the centers for disease control and prevention workgroup. J Women's Heal. 2006;15:224–9. doi:10.1089/jwh.2006.15.224. [DOI] [PubMed] [Google Scholar]
- 41. Bonalumi S, Trapanese A, Santamaria A, et al. Cytomegalovirus infection in pregnancy: review of the literature. J Prenat Med. 2011;5:1–8. http://www.ncbi.nlm.nih.gov/pubmed/22439067 [PMC free article] [PubMed] [Google Scholar]
- 42. Marsico C, Kimberlin DW. Congenital Cytomegalovirus infection: advances and challenges in diagnosis, prevention and treatment. Ital J Pediatr. 2017;43:38. doi:10.1186%2Fs13052-017-0358-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Grosse SD, Ross DS, Dollard SC. Congenital cytomegalovirus (CMV) infection as a cause of permanent bilateral hearing loss: A quantitative assessment. J Clin Virol. 2008;41:57–62.doi:10.1016/j.jcv.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 44. Lazzarotto T, Guerra B, Lanari M, et al. New advances in the diagnosis of congenital cytomegalovirus infection. J Clin Virol. 2008;41:192–7. doi:10.1016/j.jcv.2007.10.015. [DOI] [PubMed] [Google Scholar]
- 45. Kozlowski P, Knippel A, Stressig R. Individual risk of fetal loss following routine second trimester amniocentesis: A controlled study of 20,460 cases. Ultraschall der Medizin - Eur J Ultrasound. 2007;29:165–72. doi:10.1055/s-2007-963217. [DOI] [PubMed] [Google Scholar]
- 46. Boppana SB. Dried blood spot real-time polymerase chain reaction assays to screen newborns for congenital cytomegalovirus infection. JAMA. 2010;303:1375. doi:10.1001/jama.2010.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Park AH, Duval M, McVicar Set al. A diagnostic paradigm including cytomegalovirus testing for idiopathic pediatric sensorineural hearing loss. Laryngoscope. 2014;124:2624–9. doi:10.1002/lary.24752. [DOI] [PubMed] [Google Scholar]
- 48. Menlove R. H.B. 81 Cytomegalovirus Public Health Initiative. Utah State Legislature. https://le.utah.gov/∼2013/bills/static/hb0081.html. Published 2013. Accessed February 4, 2020.
- 49. Diener ML, Zick CD, McVicar SB, et al. Outcomes from a hearing-targeted cytomegalovirus screening program. Pediatrics. 2017;139:e20160789. doi:10.1542/peds.2016-0789. [DOI] [PubMed] [Google Scholar]
- 50. Congenital CMV Legislation in the United States. National CMV Foundation. https://www.nationalcmv.org/about-us/advocacy. Published2020.
- 51. Matthews T, Boehme R. Antiviral activity and mechanism of action of ganciclovir. Clin Infect Dis. 1988;10:S490–4. doi:10.1093/clinids/10.supplement_3.s490. [DOI] [PubMed] [Google Scholar]
- 52. Kimberlin DW, Lin C-Y, Sánchez PJ, et al. Effect of ganciclovir therapy on hearing in symptomatic congenital cytomegalovirus disease involving the central nervous system: a randomized, controlled trial. J Pediatr. 2003;143:16–25. doi:10.1016/s0022-3476(03)00192-6. [DOI] [PubMed] [Google Scholar]
- 53. Kimberlin DW, Jester PM, Sánchez PJ, et al. Valganciclovir for symptomatic congenital cytomegalovirus disease. N Engl J Med. 2015;372:933–43.doi:10.1056/nejmoa1404599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Pasternak Y, Ziv L, Attias J, et al. Valganciclovir is beneficial in children with congenital cytomegalovirus and isolated hearing loss. J Pediatr. 2018;199:166–70. doi:10.1016/j.jpeds.2018.02.028. [DOI] [PubMed] [Google Scholar]
- 55. Yilmaz Ciftdogan D, Vardar F. Effect on hearing of oral valganciclovir for asymptomatic congenital cytomegalovirus infection. J Trop Pediatr. 2011;57:132–4. doi:10.1093/tropej/fmq050. [DOI] [PubMed] [Google Scholar]
- 56. McCrary H, Sheng X, Greene T, et al. Long-term hearing outcomes of children with symptomatic congenital CMV treated with valganciclovir. Int J Pediatr Otorhinolaryngol. 2019;118:124–127. doi:10.1016/j.ijporl.2018.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Jansen CFM, Toet MC, Rademaker CMA, et al. Treatment of symptomatic congenital cytomegalovirus infection with valganciclovir. J Perinat Med. 2005;33::364–366. doi:10.1515/JPM.2005.065. [DOI] [PubMed] [Google Scholar]
- 58. Park A. Randomized controlled trial of valganciclovir for cytomegalovirus infected hearing impaired infants (ValEAR). ClinicalTrials.gov. https://clinicaltrials.gov/ct2/show/NCT03107871. Published 2020. Accessed February 4, 2020.
- 59. Vossen ACTM. Congenital Cytomegalovirus: Efficacy of Antiviral Treatment (CONCERT 2). ClinicalTrials.gov. https://clinicaltrials.gov/ct2/show/NCT02005822. Published 2020. Accessed February 4, 2020.
- 60. Valganciclovir Therapy in Infants and Children With Congenital CMV Infection and Hearing Loss. ClinicalTrials.gov. https://clinicaltrials.gov/ct2/show/NCT01649869. Published 2020. Accessed February 4, 2020.
- 61. Luck SE, Wieringa JW, Blázquez-Gamero D, et al. Congenital cytomegalovirus. Pediatr Infect Dis J. 2017;36:1205–13. doi:10.1097/inf.0000000000001763. [DOI] [PubMed] [Google Scholar]
- 62. Fowler KB. Congenital cytomegalovirus infection: Audiologic outcome. Clin Infect Dis. 2013;57:S182–4. doi:10.1093/cid/cit609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Peterson J, Nishimura C, Smith RJH. Genetic testing for congenital bilateral hearing loss in the context of targeted cytomegalovirus screening. Laryngoscope. January2020.doi:10.1002/lary.28536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Schroeder L. The economic costs of congenital bilateral permanent childhood hearing impairment. Pediatrics. 2006;117:1101–12. doi:10.1542/peds.2005-1335. [DOI] [PubMed] [Google Scholar]
- 65. Shearer AE, Shen J, Amr Set al. A proposal for comprehensive newborn hearing screening to improve identification of deaf and hard-of-hearing children. Genet Med. 2019;21:2614–30. doi:10.1038/s41436-019-0563-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Botkin JR, Rothwell E. Whole genome sequencing and newborn screening. Curr Genet Med Rep. 2016;4:1–6. doi:10.1007%2Fs40142-016-0084-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Dai P, Huang L-H, Wang G-J, et al. Concurrent hearing and genetic screening of 180,469 neonates with follow-up in Beijing, China. Am J Hum Genet. 2019;105:803–12. doi:10.1016/j.ajhg.2019.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Guo L, Xiang J, Sun Let al. Concurrent hearing and genetic screening in a general newborn population. Hum Genet. 2020;139:521–30. doi:10.1007/s00439-020-02118-6. [DOI] [PubMed] [Google Scholar]
- 69. Deafness Variation Database. Molecular otolaryngology & renal research lab at the University of Iowa. http://deafnessvariationdatabase.org/. Published2020.
- 70. Azaiez H, Booth KT, Ephraim SS, et al. Genomic landscape and mutational signatures of deafness-associated genes. Am J Hum Genet. 2018;103:484–97. doi:10.1016/j.ajhg.2018.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. König K, Peifer M, Fassunke J, et al. Implementation of amplicon parallel sequencing leads to improvement of diagnosis and therapy of lung cancer patients. J Thorac Oncol. 2015;10:1049–57. doi:10.1097/jto.0000000000000570. [DOI] [PubMed] [Google Scholar]
- 72. Walsh T, Lee MK, Casadei S, et al. Detection of inherited mutations for breast and ovarian cancer using genomic capture and massively parallel sequencing. Proc Natl Acad Sci. 2010;107:12629–33. doi:10.1073%2Fpnas.1007983107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Shearer A, Kolbe DL, Azaiez H, et al. Copy number variants are a common cause of non-syndromic hearing loss. Genome Med. 2014;6:37, doi:10.1186%2Fgm554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Carter NP. Methods and strategies for analyzing copy number variation using DNA microarrays. Nat Genet. 2007;39:S16–S21. doi:10.1038/ng2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Medvedev P, Stanciu M, Brudno M. Computational methods for discovering structural variation with next-generation sequencing. Nat Methods. 2009;6:S13–S20. doi:10.1038/nmeth.1374. [DOI] [PubMed] [Google Scholar]
- 76. Andermann A. Revisting Wilson and Jungner in the genomic age: a review of screening criteria over the past 40 years. Bull World Health Organ. 2008;86:317–19.doi:10.2471%2FBLT.07.050112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–23.doi:10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Hofmann B. Incidental findings of uncertain significance: To know or not to know - that is not the question. BMC Med Ethics. 2016;17:13. doi:10.1186/s12910-016-0096-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Schmidt JL, Castellanos-Brown K, Childress S, et al. The impact of false-positive newborn screening results on families: a qualitative study. Genet Med. 2012;14:76–80. doi:10.1038/gim.2011.5. [DOI] [PubMed] [Google Scholar]
- 80. Sirugo G, Williams SM, Tishkoff SA. The missing diversity in human genetic studies. Cell. 2019;177:26–31. doi:10.1016/j.cell.2019.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Suther S, Kiros G-E. Barriers to the use of genetic testing: A study of racial and ethnic disparities. Genet Med. 2009;11:655–62. doi:10.1097/gim.0b013e3181ab22aa. [DOI] [PubMed] [Google Scholar]
- 82. Karczewski K, Francioli L, MacArthur D, et al. The mutational constraint spectrum quantified from variation in 141,456 humans, Nature. 2020;; 581:434–443. doi:10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Gorlin R, Toriello H, Cohen M. Hereditary Hearing Loss and Its Syndromes. New York: Oxford University Press; 1995. [Google Scholar]
- 84. Fu S, Dong J, Wang C, Chen G. Parental attitudes toward genetic testing for prelingual deafness in China. Int J Pediatr Otorhinolaryngol. 2010;74:1122–5. doi:10.1016/j.ijporl.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 85. Brunger JW, Murray GS, O'Riordan M, et al. Parental attitudes toward genetic testing for pediatric deafness. Am J Hum Genet. 2000;67:1621–5. doi:10.1086/316901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Newton V. Benefits of an early identification and diagnosis of permanent bilateral hearing loss. Hear Balanc Commun. 2013;11:91–9. doi:10.3109/21695717.2013.820512. [Google Scholar]
- 87. Boudreault P, Baldwin EE, Fox M, et al. Deaf adults’ reasons for genetic testing depend on cultural affiliation: Results from a prospective, longitudinal genetic counseling and testing study. J Deaf Stud Deaf Educ. 2010;15:209–27. doi:10.1093/deafed/enq012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Guillemin M, Gillam L. Attitudes to genetic testing for deafness: the importance of informed choice. J Genet Couns. 2006;15:51–9. doi:10.1007/s10897-005-9003-6. [DOI] [PubMed] [Google Scholar]
- 89. Goldin-Meadow S, Brentari D. Gesture, sign, and language: The coming of age of sign language and gesture studies. Behav Brain Sci. 2017;40:e46. doi:10.1017/s0140525x15001247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Year 2019 Position Statement: Principles and Guidelines for Early Hearing Detection and Intervention Programs. J Early Hear Detect Interv, 4:1–44. doi:10.15142/fptk-b748. [Google Scholar]
- 91. Levitt H, McGarr N, Geffner D, et al. Development of language and communication skills in hearing-impaired children - The effects of hearing status of the family and age of intervention on receptive and expressive oral language skills in hearing impaired infants. ASHA Monogr. 1987;1:9–24. http://www.ncbi.nlm.nih.gov/pubmed/3509663. [PubMed] [Google Scholar]
- 92. Yoshinaga-Itano C, Sedey AL, Coulter DK, et al. Language of Early- and Later-identified children with hearing loss. Pediatrics. 1998;102:1161–71. doi:10.1542/peds.102.5.1161. [DOI] [PubMed] [Google Scholar]
- 93. Erickson FN, Van Tasell DJ. Maximum Real-Ear Gain of In-the-Ear hearing aids. J Speech, Lang Hear Res. 1991;34:351–9. doi:10.1044/jshr.3402.351. [DOI] [PubMed] [Google Scholar]
- 94. Jorgensen L, Benson E, McCreery R. Conventional amplification for children and adults with severe-to-profound hearing loss. Semin Hear. 2018;39:364–76. doi:10.1055%2Fs-0038-1670699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Eshraghi AA, Nazarian R, Telischi FFet al. The cochlear implant: Historical aspects and future prospects. Anat Rec Adv Integr Anat Evol Biol. 2012;295:1967–80. doi:10.1002%2Far.22580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Miyagawa M, Nishio S, Ikeda T, et al. Massively parallel DNA sequencing successfully identifies new causative mutations in deafness genes in patients with cochlear implantation and EAS. Kimura A, ed. PLoS One. 2013;8:e75793. doi:10.1371/journal.pone.0075793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Park JH, Kim AR, Han JH, et al. Outcome of cochlear implantation in prelingually deafened children according to molecular genetic etiology. Ear Hear. 2017;38:e316–24. doi:10.1097/AUD.0000000000000437. [DOI] [PubMed] [Google Scholar]
- 98. Shearer AE, Eppsteiner RW, Frees K, et al. Genetic variants in the peripheral auditory system significantly affect adult cochlear implant performance. Hear Res. 2017;348:138–42. doi:10.1016/j.heares.2017.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Yan Y, Li Y, Yang T, et al. The effect of GJB2 and SLC26A4 gene mutations on rehabilitative outcomes in pediatric cochlear implant patients. Eur Arch Oto-Rhino-Laryngology. 2013;270:2865–70. doi:10.1007/s00405-012-2330-y. [DOI] [PubMed] [Google Scholar]
- 100. Pecci A, Verver EJ, Schlegel N, et al. Cochlear implantation is safe and effective in patients with MYH9-related disease. Orphanet J Rare Dis. 2014;9:100. doi:10.1186%2F1750-1172-9-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Miyagawa M, Nishio S, Hattori M, et al. Mutations in the MYO15A gene are a significant cause of nonsyndromic hearing loss. Ann Otol Rhinol Laryngol. 2015;124:158S–68S. doi:10.1177%2F0003489415575058. [DOI] [PubMed] [Google Scholar]
- 102. Dror AA, Avraham KB. Hearing impairment: A panoply of genes and functions. Neuron. 2010;68:293–308. doi:10.1016/j.neuron.2010.10.011. [DOI] [PubMed] [Google Scholar]
- 103. Stamatiou GA, Stankovic KM. A comprehensive network and pathway analysis of human deafness genes. Otol Neurotol. 2013;34:961–70. doi:10.1097/MAO.0b013e3182898272. [DOI] [PubMed] [Google Scholar]
- 104. Lenz DR, Avraham KB. Hereditary hearing loss: From human mutation to mechanism. Hear Res. 2011;281:3–10. doi:10.1016/j.heares.2011.05.021. [DOI] [PubMed] [Google Scholar]
- 105. Rosenberg SA, Aebersold P, Cornetta K, et al. Gene transfer into humans — immunotherapy of patients with advanced melanoma, using tumor-infiltrating lymphocytes modified by retroviral gene transduction. N Engl J Med. 1990;323:570–8. doi:10.1056/nejm199008303230904. [DOI] [PubMed] [Google Scholar]
- 106. Omichi R, Shibata SB, Morton CC, et al. Gene therapy for hearing loss. Hum Mol Genet. 2019;28:R65–79. doi:10.1093/hmg/ddz129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Askew C, Chien WW. Adeno-associated virus gene replacement for recessive inner ear dysfunction: Progress and challenges. Hear Res. [published online ahead of print March 18, 2020],doi:10.1016/j.heares.2020.107947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Akil O, Seal RP, Burke K, et al. Restoration of hearing in the VGLUT3 knockout mouse using virally mediated gene therapy. Neuron. 2012;75:283–93. doi:10.1016%2Fj.neuron.2012.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Chien WW, Monzack EL, McDougald DS, et al. Gene therapy for sensorineural hearing loss. Ear Hear. 2015;36:1–7. doi:10.1097/aud.0000000000000088. [DOI] [PubMed] [Google Scholar]
- 110. Askew C, Rochat C, Pan B, et al. Tmc gene therapy restores auditory function in deaf mice. Sci Transl Med. 2015;7:295ra108–295ra108. doi:10.1126/scitranslmed.aab1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Isgrig K, Shteamer JW, Belyantseva IA, et al. Gene therapy restores balance and auditory functions in a mouse model of Usher syndrome. Mol Ther. 2017;25:780–91. doi:10.1016/j.ymthe.2017.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Delmaghani S, Defourny J, Aghaie A, et al. Hypervulnerability to sound exposure through impaired adaptive proliferation of peroxisomes. Cell. 2015;163:894–906. doi:10.1016/j.cell.2015.10.023. [DOI] [PubMed] [Google Scholar]
- 113. Kim M-A, Cho H-J, Bae S-H, et al. Methionine sulfoxide reductase B3-Targeted in utero gene therapy rescues hearing function in a mouse model of congenital sensorineural hearing loss. Antioxid Redox Signal. 2016;24:590–602. doi:10.1089/ars.2015.6442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. György B, Sage C, Indzhykulian AA, et al. Rescue of hearing by gene delivery to inner-ear hair cells using exosome-associated AAV. Mol Ther. 2017;25:379–91.doi:10.1016/j.ymthe.2016.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Pan B, Askew C, Galvin Aet al. Gene therapy restores auditory and vestibular function in a mouse model of Usher syndrome type 1c. Nat Biotechnol. 2017;35:264–72. doi:10.1038/nbt.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Emptoz A, Michel V, Lelli A, et al. Local gene therapy durably restores vestibular function in a mouse model of Usher syndrome type 1G. Proc Natl Acad Sci. 2017;114:9695–700. doi:10.1073/pnas.1708894114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Geng R, Omar A, Gopal SRet al. Modeling and preventing progressive hearing loss in Usher Syndrome III. Sci Rep. 2017;7:13480. doi:10.1038/s41598-017-13620-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Dulon D, Papal S, Patni P, et al. Clarin-1 gene transfer rescues auditory synaptopathy in model of Usher syndrome. J Clin Invest. 2018;128:3382–401. doi:10.1172/jci94351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. György B, Meijer EJ, Ivanchenko MV, et al. Gene transfer with AAV9-PHP.B rescues hearing in a mouse model of Usher Syndrome 3A and transduces hair cells in a non-human primate. Mol Ther - Methods Clin Dev. 2019;13:1–13. doi:10.1016/j.omtm.2018.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Kim M-A, Kim SH, Ryu N, et al. Gene therapy for hereditary hearing loss by SLC26A4 mutations in mice reveals distinct functional roles of pendrin in normal hearing. Theranostics. 2019;9:7184–99. doi:10.7150/thno.38032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Akil O, Dyka F, Calvet C, et al. Dual AAV-mediated gene therapy restores hearing in a DFNB9 mouse model. Proc Natl Acad Sci. 2019;116:4496–501. doi:10.1073/pnas.1817537116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Al-Moyed H, Cepeda AP, Jung S, et al. A dual-AAV approach restores fast exocytosis and partially rescues auditory function in deaf otoferlin knock-out mice. EMBO Mol Med. 2019;11:e9396. doi:10.15252/emmm.201809396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Wang L, Kempton JB, Brigande JV. Gene therapy in mouse models of deafness and balance dysfunction. Front Mol Neurosci. 2018;11: 300. doi:10.3389/fnmol.2018.00300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Simons EJ, Reisinger E. Compositions and methods for treating non-age-associated hearing impairment in a human subject. 2017. https://patents.google.com/patent/WO2018039375A1/en. Accessed February 10, 2020.
- 125. Shearer AE, Smith RJ. OTOF-Related Deafness, In: Adam MP, Ardinger HH, Pagon RA, et al, eds.GeneReviews®. Seattle, WA: University of Washington; 1993:Online. http://www.ncbi.nlm.nih.gov/pubmed/20301429. [PubMed] [Google Scholar]