Key Points
Innate immune and stool microbiome signatures of HEU children are region specific.
Differences in HEU immune responses were unrelated to the microbiome.
Future immune studies of HEU children must be studied across diverse settings.
Visual Abstract
Abstract
In both high- and low-income countries, HIV-negative children born to HIV-positive mothers (HIV exposed, uninfected [HEU]) are more susceptible to severe infection than HIV-unexposed, uninfected (HUU) children, with altered innate immunity hypothesized to be a cause. Both the gut microbiome and systemic innate immunity differ across biogeographically distinct settings, and the two are known to influence each other. And although the gut microbiome is influenced by HIV infection and may contribute to altered immunity, the biogeography of immune-microbiome correlations among HEU children have not been investigated. To address this, we compared the innate response and the stool microbiome of 2-y-old HEU and HUU children from Belgium, Canada, and South Africa to test the hypothesis that region-specific immune alterations directly correlate to differences in their stool microbiomes. We did not detect a universal immune or microbiome signature underlying differences between HEU versus HUU that was applicable to all children. But as hypothesized, population-specific differences in stool microbiomes were readily detected and included reduced abundances of short-chain fatty acid–producing bacteria in Canadian HEU children. Furthermore, we did not identify innate immune-microbiome associations that distinguished HEU from HUU children in any population. These findings suggest that maternal HIV infection is independently associated with differences in both innate immunity and the stool microbiome in a biogeographical population-specific way.
Introduction
Globally, over 15 million women of reproductive age are living with HIV (1). The provision of antiretroviral (ARV) treatment during pregnancy to prevent HIV transmission to their children increased from 47% in 2010 to 82% in 2018. With this increase in ARV coverage, the mother-to-child transmission of HIV has reached the global rate of 6.8% (2). Despite this success, HIV-exposed, uninfected (HEU) children are still more vulnerable to severe infection and mortality in infancy than HIV-unexposed, uninfected (HUU) children in low-, middle-, and high-income countries (3–5). The reasons for this vulnerability are not understood and could include a combination of factors ranging from socioeconomics to in utero exposure to HIV and ARV medications. A plausible biological mechanism could involve alterations in innate and adaptive immune responses. Certain ARV drugs are indeed already known to modulate aspects of host immunity (6, 7). The few studies that evaluated innate immune status in HEU compared with HUU children found that HEU children have fewer circulating innate immune cells (8, 9), have the tendency to mount higher proinflammatory responses (10, 11) and have lower antiviral responses (10, 12) compared with HUU children. Recently, early maternal ARV was associated with improved clinical outcome and lower immune activation (11). However, each of these studies involved only single-country populations. It thus remains unclear if alterations of the immune system of HEU children in early life are similar around the globe. This is a crucial aspect to delineate, as immune alterations common to all HEU child populations would suggest a common mechanism (such as HIV or ARV exposure), and thus, targeting a common pathway may be of benefit to all [e.g., based on recent findings, ARV initiation before conception may improve clinical outcomes of HEU children (11)]. If the immune differences between HEU and HUU children are not universally present but population specific, the likely culprits would not be HIV or ARVs, and avenues to improve outcomes for HEU children would have to focus on local factors.
One of the aspects known to differ dramatically among populations and with profound impact on immunity is the gut microbiome. Specifically, the microbiome is now a well-established modulator of mucosal and systemic immune responses (13, 14). There are several reasons to hypothesize that gut microbiomes of HEU differ from HUU children. HIV infection is associated with altered gut microbiomes and even differs between treated and antiretroviral therapy–naive, HIV-infected adults (15, 16). Human milk oligosaccharide composition, a key factor modifying gut microbial composition, is different in HIV-positive compared with HIV-negative mothers (17). Furthermore, formula feeding remains a recommendation for HIV-positive mothers living in some resource-rich nations to reduce risk of HIV transmission through breastmilk (18), and formula feeding itself is associated with altered gut microbiomes in infants (19, 20). However, only one study on gut microbiomes of HEU children has been published thus far, finding that the microbiomes of breastfed Haitian HEU children differed from those of HUU infants and correlated with certain human milk oligosaccharides in their mothers’ breastmilk (17).
Most immune studies to date contrasting HEU to HUU children have been performed in African populations living either in Africa or Europe (8, 10–12) and one population in Haiti (17). Yet, both microbiomes (21, 22) and immune responses (23, 24) were shown to differ from one population to another among healthy infants. Thus, to put in context the effect of HIV exposure on immune responses and microbiomes and, importantly, the associations between the two, they must be evaluated simultaneously in diverse populations to determine any population-specific and universal differences between HEU and HUU children.
To determine population-level differences between HEU and HUU children, we recruited three cohorts of HEU and HUU children from distinct regions and measured both cytokine production in response to pattern recognition receptor (PRR) ligands and stool microbiomes based on 16S amplicon sequencing. Using a stringently standardized immune assay designed to eliminate the effect of site, we previously identified site-specific immune signatures (23, 24) and microbiome composition (N. Amenyogbe, P. Dimitriu, K.K. Smolen, E.M. Brown, C.P. Shannon, S.J. Tebbutt, P.J. Cooper, A. Marchant, T. Ghoetghebuer, M. Esser, B.B. Finlay, T.R. Kollmann, and W.W. Mohn, submitted for publication) among HUU children from these cohorts. In the current study, we identified alterations in immune responses and stool microbiomes of HEU versus HUU children in three geographically distinct populations. Furthermore, using multiomic integration, we assessed whether a relationship existed between immune and microbiome data, which are both related to HIV exposure.
Materials and Methods
Ethics statement
This work was done in accordance with ethical principles of the Declaration of Helsinki. This research was approved by The University of British Columbia Institutional Ethics Board.
Study participants
HEU and HUU children were recruited between January 2011 and March 2012 at all study sites.
Both HUU and HEU children of ∼2 y of age were recruited from Vancouver, Canada; Brussels, Belgium; and Cape Town, South Africa. Study participants were only included in the study if they were considered healthy based on medical history. They were excluded if they met one or more of the following criteria: significant chronic medical condition, immune deficiency, immunosuppression by disease or medication, cancer, bone marrow or organ transplantation, receipt of blood products within 3 mo, bleeding disorder, major congenital malformation, or genetic disorder. The only difference in criteria between HUU and HEU children was that only the latter were born to HIV-positive mothers. Further details of participant recruitment were published in a previous study involving only the HUU children (24).
Innate immune phenotyping
Whole blood cytokine release assay.
Innate immune phenotyping for these cohorts was previously described and published (23, 24). Briefly, peripheral blood was drawn, and samples were kept at room temperature <4 h and stimulated with one of five PRR ligands: PAM3CYSK4 (PAM; ligand for TLR2:1, catalog no. tlrl-pms; InvivoGen) at 0.1 mg/ml, polyinosinic–polycytidylic acid [Poly(I:C), ligand TLR3, catalog no. 27473201; GE Healthcare] at 10 mg/ml, LPS (ligand TLR4, catalog no. tlrl-pelps; InvivoGen) at 1 ng/ml, peptidoglycan (PGN; ligand for TLR2:1 and nucleotide-binding oligomerization domain-containing protein 1/2 [NOD1/2], catalog no. tlrl-pgnsa; InvivoGen) at 1 mg/ml, or R848 (resiquimod, ligand for TLR7:8, catalog no. tlrl-r848; InvivoGen) at 1 mM. Unstimulated controls were not treated with any ligands. Whole blood was incubated with the ligands for 24 h at 37°C. Supernatant was then harvested after centrifugation to pellet cells and stored at −80°C until further analysis. All samples were analyzed within 1 y of collection at The University of British Columbia laboratory. The following serum cytokine concentrations were measured using the Luminex multiplex assay (Upstate/MilliporeSigma Flex Kit system; Luminex): IFN-α2, IFN-γ, CXCL10, IL-12p70, IL-12p40, IL-6, TNF-α, IL-1β, CXCL8, CCL3, CCL4, IL-10, and ELISA for IL-23.
Intracellular cytokine flow cytometry assay.
Whole blood was stimulated with PRR ligands in parallel to those described for the cytokine release assays above and subsequently analyzed via flow cytometry. Detailed methods of this assay were previously described (23). In addition to the ligands, brefeldin A (10 μg/ml) was added to each well. Whole blood diluted 1:1 in RPMI 1640 medium was added to each well. Cells were incubated for 6 h, then treated with 2 mM EDTA for 10 min at 37°C, and then suspended in BD FACS Lyse and stored at −80°C until analysis. Flow cytometric analysis was used to identify proportions of monocytes (HLA−DR+ and CD14+), conventional dendritic cells (cDCs; HLA−DR+, CD14−, CD11c+, and CD123−), plasmacytoid dendritic cells (HLA−DR+, CD14−, CD11c−, and CD123+), αβ T cells (CD3+ and γδTCR−), γδ T cells (CD3+ and γδTCR+), B cells (HLA−DR+, CD14−, CD11c−, and CD123−), and granulocytes (HLA−DR− and CD14+). Intracellular production of IL-6, IL-12, IFN-α, IFN-γ, and TNF-α were also measured via median fluorescence intensity of cytokines within the cell type of interest.
Child fecal microbiome analysis
Stool sample collection.
Stool samples were collected by the children’s parents at home, kept at 4°C, and brought to the research laboratory during the study visit within 24 h of the bowel movement. At the laboratory, samples were aliquoted into 2-ml screw-top tubes except for samples from Vancouver, which were stored in 50-ml conical tubes. All stool samples were processed and stored at −80°C within 24 h of arrival to the laboratory. Stool samples from Brussels and Cape Town were shipped to the central study site (The University of British Columbia, Vancouver, Canada) on dry ice with a temperature monitor to ensure all samples remained below −80°C during transport. Samples were stored at −80°C in Vancouver for no longer than 24 mo prior to DNA extraction. All stool samples were collected within the same month as the blood sample, with the exception of two Canadian HEU children, whose stools were obtained either 5 mo later or 1 mo prior to blood draw. Among the South African children, four HEU children provided a stool sample at a previous study visit occurring 5–7 mo prior to the collection of the blood sample.
Stool DNA extraction.
Total DNA was extracted from 180 to 220 mg stool using the QIAamp DNA Stool Mini Kit (catalog no. 51504; QIAGEN) following the manufacturer’s protocol for isolation of DNA from stool for pathogen detection. The following modifications were made: 1) all heated incubations were at 95°C, and 2) for step two of the protocol, stool samples were homogenized by adding 1.4 ml ASL buffer to each sample and shaken in a Disruptor Genie (catalog no. SI-DD238; Scientific Industries) for 2–3 min or until stool was thoroughly homogenized.
16S amplicon sequencing.
PCR amplification and sequencing were performed using previously described protocols and rationale (25). Briefly, PCR amplification targeting the V6 region of the 16S rRNA gene was performed using 2–5 ng of template DNA from each sample.
Each PCR contained 2–5 ng template DNA and GoTaq hot start 2× colorless Master Mix from Promega (catalog no. M5131). The following primers were added at 0.8 pm/μl: forward primer, 5′-ACACTCTTTCCCTACACGACGCTCTTCCGATCTnnnn[BC8mer]CWACGCGARGAACCTTACC-3′; and reverse primer, 5′-CGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATCTnnnn[BC8mer]ACRACACGAGCTGACGAC-3′.
Cycling conditions included 1-min activation of the Taq at 95°C, followed by 25 cycles of 95, 55, and 72°C at 1 min each. Amplified samples were quantified via QBit and the dsDNA reagent and then pooled at equimolar amounts. Pooled samples were purified on a PCR cleanup column. The purified library was diluted 1:100 in deionized water Illumina adapters attached using the following primers: OLJ139, 5′-AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGA-3′; and OLJ140, 5′-CAAGCAGAAGACGGCATACGAGATCGGTCTCGGCATTCCTGCTGAAC-3′.
The cycling conditions used were the same as those used for V6-16S RNA gene amplification, except with an annealing temperature of 60°C and 10 cycles. The product was purified, and 10% phiX174 DNA was added. This library was loaded onto an Illumina MiSeq instrument and sequenced with a paired-end 2 × 100 cycle format and reagents.
16S amplicon sequence analysis.
Paired-end reads were assembled using XOR-based Read Overlapper (R. J. Dickson and G. B. Gloor, manuscript posted on arXiv), and only sequences without ambiguous bases were retained for further analysis. Assembled sequences were quality filtered and binned into operational taxonomic units (OTUs) with UPARSE v7.1 (26). Briefly, assembled reads were abundance sorted and clustered into 97% similarity OTUs. Representative sequences (excluding singletons) from each OTU were further chimera filtered against the gold database (http://drive5.com/uchime/gold.fa). Reads were then mapped backed to OTUs and converted into an OTU table.
Statistical analysis
Baseline demographics.
Baseline demographics between HEU and HUU children were compared in each cohort separately. For continuous variables, the data among HEU or HUU cohorts displayed skewedness (outside the range of −1 to 1) or kurtosis (outside the range of −3 to 3) for some cohorts but not others. For consistency, we applied the nonparametric Wilcoxon rank-sum test to all continuous variables and the data presented as medians and interquartile ranges. For categorical variables, the Fisher exact test was applied. Unadjusted p values < 0.05 were considered significant.
Overview of statistical analyses
The goals of this study were to 1) identify differences in both immune responses and gut microbiomes between HEU and HUU children across diverse settings, and 2) to determine if the differences in immune responses correlated to those in the microbiome. The statistical analyses were selected to determine the global impact of cohort of origin alongside HIV exposure on cytokine responses by applying data-type specific cluster analyses (principal component analysis [PCA] for immune data and nonmetric multidimensional scaling [NMDS] for compositional microbiome data). To interrogate the effect of HIV exposure on individual features, methods based on linear regression were used with appropriate p value corrections. This univariate approach, however, failed to identify features predictive of phenotype that also covary in a multidimensional space. Hence, we used the mixOmics framework, which offers tools to extract sparse signatures from high-dimensional data either from individual datasets (sparse partial least squares–discriminant analysis [sPLS-DA]) (27) or integrated across multiple datasets (Data Integration Analysis for Biomarker Discovery using Latent Variable Approaches for Omics studies [DIABLO]) (28). Although sPLS-DA was performed to select covarying cytokine responses or microbial OTUs predictive of HIV exposure, feature selection by DIABLO was further constrained by covariance between immune cytokine and microbiome data. By comparing the classification error rate from both analyses, we addressed our second goal of determining whether differences in the gut microbiome correlated with systemic innate immune responses.
PCA of cytokine data.
Prior to PCA analysis, cytokine values were log10 transformed and scaled. The PCA was constructed with each unique data point constituting one individual stimulated with one ligand. For each ligand, variance explained by site and HIV exposure were determined and, for each ligand at each site, variance explained by HIV exposure. The variance explained in each case was determined using permutational multivariate ANOVA [PERMANOVA; adonis test on Euclidean distance in the R package vegan (29)].
Univariate analysis of cytokine response data.
The effect of HIV exposure on cytokine responses to PRR ligands was tested within each regional cohort separately. Cytokine data were log10 transformed prior to univariate analysis. For cytokine production in response to ligand, we used multiple linear regression and adjusted for levels produced in the unstimulated sample and for time of recruitment. For unstimulated cytokine production, we selected only cytokines in which at least 70% of subjects had unstimulated values above the detection limit, with a median fluorescence intensity >10. Both baseline and stimulation response analysis were adjusted for recruitment date for the Canadian cohort only, in which HEU children were recruited at a significantly different point in time compared with HUU children. Recruitment day was calculated using the number of days from the recruitment of the first to the last participant in the study. The p values were adjusted for false discovery across all ligand–cytokine combinations within each cohort using the Benjamini–Hochberg method. For all significant findings, multiple linear regression was used and included factors that were not balanced within that cohort: child sex and delivery mode in the Belgian cohort and weight-for-length z-scores (WLZ), height-for-age z-scores (HAZ), and maternal age for the Canadian cohort. Because of sample size, each demographic model was tested in a separate model. Within the Belgian cohort only, multiple linear regression was used to test whether any significant cytokines differed between HEU children whose mothers initiated ARV therapy preconception or during pregnancy.
Analysis of flow cytometry data.
Proportions of cells types in HEU and HUU children, within each regional cohort, were compared using the Wilcoxon test, and p values were adjusted within each cohort using the Benjamini–Hochberg method. Differences in the percent of cDCs, plasmacytoid dendritic cells, and monocytes producing IL-6, IL-12p40, TNF-α, IFN-α, and IFN-γ were tested using linear regression, adjusted for percent producers in the unstimulated controls. Both baseline and stimulation response analyses were adjusted for recruitment date for the Canadian cohort only, as described above for cytokine response analysis. The p values were adjusted for all ligand–cytokine combinations for each cell type separately using the Benjamini–Hochberg method.
Microbiome analysis
α Diversity.
We estimated microbial diversity with abundance-dependent (Shannon) metrics after subsampling the OTU table to account for unequal sampling depth. To test for relationships between host factors and Shannon diversity, we used the Wilcoxon test.
β Diversity.
We used the phyloseq R package (30) to compute β diversity using the Bray–Curtis index. Sample clustering was visualized using NMDS. To test whether HIV exposure explained community composition among all children or within regional cohorts, we used the PERMANOVA test [Adonis test on Bray–Curtis distance from R package vegan (29)].
Differential abundance.
Following analysis of α and β diversity, we stratified all further analyses by regional cohort. Only OTUs present in at least 6% of subjects were retained within each site. To identify differentially abundant OTUs between HEU and HUU children, we used the Wald test implemented in the R package DESeq2 (31). OTUs were considered significant if the Benjamini–Hochberg adjusted p value was <0.05.
Integrated analysis of cytokine and microbiome data
sPLS-DA.
For both microbiome and soluble cytokine datasets, we tested the ability of significantly different features to classify HEU from HUU children within each regional cohort using sPLS-DA (27), implemented in the R mixOmics package. Cytokine data were log10 transformed as per univariate analysis. OTU count data were first transformed into relative abundance, then normalized using the centered log-ratio transformation with an offset of 0.1 times the minimum value for each individual (the default offset in the mixOmics clr function). Models were first tuned to select the number of features required to achieve the minimum classification error rate, using Mfold validation with at least 10 subjects per fold and 100 iterations. The final models were run on two components, using the numbers of features selected by the tuning process. Classification error was assessed using the maximum error rate.
Cytokine and microbiome data integration.
To test whether OTU–cytokine correlations distinguished HEU from HUU children within each regional cohort, we used sparse Generalized Canonical Correlation Discriminant Analysis or DIABLO (28) implemented in the R mixOmics package. Cytokine and microbiome data were normalized using the same transformations as per sPLS-DA analysis. All data were scaled. We used the DIABLO full model, in which selected features across data types are constrained to the discriminant factor (HIV exposure) and to the features in the other data block (i.e., to maximize correlation across OTU and microbiome data). We first tuned the model to determine the maximum number of OTUs and cytokines to achieve the lowest classification error rate across two components, with a minimum of five features per dataset and a maximum equal to the number of samples. The final models used the selected number of features and error assessed using the maximum error rate.
Data accessibility.
Raw 16S amplicon sequencing data have been deposited at a publicly accessible server; the National Center for Biotechnology Information Sequence Read Archive and can be access via the following accession number: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA660015/). Data used for statistical analyses, and associated scripts, are publicly available and can be accessed on GitHub via the following url:https://github.com/nelly-amenyogbe/Global_HEU_immune_microbiome.
Results
Regional cohort characteristics
We recruited 21 HUU and 21 HEU children in Belgium, 33 HUU and 16 HEU children in Canada, and 20 HUU and 16 HEU children in South Africa. For some children, only immune or only microbiome data were available (Tables I, II, and III). HEU and HUU children differed demographically in a cohort-specific manner (Tables I, II, and III). Belgian HUU differed from HEU children in sex and delivery mode (Table I). Canadian HUU differed from HEU children in age and anthropometric measurements WLZ, HAZ, and maternal age, whereas HEU were younger and had higher WLZ but lower HAZ than HUU children. Canadian HEU children were also born to younger mothers than HUU children (Table II). South African HEU and HUU children differed for weight-for-age z-scores (WAZ), and differences in HAZ approached significance. (Table III). Maternal ARV therapy initiation differed among the three sites (Table IV). The Belgian cohort included the highest proportion of mothers starting ARV therapy prior to pregnancy. Most Canadian mothers began ARV in the second trimester, whereas most South African mothers began ARV in the third trimester. All HIV-positive mothers in the current study were advised to exclusively formula feed their children from birth to prevent HIV transmission, whereas all the HUU children in the current study were breastfed. Duration of breastfeeding was not recorded. Study participants were recruited between November 2011 and May 2012 (Supplemental Table I). In Canada only, recruitment period differed for HEU and HUU children.
Table I. Belgian cohort characteristics.
| HUU |
HEU |
|||
|---|---|---|---|---|
| n = 14 | n = 21 | p | ||
| Data available | Immune, microbiome | 7 | 15 | |
| Immune only | 5 | 6 | ||
| Microbiome | 2 | 0 | ||
| Child sex | Female | 1 (7.1) | 10 (47.6) | 0.023 |
| Male | 13 (92.9) | 11 (52.4) | ||
| Delivery mode | Caesarean | 1 (7.1) | 10 (47.6) | 0.023 |
| Vaginal | 13 (92.9) | 11 (52.4) | ||
| Age (mo) | 25.65 [21.50, 27.17] | 22.83 [20.50, 26.57] | 0.501 | |
| Missing | 1 | 0 | ||
| Gestational age (wk) | 39.00 [36.00, 40.00] | 38.00 [37.00, 39.00] | 0.298 | |
| Birthweight (g) | 3090.00 [2382.50, 3420.00] | 3050.00 [2550.00, 3240.00] | 0.796 | |
| Missing | 3 | 0 | ||
| WAZ | 0.76 [−0.05, 1.60] | 0.87 [−0.01, 1.49] | 0.804 | |
| Missing | 1 | 0 | ||
| WLZ | 0.02 [−1.29, 0.72] | −0.21 [−0.75, 0.87] | 0.632 | |
| Missing | 1 | 0 | ||
| HAZ | 1.50 [0.69, 1.86] | 1.88 [0.68, 2.49] | 0.723 | |
| Missing | 1 | 0 | ||
| Maternal age (y) | 32.00 [28.00, 37.75] | 31.00 [27.00, 33.00] | 0.438 |
Statistical analyses: categorical variables were compared using the Fisher exact test, and data presented are n (in percentages). Continuous variables were compared using the Wilcoxon rank-sum test, and data presented are median [interquartile range]. Unadjusted p values < 0.05 were considered significant.
Table II. Canadian cohort characteristics.
| HUU |
HEU |
|||
|---|---|---|---|---|
| n = 33 | n = 16 | p | ||
| Data availability | Immune, microbiome | 19 | 15 | |
| Immune only | 1 | 1 | ||
| Microbiome only | 13 | 0 | ||
| Child sex | Female | 16 (48.5) | 7 (43.8) | 1.000 |
| Male | 17 (51.5) | 9 (56.2) | ||
| Delivery mode | Caesarean | 16 (48.5) | 4 (25.0) | 0.136 |
| Vaginal | 17 (51.5) | 12 (75.0) | ||
| Age (mo) | 18.87 [18.38, 20.74] | 18.29 [18.25, 19.34] | 0.041 | |
| Gestational age (wk) | 39.00 [38.00, 40.00] | 38.50 [37.83, 39.35] | 0.110 | |
| Birth weight (g) | 3280.00 [3062.00, 3610.00] | 3215.00 [2912.50, 3381.25] | 0.176 | |
| WAZ | 0.32 [−0.30, 1.04] | 0.74 [0.43, 0.92] | 0.398 | |
| Missing | 0 | 1 | ||
| WLZ | 0.33 [−0.33, 0.80] | 1.27 [0.75, 1.56] | 0.004 | |
| Missing | 0 | 1 | ||
| HAZ | 0.27 [−0.50, 1.07] | −0.47 [−0.92, −0.27] | 0.006 | |
| Missing | 0 | 1 | ||
| Maternal age (y) | 35.00 [33.00, 38.00] | 28.00 [25.00, 32.25] | <0.001 |
Table III. South African cohort characteristics.
| HUU |
HEU |
|||
|---|---|---|---|---|
| n = 21 | n = 16 | p | ||
| Data availability | Innate, microbiome | 7 | 8 | |
| Innate only | 13 | 8 | ||
| Microbiome only | 1 | 0 | ||
| Child sex | Female | 11 (52.4) | 11 (68.8) | 0.500 |
| Male | 10 (47.6) | 5 (31.2) | ||
| Delivery mode | Caesarean | 0 (0.0) | 1 (6.2) | 0.432 |
| Vaginal | 21 (100.0) | 15 (93.8) | ||
| Age (mo) | 24.16 [23.74, 24.55] | 24.10 [23.93, 24.99] | 0.701 | |
| Gestational age (wk) | 38.00 [37.00, 40.00] | 39.00 [38.00, 40.00] | 0.546 | |
| Missing | 0 | 1 | ||
| Birth weight (g) | 3030.00 [2740.00, 3300.00] | 2990.00 [2822.50, 3200.00] | 0.794 | |
| WAZ | −0.46 [−1.16, 0.07] | 0.32 [−0.74, 1.28] | 0.052a | |
| Missing | 1 | 0 | ||
| WLZ | 0.06 [−0.56, 0.32] | 0.58 [−0.85, 1.47] | 0.474 | |
| Missing | 1 | 0 | ||
| HAZ | −1.27 [−2.07, −0.03] | −0.30 [−0.78, 0.61] | 0.048 | |
| Missing | 1 | 0 | ||
| Maternal age (y) | 25.00 [22.00, 28.00] | 26.00 [25.50, 29.50] | 0.282 | |
| Missing | 6 | 4 |
p = 0.052, approaching significance.
Table IV. Initiation of maternal ARV prophylaxis/therapy for HIV-positive mothers.
| Belgium | Canada | South Africa | |
|---|---|---|---|
| n | 21 | 14 | 16 |
| No PMTCT | 0 (0.0) | 0 (0.0) | 3 (21.4) |
| Preconception | 7 (38.9) | 3 (25.0) | 1 (7.1) |
| First trimestera | 3 (16.7) | 2 (16.7) | 0 (0.0) |
| Second trimestera | 5 (27.8) | 7 (58.3) | 2 (14.3) |
| Third trimestera | 3 (16.7) | 0 (0.0) | 8 (57.1) |
| Missing | 3 | 2 | 2 |
First trimester, 0 to under 13 wk; second trimester, 13 to under 26 wk; and third trimester, 26 wk or more. In parentheses is percent of cohort. Initiation of ARV differed significantly among cohorts (χ2 test, p < 0.001).
Global cytokine responses to PRR ligands
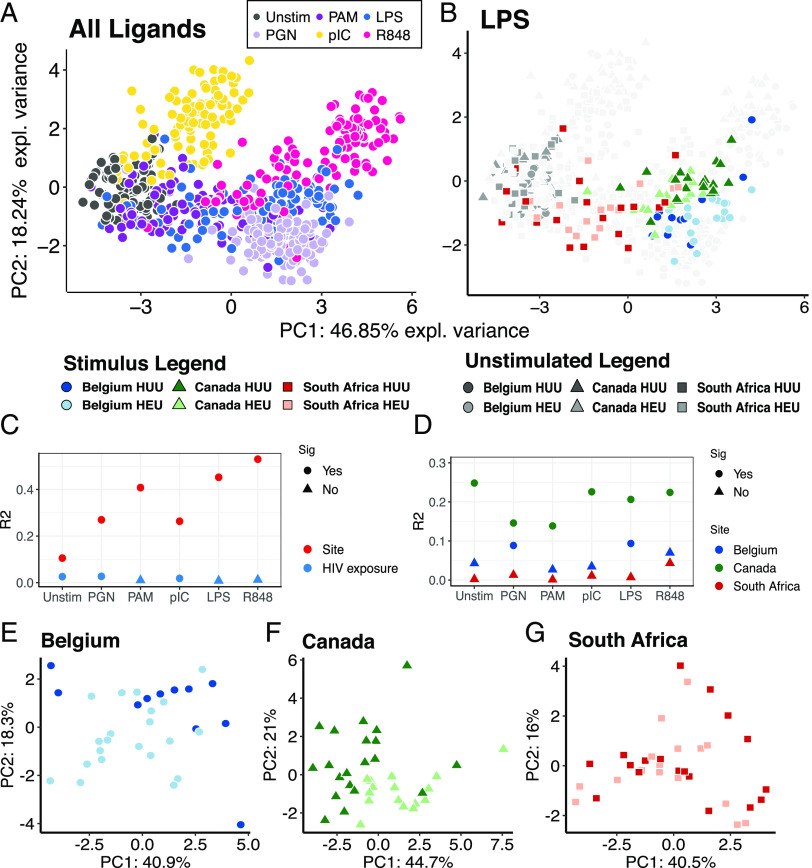
PCA of cytokine responses to PRR ligands revealed that ligands had the dominant effect on cytokine response profiles, as previously observed for HUU children (24) (Fig. 1A, Supplemental Fig. 1). For individual ligands, country of origin contributed significantly to variance of responses (Fig. 1B, 1C, Supplemental Fig. 1). For individual ligands, HIV exposure had a much smaller effect than country of origin, which was significant for only two of the six ligands and unstimulated controls (Fig. 1C). For individual countries of origin, the effect of HIV exposure on responses to each ligand was variable (Fig. 1D–G, Supplemental Fig. 2). That effect was greatest among Canadians, intermediate for Belgians, and NS for South Africans.
FIGURE 1.
PCA of whole blood cytokine response profiles. (A) Responses for each subject to each PRR ligand; ligands significantly affected cytokine responses (PERMANOVA R2 = 0.49; p = 0.001). (B) Same PCA as (A), highlighting responses to LPS. (C) Variability of cytokine responses to each stimulus explained by country of origin (site) and HIV exposure, based on PERMANOVA (adjusted p value <0.05, Bonferroni correction). (D) Variability of cytokine responses to each stimulus explained by HIV exposure for individual countries of origin, based on PERMANOVA (adjusted p value < 0.05, Bonferroni correction). (E–G) PCA comparing cytokine profiles in response to LPS in HUU versus HEU children. Sig, significant effect.
Supernatant cytokine responses in HEU versus HUU children
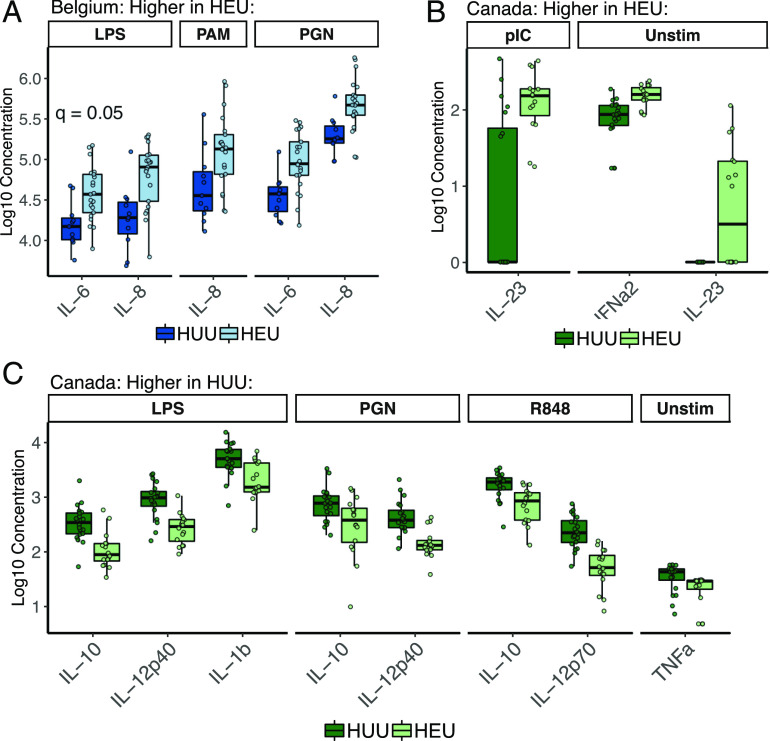
We tested the effect of HIV exposure on cytokine responses for each ligand–cytokine combination separately for each regional cohort using linear regression. Consistent with PCA analysis, HIV exposure was associated with responses to LPS and PGN in the Belgian cohort, with greater IL-6 and IL-8 production by HEU children (Fig. 2A). Timing of ARV initiation during pregnancy has been reported to account for differences between HEU and HUU children. In the Belgian cohort, seven mothers initiated ARV prior to conception and 11 during pregnancy (Table IV). We tested whether timing of ARV initiation affected IL-6 and IL-8 responses in the Belgian cohort, in which seven mothers commenced ARV therapy prior to conception and 11 after and found no significant effect. However, it is possible that we were not adequately powered to detect this. As no mothers in the Canadian cohort and only three mothers in the South African cohort commenced ARV therapy prior to conception, we did not test the effect of ARV initiation on cytokine responses in these cohorts.
FIGURE 2.
Cytokine responses to ligands within regional cohorts, which were significantly affected by HIV exposure among Belgian (A) or Canadian (B and C) children, based on linear regression (adjusted p value ≤ 0.05, corrected using the Benjamini–Hochberg method); boxplots indicate medians with first and third quartiles (25–75%). Whiskers extend no further than 1.5× interquartile range from the hinge.
Within the Canadian cohort, HEU children had higher levels than HUU children of IFN-α2 and IL-23 both at baseline (unstimulated) and in response to Poly(I:C) (Fig. 2B). Canadian HUU children had higher levels than HEU children of IL-10 in response to LPS, PGN, and R848; IL-12p40 in response to LPS and PGN; IL-1β in response to LPS only; and IL-12p70 in response to R848 only (Fig. 2C). Findings within Belgian and Canadian cohorts remained significant after adjusting for unbalanced demographic factors. Within the South African cohort, HIV exposure was not associated with responses to any ligands, in agreement with the PCA (Supplemental Fig. 2).
Intracellular cytokine responses in HEU and HUU children
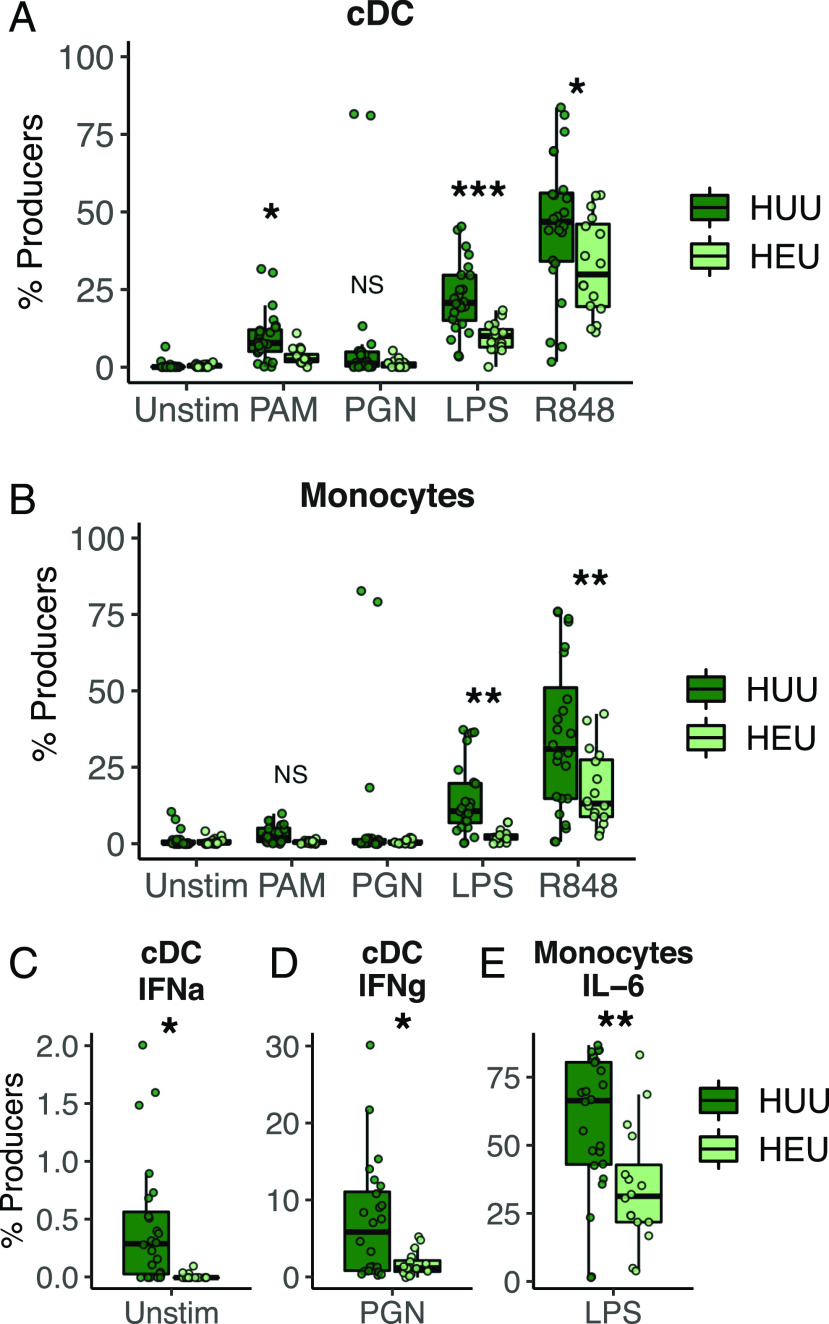
In all regional cohorts, HEU and HUU children had similar proportions of monocytes, granulocytes, dendritic cells, and T cells (Supplemental Fig. 3). Canadians had higher proportions of γδ T cells and lower proportions of B cells and T cells in HEU versus HUU children. Canadian intracellular cytokine responses supported the trends seen for IL-12p40, as HEU children had fewer cDCs producing IL-12p40 in response to PAM, LPS, and R848 and fewer monocytes producing IL-12p40 in response to LPS and R848 (Fig. 3). Few other differences in intracellular cytokine responses were found in Canadians; HEU cDCs produced less IFN-γ in response to PGN, and fewer monocytes produced IL-6 in response to LPS (Fig. 3). No differences between HEU and HUU children were found in the other two cohorts.
FIGURE 3.
(A and B) Intracellular production of IL-12p40 in response to four PRR ligands by cDCs (A) and monocytes (B) from HUU and HEU Canadian infants. (C–E) Proportions of cDCs producing IFN-α (C) and IFN-γ (D) and proportions of monocytes producing IL-6 (E) differ among HEU and HUU children. Boxplots indicate medians with first and third quartiles (25–75%). Whiskers extend no further than 1.5× interquartile range from the hinge. *p < 0.05, **p < 0.01, ***p < 0.001, for linear regression, adjusted for unstimulated values.
Microbiome differences between HEU and HUU children
α And β diversity.
Country of origin had the greatest effect on fecal microbiome community composition (Fig. 4A). α Diversity, based on Shannon index, did not differ between HEU and HUU children from any country of origin (Fig. 4B). Within each cohort individually (Fig. 4C, 4D), HIV exposure was only significantly associated with community composition among the Canadian children but only accounted for 3.5% of the variance.
FIGURE 4.
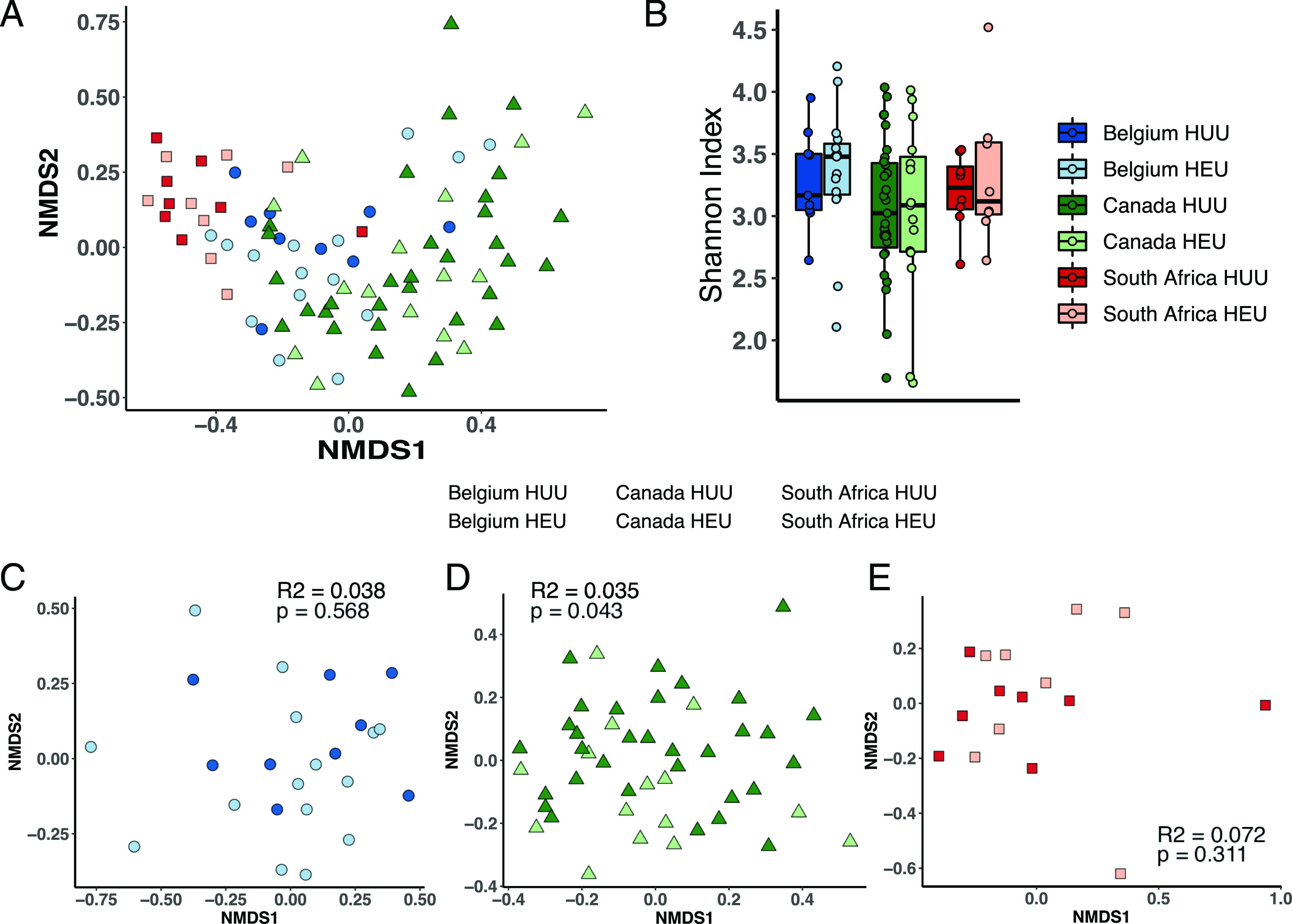
The intestinal microbiota is minimally affected by HIV exposure. (A) Microbiome subject clustering is driven by site in NMDS ordination of all samples. (B) α Diversity computed with the Shannon index is not affected by HIV exposure for any site. (C–E) NMDS ordinations showing microbiome samples for Belgian (C), Canadian (D), and South African (E) HEU and control infants with inset PERMANOVA R2 and p values for effect of HIV exposure on sample clustering.
OTUs differentially abundant between HEU and HUU children.
We identified differentially abundant OTUs between HEU and HUU children within each regional cohort (Fig. 5A–C), with 17, 63, and three OTUs from Belgium, Canada, and South Africa, respectively. Belgian HEU children were characterized by increased abundances 12 OTUs, including several Firmicutes (Eubacterium, Ruminococcus, and Blautia) and Bacteroidetes (Rikenellaceae and Odoribacter) (Fig. 5A, 5D). HUU children had a higher abundance of only five OTUs, representing Proteobacteria and Firmicutes. OTUs belonging to the Bacteroides genus had both higher and lower abundances in Belgian HEU children.
FIGURE 5.
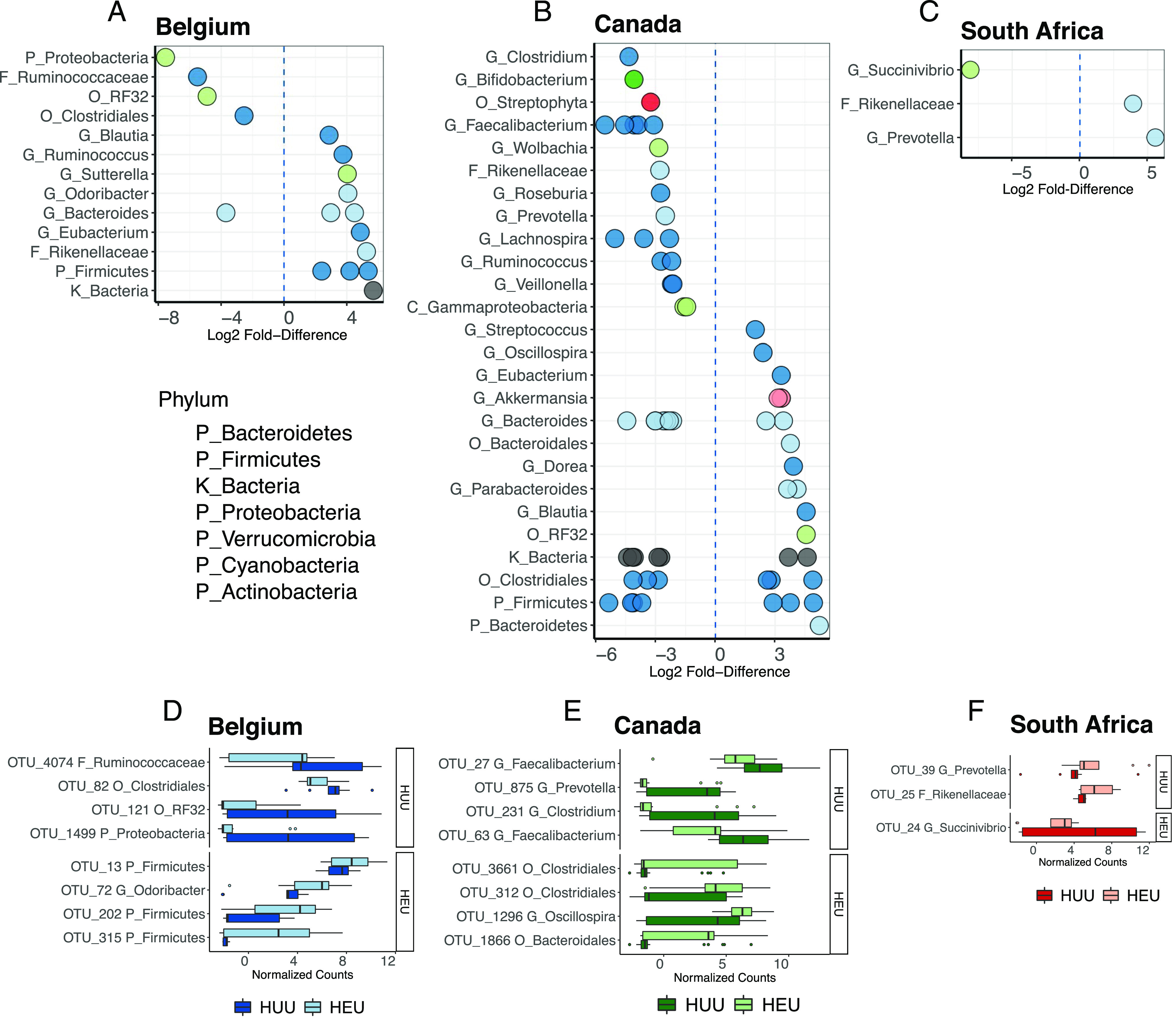
OTUs differentially abundant between HEU and HUU children from Belgium, Canada, and South Africa. (A–C) Log2 fold difference in abundance of bacterial taxa; some taxa include multiple differentially abundant OTUs (individual dots); positive values indicate enrichment in HEU children. (D–F) Relative abundance of top four OTUs classified below the kingdom level that discriminate HUU and HEU children, based on sPLS-DA. Facets separate OTUs enriched in HUU or HEU children. Boxplots indicate medians with first and third quartiles (25–75%). Whiskers extend no further than 1.5× interquartile range from the hinge.
Canadian HEU children had increased abundances of 22 OTUs (Fig. 5B, 5E). OTUs classified as Firmicutes (phylum), Clostridiales (order), and Bacteroides (genus) had both higher and lower abundances in Canadian HEU children. Ten taxa contained OTUs that were exclusively more abundant in HEU children, including the Bacteroidetes, Parabacteroides (genus) and Bacteroidales (order), and the Firmicutes, Blautia, Dorea, Eubacterium, Oscillospira, and Streptococcus (genera). Multiple Akkermansia (genus) OTUs were more abundant in HEU. Conversely, OTUs of 12 taxa were exclusively more abundant in HUU children, including the Firmicutes, Veillonella, Ruminococcus, Lachnospira, Roseburia, Faecalibacterium, and Clostridium, which are all known short-chain fatty acid (SCFA) producers. Others included members of the phyla Proteobacteria, Bacteroidetes, and Actinobacteria.
Between the South African HEU and HUU children, only three OTUs were differentially abundant, including one Rikenellaceae OTU was more abundant in HEU children, and one Succinivibrio OTU was more abundant in HUU children (Fig. 5C, 5F).
Cytokine–OTU relationships in HEU and HUU children
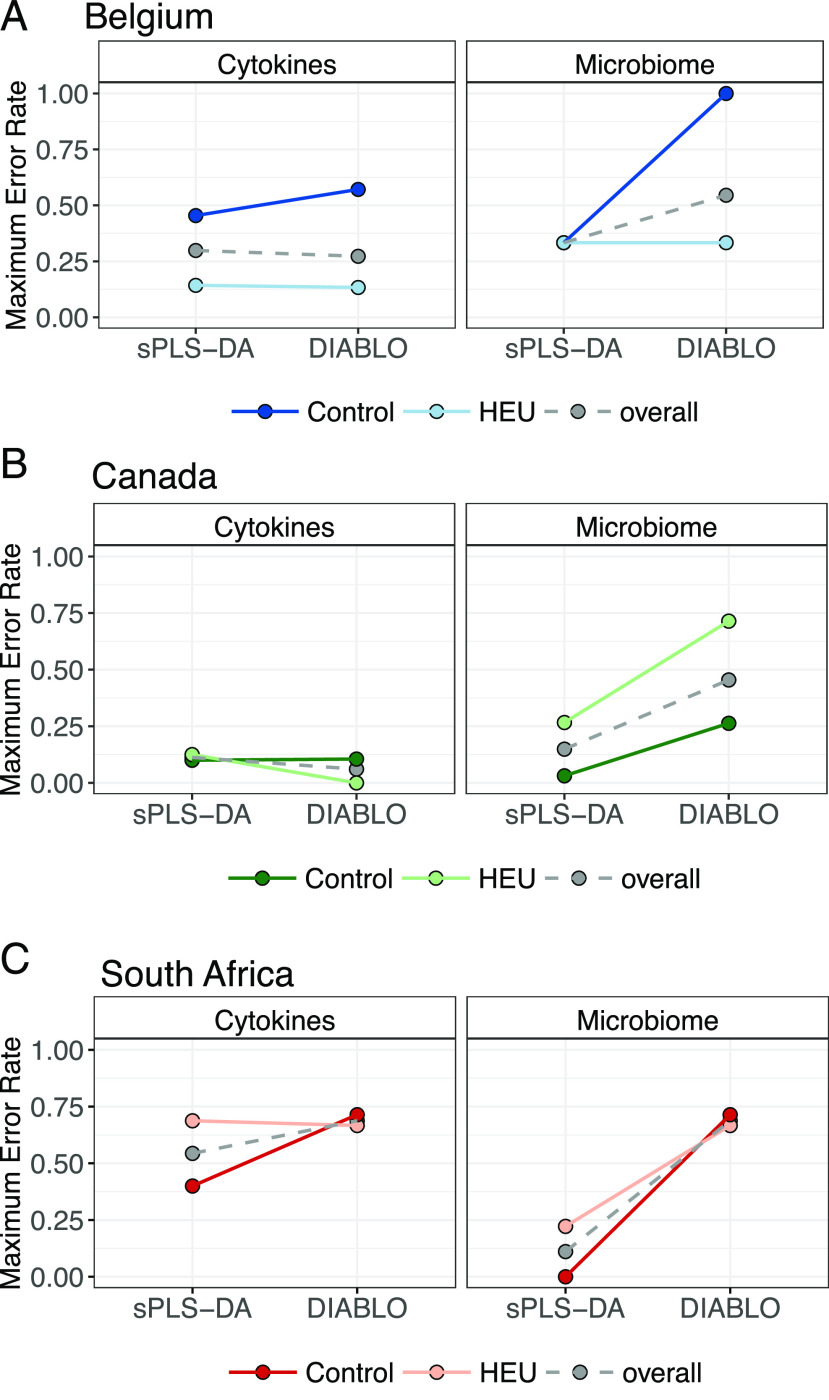
To test whether any differences in the microbiomes of HEU children correlated with their cytokine responses to PRR stimulation, we employed DIABLO, a sparse generalized canonical correlation discriminant analysis designed to select features that both covary across data types and discriminate between classes of interest (28), in this study, HEU versus HUU children across cytokine response and microbiome datasets. Because achieving a minimal classification error rate is a primary goal for this analysis, we compared classification error rates with each dataset alone using sPLS-DA and DIABLO models, both tuned to select the number of features, resulting in the lowest error rate for cytokine and microbiome data. Using sPLS-DA, cytokine data alone distinguished between Canadian HEU and HUU children with an overall error rate of 11%, compared with 30 and 55% for Belgians and South Africans, respectively. Microbiome data alone distinguished HEU from HUU children with 35, 14, and 11% error for Belgians, Canadians, and South Africans (Fig. 6).
FIGURE 6.
Maximum classification error rates for discriminating HEU from HUU children from Belgium (A), Canada (B), or South Africa (C) using single (sPLS-DA) or integrated (DIABLO) datasets.
Except for cytokine data for the Canadian cohort, DIABLO selected features with inferior classification accuracy to sPLS-DA models. With only five OTUs (the minimum allowed) selected for each DIABLO model, they discriminated HEU from HUU children with error rates of 50% or above for each cohort (Fig. 6). Thus, no OTUs were identified that both correlated with HIV exposure and discriminated between HEU and HUU children in any regional cohort.
Discussion
This study measured, for the first time, to our knowledge, immune and microbiome differences between HIV-exposed and -unexposed children across geographically diverse settings. Our findings suggest that country of origin is a stronger driver of innate immune phenotype than HIV or ARV exposure. However, an effect of HIV exposure on immune response was still evident within two of three countries. Notably, the immune differences between HEU and HUU children were site specific; immune differences in one population could not simply be extrapolated to another.
Belgian HEU children were distinguished from HUU children by higher IL-6 and IL-8 production in response to LPS and PGN and higher IL-8 production in response to TLR2:1 stimulation, and we determined that child sex and delivery mode did not drive these associations. Increased inflammatory responses in HEU children were described previously (10), including children recruited at the exact same center used in the current study (11). However, this is the first study, to our knowledge, to report an effect of HIV exposure on immune responses at 2 y of age; previous studies found the greatest effect at birth, with differences persisting for up to 1 y of age (10, 11). Increased severity to infection and altered immunity in HEU infants recruited within the same study site in Belgium were recently reported to be associated with timing of ARV initiation (11). Although the immune alterations identified in our study did not differ between HEU children whose mothers initiated ARV therapy prior to or during pregnancy, our study measured different immune parameters later in childhood and with a very small sample size. Also, we did not attempt to associate immune differences to clinical outcomes. Thus, although we were not able to support the hypothesis that ARV initiation associates with altered immunity, these important differences may account for the result.
Innate immune responses to PRR stimulation in HEU children have not been previously measured in any North American populations. This study reports that in Canada, HEU children differed from HUU children by their lower production of IL-10 and IL-12 in response to LPS, PGN, and R848. Thus, although responses to similar ligands were affected by HIV exposure in both the Belgian and Canadian cohorts, the specific cytokines affected were completely distinct. Canadian HEU and HUU children differed from one another demographically in several aspects: child age, maternal age, WLZ, and HAZ. Thus, several host factors independent of HIV infection may have contributed to the distinct immune responses. Our findings provide further evidence that HIV/ARV exposure is associated with altered immune responses across high-income countries.
The lack of immune differences between South African HEU and HUU children is consistent with previous findings from this cohort, in which different bulk and cell-specific cytokine responses to LPS and PAM were evident up to 6 mo of life, but not by 1 y (10). Notably, immune phenotypes of South African children were most distinct from those of other cohorts because of lower responses to almost all PRR ligands (24). Thus, it is possible that environment influences immune responses such that any effects of HIV or ARV exposure are no longer evident.
This study did not allow us to determine the mechanisms driving differences in cytokine responsiveness. However, there are several known mechanisms that have been demonstrated to underlie heterogeneity of in vitro cytokine responses to PRR stimulation. For example, population-level differences in TLR expression have been shown to contribute to differences in cytokine responses (32). Altered TLR expression has been observed in HIV-exposed individuals for TLR2 and TLR4, compared with healthy controls (33). Given that we detected both TLR-specific and cytokine-specific differences, potential mechanisms may involve higher-level regulation of immunity. Epigenetic programming underlies global differences in innate immune responsiveness, including TLR expression (34), and differential gene expression of epigenetic regulators has been observed among HEU children (35). Thus, future studies employing systems biology approaches may reveal more holistic differences in the host response beyond cytokine responsiveness.
To our knowledge, this is the first study to measure stool microbiome composition of HEU children at 2 y of age. Only one other study has reported stool microbiomes of HEU infants at 3 mo of age (17). Gut microbiomes in early infancy, prior to addition of solid foods, are known to be highly distinct from those in later infancy or toddlerhood, when solid foods are introduced and after weaning (20, 21). Thus, it is not surprising that taxonomic differences in the 3-mo-old infant study did not at all apply to our findings in 2-y-old children. Breast versus formula feeding may have contributed to microbiome differences between HEU and HUU children in the current study. Given that all HIV-positive mothers in the current study were advised to exclusively formula feed their infants from birth to prevent HIV transmission, whereas all the HUU children in the current study were breastfed, exclusive formula feeding may have contributed to the distinct microbial composition in HEU children.
Abundances of SCFA-producing bacteria were reduced in Canadian HEU children compared with Canadian HUU children. We found that gut microbiomes of Canadian HEU children had lower relative abundances of SCFA-producing taxa that have been isolated from breast milk and are associated with breastfeeding—Bifidobacterium and Veillonella (36, 37). As well, other prominent SCFA producers normally present in adult microbiomes, Ruminococcus, Lachnospira, Clostridia, Faecalibacterium, and Roseburia (38), were also less abundant in Canadian HEU children. Diets rich in nondigestible fibers heavily increase the relative abundance of SCFA-producing bacteria (39). However, as dietary composition was not analyzed in this study, we cannot determine whether this underlies the observed difference. Given the known roles of SCFAs in maintaining intestinal and immunological health, possible effects of reduced SCFA-producing bacteria in Canadian HEU children deserve further attention. However, this difference was not apparent in the other two cohorts. In Belgian HEU children, OTUs representing SCFA-producing Ruminococcus and Eubacterium were enriched compared with HUU children. Several bacterial classes contained both enriched and depleted OTUs in Belgian HEU children, making any functional impact of these differences difficult to infer. Only three OTUs were differentially abundant between South African HEU and HUU children, suggesting their gut microbiome was not greatly affected by HIV exposure or the differences were too subtle to detect with our sample size.
sPLS-DA and DIABLO analyses both demonstrated that although both cytokine and OTU datasets had very similar discriminatory power for Belgian and Canadian cohorts, no discriminatory multivariate relationships between the two datasets (immune and microbiome) could be identified using DIABLO, a powerful data integration tool. This suggests that systemic innate immunity and the gut microbiome were affected by independent factors associated with HIV or ARV exposure or that we were not adequately powered to detect these differences. For instance, although formula feeding may have contributed to microbial differences seen between Canadian HEU and HUU children, another aspect of HIV exposure, partly accounted for by differences in maternal age, could have contributed to differences in the observed cytokine response.
This study could not assess any relationships between immune phenotype or gut microbiome and adverse clinical outcomes, such as severe infections, associated with prenatal HIV exposure. A much larger number of children would have to be included to capture such relatively rare clinical outcomes. Further, our HEU cohorts were at reduced risk of infection because they were almost all born at full term and normal birth weight. HIV and ARV exposure are both associated with adverse birth outcomes including low birth weight and preterm birth (40, 41), which are both in turn associated with increased risk for infection in early life. Further, a limited set of participant demographic information was available for this study. Other important variables, such as socioeconomic or ethnic background of the mothers, or child diet, may have contributed to the observed phenotypes. These important variables should be considered in future works.
In conclusion, we found that the impact of maternal HIV/ARV exposure on innate immunity of the child is population specific, adding to the evidence that immune development is population specific (23, 24). This concept has important implications for immunity to pathogens and interventions across the globe. Similarly, we found that the impact of HIV exposure on the gut microbiome composition is population specific. We found no universal signature of HIV effect on immune status or stool microbiome composition. Furthermore, the microbiome differences between HEU and HUU children within each population did not correlate with their respective differences in immune status. This suggests that the microbiome does not play a major role in mediating the effect of HIV or ARV exposure on the innate immune system and, conversely, that the innate immune system does not play a major role in mediating the effect of HIV or ARV exposure on the microbiome. This notion warrants more attention in future studies powered to detect more subtle effects, alongside relevant clinical outcomes, and a more integrative assessment of the gut microbiome using metagenomics or metatranscriptomic approaches.
Supplementary Material
Acknowledgments
We thank the mothers, fathers, and infants who generously donated time to the study in Belgium, Canada, and South Africa. Without whose participation, the study would not have been possible. We thank the expert assistance provided by the staff at the Centre Hospitalier Universitaire Saint-Pierre, Brussels; Children's Infectious Diseases Clinical Research Unit, Department of Paediatrics and Child Health, Tygerberg Academic Hospital, Stellenbosch University, Cape Town, South Africa; the Oak Tree Clinic, BC Women’s Hospital; and the Vaccine Evaluation Center, BC Children’s Hospital, The University of British Columbia, Vancouver, Canada. We also thank Dr. Gregory Gloor for providing 16S amplicon sequencing data from the stool samples and the study staff that facilitated participant recruitment and sample collection. We thank the following study staff across the research centers for contribution to study participant recruitment, biological assessment, and study design: Vancouver, Dr. David Speert, Laura Gelinas, Kevin Ho, and Darren Sutherland; South Africa, Rozanne Adams, Santhashan Pillay, and Corena De Beer; and Belgium, Mustapha Chamekh.
This work was supported by grants from the Canadian Institutes of Health Research (PJT-148781 and HET-85515) and The University of British Columbia (F0906208).
The sequences presented in this article have been submitted to the National Center for Biotechnology Information Sequence Read Archive (https://www.ncbi.nlm.nih.gov/bioproject/) under accession number PRJNA660015.
The online version of this article contains supplemental material.
- ARV
- antiretroviral
- cDC
- conventional dendritic cell
- DIABLO
- Data Integration Analysis for Biomarker Discovery using Latent Variable Approaches for Omics studies
- HAZ
- height-for-age z-score
- HEU
- HIV-exposed, uninfected
- HUU
- HIV-unexposed, uninfected
- NMDS
- nonmetric multidimensional scaling
- OTU
- operational taxonomic unit
- PAM
- PAM3CYSK4
- PCA
- principal component analysis
- PERMANOVA
- permutational multivariate ANOVA
- PGN
- peptidoglycan
- PRR
- pattern recognition receptor
- SCFA
- short-chain fatty acid
- sPLS-DA
- sparse partial least squares–discriminant analysis
- WAZ
- weight-for-age z-score
- WLZ
- weight-for-length z-score.
Disclosures
A.M. is Research Director of the Fonds de la Recherche Scientifique, Belgium. The other authors have no financial conflicts of interest.
References
- 1.World Health Organization 2016. Consolidated Guidelines on the Use of Antiretroviral Drugs for Treating and Preventing HIV Infection: Recommendations for a Public Health Approach. World Health Organization, Geneva, Switzerland. [PubMed] [Google Scholar]
- 2.UNAIDS. 2019. Start free stay free AIDS free-2019 report. Available at: https://www.unaids.org/en/resources/documents/2019/20190722_UNAIDS_SFSFAF_2019. Accessed: August 15, 2019.
- 3.Slogrove A. L., Goetghebuer T., Cotton M. F., Singer J., Bettinger J. A. 2016. Pattern of infectious morbidity in HIV-exposed uninfected infants and children. Front. Immunol. 7: 164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Desmonde S., Goetghebuer T., Thorne C., Leroy V. 2016. Health and survival of HIV perinatally exposed but uninfected children born to HIV-infected mothers. Curr. Opin. HIV AIDS 11: 465–476. [DOI] [PubMed] [Google Scholar]
- 5.Goetghebuer T., Smolen K. K., Adler C., Das J., McBride T., Smits G., Lecomte S., Haelterman E., Barlow P., Piedra P. A., et al. 2019. Initiation of antiretroviral therapy before pregnancy reduces the risk of infection-related hospitalization in human immunodeficiency virus-exposed uninfected infants born in a high-income country. Clin. Infect. Dis. 68: 1193–1203. [DOI] [PubMed] [Google Scholar]
- 6.Dainiak N., Worthington M., Riordan M. A., Kreczko S., Goldman L. 1988. 3′-Azido-3′-deoxythymidine (AZT) inhibits proliferation in vitro of human haematopoietic progenitor cells. Br. J. Haematol. 69: 299–304. [DOI] [PubMed] [Google Scholar]
- 7.Pacheco S. E., McIntosh K., Lu M., Mofenson L. M., Diaz C., Foca M., Frederick M., Handelsman E., Hayani K., Shearer W. T., Women and Infants Transmission Study 2006. Effect of perinatal antiretroviral drug exposure on hematologic values in HIV-uninfected children: an analysis of the women and infants transmission study. J. Infect. Dis. 194: 1089–1097. [DOI] [PubMed] [Google Scholar]
- 8.European Collaborative Study 2004. Levels and patterns of neutrophil cell counts over the first 8 years of life in children of HIV-1-infected mothers. AIDS 18: 2009–2017. [DOI] [PubMed] [Google Scholar]
- 9.Bunders M. J., Bekker V., Scherpbier H. J., Boer K., Godfried M., Kuijpers T. W. 2005. Haematological parameters of HIV-1-uninfected infants born to HIV-1-infected mothers. Acta Paediatr 94: 1571–1577. [DOI] [PubMed] [Google Scholar]
- 10.Reikie B. A., Adams R. C. M., Leligdowicz A., Ho K., Naidoo S., Rusk C. E., de Beer C., Preiser W., Cotton M. F., Speert D. P., et al. 2014. Altered innate immune development in HIV-exposed uninfected infants. J. Acquir. Immune Defic. Syndr. 66: 245–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goetghebuer T., Smolen K. K., Adler C., Das J., McBride T., Smits G., Lecomte S., Haelterman E., Barlow P., Piedra P. A., et al. 2019. Initiation of antiretroviral therapy before pregnancy reduces the risk of infection-related hospitalization in human immunodeficiency virus-exposed uninfected infants born in a high-income country. Clin. Infect. Dis. 68: 1193–1203. [DOI] [PubMed] [Google Scholar]
- 12.Slyker J. A., Lohman-Payne B., John-Stewart G. C., Dong T., Mbori-Ngacha D., Tapia K., Atzberger A., Taylor S., Rowland-Jones S. L., Blish C. A. 2012. The impact of HIV-1 infection and exposure on natural killer (NK) cell phenotype in Kenyan infants during the first year of life. Front. Immunol. 3: 399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thaiss C. A., Zmora N., Levy M., Elinav E. 2016. The microbiome and innate immunity. Nature 535: 65–74. [DOI] [PubMed] [Google Scholar]
- 14.Sjogren Y. M., Tomicic S., Lundberg A., Bottcher M. F., Bjorksten B., Sverremark-Ekstrom E., Jenmalm M. C. 2009. Influence of early gut microbiota on the maturation of childhood mucosal and systemic immune responses. Clin. Exp. Allergy 39: 1842–1851. [DOI] [PubMed] [Google Scholar]
- 15.Lozupone C. A., Li M., Campbell T. B., Flores S. C., Linderman D., Gebert M. J., Knight R., Fontenot A. P., Palmer B. E. 2013. Alterations in the gut microbiota associated with HIV-1 infection. Cell Host Microbe 14: 329–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nowak P., Troseid M., Avershina E., Barqasho B., Neogi U., Holm K., Hov J. R., Noyan K., Vesterbacka J., Svärd J., et al. 2015. Gut microbiota diversity predicts immune status in HIV-1 infection. AIDS 29: 2409–2418. [DOI] [PubMed] [Google Scholar]
- 17.Bender J. M., Li F., Martelly S., Byrt E., Rouzier V., Leo M., Tobin N., Pannaraj P. S., Adisetiyo H., Rollie A., et al. Maternal HIV infection influences the microbiome of HIV-uninfected infants. [Published erratum appears in 2016 Sci. Transl. Med. 8: 351er6.] Sci. Transl. Med. 8: 349ra100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuhn L., Aldrovandi G. 2010. Survival and health benefits of breastfeeding versus artificial feeding in infants of HIV-infected women: developing versus developed world. Clin. Perinatol. 37: 843–862, x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bokulich N. A., Chung J., Battaglia T., Henderson N., Jay M., Li H., D Lieber A., Wu F., Perez-Perez G. I., Chen Y., et al. 2016. Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci. Transl. Med. 8: 343ra82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bäckhed F., Roswall J., Peng Y., Feng Q., Jia H., Kovatcheva-Datchary P., Li Y., Xia Y., Xie H., Zhong H., et al. 2015. Dynamics and stabilization of the human gut microbiome during the first year of life. [Published erratum appears in 2015 Cell Host Microbe 17: 852.] Cell Host Microbe 17: 690–703. [DOI] [PubMed] [Google Scholar]
- 21.Yatsunenko T., Rey F. E., Manary M. J., Trehan I., Dominguez-Bello M. G., Contreras M., Magris M., Hidalgo G., Baldassano R. N., Anokhin A. P., et al. 2012. Human gut microbiome viewed across age and geography. Nature 486: 222–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De Filippo C., Di Paola M., Ramazzotti M., Albanese D., Pieraccini G., Banci E., Miglietta F., Cavalieri D., Lionetti P. 2017. Diet, environments, and gut microbiota. A preliminary investigation in children living in rural and urban Burkina Faso and Italy. Front. Microbiol. 8: 1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smolen K. K., Cai B., Gelinas L., Fortuno E. S., 3rd, Larsen M., Speert D. P., Chamekh M., Cooper P. J., Esser M., Marchant A., Kollmann T. R. 2014. Single-cell analysis of innate cytokine responses to pattern recognition receptor stimulation in children across four continents. J. Immunol. 193: 3003–3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smolen K. K., Ruck C. E., Fortuno E. S., III, Ho K., Dimitriu P., Mohn W. W., Speert D. P., Cooper P. J., Esser M., Goetghebuer T., et al. 2014. Pattern recognition receptor-mediated cytokine response in infants across 4 continents. J. Allergy Clin. Immunol. 133: 818–826.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gloor G. B., Hummelen R., Macklaim J. M., Dickson R. J., Fernandes A. D., MacPhee R., Reid G. 2010. Microbiome profiling by illumina sequencing of combinatorial sequence-tagged PCR products. PLoS One 5: e15406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Edgar R. C. 2013. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10: 996–998. [DOI] [PubMed] [Google Scholar]
- 27.Lê Cao K. A., Boitard S., Besse P. 2011. Sparse PLS discriminant analysis: biologically relevant feature selection and graphical displays for multiclass problems. BMC Bioinformatics 12: 253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singh A., Shannon C. P., Gautier B., Rohart F., Vacher M., Tebbutt S. J., Lê Cao K. A. 2019. DIABLO: an integrative approach for identifying key molecular drivers from multi-omics assays. Bioinformatics 35: 3055–3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oksanen J., Blanchet F. G., Friendly M., Kindt R., Legendre P., McGlinn D., Minchin P. R., O’hara R., Simpson G. L., Solymos P. 2016. vegan: Community Ecology Package, R package version 2.4-3. Vienna: R Foundation for Statistical Computing. [Google Scholar]
- 30.Charlop-Powers Z., Brady S. F. 2015. phylogeo: an R package for geographic analysis and visualization of microbiome data. Bioinformatics 31: 2909–2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Love M. I., Huber W., Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van den Biggelaar A. H., Prescott S. L., Roponen M., Nadal-Sims M. A., Devitt C. J., Phuanukoonnon S., Pomat W., Tulic M. K., Lehmann D., Siba P. M., et al. 2009. Neonatal innate cytokine responses to BCG controlling T-cell development vary between populations. J. Allergy Clin. Immunol. 124: 544–550, 550.e1–2. [DOI] [PubMed] [Google Scholar]
- 33.Hernandez J. C., St Laurent G., III, Urcuqui-Inchima S. 2016. HIV-1-exposed seronegative individuals show alteration in TLR expression and pro-inflammatory cytokine production ex vivo: an innate immune quiescence status? Immunol. Res. 64: 280–290. [DOI] [PubMed] [Google Scholar]
- 34.Boosani C. S., Agrawal D. K. 2016. Epigenetic regulation of innate immunity by microRNAs. Antibodies (Basel) 5: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Musimbi Z. D., Rono M. K., Otieno J. R., Kibinge N., Ochola-Oyier L. I., de Villiers E. P., Nduati E. W. 2019. Peripheral blood mononuclear cell transcriptomes reveal an over-representation of down-regulated genes associated with immunity in HIV-exposed uninfected infants. Sci. Rep. 9: 18124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jost T., Lacroix C., Braegger C., Chassard C. 2013. Assessment of bacterial diversity in breast milk using culture-dependent and culture-independent approaches. Br. J. Nutr. 110: 1253–1262. [DOI] [PubMed] [Google Scholar]
- 37.Jiang T., Liu B., Li J., Dong X., Lin M., Zhang M., Zhao J., Dai Y., Chen L. 2018. Association between sn-2 fatty acid profiles of breast milk and development of the infant intestinal microbiome. Food Funct. 9: 1028–1037. [DOI] [PubMed] [Google Scholar]
- 38.Morrison D. J., Preston T. 2016. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes 7: 189–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trompette A., Gollwitzer E. S., Yadava K., Sichelstiel A. K., Sprenger N., Ngom-Bru C., Blanchard C., Junt T., Nicod L. P., Harris N. L., Marsland B. J. 2014. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat. Med. 20: 159–166. [DOI] [PubMed] [Google Scholar]
- 40.Santosa W. B., Staines-Urias E., Tshivuila-Matala C. O. O., Norris S. A., Hemelaar J. 2019. Perinatal outcomes associated with maternal HIV and antiretroviral therapy in pregnancies with accurate gestational age in South Africa. AIDS 33: 1623–1633. [DOI] [PubMed] [Google Scholar]
- 41.Wedi C. O., Kirtley S., Hopewell S., Corrigan R., Kennedy S. H., Hemelaar J. 2016. Perinatal outcomes associated with maternal HIV infection: a systematic review and meta-analysis. Lancet HIV 3: e33–e48. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw 16S amplicon sequencing data have been deposited at a publicly accessible server; the National Center for Biotechnology Information Sequence Read Archive and can be access via the following accession number: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA660015/). Data used for statistical analyses, and associated scripts, are publicly available and can be accessed on GitHub via the following url:https://github.com/nelly-amenyogbe/Global_HEU_immune_microbiome.