Key Points
HLA-E*01:01, HLA-E*01:03, Mamu-E, and Qa-1b have unique motifs.
Each MHC-E molecule binds alternative peptides with conserved main anchor residues.
Nonanchor residues can play an important role in peptide binding to MHC-E.
Abstract
Ag presentation via the nonclassical MHC class Ib molecule HLA-E, with nearly complete identity between the two alleles expressed in humans, HLA-E*01:01 and HLA-E*01:03, can lead to the activation of unconventional T cells in humans. Despite this virtual genetic monomorphism, differences in peptide repertoires binding to the two allelic variants have been reported. To further dissect and compare peptide binding to HLA-E*01:01 and HLA-E*01:03, we used an UV-mediated peptide exchange binding assay and an HPLC-based competition binding assay. In addition, we investigated binding of these same peptides to Mamu-E, the nonhuman primate homologue of human HLA-E, and to the HLA-E–like molecule Qa-1b in mice. We next exploited the differences and homologies in the peptide binding pockets of these four molecules to identify allele specific as well as common features of peptide binding motifs across species. Our results reveal differences in peptide binding preferences and intensities for each human HLA-E variant compared with Mamu-E and Qa-1b. Using extended peptide libraries, we identified and refined the peptide binding motifs for each of the four molecules and found that they share main anchor positions, evidenced by conserved amino acid preferences across the four HLA-E molecules studied. In addition, we also identified differences in peptide binding motifs, which could explain the observed variations in peptide binding preferences and affinities for each of the four HLA-E–like molecules. Our results could help with guiding the selection of candidate pathogen-derived peptides with the capacity to target HLA-E–restricted T cells that could be mobilized in vaccination and immunotherapeutic strategies.
Introduction
Unconventional T cells are unique in their capacity to recognize Ags, including peptides, lipids, phosphoantigens, and metabolites presented through highly conserved Ag-presenting molecules such as HLA-E, CD1, butyrophilin-3, and MR-1, respectively (1). The very limited genetic polymorphism characteristic of these Ag-presenting molecules allows the activation of T cell responses in the majority of individuals by identical ligands, leading to the moniker donor-unrestricted T cells (2).
HLA-E–restricted T cells are triggered by the presentation of peptide Ags via HLA-E, a virtually monomorphic HLA class Ib molecule, for which two coding variants are known to be expressed in humans. Their sequences contain only 1 aa difference, in position 107, located outside of the peptide binding groove (arginine in HLA-E*01:01 and glycine in HLA-E*01:03), and are therefore considered to be unlikely to affect peptide binding (3). HLA-E typically binds nonameric peptides derived from HLA class Ia signal sequences, leading to its stabilization on the cell surface and allowing recognition by CD94/NKG2A and CD94/NKG2C, which inhibit or activate NK cell activity, respectively (4, 5). In addition, HLA-E functions as an Ag presentation molecule for TCR-mediated recognition of self- or pathogen-derived peptides, triggering the adaptive immune system (6–13). Its virtual monomorphism along with its capacity to stimulate unconventional CD8+ T cells suggest HLA-E’s potential can be exploited (e.g., by vaccination) to effectively mobilize HLA-E–restricted T cells across human populations (14). A series of studies in rhesus macaques (RM) vaccinated with modified rhesus CMV (RhCMV) vectors carrying pathogen-encoded Ags showed that the vaccine-induced protection was largely, if not fully, dependent on Mamu-E (ortholog of HLA-E in RM)–restricted T cell responses in the context of Plasmodium knowlesi and SIV infections (15, 16). More recently, Mamu-E–restricted hepatitis B virus–specific CD8+ T cells induced by a modified RhCMV vector were able to recognize hepatitis B virus–infected primary hepatocytes derived from both RM and humans, further supporting the potential of these unconventional T cells as vaccine targets in humans (17).
Mamu-E and Mafa-E, the orthologs of HLA-E in RM and Mauritian cynomolgus macaques, respectively, were shown to bind the same peptides as HLA-E and to have conserved Ag presentation capacities akin to their human counterpart (18). Thus, nonhuman primates (NHP) provide a relevant model for human HLA-E immunobiology and represent one of the frequently used models for human disease pathogenesis studies and preclinical trials. In mice, Qa-1b is the functional homolog of HLA-E, which despite greater evolutionary distance to HLA-E, also shows similar peptide binding specificity, likely due to the high structural conservation of the peptide binding groove (19–23). In humans as well as in mice, a broad functional capacity for HLA-E/Qa-1b–restricted T cells, including regulatory and cytotoxic effector functions, was associated with control of intracellular mycobacterial growth in vitro and protection against severe Mycobacterium tuberculosis infection in vivo (7, 8, 24, 25).
Strong positional specificity was initially observed in nonameric peptide sequences binding to HLA-E, with preferred hydrophobic “anchor” residues at position 2 (P2) and P9, based on analyses of the highly conserved HLA class Ia signal peptide sequence (20, 26). Based on this motif, in silico prediction efforts led to the identification of M. tuberculosis–derived peptides with the capacity to stimulate effector CD8+ T cell responses in an HLA-E–restricted fashion (7, 8, 24). However, more recent peptide elution studies have shown much greater sequence and length diversity in the HLA-E peptide binding repertoire (27–30). Unexpected differences were also identified in peptide repertoires binding to HLA-E*01:01 and HLA-E*01:03 (28, 31). Structural analyses have demonstrated a broad tolerance for hydrophobic and polar amino acids in the primary pockets of HLA-E and Mamu-E molecules, supporting the notion that HLA-E has the capacity to bind more diverse peptides than previously thought (15, 32). The recently estimated density of ∼4 HLA-E epitopes per 100 aa of protein length further supports the notion that HLA-E can bind more peptides than originally anticipated (15).
In this study, we have compared the peptide binding capacities of HLA-E*01:01 and HLA-E*01:03 to dissect the precise peptide binding motifs for these two highly conserved Ag-presenting molecules, using large panels of peptides (Supplemental Table I). To optimize the future selection of peptides for in vivo and preclinical studies in animal models, we also examined and cross-validated binding of the same peptides to the HLA-E–like molecules Mamu-E and Qa-1b to compare peptide binding properties by human, NHP, and mouse HLA-E–like molecules. The resulting algorithm may help with identifying novel pathogen-derived peptides with an ability to bind MHC-E (referring to HLA-E*01:01, HLA-E*01:03, Mamu-E, and Qa-1b) and could assist in selecting pathogen-derived peptides to target MHC-E–restricted T cells in humans, NHP, and mice, with the potential to stimulate MHC-E–restricted effector T cell immunity against pathogens and tumors.
Materials and Methods
Protein production
Recombinant extracellular HLA-E*01:01, HLA-E*01:03, Mamu-E [consensus sequence obtained after alignment (33) of 22 available alleles (Supplemental Fig. 1)], and Qa-1b (Protein Data Bank: 3VJ6) H chains with a C-terminal BirA recognition site were overexpressed in Escherichia coli as described previously (7) and stored as inclusion bodies at −80°C. Human β2-microglobulin (β2m) was also overexpressed in E. coli and dissolved in 8 M urea before being prefolded by dialysis against PBS with 10 mM Tris.HCl (pH 8) and stored at −80°C in 200 μM stock solutions. Protein concentration was determined by the Bradford protein assay, and purity was assessed by SDS-PAGE.
Folding monomers for UV-mediated peptide exchange
Purified inclusion bodies containing 2.5 mg of H chain were solubilized in 8 M urea and folded for 5 d at 10°C with 1.2 mg prefolded β2m and 3 mg UV-sensitive peptide (VMAPJTLVL, where J is a photolabile 2-nitrophenylamino acid) (32, 34) in 50 ml folding buffer (400 mM l-Arginine, 0.5 mM oxidized glutathione, 5 mM reduced glutathione, 2 mM EDTA, 100 mM Tris.HCl [pH 8], glycerol 5%, and half a tablet of protease inhibitor mixture [cOmplete; Roche]). After folding, monomers were concentrated on a 30 kD filter (Amicon Ultracel 15). Mamu-E and Qa-1b monomers were biotinylated using the BirA enzyme, whereas HLA-E*01:01 and HLA-E*01:03 monomers were not biotinylated. All monomers were purified by gel filtration on a HiLoad 16/60 Superdex 75 prep grade (GE Healthcare) and stored with 16% of glycerol in PBS at −80°C in small aliquots.
UV-mediated peptide exchange reaction
UV-mediated peptide exchange was performed as described previously (32, 35, 36). Briefly UV photo cleavage and peptide exchange were performed in duplicate in polypropylene U-bottom 96-well plates (Greiner Bio-One). Each well contained a single reaction volume containing 0.5 μM of UV-sensitive MHC-E monomer in the presence of 100 μM exchange peptide in a total volume of 25 μl, which was adjusted by adding exchange buffer (20 mM Tris [pH 7.4] and 150 mM NaCl). As a positive control, pCMV was used as exchange peptide because it is known to have a high binding capacity to HLA-E, and VL9 and Qdm, canonical binders to HLA-E and Qa-1b, respectively, were used as an additional control in all assays. As a negative control, no additional peptide was added, and the reaction consisted of the UV-sensitive MHC-E monomer alone. UV-mediated peptide exchange reaction was triggered by exposure to UV light >350 nm for 60 min at 4°C.
Detection of peptide exchange by sandwich ELISA
For HLA-E*01:01 and HLA-E*01:03, 96-well half area ELISA microplates (Greiner Bio-One) were coated with 10 μg/ml of purified anti-human HLA-E Ab (clone 3D12; BioLegend). For Mamu-E and Qa-1b, 96-well Nunc MaxiSorp plates (Invitrogen) were coated with 2 μg/ml streptavidin (Invitrogen). Subsequent to the UV exposure, the resulting exchanged monomers were diluted 100-fold in blocking buffer, and 25 μl was added to plates previously blocked with 2% IgG-free BSA in PBS and washed with 0.05% Tween 20 in PBS. Detection of β2m-associated complexes was performed using 2 μg/ml HRP-conjugated anti-β2m Ab (Thermo Fisher Scientific) in blocking buffer, and the signal was amplified with a 1:15 dilution of HRP-coupled goat anti-rabbit IgG (Dako) before developing with tetramethylbenzidine substrate (Invitrogen) and terminating the reaction with H2SO4 stop solution. Absorbance readings were obtained at 450 nm using a SpectraMax i3x Reader. Sample-to-positive (s/p) ratio was calculated by normalizing values to positive control (pCMV) after subtraction of the background obtained in the absence of test peptide (no rescue): s/p ratio = (value − no rescue)/(pCMV − no rescue). Binding for all peptides was measured in at least two independent experiments, and results represent mean values.
HPLC size exclusion chromatography peptide binding assay
MHC-E H chains were dissolved in 8 M urea and stored in 50 μM stock solutions at −80°C until use. MHC-E preparations were titrated in duplicate in the presence of 100 fmol fluorescent standard peptide to determine the rMHC-E concentration at which 20–50% binding of the fluorescent standard peptide was obtained. All subsequent assays were performed at these MHC-E concentrations. For HLA-E*01:01, HLA-E*01:03, and Mamu-E, VXAPCTLLL (where X is norleucine [the use of norleucine showed reduced oxidation compared with the methionine in P2. This reduced unspecific aggregation of the proteins while maintaining the binding capacity of methionine] and C is labeled with fluorescein) was used as a fluorescent standard peptide. For the Qa-1b, VMAPCTLVL (where C is labeled with Alexa Fluor 488) was used.
Test peptides were dissolved in DMSO at a concentration of 10 mM, and binding assays were performed as previously described (7, 37). Briefly, a 10-fold titration series was made on a low protein binding U-bottom 96-well microtiter plate (polypropylene; CoStar). HLA-E*01:01, HLA-E*01:03, or Mamu-E (5 pmol) or Qa-1b H chain (25 pmol) was incubated for 24 h at room temperature in a total reaction volume of 100 μl containing 25 pmol β2m, 2 μl protease inhibitor mixture (50× solution cOmplete; Roche), 100 fmol fluorescent standard peptide, and 10 μl of test peptide dilution in binding buffer (pH 7) (100 mM phosphate, 75 mM NaCl, and 1 mM CHAPS). Samples were analyzed on a SynChroPak GPC 100 column (250 × 4.6 mm; Eprogen) using a Jasco AS-950. As HPLC running buffer, binding buffer containing 1% CH3CN was used. Fluorescence emission was detected at 516 nm using a Jasco FP 920 fluorescence monitor (B&L Systems, Maarssen, the Netherlands). The percentage of peptide bound was calculated as the amount of fluorescence bound to MHC-E divided by total fluorescence. The concentration of peptide yielding IC50 was deduced from the dose-response curve. Binding for each peptide was measured once.
9-mer combinatorial peptide library scan and analysis
The combinatorial peptide library (CPL) consists of 180 peptide pools, each of them containing a single amino acid fixed at one position, whereas the rest of the sequence is random (38, 39). Binding of CPL peptide pools was measured in the same way as with single peptides using 100 μM of each peptide pool. Two measurements were performed in the context of each MHC-E molecule, and averages were calculated for further analysis with the Warwick Systems Biology Centre Peptide Identification CPL webtool, which generated the heatmaps (40).
Results
To investigate peptide binding to MHC-E, we implemented an UV-mediated peptide exchange assay with detection by ELISA previously described for HLA class I (35) and adapted to HLA-E*01:03 (36). Based on previous observations that hinted at possible differences in peptide binding to the two human HLA-E molecules (28, 31), we adjusted the assay to test peptide binding to both HLA-E*01:01 and HLA-E*01:03 independently. In this assay, we refolded the H chains of each HLA-E molecule with human β2m in the presence of a modified VL9 peptide containing an UV-sensitive 2-nitrophenylamino acid termed J at P5, as previously described (36). To induce UV-mediated peptide exchange, the degradation of purified monomers was induced through exposure to UV light, which promoted cleavage of the UV-sensitive peptide and destabilization of the HLA-E–peptide complex. In the presence of an exchange peptide with binding capacity to HLA-E, the monomer was rescued and stabilized, whereas the absence of exchange peptide led to complex instability and loss of HLA-E/β2m binding. Efficiency of monomer degradation and peptide exchange was measured by sandwich ELISA in which complexes were captured using an HLA-E–specific Ab (Fig. 1A) (35, 36).
FIGURE 1.
Peptide binding to HLA-E*01:01, HLA-E*01:03, Mamu-E, and Qa-1b quantified with an UV-mediated peptide exchange assay and a competition-based HPLC assay. (A) Schematic representation of the UV-mediated peptide exchange assay to measure peptide binding to HLA-E*01:01 and HLA-E*01:03 by capturing rescued complexes with plates coated with a specific HLA-E Ab. (B) Schematic representation of a modified UV-mediated peptide exchange assay to measure peptide binding to Mamu-E and Qa-1b in which the capture of previously biotinylated complexes is done with streptavidin-coated plates. In both (A and B), rescued complexes are detected with an HRP-coupled β2m Ab. (C–E) Previously described MHC-E binding peptides in humans: pCMV (VLAPRTLLL, red bar) and VL9, canonical HLA-E binders; p34-p68, M. tuberculosis–derived peptides previously predicted to bind HLA-E (7); in mice, Qdm-FL9, Qa-1b binders (12); and in NHP, Gag120-Gag69, SIV-derived peptides known to bind Mamu-E (15). The sequences of these peptides are available in Supplemental Table I. Peptides are ranked according to their capacity to bind HLA-E*01:01 to facilitate comparison between alleles. (C) Absorbance represents the number of MHC-E complexes that are rescued in the presence of test peptide, the value being higher for peptides with better binding capacity. Bars show the average and SD of a representative experiment with two biological repeats and two technical replicates. (D) The s/p ratios were calculated by normalizing values to positive control (pCMV) after subtraction of background obtained in the absence of test peptide (no rescue): (value − no rescue)/(pCMV − no rescue). Bars represent calculations from one representative experiment containing two biological repeats and two technical replicates. (E) IC50 were obtained by HPLC size exclusion chromatography using recombinant MHC-E and competition of a fluorescently labeled peptide. Binding was calculated as the concentration (micromolar) of peptide required to reduce fluorescence intensity of the standard peptide by 50% (IC50), and 100 μM was the highest concentration tested. Bars represent one representative experiment.
In addition to human HLA-E, we also adapted the protocol to measure peptide binding to Mamu-E and Qa-1b as these are the most commonly used HLA-E–related animal models for future in vivo preclinical vaccination and immunotherapeutic studies. The RM genome encodes at least 30 different Mamu-E molecules with 88% amino acid sequence similarity within the peptide binding groove. Notably, residues forming the B and F pockets, which accommodate the primary anchor residues, are conserved among all except 3 Mamu-E alleles (18). Therefore, we performed a multiple alignment analysis of the 22 available Mamu-E proteins and obtained an artificial consensus sequence (93.03–99.33% identity with Mamu-E alleles, with the most similar allele being Mamu-E*02:11 with a single Gly37Glu substitution), which was genetically synthesized, produced as recombinant proteins, and used for all experiments (Supplemental Fig. 1). Peptide exchange for Mamu-E and Qa-1b was performed in the same way as for the human HLA-E molecules. However, because of the lack of a specific capture Ab, detection of exchanged complexes, which were previously biotinylated, was performed using streptavidin-coated plates (Fig. 1B). This allowed us to define the peptide binding motifs for HLA-E*01:01, HLA-E*01:03, Mamu-E, and Qa-1b.
Evaluation of peptides binding to HLA-E*01:01, HLA-E*01:03, Mamu-E, and Qa-1b
We first used the UV-mediated peptide exchange assay to test a panel of 9-mer peptides, previously described to bind HLA-E*01:03 (Mtb44, Mtb34, Mtb62, Mtb68, and Mtb55) (7), Mamu-E (Gag120 and Gag69) (15), and Qa-1b (Qdm and FL9) (12). VL9, a canonical HLA-E binder, was included as an additional positive control (41). Peptide binding intensity as indicated by absorbance at 450 nm was generally higher for HLA-E*01:01 compared with HLA-E*01:03 and slightly lower for Mamu-E and Qa-1b (Fig. 1C). However, normalization of the data relative to pCMV control after background/no rescue subtraction (s/p ratio) suggests that binding of tested peptides was similar to both human molecules, with only subtle differences, which were more evident for Qdm, Mtb34, and FL9. Although peptides Mtb44, VL9, and Qdm showed significant binding to all four molecules, other peptides showed greater differences in binding to Mamu-E and Qa-1b compared with HLA-E. Such is the case of Mtb68, which bound best to HLA-E*01:01 and HLA-E*01:03; Gag120 bound best to Mamu-E followed by HLA-E*01:01 and HLA-E*01:03 but not to Qa-1b; and FL9 bound best to Qa-1b with little to no binding to HLA-E*01:03 and Mamu-E (Fig. 1D). When comparing all four molecules, Qa-1b stood out by having an s/p ratio higher than 0 for only 7 out of 10 tested peptides. For Mamu-E, this was the case for 9 out of 10, whereas for HLA-E*01:01 and HLA-E*01:03, all 10 tested peptides had an s/p ratio above 0.
We next validated and extended these results using a second, independent HPLC-based peptide binding assay. This method was previously established for HLA-E*01:03 in our laboratory (7), and in this study, we adapted and optimized it for HLA-E*01:01, Mamu-E, and Qa-1b. This assay is based on competition of a given test peptide with a fluorescently labeled indicator binding peptide and allows relative quantitative affinity measurements (IC50). A low IC50 indicates that the test peptide is a strong binder as it effectively outcompetes the fluorescent indicator peptide at low concentrations (Fig. 1E). Using this assay, similarities and differences in peptide binding to all four MHC-E molecules, as observed with the UV-mediated peptide exchange method, were confirmed. Peptide binding to HLA-E*01:01 and HLA-E*01:03 were similar, whereas greater differences were observed in peptide binding to Mamu-E and Qa-1b. More specifically, Mtb44, VL9, Qdm, and the positive control pCMV had the lowest IC50 values for all four molecules. In addition, Gag120 showed the lowest IC50 for Mamu-E, HLA-E*01:03, and HLA-E*01:01 but showed no binding to Qa-1b, whereas FL9 had the lowest IC50 for Qa-1b. Confirming the above results, Qa-1b bound (IC50 < 100 μM) fewer peptides (8 out of 11 tested) compared with the other three molecules (10 for Mamu-E and 11 for HLA-E*01:01 and HLA-E*01:03 out of 11 tested).
Peptides bind differently to HLA-E*01:01, HLA-E*01:03, Mamu-E, and Qa-1b
To improve the definition of peptide binding motifs to all four MHC-E molecules, we tested binding of HLA-E*01:01, HLA-E*01:03, Mamu-E, and Qa-1b to a larger panel of previously described peptides that were predicted to bind HLA-E in humans (7). We additionally tested peptides previously described to be binders or nonbinders to Mamu-E in NHP (15, 42, 43) and to be immunogenic in mice (13). Heatmaps represent s/p ratio corresponding to binding of the tested peptides to each MHC-E molecule, measured with the UV-mediated peptide exchange binding assay (Fig. 2). For each allele, we obtained the SD for all peptide measurements (at least two per peptide) and subsequently estimated a threshold for defining binding peptides by calculating the mean (SD) (SD ≤ s/p ratio ≤ 2 × SD). We additionally used two times the mean SD to estimate strong binders in the context of each MHC-E allele individually (s/p ratio ≥ 2 × SD) (Supplemental Fig. 2).
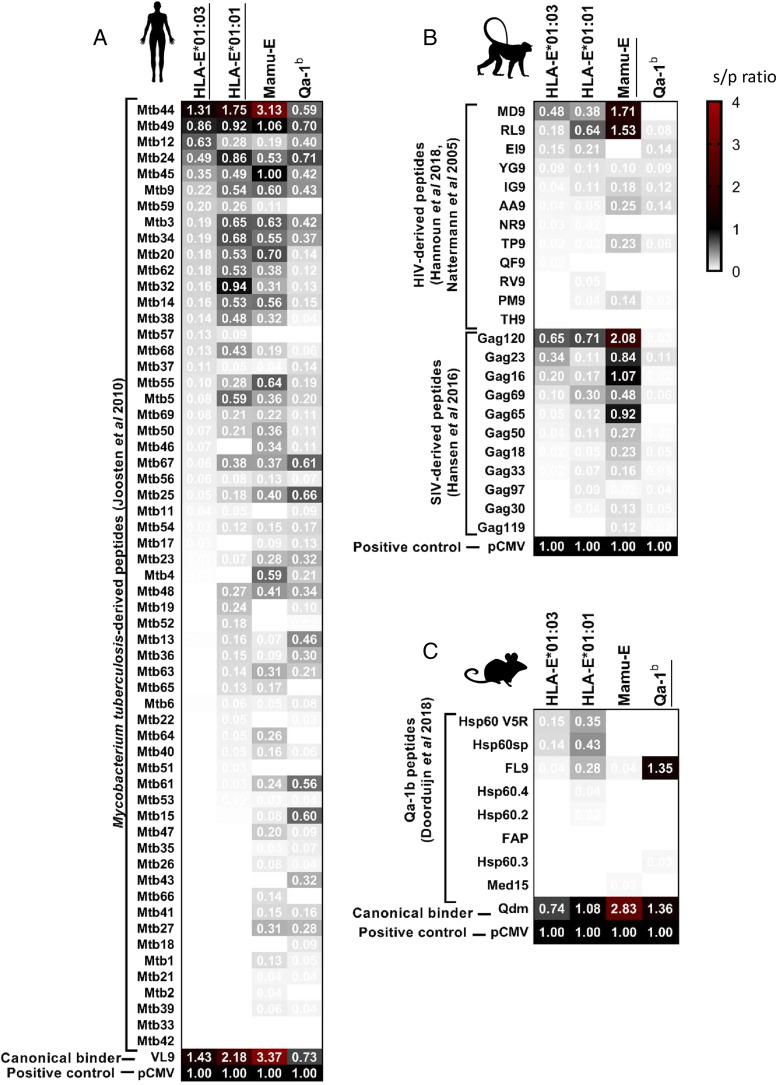
FIGURE 2.
Peptides bind differently to HLA-E*01:01, HLA-E*01:03, Mamu-E, and Qa-1b. Peptide binding was measured using the UV-mediated peptide exchange assay. Heatmaps represent mean values of at least two independent experiments and indicate s/p ratios of the tested peptides for each MHC-E molecule with white being no binding (s/p ratio = 0), black being a binding similar to positive control (s/p ratio = 1), and red being a binding higher than the positive control. Peptides were arranged in a decreasing order of binding to HLA-E*01:01, with the stronger binder at the top and canonical and control binders at the bottom. (A) M. tuberculosis–derived peptides previously predicted to bind HLA-E (7). (B) HIV- and SIV-derived peptides previously described to bind Mamu-E (15, 42, 43). (C) Peptides previously described to bind Qa-1b (13).
Surprisingly, we found important differences in peptide binding to all MHC-E molecules tested. Of the 59 M. tuberculosis–derived peptides previously predicted to bind HLA-E*01:03 in humans, 18 were binders, and 11 were strong binders to HLA-E*01:03. Among the 11 strong binders to HLA-E*01:03, 9 were also strong binders to HLA-E*01:01, 9 to Mamu-E, and 8 to Qa-1b. Only seven peptides were strong binders to HLA-E*01:01, HLA-E*01:03, Mamu-E, and Qa-1b (Mtb44, Mtb49, Mtb24, Mtb34, Mtb3, Mtb9, and Mtb45) (Fig. 2A). Among the nine peptides with less strong binding affinity to HLA-E*01:03, five were strong binders to HLA-E*01:01, two to Mamu-E, and none to Qa-1b, highlighting the differences in peptide binding to each MHC-E molecule. As observed previously, peptides generally had a higher binding intensity for HLA-E*01:01 than for HLA-E*01:03, possibly as a consequence of an overall lower stability of HLA-E*01:01 compared with HLA-E*01:03, which might facilitate peptide exchange.
Interestingly, we found that among the 31 peptides described as immunogenic in NHP or in mice, 7 were strong binders to Mamu-E, and only 1 to Qa-1b, whereas 5 were strong binders to HLA-E*01:01 and HLA-E*01:03 (Fig. 2B, 2C). None of these tested peptides was shown to have binding capacity to all four MHC-E molecules, and only three peptides were strong binders to human and NHP MHC-E (notably RL9, MD9, and Gag120). Although strong peptide binding is not a strict predictor of immunogenicity and other measurements such as stability might be a better indicator (44), peptide binding to HLA is necessary for productive recognition of the presented peptides by TCRs. The differences we observed also in peptides with moderate and low binding capacity to each of the four MHC-E molecules suggest that comparing peptide binding to human and animal MHC-E molecules might facilitate the selection of promising candidate peptides.
We next validated the peptide binding and measured relative affinities using the HPLC-based binding assay. Comparison of binding data for M. tuberculosis–derived peptides (except for Mtb34, Mtb44, Mtb55, Mtb62, and Mtb68) was only performed in the context of HLA-E*01:03 based on the results published previously (7). The analysis across the four molecules showed significant correlation between the two binding assays, indicating that the different results (binding versus relative affinity) are in agreement across alternative methods and supporting that differences exist in peptide binding to the two highly conserved HLA-E molecules (Fig. 3). The UV-mediated peptide exchange binding assay is relatively fast and high throughput, allowing the analysis of 10 times the number of peptides compared with the HPLC-based method in the same time interval. Therefore, we continued using UV-mediated peptide exchange binding assay to test peptide binding to MHC-E.
FIGURE 3.
Good correlation between UV-mediated peptide exchange and HPLC binding assays. Correlations between the s/p ratio obtained with the UV-mediated peptide exchange binding assay and the IC50 obtained with the HPLC binding assay are plotted for HLA-E*01:03, HLA-E*01:01, Mamu-E, and Qa-1b as indicated. IC50 values for peptides represented as black circles correspond to HIV- and SIV-derived peptides previously described to bind Mamu-E (15, 42, 43) and peptides previously described to bind Qa-1b (13). IC50 values for peptides represented as gray triangles correspond to those previously published in the context of only HLA-E*01:03 (7). Dotted lines represent the thresholds indicating binders and strong binders to each MHC-E molecule, according to the UV-mediated peptide exchange binding assay. The s/p ratios represent mean values from at least two independent experiments with two biological replicates and two technical replicates. IC50 values represent one experiment. Statistical significance was tested by nonparametric Spearman correlation analysis.
Importance of anchor residues for 9-mer peptides binding to HLA-E*01:01, HLA-E*01:03, Mamu-E, and Qa-1b
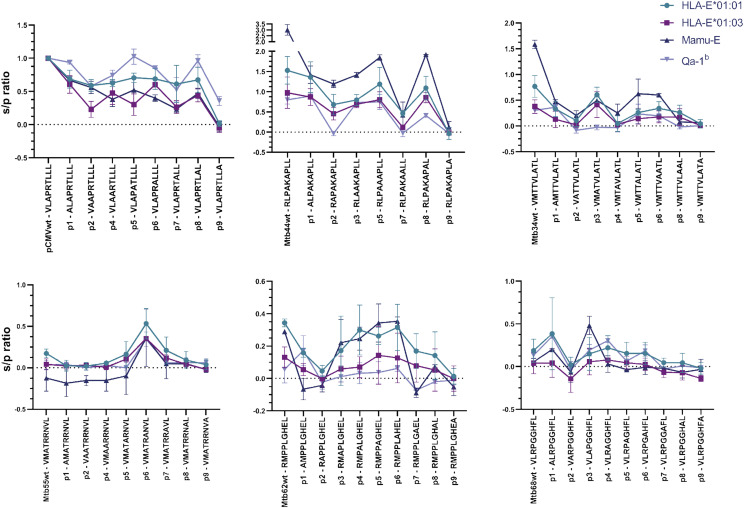
To study the influence of each amino acid position on peptide binding to MHC-E, we substituted each residue by alanine (Ala), one of the smallest natural amino acids. This replacement is expected to reduce the binding capacity when substituting anchor wild-type residues as its side-chain chemical properties will be lost and additionally may also reveal gain-of-function substitutions improving binding. We synthesized Ala substitutions of six M. tuberculosis–derived HLA-E binding peptides, previously shown to be recognized in an HLA-E–specific manner (7). The substitutions prompted changes in peptide binding to all four MHC-E molecules; when replacing an apparently critical anchor residue, we observed reduction or complete elimination of peptide binding. In particular, we observed a complete or almost complete loss of binding to HLA-E*01:01, HLA-E*01:03, Mamu-E, and Qa-1b when P7-Ala or P9-Ala substitutions were introduced in pCMV and Mtb44. In addition, P2-Ala substitution also showed a considerable loss of binding to all four molecules (Fig. 4, top left and middle). Ala substitutions in all positions of Mtb34 resulted in loss of binding, except for P3-Ala (Fig. 4, top right). Considering the relatively low HLA-E binding affinity of Mtb34 compared with pCMV and Mtb44, this might suggest that a combination of optimal anchor residues is essential for effective peptide binding to HLA-E in certain cases. The wild-type sequence of Mtb34 contains a P7-Ala possibly already reducing its binding capacity as was seen for pCMV and Mtb44 Ala substitutions at P7. Moreover, substitution of nonanchor positions sometimes led to increased peptide binding (Fig. 4). Mtb34 containing P3-Ala and Mtb68 containing P3-Ala or P1-Ala had increased binding to certain MHC-E molecules, suggesting that the presence of a small residue adjacent to a primary anchor residue (P2) might allow more space or less interference to favor its anchorage within the primary binding pocket (Fig. 4, top and bottom right). Mtb62, as with higher binders, had reduced binding when P2-Ala, P7-Ala, or P9-Ala substitutions were tested, and binding was not affected when P4-Ala, P5-Ala, or P6-Ala substitutions were used, suggesting that these positions are less crucial for peptide binding to MHC-E (Fig. 4, bottom middle). However, for Mtb55, the peptide with lowest binding to MHC-E among the six tested, there was a remarkable increase in peptide binding to all four MHC-E molecules when the P6-Ala substitution was tested (Fig. 4, bottom left).
FIGURE 4.
Binding of alanine substitutions reveals the importance of anchor positions for binding HLA-E*01:01, HLA-E*01:03, Mamu-E, and Qa-1b. Peptides were modified by an alanine substitution at each position and tested for binding to HLA-E*01:01, HLA-E*01:03, Mamu-E, and Qa-1b with the UV-mediated peptide exchange binding assay. Each graph represents peptide binding as s/p ratio for a peptide and its alanine substitutions in the context of the four MHC-E molecules. The graphs show the mean and SD of at least two independent experiments with two biological replicates and two technical replicates each.
Altogether, these results support that P2 and P9 are the most critical anchor positions for peptide binding to MHC-E in humans, RM, and mice and further underscore the importance of P7 given the level of disruption in peptide binding upon Ala substitution. Moreover, these results suggest that there might be an advantage for peptide binding when a residue adjacent to an anchor residue is small, such as Ala, allowing an optimal docking of the anchor residue within the binding pocket. Overall, the four MHC-E molecules investigated behaved similarly when binding these six peptides and their corresponding Ala substitutions, with Qa-1b being somewhat more distinct compared with HLA-E*01:01, HLA-E*01:03, and Mamu-E, which behaved more similar.
Binding of CPL to HLA-E*01:01, HLA-E*01:03, Mamu-E, and Qa-1b
For a comprehensive, systematic, and unbiased analysis of 9-mer peptide sequences with a capacity to bind to MHC-E and to assess particular amino acid preferences for each anchor position, we next used a CPL scanning approach in combination with the UV-mediated peptide exchange binding assay. The CPL consists of 180 peptide pools, each of them containing a single amino acid fixed at one position, whereas the rest of the sequence is random, and therefore, this approach can reveal the specific contribution of each residue at each position (38, 39). Results are depicted in heatmaps representing the MHC-E binding obtained when each amino acid (columns) is fixed at each position (rows) (Fig. 5A). In line with our results above, HLA-E*01:01, HLA-E*01:03, and Mamu-E showed a more similar peptide binding signature compared with Qa-1b. In addition to the canonical anchor residues Met and Leu at P2 and P9, respectively, we observed a certain flexibility in the amino acids present at key anchor positions of binding peptides. HLA-E*01:01 and Qa-1b bound, to some extent, peptides containing a P2-Phe, whereas Mamu-E and Qa-1b also bound peptides with P2-Thr. Similarly, binding of peptides with P9-Phe was possible in the case of HLA-E*01:01 and HLA-E*01:03, whereas Qa-1b also bound peptides with P9-Met and P9-Lys. Interestingly, large hydrophobic residues such as Phe, Trp, or Tyr at secondary anchor position P7 and nonanchor position P5 favored peptide binding to all four molecules and to Mamu-E and Qa-1b when they are present at P8. The presence of these large residues at these commonly bulging out peptide positions (P5 and P8) might favor peptide binding by increasing the capacity of the peptides to bulge out of the binding groove at these positions, facilitating that anchor residues could reach deeper into the primary binding pockets (32). Alternatively, small residues at P5 could result in peptide binding as a result of low interference as could be seen for HLA-E*01:01 and HLA-E*01:03 but not Mamu-E and Qa-1b. Qa-1b additionally showed preference for peptides with P7-Pro. Finally, an apparent characteristic of peptides binding to HLA-E*01:01 and HLA-E*01:03, but not Mamu-E and Qa-1b, was the preference for small residues such as Ala, Pro, or Thr in secondary anchor position P6. Although our Ala substitution analysis showed a similar effect of P6-Ala in all four MHC-E molecules, the CPL binding analysis showed that P6 was less important for peptide binding to Mamu-E, whereas P6-Thr and P6-Met increased peptide binding to Qa-1b. Overall, HLA-E*01:03 and Mamu-E allowed fewer variations in peptide amino acid sequences able to anchor to its peptide binding groove, which might be a reflection of higher conformational rigidity of these molecules compared with the other two.
FIGURE 5.
New insights into amino acid preferences for peptide binding to HLA-E*01:01, HLA-E*01:03, Mamu-E, and Qa-1b. (A) Heatmap representation of CPL binding intensity to HLA-E*01:01, HLA-E*01:03, Mamu-E, and Qa-1b, indicating which amino acids (columns) are preferred at each peptide position (row) for the peptide to bind each MHC-E molecule. Data represent mean values of two independent experiments with two biological replicates and are normalized for each position with values ranging from high binding in red to low binding in blue. Highlighted with black squares are the residues and positions that resulted in preferred binding to each molecule. (B) Amino acid substitutions on pCMV and Mtb44 were tested based on the results above to confirm flexibility or strictly preferred amino acids in anchor positions. Bar graphs show mean and SD from at least two independent experiments. The s/p ratios represent binding affinity of the indicated peptides to HLA-E*01:01, HLA-E*01:03, Mamu-E, and Qa-1b.
Amino acid substitutions testing the flexibility of main anchor positions
Given the diversity of peptide repertoires previously reported to bind MHC-E and based on the results we obtained from the analysis of CPL binding to all four MHC-E molecules, we decided to evaluate MHC-E binding capacity after introducing certain amino acid substitutions at the main anchor positions of the strong binders pCMV and Mtb44. (Fig. 5B). We examined Leu2Met (substitution of Leu by Met at P2) and observed that the presence of any of the two hydrophobic residues led to strong binding of the peptides to the four MHC-E molecules. Substitutions at P7 of pCMV with very large hydrophobic residues such as Leu7Phe and Leu7Trp, resulted in decreased peptide binding capacity, which was more prominent for Mamu-E and Qa-1b compared with HLA-E*01:01 and HLA-E*01:03, although these peptides continued to have strong binding, in line with the above CPL binding data. However, Leu7Pro led to an increase in peptide binding capacity to all four molecules, suggesting that the presence of the relatively rigid Pro in P7, which may restrict peptide structural flexibility, could have an advantage for peptide binding to MHC-E. The strong binder Mtb44 has a Pro in P7 in its wild-type form, and Pro7Phe and Pro7Trp substitutions led to a marked reduction in peptide binding to HLA-E*01:01 and Mamu-E, further supporting the importance of Pro in P7 for peptide binding to MHC-E. This reduction was less evident in the context of HLA-E*01:03 and was only seen for Pro7Trp, but not Pro7Phe, in the context of Qa-1b. Based on our Ala substitution analyses in which we found that P6-Ala could result in an increase in peptide binding to MHC-E, we further studied the effect of Ala6Thr substitution, as P6-Thr is present in pCMV but observed no apparent changes in peptide binding to the four MHC-E molecules, suggesting that other small residues at P6 can also facilitate effective peptide binding to MHC-E, in line with findings from the CPL binding analysis. Because P9 is one of the most important anchor positions, we additionally studied the influence of amino acid substitutions at this final position of the 9-mers. A Leu9Phe substitution maintained strong binding to MHC-E, with the exception of Mtb44 Leu9Phe, which led to reduced binding to Mamu-E and Qa-1b. However, Leu9Tyr and Leu9Trp substitutions led to a marked reduction in peptide binding to all four MHC-E molecules, suggesting that peptide binding to MHC-E strictly needs a large and hydrophobic anchor residue in P9.
Altogether, our findings suggest that peptides binding HLA-E*01:01 and HLA-E*01:03 have a strict preference for large hydrophobic residues at main anchor P2 and P9. Binding to these two molecules was also higher for peptides containing large hydrophobic residues at main anchor P7, although peptides carrying a Pro at this position were also binders. In addition, we identified a potential preference for small amino acids adjacent to main anchor P2 (P1-Ala/Thr) and P7 (P6-Ala/Thr) as well as increased binding of peptides containing either very large or small residues at P5. Overall, peptide binding motifs to HLA-E*01:01 and HLA-E*01:03 were highly overlapping, the main difference being the apparently higher permissive binding to HLA-E*01:01 of peptides containing alternative residues at P7 (Fig. 6).
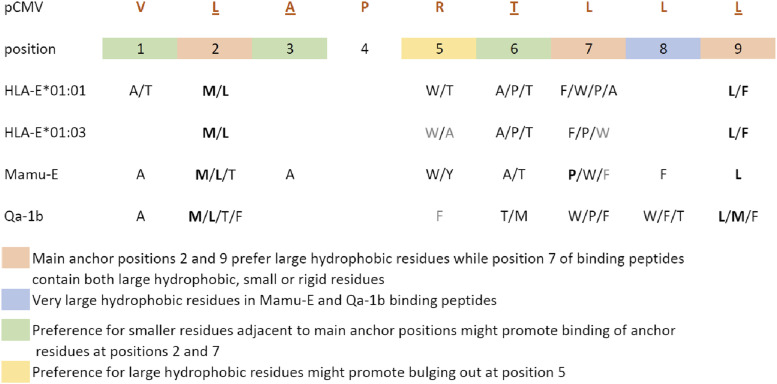
FIGURE 6.
Binding motifs to HLA-E*01:01, HLA-E*01:03, Mamu-E, and Qa-1b. New insights into the residue preferences at each peptide position allowing peptide binding to HLA-E*01:01, HLA-E*01:03, Mamu-E, and Qa-1b are indicated. The figure represents a summary of the peptide binding motifs for each MHC-E molecule based on all the data obtained in this study. The canonical sequence of pCMV is indicated with the underlined positions following the motifs. Residues in bold highlight stronger preferences, whereas those in gray are less strongly preferred. Primary anchor positions are highlighted in red, and secondary anchor positions are highlighted in green. Positions with newly defined residue preferences are highlighted in yellow and blue.
Similarly to the human molecules, peptides binding Mamu-E and Qa-1b showed a preference for large hydrophobic residues at main anchor P2 and P9, with more alternative binding residues compared with HLA-E*01:01 and HLA-E*01:03. In addition, P7 in peptides binding to Mamu-E and Qa-1b permits both large hydrophobic as well as small residues, and peptides with small residues at P1, P3, and P6 adjacent to main anchor positions show better binding. In contrast to the human molecules, Mamu-E and Qa-1b bound peptides with only large residues at P5 and showed preference for large hydrophobic amino acids also at P8 (Fig. 6).
Discussion
In this study, we modified and used a novel high-throughput UV-mediated peptide exchange binding assay to gain insight into the peptide binding requirements and resulting motifs to HLA-E*01:01, HLA-E*01:03, Mamu-E, and Qa-1b. Altogether, our results demonstrate that differences exist in peptide binding to HLA-E*01:01, HLA-E*01:03, Mamu-E, and Qa-1b, which might in part be explained by variations in MHC-E/peptide binding motifs across species (Fig. 6). We observed that for all four MHC-E molecules, the anchor P2, P7, and P9 generally preferred large hydrophobic residues, such as Met, Leu, Trp, and Phe, whereas anchor P7 showed additional preference for small Ala or Pro. Binding peptide positions adjacent to anchor positions, such as P6 and P1, showed a preference for smaller Ala or Thr residues, whereas P5 preferred large hydrophobic Trp, Tyr, or Phe residues. We identified allele-specific preferences such as binding of peptides with small P1-Ala or P1-Thr as well as the more permissive binding of peptides with P7-Ala for HLA-E*01:01 but not HLA-E*01:03. Moreover, Mamu-E and Qa-1b bound peptides with P2-Thr or P2-Phe and, contrary to the human HLA-E molecules, showed preference for binding peptides with large hydrophobic residues Phe or Trp at P8. These results suggest that although main anchor positions are conserved, the diversity in residues not only at anchor but also nonanchor positions could individually influence the docking of peptides within the binding groove of each of the four MHC-E molecules.
Despite both HLA-E alleles being equally prevalent in the human population (45, 46), HLA-E*01:01 and HLA-E*01:03 homozygosity have been associated with increased risk to or protection from infections, cancer, and autoimmune disorders, suggesting distinct roles in certain pathological conditions (47). Differences in peptide binding and surface stability, which may lead to functional differences between HLA-E*01:01 and HLA-E*01:03, support the importance of defining peptide binding motifs to each human HLA-E molecule (31, 48, 49). In the side-by-side comparison performed in this study, we found an important overlap between the peptide binding motifs for both human molecules, confirming previously defined primary anchor residues at P2 and P9, with a preference for large hydrophobic residues Met or Leu and Leu or Phe, respectively, in both HLA-E*01:01 and HLA-E*01:03 (20, 50). In addition, secondary anchor P6 had a clear preference for small residues in binding peptides, which could favor a less kinked out conformation of the HLA-E bound peptide, possibly increasing its stability within the groove (32). In line with the structure of the HLA-E*01:03/Mtb44 complex, we observed a preference for large hydrophobic residues such as Trp at P5, which might promote the burying of P6 and even P7 deep into the groove. Smaller residues such as Ala and Thr can also be present at P5 of HLA-E binding peptides, which could facilitate binding of peptides in which P6 and P7 are shallower, as is the case of the HLA-E*01:03/RL9 complex (32). Taken together, these data suggest that HLA-E and peptides can use alternative conformations and anchor interactions in the context of diverse binding sequences.
The peptide binding groove sequences of the two human HLA-E molecules are identical (3). However, it is possible that the hydrophobic and positive charges around the single nucleotide polymorphism encoded in residue 107 might lead to a reduced stability in the context of an arginine in HLA-E*01:01 versus a higher stability with a glycine in HLA-E*01:03. Indeed, HLA-E*01:03 molecules were previously shown to have higher thermal stability and higher relative affinity to peptide compared with HLA-E*01:01 (43, 49). Our results showing lower binding values for HLA-E*01:03 complexes compared with HLA-E*01:01 after an UV-mediated exchange reaction could therefore be a consequence of the relatively higher stability of HLA-E*01:03, which might partially inhibit peptide exchange. In support of this, IC50 values obtained with the HPLC-based binding assay, in which differences in complex stability are not of influence as refolding takes place in the presence of the peptide being tested, showed more comparable results for the two human HLA-E molecules. In addition, peptides such as Mtb12 and MD9 showed higher binding to HLA-E*01:03 than to HLA-E*01:01, and Gag23 bound HLA-E*01:03 but not HLA-E*01:01 (Fig. 2). This suggests that differences in peptide binding exist that cannot solely be explained by possible variations in stability but might instead be explained by variations in peptide binding motifs for each HLA-E molecule. We observed that HLA-E*01:01, but not HLA-E*01:03, showed a preference for binding peptides with small residues such as Ala and Thr at P1. In addition, the lower promiscuity in peptide binding to HLA-E*01:03 compared with HLA-E*01:01 may be explained by a higher P7 permissive binding in HLA-E*01:01 compared with HLA-E*01:03. These results support an important anchoring role for P7, which is well conserved among known consensus sequences and some alternative HLA-E binding peptides (42). These differences between the two HLA-E human molecules should be considered in the selection of vaccine peptides to target HLA-E–restricted cytotoxic T cells (14).
Like HLA-E, Qa-1b and Mamu-E can bind a wide array of peptides and have the capacity to present self- or pathogen-derived peptides for recognition by T cells (15, 19, 51). Despite Mamu-E and HLA-E showing a conserved functionality, differences were suggested in peptide binding intensities to each molecule (18). By directly comparing a large number of peptide sequences, we identified similitudes in peptide binding motifs to HLA-E, Mamu-E, and Qa-1b. As with human HLA-E molecules, Mamu-E and Qa-1b preferred binding peptides with small amino acids at P1, P2, and P6, adjacent to the main anchor positions, which might represent an advantage for main anchor residues to bind with little interference from the surrounding residues. Similar to human HLA-E, Mamu-E showed a preference for large residues at P5, which was less evident for Qa-1b. Interestingly, we also identified differences, such as Mamu-E and Qa-1b binding to peptides with greater variation in anchor P2 residues with additional preference for Thr or Phe compared with only Met and Leu for HLA-E in humans. In contrast to human HLA-E, the preference of Mamu-E and Qa-1b for large hydrophobic residues at P8 might allow a peptide conformation that enhances binding at P7 and P9. At anchor P9, Mamu-E showed higher constraints for binding peptides with Leu, whereas Qa-1b bound peptides with Met in addition to Leu and Phe, the two residues preferred for binding to HLA-E. These differences in the main anchor and nonanchor residues across all four MHC-E molecules might account for the observed variations in peptide binding between HLA-E, Mamu-E, and Qa-1b. Even though MHC-E is highly conserved in humans, mice, and RM, differences in peptide binding motifs might be explained by variations in the residues forming the binding pockets of each species (26, 32). Therefore, the single difference I73T within the C pocket between human HLA-E and Mamu-E might explain anchoring preferences for P6. Similarly, H9T within the B pocket, T70M and I73N within the C pocket, and F117E within the C, E, and F pockets of human HLA-E versus Qa-1b might explain the even larger differences in peptide binding motifs between these two molecules (Supplemental Fig. 3). Further direct structural analyses comparing the four MHC-E molecules are needed to elucidate differences in pocket occupancy by the same peptides and its functional consequences.
Remarkably, Gag97, which was recognized by CD8+ T cells from RM vaccinated with a rhCMV vector carrying SIV Ags (15), or Med15, an immunogenic T cell epitope in the context of Qa-1b (13), are low MHC-E binding epitopes. Plasticity in TCR–peptide–HLA interactions, as demonstrated in the context of melanoma Ag-specific T cells (52), might result in the context of HLA-E, in conformational changes after initial recognition and binding of TCR to a low binding HLA-E/peptide complex, leading to increased binding, complex stability, and productive TCR signaling. This mechanism might be particularly relevant in the context of peptides that bind HLA-E with a bulging out, less stable conformation (53). Besides aiming for strong HLA-E/peptide binding affinity and stability for the identification of most immunogenic peptides (54), weaker HLA-E binding peptides could also have an important role in priming T cell and driving T cell differentiation and effector function (55). Alternatively, the low correlation observed between HLA-E/peptide binding affinity and T cell recognition (7) might be a consequence of a limitation in the assay that does not represent the cellular processes involved in Ag processing and peptide presentation and also did not measure the stability and off-rates of HLA-E bound peptides. Therefore, possible unknown mechanisms or molecular interactions with coreceptors or other factors that promote peptide binding and complex stability could be involved, requiring further study (36).
The UV-mediated peptide exchange binding assay, as opposed to the HPLC-based binding assay, allows the high-throughput identification of binding peptides with alternative anchor residues across species. The definition of alternative MHC-E/peptide binding motifs will allow the improvement of prediction algorithms for the discovery of novel HLA-E binding peptides derived from persisting as well as emerging human pathogens, which could be relevant for the development of vaccines targeting the nonpolymorphic HLA-E molecule (14).
Supplementary Material
Acknowledgments
We thank Prof. Thorbald van Hall (Department of Medical Oncology, Leiden University Medical Center) for providing us with Qa-1b peptides and reviewing the manuscript. We also thank Linda Voogd (Department of Infectious Diseases, Leiden University Medical Center) for reviewing the manuscript. We thank Lucy Walters (Nuffield Department of Medicine, University of Oxford, Oxford, U.K.) for sharing protocols and expertise on the UV-mediated peptide exchange method and detection of peptide exchange by sandwich ELISA ahead of publication.
This work was supported by the European Union’s Horizon 2020 research and innovation program under Marie Skłodowska-Curie Actions Grants 707404 and 793027. Research reported in this study was also supported by the National Institute of Allergy and Infectious Diseases, National Institutes of Health under Awards R21AI127133 and R01AI141315.
The online version of this article contains supplemental material.
- CPL
- combinatorial peptide library
- β2m
- β2-microglobulin
- NHP
- nonhuman primate
- P2
- position 2
- RhCMV
- rhesus CMV
- RM
- rhesus macaque
- s/p
- sample-to-positive.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.D’Souza M. P., Adams E., Altman J. D., Birnbaum M. E., Boggiano C., Casorati G., Chien Y. H., Conley A., Eckle S. B. G., Früh K., et al. 2019. Casting a wider net: immunosurveillance by nonclassical MHC molecules. PLoS Pathog. 15: e1007567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Joosten S. A., Ottenhoff T. H. M., Lewinsohn D. M., Hoft D. F., Moody D. B., Seshadri C., Collaboration for Tuberculosis Vaccine Discovery - Donor-Unrestricted T-cells Working Group, Bill and Melinda Gates Foundation 2019. Harnessing donor unrestricted T-cells for new vaccines against tuberculosis. Vaccine 37: 3022–3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Geraghty D. E., Stockschleader M., Ishitani A., Hansen J. A. 1992. Polymorphism at the HLA-E locus predates most HLA-A and -B polymorphism. Hum. Immunol. 33: 174–184. [DOI] [PubMed] [Google Scholar]
- 4.Braud V. M., Allan D. S., O’Callaghan C. A., Söderström K., D’Andrea A., Ogg G. S., Lazetic S., Young N. T., Bell J. I., Phillips J. H., et al. 1998. HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature 391: 795–799. [DOI] [PubMed] [Google Scholar]
- 5.Lee N., Llano M., Carretero M., Ishitani A., Navarro F., López-Botet M., Geraghty D. E. 1998. HLA-E is a major ligand for the natural killer inhibitory receptor CD94/NKG2A. Proc. Natl. Acad. Sci. USA 95: 5199–5204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pietra G., Romagnani C., Falco M., Vitale M., Castriconi R., Pende D., Millo E., Anfossi S., Biassoni R., Moretta L., Mingari M. C. 2001. The analysis of the natural killer-like activity of human cytolytic T lymphocytes revealed HLA-E as a novel target for TCR α/β-mediated recognition. Eur. J. Immunol. 31: 3687–3693. [DOI] [PubMed] [Google Scholar]
- 7.Joosten S. A., van Meijgaarden K. E., van Weeren P. C., Kazi F., Geluk A., Savage N. D. L., Drijfhout J. W., Flower D. R., Hanekom W. A., Klein M. R., Ottenhoff T. H. M. 2010. Mycobacterium tuberculosis peptides presented by HLA-E molecules are targets for human CD8 T-cells with cytotoxic as well as regulatory activity. PLoS Pathog. 6: e1000782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Meijgaarden K. E., Haks M. C., Caccamo N., Dieli F., Ottenhoff T. H. M., Joosten S. A. 2015. Human CD8+ T-cells recognizing peptides from Mycobacterium tuberculosis (Mtb) presented by HLA-E have an unorthodox Th2-like, multifunctional, Mtb inhibitory phenotype and represent a novel human T-cell subset. PLoS Pathog. 11: e1004671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heinzel A. S., Grotzke J. E., Lines R. A., Lewinsohn D. A., McNabb A. L., Streblow D. N., Braud V. M., Grieser H. J., Belisle J. T., Lewinsohn D. M. 2002. HLA-E-dependent presentation of Mtb-derived antigen to human CD8+ T cells. J. Exp. Med. 196: 1473–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pietra G., Romagnani C., Mazzarino P., Falco M., Millo E., Moretta A., Moretta L., Mingari M. C. 2003. HLA-E-restricted recognition of cytomegalovirus-derived peptides by human CD8+ cytolytic T lymphocytes. Proc. Natl. Acad. Sci. USA 100: 10896–10901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoare H. L., Sullivan L. C., Pietra G., Clements C. S., Lee E. J., Ely L. K., Beddoe T., Falco M., Kjer-Nielsen L., Reid H. H., et al. 2006. Structural basis for a major histocompatibility complex class Ib-restricted T cell response. Nat. Immunol. 7: 256–264. [DOI] [PubMed] [Google Scholar]
- 12.Nagarajan N. A., Gonzalez F., Shastri N. 2012. Nonclassical MHC class Ib-restricted cytotoxic T cells monitor antigen processing in the endoplasmic reticulum. Nat. Immunol. 13: 579–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doorduijn E. M., Sluijter M., Querido B. J., Seidel U. J. E., Oliveira C. C., van der Burg S. H., van Hall T. 2018. T cells engaging the conserved MHC class Ib molecule Qa-1 b with TAP-independent peptides are semi-invariant lymphocytes. Front. Immunol. 9: 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ottenhoff T. H. M., Joosten S. A. 2019. Mobilizing unconventional T cells. Science 366: 302–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hansen S. G., Wu H. L., Burwitz B. J., Hughes C. M., Hammond K. B., Ventura A. B., Reed J. S., Gilbride R. M., Ainslie E., Morrow D. W., et al. 2016. Broadly targeted CD8+ T cell responses restricted by major histocompatibility complex E. Science 351: 714–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hansen S. G., Womack J., Scholz I., Renner A., Edgel K. A., Xu G., Ford J. C., Grey M., St Laurent B., Turner J. M., et al. 2019. Cytomegalovirus vectors expressing Plasmodium knowlesi antigens induce immune responses that delay parasitemia upon sporozoite challenge. PLoS One 14: e0210252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burwitz B. J., Hashiguchi P. K., Mansouri M., Meyer C., Gilbride R. M., Biswas S., Womack J. L., Reed J. S., Wu H. L., Axthelm M. K., et al. 2020. MHC-E-restricted CD8+ T cells target hepatitis B virus-infected human hepatocytes. J. Immunol. 204: 2169–2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu H. L., Wiseman R. W., Hughes C. M., Webb G. M., Abdulhaqq S. A., Bimber B. N., Hammond K. B., Reed J. S., Gao L., Burwitz B. J., et al. 2018. The role of MHC-E in T cell immunity is conserved among humans, rhesus macaques, and cynomolgus macaques. J. Immunol. 200: 49–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kraft J. R., Vance R. E., Pohl J., Martin A. M., Raulet D. H., Jensen P. E. 2000. Analysis of Qa-1(b) peptide binding specificity and the capacity of CD94/NKG2A to discriminate between Qa-1-peptide complexes. J. Exp. Med. 192: 613–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller J. D., Weber D. A., Ibegbu C., Pohl J., Altman J. D., Jensen P. E. 2003. Analysis of HLA-E peptide-binding specificity and contact residues in bound peptide required for recognition by CD94/NKG2. J. Immunol. 171: 1369–1375. [DOI] [PubMed] [Google Scholar]
- 21.Jensen P. E., Sullivan B. A., Reed-Loisel L. M., Weber D. A. 2004. Qa-1, a nonclassical class I histocompatibility molecule with roles in innate and adaptive immunity. Immunol. Res. 29: 81–92. [DOI] [PubMed] [Google Scholar]
- 22.Ying G., Wang J., Kumar V., Zajonc D. M. 2017. Crystal structure of Qa-1a with bound Qa-1 determinant modifier peptide. PLoS One 12: e0182296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zeng L., Sullivan L. C., Vivian J. P., Walpole N. G., Harpur C. M., Rossjohn J., Clements C. S., Brooks A. G. 2012. A structural basis for antigen presentation by the MHC class Ib molecule, Qa-1b. J. Immunol. 188: 302–310. [DOI] [PubMed] [Google Scholar]
- 24.Prezzemolo T., van Meijgaarden K. E., Franken K. L. M. C., Caccamo N., Dieli F., Ottenhoff T. H. M., Joosten S. A. 2018. Detailed characterization of human Mycobacterium tuberculosis specific HLA-E restricted CD8+ T cells. Eur. J. Immunol. 48: 293–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bian Y., Shang S., Siddiqui S., Zhao J., Joosten S. A., Ottenhoff T. H. M., Cantor H., Wang C. R. 2017. MHC Ib molecule Qa-1 presents Mycobacterium tuberculosis peptide antigens to CD8+ T cells and contributes to protection against infection. PLoS Pathog. 13: e1006384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O’Callaghan C. A., Tormo J., Willcox B. E., Braud V. M., Jakobsen B. K., Stuart D. I., McMichael A. J., Bell J. I., Jones E. Y. 1998. Structural features impose tight peptide binding specificity in the nonclassical MHC molecule HLA-E. Mol. Cell 1: 531–541. [DOI] [PubMed] [Google Scholar]
- 27.Lampen M. H., Hassan C., Sluijter M., Geluk A., Dijkman K., Tjon J. M., de Ru A. H., van der Burg S. H., van Veelen P. A., van Hall T. 2013. Alternative peptide repertoire of HLA-E reveals a binding motif that is strikingly similar to HLA-A2. Mol. Immunol. 53: 126–131. [DOI] [PubMed] [Google Scholar]
- 28.Kraemer T., Celik A. A., Huyton T., Kunze-Schumacher H., Blasczyk R., Bade-Döding C. 2015. HLA-E: presentation of a broader peptide repertoire impacts the cellular immune response-implications on HSCT outcome. Stem Cells Int. 2015: 346714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harriff M. J., Wolfe L. M., Swarbrick G., Null M., Cansler M. E., Canfield E. T., Vogt T., Toren K. G., Li W., Jackson M., et al. 2017. HLA-E presents glycopeptides from the Mycobacterium tuberculosis protein MPT32 to human CD8+ T cells. Sci. Rep. 7: 4622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McMurtrey C., Harriff M. J., Swarbrick G. M., Duncan A., Cansler M., Null M., Bardet W., Jackson K. W., Lewinsohn D. A., Hildebrand W., Lewinsohn D. M. 2017. T cell recognition of Mycobacterium tuberculosis peptides presented by HLA-E derived from infected human cells. PLoS One 12: e0188288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Celik A. A., Kraemer T., Huyton T., Blasczyk R., Bade-Döding C. 2016. The diversity of the HLA-E-restricted peptide repertoire explains the immunological impact of the Arg107Gly mismatch. Immunogenetics 68: 29–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walters L. C., Harlos K., Brackenridge S., Rozbesky D., Barrett J. R., Jain V., Walter T. S., O’Callaghan C. A., Borrow P., Toebes M., et al. 2018. Pathogen-derived HLA-E bound epitopes reveal broad primary anchor pocket tolerability and conformationally malleable peptide binding. [Published erratum appears in 2018 Nat. Commun. 9: 4833] Nat. Commun. 9: 3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Combet C., Blanchet C., Geourjon C., Deléage G. 2000. NPS@: network protein sequence analysis. Trends Biochem. Sci. 25: 147–150. [DOI] [PubMed] [Google Scholar]
- 34.Toebes M., Rodenko B., Ovaa H., Schumacher T. N. M. 2009. Generation of peptide MHC class I monomers and multimers through ligand exchange. Curr. Protoc. Immunol. Chapter 18: Unit 18.16. [DOI] [PubMed] [Google Scholar]
- 35.Rodenko B., Toebes M., Hadrup S. R., van Esch W. J. E., Molenaar A. M., Schumacher T. N. M., Ovaa H. 2006. Generation of peptide-MHC class I complexes through UV-mediated ligand exchange. Nat. Protoc. 1: 1120–1132. [DOI] [PubMed] [Google Scholar]
- 36.Walters, L. C., A. J. McMichael, and G. M. Gillespie. 2020. Detailed and atypical HLA‐E peptide binding motifs revealed by a novel peptide exchange binding assay. Eur. J. Immunol. DOI: 10.1002/eji.202048719. [DOI] [PubMed]
- 37.Tan T. L. R., Geluk A., Toebes M., Ottenhoff T. H. M., Drijfhout J. W. 1997. A novel, highly efficient peptide-HLA class I binding assay using unfolded heavy chain molecules: identification of HIV-1 derived peptides that bind to HLA-A*0201 and HLA-A*0301. J. Immunol. Methods 205: 201–209. [DOI] [PubMed] [Google Scholar]
- 38.Wooldridge L., Laugel B., Ekeruche J., Clement M., van den Berg H. A., Price D. A., Sewell A. K. 2010. CD8 controls T cell cross-reactivity. J. Immunol. 185: 4625–4632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bijen H. M., van der Steen D. M., Hagedoorn R. S., Wouters A. K., Wooldridge L., Falkenburg J. H. F., Heemskerk M. H. M. 2018. Preclinical strategies to identify off-target toxicity of high-affinity TCRs. Mol. Ther. 26: 1206–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Szomolay B., Liu J., Brown P. E., Miles J. J., Clement M., Llewellyn-Lacey S., Dolton G., Ekeruche-Makinde J., Lissina A., Schauenburg A. J., et al. 2016. Identification of human viral protein-derived ligands recognized by individual MHCI-restricted T-cell receptors. Immunol. Cell Biol. 94: 573–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee N., Goodlett D. R., Ishitani A., Marquardt H., Geraghty D. E. 1998. HLA-E surface expression depends on binding of TAP-dependent peptides derived from certain HLA class I signal sequences. J. Immunol. 160: 4951–4960. [PubMed] [Google Scholar]
- 42.Nattermann J., Nischalke H. D., Hofmeister V., Kupfer B., Ahlenstiel G., Feldmann G., Rockstroh J., Weiss E. H., Sauerbruch T., Spengler U. 2005. HIV-1 infection leads to increased HLA-E expression resulting in impaired function of natural killer cells. Antivir. Ther. (Lond.) 10: 95–107. [DOI] [PubMed] [Google Scholar]
- 43.Hannoun Z., Lin Z., Brackenridge S., Kuse N., Akahoshi T., Borthwick N., McMichael A., Murakoshi H., Takiguchi M., Hanke T. 2018. Identification of novel HIV-1-derived HLA-E-binding peptides. Immunol. Lett. 202: 65–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harndahl M., Rasmussen M., Roder G., Dalgaard Pedersen I., Sørensen M., Nielsen M., Buus S. 2012. Peptide-MHC class I stability is a better predictor than peptide affinity of CTL immunogenicity. Eur. J. Immunol. 42: 1405–1416. [DOI] [PubMed] [Google Scholar]
- 45.Grimsley C., Ober C. 1997. Population genetic studies of HLA-E: evidence for selection. Hum. Immunol. 52: 33–40. [DOI] [PubMed] [Google Scholar]
- 46.Matte C., Lacaille J., Zijenah L., Ward B., Roger M. 2000. HLA-G and HLA-E polymorphisms in an indigenous African population. The ZVITAMBO Study Group. Hum. Immunol. 61: 1150–1156. [DOI] [PubMed] [Google Scholar]
- 47.Kanevskiy L., Erokhina S., Kobyzeva P., Streltsova M., Sapozhnikov A., Kovalenko E. 2019. Dimorphism of HLA-E and its disease association. Int. J. Mol. Sci. 20: 5496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maier S., Grzeschik M., Weiss E. H., Ulbrecht M. 2000. Implications of HLA-E allele expression and different HLA-E ligand diversity for the regulation of NK cells. Hum. Immunol. 61: 1059–1065. [DOI] [PubMed] [Google Scholar]
- 49.Strong R. K., Holmes M. A., Li P., Braun L., Lee N., Geraghty D. E. 2003. HLA-E allelic variants. Correlating differential expression, peptide affinities, crystal structures, and thermal stabilities. J. Biol. Chem. 278: 5082–5090. [DOI] [PubMed] [Google Scholar]
- 50.Braud V., Jones E. Y., McMichael A. 1997. The human major histocompatibility complex class Ib molecule HLA-E binds signal sequence-derived peptides with primary anchor residues at positions 2 and 9. Eur. J. Immunol. 27: 1164–1169. [DOI] [PubMed] [Google Scholar]
- 51.van Hall T., Oliveira C. C., Joosten S. A., Ottenhoff T. H. M. 2010. The other Janus face of Qa-1 and HLA-E: diverse peptide repertoires in times of stress. Microbes Infect. 12: 910–918. [DOI] [PubMed] [Google Scholar]
- 52.Madura F., Rizkallah P. J., Legut M., Holland C. J., Fuller A., Bulek A., Schauenburg A. J., Trimby A., Hopkins J. R., Wells S. A., et al. 2019. TCR-induced alteration of primary MHC peptide anchor residue. Eur. J. Immunol. 49: 1052–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tynan F. E., Reid H. H., Kjer-Nielsen L., Miles J. J., Wilce M. C. J., Kostenko L., Borg N. A., Williamson N. A., Beddoe T., Purcell A. W., et al. 2007. A T cell receptor flattens a bulged antigenic peptide presented by a major histocompatibility complex class I molecule. Nat. Immunol. 8: 268–276. [DOI] [PubMed] [Google Scholar]
- 54.Borbulevych O. Y., Baxter T. K., Yu Z., Restifo N. P., Baker B. M. 2005. Increased immunogenicity of an anchor-modified tumor-associated antigen is due to the enhanced stability of the peptide/MHC complex: implications for vaccine design. J. Immunol. 174: 4812–4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Clancy-Thompson E., Devlin C. A., Tyler P. M., Servos M. M., Ali L. R., Ventre K. S., Bhuiyan M. A., Bruck P. T., Birnbaum M. E., Dougan S. K. 2018. Altered binding of tumor antigenic peptides to MHC class I affects CD8+ T cell-effector responses. Cancer Immunol. Res. 6: 1524–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.