Abstract
Aberrant expression of the simple mucin‐type carbohydrate antigens such as T, Tn, sialyl‐T and sialyl‐Tn is associated with poor prognosis in several cancers. β1,3‐N‐acetylglucosaminyltransferase‐3 (B3GNT3), a member of the β3GlcNAcT family, is responsible for forming extended core 1 (T antigen) oligosaccharides. The role of B3GNT3, which is expressed in various tissues including human fetal brain, in regulating neuroblastoma (NB) formation and cell behaviors remains unclear. Here, we showed that increased B3GNT3 expression evaluated using immunohistochemistry in NB tumor tissues correlated well with the histological grade of differentiation as well as a favorable Shimada's subset of pathology. Univariate and multivariate analyses revealed that positive B3GNT3 expression in tumor tissues predicted a favorable prognosis in NB patients independent of other prognostic markers. B3GNT3 overexpression suppresses T antigen formation and malignant phenotypes including migration and invasion of SK‐N‐SH cells, whereas B3GNT3 knockdown enhances these phenotypes of SK‐N‐SH cells. Moreover, B3GNT3 expression decreased phosphorylation of focal adhesion kinase (FAK), Src, paxillin, Akt and ERK1/2. We conclude that B3GNT3 predicts a favorable cancer behavior of NB and suppresses malignant phenotypes by modulating mucin‐type O‐glycosylation and signaling in NB cells.
B3GNT3 expression suppresses cell migration and invasion and predicts favorable outcomes in neuroblastoma.
Neuroblastoma (NB) is the most common extracranial solid tumor in childhood, accounting for 8–10% of all pediatric malignancies. This tumor arises from primordial neuroepithelial cells of the neural crest.1, 2 The behavior of NB is markedly heterogeneous, ranging from spontaneous differentiation or regression into ganglioneuroblastoma (GNB) or ganglioneuroma with a favorable prognosis to highly undifferentiated tumors with a rapid progression and very poor outcomes.3 Metastasis, a NB staging factor, is found in 50–60% of all NB cases, and advanced NB typically metastasize to distant lymph nodes, bone marrow, bone, liver or other organs. Although the overall prognosis for NB patients has improved remarkably with recent advances in therapies, the long‐term survival of patients with high‐risk NB remains poor despite intensive multimodal therapy.1, 2 Thus, finding new prognostic factors is required to further understand NB pathogenesis and to tailor therapies for improving treatment outcomes of patients with unfavorable NB.
Glycosylation is one of the most important processes in posttranslational modification of proteins. It is commonly found that oligosaccharide structures of glycoproteins are changed during malignant transformations.4 Because the sugar chains of glycoproteins are essential for maintaining ordered intercellular interactions among differentiated cells in multicellular organisms, alterations in the sugar chains constitute the molecular basis of abnormal intercellular behaviors in tumor cells, such as invasion into the surrounding tissues and metastasis.5
There are two major types of protein glycosylation in mammalian cells, namely N‐linked and O‐linked. Mucin‐type O‐glycans contain N‐acetylgalactosamine residues linked to serine or threonine residues (GalNAcα1‐O‐Ser/Thr, Tn antigen) in a polypeptide. T synthase galactosylates Tn to form core 1 (Galβ1→3GalNAc, T antigen).6 Aberrant expression of the simple mucin‐type antigens such as T, Tn, sialyl‐T and sialyl‐Tn is associated with poor prognosis in several cancers.7
β1,3‐N‐acetylglucosaminyltransferase‐3 (B3GNT3), formerly called core 1 β3GlcNAcT, is responsible for adding GlcNAc to core 1 (T antigen) in a β1,3 linkage, forming extended core 1 oligosaccharides.8, 9 B3GNT3 belongs to the β3GlcNAcT gene family, which consists of at least eight different β3GlcNAcTs.9 Many B3GNT family members are also associated with malignant transformation. Premature translation termination of the putative B3GNT1 within the Large locus in mice results in myodystrophy,10 whereas the LARGE locus is always deleted in human patients with meningioma, a meningeal tumor of the central nervous system.11 B3GNT8 is involved in malignancy through synthesis of poly‐N‐acetyllactosamine (polyLacNAc) on β1–6 branched N‐glycans in colon cancer.12 Glycoconjugates synthesized by B3GNT7 might also serve to diminish the motility of lung cancer cell lines.13
B3GNT3 transcripts are expressed in the small intestine, colon, placenta, liver, kidney, pancreas, prostate, neutrophils, lymphocytes and fetal brain.14 However, the role of B3GNT3 in cell behaviors and clinical significance of NB remains unclear. To address these issues, we demonstrated the expression of B3GNT3 in NB tissues having varying differentiation status. The clinicopathological factors and survival outcomes with respect to B3GNT3 expression were also analyzed. Parallel experiments in a NB cell line were designed to analyze the effects of B3GNT3 on NB cell behaviors. Possible mechanisms by which B3GNT3 affects NB cells were further investigated.
Materials and Methods
Patients and treatment
Tissue samples were collected from 102 NB patients receiving treatment at the National Taiwan University Hospital between 1 December 1990 and 31 December 2007. The use of human tissues for the present study was approved by the National Taiwan University Hospital Ethics Committee and written consent was obtained from patients before sample collection. Of these 102 patients with complete follow up, 87 were enrolled in the present study. The median age at diagnosis was 2.5 years (range, 0–13.5 years). Male patients were slightly predominant, with a male/female ratio of 49:38. Most tumors (52 cases) originated primarily from the adrenal gland, followed by the retroperitoneum (19 cases), mediastinum (seven cases), neck (five cases) and pelvis (four cases). Tumor histology was categorized into four types according to the criteria of the International Neuroblastoma Pathology Classification:15, 16 (i) NB (including undifferentiated, poorly differentiated and differentiating subtypes); (ii) GNB, intermixed; (iii) GNB, nodular; and (iv) GNB, maturing subtype. GNB, maturing subtype, is a benign lesion and was not included in this study. To simplify the description of the correlation between B3GNT3 expression and histological features of NB tissues, we defined NB as undifferentiated NB (UNB), poorly differentiated NB (PDNB), differentiating NB (DNB) or GNB, intermixed. The GNB, nodular subtype, was classified into UNB, PDNB or DNB according to the morphological features of NB nodules because the tumor behavior of this subtype depends mainly on the NB nodules. For prognostic analysis, GNB, intermixed was classified as a favorable histological type; UNB, PDNB and DNB were classified as either favorable or unfavorable according to the mitosis–karyorrhexis index and patient age at diagnosis based on the criteria of the International Neuroblastoma Pathology Classification.15, 16 Tumor staging was based on the International NB Staging System.17 MYCN status of each tumor tissue was evaluated using fluorescence in situ hybridization analysis of formalin‐fixed paraffin‐embedded tissues or fresh single tumor cells.18, 19 Patients were treated with surgery only or a combination of multiple modalities including chemotherapy, radiotherapy and/or autologous bone marrow transplantation according to the patient's risk grouping based on the risk classification of the Children's Cancer Group.20 The median follow up after diagnosis was 64 months with a range of 1–258 months, and the overall predictive 5‐year survival rate for this cohort was 55%.
Immunohistochemical analysis
Paraffin‐embedded tissue sections were deparaffinized in xylene and rehydrated in a series of graded alcohols. After quenching the activity of endogenous peroxidase with 1% H2O2 in PBS for 10 min, the sections were rinsed three times with PBS and then incubated with 5% non‐fat milk/PBS for 30 min to reduce non‐specific binding. Sections were incubated with an anti‐B3GNT3 polyclonal antibody (Sigma‐Aldrich, St Louis, MO, USA) diluted with 5% non‐fat milk/PBS for 16 h at 4°C. After rinsing twice with PBS, the Super Sensitive Link‐Label Immunohistochemistry Detection System (BioGenex, San Ramon, CA, USA) was applied to tissue sections. The specific immunostaining was then visualized with the 3,3‐Diaminobenzidine Liquid Substrate System (Sigma‐Aldrich). All sections were counterstained with hematoxylin and mounted with UltraKitt (J.T. Baker, Deventer, Holland). Negative controls were generated by replacing primary antibodies with a control non‐immune IgG at the same concentration. To confirm the specificity of staining, B3GNT3 peptide (10 μg/mL) was used to block binding of anti‐B3GNT3 antibody to tissues.
Cell culture and transfection
The NB cell line, SK‐N‐SH, from American Type Culture Collection (ATCC, Manassas, VA, USA) was maintained in Dulbecco's modified Eagle's medium (DMEM; JRH Biosciences, Lenexa, KS, USA) containing 10% FBS (PAA Laboratories, Pasching, Austria) in a humidified tissue culture incubator at 37°C in a 5% CO2 atmosphere. For stable transfection, SK‐N‐SH cells were transfected with B3GNT3/pcDNA3.1A or pcDNA3.1A/myc‐His (Invitrogen, Life Technologies Inc., Grand Island, NY, USA) using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). The transfected cells were selected with 400 μg/mL G418 (Calbiochem, Darmstadt, Germany) for 14 days and pooled for further studies. B3GNT3 expression was analyzed using RT‐PCR and immunoblotting.
Knockdown of B3GNT3 expression
Specific si‐RNA against B3GNT3 and non‐targeting control si‐RNA (Thermo Scientific, Bannockburn, IL, USA) were used for further experiments. SK‐N‐SH cells were transfected with 20 nM si‐RNA using Lipofectamine RNAiMAX (Invitrogen, Life Technologies Inc.) for 2 days.
Reverse transcription‐polymerase chain reaction (RT‐PCR) and real‐time PCR
Total cellular RNA was isolated from cells grown to 70% confluency using TRIzol reagent (Invitrogen, Life Technologies Inc.). For cDNA synthesis, 2 μg of total RNA was used as a template in a 25‐μL reverse‐transcription reaction. The PCR reactions were incubated for 5 min at 95°C, followed by 35 cycles with 30 s of denaturation at 94°C, 30 s of annealing at 60°C and 30 s of extension at 72°C. For real‐time PCR, the quantitative PCR system Mx3000P (Stratagene, La Jolla, CA, USA) was used to analyze gene expression. Briefly, 25‐μL reactions contained 2 μL of cDNA, 400 nmol/L each of sense and antisense primers and 12.5 μL of Brilliant SYBR Green QPCR Master Mix (Stratagene). To detect glyceraldehyde‐3‐phosphate dehydrogenase (GAPDH) expression, the sense and antisense primers were 5′‐ACAGTCAGC CGCATCT TCTT‐3′ and 5′‐GACAAGCTTCCCGTTCTCAG‐3′, respectively, generating a 259‐bp fragment. For B3GNT3 detection, sense and antisense primers were 5′‐TATGTGT CTGGAGCTTGAGG‐3′ and 5′‐AAGGATGTGTAGGAGTTC GC‐3′, respectively, generating a 382‐bp fragment. The PCR products were confirmed to be correct using DNA sequencing.
Immunoblotting and immunoprecipitation
A biotinylated lectin, peanut agglutinin (PNA; Vector Laboratories, Burlingame, CA, USA), was used to detect the Galβ1‐3GalNAcα1‐Ser/Thr (T antigen) structure in primary tumors. The PNA recognizes and binds preferentially to the T antigen structure. The extension of core 1 (T antigen) interferes with the binding affinity of PNA to glycoconjugates. To detect the T antigen structure on cell surface proteins, intact cells were surface‐biotinylated with sulfosuccinimidyl biotin (Pierce Chemical Co., Rockford, IL, USA), lysed and then pulled down with PNA‐agarose beads (Vector Laboratories). The pulled‐down proteins were detected using horseradish peroxidase–conjugated streptavidin (Santa Cruz Biotechnology, Santa Cruz, CA, USA). For cell signaling analyses, we used rabbit anti‐FAK pY397 polyclonal antibody (Biosource, Nivelles, Belgium), rabbit anti‐FAK polyclonal antibody (C‐20; Santa Cruz Biotechnology), anti‐paxillin pY118 (BD Transduction Laboratories, Heidelberg, Germany), anti‐paxillin polyclonal antibody (BD Transduction Laboratories), rabbit anti‐pERK1/2 monoclonal antibody, rabbit anti‐pAkt and mouse anti‐pan Akt monoclonal antibody (Cell Signaling Technology Inc., Beverly, MA, USA), rabbit anti‐ERK1/2 antibody (Santa Cruz Biotechnology), rabbit anti‐Src pY418 antibody (Invitrogen) and rabbit anti‐c‐Src antibody (Invitrogen). Immunoblotted membranes were then incubated with horseradish peroxidase–conjugated streptavidin, horseradish peroxidase–conjugated anti‐rabbit IgG or anti‐mouse IgG (Santa Cruz Biotechnology). Signals were visualized using ECL reagents (Amersham Biosciences, Piscataway, NJ, USA) and images were quantified using ImageQuant 5.1 (Amersham Biosciences). For immunoprecipitation, 1 mg of cell tracts were incubated with PNA‐agarose beads (Vector Laboratories) or protein G Sepharose 4 Fast Flow (GE Healthcare UK Ltd, Little Chalfont, UK) conjugated with 2 μg of antibody overnight at 4°C.
Cell growth analysis
Cells were plated in triplicate wells of 96‐well plates at a density of 3 × 103 cells per well. After incubation for 24 or 48 h, 3‐(4,5‐dimethyl‐2‐thiazolyl)‐2,5‐ diphenyl‐2H‐tetrazolium bromide solution (MTT; Sigma‐Aldrich) was added to each well at a final concentration of 0.5 mg/mL and was incubated for 4 h to allow MTT reduction. Formazan crystals were dissolved in a solution containing 0.01 mol/L HCl and 10% (w/v) SDS, and absorbance was measured at the dual wavelengths of 570 and 630 nm using a spectrophotometer.
Anchorage‐independent growth in soft agar
Cells (1 × 104) in 0.3% (w/v) Bacto Agar (Sigma‐Aldrich) in DMEM supplemented with 10% FBS were overlaid on a base of 0.6% Bacto Agar in DMEM supplemented with 10% FBS in six‐well plates. Cells were incubated at 37°C in a 5% CO2 atmosphere. Triplicate wells were used for the cell line and three independent experiments were performed. The number of colonies with a diameter >30 μm was counted on day 7.
Migration and invasion assays
Transwell motility assays were performed using 8‐μm pore, 6.5‐mm polycarbonate Transwell filters (Corning Costar Corp., Cambridge, MA, USA). Single‐cell suspensions (3 × 105 cells) were seeded in serum‐free DMEM on the upper surface of the filters and were allowed to migrate toward 10% FBS/DMEM or 1 μg/mL of extracellular matrix proteins (collagen IV, fibronectin and laminin) (Sigma‐Aldrich). After a 48‐h incubation period, cells that had migrated to the underside of the filter were fixed, stained with 0.5% crystal violet (Sigma–Aldrich) and counted under a microscope at ×200 in five random fields.
Cell invasion assays were performed in BioCoat Matrigel invasion chambers (Becton‐Dickinson, Bedford, MA, USA). Briefly, 500 μL of DMEM containing 10% FBS was loaded in the lower part of the chamber and 3 × 105 cells in 500 μL of serum‐free DMEM were seeded to the upper part. Cells were allowed to invade the Matrigel for 48 h in a humidified tissue culture incubator at 37°C in a 5% CO2 atmosphere. Non‐invading cells on the upper surface of the membrane were removed from the chamber and the invading cells on the lower surface of the membrane were fixed with 100% methanol and stained with 0.5% crystal violet. Invading cells were counted in each well under a phase‐contrast microscope. The mean ± SD values were calculated from the numbers of invading cells observed under the microscope in three independent experiments.
Statistical analysis
spss 10.0 for Windows software (SPSS Inc., Chicago, IL, USA) was used for statistical analyses. Pairs of categorical variables were compared using Pearson's χ2 test. Variations between the data resulting from different treatments were assessed using the Student's t‐test. The Kaplan–Meier method was used to estimate survival probabilities in various subgroups and significant differences between groups were established using log‐rank tests. Each variable that could possibly affect patient survival was further evaluated using univariate and multivariate Cox proportional hazards model analysis. All statistical tests were two sided and those with P < 0.05 were considered significant.
Results
B3GNT3 expression correlates with tumor histology and survival probability of NB patients
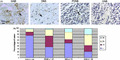
To investigate the clinical importance of B3GNT3 and its correlation with clinicopathological factors in NB, we examined B3GNT3 expression in NB tumors using immunohistochemical staining. Positive B3GNT3 staining was observed specifically in the cytoplasm of ganglion cells of GNB tumor tissues (Fig. 1a) but not in Schwannian stromal cells. In addition to the mature ganglion cells, neuroblastic cells showing various differentiated states in GNB, DNB or PDNB also had positive cytoplasmic staining (Fig. 1a), whereas undifferentiated neuroblastic cells usually had negative staining (Fig. 1a). Therefore, B3GNT3 expression correlated positively with the differentiation status of human NB.
Figure 1.
Immunohistochemical analysis of B3GNT3 in human neuroblastoma (NB) cells. (a) B3GNT3 expression in ganglioneuroblastoma (GNB), differentiating NB (DNB), poorly differentiated NB (PDNB) and undifferentiated NB (UNB). The arrows in GNB, DNB and PDNB indicate positive staining of neuroblastic cells in various states of differentiation. The UNB shows negative staining. Scale bar = 100 μm. Original magnification, ×400. (b) Percentage distribution of B3GNT3 expression in tumors with UNB, PDNB, DNB or GNB histology.
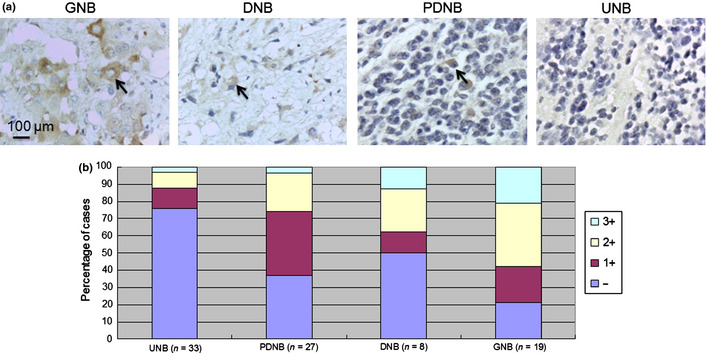
The immunoreactivity of B3GNT3 in NB tumors was categorized into four groups: –, no expression (no stained cells or only isolated single stained cells seen); 1+, weak expression (expression in ~10–35% of neuroblastic cells); 2+, moderate expression (expression in ~35–70% of neuroblastic cells); and 3+, strong expression (expression in >70% of neuroblastic cells). B3GNT3 expression (1+, 2+ and 3+) was observed in most PDNB (63.0%), DNB (50.0%) and GNB tumors (78.9%) and was observed much less in UNB tumors (24.2%), indicating that the intensity and percentage of positive B3GNT3 immunostaining correlated with the histological grade of differentiation (Fig. 1b, Table 1; P = 0.001, χ2 test). For further analyses, NB tumors were assigned to negative B3GNT3 expression (– in immunoreactivity) and positive B3GNT3 expression (1+, 2+ or 3+ in immunoreactivity). The immunohistochemical staining revealed B3GNT3‐positive expression (1+ to 3+) in 50.6% (44/87) of NB tumors. Moreover, Kaplan–Meier analysis showed that patients with B3GNT3‐positive expression had a higher predictive 5‐year survival rate than those with B3GNT3‐negative expression (Fig. 2a; P < 0.001, log‐rank test). These data suggest that B3GNT3 expression predicts more differentiated histology and a higher predictive 5‐year survival rate for NB patients.
Table 1.
B3GNT3 expression and clinicopathological and biological characteristics of neuroblastoma
| Variable | No. cases | Positive B3GNT3 expression (%) | P‐valuea |
|---|---|---|---|
| Age at diagnosis | |||
| ≤1 year | 29 | 13 (44.8) | 0.448 |
| >1 year | 58 | 31 (53.4) | |
| Gender | |||
| Male | 49 | 22 (44.9) | 0.229 |
| Female | 38 | 22 (57.9) | |
| Clinical stage | |||
| 1, 2 and 4S | 30 | 17 (56.7) | 0.410 |
| 3 and 4 | 57 | 27 (47.4) | |
| Primary tumor site | |||
| Adrenal | 52 | 25 (48.1) | 0.570 |
| Extra‐adrenal | 35 | 19 (54.3) | |
| INPC histology | |||
| UNB | 33 | 8 (24.2) | 0.001 |
| PDNB | 27 | 17 (63.0) | |
| DNB | 8 | 4 (50.0) | |
| GNB | 19 | 15 (78.9) | |
| Shimada histology | |||
| Favorable | 45 | 32 (71.1) | <0.001 |
| Unfavorable | 42 | 12 (28.6) | |
| MYCN | |||
| Amplified | 23 | 8 (34.8) | 0.077 |
| Non‐amplified | 64 | 36 (56.3) | |
| Outcome | |||
| Alive | 43 | 30 (69.8) | <0.001 |
| Dead | 44 | 14 (31.8) | |
χ2 test. DNB, differentiating neuroblastoma; GNB, ganglioneuroblastoma; INPC, International Neuroblastoma Pathology Classification; PDNB, poorly differentiated neuroblastoma; UNB, undifferentiated neuroblastoma.
Figure 2.
(a) Kaplan–Meier survival analysis according to the expression of B3GNT3 in 87 neuroblastoma (NB) patients. P values were calculated using the log‐rank test. (b) Kaplan–Meier survival analysis according to the expression of B3GNT3 in NB patients with normal MYCN status. (c) Kaplan–Meier survival analysis according to the expression of B3GNT3 in NB patients at an early stage. (d) Kaplan–Meier survival analysis according to the expression of B3GNT3 in NB patients at an advanced stage.
B3GNT3 expression and clinicopathological and biological factors of NB tumors
We further analyzed the association between B3GNT3 expression and other clinicopathological and biological variables of NB tumors. B3GNT3 expression significantly correlated with favorable Shimada histology and better survival outcomes (both P < 0.001, χ2 test; Table 1). Furthermore, univariate analysis showed that in addition to the absence of B3GNT3 expression, older age at diagnosis (>1 year), advanced clinical stage (stages 3 and 4), MYCN amplification and unfavorable Shimada histology strongly correlated with poor survival (Table 2). Multivariate analysis revealed that advanced clinical stage, MYCN amplification and negative B3GNT3 expression remained independent prognostic factors for poor survival (Table 2).
Table 2.
Clinicopathological and biological factors affecting the 5‐year predictive survival rate
| Variable | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
| RR | 95% CI | P‐value | RR | 95% CI | P value | |
| Age at diagnosis | ||||||
| >1 year vs ≤1 year | 4.145 | 1.965–8.746 | <0.001 | 2.673 | 0.922–7.747 | 0.070 |
| Clinical stage | ||||||
| Advanced (3 and 4) vs early (1, 2 and 4S) | 11.386 | 4.099–31.627 | <0.001 | 4.741 | 1.189–18.905 | 0.027 |
| MYCN | ||||||
| Amplified vs non‐amplified | 3.427 | 2.039–5.758 | <0.001 | 2.112 | 1.114–4.007 | 0.022 |
| B3GNT3 expression | ||||||
| Negative vs positive | 3.021 | 1.598–5.710 | 0.001 | 2.248 | 1.102–4.585 | 0.026 |
| Shimada histology | ||||||
| Unfavorable vs favorable | 3.862 | 2.202–6.771 | <0.001 | 1.578 | 0.788–3.159 | 0.198 |
CI, confidence interval; RR, risk ratio.
To further evaluate the significance of B3GNT3 expression in prognostic discrimination, the impact of B3GNT3 expression on survival rate was analyzed according to the other two independent prognostic factors of clinical stage and MYCN status. The result show that, except for patients with MYCN amplification who have a very poor prognosis, positive B3GNT3 expression predicted higher survival probability in patients with normal MYCN status and in those with either early or advanced‐stage disease (Fig. 2b–d). These results suggest that B3GNT3 expression is an independent prognostic factor of better survival outcome for NB patients, probably by affecting tumor cell behaviors in vivo.
B3GNT3 expression in transfected SK‐N‐SH cells
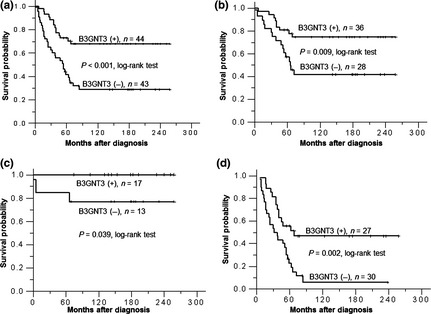
To demonstrate the effects of B3GNT3 on NB cells, SK‐N‐SH cells were transfected with pcDNA3.1 containing B3GNT3. We tried to isolate single clones using G418 selection, but these clones were not stable. Therefore, we pooled the mock‐ or B3GNT3‐transfected cells for further experiments after selection with 400 μg G418 for 10 days. Transfection efficiency was nearly 70% and the pooled B3GNT3‐transfected cells could be grown in vitro for 1 month. B3GNT3 overexpression in SK‐N‐SH cells was demonstrated using RT‐PCR and immunoblotting (Fig. 3a). B3GNT3 expression decreased PNA binding to glycoproteins, suggesting that it had intact enzymatic activity to promote the formation of extended core 1 oligosaccharides in NB cells (Fig. 3b).
Figure 3.
B3GNT3 expression in SK‐N‐SH cells and the effects of B3GNT3 on SK‐N‐SH cells. (a) Forced expression of B3GNT3 in SK‐N‐SH cells. SK‐N‐SH cells were transfected with pcDNA3.1 (mock) or pcDNA3.1/B3GNT3 and selected with G418 for 14 days. The G418‐resistent cells were pooled and overexpression of B3GNT3 mRNA and protein was demonstrated using RT‐PCR (top panel) and Western blotting (bottom panel), respectively. (b) B3GNT3 expression modifies glycosylation and decreases peanut agglutinin (PNA) binding to glycoconjugates, as demonstrated using Western blotting with PNA. Molecular weight markers (kDa) are shown on the left. IP, immunoprecipitated. In panels (a) and (b), GAPDH was used as a loading control. (c) B3GNT3 expression does not significantly affect cell growth as shown using MTT assays conducted over 5 days. Cells were cultured in DMEM containing 10% FBS and a MTT reagent was applied to the cells at the indicated times. The results were standardized by setting the value at day 0 to 1.0. Error bar = SD. (d) B3GNT3 suppresses anchorage‐independent cell growth of SK‐N‐SH cells. Mock and B3GNT3 transfectants were seeded into soft agar for 14 days and colony numbers (colony diameter >50 μm) were counted under a microscope. *P < 0.01. (e) B3GNT3 inhibits SK‐N‐SH cell migration as assessed using Transwell migration assays. Mock or B3GNT3 transfectants of SK‐N‐SH cells were seeded onto the upper chamber and 10% FBS was used as a chemoattractant in the lower chamber. After 48 h, migrated cells in the lower chamber were stained, and representative images are shown (top panel). Original magnification, ×200. The relative migration of mock and B3GNT3 cells is shown (bottom panel). Data represent the mean ± SD of three independent experiments. *P < 0.01. (f) B3GNT3 suppresses SK‐N‐SH cell invasion as assessed using Matrigel invasion assays. Mock or B3GNT3 transfectants of SK‐N‐SH cells were seeded in each chamber and cultured for 48 h. The chemoattractant in the lower chamber was 10% FBS. Invaded cells were fixed and stained with crystal violet and representative images are shown (top panel). Original magnification, ×200. The relative invasion of mock and B3GNT3 cells is shown (bottom panel). Data represent the mean ± SD of three independent experiments. *P < 0.01.
B3GNT3 expression inhibits anchorage‐independent cell growth
To investigate whether B3GNT3 expression influenced anchorage‐dependent cell growth, cell viability was analyzed using MTT assays conducted over 5 days. B3GNT3 expression did not significantly affect cell growth (Fig. 3c). Next, we examined the effect of B3GNT3 on anchorage‐independent growth of SK‐N‐SH cells using colony formation assays in soft agar. The number of colonies for B3GNT3 transfectants was 3.3 ± 0.6 compared with 19 ± 2.6 for mock transfectants (Fig. 3d), suggesting that B3GNT3 expression decreases colony formation in SK‐N‐SH cells.
B3GNT3 suppresses cell migration and invasion
To determine whether B3GNT3 expression can regulate cell migration, Transwell migration assays were performed. SK‐N‐SH cell migration for B3GNT3 transfectants was 70.8 ± 7.5% compared with mock transfectants (Fig. 3e). Next, we examined the effect of B3GNT3 on SK‐N‐SH cell invasion using a Matrigel invasion assay that mimics active transmigration of cancer cells across a reconstituted basement membrane. The relative invading ability of B3GNT3 transfectants was 65.9 ± 10.2% compared with mock transfectants (Fig. 3f). These results suggest that expression of B3GNT3 suppresses the migration and invasion potential of SK‐N‐SH cells.
Effects of B3GNT3 knockdown on malignant phenotypes of NB cells
To further confirm the effects of B3GNT3 on NB cells, we knocked down B3GNT3 expression in SK‐N‐SH cells, which was demonstrated using RT‐PCR (Fig. 4a). The results show that B3GNT3 knockdown did not significantly affect cell growth (Fig. 4b). However, B3GNT3 knockdown significantly enhanced migration (Fig. 4c) and invasion (Fig. 4d) in SK‐N‐SH cells. These results further confirmed the role of B3GNT3 in regulating malignant properties, including migration and invasion abilities, in SK‐N‐SH cells.
Figure 4.
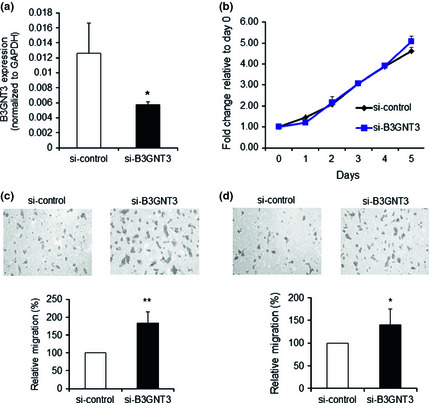
Effects of B3GNT3 knockdown on malignant phenotypes of SK‐N‐SH cells. (a) B3GNT3 knockdown with siRNA in SK‐N‐SH cells. B3GNT3 expression was knocked down by B3GNT3 siRNA (si‐B3GNT3) compared with the control siRNA (si‐control), which was demonstrated using RT‐PCR. (b–d) Effects of B3GNT3 knockdown on malignant phenotypes, including cell growth (b), cell migration (c) and cell invasion (d). Data represent the mean ± SD of three independent experiments. Original magnification, ×200. *P < 0.05. **P < 0.01.
Inhibition of Akt and ERK activities is required for B3GNT3 to suppress NB cell migration and invasion
To elucidate the signaling changes modulated by B3GNT3 expression, the phosphorylation of FAK, Src and paxillin was examined because these signaling molecules are crucial for cell migration and invasion. Phosphorylation levels of FAK at Y397, Src at Y418 and paxillin at Y118 were lower in B3GNT3 transfectants compared with mock transfectants. Furthermore, we found that the expression of FAK and Src protein was also inhibited by B3GNT3 expression (Fig. 5a).
Figure 5.
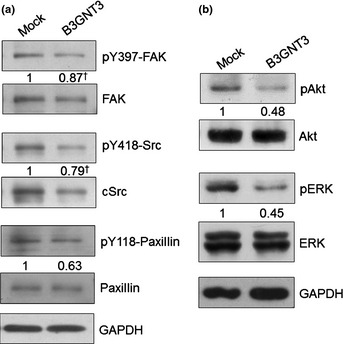
Signaling molecules affected by B3GNT3 expression. (a) B3GNT3 decreases tyrosine phosphorylation (indicated by lowercase ‘p’) of FAK, Src and paxillin in SK‐N‐SH cells. The band intensity of pY118‐paxillin was normalized to the total protein. †The band intensity of pY397‐FAK and pY418‐Src was normalized to the ratio of their total protein to GAPDH, respectively. The results are representative of three independent experiments. The signals were quantified using ImageQuant 5.1 (Amersham Biosciences, Piscataway, NJ, USA). (b) B3GNT3 suppresses phosphorylation of Akt and ERK1/2 in SK‐N‐SH cells. The results are representative of three independent experiments. The signals were quantified using ImageQuant 5.1 (Amersham Biosciences) and were normalized to their controls.
ERK and Akt are important downstream signaling molecules for FAK and their aberrant expression is closely associated with malignant cell behaviors. Thus, we examined the effect of B3GNT3 expression on the phosphorylation of ERK and Akt proteins. Overexpression of B3GNT3 significantly suppressed Akt and ERK1/2 phosphorylation, whereas the expression levels of total Akt and ERK1/2 remained unchanged (Fig. 5b). These results suggest that suppression of Akt and ERK activation might be required for B3GNT3 to inhibit NB cell migration and invasion.
Discussion
Pan‐carcinoma antigens such as T, Tn, sialyl‐T and sialyl‐Tn result from poor glycosylation of glycoproteins, leading to the expression of truncated O‐glycans on the cell surface. Mechanisms for cancer‐associated expression of truncated O‐glycans include disorganization of secretory pathway organelles (endoplasmic reticulum or Golgi) in cancer cells and, more commonly, altered expression of glycosyltransferases, which are responsible for the synthesis of core structures used as substrates for chain elongation.6, 21 B3GNT3 transcripts are expressed in various tissues including human fetal brain;14 however, the role of B3GNT3 in regulating NB formation and cell behaviors remains unclear. In the present study, we demonstrated that B3GNT3 was highly expressed in either differentiated NB or mature ganglion cells. In contrast, its expression was observed much less in UNB cells. In addition to early clinical stage and non‐amplified MYCN status, B3GNT3 expression predicts a better survival outcome for NB patients. Expression of B3GNT3 in NB cells resulted in a reduction of malignant phenotypes, including migration and invasion. Moreover, these results suggest that the phenotypic changes caused by B3GNT3 expression are likely mediated by attenuation of Akt and ERK activation. Therefore, B3GNT3 might play a critical role in suppressing the malignant properties of NB and its altered expression might contribute to the pathogenesis of NB.
Because NB is a heterogeneous tumor, treatment is based on risk stratification depending on several clinical and biological features. Among them, clinical stage and MYCN status are two well‐established prognostic factors of NB. The present study suggests that assessing B3GNT3 expression in NB tumors could provide prognostic information that complements the clinical stage and MYCN status, allowing clinicians to devise an individualized treatment strategy for each patient, such as a reduction in treatment intensity for children who have low‐risk tumors and continued increased treatment intensity and the addition of new agents for treatment of children who have high‐risk NB.
When analyzing the clinicopathological and biological characteristics with respect to B3GNT3 expression in NB, we found that B3GNT3 expression strongly correlated with more differentiated histology and favorable Shimada histology. However, it was B3GNT3 expression but not the tumor histological profile that predicted survival outcomes when analyzed using multivariate survival analysis. These results suggest that B3GNT3 affects NB cell malignant phenotypes and subsequent survival outcomes by alternative mechanisms aside from cell differentiation status. Indeed, we showed that in NB cells B3GNT3 overexpression suppresses malignant phenotypes including migration and invasion. Conversely, B3GNT3 knockdown in NB cells enhances these phenotypes.
Despite the absence of evidence for a role of altered glycosylation in cancer initiation and the limited information about the mechanisms that generate abnormal glycosylation, it is well established that altered glycosylation contributes temporally from early stages to invasion and metastasis. Two of the most common cancer‐associated modifications are the generation of truncated versions of normal oligosaccharides and the generation of unusual forms of terminal structures.7 N‐acetylglucosaminyltransferase V (GnT‐V) catalyzes the formation of β1,6GlcNAc branches on N‐glycans that are elongated with polyLacNAc and is able to enhance tumor growth and metastasis.22 N‐acetylglucosaminyltransferase III (GnT‐III) catalyzes the attachment of a GlcNAc to a core mannose of N‐glycan via a β1,4‐linkage to form the bisecting GlcNAc structure and this enzyme has been proposed to antagonize GnT‐V, thereby contributing to the suppression of cancer metastasis.23 In the present study, we found that B3GNT3 expression promoted the formation of extended core 1 oligosaccharides and prevented PNA lectin binding to T antigen (Fig. 3b). Therefore, we suggest that B3GNT3 might play an important role in suppressing the malignant phenotypes of NB cells by altering glycosylation of the cell‐surface molecules of NB. However, the acceptor substrates for B3GNT3 on the cell surface of NB and the signaling pathways following B3GNT3‐mediated altered glycosylation of these acceptor substrates require further investigation.
Integrins can alter cellular behavior through the recruitment and activation of signaling proteins such as non‐receptor tyrosine kinases including FAK and c‐Src, which form a dual kinase complex. The FAK‐Src complex is activated in many tumor cells and generates signals leading to tumor growth and metastasis.24 Data from the present study showed that B3GNT3 expression inhibited phosphorylation of FAK, Src and paxillin. The FAK‐Src complex binds to and phosphorylates various adaptor proteins including paxillin and thus serves as a platform to recruit numerous regulatory and structural proteins that together control the dynamic changes in cytoskeletal reorganization and gene expression that are necessary for cell migration and survival.25 Moreover, the paxillin–ERK complex plays a role in cell survival and motility.26 Expression of mutant Y397F FAK inhibits FAK‐to‐ERK/MAP kinase signaling through which FAK functions to promote cell proliferation. FAK‐enhanced activation of ERK can also promote transcription‐associated increases in protease expression, which is essential for cell invasion.27 Additionally, FAK can promote survival by enhancing phosphatidylinositol 3‐kinase–mediated activation of Akt.28 Activation of Akt protects neuroblastoma cells against apoptosis induced by tumor necrosis factor‐related apoptosis‐inducing ligand or chemotherapy.29 Data from the present study showed that decreased ERK and Akt signaling in NB might be essential for the phenotypic changes caused by B3GNT3 expression. ERK and Akt are important downstream signaling molecules for both integrins and receptor tyrosine kinases stimulated by growth factors such as insulin‐like growth factor, epidermal growth factor or brain‐derived neurotrophic factor.29, 30, 31, 32, 33, 34 In addition, integrins and receptor tyrosine kinases have been found to carry O‐glycans. For example, ST6GalNac‐I and ST6Gal‐I modify carbohydrate structures on β1 integrin, leading to altered cell morphological features and behavior by changing N‐glycosylation or O‐glycosylation of β1 integrin.35, 36, 37, 38 N‐acetylgalactosaminyl‐transferase 2 (GALNT2) modifies EGFR O‐glycosylation and internalization and thereby regulates its downstream signaling.39 Therefore, it is possible that B3GNT3 modifies O‐glycans of integrins and receptor tyrosine kinases on NB cells by forming extended core 1 oligosaccharides and thereby suppressing malignant phenotypes of NB cells.
In summary, we have shown that expression of B3GNT3 correlates positively with the histological grade of differentiation as well as favorable Shimada histology and might predict favorable outcomes of NB. B3GNT3 alters O‐glycosylation and suppresses migration and invasion of NB cells. The present study also suggests that suppression of FAK, Akt and ERK, important downstream signaling molecules for integrins and numerous growth factor receptors, might be required for B3GNT3 to inhibit malignant phenotypes of NB cells.
Disclosure Statement
The authors have no conflict of interest.
Acknowledgments
This study was supported by Shin Kong Wu Ho‐Su Memorial Hospital (SKH‐8302‐101‐DR‐18 to W‐L.H.), National Taiwan University (NTU.101‐R7808 to M‐C.H.), National Science Council, Taiwan (NSC 101‐2320‐B‐002‐007‐MY3 to M‐C.H.; NSC 99‐2628‐B‐002‐056‐MY3 to W‐M.H.) and National Taiwan University Hospital (NTUH.101‐S1787 to W‐M.H.).
(Cancer Sci 2013; 104: 1600–1608)
References
- 1. Kamijo T, Nakagawara A. Molecular and genetic bases of neuroblastoma. Int J Clin Oncol 2012; 17: 190–5. [DOI] [PubMed] [Google Scholar]
- 2. Maris JM, Hogarty MD, Bagatell R, Cohn SL. Neuroblastoma. Lancet 2007; 369: 2106–20. [DOI] [PubMed] [Google Scholar]
- 3. Brodeur GM. Neuroblastoma: biological insights into a clinical enigma. Nat Rev Cancer 2003; 3: 203–16. [DOI] [PubMed] [Google Scholar]
- 4. Hakomori S. Aberrant glycosylation in tumors and tumor‐associated carbohydrate antigens. Adv Cancer Res 1989; 52: 257–331. [DOI] [PubMed] [Google Scholar]
- 5. Kobata A, Amano J. Altered glycosylation of proteins produced by malignant cells, and application for the diagnosis and immunotherapy of tumours. Immunol Cell Biol 2005; 83: 429–39. [DOI] [PubMed] [Google Scholar]
- 6. Cao Y, Stosiek P, Springer GF, Karsten U. Thomsen‐Friedenreich‐related carbohydrate antigens in normal adult human tissues: a systematic and comparative study. Histochem Cell Biol 1996; 106: 197–207. [DOI] [PubMed] [Google Scholar]
- 7. Reis CA, Osorio H, Silva L, Gomes C, David L. Alterations in glycosylation as biomarkers for cancer detection. J Clin Pathol 2010; 63: 322–9. [DOI] [PubMed] [Google Scholar]
- 8. Yeh JC, Hiraoka N, Petryniak B et al Novel sulfated lymphocyte homing receptors and their control by a Core1 extension beta 1,3‐N‐acetylglucosaminyltransferase. Cell 2001; 105: 957–69. [DOI] [PubMed] [Google Scholar]
- 9. Mitoma J, Petryniak B, Hiraoka N, Yeh JC, Lowe JB, Fukuda M. Extended core 1 and core 2 branched O‐glycans differentially modulate sialyl Lewis X‐type L‐selectin ligand activity. J Biol Chem 2003; 278: 9953–61. [DOI] [PubMed] [Google Scholar]
- 10. Grewal PK, Holzfeind PJ, Bittner RE, Hewitt JE. Mutant glycosyltransferase and altered glycosylation of alpha‐dystroglycan in the myodystrophy mouse. Nat Genet 2001; 28: 151–4. [DOI] [PubMed] [Google Scholar]
- 11. Peyrard M, Seroussi E, Sandberg‐Nordqvist AC et al The human LARGE gene from 22q12.3‐q13.1 is a new, distinct member of the glycosyltransferase gene family. Proc Natl Acad Sci U S A 1999; 96: 598–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ishida H, Togayachi A, Sakai T et al A novel beta1,3‐N‐acetylglucosaminyltransferase (beta3Gn‐T8), which synthesizes poly‐N‐acetyllactosamine, is dramatically upregulated in colon cancer. FEBS Lett 2005; 579: 71–8. [DOI] [PubMed] [Google Scholar]
- 13. Kataoka K, Huh NH. A novel beta1,3‐N‐acetylglucosaminyltransferase involved in invasion of cancer cells as assayed in vitro . Biochem Biophys Res Commun 2002; 294: 843–8. [DOI] [PubMed] [Google Scholar]
- 14. Shiraishi N, Natsume A, Togayachi A et al Identification and characterization of three novel beta 1,3‐N‐acetylglucosaminyltransferases structurally related to the beta 1,3‐galactosyltransferase family. J Biol Chem 2001; 276: 3498–507. [DOI] [PubMed] [Google Scholar]
- 15. Shimada H, Ambros IM, Dehner LP et al The international neuroblastoma pathology classification (the Shimada system). Cancer 1999; 86: 364–72. [PubMed] [Google Scholar]
- 16. Shimada H, Ambros IM, Dehner LP, Hata J, Joshi VV, Roald B. Terminology and morphologic criteria of neuroblastic tumors: recommendations by the International Neuroblastoma Pathology Committee. Cancer 1999; 86: 349–63. [PubMed] [Google Scholar]
- 17. Brodeur GM, Pritchard J, Berthold F et al Revisions of the international criteria for neuroblastoma diagnosis, staging, and response to treatment. J Clin Oncol 1993; 11: 1466–77. [DOI] [PubMed] [Google Scholar]
- 18. Tajiri T, Shono K, Fujii Y et al Highly sensitive analysis for N‐myc amplification in neuroblastoma based on fluorescence in situ hybridization. J Pediatr Surg 1999; 34: 1615–9. [DOI] [PubMed] [Google Scholar]
- 19. Kopf I, Hanson C, Delle U, Verbiene I, Weimarck A. A rapid and simplified technique for analysis of archival formalin‐fixed, paraffin‐embedded tissue by fluorescence in situ hybridization (FISH). Anticancer Res 1996; 16: 2533–6. [PubMed] [Google Scholar]
- 20. Castleberry RP. Neuroblastoma. Eur J Cancer 1997; 33: 1430–7; discussion 7–8. [DOI] [PubMed] [Google Scholar]
- 21. Kjeldsen T, Clausen H, Hirohashi S, Ogawa T, Iijima H, Hakomori S. Preparation and characterization of monoclonal antibodies directed to the tumor‐associated O‐linked sialosyl‐2—6 alpha‐N‐acetylgalactosaminyl (sialosyl‐Tn) epitope. Cancer Res 1988; 48: 2214–20. [PubMed] [Google Scholar]
- 22. Dennis JW, Pawling J, Cheung P, Partridge E, Demetriou M. UDP‐N‐acetylglucosamine:alpha‐6‐D‐mannoside beta1,6 N‐acetylglucosaminyltransferase V (Mgat5) deficient mice. Biochim Biophys Acta 2002; 1573: 414–22. [DOI] [PubMed] [Google Scholar]
- 23. Gu J, Sato Y, Kariya Y, Isaji T, Taniguchi N, Fukuda T. A mutual regulation between cell–cell adhesion and N‐glycosylation: implication of the bisecting GlcNAc for biological functions. J Proteome Res 2009; 8: 431–5. [DOI] [PubMed] [Google Scholar]
- 24. Mitra SK, Schlaepfer DD. Integrin‐regulated FAK‐Src signaling in normal and cancer cells. Curr Opin Cell Biol 2006; 18: 516–23. [DOI] [PubMed] [Google Scholar]
- 25. Deakin NO, Turner CE. Paxillin comes of age. J Cell Sci 2008; 121: 2435–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ishibe S, Joly D, Liu ZX, Cantley LG. Paxillin serves as an ERK‐regulated scaffold for coordinating FAK and Rac activation in epithelial morphogenesis. Mol Cell 2004; 16: 257–67. [DOI] [PubMed] [Google Scholar]
- 27. Hu B, Jarzynka MJ, Guo P, Imanishi Y, Schlaepfer DD, Cheng SY. Angiopoietin 2 induces glioma cell invasion by stimulating matrix metalloprotease 2 expression through the alphavbeta1 integrin and focal adhesion kinase signaling pathway. Cancer Res 2006; 66: 775–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schlaepfer DD, Mitra SK, Ilic D. Control of motile and invasive cell phenotypes by focal adhesion kinase. Biochim Biophys Acta 2004; 1692: 77–102. [DOI] [PubMed] [Google Scholar]
- 29. Opel D, Poremba C, Simon T, Debatin KM, Fulda S. Activation of Akt predicts poor outcome in neuroblastoma. Cancer Res 2007; 67: 735–45. [DOI] [PubMed] [Google Scholar]
- 30. Sartelet H, Oligny LL, Vassal G. AKT pathway in neuroblastoma and its therapeutic implication. Expert Rev Anticancer Ther 2008; 8: 757–69. [DOI] [PubMed] [Google Scholar]
- 31. Li Z, Jaboin J, Dennis PA, Thiele CJ. Genetic and pharmacologic identification of Akt as a mediator of brain‐derived neurotrophic factor/TrkB rescue of neuroblastoma cells from chemotherapy‐induced cell death. Cancer Res 2005; 65: 2070–5. [DOI] [PubMed] [Google Scholar]
- 32. Li Z, Thiele CJ. Targeting Akt to increase the sensitivity of neuroblastoma to chemotherapy: lessons learned from the brain‐derived neurotrophic factor/TrkB signal transduction pathway. Expert Opin Ther Targets 2007; 11: 1611–21. [DOI] [PubMed] [Google Scholar]
- 33. Ho R, Minturn JE, Hishiki T et al Proliferation of human neuroblastomas mediated by the epidermal growth factor receptor. Cancer Res 2005; 65: 9868–75. [DOI] [PubMed] [Google Scholar]
- 34. Evangelopoulos ME, Weis J, Kruttgen A. Signalling pathways leading to neuroblastoma differentiation after serum withdrawal: HDL blocks neuroblastoma differentiation by inhibition of EGFR. Oncogene 2005; 24: 3309–18. [DOI] [PubMed] [Google Scholar]
- 35. Seales EC, Jurado GA, Brunson BA, Wakefield JK, Frost AR, Bellis SL. Hypersialylation of beta1 integrins, observed in colon adenocarcinoma, may contribute to cancer progression by up‐regulating cell motility. Cancer Res 2005; 65: 4645–52. [DOI] [PubMed] [Google Scholar]
- 36. Isaji T, Gu J, Nishiuchi R et al Introduction of bisecting GlcNAc into integrin alpha5beta1 reduces ligand binding and down‐regulates cell adhesion and cell migration. J Biol Chem 2004; 279: 19747–54. [DOI] [PubMed] [Google Scholar]
- 37. Guo HB, Lee I, Kamar M, Akiyama SK, Pierce M. Aberrant N‐glycosylation of beta1 integrin causes reduced alpha5beta1 integrin clustering and stimulates cell migration. Cancer Res 2002; 62: 6837–45. [PubMed] [Google Scholar]
- 38. Clement M, Rocher J, Loirand G, Le Pendu J. Expression of sialyl‐Tn epitopes on beta1 integrin alters epithelial cell phenotype, proliferation and haptotaxis. J Cell Sci 2004; 117: 5059–69. [DOI] [PubMed] [Google Scholar]
- 39. Wu YM, Liu CH, Hu RH et al Mucin glycosylating enzyme GALNT2 regulates the malignant character of hepatocellular carcinoma by modifying the EGF receptor. Cancer Res 2011; 71: 7270–9. [DOI] [PubMed] [Google Scholar]


