Abstract
Human papillomavirus (HPV)‐associated oropharyngeal squamous cell carcinomas (OPSCCs) frequently show different clinical and pathological features, which tend to be younger age, better performance status, less tobacco and alcohol consumption, more poorly differentiated histopathology, but usually with better treatment response and prognosis compared with HPV‐negative OPSCCs. In tumor tissue, HPV infection is closely correlated with p16INK4A expression, which has been suggested to be a surrogate biomarker of HPV infection. However, there is diversity of sensitivity and specificity about p16INK4A in surrogate detection of HPV status. Herein, we summarize the current knowledge and note some aspects for consideration concerning p16INK4A as a surrogate biomarker for HPV‐associated OPSCC.
Human papillomavirus‐associated oropharyngeal carcinomas have been extensively recognized a distinct entity. P16INK4A is evidenced to be a surrogate biomarker of HPV status. However, there is diversity of sensitivity and specificity about p16 in surrogate detection of HPV infection or transactivation. We summarized the current knowledge to explain the discrepancy between p16INK4A expression and HPV infection, and recommended suitable method of HPV status detection for clinical use. In addition, we reviewed the prognosis of p16INK4A or HPV associated entity and pointed out the cautious perspective for deintensified treatment.

As one of the cyclin‐dependent kinase inhibitors that inhibit cyclin‐dependent kinases 4 and 6, p16INK4A is encoded by the tumor suppressor gene CDKN2A. In non‐human papillomavirus (HPV) infected carcinomas, p16INK4A frequently showed in low levels or loss for epigenetic alteration and gene mutation.1 However, in HPV‐related cervical lesions and head and neck squamous cell carcinomas (HNSCCs), oncoprotein E7 could combine with Rb, and cause the dysfunction of Rb. The functional inactivation of Rb by E7 therefore results in the release of the transcriptional factor E2F from the Rb–E2F protein complex and the promotion of cell cycle progression, and also leads to release of the p16 INK4A gene from its transcriptional inhibition, causing p16INK4A to be expressed at a high level.2, 3, 4 Because of this molecular event, the fact that p16INK4A turned out to be substantially overexpressed in virtually all HPV‐transformed cells in cervical lesions, p16INK4A expression has been used for distinguishing high‐risk from low‐risk HPV infection,5 for ancillary confirmation and grading of histological diagnosis of cervical intraepithelial neoplasia,6 and for predicting progression or regression of low‐grade cervical intraepithelial neoplasia.7, 8 Considering the etiological link with HPV in HNSCCs, p16INK4A expression has been detected in many HPV‐related HNSCCs.9, 10, 11, 12, 13, 14 Those investigations suggest that overexpression of p16INK4A is caused by HPV infection, induced by dysfunction of the Rb tumor suppressor gene; also, theoretically, tumors with a high level of p16INK4A are the result of HPV infection.15 However, it is arbitrary to consider p16INK4A as a surrogate biomarker of HPV infection or HPV‐related carcinomas solely according to the theory mentioned above. First, the definition of “surrogate” should be identified. Does the high expression of p16INK4A indicate HPV infection, or the initiation of carcinogenesis induced by HPV, or tumors originating from HPV? Second, if p16INK4A serves as a surrogate biomarker, it should represent the biological and clinical features of HPV‐related carcinomas. If p16INK4A expression can represent or partially represent the features of HPV‐related carcinomas, it would be valuable as a surrogate biomarker.
Expression of p16INK4A and HPV Infection in Oropharyngeal Carcinomas
Human papillomavirus infection was found in 25.9% of head and neck cancers and 35.6% of oropharyngeal squamous cell carcinomas (OPSCCs).16 Patients with HPV‐associated OPSCC are frequently of a younger age at the time of diagnosis, with better performance status, less tobacco and alcohol exposure,17 with oral–genital sexual behavior,18, 19 poorly differentiated disease, and better prognosis than OPSCC patients without HPV infection.13, 20 We collected published reports on OPSCCs with p16INK4A expression and simultaneous HPV DNA detection (PCR and/or in situ hybridization [ISH]) in the past 5 years in the PubMed medical literature database,9, 21, 22, 23, 24, 25, 26, 27, 28, 29 and found that the sensitivity of p16INK4A for HPV DNA detection varied from 46% to 98%. Although it was recognized that overexpression of p16INK4A was closely correlated with HPV infection in OPSCCs, 3–51% of HPV DNA‐negative OPSCCs were found to express p16INK4A, and 2–54% of HPV DNA‐positive OPSCCs were found to display negative expression of p16INK4A (Table 1). In fact, regarding HPV DNA ISH, p16INK4A expression immunohistochemistry (IHC) detection shows excellent sensitivity, but there were too many p16INK4A(+)/HPV(−) cases (18–51%).9, 22, 27, 29 Similarly, regarding HPV DNA PCR, there were many p16INK4A(−)/HPV(+) cases (25–54%).26, 28, 30 We can ascribe the discrepancy between the p16INK4A IHC and HPV DNA tests to the excessive sensitivity of the PCR test and the specificity of the ISH test. Additionally, the pathology grade of tumors and the cut‐off point of p16INK4A interpretation must be considered. Data from Thavaraj et al.24 (Table 1) showed excellent concordance between the p16INK4A IHC and HPV DNA tests, partially due to the majority of poorly differentiated samples and suitably stringent criteria for p16INK4A staining (scored as positive if there was strong and diffuse staining present in >70% of the malignant cells). A published guide for interpretation of p16INK4A expression31 provides much instruction for p16INK4A expression as a surrogate biomarker for HPV‐associated OPSCCs, in which the pathology grade and stringent criteria were also emphasized.
Table 1.
Expression of p16INK4A in oropharyngeal carcinomas with different human papillomavirus (HPV) DNA status, as reported in research published 2009–2013
| Study | Total cases | P16+/HPV+ (sensitivity, %) | P16+ /HPV− (false positivity, %) | P16−/HPV+, false (negativity, %) | P16−/HPV−, (specificity, %) | Pathology grade | HPV DNA detection | P16 IHC expression pattern† |
|---|---|---|---|---|---|---|---|---|
| Shi et al.9 | 111 | 61/62 (98) | 8/44 (18) | 2 | 82 | No data | ISH | Neu and cyto a |
| Gao et al.29 | 150 | 54/55 (98) | 22/43 (51) | 2 | 49 | No data | ISH | Neu and cyto b |
| Thavanaj et al.24 | 142 | 88/90 (98) | 2/52 (4) | 2 | 96 | Grade I, 0; II, 71; III, 71 | ISH or PCR‡ | Neu and cyto c |
| Lewis et al.22 | 239 | 158/163 (97) | 29/76 (38) | 3 | 62 | K, 54; H*, 50; NK, 126 | ISH or PCR‡ | Neu and/or cyto d |
| Jordan et al.27 | 235 | 138/143 (97) | 27/89 (30) | 3 | 70 | No data | ISH | No data e |
| Evans et al.25 | 30 | 20/22 (91) | 2/4 (50) | 9 | 50 | No data | PCR | No data f |
| Nasman et al.21 | 175 | 119/136 (88) | 4/35 (11) | 12 | 89 | High, 5; medium, 54; low, 91 | PCR | No data g |
| Hong et al.23 | 198 | 62/83 (75) | 3/115 (3) | 25 | 97 | Grade I & II, 118; III, 77 | PCR | Neu and cyto h |
| Junor et al.26 | 254 | 77/133 (58) | 4/51 (8) | 42 | 92 | HPV+ and p16+ are likely to be grade III | PCR | Neu and/or cyto i |
| Holzinger et al.28 | 199 | 42/92 (46) | 12/85 (14) | 54 | 86 | No differential impact of p16 overexpression | PCR | Neu and cyto j |
†Criteria for p16INK4A expression: aA tumor was considered positive when strong signals were detected in both the tumor nuclei (Neu) and the cytoplasm (cyto). bClassified in a binary manner as positive when >50% of the cells showed nuclear and cytoplasmic staining. cScored as positive if there was strong and diffuse staining present in >70% of the malignant cells, other staining patterns were scored as negative. dGraded in a quartile manner for its extent: 0, negative; 1+, 1–25% of cells positive; 2+, 26–50%; 3+, 51–75%; 4+, 76–100%. Cases were also divided into positive (1–4 + ) and negative (0) groups. eSignificant differences in AUC were observed for both intensity score and percentage staining. A p16INK4A intensity score cut‐off point of 2 on a scale of 0–3 was most sensitive, and a percentage staining cut‐off point of 35% on a scale of 0–100% was most specific. *A hybrid (H) score cut‐off point of 60 on a scale of 0–300 yielded an average sensitivity of 91.6% and specificity of 90.4%. fp16INK4A was positive in diffuse staining and patchy staining. gNo detailed data were found. hWeak focal staining was recorded as negative. iStaining was scored as negative, focal positive, and positive based on both nuclear and cytoplasmic staining. Diffuse and continuous cytoplasmic and nuclear staining was considered a positive reaction. jScoring p16INK4A‐high required strong nuclear and cytoplasmic staining in the proliferating tumor cells. Patchy and negative staining was recorded as p16INK4A‐low. ‡Positive result, when either PCR or in situ hybridization (ISH) test was positive; negative result, when both PCR and ISH were negative. K, keratinizing; NK, non‐keratinizing.
Apart from the detection techniques that can disturb the accuracy of p16INK4A staining for HPV status detection, we cannot deny the theoretical possibility that there are the p16INK4A(+)/HPV(−) and p16INK4A(−)/HPV(+) cases (Fig. 1). Results regarding aberrant expression of p16INK4A had been reported in breast cancer32 and small‐cell lung cancer.33 Klingenberg et al.34 previously found p16INK4A overexpression was frequently detected in tumor‐free tonsil tissue without association with HPV infection (detected by PCR and FISH analysis). Therefore, mechanisms other than HPV infection are implicated in p16INK4A upregulation. Little is known concerning the topic, and further investigation is warranted. Of course, we can improve the criteria of p16INK4A interpretation to decrease the incidence of p16INK4A(+)/HPV(−) cases, however, which would take risk of increasing p16INK4A(−)/HPV(+) cases. Junor et al.26 excessively restricted the criteria of p16INK4A staining (Table 1), resulting in a p16INK4A(−)/HPV(+) set whose survival was much closer to that of p16INK4A(+)/HPV(+) patients than p16INK4A(−)/HPV(−) patients. Hoffmann et al.35 found that p16INK4A expression was closely correlated to HPV E6/E7 mRNA expression. In the HPV‐associated OPSCCs, HPV DNA‐negative sets and HPV DNA(+)/mRNA(−) sets showed similar survival curves.36 Human papillomavirus DNA detection can reflect the status of existing HPV infection but may be insufficient to indicate whether HPV is transformally active or not. No impact on survival was reported when the presence of HPV DNA was focused as a single factor, but HPV E6/E7 mRNA expression, p16INK4A overexpression, or the HPV DNA/p16INK4A combined test clearly showed statistical significance for better overall survival.35 Overexpression of p16INK4A or HPV E6/E7 mRNA expression were thought to be the parameter that described an activity of viral oncogenes, a finding that exactly explained that the p16INK4A(−)/HPV DNA(+) events were the results of HPV inactive infection. Consequently, HPV status was decided by HPV infection and transactivation (Fig. 1). The HPV DNA PCR test can detect HPV infection but cannot detect its activation. Although p16INK4A expression and HPV DNA infection are correlated with HPV‐associated OPSCCs, neither of the tests alone is the optimal method for HPV status detection.
Figure 1.
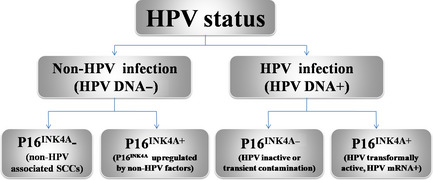
Human papillomavirus (HPV) status impacted by HPV infection and p16INK4A expression. SCC, squamous cell carcinoma.
Detection of HPV‐Associated OPSCC
Gold standard for HPV status detection
Although p16INK4A staining or the HPV DNA test can well reflect the HPV status of OPSCCs, inevitable discrepancy was found between p16INK4A staining and the HPV DNA test in a few cases. The use of HPV E6/E7 mRNA RT‐PCR detection can directly provide the present level of HPV oncoproteins in existing clinical samples (e.g., formalin‐fixed paraffin‐embedded tumors),37 and it can remove the situation of HPV‐inactive status and transient HPV infection or contamination. Survival analysis showed that the HPV mRNA RT‐PCR test can well stratify survival9, 29 and was superior to the HPV DNA test.36, 38 Detection HPV E6/E7 mRNA by RT‐PCR has been considered the gold standard for meaningful HPV infection. Another HPV RNA detection method is RNA ISH, which can also be used for detecting transcriptionally active HPV infection in formalin‐fixed paraffin‐embedded samples. Currently, the application of HPV RNA ISH for clinical detection is rare; Gao et al.29 has reported a perfect correlation (100% sensitivity and specificity) with HPV mRNA real‐time quantitative PCR. In contrast to the gold standard HPV mRNA RT‐PCR, RNA ISH is not quantitative and requires positive and negative controls. However, because HPV RNA ISH is slide based, it is convenient for clinical use. In addition, HPV RNA ISH requires less tissue and allows for visualization of viral transcripts directly in tumor cells. The HPV RNA ISH technique has shown perfect concordance with HPV mRNA RT‐PCR,29 HPV DNA ISH, p16INK4A staining, and survival.39, 40 Thus, HPV RNA ISH is thought to be an ideal platform for HPV detection, but further supportive data is needed.
Widely used methods for HPV status detection
Other widely used methods for HPV status detection include HPV DNA consensus PCR detection and type, HPV DNA ISH/FISH, HPV DNA load real‐time PCR, and indirect detections such as the p16INK4A expression IHC test and serum HPV L1/E6/E7 antibody tests. Table 2 shows the different sensitivities and specificities of each method according to the HPV E6 and/or E7 mRNA test (RT‐PCR or RNA ISH) as the gold standard.
Table 2.
Sensitivities and specificities of detection methods for human papillomavirus (HPV) status in oropharyngeal squamous cell carcinomas with HPV E6 or E7 mRNA (RT‐PCR or RNA in situ hybridization [ISH]) as the gold standard
| Study | Total cases | Tumor site | HPV DNA PCR, % | ISH/FISH, % | Viral load, %† | P16INK4A IHC, %‡ | P16 IHC interpretation | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sens. | Spec. | Sens. | Spec. | Sens. | Spec. | Sens. | Spec. | Intensity | % | |||
| Smeets et al.11 | 48 | Oropharynx, 18 Oral, 30 | 100 | 89 | 83 | 100 | 92 | 97 a | 100 | 79 d | ≥1+ Scale, 0–3 | >10 |
| Jordon et al.27 | 235 | Oropharynx | 100 | 66 | 88 | 94 | 91 | 96 b | 97 | 84 e | ≥2+ Scale, 0–3 | >35 |
| Rotnaglova et al.41 | 109 | Tonsillar | 100 | 89 | – | – | – | – | 96 | 94 f | None | >50 |
| Gao et al.29 | 150 | Oropharynx | – | – | 69 | 95 | – | – | 95 | 90 g | None | >50 |
| Schache et al.38 | 108 | Oropharynx | 97 | 87 | 88 | 88 | – | – | 94 | 82 h | None | >70 |
| Schlecht et al.43 | 110 | Oropharynx, 30 | – | – | Ventana 67 | 30 | – | – | 90 | 100 i | ≥2+ Scale, 0–3 | >75 |
| Dako 38 | 100 | |||||||||||
| Shi et al.9 | 111 | Oropharynx | – | – | 84 | 92 | – | – | 89 | 81 j | Strong | None |
| Jung et al.36 | 231 | Oropharynx | 100 | 91 | – | – | 100 | 100 c | – | – | – | – |
†Criteria for viral load as follows. aTumors with >0.5 copies per cell were scored as positive. bNo data found. cLow viral loads <1 copy/diploid genome equivalent, higher HPV16 loads >1 copy/diploid genome equivalent. ‡Criteria for p16INK4A immunohistochemical (IHC) staining as follows: dStaining intensity (graded 0–3 proportional to staining intensity) and the percentage of the tumor cells positively stained per slide were assessed, positive defined as intensity >1 and percentage >10%. eSignificant differences in AUC were observed for both intensity score and percentage staining. A p16INK4A intensity score cut‐off point of 2 on a scale of 0–3 was most sensitive, and a percentage staining cut‐off point of 35% on a scale of 0–100% was most specific. An H score cut‐off point of 60 on a scale of 0–300 yielded an average sensitivity of 91.6% and specificity of 90.4%. fPositive for p16INK4A expression had to show more than 50% of positive cells and reveal nuclear and/or cytoplasmic staining. gClassified in a binary manner as positive when >50% of the cells showed nuclear and cytoplasmic staining. hP16INK4A IHC was scored as positive if there was strong and diffuse nuclear and cytoplasmic staining present in >70% of the malignant cells. iP16INK4A positivity was defined by a mean intensity cut‐off >2 and diffuse (>75%) staining distribution in either the nuclei or cytoplasm. jA tumor was considered positive when strong signals were detected in both the tumor nuclei as well as the cytoplasm. –, no data; Sens., sensitivity; Spec., specificity.
In OPSCCs, the HPV DNA PCR assay was used early and widely in HPV status detection with a high sensitivity range from 97% to 100% and a relative low specificity of 66–91%.11, 27, 36, 38, 41 Human papillomavirus DNA was usually amplified with the L1 consensus HPV MY09/MY11 or GP5+/6+ primer set, or HPV E6/E7 specific primers. HPV DNA PCR assay usually overestimates the results for inactive HPV infection. In addition, the possibility of a false‐negative HPV L1 consensus PCR assay exists because of the presence of integrated virus with loss or disruption of the L1 ORF.42, 43 Additionally, HPV DNA PCR assay does not distinguish the integrated form from the episomal form of the virus, a finding that argues against the use of PCR alone for classification of HPV status.
Another commonly used method is the type‐specific HPV DNA detection by ISH or FISH assays. Usually punctuated nuclear (F)ISH signals indicate HPV DNA integrated into the host genome, and areas with diffuse nuclear (F)ISH staining indicate episomal HPV DNA. These assays allow visual confirmation of HPV DNA within individual tumor cell nuclei,37 making excellent specificity for HPV status detection, at approximately 88–100%.9, 27, 29, 38, 43 However, the interpretation of staining is subjective, leading to ambiguous interpretation for non‐specific staining in situ.43
The assay with HPV DNA copy number has not yet formed a standardized protocol for HPV transcriptional activity, and it is very labor‐intensive as the neoplastic cells need to be enriched by microdissection.44 Jung et al.36 showed that all the HPV16 transcriptionally active tumors had elevated viral load values, with a cut‐off point at 1 copy per diploid genome equivalent. Considering some normal tissue contamination in the microdissected tumor samples, Smeets et al.11 scored tumors with >0.5 copies per cell as positive and showed a high sensitivity and specificity of 92% and 97%, respectively, for HPV status detection.
Detection of the antibody to HPV E6 and E7 in serum was investigated in several large clinical studies concerning cervical cancer patients.45, 46 Seropositivity validated by HPV 16 L1 virus‐like particles was associated with a significantly increased risk of oropharyngeal cancer.47 Smeets et al. detected antibodies against the proteins HPV 16 L1, E6, and E7, indirectly reflecting HPV 16 infection, and the highest sensitivity was reached when positive serology was defined with any of the three antibodies (91%), but the specificity was then limited (74%).11 Better results from Rotnaglova et al.41 showed a high correlation between HPV DNA/RNA status and seropositivity of E6/E7 oncoproteins, and indicated that antibodies against HPV 16 E6 and E7 oncoproteins reached high sensitivity (96%) and specificity (89%) for detection of HPV‐associated tonsillar cancer. Serological testing is simple, convenient, and cheap, but the value of clinical application of HPV L1/E6/E7 and other antigens requires further investigations.
Immunohistochemical staining of p16INK4A for HPV status detection in OPSCCs
Apart from HPV status mentioned above, p16INK4A expression can also be regulated through epigenetic control and multiple transcription factors, such as PRC1, PRC2, YY1, Id1, CTCF, Sp1, Ets, and HBP‐1.48 Frequently, HPV‐positive OPSCCs are less likely to carry genetic alterations compared with HPV‐negative ones, including chromosomal aberration,49 gene mutation,50 and transcriptional expression.36 The difference of gene profiling would indirectly lead to diametrically opposed expression of p16INK4A, for example, 11q is frequently found lost in HPV‐positive but gained in HPV‐negative OPSCCs, on which Ets (a protein that can raise the level of p16INK4A) is located. The different genetic landscapes associated with transcriptionally active HPV are consistent with epidemiologic and clinical features (e.g., age, tobacco and alcohol exposure, tumor stage, grade, and response to treatment). Selection of the patients with different features can in turn reflect the initiation and genotype of tumors, which could impact p16INK4A expression in OPSCCs with different HPV status. Additionally, there were different interpretations for p16INK4A IHC staining between different investigators. The results of IHC staining usually depended on comprehensive elements, including the expression pattern (nuclear and/or cytoplasmic), intensity of staining, and percentage of stained tumor cells. The selection of the experimental reagents and interpretation of the staining may lead to different results. Considering the reasons mentioned above, controversy appeared regarding the capacity of p16INK4A to indicate HPV status.
The interpretation of p16INK4A IHC staining likely contributes to most of the discrepancy regarding p16INK4A detection and is the only element we can easily control. Cut‐off points for the intensity and percentage of tumor cell staining are equally important. A single limitation of the cut‐off point for either intensity9, 27 or percentage38 of the staining will certainly result in increased sensitivity and decreased specificity (Table 2). Suitably restricting both the cut‐off points can eliminate the false‐positivity of p16INK4A expression induced by low‐risk HPV infection and non‐HPV factors, and even cause the specificity of p16INK4A detection to surpass its sensitivity.43 Easy to carry out, p16INK4A IHC detection is also low cost and has a high sensitivity. However, there is a tendency toward false‐positive results in p16INK4A staining, and there is a lack of a direct and exclusive mechanistic link between HPV DNA integration and p16INK4A expression. Cautious interpretation and stringent criteria for p16INK4A IHC staining with attached information of various histologic, anatomic, clinical, and technical considerations were advocated.31 And further HPV testing was suggested when p16INK4A staining was absent/weak or when keratinizing squamous cell carcinoma staining was present.
Further HPV testing, for instance, p16INK4A IHC in combination with HPV DNA PCR assays or ISH assays, was frequently used in studies and in clinics. Table 3 shows the comparison of the detection of p16INK4A combined with HPV DNA PCR or ISH assays based on HPV mRNA detection as the gold standard.11, 27, 29, 38, 41 In the combined detection using the p16INK4A IHC and HPV DNA ISH assays, if results set both positive as a positive test and all others as a negative test, we can obtain a perfect specificity but with decreased sensitivity; if results set either positive as a positive test and both negative as a negative test, we can scarcely obtain any enhancement in sensitivity or specificity (Table 3). Unlike the combination with the ISH assay, HPV IHC combined with PCR can conspicuously improve the specificity on the premise of continuously high sensitivity (Table 3). Using HPV DNA PCR combined with p16INK4A IHC for HPV status detection, Hong et al. and Heath et al.30, 51both showed that the HPV DNA(+)/p16INK4A(+) groups had better survival than the HPV DNA(+)/p16INK4A(−) and HPV DNA(−)/p16INK4A(−) groups. Clearly, the combined detection of p16INK4A IHC and HPV DNA PCR can not only eliminate inactive infection and transient contamination but can also omit the contingently elevated p16INK4A expression by non‐viral related alterations. The p16INK4A IHC/HPV DNA PCR combination test offers a valuable alternative to RNA analysis, with excellent sensitivity/specificity and prognostic value.38, 41 Additionally, p16INK4A combined with other cellular proteins has been reported as a feasible biomarker to identify OPSCCs with active HPV, for example, combined with pRb (sensitivity 78%, specificity 93%), with p53 (sensitivity 67%, specificity 95%), or with cyclin D1 (sensitivity 78%, specificity 90%).52 In the combined detection of p16INK4A/p53 IHC staining, active HPV infection is inversely associated with p53 mutation. The HPV‐associated OPSCCs with wild‐type p53 gene always show a low level (“negative” in standard IHC and “normal low” in TSA‐IHC staining) of p53 protein due to ubiquitination and degradation through viral E6 protein. Intriguingly, HPV‐negative OPSCCs (inclined to p53 mutation) always show “absent” or “high” p53 protein level (detected with standard IHC and TSA‐IHC)52 because absent or high p53 staining was correlated to nonsense or missense p53 mutations,53 respectively, and mutant p53 protein was more stable and had heavier staining than wild‐type p53 protein. Mannweiler et al.54 reported completely consistent p53 staining with p16INK4A/HPV DNA combined detection in penile lesions and suggested p53 expression along with p16INK4A negativity to identify HPV‐negative cancers. However, the method of p53 detection was questionable and normal p53 and nonsense mutations were not taken into consideration.
Table 3.
Human papillomavirus (HPV) status detection using p16INK4A immunohistochemical (IHC) staining in combination with HPV DNA PCR or in situ hybridization (ISH) assay
| Study | Cases | Gold standard | Combined with PCR, % | Combined with ISH, % | Compared with single p16INK4A IHC staining, %† | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Both positive | Both positive | Either positive | ||||||||
| Sens. | Spec. | Sens. | Spec. | Sens. | Spec. | Sens. | Spec. | |||
| Smeets et al.11 | 48 | HPV E6/E7 mRNA | 100 | 100 | – | – | – | – | 100 | 79 a |
| Rotnaglova et al.41 | 109 | HPV E6*I mRNA | 100 | 88 | – | – | – | – | 96 | 94 b |
| Schache et al.38 | 108 | HPV E6 mRNA | 97 | 94 | 88 | 90 | – | – | 94 | 82 c |
| Jordon et al.27 | 232 | HPV E6/E7 mRNA | – | – | 86.1 | 97.3 | 98.7 | 81.1 | 97 | 84 d |
| Gao et al.29 | 150 | HPV E6/E7 mRNA ISH | – | – | 69 | 100 | 95 | 85 | 95 | 90 e |
†Criteria for p16INK4A immunohistochemical (IHC) staining as follows. aStaining intensity (graded 0–3 proportional to staining intensity) and the percentage of the tumor cells positively stained per slide were assessed, positive defined as intensity >1 and percentage >10%. bPositive for p16INK4A expression had to show more than 50% of positive cells and reveal nuclear and/or cytoplasmic staining. cP16INK4A IHC was scored as positive if there was strong and diffuse nuclear and cytoplasmic staining present in >70% of the malignant cells. dSignificant differences in AUC were observed for both intensity score and percentage staining. A p16INK4A intensity score cut‐off point of 2 on a scale of 0–3 was most sensitive, and a percentage staining cut‐off point of 35% on a scale of 0–100% was most specific. An H score cut‐off point of 60 on a scale of 0–300 yielded an average sensitivity of 91.6% and specificity of 90.4%. eClassified in a binary manner as positive when >50% of the cells showed nuclear and cytoplasmic staining. –, no data; Sens., sensitivity; Spec., specificity.
Significance of p16INK4A for Prognosis and Treatment in HPV‐Associated OPSCCs
Despite the controversy concerning the significance of HPV status at other sites of HNSCCs, its particular meaning for OPSCCs has been extensively recognized.55 The HPV‐associated HNSCCs, particularly OPSCCs, have been defined as a distinct entity with different epidemiology, etiology, pathogenesis, pathology and molecular pathology, clinical manifestations, treatment response, and prognosis. Most interestingly, despite the poor differentiation, and early cervical metastasis, HPV‐associated OPSCCs usually demonstrate a better treatment response and prognosis than HPV‐negative OPSCCs.
Immunohistochemical staining for p16INK4A can not only represent HPV status but can also indicate the prognosis of HPV‐associated OPSCCs.56, 57 P16INK4A and HPV status is a strong and consistent determinant of superior survival, regardless of treatment strategy, such as surgery,12 radiotherapy,58 chemoradiotherapy,56 or induction chemotherapy plus chemoradiotherapy.13 Of course, most of the existing clinical trials reflecting advantageous prognosis were related to radiotherapy and/or chemotherapy. Parts of the studies investigated the effect of radical surgery on HPV‐associated OPSCCs; however, either most of the included patients received postoperative radiotherapy12 or no statistically significant differences were found between HPV‐positive and ‐negative groups.59 Lassen reviewed the clinical data, addressing the impact of HPV on radiotherapy, including conventionally fractionated radiotherapy, accelerated fractionated radiotherapy, hypoxic modification in radiotherapy, and chemoradiotherapy.60 Human papillomavirus‐positive or p16INK4A‐positive tumors support a better prognosis in various schedules of radiotherapy and chemotherapy except the hypoxic modification in radiotherapy.61 The HPV‐associated OPSCCs have been considered to show an excellent sensitivity to radiotherapy and chemotherapy. Given that HPV‐associated OPSCCs are distinct from HPV‐negative carcinomas in treatment response and prognosis, numerous clinical trials that deintensify treatment for HPV‐associated carcinomas are underway, with the aim of reducing treatment toxicity and improving the quality of life. A review authored by Mehra et al.62 summarized the ongoing clinical trials.
Reduced‐intensity therapy mostly focuses on reducing the radiation dose and replacing concurrent chemotherapy with cetuximab. However, little is known concerning the mechanism of enhanced sensitivity to radiotherapy and chemotherapy in HPV‐associated OPSCCs. The HPV‐associated OPSCCs possess fewer p53 mutations and lower EGFR expression, which may play a role in better prognosis, simultaneously questioning the replacement of treatment with cetuximab. Human papillomavirus E7 can bind to the catalytic and structural subunits of protein phosphatase 2A and inhibit their interaction with Akt, thereby maintaining PKB/Akt signaling by inhibiting its dephosphorylation.63 The activated PI3K/Akt pathway is known to be a potent inducer of radiation resistance in cervical carcinoma, whereas little is known regarding its role in head and neck carcinoma. In terms of radiation, hypoxic cells in tumors are resistant to treatment. It has been reported that hypoxic modification improved the outcome in HPV‐ or p16INK4A‐negative tumors but was of no significant benefit in HPV‐ or p16INK4A‐positive tumors.61 The hypothesis supported that the extent of hypoxia may be more pronounced in p16INK4A‐negative tumors compared with p16INK4A‐positive tumors.64 Additionally, elevated p16INK4A is induced by functional inactivation of the tumor suppressor gene Rb, which may also contribute to p16INK4A‐positive tumors' sensitivity to chemo‐ or radiotherapy. Conclusions regarding the mechanism to explain why p16INK4A‐positive/HPV‐associated OPSCCs possess a superior prognosis are difficult to draw from inconsistent data. With the current findings of molecular biology and clinicopathology of HPV‐associated OPSCCs, we hypothesize that increased sensitivity to radiotherapy and/or chemotherapy is the aggregate result of poor differentiation, continuous proliferation, abrogation of the inhibition of DNA synthesis induced by radiation,65 slight hypoxia status, non‐mutation but dysfunctional p53, and its own genomic instability.66
Oropharyngeal SCC is a distinct entity frequently associated with younger age, better performance status, less tobacco and alcohol consumption, improved adaptive immunity67 and different patterns of gene‐expression profiles.68 Additionally, the mechanism of superior prognosis is unclear and needs further investigation. As described above, the excellent prognosis of the HPV‐positive entity is the result of the comprehensive effect of multiple factors. Additionally, change to any single factor may cause alteration of the outcome, for instance, tobacco consumption.56 Likewise, no single factor alone can reflect all the characteristics of an individual patient, particularly regarding treatment response and prognosis. Although a series of clinical trials on deintensification for reducing treatment toxicity are underway, we believe the deintensification should proceed with caution when considering the potential cost of treatment efficacy, particularly the formulation and mastery of the indications for deintensified treatment. By contrast, HPV‐associated OPSCCs show an excellent radiotherapy or chemoradiotherapy response. Thus, the following questions arise: why should these effective adjuvant treatments be discarded when radical surgery is the primary treatment algorithm, and could HPV‐positive and/or p16INK4A expression be a clinical indication for postoperative radiotherapy or chemoradiotherapy? There is a lack of research regarding this subject area. With different treatment intensifications, as well as different efficacies and toxicities, which is the key point is determined by the patient's choice.
Conclusions and Future Perspectives
The tumor suppressor p16INK4A plays an important role in cell cycle regulation. When p16INK4A is expressed in HNSCCs, particularly in OPSCCs, it is associated with a new implication: HPV status and superior prognosis. Immunohistochemical staining of p16INK4A does not exactly match the HPV DNA test, and two inconsistent patterns of p16INK4A(+)/HPV DNA(−) and p16INK4A(−)/HPV DNA(+) cases are evidence. Restricting the cut‐off point of criteria can improve the specificity of p16INK4A IHC staining as a surrogate biomarker for HPV‐associated OPSCC detection. The combination of p16INK4A staining with the HPV DNA PCR test can produce almost perfect sensitivity and specificity with HPV E6 or E7 mRNA (RT‐PCR or RNA ISH) as a gold standard. Granted, p16INK4A can stratify prognosis in OPSCCs, but the mechanism for better survival remains unclear, warranting further investigation. Despite the advantageous treatment response and prognosis of HPV‐associated carcinomas, decreasing the treatment intensification would lead to a potential risk of reducing treatment efficacy. Additionally, the clinical indication and implementation of deintensification should be done with caution. Finally, we would like to reiterate the features of HPV‐associated OPSCCs: they are not only carcinomas associated with HPV infection but are OPSCCs associated with high‐risk HPV infection and activation of malignant transformation in carcinogenesis, with distinct characteristics in epidemiology, etiology and pathogenesis, clinical manifestations, pathology and molecular phenotype, treatment response and survival.
Disclosure Statement
The authors have no conflict of interest.
(Cancer Sci 2013; 104: 1553–1559)
References
- 1. Nobori T, Miura K, Wu DJ et al Deletions of the cyclin‐dependent kinase‐4 inhibitor gene in multiple human cancers. Nature 1994; 368: 753–6. [DOI] [PubMed] [Google Scholar]
- 2. Khleif SN, DeGregori J, Yee CL et al Inhibition of cyclin D‐CDK4/CDK6 activity is associated with an E2F‐mediated induction of cyclin kinase inhibitor activity. Proc Natl Acad Sci U S A 1996; 93: 4350–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dyson N, Howley PM, Munger K et al The human papilloma virus‐16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science 1989; 243: 934–7. [DOI] [PubMed] [Google Scholar]
- 4. Munger K, Werness BA, Dyson N et al Complex formation of human papillomavirus E7 proteins with the retinoblastoma tumor suppressor gene product. EMBO J 1989; 8: 4099–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sano T, Oyama T, Kashiwabara K et al Expression status of p16 protein is associated with human papillomavirus oncogenic potential in cervical and genital lesions. Am J Pathol 1998; 153: 1741–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mulvany NJ, Allen DG, Wilson SM. Diagnostic utility of p16INK4a: a reappraisal of its use in cervical biopsies. Pathology 2008; 40: 335–44. [DOI] [PubMed] [Google Scholar]
- 7. Carozzi F, Gillio‐Tos A, Confortini M et al Risk of high‐grade cervical intraepithelial neoplasia during follow‐up in HPV‐positive women according to baseline p16‐INK4A results: a prospective analysis of a nested substudy of the NTCC randomised controlled trial. Lancet Oncol 2013; 14: 168–76. [DOI] [PubMed] [Google Scholar]
- 8. Nishio S, Fujii T, Nishio H et al p16(INK4a) immunohistochemistry is a promising biomarker to predict the outcome of low grade cervical intraepithelial neoplasia: comparison study with HPV genotyping. J Gynecol Oncol 2013; 24: 215–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shi W, Kato H, Perez‐Ordonez B et al Comparative prognostic value of HPV16 E6 mRNA compared with in situ hybridization for human oropharyngeal squamous carcinoma. J Clin Oncol 2009; 27: 6213–21. [DOI] [PubMed] [Google Scholar]
- 10. Reimers N, Kasper HU, Weissenborn SJ et al Combined analysis of HPV‐DNA, p16 and EGFR expression to predict prognosis in oropharyngeal cancer. Int J Cancer 2007; 120: 1731–8. [DOI] [PubMed] [Google Scholar]
- 11. Smeets SJ, Hesselink AT, Speel EJ et al A novel algorithm for reliable detection of human papillomavirus in paraffin embedded head and neck cancer specimen. Int J Cancer 2007; 121: 2465–72. [DOI] [PubMed] [Google Scholar]
- 12. Licitra L, Perrone F, Bossi P et al High‐risk human papillomavirus affects prognosis in patients with surgically treated oropharyngeal squamous cell carcinoma. J Clin Oncol 2006; 24: 5630–6. [DOI] [PubMed] [Google Scholar]
- 13. Fakhry C, Westra WH, Li S et al Improved survival of patients with human papillomavirus‐positive head and neck squamous cell carcinoma in a prospective clinical trial. J Natl Cancer Inst 2008; 100: 261–9. [DOI] [PubMed] [Google Scholar]
- 14. Weinberger PM, Yu Z, Haffty BG et al Molecular classification identifies a subset of human papillomavirus—associated oropharyngeal cancers with favorable prognosis. J Clin Oncol 2006; 24: 736–47. [DOI] [PubMed] [Google Scholar]
- 15. von Knebel DM. New markers for cervical dysplasia to visualise the genomic chaos created by aberrant oncogenic papillomavirus infections. Eur J Cancer 2002; 38: 2229–42. [DOI] [PubMed] [Google Scholar]
- 16. Kreimer AR, Clifford GM, Boyle P et al Human papillomavirus types in head and neck squamous cell carcinomas worldwide: a systematic review. Cancer Epidemiol Biomarkers Prev 2005; 14: 467–75. [DOI] [PubMed] [Google Scholar]
- 17. Marur S, D'Souza G, Westra WH et al HPV‐associated head and neck cancer: a virus‐related cancer epidemic. Lancet Oncol 2010; 11: 781–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gillison ML, D'Souza G, Westra W et al Distinct risk factor profiles for human papillomavirus type 16‐positive and human papillomavirus type 16‐negative head and neck cancers. J Natl Cancer Inst 2008; 100: 407–20. [DOI] [PubMed] [Google Scholar]
- 19. Smith EM, Ritchie JM, Summersgill KF et al Age, sexual behavior and human papillomavirus infection in oral cavity and oropharyngeal cancers. Int J Cancer 2004; 108: 766–72. [DOI] [PubMed] [Google Scholar]
- 20. Gillison ML, Koch WM, Capone RB et al Evidence for a causal association between human papillomavirus and a subset of head and neck cancers. J Natl Cancer Inst 2000; 92: 709–20. [DOI] [PubMed] [Google Scholar]
- 21. Nasman A, Andersson E, Nordfors C et al MHC class I expression in HPV positive and negative tonsillar squamous cell carcinoma in correlation to clinical outcome. Int J Cancer 2013; 132: 72–81. [DOI] [PubMed] [Google Scholar]
- 22. Lewis JJ, Thorstad WL, Chernock RD et al p16 positive oropharyngeal squamous cell carcinoma:an entity with a favorable prognosis regardless of tumor HPV status. Am J Surg Pathol 2010; 34: 1088–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hong AM, Dobbins TA, Lee CS et al Human papillomavirus predicts outcome in oropharyngeal cancer in patients treated primarily with surgery or radiation therapy. Br J Cancer 2010; 103: 1510–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Thavaraj S, Stokes A, Guerra E et al Evaluation of human papillomavirus testing for squamous cell carcinoma of the tonsil in clinical practice. J Clin Pathol 2011; 64: 308–12. [DOI] [PubMed] [Google Scholar]
- 25. Evans MF, Matthews A, Kandil D et al Discrimination of ‘driver’ and ‘passenger’ HPV in tonsillar carcinomas by the polymerase chain reaction, chromogenic in situ hybridization, and p16(INK4a) immunohistochemistry. Head Neck Pathol 2011; 5: 344–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Junor E, Kerr G, Oniscu A et al Benefit of chemotherapy as part of treatment for HPV DNA‐positive but p16‐negative squamous cell carcinoma of the oropharynx. Br J Cancer 2012; 106: 358–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jordan RC, Lingen MW, Perez‐Ordonez B et al Validation of methods for oropharyngeal cancer HPV status determination in US cooperative group trials. Am J Surg Pathol 2012; 36: 945–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Holzinger D, Schmitt M, Dyckhoff G et al Viral RNA patterns and high viral load reliably define oropharynx carcinomas with active HPV16 involvement. Cancer Res 2012; 72: 4993–5003. [DOI] [PubMed] [Google Scholar]
- 29. Gao G, Chernock RD, Gay HA et al A novel RT‐PCR method for quantification of human papillomavirus transcripts in archived tissues and its application in oropharyngeal cancer prognosis. Int J Cancer 2013; 132: 882–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hong A, Jones D, Chatfield M et al HPV status of oropharyngeal cancer by combination HPV DNA/p16 testing: biological relevance of discordant results. Ann Surg Oncol 2012; doi: 10.1245/s10434-012-2778-4. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 31. El‐Naggar AK, Westra WH. p16 expression as a surrogate marker for HPV‐related oropharyngeal carcinoma: a guide for interpretative relevance and consistency. Head Neck 2012; 34: 459–61. [DOI] [PubMed] [Google Scholar]
- 32. Cui SP, Wang HL, Peng W et al Aberrant expression and correlative analysis of P16 in breast cancers. Beijing Da Xue Xue Bao 2012; 44: 755–9. [PubMed] [Google Scholar]
- 33. Yuan J, Knorr J, Altmannsberger M et al Expression of p16 and lack of pRB in primary small cell lung cancer. J Pathol 1999; 189: 358–62. [DOI] [PubMed] [Google Scholar]
- 34. Klingenberg B, Hafkamp HC, Haesevoets A et al p16 INK4A overexpression is frequently detected in tumour‐free tonsil tissue without association with HPV. Histopathology 2010; 56: 957–67. [DOI] [PubMed] [Google Scholar]
- 35. Hoffmann M, Ihloff AS, Gorogh T et al p16(INK4a) overexpression predicts translational active human papillomavirus infection in tonsillar cancer. Int J Cancer 2010; 127: 1595–602. [DOI] [PubMed] [Google Scholar]
- 36. Jung AC, Briolat J, Millon R et al Biological and clinical relevance of transcriptionally active human papillomavirus (HPV) infection in oropharynx squamous cell carcinoma. Int J Cancer 2010; 126: 1882–94. [DOI] [PubMed] [Google Scholar]
- 37. Chung CH, Gillison ML. Human papillomavirus in head and neck cancer: its role in pathogenesis and clinical implications. Clin Cancer Res 2009; 15: 6758–62. [DOI] [PubMed] [Google Scholar]
- 38. Schache AG, Liloglou T, Risk JM et al Evaluation of human papilloma virus diagnostic testing in oropharyngeal squamous cell carcinoma: sensitivity, specificity, and prognostic discrimination. Clin Cancer Res 2011; 17: 6262–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bishop JA, Ma XJ, Wang H et al Detection of transcriptionally active high‐risk HPV in patients with head and neck squamous cell carcinoma as visualized by a novel E6/E7 mRNA in situ hybridization method. Am J Surg Pathol 2012; 36: 1874–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ukpo OC, Flanagan JJ, Ma XJ et al High‐risk human papillomavirus E6/E7 mRNA detection by a novel in situ hybridization assay strongly correlates with p16 expression and patient outcomes in oropharyngeal squamous cell carcinoma. Am J Surg Pathol 2011; 35: 1343–50. [DOI] [PubMed] [Google Scholar]
- 41. Rotnaglova E, Tachezy R, Salakova M et al HPV involvement in tonsillar cancer: prognostic significance and clinically relevant markers. Int J Cancer 2011; 129: 101–10. [DOI] [PubMed] [Google Scholar]
- 42. Duray A, Descamps G, Arafa M et al High incidence of high‐risk HPV in benign and malignant lesions of the larynx. Int J Oncol 2011; 39: 51–9. [DOI] [PubMed] [Google Scholar]
- 43. Schlecht NF, Brandwein‐Gensler M, Nuovo GJ et al A comparison of clinically utilized human papillomavirus detection methods in head and neck cancer. Mod Pathol 2011; 24: 1295–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ha PK, Pai SI, Westra WH et al Real‐time quantitative PCR demonstrates low prevalence of human papillomavirus type 16 in premalignant and malignant lesions of the oral cavity. Clin Cancer Res 2002; 8: 1203–9. [PubMed] [Google Scholar]
- 45. Combita AL, Bravo MM, Touze A et al Serologic response to human oncogenic papillomavirus types 16, 18, 31, 33, 39, 58 and 59 virus‐like particles in colombian women with invasive cervical cancer. Int J Cancer 2002; 97: 796–803. [DOI] [PubMed] [Google Scholar]
- 46. Mork J, Lie AK, Glattre E et al Human papillomavirus infection as a risk factor for squamous‐cell carcinoma of the head and neck. N Engl J Med 2001; 344: 1125–31. [DOI] [PubMed] [Google Scholar]
- 47. Dahlstrom KR, Adler‐Storthz K, Etzel CJ, et al Human papillomavirus type 16 infection and squamous cell carcinoma of the head and neck in never‐smokers: a matched pair analysis. Clin Cancer Res 2003; 9: 2620–6. [PubMed] [Google Scholar]
- 48. Rayess H, Wang MB, Srivatsan ES. Cellular senescence and tumor suppressor gene p16. Int J Cancer 2012; 130: 1715–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Dahlgren L, Mellin H, Wangsa D et al Comparative genomic hybridization analysis of tonsillar cancer reveals a different pattern of genomic imbalances in human papillomavirus‐positive and ‐negative tumors. Int J Cancer 2003; 107: 244–9. [DOI] [PubMed] [Google Scholar]
- 50. Agrawal N, Frederick MJ, Pickering CR et al Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science 2011; 333: 1154–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Heath S, Willis V, Allan K et al Clinically significant human papilloma virus in squamous cell carcinoma of the head and neck in UK practice. Clin Oncol (R Coll Radiol) 2012; 24: e18–23. [DOI] [PubMed] [Google Scholar]
- 52. Holzinger D, Flechtenmacher C, Henfling N et al Identification of oropharyngeal squamous cell carcinomas with active HPV16 involvement by immunohistochemical analysis of the retinoblastoma protein pathway. Int J Cancer 2013; 133: 1389–99. [DOI] [PubMed] [Google Scholar]
- 53. Bosch FX, Ritter D, Enders C et al Head and neck tumor sites differ in prevalence and spectrum of p53 alterations but these have limited prognostic value. Int J Cancer 2004; 111: 530–8. [DOI] [PubMed] [Google Scholar]
- 54. Mannweiler S, Sygulla S, Winter E et al Two major pathways of penile carcinogenesis: HPV‐induced penile cancers overexpress p16ink4a, HPV‐negative cancers associated with dermatoses express p53, but lack p16ink4a overexpression. J Am Acad Dermatol 2013; 69: 73–81. [DOI] [PubMed] [Google Scholar]
- 55. Rainsbury JW, Ahmed W, Williams HK et al Prognostic biomarkers of survival in oropharyngeal squamous cell carcinoma: systematic review and meta‐analysis. Head Neck 2013; 35: 1048–55. [DOI] [PubMed] [Google Scholar]
- 56. Ang KK, Harris J, Wheeler R et al Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med 2010; 363: 24–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Rischin D, Young RJ, Fisher R et al Prognostic significance of p16INK4A and human papillomavirus in patients with oropharyngeal cancer treated on TROG 02.02 phase III trial. J Clin Oncol 2010; 28: 4142–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lassen P, Eriksen JG, Hamilton‐Dutoit S et al Effect of HPV‐associated p16INK4A expression on response to radiotherapy and survival in squamous cell carcinoma of the head and neck. J Clin Oncol 2009; 27: 1992–8. [DOI] [PubMed] [Google Scholar]
- 59. Cohen MA, Weinstein GS, O'Malley BJ et al Transoral robotic surgery and human papillomavirus status: Oncologic results. Head Neck 2011; 33: 573–80. [DOI] [PubMed] [Google Scholar]
- 60. Lassen P. The role of human papillomavirus in head and neck cancer and the impact on radiotherapy outcome. Radiother Oncol 2010; 95: 371–80. [DOI] [PubMed] [Google Scholar]
- 61. Lassen P, Eriksen JG, Hamilton‐Dutoit S et al HPV‐associated p16‐expression and response to hypoxic modification of radiotherapy in head and neck cancer. Radiother Oncol 2010; 94: 30–5. [DOI] [PubMed] [Google Scholar]
- 62. Mehra R, Ang KK, Burtness B. Management of human papillomavirus‐positive and human papillomavirus‐negative head and neck cancer. Semin Radiat Oncol 2012; 22: 194–7. [DOI] [PubMed] [Google Scholar]
- 63. Pim D, Massimi P, Dilworth SM et al Activation of the protein kinase B pathway by the HPV‐16 E7 oncoprotein occurs through a mechanism involving interaction with PP2A. Oncogene 2005; 24: 7830–8. [DOI] [PubMed] [Google Scholar]
- 64. Overgaard J, Eriksen JG, Nordsmark M et al Plasma osteopontin, hypoxia, and response to the hypoxia sensitiser nimorazole in radiotherapy of head and neck cancer: results from the DAHANCA 5 randomised double‐blind placebo‐controlled trial. Lancet Oncol 2005; 6: 757–64. [DOI] [PubMed] [Google Scholar]
- 65. Song S, Gulliver GA, Lambert PF. Human papillomavirus type 16 E6 and E7 oncogenes abrogate radiation‐induced DNA damage responses in vivo through p53‐dependent and p53‐independent pathways. Proc Natl Acad Sci U S A 1998; 95: 2290–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Duensing S, Munger K. Mechanisms of genomic instability in human cancer: insights from studies with human papillomavirus oncoproteins. Int J Cancer 2004; 109: 157–62. [DOI] [PubMed] [Google Scholar]
- 67. Wansom D, Light E, Worden F et al Correlation of cellular immunity with human papillomavirus 16 status and outcome in patients with advanced oropharyngeal cancer. Arch Otolaryngol Head Neck Surg 2010; 136: 1267–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Martinez I, Wang J, Hobson KF et al Identification of differentially expressed genes in HPV‐positive and HPV‐negative oropharyngeal squamous cell carcinomas. Eur J Cancer 2007; 43: 415–32. [DOI] [PMC free article] [PubMed] [Google Scholar]


