Abstract
The estrogen receptor (ER) is a key molecule for growth of breast cancers. It has been a successful target for treatment of breast cancers. Elucidation of the ER expression mechanism is of importance for designing therapeutics for ER‐positive breast cancers. However, the detailed mechanism of ER stability is still unclear. Here, we report that histone acetyltransferase Hbo1 promotes destabilization of estrogen receptor α (ERα) in breast cancers through lysine 48‐linked ubiquitination. The acetyltransferase activity of Hbo1 is linked to its activity for ERα ubiquitination. Depletion of Hbo1 and anti‐estrogen treatment displayed a potent growth suppression of breast cancer cell line. Hbo1 modulated transcription by ERα. Mutually exclusive expression of Hbo1 and ERα was observed in roughly half of the human breast tumors examined in the present study. Modulation of ER stability by Hbo1 in breast cancers may provide a novel therapeutic possibility.
Approximately two‐thirds of breast cancers are dependent for proliferation and they respond to anti‐hormone therapy. However, some breast cancers show resistance against anti‐hormone therapy and recurrence occurs. The molecular mechanism of the resistance is largely unknown. Iizuka et al. uncovered a novel regulation of stability of estrogen receptor alpha, a central player for the estrogen‐dependent growth of breast cancers. They found that DNA replication‐associated histone acetyltransferase, Hbo1, promotes degradation of estrogen receptor alpha by ubiquitination.
Breast cancer is the most common cancer among women and causes 458 000 deaths per year worldwide.1 The estrogen receptor (ER), primarily ERα, has a central role in the proliferation of breast cancer. Approximately two‐thirds of human breast cancers are positive for ERα, and, therefore, are dependent on estrogen for proliferation and respond to anti‐estrogen therapy.2 However, a substantial number of ERα‐positive breast cancers become resistant to anti‐estrogens and recurrence occurs. Studies over the past decade have revealed that splicing variants of ERα, mutations of the ERα gene and alteration in the stability of the ERα protein are responsible for the resistance to anti‐estrogens.3, 4, 5
Ubiquitination of ERα is one of the mechanisms for ERα degradation. Several E3 ligases for ERα have been characterized. Mouse double minute 2 homolog (Mdm2) physically interacts with ERα, and enhances ubiquitination of ERα in vivo.6 The breast cancer susceptibility gene 1/Brca1‐associated RING domain protein 1 (BRCA1/BARD1) heterodimer ubiquitinates ERα and appears to inhibit transcriptional activity of ERα.7 The estrogen‐responsive finger protein (EFP), a member of the RING finger protein family, polyubiquitinates ERα through lysine 48 within the ubiquitin molecule and promotes degradation of ERα, although EFP contributes to ERα‐mediated transcription.8 The carboxyl‐terminus of heat shock protein 70 (Hsc70)‐interacting protein (CHIP), a ubiquitin ligase with U‐box domain, promotes ERα degradation in the nucleus.9
Hbo1, a histone acetyltransferase binding to origin recognition complex 1 (ORC1),10 regulates histone acetylation11, 12, 13, 14 and is involved in replicational licensing.15, 16 Hbo1 is required for adipogenesis,17 embryonic development13 and survival of erythroblasts in fetal liver.14 The Hbo1 protein complex includes inhibitor of growth 4 (ING4) or inhibitor of growth 5 (ING5), and gene for apoptosis and differentiation‐1 (Jade‐1).11 Tumor suppressor p53 inhibits histone acetyltransferase (HAT) activity of Hbo118 and Hbo1 is overexpressed in some human primary cancers,12 suggesting a link between Hbo1 and cancers. Indeed, overexpression of Hbo1 causes an increase in colony formation on soft agar in breast cancer cell lines.19 However, the molecular basis for growth control by Hbo1 in cancers is unclear. Here we report that Hbo1 promotes the degradation of ERα through ubiquitination. Hbo1 modulates transcription by ERα and growth of tamoxifen‐treated breast cancers.
Materials and Method
Reverse‐transcription and real‐time quantitative PCR
Total RNA was isolated with Trizol (Invitrogen, Carlsbad, CA, USA) or RNA iso‐Plus (TaKaRa Biotechnology, Otsu, Japan). Total RNA (0.5 μg) was reverse transcribed using random hexamers, oligo(dT) primer, and Prime‐Script RT regent kit (TaKaRa Biotechnology). Quantitative real‐time PCR analysis was performed using the Thermal Cycler Dice TM TP860 (TaKaRa Biotechnology) and the SYBR Premix EXTaqII (TaKaRa Biotechnology). Relative gene expression was determined following the δCT method. The data were normalized to GAPDH expression. Experiments were performed at least in triplicate. Synthetic oligonucleotides for PCR were purchased from Greiner Japan. Primer sequences for PCR are listed in Supplementary Table S1.
Plasmids, chemicals and antibodies
Mammalian expression plasmids for FLAG‐tagged ING4 and ING520 were gifts from Dr Curtis Harris. Expression plasmids for HA‐tagged wild‐type, K48R and K0 ubiquitins21 were donated by Dr Yuichi Machida. A mammalian expression plasmid for human ERα was made by inserting human ERα cDNA (a gift from Dr Shigeaki Kato) into pFLAG‐CMV‐2 plasmid. The Jade‐1 expression plasmid was an IMAGE clone, MGC40386. Cycloheximide and 17‐β‐estradiol (E2) were from Sigma–Aldrich (St. Louis, MO, USA). MG‐132 was from Biomol. The recombinant full‐length ERα protein was from Invitrogen. The His‐tagged Hbo1 recombinant protein was prepared as reported previously.18 The following commercial antibodies were used: anti‐α‐tubulin monoclonal and anti‐FLAG monoclonal (Sigma‐Aldrich); anti‐ERα and anti‐progesterone receptor (PgR) (Santa‐Cruz, Dallas, TX, USA); anti‐HA monoclonal (Roche, Basel, Switzerland); and anti‐acetylated histone H4 polyclonal (H4KAc) (Millipore, Billerica, MA, USA). Generation of anti‐Hbo1 antibody was documented.10, 12 Anti‐Jade‐1 antibody was generated by immunizing rabbits with a synthetic peptide LFTHLRQDLERVMIDTDTL. Two different siRNAs targeting nucleotide 92 (#1) and 1313 (#2) of the Hbo1 coding region were used. Knocking down by siRNA was undertaken as reported previously.18 Antibodies for immunohistochemistry are listed in Supplementary Table S2.
In vitro acetylation assay
Hbo1 or its mutant18 (1 μM) and recombinant ERα (0.5 μM) proteins were incubated in 20 μL of protein acetylation buffer (50 mM Tris, pH 8.0, 50 mM KCl, 5% glycerol and 1 mM EDTA) with 1 μL (0.02 μCi) of [14C]‐acetyl‐CoA (Perkin Elmer) for 1 h at 30°C. The proteins were separated on SDS‐PAGE, and transferred onto membrane, followed by fluorography.
In vivo ubiquitination assay
The in vivo ubiquitination assay was performed according to a previous report.22 In brief, 293T cells were transfected using the calcium‐phosphate‐mediated gene transfer method.23 Forty‐eight hours post‐transfection, the cells were treated with MG‐132 for 6 h, lysed with pre‐boiled buffer (50 mM Tis‐HCl, pH 7.5, 0.5 mM EDTA, 1% SDS, 1 mM DTT) and boiled for 10 min. After centrifugation, supernatant was diluted with buffer A (50 mM Tris‐Cl, 0.5% NP‐40, 150 mM NaCl, 50 mM NaF, 1 mM DTT, 1 mM NaVO3) with proteinase inhibitors, and incubated with the primary antibody and protein A beads. The immunoprecipitates were washed three times with buffer A, separated on SDS‐PAGE, transferred onto the membrane and subjected to western blotting.
Immunohistochemistry
A formalin‐fixed paraffin‐embedded section of a tissue microarray containing 24 breast tumors was immunostained using the EnVision FLEX system (DakoCytomation, Grostrup, Denmark) or the Histofine Simple Stain MAX‐PO kit (Nichirei, Tokyo, Japan) according to the instruction manuals. The results were evaluated as follows: negative, <10% of tumor cells stained; weakly posivite, 10–50% of tumor cells stained; and strongly positive, >50% of tumor cells stained.
Statistical analysis
Statistical data analysis of the mRNA expression levels was conducted using a one‐factor anova and the Tukey–Kramer test. The differences were considered significant at P < 0.05. The relationship between protein levels in the immunohistochemistry study was analyzed using cross‐tabulation and Pearson's χ2.
Results
Hbo1 promotes destabilization of estrogen receptor α protein
Hbo1 modulates ER‐dependent transcription.24 However, it remained a question whether Hbo1 physically interacts with ERα. We were unable to demonstrate interaction between endogenous Hbo1 and ERα proteins by coimmunoprecipitation experiments under multiple conditions, suggesting that the interaction might be weak or dynamic. To circumvent the difficulty, we lysed 293T cells transfected with Hbo1 and ERα, and immunoprecipitated the whole cell extracts with anti‐Hbo1 antibody. The immunoprecipitates were assayed for ERα protein by immunoblotting (Fig. 1a). ERα was coimmunoprecipitated with Hbo1 (lane 4), whereas no ERα signal was detected in the control reaction performed by immunoprecipitaion with the purified rabbit IgG (lane 8), indicating that Hbo1 and ERα are in the same protein complex. To test whether Hbo1 acetylates ERα in vitro, a recombinant wild‐type Hbo1 protein and a catalytically‐inactive mutant (G485A) of Hbo1 were incubated with the recombinant ERα protein in the presence of 14C‐labelled acetyl‐CoA, and acetylation was detected by fluorography (Fig. 1b). The wild‐type Hbo1 acetylated ERα protein (lane 1) in addition to Hbo1 itself, whereas the catalytically‐inactive mutant Hbo1 did not (lane 2), suggesting that Hbo1 acetylates ERα in vitro. To explore the biological significance of physical interaction between Hbo1 and ERα, we assayed protein and mRNA amounts of ERα by immunoblotting (Fig. 1c) and RT‐PCR analysis (Fig. 1d), respectively, after knocking down Hbo1 expression in a breast cancer cell line, MCF‐7. Depletion of Hbo1 caused an increase in ERα protein (second panel). An equivalent amount of protein was loaded onto each lane judged by blotting with anti‐α‐tubulin antibody (third panel). Decrease in histone H4 acetylation upon Hbo1 depletion (fourth panel) has been reported previously.12 In contrast, knockdown of Hbo1 expression caused a decrease in ERα mRNA (Fig. 1d), suggesting that the increase in ERα protein levels upon Hbo1 knockdown was not due to transcriptional activation of the ERα gene. Therefore, we tested whether Hbo1 affected the stability of the ERα protein. Following knockdown of Hbo1, the MCF‐7 cells were treated with cycloheximide, an inhibitor of protein synthesis, and were assayed for ERα expression (Fig. 1e). The ERα protein started to degrade 30 min after cycloheximde treatment in the control siRNA‐transfected cells (lanes 1–4), whereas the ERα protein was stable in the Hbo1 knocked‐down cells for up to 90 min (lanes 5–8). This result suggests that Hbo1 negatively regulates the stability of the ERα protein.
Figure 1.
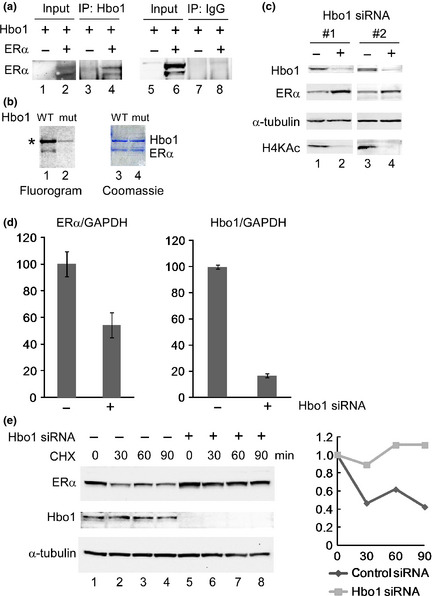
Hbo1 contributes to destabilization of estrogen receptor α (ERα) protein. (a) 293T cells were transfected with Hbo1 and/or ERα, lysed, and immunoprecipitated with anti‐Hbo1 antibody (lanes 3 and 4) and purified rabbit IgG (lanes 7 and 8). The immunoprecipitates were separated on SDS‐PAGE, transferred onto membrane, and probed with anti‐ERα antibody. Transfection of ERα into 293T cells reproducibly gave two bands corresponding to ERα, presumably due to post‐translational modifications. Ten percent of the input was loaded. (b) Recombinant Hbo1 proteins, wild‐type (WT) (lanes 1 and 3) and catalytically‐inactive mutant (mut) (lanes 2 and 4), were incubated with recombinant ERα in the presence of 14C‐ acetyl‐CoA, separated on SDS‐PAGE, followed by fluorography and Coomassie staining (Sigma‐Aldrich). *Autoacetylation of Hbo1. (c) MCF‐7 whole cell extracts transfected with control (−) (lanes 1 and 3) and two different Hbo1 (+) (lanes 2 and 4) siRNA oligonucleotides were probed with the listed antibodies. (d) MCF‐7 cells were transfected with Hbo1 or control siRNA and the expression of ERα mRNA was analyzed by qRT‐PCR. A ratio of the ERα or Hbo1 mRNA to GAPDH mRNA in the control siRNA‐treated cells was arbitrarily set to 100. Each value is represented as a mean ± SD. (e) Three days post‐transfection of Hbo1 siRNAs, MCF‐7 cells were treated with cycloheximide (CHX) for indicated time, lysed and immunoblotted with the listed antibodies (left). The ratio of intensity of the ERα to α‐tubulin was plotted with the time of CHX treatment (right).
Hbo1 is ubiquitinated
Treatment of MCF‐7 cells with MG‐132 caused an increased signal of ERα protein in the absence of the ligand, indicating that ERα stability is regulated in a ubiquitin‐dependent fashion.25 Moreover, several histone acetyltransferases, including cAMP response element binding protein‐binding protein (CBP), p300, P300/CBP‐associated factor (PCAF) and TATA box binding protein (TBP)‐associated factor 250 kDa (TAFII250) and one histone deacetylase (HDAC), HDAC6, have ubiquitin‐associated capabilities.26 These findings suggest that Hbo1 contributes to ubiquitin‐dependent ERα degradation. We examined whether Hbo1 was ubiquitinated. We transfected 293T cells with Hbo1 and HA‐tagged ubiquitin, and immunoprecipitated under denaturing conditions with anti‐Hbo1 antibody, and the immunoprecipitates were assayed for HA‐ubiquitin by immunoblotting (Fig. 2a). The slow‐migrating bands above the size of Hbo1 (approximately 80 kDa) solely in the cells with overexpressed Hbo1 (lane 6) but not in the cells transfected with the empty vector (lane 5) suggested that the Hbo1 protein was ubiquitinated.
Figure 2.
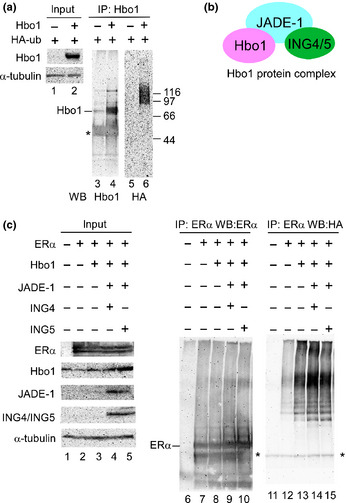
Ubiquitination of Hbo1 and promotion of estrogen receptor α (ERα) ubiquitination by Hbo1. (a) Hbo1 (lanes 2, 4 and 6) and the empty vector (lanes 1, 3 and 5), together with HA‐tagged ubiquitin, were transfected into 293T cells. Two days post‐transfection, the cells were lysed under denaturing conditions and immunoprecipitated with anti‐Hbo1 antibody. The immunoprecipitates were assayed for Hbo1 (lanes 3 and 4) and HA peptide (lanes 5 and 6). Ten percent of the input was analyzed. *Immunoglobulin heavy chain. (b) Components of the Hbo1 protein complex. (c) 293T cells were transfected with ERα, Hbo1, Jade‐1, ING4 or ING5, with HA‐tagged ubiquitin by the listed combination. Immunoprecipitation with anti‐ERα antibody was done similarly to Figure 2(a). The immunoprecipitates were probed with anti‐ERα and anti‐HA antibodies. Ten percent of the input was analyzed. *Immunoglobulin heavy chain.
The Hbo1 protein complex promotes estrogen receptor α ubiquitination in vivo
To examine whether Hbo1 promotes ERα ubiquitination, we overexpressed Hbo1, ING4 or ING5, and Jade‐1, which are components of the Hbo1 protein complex (Fig. 2b),11 along with HA‐ubiquitin and ERα, and immunoprecipitated the whole cell extracts with anti‐ERα antibody under denaturing conditions. The immunoprecipitates were assayed for ubiquitin (Fig. 2c). An equivalent amount of ERα was precipitated among the cells transfected with ERα (lanes 7–10). Remarkable slow‐migrating poly‐ubiquitin signals were detected in the Hbo1, Hbo1/ING4/Jade‐1 or Hbo1/ING5/Jade‐1 transfectants (lanes 13–15), whereas the ERα‐only transfectant showed a weak intensity of poly‐ubiquitination (lane 12). The protein complexes containing Hbo1/ING4/Jade‐1 and Hbo1/ING5/Jade‐1 both appeared to have an equivalent level of ubiquitinating activity for ERα (lanes 14 and 15). Thus, Hbo1 contributes to ERα ubiquitination in vivo.
The HAT activity of Hbo1 is linked with its ubiquitinating activity
The HAT activity of Hbo1 is linked to global histone acetylation12 and replicational licensing.18, 27 To check whether the Hbo1 HAT activity is associated with its ability to ubiquitinate ERα, we transfected Hbo1 (G485A), which is deficient in HAT activity,12 with ING4 or ING5 and Jade‐1, and assayed its ability to ubiquitinate ERα in vivo (Fig. 3a). The Hbo1 mutant showed a marked decrease in ERα ubiquitination compared to the wild‐type, irrespective of ING4 or ING5 transfected (lanes 9 vs 10 and lanes 11 vs 12), suggesting that the HAT activity of Hbo1 is linked to its ability to ubiquitinate the ERα protein.
Figure 3.
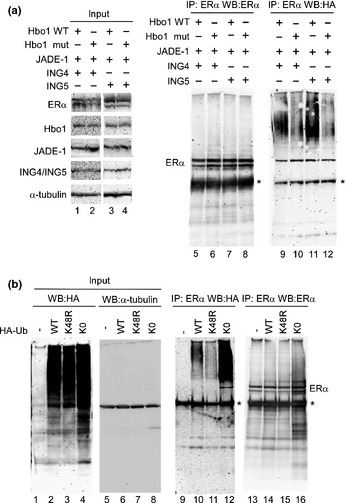
Poly‐ubiquitination of estrogen receptor α (ERα) by Hbo1 is dependent on the histone acetyltransferase (HAT) activity of Hbo1 and linked through lysine 48 of the ubiquitin. (a) Hbo1 wild‐type (Hbo1 WT) and the catalytically‐deficient mutant, Hbo1 (G485A) (Hbo1 mut), were transfected into 293T cells with the other Hbo1 complex components, and ERα and HA‐ubiquitin, and analyzed similarly to Figure 2(a). Ten percent of the input was loaded. (b) The ING4‐containing components of Hbo1 protein complex, and ERα were transfected with the empty vector, wild‐type ubiquitin, K48R mutant, or K0 mutant tagged with the HA epitope. The transfectants were handled in a similar fashion to Figure 2(a). *Immunoglobulin heavy chain.
Poly‐ubiquitination of estrogen receptor α by Hbo1 through lysine 48 of ubiquitin
The specific lysine position within the ubiquitin molecule is used to connect multiple ubiquitin moieties and the position is closely linked to the biological fate of the proteins tagged with ubiquitin. For instance, proteins modified with multiple ubiquitins through lysine 48 are destined for their degradation by the proteasome. In contrast, being tagged with ubiquitins through lysine 63 is associated with other biological events including DNA repair and signal transduction.28, 29 As Hbo1 knockdown caused increased stability of the ERα protein (Fig. 1c,e), the ubiquitination of the ERα protein by Hbo1 through the lysine at position 48 was predicted. To examine the ubiquitin linkage, we transfected into 293T cells a ubiquitin mutant (K48R), whose lysine at position 48 is substituted to arginine, with Hbo1, ING4, Jade‐1 and ERα, and analyzed the ubiquitination of the ERα protein (Fig. 3b). As a negative control, we transfected ubiquitin K0 mutant where all the lysines are substituted to arginine (lanes 4, 8, 12 and 16). With an equivalent amount of ERα pulled down among the transfectants (lanes 13–16), the wild‐type ubiquitin produced a poly‐ubiquitination (lane 10), whereas the K48R mutant displayed a marked decrease of poly‐ubiquitination (lane 11), suggesting that the poly‐ubiquitination of ERα by Hbo1 was through lysine 48 of the ubiquitin molecule. The ubiquitin K0 mutant gave a robust signal of ERα ubiquitination (lane 12), in accordance with the previous study using the K0 mutant.21 Presumably, expression of the K0 mutant was not sufficient to suppress ubiquitin elongation of ERα due to an abundance of endogenous ubiquitin.
Hbo1 depletion potentiated growth suppression of anti‐estrogen‐treated breast cancer cells
Tamoxifen, a competitive inhibitor of estradiol binding to the ER, has been effective for the therapy of ER‐positive breast cancer.2 In contrast, Hbo1 depletion causes inhibition of proliferation of 293T cells.11 To test whether Hbo1 depletion affects the growth of breast cancer cells treated with tamoxifen, we examined growth suppression of breast cancer cell line MCF‐7 by tamoxifen following knocking down Hbo1 expression (Fig. 4). Tamoxifen treatment per se (22% reduction in growth compared to the non‐treated control) and Hbo1 depletion per se (20% reduction in growth for siRNA#1 and 34% reduction for siRNA#2, compared to the siRNA control) 1 had a comparable level of growth suppression. Hbo1 knockdown plus tamoxifen treatment had the more growth suppression (36% reduction in growth for #1 and 65% reduction in growth for #2).
Figure 4.
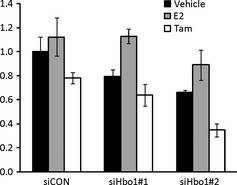
Hbo1 influences growth suppression of anti‐estrogen‐treated breast cancer cell line. MCF‐7 cells were transfected with control (siCON) or Hbo1 (siHbo1 #1, #2) siRNA oligonucleotides. Twenty‐four hours post‐transfection, the cells were treated with vehicle (black), E2 (10−7 M) (grey) or tamoxifen (10−7 M) (white) for 72 h. Cell numbers were counted. The average number of cells treated with control siRNA and vehicle was arbitrarily set to 1.
Hbo1 modulates transcription by estrogen receptor α
By binding to a ligand, ERα modulates the transcription of downstream genes.30 As Hbo1 contributes to the destabilization of the ERα protein, we predicted that knockdown of Hbo1 caused an increase in expression of the ERα protein, leading to potentiating estrogen‐dependent transcription of the downstream genes. We depleted Hbo1 expression by siRNA, challenged with E2, and analyzed the expression of 15 genes whose expression is known to be activated in an ERα‐dependent fashion (Fig. 5).31, 32 The effect of Hbo1 depletion was categorized into three groups: repression (4 genes), activation (3 genes), and no change (8 genes) of the E2‐dependent transcription. Unexpectedly, expression of four genes, E2F1, ribonucleotide reductase M2 (RRM2), cathepsin D (CTSD) and NRGN, was repressed by Hbo1 depletion (Fig. 5a). The decrease in transcription was probably due to reduced acetylation around the promoters because Hbo1 is bound to some of the active gene promoters.33 In contrast, three genes, PgR, SDF1 and interleukin‐24 (IL24), were activated by Hbo1 depletion (Fig. 5b), in agreement with the idea that Hbo1 promotes ubiquitin‐dependent degradation of ERα.
Figure 5.
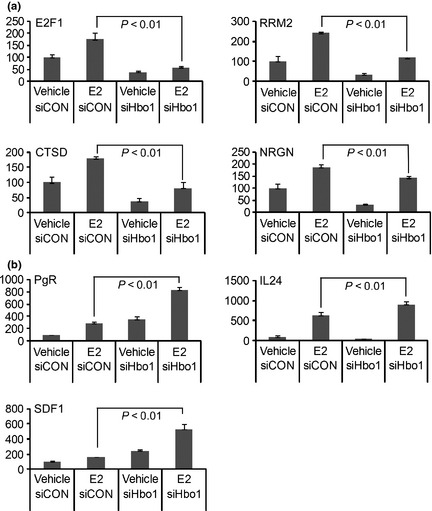
Hbo1 activates (a) and represses (b) transcription by estrogen receptor α (ERα). MCF‐7 cells were transfected with siRNA oligonucleotides for control (−) (siCON) or Hbo1 (+) (siHbo1) for 48 h followed by treatment with E2 (2 × 10−8 M) for 24 h. Amount of mRNA was quantified by qRT‐PCR. GAPDH mRNA was used as an expression standard. A ratio of mRNA to GAPDH in the vehicle‐treated cells transfected with the control siRNA was set to 100.
Immunohistochemistry study of Hbo1, ING4, ING5 and Jade‐1
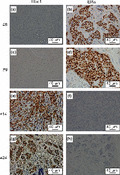
To explore the in vivo biological role of the Hbo1 complex in breast cancers, we examined the expression of Hbo1, ING4, ING5 and Jade‐1 in 24 primary breast cancers by immunohistochemistry. The breast cancers were composed of 3 non‐invasive carcinomas and 21 invasive carcinomas (1 well differentiated, 10 moderately differentiated and 9 poorly differentiated carcinomas) (Fig. 6, Table 1, Suppl. Table S3). Staining of Hbo1, ING4, ING5 and Jade‐1 was nuclear, nuclear and/or cytoplasmic, nuclear and/or cytoplasmic, and cytoplasmic, respectively. Cytoplasmic staining of ING4,34 ING535 and Jade‐136 by immunohistochemistry has been described previously. Out of the 24 cancers, 5 (21%) were positive for Hbo1 and 9 were positive for Jade‐1 (38%). In agreement with the destabilization of ERα by Hbo1, 11 breast tumors (46%) showed mutually exclusive expression of Hbo1 and ERα (Patients 1, 5, 6, 8, 9, 14, 16, 18, 19, 20 and 24; Table 1, Fig. 7), whereas the remaining 13 cases did not. We tested whether this mutually exclusive expression was statistically significant. We examined expression of ERα, PgR and HER2 by immunohistochemistry to see whether the staining pattern of Hbo1 was correlated with that of those factors important for expression profiles of breast cancers37 by pair‐wise χ2 analysis (Suppl. Table S4). As progesterone receptor is highly E2‐sensitive,31 there was a strong correlation between expression of ER and progesterone receptor (P < 0.01). However, Hbo1 expression had no statistically significant correlation with ERα expression.
Figure 6.
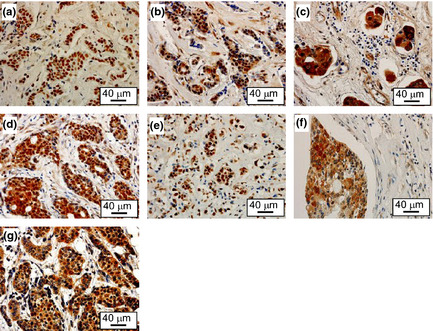
Overexpression of Hbo1, ING4, ING5 and Jade‐1 in human primary breast tumors. The immunohistochemical study showing staining of Hbo1 (a), nuclear staining of ING4 (b), cytoplasmic staining of ING4 (c), nuclear and cytoplasmic staining of ING4 (d), nuclear staining of ING5 (e), cytoplasmic staining of ING5 (f), and cytoplasmic staining of Jade‐1 (g). Magnification: ×400.
Table 1.
Pathological features and staining pattern of 24 breast tumors
| Patient | Invasiveness | Differentiation | Age | Hbo1 | ING4 | ING5 | Jade‐1 | ER | PgR | HER2 |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Non | N/A | 43 | – | sNC | wN | s | w | s | – |
| 2 | Non | N/A | 42 | – | wC | wNC | – | – | – | – |
| 3 | Non | N/A | 54 | – | sC | sC | – | – | – | s |
| 4 | Non | N/A | 80 | – | sC | – | – | – | – | – |
| 5 | Inv | well | 44 | – | sNC | sN | s | s | s | – |
| 6 | Inv | mod | 42 | – | sC | – | w | w | s | – |
| 7 | Inv | mod | 69 | – | sC | wC | s | – | – | w |
| 8 | Inv | mod | 64 | – | sC | – | – | s | s | – |
| 9 | Inv | mod | 82 | – | sNC | – | s | s | s | – |
| 10 | Inv | mod | 71 | – | sC | wC | s | – | – | s |
| 11 | Inv | mod | 68 | – | sC | wC | s | – | s | – |
| 12 | Inv | mod | 51 | s | sNC | wNC | s | s | – | s |
| 13 | Inv | mod | 50 | – | wC | – | – | – | – | s |
| 14 | Inv | mod | 57 | s | sC | – | – | – | – | – |
| 15 | Inv | mod | 29 | – | sC | – | – | – | – | – |
| 16 | Inv | poorly | 39 | – | sC | wC | – | s | s | – |
| 17 | Inv | poorly | 63 | s | sN | sN | – | s | s | w |
| 18 | Inv | poorly | 52 | – | sC | – | w | s | s | – |
| 19 | Inv | poorly | 61 | – | wC | – | – | w | – | – |
| 20 | Inv | poorly | 71 | – | wC | sNC | – | s | w | w |
| 21 | Inv | poorly | 35 | – | sC | wC | – | – | – | – |
| 22 | Inv | poorly | 65 | – | – | – | – | – | – | – |
| 23 | Inv | poorly | 41 | s | sC | sN | – | s | w | w |
| 24 | Inv | poorly | 42 | w | sC | – | – | – | w | – |
–, negative; C, cytoplasmic; Inv, invasive; mod, moderately; N, nuclear; N/A, not applied; Non, non‐invasive; s, strong; w, weak.
Figure 7.
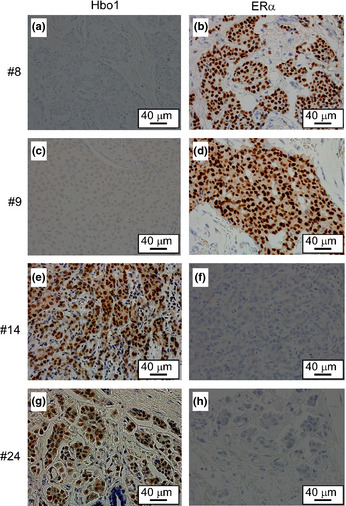
Mutually exclusive expression of Hbo1 and estrogen receptor α (ERα) in human primary breast tumors. The immunohistochimestry staining of the tumors from the patients 8, 9, 14 and 24 with anti‐Hbo1 (a, c, e and g) and anti‐ERα (b, d, f and g) antibodies. Magnification: ×400.
Discussion
Here we report a novel function of Hbo1: promotion of ERα degradation through ubiquitination. Two possible mechanisms for ERα degradation by Hbo1 are conceivable. The first is: as Hbo1 regulates global acetylation of core histones,11, 12, 13, 14 Hbo1 might activate the transcription of ubiquitinating enzyme(s) for ERα through changes in the chromatin structure by altering acetylation levels of core histones. However, even a large input of exogenous Hbo1, which induced ERα ubiquitination (Fig. 2c), did not cause changes in core histone acetylation.38 Therefore, it is unlikely that ERα ubiquitination by transfection of Hbo1 (Fig. 2c) is caused by alteration of global histone acetylation. A second possibility is that aceylation of ERα by Hbo1 facilitates ubiquitination and proteasomal degradation of ERα. Acetylation‐induced ubiquitination/proteasomal degradation have been described for some proteins, including histone H2A.X,39 DNA methyltransferase 1 (DNMT1),40 phosphoenolpyruvate carboxylase 1 (PCK1),41 p2742 and core histones.43 This possibility is consistent with the in vitro acetylation of ERα by Hbo1 (Fig. 1b) and the close link between the Hbo1 HAT activity and ERα ubiquitination (Fig. 3a).
Hbo1 activated and suppressed transcription by ERα (Fig. 5). Hbo1 activated the basal level of E2F1, RRM2 and CTSD genes and suppressed IL24 transcription. E2F1 can stimulate proliferation.44 RRM2 plays a direct role in tumor progression.45 CTSD, a proteolytic enzyme, is abundant in cancers and is potential tumor marker.46 IL24 can induce apoptosis in cancer cells.47 Thus, Hbo1 might coordinately alter the expression profile through changes in histone acetylation and ERα stability to link cellular proliferation with initiation of DNA replication. In this context, it is of interest that Hbo1 depletion enhanced the growth suppression of tamoxifen‐treated MCF‐7 cells (Fig. 4). As ING5 depletion causes tamoxifen sensitivity,48 we speculate that Hbo1, as a protein complex, modulates the survival of tamoxifen‐treated breast cancer cells.
Since the identification of protein complex including Hbo1, Jade‐1, and ING4/5,11 it has been enigmatic that the proteins with contrasting functions are in the same protein complex, because Hbo1 acts as oncogene, whereas ING4/5 and Jade‐1 behave as anti‐oncogenes. Positive regulation of DNA replication by Hbo115 and other findings11, 12, 18, 19 indicate that Hbo1 is an oncogene. In contrast, ING4 and ING5 are tumor suppressors.20, 49, 50, 51 Moreover, Jade‐1 contributes to apoptosis and tumor suppression.52, 53 The integrity of Hbo1/Jade‐1/ING protein complex appears to be important in the suppression of cellular proliferation.33, 54 However, the antagonism between Hbo1 and Jade‐1 does not explain why the transcriptional profiles are not similar between the Hbo1‐depleted cells and the Jade‐1L overexpressing cells.33 Havasi et al. (2013) report that Jade‐1 depletion inhibits DNA replication and cell proliferation,55 indicating a possibility that Jade‐1 is oncogenic. ERα ubiquitination promoted by Hbo1 protein complex as well as Hbo1 alone (Fig. 2c) supports the notion.
In sum, we report that Hbo1 contributes to degradation of ERα by ubiquitination through lysine 48. As this control of ERα ubiquitination by Hbo1 is linked to histone acetyltransferase activity of Hbo1 (Fig. 3) and Hbo1 acetylase activity is important for replicational licensing,18, 27 Hbo1 might be a molecule linking activated DNA replication, enhanced ubiquitin‐dependent proteasomal degradation of ERα and resistance to anti‐estrogen in breast cancers.
Disclosure Statement
The authors have no conflict of interest.
Supporting information
Table S1. Sequences for PCR primers.
Table S2. Antibodies for immunohistochemistry.
Table S3. Pathological details of 24 breast tumors.
Table S4. Correlation of expression of Hbo1 in relation to ER, PgR and HER2 in breast cancers.
Acknowledgments
This work was supported by a grant from the Science Research Promotion Fund from the Promotion and Mutual Aid Corporation for Private Schools of Japan and a grant from the Ministry of Education, Science, Sports and Culture, Japan. We thank Masato Watanabe for technical assistance and Drs Curtis Harris, Shigeaki Kato, Yuichi Machida and Tomohiko Ohta for reagents. We also thank Dr Koji Okamoto for critical reading of the manuscript.
(Cancer Sci 2013; 104: 1647–1655)
References
- 1. Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer 2010; 127: 2893–917. [DOI] [PubMed] [Google Scholar]
- 2. Brunton LL, Chabner BA, Knollmann BC. Goodman and Gilman's The Pharmacological Basis of Therapeutics, 12th edn. New York: McGraw‐Hill Professional, 2010. [Google Scholar]
- 3. Weinberg OK, Marquez‐Garban DC, Pietras RJ. New approaches to reverse resistance to hormonal therapy in human breast cancer. Drug Resist Updat 2005; 8: 219–33. [DOI] [PubMed] [Google Scholar]
- 4. Billam M, Witt AE, Davidson NE. The silent estrogen receptor–Can we make it speak? Cancer Biol Ther 2009; 8: 485–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Barone I, Brusco L, Fuqua SA. Estrogen receptor mutations and changes in downstream gene expression and signaling. Clin Cancer Res 2010; 16: 2702–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Duong V, Boulle N, Daujat S et al Differential regulation of estrogen receptor alpha turnover and transactivation by Mdm2 and stress‐inducing agents. Cancer Res 2007; 67: 5513–21. [DOI] [PubMed] [Google Scholar]
- 7. Ma Y, Fan S, Hu C et al BRCA1 regulates acetylation and ubiquitination of estrogen receptor‐alpha. Mol Endocrinol 2010; 24: 76–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nakajima A, Maruyama S, Bohgaki M et al Ligand‐dependent transcription of estrogen receptor alpha is mediated by the ubiquitin ligase EFP. Biochem Biophys Res Commun 2007; 357: 245–51. [DOI] [PubMed] [Google Scholar]
- 9. Fan M, Park A, Nephew KP. CHIP (carboxyl terminus of Hsc70‐interacting protein) promotes basal and geldanamycin‐induced degradation of estrogen receptor‐alpha. Mol Endocrinol 2005; 19: 2901–14. [DOI] [PubMed] [Google Scholar]
- 10. Iizuka M, Stillman B. Histone acetyltransferase HBO1 interacts with the ORC1 subunit of the human initiator protein. J Biol Chem 1999; 274: 23027–34. [DOI] [PubMed] [Google Scholar]
- 11. Doyon Y, Cayrou C, Ullah M et al ING tumor suppressor proteins are critical regulators of chromatin acetylation required for genome expression and perpetuation. Mol Cell 2006; 21: 51–64. [DOI] [PubMed] [Google Scholar]
- 12. Iizuka M, Takahashi Y, Mizzen CA et al Histone acetyltransferase Hbo1: catalytic activity, cellular abundance, and links to primary cancers. Gene 2009; 436: 108–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kueh AJ, Dixon MP, Voss AK, Thomas T. HBO1 is required for H3K14 acetylation and normal transcriptional activity during embryonic development. Mol Cell Biol 2011; 31: 845–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mishima Y, Miyagi S, Saraya A et al The Hbo1‐Brd1/Brpf2 complex is responsible for global acetylation of H3K14 and required for fetal liver erythropoiesis. Blood 2011; 118: 2443–53. [DOI] [PubMed] [Google Scholar]
- 15. Iizuka M, Matsui T, Takisawa H, Smith MM. Regulation of replication licensing by acetyltransferase Hbo1. Mol Cell Biol 2006; 26: 1098–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Miotto B, Struhl K. HBO1 histone acetylase is a coactivator of the replication licensing factor Cdt1. Genes Dev 2008; 22: 2633–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Johmura Y, Osada S, Nishizuka M, Imagawa M. FAD24 acts in concert with histone acetyltransferase HBO1 to promote adipogenesis by controlling DNA replication. J Biol Chem 2008; 283: 2265–74. [DOI] [PubMed] [Google Scholar]
- 18. Iizuka M, Sarmento OF, Sekiya T, Scrable H, Allis CD, Smith MM. Hbo1 Links p53‐dependent stress signaling to DNA replication licensing. Mol Cell Biol 2008; 28: 140–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hu X, Stern HM, Ge L et al Genetic alterations and oncogenic pathways associated with breast cancer subtypes. Mol Cancer Res 2009; 7: 511–22. [DOI] [PubMed] [Google Scholar]
- 20. Shiseki M, Nagashima M, Pedeux RM et al p29ING4 and p28ING5 bind to p53 and p300, and enhance p53 activity. Cancer Res 2003; 63: 2373–8. [PubMed] [Google Scholar]
- 21. Machida YJ, Machida Y, Vashisht AA, Wohlschlegel JA, Dutta A. The deubiquitinating enzyme BAP1 regulates cell growth via interaction with HCF‐1. J Biol Chem 2009; 284: 34179–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nishikawa H, Ooka S, Sato K et al Mass spectrometric and mutational analyses reveal Lys‐6‐linked polyubiquitin chains catalyzed by BRCA1‐BARD1 ubiquitin ligase. J Biol Chem 2004; 279: 3916–24. [DOI] [PubMed] [Google Scholar]
- 23. Green MR, Sambrook J. Molecular Cloning, A Laboratory Manual, 4th edn. New York: Cold Spring Harbor, 2012. [Google Scholar]
- 24. Georgiakaki M, Chabbert‐Buffet N, Dasen B et al Ligand‐controlled interaction of histone acetyltransferase binding to ORC‐1 (HBO1) with the N‐terminal transactivating domain of progesterone receptor induces steroid receptor coactivator 1‐dependent coactivation of transcription. Mol Endocrinol 2006; 20: 2122–40. [DOI] [PubMed] [Google Scholar]
- 25. Tateishi Y, Kawabe Y, Chiba T et al Ligand‐dependent switching of ubiquitin‐proteasome pathways for estrogen receptor. EMBO J 2004; 23: 4813–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sadoul K, Boyault C, Pabion M, Khochbin S. Regulation of protein turnover by acetyltransferases and deacetylases. Biochimie 2008; 90: 306–12. [DOI] [PubMed] [Google Scholar]
- 27. Miotto B, Struhl K. HBO1 histone acetylase activity is essential for DNA replication licensing and inhibited by Geminin. Mol Cell 2010; 37: 57–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Haglund K, Dikic I. Ubiquitylation and cell signaling. EMBO J 2005; 24: 3353–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Skaug B, Jiang X, Chen ZJ. The role of ubiquitin in NF‐kappaB regulatory pathways. Annu Rev Biochem 2009; 78: 769–96. [DOI] [PubMed] [Google Scholar]
- 30. Ikeda K, Inoue S. Estrogen receptors and their downstream targets in cancer. Arch Histol Cytol 2004; 67: 435–42. [DOI] [PubMed] [Google Scholar]
- 31. Coser KR, Chesnes J, Hur J, Ray S, Isselbacher KJ, Shioda T. Global analysis of ligand sensitivity of estrogen inducible and suppressible genes in MCF7/BUS breast cancer cells by DNA microarray. Proc Natl Acad Sci U S A 2003; 100: 13994–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hall JM, Korach KS. Stromal cell‐derived factor 1, a novel target of estrogen receptor action, mediates the mitogenic effects of estradiol in ovarian and breast cancer cells. Mol Endocrinol 2003; 17: 792–803. [DOI] [PubMed] [Google Scholar]
- 33. Avvakumov N, Lalonde ME, Saksouk N et al Conserved molecular interactions within the HBO1 acetyltransferase complexes regulate cell proliferation. Mol Cell Biol 2012; 32: 689–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang QS, Li M, Zhang LY et al Down‐regulation of ING4 is associated with initiation and progression of lung cancer. Histopathology 2010; 57: 271–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zheng HC, Xia P, Xu XY, Takahashi H, Takano Y. The nuclear to cytoplasmic shift of ING5 protein during colorectal carcinogenesis with their distinct links to pathologic behaviors of carcinomas. Hum Pathol 2011; 42: 424–33. [DOI] [PubMed] [Google Scholar]
- 36. Lian X, Duan X, Wu X et al Expression and clinical significance of von Hippel‐Lindau downstream genes: Jade‐1 and beta‐catenin related to renal cell carcinoma. Urology 2012; 80(485): e7–13. [DOI] [PubMed] [Google Scholar]
- 37. Perou CM, Sorlie T, Eisen MB et al Molecular portraits of human breast tumours. Nature 2000; 406: 747–52. [DOI] [PubMed] [Google Scholar]
- 38. Foy RL, Song IY, Chitalia VC et al Role of Jade‐1 in the histone acetyltransferase (HAT) HBO1 complex. J Biol Chem 2008; 283: 28817–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ikura T, Tashiro S, Kakino A et al DNA damage‐dependent acetylation and ubiquitination of H2AX enhances chromatin dynamics. Mol Cell Biol 2007; 27: 7028–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Du Z, Song J, Wang Y et al DNMT1 stability is regulated by proteins coordinating deubiquitination and acetylation‐driven ubiquitination. Sci Signal 2010; 3: ra80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jiang W, Wang S, Xiao M et al Acetylation regulates gluconeogenesis by promoting PEPCK1 degradation via recruiting the UBR5 ubiquitin ligase. Mol Cell 2011; 43: 33–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Perez‐Luna M, Aguasca M, Perearnau A et al PCAF regulates the stability of the transcriptional regulator and cyclin‐dependent kinase inhibitor p27 Kip1. Nucleic Acids Res 2012; 40: 6520–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Qian MX, Pang Y, Liu CH et al Acetylation‐mediated proteasomal degradation of core histones during DNA repair and spermatogenesis. Cell 2013; 153: 1012–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Polager S, Ginsberg D. E2F – at the crossroads of life and death. Trends Cell Biol 2008; 18: 528–35. [DOI] [PubMed] [Google Scholar]
- 45. Furuta E, Okuda H, Kobayashi A, Watabe K. Metabolic genes in cancer: their roles in tumor progression and clinical implications. Biochim Biophys Acta 2010; 1805: 141–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Benes P, Vetvicka V, Fusek M. Cathepsin D–many functions of one aspartic protease. Crit Rev Oncol Hematol 2008; 68: 12–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tahara I, Miyake K, Hanawa H et al Systemic cancer gene therapy using adeno‐associated virus type 1 vector expressing MDA‐7/IL24. Mol Ther 2007; 15: 1805–11. [DOI] [PubMed] [Google Scholar]
- 48. Mendes‐Pereira AM, Sims D, Dexter T et al Genome‐wide functional screen identifies a compendium of genes affecting sensitivity to tamoxifen. Proc Natl Acad Sci U S A 2012; 109: 2730–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Garkavtsev I, Kozin SV, Chernova O et al The candidate tumour suppressor protein ING4 regulates brain tumour growth and angiogenesis. Nature 2004; 428: 328–32. [DOI] [PubMed] [Google Scholar]
- 50. Kim S, Chin K, Gray JW, Bishop JM. A screen for genes that suppress loss of contact inhibition: identification of ING4 as a candidate tumor suppressor gene in human cancer. Proc Natl Acad Sci U S A 2004; 101: 16251–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Shah S, Smith H, Feng X, Rancourt DE, Riabowol K. ING function in apoptosis in diverse model systems. Biochem Cell Biol 2009; 87: 117–25. [DOI] [PubMed] [Google Scholar]
- 52. Zhou MI, Foy RL, Chitalia VC et al Jade‐1, a candidate renal tumor suppressor that promotes apoptosis. Proc Natl Acad Sci U S A 2005; 102: 11035–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chitalia VC, Foy RL, Bachschmid MM et al Jade‐1 inhibits Wnt signalling by ubiquitylating beta‐catenin and mediates Wnt pathway inhibition by pVHL. Nat Cell Biol 2008; 10: 1208–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hung T, Binda O, Champagne KS et al ING4 mediates crosstalk between histone H3 K4 trimethylation and H3 acetylation to attenuate cellular transformation. Mol Cell 2009; 33: 248–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Havasi A, Haegele JA, Gall JM et al Histone acetyl transferase (HAT) HBO1 and JADE1 in epithelial cell regeneration. Am J Pathol 2013; 182: 152–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Sequences for PCR primers.
Table S2. Antibodies for immunohistochemistry.
Table S3. Pathological details of 24 breast tumors.
Table S4. Correlation of expression of Hbo1 in relation to ER, PgR and HER2 in breast cancers.


