Abstract
Inorganic arsenic is known to be a human carcinogen. Previous studies have reported that DNA methylation changes are involved in arsenic‐induced carcinogenesis, therefore, DNA methylation changes that are specific to arsenic‐induced tumors would be useful to distinguish tumors induced by arsenic from tumors caused by other factors and to dissect arsenic carcinogenesis. Previous studies have shown that gestational arsenic exposure of C3H mice, which tend to spontaneously develop liver tumors, increases the incidence of tumors in male offspring. In this study we used the same experimental protocol as in those previous studies and searched for DNA regions where methylation status was specifically altered in the liver tumors of arsenic‐exposed offspring by using methylated DNA immunoprecipitation–CpG island microarrays. The methylation levels of the DNA regions selected were measured by quantitative methylation‐specific PCR and bisulfite sequencing. The results of this study clarified a number of regions where DNA methylation status was altered in the liver tumors in the C3H mice compared to normal liver tissues. Among such regions, we showed that a gene body region of the oncogene Fosb underwent alteration in DNA methylation by gestational arsenic exposure. We also showed that Fosb expression significantly increased corresponding to the DNA methylation level of the gene body in the arsenic‐exposed group. These findings suggest that the DNA methylation status can be used to identify tumors increased by gestational arsenic exposure.
This study further revealed that the gene body region of the oncogene Fosb is highly methylated and its expression is greatly increased in the tumors of gestationally arsenic‐exposed mice in comparison with the tumors of control mice. These results suggest that DNA methylation changes are involved in the alteration in tumor phenotype by gestational arsenic exposure.

Naturally occurring inorganic arsenic is known to cause serious health problems.1, 2 In many areas in the world, exposure to high concentrations of inorganic arsenic in drinking water has been found to increase the risk of cancer in the skin and several other organs, including the urinary bladder, lung, and liver.1 Epidemiological studies have also indicated that gestational arsenic exposure is associated with increased incidence of cancers in several organs, including the bladder and liver, in adulthood.3, 4 Despite its high carcinogenicity, arsenic has been documented to be a relatively weak mutagen.5 Consistent with the notion of arsenic being a weak mutagen, many studies have shown that arsenic induces aberrant epigenetic changes, mainly in DNA methylation and histone modification, and the arsenic‐induced epigenetic effects have been implicated in carcinogenesis.6, 7 However, consensus on the epigenetic effects of arsenic remains to be established, as some of these studies are still contradictory and mostly descriptive.6
The most studied epigenetic changes in cancer have included global DNA hypomethylation and site‐specific DNA hypermethylation.8 Global DNA hypomethylation is thought to play a role in inducing genome instability and thereby promoting carcinogenesis.9 Hypermethylation of CpG islands in promoter regions generally results in the transcriptional silencing of genes, and many different tumor suppressor genes have been found to be silenced by promoter hypermethylation in many types of cancers.8 Alterations in DNA methylation have already been applied as biomarkers that predict patient outcome and to choose treatments that take advantage of many unique features that are different from mutations.10, 11 If there exist alterations in DNA methylation that are specific to arsenic‐induced cancers, they should also be useful markers to distinguish arsenic‐induced cancers, and provide important clues to the selection of suitable treatments.
The C3H mouse has a high incidence of spontaneous liver tumors and chemically induced liver cancers,12 so this strain is often used in studies of induction of liver carcinogenesis by chemicals, including arsenic.12 Giving pregnant mice water containing a 42.5 or 85 ppm concentration of sodium arsenite to drink from days 8–18 of gestation has been found to dose‐dependently increase the incidence and numbers of liver tumors in their male offspring when examined at 74 weeks of age.13 Hypomethylation of the promoter region of ERα and upregulation of ERα expression have been found in the normal liver tissues of tumor‐bearing livers in a gestationally arsenic‐exposed group of C3H mice in comparison with the normal liver tissues of control mice.14 In a study in which we used the same experimental protocol we also detected an increase in liver tumors in C3H male mice,15 but we did not detect hypomethylation of the promoter region of ERα or upregulation of ERα expression.15 These findings indicated that changes in DNA methylation and ERα expression are not necessary for liver tumors to increase as a result of gestational arsenic exposure. Thus, it remains to be clarified whether there is an association between DNA methylation changes and increases in liver tumors after gestational arsenic exposure.
In this study we used MeDIP–CpG island microarray assays to identify DNA regions where methylation status is altered in the liver tumors of C3H mice in an experimental model of liver tumor induction by gestational arsenic exposure.13, 14, 15 We then searched for DNA regions where methylation status was specifically altered in the liver tumors of arsenic‐exposed C3H mice compared to the liver tumors of control mice by MSP and bisulfite sequencing. We also investigated the expression of genes that contained regions where DNA methylation was altered in the liver tumors of arsenic‐exposed C3H mice.
Materials and Methods
Sample preparation
The normal tissues and tumor tissues were obtained in our previous study15) from the livers of male 74–84‐week‐old offspring born to C3H mice mothers given drinking water or water containing 85 ppm sodium arsenite ad libitum from day 8 to 18 of gestation. The incidence of liver tumors in the offspring in the control group and arsenic‐exposed group was 51% and 41%, respectively. The predominant histological type of tumor was adenoma in both groups.15 The previous study revealed that the liver tumors in the arsenic‐exposed group had a higher rate of Ha‐ras mutation (71%) compared to the liver tumors in the control group (47%).15 The mice were handled in a humane manner in accordance with the National Institute for Environmental Studies (Tsukuba, Japan) guidelines for animal experiments.
Genomic DNA extraction
The liver tissues were lysed in lysis buffer (50 mM Tris‐HCl (pH 8.0), 0.1 M NaCl, 20 mM EDTA, 1% SDS, 0.3 mg/mL proteinase K). After RNase treatment, genomic DNA was purified with phenol chloroform mixture, precipitated with ethanol, dried, and resuspended in buffer containing 10 mM Tris‐HCl and 1 mM EDTA, pH 8.0.
Methylated DNA immunoprecipitation–CpG island microarray analysis
The MeDIP‐CpG island microarray analysis was carried out as previously described.16 Briefly, genome DNAs were prepared from pooled normal liver homogenates of three control mice and pooled tumor tissue homogenates of three arsenic‐exposed mice. Five micrograms of genome DNA of each group was immunoprecipitated with an anti‐5‐methylcytidine antibody (Diagenode, Liége, Belgium), and the precipitated DNA and the input DNA were labeled with Cy5 and Cy3, respectively. A mouse CpG island microarray that contains 97 652 probes covering 12 573 genes (Agilent Technologies, Santa Clara, CA, USA) was hybridized with the labeled probes and scanned with an Agilent G2565BA microarray scanner (Agilent Technologies). Scanned data were processed with Feature Extraction version 9.1 and Agilent G4477AA ChIP Analytics 1.3 software (both Agilent Technologies). A signal of a probe was converted into a ‘‘Me‐value’’ which represented the methylation level as a value from 0 (unmethylated) to 1 (methylated).16 Differentially methylated regions were detected by comparison of the Me‐values of the two groups.
Methylated control DNA and unmethylated control DNA
Fully methylated control DNA (CpG methylated NIH3T3 genomic DNA) was purchased from New England Biolabs (Ipswich, MA, USA), and fully unmethylated control DNA was prepared by amplifying NIH3T3 genomic DNA using GenomiPhi DNA Amplification Kit (GE Healthcare Bio‐Sciences, Little Chalfont, UK).
Bisulfite treatment
Bisulfite treatment was carried out as previously described.17 Briefly, genomic DNA, methylated control DNA, and unmethylated control DNA, prepared as described above, were digested with EcoRI and incubated with freshly prepared 0.3 M NaOH for 15 min. To this solution was added sodium metabisulfite, pH 5.0, and hydroquinone to give final concentrations of 2.0 M and 0.5 mM, respectively. The mixture was incubated for 16 h at 50°C in the dark. The samples were desalted with the Wizard DNA Clean‐up System (Promega, Madison, WI, USA), and the bisulfite reaction was terminated by adding NaOH to give a final concentration of 0.3 M and incubating for 15 min at 37°C. The DNA was then precipitated with ethanol, dried, and resuspended in buffer containing 10 mM Tris‐HCl and 1 mM EDTA, pH 8.0.
Methylation‐specific PCR
Methylation‐specific PCR was carried out by using bisulfite‐treated DNA and specific primers. The primer sequences and annealing temperatures used for MSP are shown in Table 1. The gel images were captured and visualized as described previously.18
Table 1.
Oligonucleotide primers for methylation‐specific PCR, bisulfite sequencing, and real‐time RT‐PCR
| Gene symbol | Status | Forward primer (5′–3′) | Reverse primer (5′–3′) | Annealing temp. (°C) |
|---|---|---|---|---|
| Fosb | M | tagagttggagtcggagatcgtc | gaccaacgaacctaaccccg | 60 |
| U | tagagttggagttggagattg | ccaaaaccatcttccttaaca | 60 | |
| BS | ttgttgggatttattaggaaattga | aaacttaaacttcactatatataaaaaaaa | 52 | |
| RT | gtgcgagcttccttgttttc | gtctccaacagccagaggag | 60 | |
| Btd | M | aagttttagcgttaaatatatc | aacaaactcgatatttacctcg | 56 |
| U | gaattgtgtttaaaatttagttgt | aaaacaaactcaatatttacctca | 56 | |
| BS | agggagagagttttttatggttgtt | aactaaacttcaaaacatttccaaaac | 56 | |
| Mab21l2 | M | atatgtttcgttttcgtttatc | ataatctcgcccaccgaattcg | 60 |
| U | agatatgttttgtttttgtttatt | aaataatctcacccaccaaattca | 60 | |
| BS | tttgggttattagtgtttgg | caaactaatttaccaactcaataaa | 52 | |
| Fam43a | M | agatgaagttgatcgtgagtgc | ctcataccgatatacccaaacg | 56 |
| U | ttaaagtttttgtttgggtatatt | aaacatttaaactctaccaaaaca | 56 | |
| Hspa2 | M | agagtattaatttcgacgaggc | cataacaccgccaactatttcg | 56 |
| U | taagagtattaattttgatgaggt | atcataacaccaccaactatttca | 56 | |
| Il1r1 | M | ttcggttgttttcggtttgaac | ttctcctacctaaacgtacccg | 56 |
| U | gttttggttgtttttggtttgaat | ttttctcctacctaaacataccca | 56 | |
| Cyp26a1 | M | tgcggagtcggagtaggggaac | actctataacctaaaaccgacg | 56 |
| U | gttggagtaggggaatgtaagttt | aacctaaaaccaacaaatcttaaca | 56 | |
| Adam11 | M | tattgggaggcgttcgagtttc | taaataaaaaacgtcgcttacg | 52 |
| U | gttattgggaggtgtttgagtttt | aataaataaaaaacatcacttaca | 56 | |
| B4galt6 | M | attagagagcgattggagattc | ttaaaacctaaaccgtaacccg | 56 |
| U | taattagagagtgattggagattt | atttaaaacctaaaccataaccca | 56 | |
| Cdh8 | M | gtggagtttggtgcggggatgc | ataaacctaaacgtcaacaccg | 60 |
| U | ttgtggagtttggtgtggggatgt | acataaacctaaacatcaacacca | 60 | |
| Tram1l1 | M | ttttgattgatcggtcggtagc | aattcctaacgttcttcttacg | 52 |
| U | gtttttgattgattggttggtagt | aaaattcctaacattcttcttaca | 52 | |
| 18S rRNA | RT | taccacatccaaggaaggcag | tgccctccaatggatcctc | 64 |
BS, primers for bisulfite sequencing; M, primers specific to methylated DNA; RT, primers for real‐time RT‐PCR; temp., temperature; U, primers specific to unmethylated DNA.
Real‐time MSP
Real‐time MSP was carried out by using bisulfite‐treated DNA and specific primers. The primers were designed for the gene body region in which Me‐value changes were detected by MeDIP–CpG island array assays. The approximate positions from transcription start site of the primers of Fosb, Btd, and Mab21l2 are indicated by arrows in Figures 2–4. The primer sequences and annealing temperatures used for real‐time MSP are shown in Table 1. LightCycler 480 software, version 1.5 (Roche Diagnostic, Basel, Switzerland) was used to compare amplification in bisulfite‐treated tissue samples during the log‐linear phase of the standard curve based on the results obtained in a dilution series of bisulfite‐treated control DNA. The PCR efficiencies were calculated from measurements of bisulfite‐treated fully methylated DNA by using M primers, or measurements of bisulfite‐treated fully unmethylated DNA by using U primers. All of the primers yielded similar efficiencies. DNA methylation levels were calculated as percentages as follows: (amount of DNA amplified with M primers) / [(amount of DNA amplified with M primers) + (amount of DNA amplified with U primers)] × 100.
Bisulfite sequencing analysis
Bisulfite sequencing was carried out as previously described.17 Briefly, the bisulfite‐treated DNA was amplified with the primers shown in Table 1. The PCR products were cloned into pGEM‐T Easy vector (Promega). Fourteen to 16 clones from each sample were cycle sequenced with M13RV primers and a BigDye Terminator version 3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA), and analyzed with an Applied Biosystems 3730 DNA analyzer (Applied Biosystems).
Preparation of cDNA and real‐time PCR
Total RNA of livers was prepared with an RNeasy Mini Kit (Qiagen, Valencia, CA, USA). The purity of RNA was checked by confirming no amplification of target DNA occurs before reverse transcription. Reverse transcription reaction was carried out using the TaKaRa RNA PCR Kit (AMV) version 3.0 (TaKaRa Bio, Shiga, Japan). Quantitative real‐time PCR analysis was carried out on a LightCycler 480 instrument (Roche Diagnostics, Basel, Switzerland) as described previously.19 The primer sequences and annealing temperatures used for real‐time PCR are shown in Table 1.
Global DNA methylation analysis
Global DNA methylation analysis was carried out as previously described.20 Genomic DNA was denatured by heating at 98°C for 3 min. The solution was mixed with 2 U nuclease P1 (Wako, Osaka, Japan) and 100 mM ammonium acetate to a final concentration of 10 mM and incubated at 45°C for 2 h. Next, the solution was supplemented with 0.002 U phosphodiesterase I (Worthington Biochemical, Lakewood, NJ, USA) and 1 M ammonium bicarbonate to a final concentration of 100 mM and incubated at 37°C for 2 h and then 0.5 U alkaline phosphatase (Promega) was added and incubation was continued at 37°C for an additional 1 h. A 100 mM ammonium bicarbonate solution was added to make a total volume of 100 μL, and the mixture was filtered with a Microcon Ultracel YM‐10 cartridge (Millipore, Billerica, MA, USA). The LC–MS analyses were carried out by using an LC/MS‐2010A mass spectrometer and ESI ionization probe (Shimadzu, Kyoto, Japan). Deoxycitidine and 5medC were separated by using a reversed‐phase column (Atlantis dC18, 2.1 × 150 mm, 5 μm; Waters, Milford, MA, USA) in isocratic mode with a mobile phase of methanol : 10 mM ammonium acetate (2:98, v/v) at a flow rate of 0.2 mL/min. The prepared internal standards (13C9,15N3‐2′‐deoxycytidine, 13C10,15N2‐5‐ methyl‐2′‐deoxycytidine, 100 ng each) were added to 15 μL of the hydrolyzed sample, and the mixture was diluted to 500 μL with H2O. A 10 μL volume of the mixture was injected into HPLC–MS, and dC and 5medC were analyzed on a SIM mode.
Statistical analysis
Comparisons were carried out using a two‐tailed paired Student's t‐test. A P‐value <0.05 was considered to be statistically significant. A ROC curve was plotted for detection of liver tumors that were affected by gestational arsenic exposure, and Youden index (sensitivity + specificity−1) was calculated.21
Results
Microarray analysis of DNA regions where methylation status was altered in liver tumors of C3H mice
Although DNA methylation patterns are generally fairly stable,10 DNA methylation changes have been shown to be involved in carcinogenesis and tumorigenesis.8, 9, 10, 11 We assumed that arsenic exposure may target, by changing the status of tumor, the DNA methylation regions that are affected by tumorigenesis. Thus, we first detected DNA regions where methylation status was altered in the liver tumors by using MeDIP–CpG island microarray analysis. To do so, we compared the DNA methylation status of genome DNAs prepared from pooled normal liver homogenates of three control mice and pooled tumor tissues homogenates of three arsenic‐exposed mice. The Me‐values were used to represent methylation levels.
We selected regions where two or more consecutive probes in the control group or arsenic group yielded Me‐values of 0.20 or less and where the differences in the Me‐values of corresponding probes in the two groups were 0.30 or more. According to the criteria, 16 regions were selected, as shown in Table 2. We found that the DNA methylation level in 12 regions was higher, and the level in four regions was lower, in the tumors of the arsenic group than in normal livers of the control group. The results also showed that DNA methylation was mainly altered in the gene body (annotated as “INSIDE” in Table 2).
Table 2.
List of primary annotation identified by methylated DNA immunoprecipitation–CpG island microarray as having different methylation status between normal livers of mice in control group and liver tumors of mice in arsenic‐exposed group
| Location | Primary annotation | Primary annotation type | Average Me‐value | |||
|---|---|---|---|---|---|---|
| Chr | nt number | Tumor tissue | Normal tissue | Difference | ||
| 1 | 111810301–111810709 | Cdh7 | PROMOTER | 0.52 | 0.19 | 0.33 |
| 1 | 040211362–040211474 | Il1r1 | INSIDE | 0.18 | 0.54 | −0.35 |
| 3 | 086633247–086633682 | Mab21l2 | INSIDE | 0.64 | 0.17 | 0.47 |
| 3 | 124313191–124313473 | Tram1l1 | INSIDE | 0.55 | 0.19 | 0.36 |
| 6 | 118559274–118559490 | ENSMUST00000078320.4 | Unknown | 0.49 | 0.16 | 0.34 |
| 6 | 030889400–030889519 | ENS MUST00000101589.1 | Unknown | 0.54 | 0.12 | 0.41 |
| 6 | 110611869–110612025 | Grm7 | INSIDE | 0.48 | 0.17 | 0.32 |
| 7 | 018463460–018463833 | Fosb | INSIDE | 0.60 | 0.18 | 0.42 |
| 8 | 102305418–102305602 | Cdh8 | INSIDE | 0.50 | 0.16 | 0.34 |
| 11 | 102590006–102590400 | Adam11 | INSIDE | 0.18 | 0.71 | −0.54 |
| 11 | 033413037–033413188 | Ranbp17 | INSIDE | 0.48 | 0.18 | 0.31 |
| 12 | 077324539–077324770 | Hspa2 | INSIDE | 0.53 | 0.17 | 0.36 |
| 14 | 030490675–030491319 | Btd | INSIDE | 0.62 | 0.18 | 0.44 |
| 16 | 030520656–030520955 | Fam43a | INSIDE | 0.20 | 0.69 | −0.49 |
| 18 | 020888422–020888579 | B4galt6 | INSIDE | 0.17 | 0.51 | −0.34 |
| 19 | 037764225–037764481 | Cyp26a1 | INSIDE | 0.54 | 0.18 | 0.35 |
Chr, chromosome number; INSIDE, probe position is downstream within 10 kb of transcription start site and in translated region; Normal liver, normal livers of mice in control group; nt number, nucleotide number in the NCBI database (NCBI36/mm8); PROMOTER, probe position is within 10 kb upstream of transcription start site; Tumor tissue, tumor tissue from tumor‐bearing livers of mice in arsenic‐exposed group.
In order to validate the data obtained by the MeDIP–CpG island microarray analyses and to measure the methylation levels of those regions in each liver tissue, we established real‐time MSP primers for seven regions (Il1r1, Mab21l2, Fosb, Hspa2, Btd, Fam43a, and Cyp26a1) and measured the methylation levels of individual genome samples. The DNA methylation levels were calculated as described in “Materials and Methods” and are summarized in Figure 1. The results showed that the methylation levels of the seven DNA regions were consistent with the data obtained by the MeDIP–CpG island microarray analysis (Fig. 1a). We also obtained ordinary MSP primers for four additional regions and analyzed the MSP products by electrophoresis (Fig. 1b). The results also supported those of the MeDIP–CpG island microarray analyses.
Figure 1.
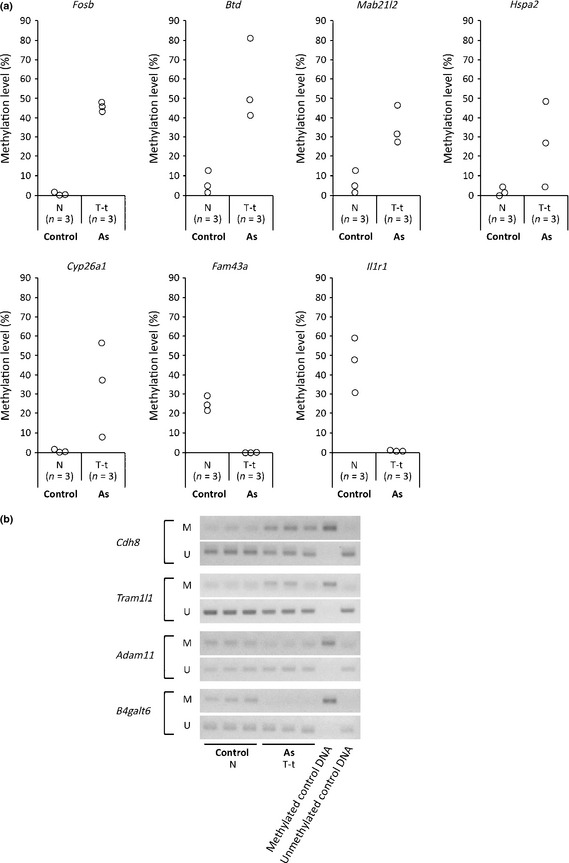
Validation of data obtained by methylated DNA immunoprecipitation–CpG island microarray analysis by real‐time methylation‐specific PCR (MSP). Individual genome samples that were used for the microarray analyses (obtained from normal livers of three control mice [Control‐N] and tumor tissues from tumor‐bearing livers of three gestationally arsenic‐exposed mice [As T‐t]) were examined by real‐time MSP. (a) Methylation levels of DNA regions were measured by real‐time MSP. (b) Methylation changes of DNA regions were measured by MSP. Electrophoresis patterns of each DNA region are shown. M, M primer set; U, U primer set.
Real‐time MSP analyses of individual samples
We then analyzed the DNA methylation levels of individual genomic samples obtained from the normal livers and the normal tissues and the tumor tissues from tumor‐bearing livers of the control group and arsenic group by real‐time MSP. We investigated the DNA regions of Fosb, Btd, Mab21l2, and Il1r1, where MeDIP–CpG island microarray analyses showed higher Me‐values in the tumor tissues than in the normal tissues, and the region of Il1r1, where the microarray analysis showed lower Me‐values in the tumor tissues than in the normal tissues (Fig. 2). The comparison between the normal tissues and the tumor tissues in tumor‐bearing livers showed that the DNA methylation levels of the Fosb and Mab21l2 regions in the arsenic group and Il1r1 region in both the control and arsenic groups were significantly different (Fig. 2).
Figure 2.
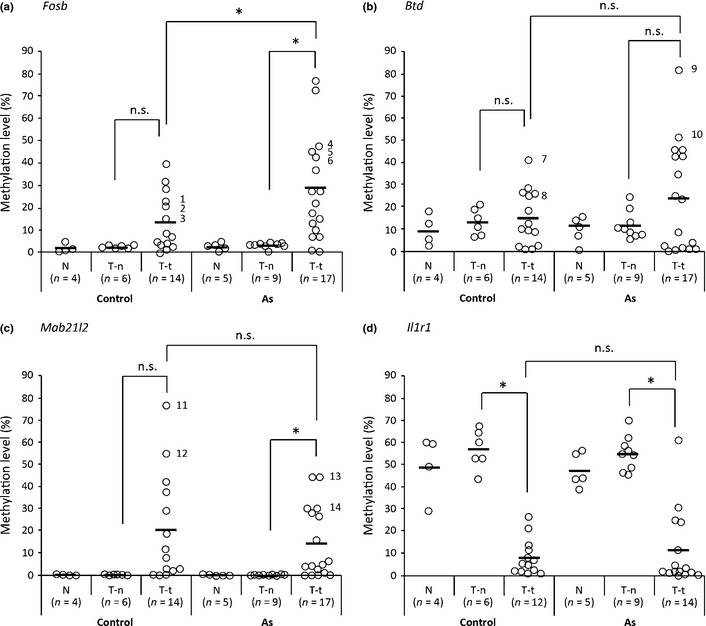
Methylation levels of DNA regions in liver tissues from control mice and gestationally arsenic‐exposed mice. Methylation levels of DNA regions of Fosb, Btd, Mab21l2, and Il1r1 in tissues from normal livers (Control‐N, As‐N), normal tissues from tumor‐bearing livers (Control T‐n, As T‐n), and tumor tissues from tumor‐bearing livers (Control T‐t, As T‐t) were measured by real‐time methylation‐specific PCR. The distributions of the methylation levels of the DNA region of Fosb (a), Btd (b), Mab21l2 (c), and Il1r1 (d) are shown, and the horizontal lines in the chart represent the average methylation level of the liver tissues from each source. Statistically significant differences between the two groups (Control T‐n vs. Control T‐t, As T‐n vs. As T‐t, or Control T‐t vs. As T‐t) were analyzed by Student's t‐test. *P < 0.05 was considered statistically significant. n.s., not significant. 1–14, sample numbers.
Comparison of the DNA methylation level of tumor tissues between the control group and the arsenic‐exposed group for the seven regions showed that the DNA methylation level of the Fosb region was significantly different in the two groups (Figs. 2,S1). We further plotted the ROC curve for detection of liver tumors that were affected by gestational arsenic exposure. The area under the ROC curve was 0.69, and the cut‐off value at which the Youden index showed maximum (sensitivity = 0.65, specificity = 0.64, and Youden index = 0.29) was 0.15.
Analysis of DNA methylation status by bisulfite sequencing
Additionally, we carried out bisulfite sequencing to evaluate the methylation pattern of the DNA regions of Fosb, Btd, and Mab21l2. The results showed that the DNA regions of Fosb (Fig. 3) and Mab21l2 (Fig. 5) exhibited a mosaic pattern of DNA methylation and that the DNA region of Btd consisted of densely methylated DNA and unmethylated DNA (Fig. 4). The DNA methylation levels measured by real‐time MSP almost completely paralleled the DNA methylation levels calculated by bisulfite sequencing (Figs 2, 3, 4, 5).
Figure 3.
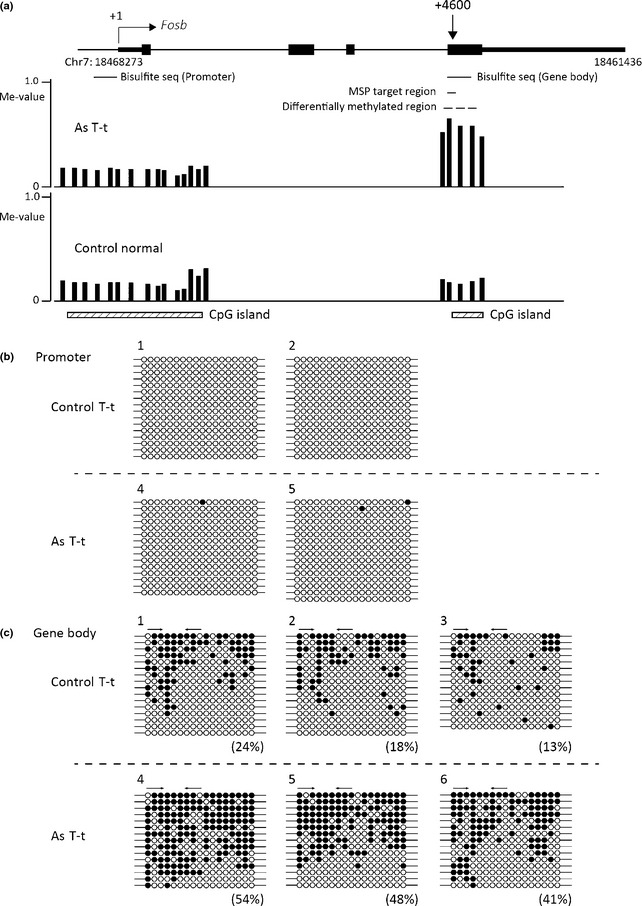
Bisulfite sequencing (seq) analysis of the Fosb region in tumor tissues from tumor‐bearing livers of control mice (Control T‐t) and gestationally arsenic‐exposed mice (As T‐t). (a) Schematic representation of the Fosb locus. The vertical arrow indicates the approximate position of methylation‐specific PCR (MSP) primers from the transcription start site (+1). The black boxes indicate the transcribed region of Fosb. Vertical bars show methylation levels (Me‐values) of each probe. Dashed lines show the areas where two or more consecutive probes in the control group or arsenic group yielded Me‐values of 0.20 or less and where the differences in the Me‐values of corresponding probes in the two groups were 0.30 or more. The MSP primers were designed in the area shown by a black line. This figure was drawn by the UCSC Genome Browser (http://genome.ucsc.edu/) on NCBI36/mm8 assembly that shows the Fosb locus on chromosome 7 (Chr7). The bisulfite sequencing data of samples 1, 2, 4, and 5 in the promoter region (b) and samples 1–6 in the gene body region (c) are shown. The percentages in parentheses are the DNA methylation levels of each MSP primer region calculated from the data of bisulfite sequencing. The arrows indicate the positions of the MSP primers. ○, Unmethylated cytosine; ●, methylated cytosine.
Figure 5.
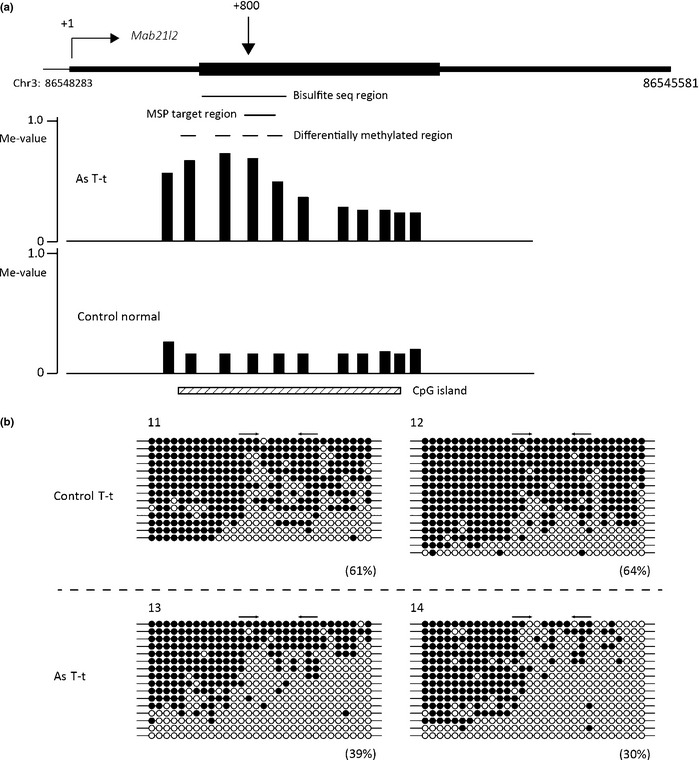
Bisulfite sequencing (seq) analysis of the Mab21l2 region of tumor tissues from the tumor‐bearing livers of control mice (Control T‐t) and gestationally arsenic‐exposed mice (As T‐t). (a) Schematic representation of the Mab21l2 locus. The vertical arrow indicates the approximate position of methylation‐specific PCR (MSP) primers from the transcription start site (+1). The black boxes indicate the transcribed region of Mab21l2. Vertical bars show methylation levels (Me‐values) of each probe. Dashed lines show the areas where two or more consecutive probes in the control group or arsenic group yielded Me‐values of 0.20 or less and where the differences in the Me‐values of corresponding probes in the two groups were 0.30 or more. The MSP primers were designed in the area shown by a black line. This figure was drawn by the UCSC Genome Browser (http://genome.ucsc.edu/) on NCBI36/mm8 assembly that shows the Mab21l2 locus on chromosome 3 (Chr3). (b) Bisulfite sequencing data of samples 11–14. The percentages in parentheses are the DNA methylation levels of each MSP primer region calculated from the data of bisulfite sequencing. The arrows indicate the positions of the MSP primers. ○, Unmethylated cytosine; ●, methylated cytosine.
Figure 4.
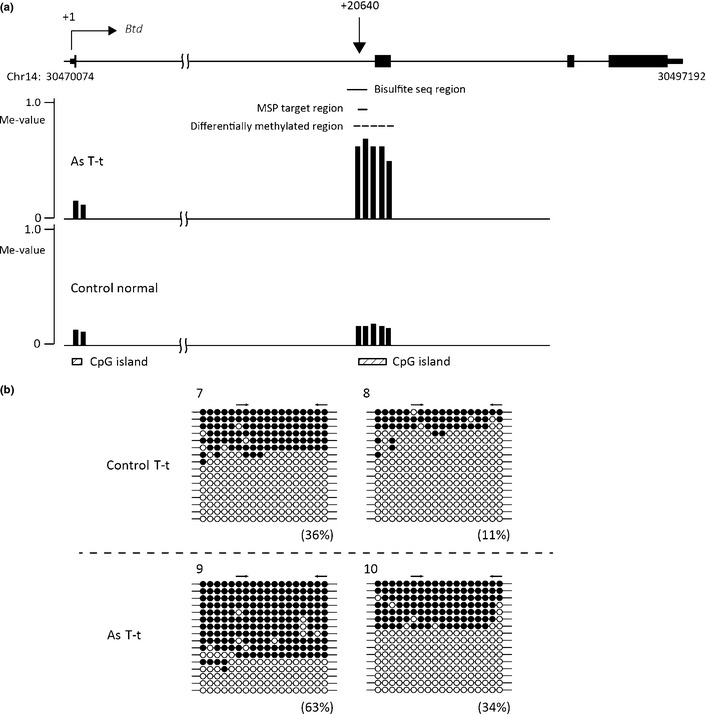
Bisulfite sequencing (seq) analysis of the Btd region of tumor tissues from tumor‐bearing livers of control mice (Control T‐t) and gestationally arsenic‐exposed mice (As T‐t). (a) Schematic representation of the Btd locus. The vertical arrow indicates the approximate position of methylation‐specific PCR (MSP) primers from the transcription start site (+1). The black boxes indicate the transcribed region of Btd. Vertical bars show methylation levels (Me‐values) of each probe. Dashed lines show the areas where two or more consecutive probes in the control group or arsenic group yielded Me‐values of 0.20 or less and where the differences in the Me‐values of corresponding probes in the two groups were 0.30 or more. The MSP primers were designed in the area shown by a black line. This figure was drawn by the UCSC Genome Browser (http://genome.ucsc.edu/) on NCBI36/mm8 assembly that shows the Fosb locus on chromosome 14 (Chr14). (b) Bisulfite sequencing data of samples 7–10. The percentages in parentheses are the DNA methylation levels of each MSP primer region calculated from the data of bisulfite sequencing. The arrows indicate the positions of the MSP primers. ○, Unmethylated cytosine; ●, methylated cytosine.
Upregulation of Fosb mRNA in liver tumors of gestationally arsenic‐exposed mice
As the DNA methylation level of Fosb was found to be altered between the tumor tissues in the control group and arsenic group, we measured the expression of Fosb by real‐time RT‐PCR. The results showed that the mRNA level of Fosb in tumors of the arsenic‐exposed mice was approximately 10 times higher than in other tissues of the control and arsenic groups (Fig. 6).
Figure 6.
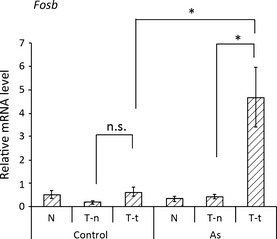
Increased Fosb expression in the liver tumors of gestationally arsenic‐exposed mice. Expression of Fosb in tissues from normal livers (Control‐N, As‐N), normal tissues from tumor‐bearing livers (Control T‐n, As T‐n), and tumor tissues from tumor‐bearing livers (Control T‐t, As T‐t) was measured by real‐time PCR. The Fosb expression levels were normalized to the levels of 18S rRNA expression. Results are reported as means ± SE (n = 6). Statistical significance between the two groups (Control T‐n vs. Control T‐t, As T‐n vs. As T‐t, or Control T‐t vs. As T‐t) was analyzed by Student's t‐test. *P < 0.05 was considered statistically significant. n.s., not significant.
It was recently reported that gene body DNA methylation corresponded to upregulation of gene expression.22, 23, 24 In this study, the increase in DNA methylation in the gene body of Fosb was accompanied by an increase in expression of Fosb mRNA in the liver tumors of the gestationally arsenic‐exposed mice (Figs 3, 6).
Global DNA methylation analysis based on LC/ESI–MS measurements
Aberrant DNA methylations, such as global DNA hypomethylation (lower percentage of 5‐methylcytosine) and region‐specific hypermethylation, are often associated with cancer.8 To determine the global DNA methylation status of liver tissues, we precisely measured the level of 5medC in liver genome DNA by LC/ESI–MS and calculated 5 methylcytosine (5meC) content as a percentage of total cytosine. The results showed a significantly lower percentage of 5meC in the tumor tissues than in the tissues from normal livers and the normal tissues from tumor‐bearing livers in the control group (Table 3), but we did not observe any significant difference among those tissues in the arsenic group (Table 3).
Table 3.
Proportion (%) of 5‐methylcytosine in total cytosine (cytosine + 5‐methylcytosine) from normal livers (Normal), normal tissues from tumor‐bearing livers (T‐n), and tumor tissues from tumor‐bearing livers (T‐t) in control and gestationally arsenic‐exposed mice (As)
| Normal | T‐n | T‐t | |
|---|---|---|---|
| Control (%) | 4.63 ± 0.04 (100) | 4.65 ± 0.07 (100.4) | 4.29 ± 0.09* (92.7) |
| As (%) | 4.61 ± 0.06 (100) | 4.62 ± 0.06 (100.2) | 4.46 ± 0.07 (96.7) |
*Significant differences from Control normal and Control T‐n at *P < 0.01. The number in the parenthesis indicates the value of each sample compared to the value in the Normal groups, in which it was set as 100 for both Control and As mice. Results are reported as means ± SE (n = 6).
Discussion
In this study the results of MeDIP–CpG island microarray analyses and real‐time MSP indicated that there exist several DNA regions where methylation levels were significantly different between the non‐tumor tissues and tumor tissues in control C3H mice and/or C3H mice gestationally exposed to arsenic (Table 2, Fig. 2). This is the first report on the genome‐wide DNA methylation analysis of liver tumors in C3H mice. A study by Cui et al. reported that chronic exposure of male A/J mice to inorganic arsenic increased their incidence of lung tumors compared with control mice.25 They also reported that MSP analyses of the promoter region of the tumor suppressor genes p16 Ink4a and RASSF1A showed higher methylation in lung tumors of the arsenic‐exposed group than in the control group.25 In this study, we newly clarified that the methylation level of the gene body region of Fosb was significantly higher in the liver tumor tissues of C3H mice gestationally exposed to arsenic than in the tumor tissues of control mice (Fig. 2a).
Real‐time PCR analyses of tumor tissue revealed significantly increased expression of the oncogene Fosb in gestationally arsenic‐exposed mice (Fig. 6). Hypermethylation of CpG islands in promoter regions generally results in transcriptional silencing, however, increased DNA methylation of the CpG island of gene bodies is reported to be associated with increased gene expression.22, 23, 24 In this study, the increase in DNA methylation in the gene body of Fosb was found to be associated with increased expression of Fosb in tumor tissues of the arsenic‐exposed group (Fig. 2). In contrast, the CpG island of the Fosb promoter region was completely unmethylated in normal tissues from the tumor‐bearing livers and tumor tissues of both the control group and arsenic group (Fig. 3 and data not shown). Thus, the upregulation of Fosb in the tumor tissues of the arsenic group may be associated with increased DNA methylation in the gene body. Previous studies suggested several explanations for the linkage between increased DNA methylation in the gene body and increased gene expression. An interpretation is that upregulated transcription may facilitate de novo DNA methylation in the gene body, and an alternative explanation is that gene body methylation represses the expression of antisense transcripts that would downregulate the expression of sense transcripts.23, 24, 26 However, experimental evidence is yet to be obtained.
Fosb is a member of the Fos family of proteins, which constitutes transcription factor complex AP1. AP1 is activated by ERK1/2‐mediated phosphorylation and is involved in tumor formation.27 Our recent study, carried out in the same model as used in this study, revealed that liver tumors in mice gestationally exposed to arsenic harbored a higher rate of Ha‐ras mutation than tumors of control mice.15 Ha‐ras is activated on mutation and in turn activates the Raf–ERK signaling pathway,28 which leads to AP1 activation.29 The upregulation of Fosb seems to be a downstream event of Ha‐ras mutation and to be involved in the increase in liver tumors as a result of gestational arsenic exposure. As the area under the ROC curve for the DNA methylation status of the Fosb region in tumors was not large, that region alone may not be a potent marker to distinguish tumors affected by arsenic. However, the gene body methylation status in conjunction with the gene expression of Fosb would give a promising clue to diagnose those tumors affected by gestational arsenic exposure.
In the present study, we also measured the contents of 5meC in the liver tissues. Although the tumor tissues of the control group showed significantly lower levels of 5meC compared to normal tissues, as reported in previous studies,8 the 5meC level of tumor tissues in the arsenic group was not significantly reduced. These results may be attributable to the effects of arsenic on the function of machineries involved in the maintenance of DNA methylation. Another explanation would be that arsenic affects the status of tumor, which indirectly leads to the alteration in the level of 5meC.
In summary, we carried out MeDIP–CpG island microarray analyses to dissect the DNA methylation status in liver tumors of C3H mice gestationally exposed to arsenic. Validation of the results of microarray analyses by real‐time MSP indicated that the microarray method correctly depicted the changes in DNA methylation in the genome. As a result of the analyses, we revealed that the gene body region of the oncogene Fosb is highly methylated and its expression is greatly increased in the tumors of gestationally arsenic‐exposed mice, compared with the tumors of control mice. These results suggest that DNA methylation changes are involved in the alteration in tumor phenotype by gestational arsenic exposure.
The study first compared the DNA methylation statuses of normal tissues from the control group and tumor tissues from the arsenic group by MeDIP–CpG island microarray analyses. However, the combination may not have identified the DNA regions where the methylation statuses were largely altered in tumors from the control group. Thus, further precise study would potentially clarify additional DNA methylation regions that discriminate arsenic‐induced tumors from other tumors. Application of next generation sequencing‐based methods, such as reduced representation bisulfite sequencing30 and whole genome bisulfite sequencing,31 would be useful to improve the resolving power and sensitivity of analysis of the present study.
Disclosure Statement
The authors have no conflict of interest.
Abbreviations
- 5meC
5‐methylcytosine
- 5medC
5‐methyldeoxycytidine
- Btd
biotinidase
- Cyp26a1
cytochrome P450, family 26, subfamily a, polypeptide 1
- dC
deoxycytidine
- ERα
estrogen receptor α
- ESI
electrospray ionization
- Fam43a
family with sequence similarity 43, member A
- Fosb
FBJ osteosarcoma oncogene B
- Hspa2
heat shock protein 2
- Il1r1
interleukin 1 receptor, type I
- LC
liquid chromatography
- Mab21l2
Mab‐21‐like 2
- MeDIP
methylated DNA immunoprecipitation
- MS
mass spectrometry
- MSP
methylation‐specific PCR
- ROC
receiver operating characteristic
Supporting information
Fig. S1. Methylation levels of the DNA region of Hspa2, Fam43a, and Cyp26a1 in liver tumor tissues from control mice and gestationally arsenic‐exposed mice.
Acknowledgments
We wish to thank Dr. A. Takeuchi (National Institute for Environmental Studies, Tsukuba, Japan) for her useful advice on statistical analysis, H. Murai and M. Matsumoto for their excellent technical assistance, and S. Itaki for her helpful secretarial assistance. This study was partly supported by the National Institute for Environmental Studies (0710AG333 to K.N.), a Grant‐in‐Aid for Young Scientists (B) (No. 23790680 to T.S.), and a Grant‐in‐Aid for Scientific Research (B) (No. 23390166 to K.N.) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
(Cancer Sci 2013; 104: 1575–1585)
References
- 1. Hughes MF, Beck BD, Chen Y, Lewis AS, Thomas DJ. Arsenic exposure and toxicology: a historical perspective. Toxico Sci 2011; 123: 305–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jomova K, Jenisova Z, Feszterova M et al Arsenic: toxicity, oxidative stress and human disease. J Appl Toxicol 2011; 31: 95–107. [DOI] [PubMed] [Google Scholar]
- 3. Yuan Y, Marshall G, Ferreccio C et al Kidney cancer mortality: fifty‐year latency patterns related to arsenic exposure. Epidemiology 2010; 21: 103–8. [DOI] [PubMed] [Google Scholar]
- 4. Smith AH, Marshall G, Liaw J, Yuan Y, Ferreccio C, Steinmaus C. Mortality in young adults following in utero and childhood exposure to arsenic in drinking water. Environ Health Perspect 2012; 120: 1527–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rossman TG. Mechanism of arsenic carcinogenesis: an integrated approach. Mutat Res 2003; 533: 37–65. [DOI] [PubMed] [Google Scholar]
- 6. Reichard JF, Puga A. Effects of arsenic exposure on DNA methylation and epigenetic gene regulation. Epigenomics 2010; 2: 87–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ren X, McHale CM, Skibola CF, Smith AH, Smith MT, Zhang L. An emerging role for epigenetic dysregulation in arsenic toxicity and carcinogenesis. Environ Health Perspect 2011; 119: 11–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer 2004; 4: 143–53. [DOI] [PubMed] [Google Scholar]
- 9. Eden A, Gaudet F, Waghmare A, Jaenisch R. Chromosomal instability and tumors promoted by DNA hypomethylation. Science 2003; 300: 455. [DOI] [PubMed] [Google Scholar]
- 10. Laird PW. The power and the promise of DNA methylation markers. Nat Rev Cancer 2003; 3: 253–66. [DOI] [PubMed] [Google Scholar]
- 11. Ushijima T, Asada K. Aberrant DNA methylation in contrast with mutations. Cancer Sci 2010; 101: 300–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Köhle C, Schwarz M, Bock KW. Promotion of hepatocarcinogenesis in humans and animal models. Arch Toxicol 2008; 82: 623–31. [DOI] [PubMed] [Google Scholar]
- 13. Waalkes MP, Ward JM, Liu J, Diwan BA. Transplacental carcinogenicity of inorganic arsenic in the drinking water: induction of hepatic, ovarian, pulmonary, and adrenal tumors in mice. Toxicol Appl Pharmacol 2003; 186: 7–17. [DOI] [PubMed] [Google Scholar]
- 14. Waalkes MP, Liu J, Chen H et al BA Estrogen signaling in livers of male mice with hepatocellular carcinoma induced by exposure to arsenic in utero. J Natl Cancer Inst 2004; 96: 466–74. [DOI] [PubMed] [Google Scholar]
- 15. Nohara K, Tateishi Y, Suzuki T et al Late‐onset increases in oxidative stress and other tumorigenic activities and tumors with a Ha‐ras mutation in the liver of adult male C3H mice gestationally exposed to arsenic. Toxicol Sci 2012; 129: 293–304. [DOI] [PubMed] [Google Scholar]
- 16. Yamashita S, Hosoya K, Gyobu K, Takeshima H, Ushijima T. Development of a novel output value for quantitative assessment in methylated DNA immunoprecipitation‐CpG island microarray analysis. DNA Res 2009; 16: 275–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Suzuki T, Nohara K. Long‐term arsenic exposure induces histone H Lys9 dimethylation without altering DNA methylation in the promoter region of p16(INK4a) and down‐regulates its expression in the liver of mice. J Appl Toxicol 2013; 33: 951–8. [DOI] [PubMed] [Google Scholar]
- 18. Nohara K, Pan X, Tsukumo S et al Constitutively active aryl hydrocarbon receptor expressed specifically in T‐lineage cells causes thymus involution and suppresses the immunization‐induced increase in splenocytes. J Immunol 2005; 174: 2770–7. [DOI] [PubMed] [Google Scholar]
- 19. Nohara K, Ao K, Miyamoto Y et al Comparison of the 2,3,7,8‐tetrachlorodibenzo‐p‐dioxin (TCDD)‐induced CYP1A1 gene expression profile in lymphocytes from mice, rats, and humans: most potent induction in humans. Toxicology 2006; 225: 204–13. [DOI] [PubMed] [Google Scholar]
- 20. Nohara K, Baba T, Murai H et al Global DNA methylation in the mouse liver is affected by methyl deficiency and arsenic in a sex‐dependent manner. Arch Toxicol 2011; 85: 653–61. [DOI] [PubMed] [Google Scholar]
- 21. Akobeng AK. Understanding diagnostic tests 3: receiver operating characteristic curves. Acta Paediatr 2007; 96: 644–7. [DOI] [PubMed] [Google Scholar]
- 22. Hellman A, Chess A. Gene body‐specific methylation on the active X chromosome. Science 2007; 315: 1141–3. [DOI] [PubMed] [Google Scholar]
- 23. Ball MP, Li JB, Gao Y et al Targeted and genome‐scale strategies reveal gene‐body methylation signatures in human cells. Nat Biotechnol 2009; 27: 361–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rauch TA, Wu X, Zhong X, Riggs AD, Pfeifer GP. A human B cell methylome at 100‐base pair resolution. Proc Natl Acad Sci USA 2009; 106: 671–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cui X, Wakai T, Shirai Y, Hatakeyama K, Hirano S. Chronic oral exposure to inorganic arsenate interferes with methylation status of p16INK4a and RASSF1A and induces lung cancer in A/J mice. Toxicol Sci 2006; 91: 372–81. [DOI] [PubMed] [Google Scholar]
- 26. Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet 2008; 9: 465–76. [DOI] [PubMed] [Google Scholar]
- 27. Milde‐Langosch K. The Fos family of transcription factors and their role in tumourigenesis. Eur J Cancer 2005; 41: 2449–61. [DOI] [PubMed] [Google Scholar]
- 28. Pylayeva‐Gupta Y, Grabocka E, Bar‐Sagi D. RAS oncogenes: weaving a tumorigenic web. Nat Rev Cancer 2011; 11: 761–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Frost JA, Geppert TD, Cobb MH, Feramisco JR. A requirement for extracellular signal‐regulated kinase (ERK) function in the activation of AP‐1 by Ha‐Ras, phorbol 12‐myristate 13‐acetate, and serum. Proc Natl Acad Sci USA 1994; 91: 3844–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Meissner A, Gnirke A, Bell GW, Ramsahoye B, Lander ES, Jaenisch R. Reduced representation bisulfite sequencing for comparative high‐resolution DNA methylation analysis. Nucleic Acids Res 2005; 33: 5868–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lister R, Pelizzola M, Dowen RH et al Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 2009; 462: 315–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Methylation levels of the DNA region of Hspa2, Fam43a, and Cyp26a1 in liver tumor tissues from control mice and gestationally arsenic‐exposed mice.


