Abstract
Background: Head and neck squamous cell carcinoma (HNSCC) is the fifth most common malignancy in the world. The 5-methylcytosine (m5C) plays vital roles in pathological conditions, such as cancer. Methods: This study investigated The Cancer Genome Atlas (TCGA) database for patients with HNSCC. We characterized the mutations and copy number variations (CNVs) in m5C-regulatory genes, in addition to analyzing their mRNA expressions. Gene set enrichment analysis (GSEA) and protein-protein interaction (PPI) analyses were used to explore relevant functional annotations of m5C-regulatory genes. Results: Alterations in m5C-regulatory genes were closely associated with patient clinicopathological characteristics. The expression of ten m5C-regulatory genes was significantly correlated with CNV patterns, indicating that m5C-regulatory genes have important regulatory effects. There was increased expression of m5C-regulatory genes, particularly ALYREF and NSUN5, during the tumor, node, and metastasis stages. Cox regression analysis revealed that the expression of DNMT1, TET2, and NSUN6 correlated with HNSCC prognoses. Furthermore, the expression of DNMT1 and ALYREF could effectively predict HNSCC risk in patients. In addition, the high expression levels of ALYREF correlated with mitochondrial function, and the elevated DNMT1 expression was associated with peptide cross-linking and humoral immunity. These results provide promising insight into the roles of m5C genes in tumor energy-metabolism and protein synthesis. Conclusions: Collectively, the results indicate that m5C plays critical roles in HNSCC progression, and is also a potential HNSCC prognostic marker.
Keywords: 5-methylcytosine, head and neck squamous cell carcinoma, the cancer genome atlas, prognosis
Introduction
Head and neck squamous cell carcinoma (HNSCC) is a major contributor to the global cancer burden [1]. Tobacco consumption and alcohol abuse contribute to approximately 75% of all HNSCC risk factors, with both of these factors exerting synergistic effects [2]. Surgery and radiotherapy remain the two primary modalities for HNSCC management, for both early and advanced cases [3]. Notably, the majority of patients with HNSCC present with an advanced stage of disease [4]. Despite aggressive, site-specific, multimodal therapies, a significant proportion of patients have recurring tumors and/or metastases. This often plays a role in reducing a patient’s belief in curative therapy, which results in high morbidity and dismal survival rates [5]. Novel prognostic clinical factors and biomarkers are needed to improve the prognosis of HNSCC.
RNA from all living organisms can be post-transcriptionally modified in more than 100 distinct ways [6,7]. Among these modifications, 5-methylcytosine (m5C), which was discovered in 1925, is a widespread mRNA modification [8], and localizes to the untranslated regions (UTRs) of mRNA transcripts [9]. At the posttranscriptional level, m5C RNA modifications can be dynamically regulated by a series of significant mediator proteins known as “writers, erasers, and readers” [10]. The “writers” are the RNA-methylases NSUN1-7, DNMT1, DNMT2, DNMT3A, and DNMT3B [11]. The main “eraser” is TET2, which acts as a demethylase [12], and ALYREF is an important “reader” protein, which recognizes and binds to m5C sites on mRNA [13].
Previous reports suggest that m5C is an important regulator of many aspects of gene expression, including RNA export, ribosome assembly, translation, and RNA stability [10,14,15], and recent discoveries have indicated that m5C also plays vital roles in pathological conditions, such as cancers [16]. One recent study indicated that m5C promotes the pathogenesis of human bladder urothelial carcinoma by stabilizing oncogene-mRNAs through m5C-modified sites in their 3’ UTRs [17]. However, our current knowledge of how m5C levels are controlled, and how m5C gene levels are regulated in HNSCC, remains unclear.
The Cancer Genome Atlas (TCGA) dataset contains the largest number of publicly-available HNSCC samples, and serves as a cornerstone for future research [18,19]. In this study, we first selected m5C-regulated genes from the TCGA database related to HNSCC, and performed in-depth sequence analyses. We then further analyzed the relationships between m5C-regulated gene alterations, and HNSCC clinicopathological features to determine the impact of genetic changes on survival. We found that the mRNA expressions of m5C-regulatory genes correlated with copy number variation (CNV) patterns. These results then prompted a gene set enrichment analysis (GSEA) to analyze the biological functions of DNMT1 and ALYREF. We determined that DNMT1 and ALYREF expression levels effectively predicted risk factors (morbidity and mortality) for patients with HNSCC. These results also led to our study of m5C modifications during HNSCC progression. Overall, the results indicate that m5C is crucial for regulating the progression of HNSCC.
Materials and methods
Datasets and samples
The data relevant to patients with HNSCC were collected using the TCGA-Assembler from the atlas website (https://gdc.cancer.gov), including clinical data, CNVs, mutations, and mRNA expression levels, as described previously [20]. To further validate the results, The Cancer Genome Atlas Cervical Squamous Cell Carcinoma and Endocervical Adenocarcinoma (TCGA-CESC) database was also searched and the results were downloaded from the website in the same manner.
For the HNSCC clinical data, we collected 527 samples. The inclusion criteria were as follows: HNSCC samples with complete CNV data, single nucleotide variation (SNV) data, transcriptome data, and clinical information. A total of 490 samples were used for the survival analysis. The total number of samples was determined after integrating the data and excluding samples with incomplete clinical information and those with survival times of less than 30 days. A total of 521 HNSCC samples with CNV data were retrieved and 508 HNSCC samples with SNV data were used for analyses. For our mRNA-expression dataset, we obtained 500 HNSCC samples.
Gene set enrichment analysis and gene set variation analysis (GSVA)
GSEA was performed with the GSEA v2.0.13 tool (http://www.broad.mit.edu/gsea/), as described previously [21]. The mRNA expression profiles of ALYREF and DNMT1 were extracted from the TCGA database to explore their potential biological functions. We used GSEA to identify the potential influences of different expression levels of ALYREF and DNMT1 on HNSCC pathogenesis. Additionally, we used GSVA, an unsupervised method without parametric values, to estimate variations in enrichment throughout the gene expression data set.
Protein-protein interaction (PPI) analysis
PPI information was extracted from the STRING database (https://string-db.org/), as described previously [22]. We performed PPI analysis on 11 m5C regulators to detect potentially related proteins and interactions between them.
Statistical analysis
Statistical analyses were performed using SPSS 21.0 (IBM, Chicago, IL, USA) and GraphPad Prism 8.0 (GraphPad Software, La Jolla, CA, USA). Survival analyses were performed using the Kaplan-Meier method. Associations between clinical characteristics and alterations in m5C regulatory gene were evaluated using univariate Cox regression analyses. Multivariate Cox regression analysis was used to investigate the effects of m5C regulatory genes on patient prognosis. The chi-square test was used to analyze any correlations between alterations in m5C-regulatory genes and driver-gene variations. A least absolute shrinkage and selection operator (LASSO) regression analysis was also used to identify potential HNSCC prognostic markers, as described previously [23]. A P < 0.05 was considered statistically significant.
Results
Mutations and CNVs in m5C-regulatory genes in patients with HNSCC
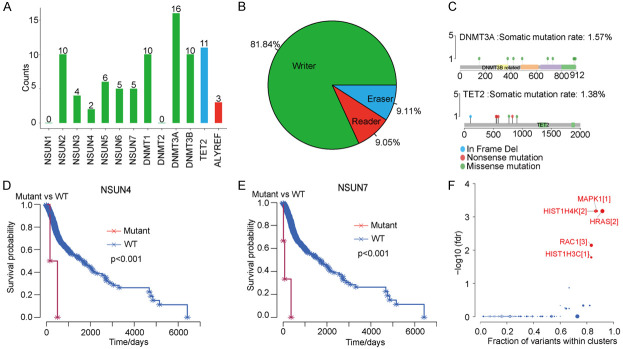
To obtain an overall understanding of the alterations in m5C-regulatory genes related to HNSCC, SNV data from 508 HNSCC samples and CNV data from 521 HNSCC samples were downloaded from the online atlas database. We identified mutations in m5C-regulatory genes in 82 independent samples. Among them, an m5C “writer” gene, DNMT3A, had the highest frequency of mutations (19.51%, 16/82) when compared to the “eraser” TET2 gene (13.41%, 11/82) (Figure 1A). At the same time, the “writer” genes NSUN2, DNMT1, and DNMT3A each had equal mutation frequencies (11.90%, 10/84). From the perspective of SNVs, single nucleotide alterations in the known functional regions of annotated m5C-regulatory genes occurred in 43 samples. Among the 13 m5C-regulatory genes, 11 genes were annotated with functional changes, and all of these had missense mutations (Supplementary Table 1). In addition to SNVs, we detected a high frequency of CNVs in m5C-regulatory genes (Figure 1B). The highest frequency of CNVs (61.65%) occurred in the “writer” gene NSUN2, followed by the “writer” gene NSUN3 (61.09%). The “writer” gene DNMT3A had the lowest CNV frequency (11.09%) (Supplementary Table 2).
Figure 1.
Mutations and CNVs in m5C-regulatory genes in patients with HNSCC. (A) The mutation frequencies of different m5C-regulatory genes. (B) The frequency of CNVs in m5C-regulatory genes. (C) The mutation sites in DNMT3A and TET2. (D, E) Kaplan-Meier analysis displays of significant associations between the mutations in (D) NSUN4 and (E) HNSCC, and survival probability (P < 0.001). (F) HNSCC driver-gene predictions by the oncodrive function of maftools.
Next, we explored the mutation sites for DNMT3A and TET2, which had the highest mutation frequencies (Figure 1C). The somatic mutation rates for DNMT3A and TET2 were 1.57% and 1.38%, respectively. Considering that mutations in m5C genes could alter HNSCC phenotypes, we performed patient-survival analyses for the 11 genes with functional changes to explore their relationships to HNSCC patient prognosis. We found a significant association between NSUN4 and NSUN7 mutations and prognosis; patients with these mutations had worse prognoses (Figure 1D, 1E). Thus, mutations and CNVs were common in these m5C-regulatory genes in patients with HNSCC, and may serve important markers for patient prognosis.
Alterations in m5C-regulatory genes and clinicopathological and molecular characteristics
We assessed relationships between alterations (CNVs and/or mutations) in m5C-regulatory genes and HSNCC patients’ clinicopathological characteristics. Univariate Cox regression analyses demonstrated that the survival of patients with HNSCC was significantly associated with age and cancer stage, but not with the presence of SNVs or CNVs (Table 1). To explore the correlations between changes in m5C-regulatory genes and pathogenic molecules in HNSCC, we first conducted driver-gene predictions using the oncodrive function of maftools. MAPK1, HRAS, RAC1, HIST1H4K, and HIST1H3C were identified as potential driver genes in HNSCC (Figure 1F). We further evaluated whether mutations in m5C-regulatory genes were associated with changes in these five driver genes. Alterations in m5C-regulatory genes were not significantly correlated with variations in MAPK1, HIST1H4K, or HIST1H3C, but were significantly associated with variations in HRAS and RAC1 (Table 2). In particular, alterations in m5C-regulatory genes were seen in 14 of the 27 patients with HRAS variations.
Table 1.
Univariate Cox analysis of clinical features and HNSCC patient survival
| Genes | beta | HR (95% CI for HR) | wald.test | p-value |
|---|---|---|---|---|
| age | 0.022 | 1 (1-1) | 12 | 0.00054 |
| TNM stage | 0.55 | 1.7 (1.2-2.5) | 8.5 | 0.0036 |
| gender | -0.27 | 0.76 (0.57-1) | 3.3 | 0.069 |
| N stage | 0.27 | 1.3 (0.98-1.7) | 3.3 | 0.07 |
| T stage | 0.21 | 1.2 (0.92-1.7) | 2 | 0.16 |
| SNV | 0.2 | 1.2 (0.76-2) | 0.68 | 0.41 |
| grade | -0.1 | 0.9 (0.67-1.2) | 0.46 | 0.5 |
| M stage | -0.17 | 0.84 (0.35-2) | 0.15 | 0.7 |
| CNV or SNV | -0.056 | 0.95 (0.66-1.3) | 0.1 | 0.76 |
| CNV | -0.042 | 0.96 (0.68-1.4) | 0.05 | 0.82 |
Abbreviations: HR = hazard ratio; CI = confidence interval; CNV = copy number variant; SNV = single nucleotide variation.
Table 2.
The relationship between alterations in m5C-regulatory genes and HNSCC-related driver genes
| without SNV and CNV | with SNV or CNV | X2 | P | |||
|---|---|---|---|---|---|---|
| HRAS | n = 490 | Wild type | 66 | 397 | 19.2381575 | 1.1538E-05 |
| alternation | 13 | 14 | ||||
| MAPK1 | n = 490 | Wild type | 77 | 406 | 0.14785117 | 0.70059737 |
| alternation | 2 | 5 | ||||
| RAC1 | n = 490 | Wild type | 71 | 408 | 22.5514412 | 2.0459E-06 |
| alternation | 8 | 3 | ||||
| HIST1H4K | n = 490 | Wild type | 79 | 405 | 0.27252197 | 0.60164501 |
| alternation | 0 | 6 | ||||
| HIST1H3C | n = 490 | Wild type | 78 | 406 | 0.27252197 | 0.60164501 |
| alternation | 1 | 5 |
Abbreviations: CNV = copy number variant; SNV = single nucleotide variation.
mRNA expressions of m5C-regulatory genes
We then evaluated the effects of m5C-regulatory gene alterations on their mRNA expressions. For the 500 HNSCC samples tested, 2 of the 13 identified m5C-regulatory genes were not detected (NSUN1 and DNMT2). After excluding these two genes, the mRNA expression profiles for 10 of the 11 m5C-regulatory genes were found to be significantly correlated with different CNV patterns (Figure 2A-J). For these 10 regulatory genes, increases in gene copy number were associated with higher mRNA expression levels, whereas gene-copy deletions were associated with decreased levels of mRNA expression. These 10 genes were distributed throughout the m5C-regulation process. NSUN7 was the only regulator gene whose expression level did not correlate with CNVs (Figure 2K), suggesting that CNVs may have a central role in controlling the expression of m5C regulators.
Figure 2.
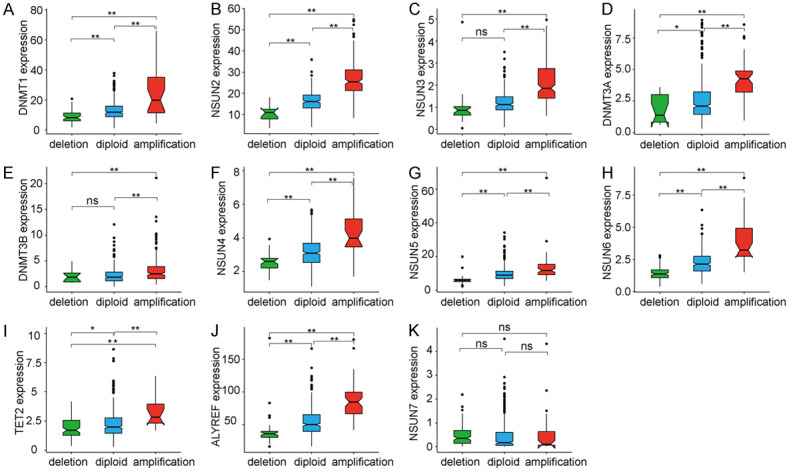
The effect of m5C-regulatory gene alterations on mRNA expression. A-J. The mRNA expressions of 10 m5C-regulatory genes were significantly correlated with different CNV patterns. K. No significant correlation existed between CNVs and the mRNA expression level of NSUN7. *P < 0.05, **P < 0.01.
Associations between m5C-regulatory gene expression and tumor, node, and metastasis (TNM) staging in HNSCC
Our analyses showed a significant association between TNM staging and the prognoses of patients with HNSCC. We defined TNM stages I/II as low and stages III/IV as high. Patients with low TNM stages demonstrated better survival rates (Figure 3A). Thus, we used cluster analysis to examine the expression of m5C-regulatory genes according to TNM staging; m5C-regulatory genes tended to be highly expressed in high-stage TNM samples (Figure 3B). Next, we analyzed the expression of m5C-regulatory genes in patients according to TNM staging. The results indicated that the expression of ALYREF and NSUN5 positively correlated with specific TNM stages (Figure 4A, 4B), while no significant association between TNM stage and any other m5C-regulator gene was observed (Figure 4C-K). In combination with the aforementioned analyses, there was a significant association between TNM staging and patient prognoses, suggesting that m5C-regulatory gene expression was related to the prognoses of patients with HNSCC.
Figure 3.
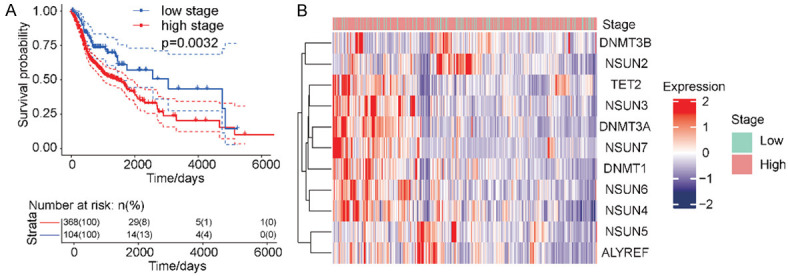
Associations between m5C-regulatory gene expression and TNM staging in HNSCC. A. Kaplan-Meier analysis revealed that patients with low TNM stages had better survival rates (P = 0.0032). B. The heat map showed that m5C-regulatory genes tended to have increased expression during later TNM stages.
Figure 4.
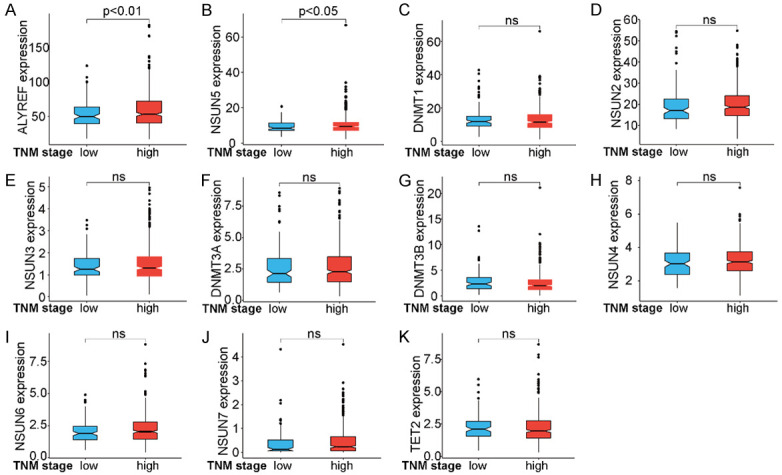
Differences in m5C-regulatory gene expression in patients at different TNM stages. (A, B) The expressions of (A) ALYREF and (B) NSUN5 were positively correlated with TNM staging. (C-K) No significant associations were observed between nine m5C-regulator genes and TNM staging.
Associations between m5C-regulatory gene expression levels and the survival of patients with HNSCC
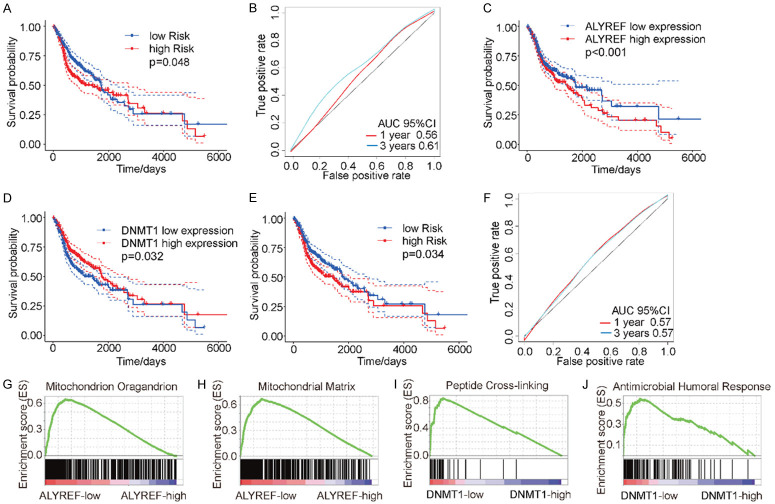
To further explore the relationship between different levels of m5C-regulatory gene expression and patient prognoses, we used a univariate Cox regression analysis. The results demonstrated that the expression levels of DNMT1, TET2, and NSUN6 significantly correlated with patient prognoses (Supplementary Table 3). The expression of these three genes also significantly correlated with CNV patterns. When we explored the influence of 11 m5C-regulatory genes on patient prognoses by multivariate Cox regression analysis, we observed that their expression significantly predicted patient risk factors (Figure 5A). For these results, the area under the receiver operating characteristic (ROC) curve for 1- and 3-years post HNSCC was > 0.55, this indicated that m5C-regulatory gene expression may be used as a potential prognostic marker for HNSCC (Figure 5B).
Figure 5.
The relationships between m5C-regulatory gene expression and patient prognoses. A. The expression of m5C-regulatory genes significantly predicted patient mortality risks (P = 0.048). B. The area under the curve (AUC) values at 1- and 3-years were greater than 0.55, indicating a strong predictive value. C. Kaplan-Meier analysis indicated that elevated ALYREF expression was associated with poor survival probability (P < 0.001). D. Kaplan-Meier analysis demonstrated that low DNMT1 expression predicted poor survival probability (P = 0.032). E. A Cox regression analysis indicated that the expressions of DNMT1 and ALYREF effectively predicted the risk of HNSCC patient mortality (P = 0.034). F. The 1- and 3-year AUCs for the DNMT1 and ALYREF m5C-regulatory genes were 0.57. G and H. Gene set enrichment analysis of DNMT1 expression levels. I and J. Gene set enrichment analysis of ALYREF expression levels.
Expressions of DNMT1 and ALYREF in patients with HNSCC
To obtain accurate and reliable prognostic markers for HNSCC, we performed a LASSO analysis on the 11 identified m5C-regulatory genes. Two genes, DNMT1 and ALYREF, were selected by combining the results of 1000 LASSO regressions. DNMT1, as a “writer” gene for m5C, and ALYREF, as a “reader” gene for m5C, are both important for m5C regulation. First, we analyzed the relationships between DNMT1 expression and ALYREF expression with the prognoses of patients with HNSCC. The results indicated that high ALYREF expression was associated with poor survival rates (Figure 5C), whereas the same outcome was associated with low DNMT1 expression (Figure 5D). Next, we performed Cox regression analyses for both DNMT1 and ALYREF to calculate the associated risks of patient death. We observed that DNMT1 and ALYREF both effectively predicted the death risk in patients with HNSCC (P = 0.034) (Figure 5E). With respect to these two m5C-regulatory genes, the 1- and 3-year areas under the ROC curves were calculated as 0.57, indicating the predictive strength of these two genes (Figure 5F). Thus, DNMT1 and ALYREF expression levels can effectively predict the risk of mortality in patients with HNSCC.
Functional enrichment analyses of DNMT1 and ALYREF gene-expression levels
Because DNMT1 and ALYREF are important genes in the m5C-methylation process, we next assessed their possible roles in the pathogenesis of HNSCC by examining enriched-gene sets from samples with different DNMT1 and ALYREF mRNA expression levels. GSEA indicated that high expression levels of ALYREF were related to a variety of mitochondrial functions (Figure 5G, 5H), whereas high DNMT1 expression levels were associated with peptide cross-linking and humoral immunity (Figure 5I, 5J). Elevated ALYREF expression was associated with increased mitochondrial activity, suggesting that ALYREF promotes intracellular energy metabolism to maintain continuous energy supplies, which is of great significance for tumor-cell growth. The relationship between DNMT1 expression and immunity requires further exploration due to the possibility that DNMT1 could be a target for immunotherapy against HNSCC.
Patient survival and validations of DNMT1 and ALYREF
To verify the relationships between the expressions of DNMT1 and ALYREF and the survival rates of patients with HNSCC, we conducted further analyses using a validation dataset. Because of few specific HNSCC studies, we used a validation dataset containing other homologous cancers to squamous cell carcinoma. Among the squamous cell carcinomas represented in TCGA, we chose cervical squamous cell carcinoma (CESC) as the validation dataset because CESC is similar to HNSCC in cell origin and tumor tissue-structure. Using Cox regression analyses, DNMT1 and ALYREF gene expressions and risk values were used to calculate the risk values of the samples. We found that these two genes had the ability to predict risk in the validation dataset (Figure 6A) and that the areas under the ROC curves were nearly 0.7 (Figure 6B). As before, low DNMT1 expression levels were also associated with poor prognoses (Figure 6C).
Figure 6.
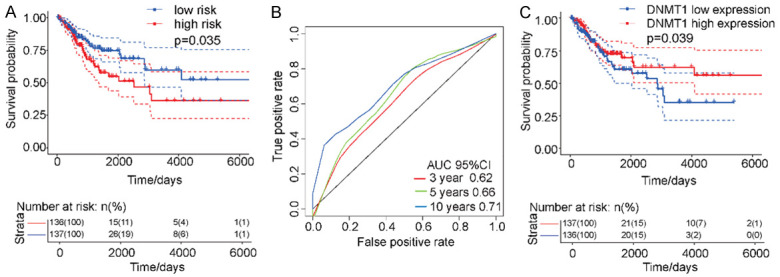
Validation of DNMT1 and ALYREF as important target molecules for HNSCC using a validation dataset (TCGA-CESC). A. Both DNMT1 and ALYREF were able to predict risks in the validation dataset. B. The AUC values were near 0.7. C. In the validation dataset, a lower DNMT1 expression was associated with poorer prognoses (P = 0.039).
Potential pathways for 5-methylcytosine regulators
To detect potential pathways for m5C regulators, we also performed a gene set variation analysis (GSVA). We assessed the correlation coefficients for m5C regulators and pathways and set a threshold of ± 0.5 for significantly related pathways. DNMT1 was found to be positively related to both mismatch repair and to the cell cycle (Figure 7A). In addition, we used m5C regulators and cancer pathways to perform a protein-protein interaction (PPI) network analysis. The m5C regulators, especially DNMT1, frequently interacted with each other (Figure 7B). These findings strongly suggest that DNMT1 is associated with cell-cycle pathways and regulates the function of m5C methylation during HNSCC.
Figure 7.
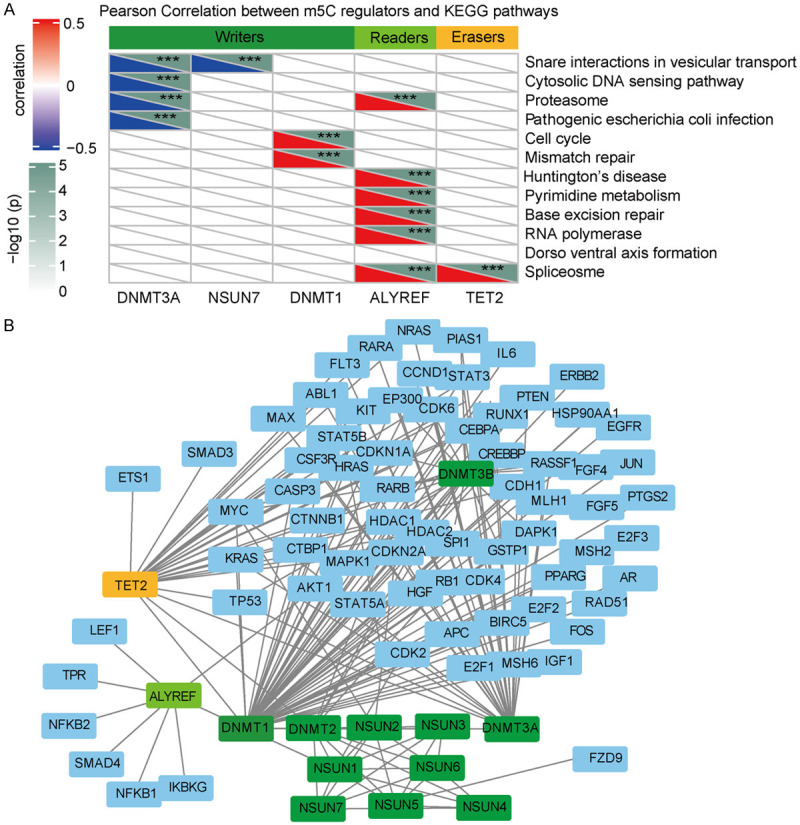
Functional analyses demonstrating that m5C regulators were closely related to cell-cycle pathways. A. GSVA analysis of m5C regulators. DNMT1 was positively correlated with mismatch-repair pathways and with cell-cycle pathways. The red color represents positive correlations, and *** denotes P < 0.001. B. Correlations between m5C regulators and cancer-pathway candidate genes demonstrated by a PPI network analysis; DNMT1 frequently interacted with candidate genes.
Discussion
The m5C is the most common methylation modification in eukaryotic mRNAs and plays essential roles in a variety of cellular, biological, and pathological processes. The “writers”, “erasers”, and “readers” are genes that regulate the methylation of cytosine. Levels of m5C methylation can be upregulated by the actions of “writers”, and reversed by “erasers”. “Readers” are effectors that bind to an m5C-site and modulate differential biological signals. Groundbreaking research has provided insights into the complex mechanisms of the reversible modifications regulated by the m5C-regulatory genes for multiple cancers [24]. Even so, the biological functions of these m5C-related genes, and how they regulate HNSCC disease development and progression, have not yet been determined.
Our TCGA dataset analyses suggest that m5C has many important regulatory effects, and that m5C levels are significantly associated with the expression levels of intracellular “writer” and “eraser” genes. The “readers” are also capable of regulating biological functions by binding to the methylation sites of m5C genes. The roles of m5C RNA methyltransferases in disease pathogenesis have also been widely studied. Mutations in the genes that encode m5C methyltransferases are linked to a variety of human diseases, and changes in m5C methyltransferase expression levels have also been observed in multiple cancers. Previous research has also indicated that functional mutations in NSUN2 are the basis for several neurodevelopmental disorders [25]. For example, Ma et al. [26] found that the NSUN2 mutation can cause the substitution of glycine 679 to arginine (p.Gly679Arg) in individuals with autosomal-recessive intellectual disabilities. In our research, mutations and CNVs in m5C-regulatory genes were observed in patients with HNSCC, and especially in the m5C “reader” gene DNMT3A, which had the highest frequency of mutations in our study. However, the molecular mechanisms linking HNSCC progression with regulatory-gene mutations require further exploration.
Alterations in m5C-regulatory genes may also be related to variations in HNSCC pathogenic molecules. Given that the control mechanisms for HNSCC pathogenesis are not clear, we employed the oncodrive function of maftools to predict driver genes. In doing so, we identified MAPK, HRAS, RAC1, HIST1H4K, and HIST1H3C. Moreover, variations in HRAS and RAC1 were significantly correlated with alterations in m5C regulatory genes. As an oncogene, HRAS mutations frequently occur in a variety of tumors, including pancreatic cancer [27], thyroid cancer [28], and bladder cancer [29]. A recent study identified associations between HRAS mutations and elevated antitumor immune signatures in HNSCC, demonstrating that HRAS may have a critical role in the immunoregulation of HNSCC [30]. RAC1, a member of the Rho family of ATP-dependent helicases, is also known to regulate the cell cycle, adhesion, motility, and epithelial-mesenchymal transitions [31]. In addition, RAC1 has many important functions in cancer, and may be a therapeutic target for many of them [32,33]. In particular, it has been shown to be a potential therapeutic target for HNSCC that is insensitive to chemoradiotherapy [34]. Thus, HRAS and RAC1 are both promising research targets to help expand our current understanding of HNSCC.
Our study concentrated on the gene signatures and prognostic values of m5C regulators in relation to HNSCC. Recently, Chen et al. [17] reported that the high co-expression of NUSN2, heparin binding growth factor (HBGF), and Y-box binding protein 1 (YBX1) could predict poor overall survival in patients with urothelial carcinoma of the bladder (UCB). In their study, NSUN2 (an m5C “writer”) and YBX1 (a m5C “reader”), drove tumor progression via an mRNA 3’-UTR m5C-methylation site in HBGF, this promoted m5C-regulated oncogene activation in UCB. With respect to HNSCC, the application of prognostic biomarkers is limited, and more predictive biomarkers are needed. Based on our research, the expressions of ALYREF and NSUN5 were positively correlated with patient clinical staging, indicating that the expression of m5C-regulatory genes may be used as prognostic markers for HNSCC. Moreover, a more comprehensive understanding of the biological, genetic, and epigenetic changes in HNSCC will contribute to the establishment of new prognostic biomarkers.
Importantly, two additional m5C-regulatory genes, DNMT1 and ALYREF, were identified by the LASSO regression analysis as strong predictors for HNSCC prognoses. These results demonstrated that high ALYREF expression and low DNMT1 expression were both associated with poor HNSCC prognosis. We found that the expression of these two genes effectively predicted the risk of mortality in HNSCC patients. Because the cell-origins and tumor tissue-structure in CESC are quite similar to that of HNSCC, data from TCGA-CESC was selected for validation of the relationship between patient survival and the expressions of DNMT1 and ALYREF. The Cox regression analysis indicated that DNMT1 and ALYREF also had strong predictive abilities using this validation dataset, and confirmed that m5C-regulatory genes are vitally important in HNSCC. Moreover, GSEA demonstrated that high ALYREF expression levels were associated with mitochondrial functions in HNSCC, and high DNMT1 levels were likely to be connected to peptide cross-linking and humoral immunity. Previous studies have reported that mitochondrial dysfunction promotes malignant tumor processes, and that the development of cancers depends on the precise regulation of mitochondrial function. Therefore, a meaningful and in-depth exploration of whether ALYREF influences energy metabolism during HNSCC is also warranted. Immunotherapies have made significant progress toward improving survival in patients with recurring or metastatic HNSCC [35]. Given that DNMT1 is related to humoral immunity, the potential value of DNMT1 use in immunotherapy also deserves further exploration.
Conclusions
The present findings have systematically demonstrated that gene alterations, expression levels, prognostic value, and other functional roles for m5C RNA methylation may exist in HNSCC patients. We identified, for the first time, genetic alterations in m5C-regulatory genes for HNSCC. We also found significant relationships between alterations in DNMT1 and ALYREF with increased mortality rates. Moreover, it is plausible that DNMT1 and ALYREF play essential biological roles during the malignant processes of HNSCC. Future cellular and molecular studies are required to further validate our findings and to better understand the genes targeted for m5C modifications during HNSCC initiation and progression.
Acknowledgements
We thank the patients and investigators who participated in TCGA for providing the data analyzed in this study. This study was supported by the Science and Technology Research Project of Henan Province (202102310074).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Marur S, Forastiere AA. Head and neck squamous cell carcinoma: update on epidemiology, diagnosis, and treatment. Mayo Clin Proc. 2016;91:386–396. doi: 10.1016/j.mayocp.2015.12.017. [DOI] [PubMed] [Google Scholar]
- 2.Blot WJ, McLaughlin JK, Winn DM, Austin DF, Greenberg RS, Preston-Martin S, Bernstein L, Schoenberg JB, Stemhagen A, Fraumeni JF Jr. Smoking and drinking in relation to oral and pharyngeal cancer. Cancer Res. 1988;48:3282–3287. [PubMed] [Google Scholar]
- 3.Argiris A, Karamouzis MV, Raben D, Ferris RL. Head and neck cancer. Lancet (London, England) 2008;371:1695–1709. doi: 10.1016/S0140-6736(08)60728-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sanderson RJ, Ironside JA. Squamous cell carcinomas of the head and neck. BMJ. 2002;325:822–827. doi: 10.1136/bmj.325.7368.822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coca-Pelaz A, Takes RP, Hutcheson K, Saba NF, Haigentz M Jr, Bradford CR, de Bree R, Strojan P, Lund VJ, Mendenhall WM, Nixon IJ, Quer M, Rinaldo A, Ferlito A. Head and neck cancer: a review of the impact of treatment delay on outcome. Adv Ther. 2018;35:153–160. doi: 10.1007/s12325-018-0663-7. [DOI] [PubMed] [Google Scholar]
- 6.Zhao BS, Roundtree IA, He C. Post-transcriptional gene regulation by mRNA modifications. Nat Rev Mol Cell Biol. 2017;18:31–42. doi: 10.1038/nrm.2016.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boccaletto P, Machnicka MA, Purta E, Piatkowski P, Baginski B, Wirecki TK, de Crécy-Lagard V, Ross R, Limbach PA, Kotter A, Helm M, Bujnicki JM. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018;46:D303–D307. doi: 10.1093/nar/gkx1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.WYATT GR. Occurrence of 5-methylcytosine in nucleic acids. Nature. 1950;166:237–238. doi: 10.1038/166237b0. [DOI] [PubMed] [Google Scholar]
- 9.Huang T, Chen W, Liu J, Gu N, Zhang R. Genome-wide identification of mRNA 5-methylcytosine in mammals. Nat Struct Mol Biol. 2019;26:380–388. doi: 10.1038/s41594-019-0218-x. [DOI] [PubMed] [Google Scholar]
- 10.Trixl L, Lusser A. The dynamic RNA modification 5-methylcytosine and its emerging role as an epitranscriptomic mark. Wiley Interdiscip Rev RNA. 2019;10:e1510. doi: 10.1002/wrna.1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reid R, Greene PJ, Santi DV. Exposition of a family of RNA m(5)C methyltransferases from searching genomic and proteomic sequences. Nucleic Acids Res. 1999;27:3138–3145. doi: 10.1093/nar/27.15.3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu H, Zhang Y. Mechanisms and functions of Tet protein-mediated 5-methylcytosine oxidation. Genes Dev. 2011;25:2436–2452. doi: 10.1101/gad.179184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang X, Yang Y, Sun BF, Chen YS, Xu JW, Lai WY, Li A, Wang X, Bhattarai DP, Xiao W, Sun HY, Zhu Q, Ma HL, Adhikari S, Sun M, Hao YJ, Zhang B, Huang CM, Huang N, Jiang GB, Zhao YL, Wang HL, Sun YP, Yang YG. 5-methylcytosine promotes mRNA export-NSUN2 as the methyltransferase and ALYREF as an mC reader. Cell Res. 2017;27:606–625. doi: 10.1038/cr.2017.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dominissini D, Rechavi G. 5-methylcytosine mediates nuclear export of mRNA. Cell Res. 2017;27:717–719. doi: 10.1038/cr.2017.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gilbert WV, Bell TA, Schaening C. Messenger RNA modifications: form, distribution, and function. Science. 2016;352:1408–1412. doi: 10.1126/science.aad8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bohnsack KE, Höbartner C, Bohnsack MT. Eukaryotic 5-methylcytosine (m5C) RNA methyltransferases: mechanisms, cellular functions, and links to disease. Genes. 2019;10:102. doi: 10.3390/genes10020102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen X, Li A, Sun BF, Yang Y, Han YN, Yuan X, Chen RX, Wei WS, Liu Y, Gao CC, Chen YS, Zhang M, Ma XD, Liu ZW, Luo JH, Lyu C, Wang HL, Ma J, Zhao YL, Zhou FJ, Huang Y, Xie D, Yang YG. 5-methylcytosine promotes pathogenesis of bladder cancer through stabilizing mRNAs. Nat Cell Biol. 2019;21:978–990. doi: 10.1038/s41556-019-0361-y. [DOI] [PubMed] [Google Scholar]
- 18.Pérez Sayáns M, Chamorro Petronacci CM, Lorenzo Pouso AI, Padín Iruegas E, Blanco Carrión A, Suárez Peñaranda JM, García García A. Comprehensive genomic review of TCGA head and neck squamous cell carcinomas (HNSCC) J Clin Med. 2019;8:1896. doi: 10.3390/jcm8111896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li J, Liu K, Zhang T, Yang Z, Wang R, Chen G, Kang M. A comprehensive investigation using meta-analysis and bioinformatics on miR-34a-5p expression and its potential role in head and neck squamous cell carcinoma. Am J Transl Res. 2018;10:2246–2263. [PMC free article] [PubMed] [Google Scholar]
- 20.He Y, Xue C, Yu Y, Chen J, Chen X, Ren F, Ren Z, Cui G, Sun R. CD44 is overexpressed and correlated with tumor progression in gallbladder cancer. Cancer Manag Res. 2018;10:3857–3865. doi: 10.2147/CMAR.S175681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xue C, He Y, Zhu W, Chen X, Yu Y, Hu Q, Chen J, Liu L, Ren F, Ren Z, Cui G, Sun R. Low expression of LACTB promotes tumor progression and predicts poor prognosis in hepatocellular carcinoma. Am J Transl Res. 2018;10:4152–4162. [PMC free article] [PubMed] [Google Scholar]
- 22.Ramly B, Afiqah-Aleng N, Mohamed-Hussein ZA. Protein-protein interaction network analysis reveals several diseases highly associated with polycystic ovarian syndrome. Int J Mol Sci. 2019;20:2959. doi: 10.3390/ijms20122959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He Y, Yu X, Li J, Zhang Q, Zheng Q, Guo W. Role of mC-related regulatory genes in the diagnosis and prognosis of hepatocellular carcinoma. Am J Transl Res. 2020;12:912–922. [PMC free article] [PubMed] [Google Scholar]
- 24.Delaunay S, Frye M. RNA modifications regulating cell fate in cancer. Nat Cell Biol. 2019;21:552–559. doi: 10.1038/s41556-019-0319-0. [DOI] [PubMed] [Google Scholar]
- 25.Flores JV, Cordero-Espinoza L, Oeztuerk-Winder F, Andersson-Rolf A, Selmi T, Blanco S, Tailor J, Dietmann S, Frye M. Cytosine-5 RNA methylation regulates neural stem cell differentiation and motility. Stem Cell Rep. 2017;8:112–124. doi: 10.1016/j.stemcr.2016.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khan MA, Rafiq MA, Noor A, Hussain S, Flores JV, Rupp V, Vincent AK, Malli R, Ali G, Khan FS, Ishak GE, Doherty D, Weksberg R, Ayub M, Windpassinger C, Ibrahim S, Frye M, Ansar M, Vincent JB. Mutation in NSUN2, which encodes an RNA methyltransferase, causes autosomal-recessive intellectual disability. Am J Hum Genet. 2012;90:856–863. doi: 10.1016/j.ajhg.2012.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weyandt J. Activation of wild-type hras suppresses the earliest stages of pancreatic cancer. Redox Biol. 2015;5:414. doi: 10.1016/j.redox.2015.09.014. [DOI] [PubMed] [Google Scholar]
- 28.Untch BR, Dos Anjos V, Garcia-Rendueles MER, Knauf JA, Krishnamoorthy GP, Saqcena M, Bhanot UK, Socci ND, Ho AL, Ghossein R, Fagin JA. Tipifarnib inhibits HRAS-driven dedifferentiated thyroid cancers. Cancer Res. 2018;78:4642–4657. doi: 10.1158/0008-5472.CAN-17-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sugita S, Enokida H, Yoshino H, Miyamoto K, Yonemori M, Sakaguchi T, Osako Y, Nakagawa M. HRAS as a potential therapeutic target of salirasib RAS inhibitor in bladder cancer. Int J Oncol. 2018;53:725–736. doi: 10.3892/ijo.2018.4435. [DOI] [PubMed] [Google Scholar]
- 30.Lyu H, Li M, Jiang Z, Liu Z, Wang X. TP53 correlate the mutation and the mutation with immune signatures in head and neck squamous cell cancer. Comput Struct Biotechnol J. 2019;17:1020–1030. doi: 10.1016/j.csbj.2019.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Payapilly A, Malliri A. Compartmentalisation of RAC1 signalling. Curr Opin Cell Biol. 2018;54:50–56. doi: 10.1016/j.ceb.2018.04.009. [DOI] [PubMed] [Google Scholar]
- 32.De P, Aske JC, Dey N. RAC1 takes the lead in solid tumors. Cells. 2019;8:382. doi: 10.3390/cells8050382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bid HK, Roberts RD, Manchanda PK, Houghton PJ. RAC1: an emerging therapeutic option for targeting cancer angiogenesis and metastasis. Mol Cancer Ther. 2013;12:1925–1934. doi: 10.1158/1535-7163.MCT-13-0164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Skvortsov S, Dudás J, Eichberger P, Witsch-Baumgartner M, Loeffler-Ragg J, Pritz C, Schartinger VH, Maier H, Hall J, Debbage P, Riechelmann H, Lukas P, Skvortsova I. Rac1 as a potential therapeutic target for chemo-radioresistant head and neck squamous cell carcinomas (HNSCC) Br J Cancer. 2014;110:2677–2687. doi: 10.1038/bjc.2014.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cohen EEW, Bell RB, Bifulco CB, Burtness B, Gillison ML, Harrington KJ, Le QT, Lee NY, Leidner R, Lewis RL, Licitra L, Mehanna H, Mell LK, Raben A, Sikora AG, Uppaluri R, Whitworth F, Zandberg DP, Ferris RL. The society for immunotherapy of cancer consensus statement on immunotherapy for the treatment of squamous cell carcinoma of the head and neck (HNSCC) J Immunother Cancer. 2019;7:184. doi: 10.1186/s40425-019-0662-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.