Abstract
Circular RNA (circRNA) is a special type of endogenous noncoding RNAs (ncRNAs), and are characterized by a covalently closed loop structure without a 5’ cap and poly-adenylated tails. Abnormal expression of circRNAs has been implicated in a wide range of human cancers, where they function as either tumor suppressor genes or oncogenes. CircHIPK3, circRNA homeodomain-interacting protein kinase 3, is associated with human cancers such as lung cancer, bladder cancer, hepatocellular carcinoma, colorectal cancer, osteosarcoma, glioma and prostate cancer, et al. Numerous studies have indicated that circHIPK3 functions as a miRNA sponge to regulate the target genes and exert specific biological effects, including regulation of cell proliferation, invasion, and migration. Furthermore, circHIPK3 is thought to be a novel diagnostic biomarker, therapeutic target, and prognostic biomarker in different cancer types. Here, we reviewed the recent progress of the mechanism and functions of circHIPK3 during the evolution of malignancies.
Keywords: Circular RNA, HIPK3, cancer, microRNA sponge
Introduction
Despite the remarkable progress made in cancer diagnosis and treatment over the past decades, cancer-related deaths are rising worldwide [1]. Oncogenesis is a multistep process mediated by a variety of factors and involves mutations in cellular genes and interaction of various signaling molecules and pathways, resulting in uncontrolled cell division, invasion, and metastasis [2]. Aberrant expression of RNA is associated with carcinogenesis [3]. Only a small minority of the total human genome sequence encodes proteins, about 98% of the genome is transcribed into noncoding RNAs, like long noncoding RNAs (lncRNAs), circular RNAs (circRNAs), miRNAs, and piwiRNAs [4,5].
CircRNA was found in RNA viruses by investigators for the first time when using an electron microscope in the 1970s and were initially thought to be the byproducts of alternative splicing. However, with the recent advancement of transcriptome sequencing technology, substantial circRNAs have been identified. More importantly, studies on circRNA are attracting more and more attention (Figure 1). Unlike linear RNAs, the structure of covalently closed-loop without a 5’ cap and poly-adenylated tail is the important feature of circRNA [6]. The majority of circRNAs are universal, conserved, stable, and particularly expressed in distinct tissues and developmental stages [7]. Its excellent stability that is not susceptible to degradation by RNase R and RNA exonuclease benefits from the covalently cyclic structure, since the half-life of circRNA is longer than that of linear mRNA. Besides regulating transcription and alternative splicing, and interacting with RNA binding proteins, miRNA sponges is one of the most frequently studied functions in circRNAs [8]. CircRNAs containing many miRNA response elements (MREs) competitively bind to miRNAs based on the principle of complementary base pairing, resulting in the reduction of the functional miRNAs and increased expression of target genes [8,9].
Figure 1.
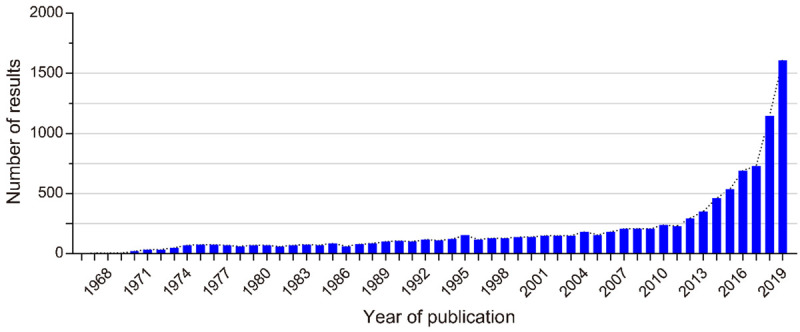
The increasing trend of the number of studies on circRNAs. A search was performed on PubMed using keywords “circular RNA” or “circRNA” (https://www.ncbi.nlm.nih.gov/pubmed, as of 2019).
Accumulating evidence indicates that circRNAs exert specific biological functions and participate in various physiological and pathophysiological processes in neoplastic cells, including proliferation, invasion, and migration [7,10]. Homeodomain-interacting protein kinase 3 (HIPK3 gene, alias PKY or FIST3) is a human protein kinase homolog of the yeast gene YAK1, cloned from a multidrug-resistant cell line (KB-V1) and functions as a co-repressor of homeodomain transcription factors [11]. CircRNA HIPK3 (circHIPK3) (circRNA ID: hsa_circ_0000284) is composed exclusively of a large second exon (1,099 nt) from the HIPK3 gene flanked on either side by long introns (Figure 2) [8,12,13]. Analysis of the flanking introns of the HIPK3 shows highly complementary Alu repeats with 51 short scattered elements downstream from Exon3 and 28 short interspersed elements in the intron upstream of HIPK3 Exon2 [13]. These short intronic repeat sequences promote the circularization of HIPK3 and are essential for the production of circHIPK3 [13,14]. Like most circRNAs, circHIPK3 is preferentially localized in the cytoplasm [12,13]. Moreover, circHIPK3 is most abundantly expressed in the brain, particularly in the cerebellum [14]. Recent studies indicate that circHIPK3 plays a vital role in the onset and progression of multiple cancers, and may act as a promising diagnostic biomarker or therapeutic target of cancer. We survey recent progress on the expression patterns and roles of circHIPK3 and expounds on the function and mechanisms of circHIPK3 in multiple malignancies.
Figure 2.
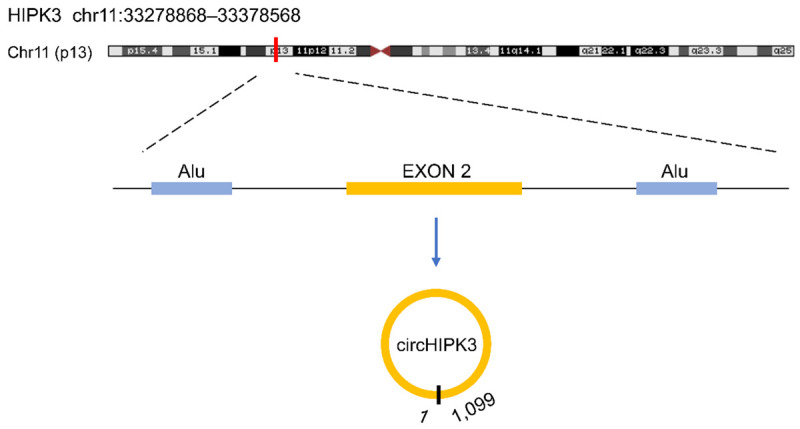
Schematic illustration of the formation of circHIPK3.
CircHIPK3 in malignancies of the respiratory system
Non-small cell lung cancer (NSCLC)
NSCLC takes up more than eighty percent of all lung cancer cases and causes over one million human mortalities each year [15]. Yu et al. found that circHIPK3 was abundant in human lung cancer tissues [16]. CircHIPK3 could facilitate cell survival and proliferation, and serve as a miR-124 sponge to modulate the level of downstream targets such as SphK1, CDK4, and STAT3, in NSCLC cells [16]. Chen et al. revealed that the quiescence of circHIPK3 effectively dampened cell proliferation, invasion, migration, and induced cell autophagy [17]. Mechanistically, they discovered that autophagy was induced through the miR-124-3p/STAT3/PRKAA/AMPKa pathway in mutant lung cancer cell lines (H838 and A549) [17]. Between circHIPK3 and linear HIPK3, it has been shown to have an antagonistic effect on autophagy. The ratio between circHIPK3 and linear HIPK3 (C/L value) reflects the levels of autophagy to some extent and a high C/L value (> 0.49) is an index of poor prognosis, particularly in advanced-stage patients [17]. Lu et al. discovered that circHIPK3 was involved in the regulation of the proliferation, invasion, and migration of NSCLC cells through the miR-149-mediated FOXM1 expression. MiR-149 could interact with FOXM1 mRNA via binding to its 3’-untranslated region [18]. These findings suggest circHIPK3 acts as an autophagy modulator and oncogene.
Nasopharyngeal carcinoma (NPC)
NPC is one of the most common head and neck cancers in Southeast Asia [19]. Ke and colleagues discovered that high expression level of circHIPK3 in NPC tissues [20]. NPC patients with a higher level of circHIPK3 indicated a lower overall survival (OS) and metastasis-free survival (MFS), meaning that it may be a prognostic indicator in NPC patients [20]. Besides, the decrease in circHIPK3 significantly impaired cell proliferation, migration, and invasion by acting as a molecular sponge of miR-4288 that targeted ELF3 in NPC cells. These results suggest that circHIPK3 promotes NPC progression through preventing ELF3 from miR-4288-mediated silencing [20].
CircHIPK3 in malignancies of the digestive system
Hepatocellular carcinoma (HCC)
HCC is the second leading cause of cancer-related death around the world [21]. The prognosis of HCC patients has hardly improved in recent years [22]. Chen and colleagues demonstrated that circHIPK3 was highly expressed in HCC tissues and positively associated with the level of aquaporin 3 (AQP3). CircHIPK3 silencing could inhibit malignant behavior of cells through decreasing the expression of AQP3 in HCC [23]. Xenograft tumor experiments indicated that circHIPK3 promoted tumor growth through the miR-124/AQP3 axis. Moreover, the upregulation of circHIPK3 was highly related to TNM stage, tumor differentiation, the presence of liver cirrhosis, and HBV-DNA copy numbers in patients with HCC [23].
Gallbladder cancer (GBC)
GBC is the most common tumor of the bile duct and a highly fatal and aggressive disease with poor prognosis [24]. Ding et al. demonstrated that the level of circHIPK3 was notably upregulated in GBC cells compared with that in gallbladder epithelial cells [25]. The elevated circHIPK3 effectively suppressed cell apoptosis and fostered cell proliferation in GBC by sequestering the tumor-suppressive miR-124 and resulting in enhanced level of downstream genes, including ROCK1 and CDK6 [25]. Taken together, circHIPK3 may be a potential biomarker of therapeutic and diagnosis target of human GBC.
Cholangiocarcinoma (CCA)
CCA is the second most common hepatic cancer and its overall incidence has gradually increased worldwide over the past decades [26]. Louis et al. found that the expression level of circHIPK3 was prominently enhanced in CCA cell lines, tumor tissues, and plasma exosomes [27]. Prediction analysis and functional assays have demonstrated that upregulated circHIPK3 enhances cell growth, invasion, and migration by sponging miR-637 that targets lymphocyte antigen-6 E (LY6E) in CCA [27]. These studies not only highlight the role of circHIPK3 as a promoter in the pathogenesis of CCA but also as a potential molecule for innovative non-invasive biomarker, since circHIPK3 is stable and enriched in exosomes.
Gastric cancer (GC)
Gastric cancer is a lethal disease and the fifth most common human cancer [28]. Cheng and colleagues showed that the level of circHIPK3 was remarkably upregulated in the GC tissues compared to para-cancerous tissues, and higher expression was observed in infiltrative-type GC cells than in expanding-type GC cells [29]. CircHIPK3 promoted cell proliferation by negatively regulating the level of miR-29b/miR-124 [29]. Bioinformatics analysis demonstrated that targets expression of the circHIPK3/miR-29b/miR-124 axis in tumor-related pathways were related to survival time in patients with GC, and even could forecast the GC status [29]. Besides, Liu et al. revealed that upregulated circHIPK3 indicated poor prognosis of patients, and the depletion of circHIPK3 inhibited the proliferation and migration of cancer cells [30]. CircHIPK3 accumulation accelerated the development of GC by regulating the Wnt/β-catenin pathway [30]. However, Ghasemi and colleagues demonstrated that the level of circHIPK3 was significantly downregulated in GC and its expression was correlated with age and M classification [31].
Colorectal cancer (CRC)
CRC is the third most common malignancy and about 50% of the patients are already at an advanced stage [32]. Zeng et al. found that c-Myb increased the level of circHIPK3 through directly interacting with its promoter region [33]. Knockdown of circHIPK3 impaired the malignant phenotypes of tumor cells in vitro, and impeded growth and metastasis of CRC in xenograft tumor model. Overexpression of circHIPK3 reversed miR-7-induced alleviation of malignant biological behavior of CRC cells via elevating the level of miR-7 targeting downstream regulatory molecules such as IGF1R, FAK, YY1, and EGFR [33]. Furthermore, it was found that circHIPK3 was remarkably enhanced in CRC, and positively related to clinical stage and distant metastasis. Multivariate Cox’s analysis indicated that increased circHIPK3 was an independent prognostic factor of poor overall survival in CRC [33]. Formin like 2 (FMNL2) was highly correlated with various cancers metastasis through the generation of protruding actin structures at the leading edge of a migrating cell [34]. CircHIPK3 could boost CRC cell growth and metastasis by regulating the miR-1207-5p/FMNL2 axis [35].
Barbagallo et al. investigated the level of circRNAs in serum exosomes from CRC patients and revealed that circHIPK3 was upregulated in serum exosomes of CRC patients relative to healthy controls [36]. Zhang et al. discovered that the expression of circHIPK3 was significantly increased in chemo-resistant CRC patients [37]. CircHIPK3 promoted oxaliplatin resistance through the suppression of autophagy. Functionally, circHIPK3 sponged miR-637 to facilitate the expression of a key gene STAT3, thus stimulating the downstream Bcl-2/beclin1 pathway [37]. CircHIPK3 acts as a chemo-resistant gene in CRC, and a promising prognostic indicator for patients with CRC, especially those receiving oxaliplatin-based chemotherapy.
CircHIPK3 in malignancies of the urinary system
Renal carcinoma (RC)
Statistics demonstrate that approximately one fifth of patients with RC have cancer metastasis at first diagnosis and the 5-year survival of metastatic RC is not optimistic [38]. CircHIPK3 boosted metastasis and suppressed apoptosis of RC cells by mediating miR-485-3p. Mechanistically, circHIPK3 was associated with the effect of miR-485-3p by partially reversing the upregulated levels of E-Cadherin, Bax, clever caspase-3 and downregulated levels of Vimentin, N-Cadherin and Bcl-2, which were epithelial-mesenchymal transition (EMT) and apoptosis-related proteins [39]. These observations offer a footstone for molecular-targeted treatment for RC patients.
Bladder cancer (BC)
BC is the mostly diagnosed malignancy of the urinary system [40]. Li et al. showed that the expression of circHIPK3 was dramatically reduced in BC tissues, and negatively related to tumor invasion, grade, and lymph node metastasis in patients with BC [41]. Accumulation of circHIPK3 was reported to significantly impair invasion, migration, and angiogenesis of BC cells in vitro, and inhibit cell growth and metastasis in vivo [41]. Furthermore, circHIPK3 harbored two important MREs for the miR-558 and acted as a molecular sponge to inhibit the expression of heparanase (HPSE) [41]. Okholm and colleagues analyzed the profiling of circRNAs in BC and found that circHIPK3 was strongly associated with aggressiveness in early-stage BC [42]. Therefore, circHIPK3 might be a potentially prognostic factor for BC patients.
CircHIPK3 in malignancies of the reproductive system
Epithelial ovarian cancer (EOC)
EOC is the lethal gynecologic cancer. It accounts for about 90% of ovarian cancer and the 5-year survival for patients is less than 40% [43,44]. Liu et al. found that the level of circHIPK3 was markedly enhanced in EOC tissues and cell lines, and potently associated with lymph node invasion and FIGO stage. Upregulation of circHIPK3 was linked to worse overall survival and disease-free survival of EOC patients [45]. Besides, multivariate Cox’s analysis revealed that circHIPK3 was an independent prognostic factor for EOC patients [45].
However, Teng at al. showed that circHIPK3 was remarkedly reduced in EOC [46]. CircHIPK3 silencing promoted cell proliferation, invasion, migration, and suppressed apoptosis of EOC cells by sponging a series of miRNAs, including miR-10a, miR-106a and miR-148b [46]. Therefore, circHIPK3 may be a crucial modulator of EOC progression.
Cervical cancer (CC)
CC is the one of most common cancer in women and the 5-year overall survival is less than 40% [1,47]. Hypoxia is a common feature of solid tumors in middle or advanced stages and exerts an important function in tumorigenesis and progression by propelling the generation of a tumor microenvironment. HIF-1α is widely expressed in various cells and regarded as the most significant modulator of oxygen homeostasis [48]. Qian et al. revealed that circHIPK3 acted as a sponge molecule of miR-338-3p to facilitate cell growth and metastasis in vivo, via modulating HIF-1α mediated EMT [49].
Prostate cancer (PCa)
PCa is common cancer in the urinary system and one of the leading causes of cancer-related death in men [1]. Chen et al. showed that circHIPK3 was significantly upregulated in PCa and an important prognostic factor in PCa patients [50]. Besides, circHIPK3 served as a competing endogenous RNA for miR-193a-3p to suppress the role on a proto-oncogene MCL1 and facilitated cell growth and metastasis of PCa [50]. Cai et al. revealed that overexpression of circHIPK3 enhanced the proliferative and invasive abilities of PCa cells through targeting miRNA-338-3p to modulate the expression of ADAM17, and thus accelerated the malignant progression of PCa [51]. Furthermore, circHIPK3 facilitated G2/M transition to stimulating PCa cell growth through controlling miR-338-3p to potentiate the levels of target gene Cdc2 and Cdc25B [52].
CircHIPK3 in malignancies of the nervous system
Glioma
Glioma the deadly disease of the central nervous system and approximately 80% of glioma tumors are characterized by invasiveness and a high recurrence rate [53]. Jin et al. discovered that circHIPK3 was highly expressed in glioma, and related to poor prognosis [54]. Functionally, circHIPK3 promoted the progression of glioma, including proliferation and invasion, by targeting miR-654 from IGF2BP3 [54]. These findings indicate that circHIPK3 might be an effective therapeutic target for glioma. Consistent with these findings, Hu et al. observed that the expression of circHIPK3 and STAT3 were upregulated, while miR-124-3p was downregulated in glioma cells [55]. Another study found that circHIPK3 promoted the growth and invasion of glioma cells via increasing the level of STAT3 after sponging miR-124-3p [55]. Besides, recent studies have found that circHIPK3 was also present in exosomes secreted by tumor cells. Exosomal circHIPK3 promoted cancer development and temozolomide resistance through controlling the miR-421/ZIC5 axis [56].
CircHIPK3 in malignancies of the hematopoietic system
Chronic myeloid leukemia (CML)
CML is a clonal myeloproliferative disease of pluripotent hematopoietic stem cells and takes up about 15% of leukemia cases [57]. Feng et al. demonstrated that circHIPK3 was significantly increased in serum samples from CML and peripheral blood mononuclear cells (PBMC) in comparison to healthy controls [58]. The Kaplan-Meier analysis demonstrated that patients containing high levels of circHIPK3 had a lower survival rate and predicted a poor outcome in CML patients [58]. The loss-function experiments indicated that circHIPK3 impaired cell proliferation and induced cell apoptosis, revealing an oncogenic role of circHIPK3 in CML [58]. These results provide a novel insight into the role of circHIPK3 in the progression and treatment of CML.
CircHIPK3 in malignancies of the skeletal-muscular system
Osteosarcoma (OS)
OS is the most prevalent bone carcinoma in children and young adolescents [59]. Due to the lack of an effective biomarker for early diagnosis, the prognosis of OS is poor, Ma et al. found that circHIPK3 could promote the malignant phenotypes of OS cells [60]. Clinically, the level of circHIPK3 was significantly downregulated in the OS tissues, plasmas, and cell lines, and highly correlated with lung metastasis and Enneking stage. Lower expression of circHIPK3 was associated with shorter overall survival time and poor prognosis in patients with OS. In addition, ROC analysis showed that the specificity and sensitivity were 0.84 and 0.56, respectively, and the area under the ROC curve was 0.783 [60].
Conclusions
CircHIPK3 has been shown to function as a molecule sponge to competitively “absorb” miRNAs and exert its different functions (Figure 3). Numerous studies have indicated that the biological role of circHIPK3 is cancer type-specificity, and it has been verified to be a carcinogenic or tumor suppressive factor in distinct types of cancer (Figure 4 and Table 1). Pancreatic cancer is one of the most devastating human cancers, with a rising incidence, however, till now, the roles and mechanisms of circHIPK3 in pancreatic cancer is still lacking, which should be further explored. As a miRNA sponge, circHIPK3 makes the circRNA-miRNA-mRNA regulatory network more abundant. Nevertheless, now the exploration of circHIPK3 mainly stays on the mechanism of miRNA sponge regarding tumor research, and it does not stand for all the functional patterns of circHIPK3. In the future, it is important to investigate other new functions or mechanisms of circHIPK3 in cancers.
Figure 3.
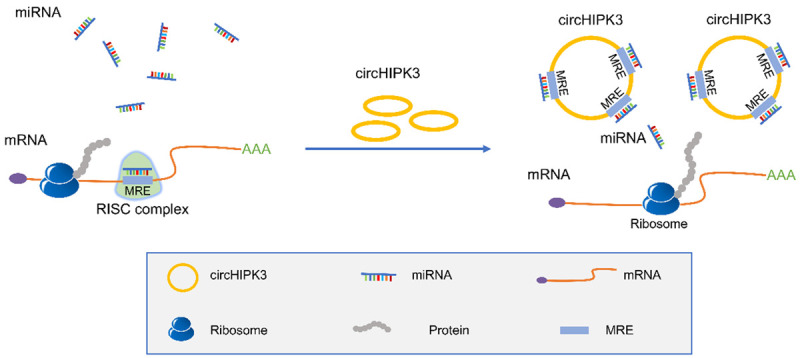
Schematic diagram showing the mechanism by which circHIPK3 acts as miRNA sponges. MiRNAs can directly bind to the matched regions of mRNAs through the seed region in a base-pairing manner, and trigger cleavage of mRNAs to inhibit the translational process of mRNAs by forming RNA-induced silencing complex (RISC). CircHIPK3 harbors multiple miRNA response elements (MREs) that contain complementary miRNA binding sites which bind to miRNAs competitively. As a consequence, circHIPK3 can indirectly regulate the expression of mRNAs and exert its biological functions by sequestering functional miRNA molecules.
Figure 4.
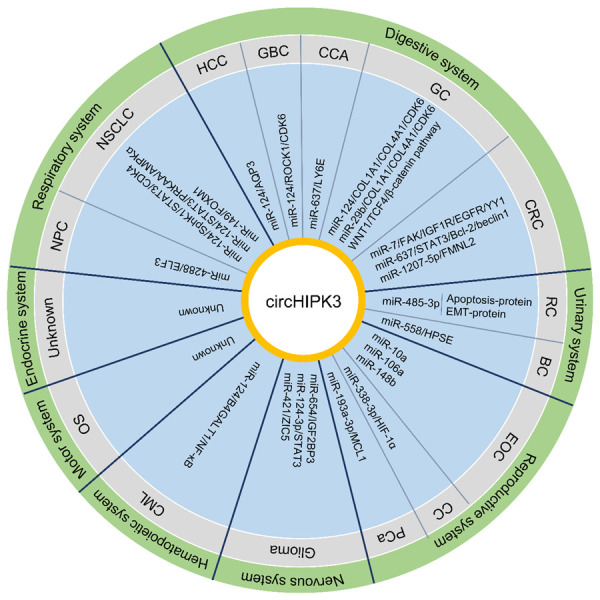
The schematic illustration showing the molecular mechanisms of circHIPK3 in multiple cancers. BC, bladder cancer; CC, cervical cancer; CCA, cholangiocarcinoma; CML, chronic myeloid leukemia; CRC, colorectal cancer; EOC, epithelial ovarian cancer; GBC, gallbladder cancer; GC, gastric cancer; HCC, Hepatocellular carcinoma; NPC, nasopharyngeal carcinoma; NSCLC, non-small cell lung cancer; OS, osteosarcoma; PCa, prostate cancer; RC, renal carcinoma.
Table 1.
Expression, role, mechanism, function and clinical significance concerning circHIPK3 in various cancers
| Cancer | Expression | Role | Mechanism (miRNA/target gene) | Function | Clinical significance | Reference |
|---|---|---|---|---|---|---|
| NSCLC | Up | O | MiR-124/SphK1/STAT3/CDK4 | Cell proliferation, apoptosis, migration, invasion, autophagy | - | [16-18] |
| MiR-124/STAT3/PRKAA/AMPKα | ||||||
| MiR-149/FOXM1 | ||||||
| NPC | Up | O | MiR-4288/ELF3 | Cell proliferation, migration, invasion | Clinical stage, distant metastasis, prognosis | [20] |
| HCC | Up | O | MiR-124/AQP3 | Cell proliferation, migration | Differentiation, TNM stage, cirrhosis, HBV-DNA copy numbers | [23] |
| GBC | Up | O | MiR-124/ROCK1/CDK6 | Cell proliferation | - | [25] |
| CCA | Up | O | MiR-637/LY6E | Cell proliferation, migration, and invasion | - | [27] |
| GC | Up/Down | O/S | MiR-124/miR-29b/COL1A1/COL4A1/CDK6 | Cell proliferation, migration | Age, T stage, Ming’s classification, prognosis | [29-31] |
| WNT1/TCF4/β-catenin pathway | ||||||
| CRC | Up | O | MiR-7/FAK/IGF1R/EGFR/YY1 | Cell proliferation, apoptosis, migration, invasion, autophagy | Tumor size, lymph node metastasis, distant metastasis, TNM stage, chemoresistance, prognosis | [33,35,37] |
| MiR-637/STAT3/Bcl-2/beclin1 | ||||||
| MiR-1207-5p/FMNL2 | ||||||
| RC | Up | O | MiR-485-3p/Clever caspase-3/Bax/Bcl-2/E-Cadherin/N-Cadherin/Vimentin/Ki-67 | Cell proliferation, migration, and invasion | Prognosis | [39] |
| BC | Down | S | MiR-558/HPSE | Cell proliferation, invasion, migration, angiogenesis | Grade, invasion, lymph node metastasis, prognosis | [41] |
| EOC | Up/Down | O/S | MiR-10a, miR-106a, miR-148b | Cell proliferation, apoptosis, migration, invasion | Lymph node invasion, FIGO stage, prognosis | [45,46] |
| CC | Up | O | MiR-338-3p/HIF-1α | Cell proliferation, apoptosis, migration, invasion | - | [49] |
| PCa | Up | O | MiR-193a-3p/MCL1 | Cell cycle, apoptosis, proliferation, migration, and invasion | Gleason score, tumor stage, prognosis | [50-52] |
| Glioma | Up | O | MiR-654/IGF2BP3 | Cell proliferation, cell cycle, apoptosis invasion | Prognosis | [54,55] |
| MiR-124-3p/STAT3 | ||||||
| MiR-421/ZIC5 | ||||||
| CML | Up | O | MiR-124/B4GALT1/NF-κB | Cell proliferation, apoptosis | Sokal relative risk, prognosis | [58] |
| OS | Down | S | - | Cell proliferation, migration and invasion | Enneking stage, lung metastasis, prognosis | [60] |
Abbreviations: Up, upregulated in cancer; Down, downregulated in cancer; O, oncogene role; S, suppressor role; HCC, Hepatocellular carcinoma; GBC, gallbladder cancer; CCA, cholangiocarcinoma; GC, gastric cancer; CRC, colorectal cancer; RC, renal carcinoma; BC, bladder cancer; EOC, epithelial ovarian cancer; CC, cervical cancer; PCa, prostate cancer; NSCLC, non-small cell lung cancer; OS, osteosarcoma; NPC, nasopharyngeal carcinoma; CML, chronic myeloid leukemia.
One of the major features of circRNA compared to linear RNA molecules is that they are more stable and not susceptible to degradation. CircHIPK3 can accumulate in cells and sustain its level and role for a considerably long time on account of its high stability, which is one of the prerequisites of a clinical biomarker. Therefore, circHIPK3 is regarded as a new diagnostic or prognostic indicator and a promising therapeutic target for certain cancer types. Of note, the combined detection using circHIPK3 and traditional tumor markers can increase the specificity and sensitivity of diagnosis.
This systematic analysis of the circHIPK3 significantly deepens our understanding of cancer pathogenesis in different cancer types. Moreover, circHIPK3 presents an extremely potential value of clinical application in diagnosis, therapy, and prognosis of tumor. Despite the accurate mechanisms of circ-HIPK3 in cancers remain limited, we believe the mystery of circHIPK3 will eventually be disclosed with the development of sequencing technologies and biological methods.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (81673023, 81272573, 81872501, and 81502068), Beijing Natural Science Foundation (7172177).
Disclosure of conflict of interest
None.
Abbreviations
- BC
bladder cancer
- CC
cervical cancer
- CCA
cholangiocarcinoma
- CircHIPK3
circRNA homeodomain-interacting protein kinase 3
- CML
chronic myeloid leukemia
- CRC
colorectal cancer
- EMT
epithelial-mesenchymal transition
- EOC
epithelial ovarian cancer
- GBC
gallbladder cancer
- GC
gastric cancer
- HCC
Hepatocellular carcinoma
- miRNA
microRNA
- MREs
miRNA response elements
- NPC
nasopharyngeal carcinoma
- NSCLC
non-small cell lung cancer
- OS
osteosarcoma
- PCa
prostate cancer
- RC
renal carcinoma
- UTR
untranslated region
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Puisieux A, Pommier RM, Morel AP, Lavial F. Cellular pliancy and the multistep process of tumorigenesis. Cancer Cell. 2018;33:164–172. doi: 10.1016/j.ccell.2018.01.007. [DOI] [PubMed] [Google Scholar]
- 3.Obeng EA, Stewart C, Abdel-Wahab O. Altered RNA processing in cancer pathogenesis and therapy. Cancer Discov. 2019;9:1493–1510. doi: 10.1158/2159-8290.CD-19-0399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lorenzi L, Avila Cobos F, Decock A, Everaert C, Helsmoortel H, Lefever S, Verboom K, Volders PJ, Speleman F, Vandesompele J, Mestdagh P. Long noncoding RNA expression profiling in cancer: challenges and opportunities. Genes Chromosomes Cancer. 2019;58:191–199. doi: 10.1002/gcc.22709. [DOI] [PubMed] [Google Scholar]
- 5.Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12:381–388. doi: 10.1080/15476286.2015.1020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geng Y, Jiang J, Wu C. Function and clinical significance of circRNAs in solid tumors. J Hematol Oncol. 2018;11:98. doi: 10.1186/s13045-018-0643-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, le Noble F, Rajewsky N. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 9.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 10.Zhang ZR, Yang TT, Xiao JJ. Circular RNAs: promising biomarkers for human diseases. Ebiomedicine. 2018;34:267–274. doi: 10.1016/j.ebiom.2018.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim YH, Choi CY, Lee SJ, Conti MA, Kim Y. Homeodomain-interacting protein kinases, a novel family of co-repressors for homeodomain transcription factors. J Biol Chem. 1998;273:25875–25879. doi: 10.1074/jbc.273.40.25875. [DOI] [PubMed] [Google Scholar]
- 12.Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, Luo Y, Lyu D, Li Y, Shi G, Liang L, Gu J, He X, Huang S. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215. doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liang D, Wilusz JE. Short intronic repeat sequences facilitate circular RNA production. Genes Dev. 2014;28:2233–2247. doi: 10.1101/gad.251926.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 16.Yu H, Chen Y, Jiang P. Circular RNA HIPK3 exerts oncogenic properties through suppression of miR-124 in lung cancer. Biochem Biophys Res Commun. 2018;506:455–462. doi: 10.1016/j.bbrc.2018.10.087. [DOI] [PubMed] [Google Scholar]
- 17.Chen X, Mao R, Su W, Yang X, Geng Q, Guo C, Wang Z, Wang J, Kresty LA, Beer DG, Chang AC, Chen G. Circular RNA circHIPK3 modulates autophagy via MIR124-3p-STAT3-PRKAA/AMPKalpha signaling in STK11 mutant lung cancer. Autophagy. 2020;16:659–671. doi: 10.1080/15548627.2019.1634945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu H, Han X, Ren J, Ren K, Li Z, Sun Z. Circular RNA HIPK3 induces cell proliferation and inhibits apoptosis in non-small cell lung cancer through sponging miR-149. Cancer Biol Ther. 2020;21:113–121. doi: 10.1080/15384047.2019.1669995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chua MLK, Wee JTS, Hui EP, Chan ATC. Nasopharyngeal carcinoma. Lancet. 2016;387:1012–1024. doi: 10.1016/S0140-6736(15)00055-0. [DOI] [PubMed] [Google Scholar]
- 20.Ke Z, Xie F, Zheng C, Chen D. CircHIPK3 promotes proliferation and invasion in nasopharyngeal carcinoma by abrogating miR-4288-induced ELF3 inhibition. J Cell Physiol. 2019;234:1699–1706. doi: 10.1002/jcp.27041. [DOI] [PubMed] [Google Scholar]
- 21.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 22.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 23.Chen G, Shi Y, Liu M, Sun J. circHIPK3 regulates cell proliferation and migration by sponging miR-124 and regulating AQP3 expression in hepatocellular carcinoma. Cell Death Dis. 2018;9:175. doi: 10.1038/s41419-017-0204-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sharma A, Sharma KL, Gupta A, Yadav A, Kumar A. Gallbladder cancer epidemiology, pathogenesis and molecular genetics: recent update. World J Gastroenterol. 2017;23:3978–3998. doi: 10.3748/wjg.v23.i22.3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kai D, Yannian L, Yitian C, Dinghao G, Xin Z, Wu J. Circular RNA HIPK3 promotes gallbladder cancer cell growth by sponging microRNA-124. Biochem Biophys Res Commun. 2018;503:863–869. doi: 10.1016/j.bbrc.2018.06.088. [DOI] [PubMed] [Google Scholar]
- 26.Saha SK, Zhu AX, Fuchs CS, Brooks GA. Forty-year trends in cholangiocarcinoma incidence in the U. S.: intrahepatic disease on the rise. Oncologist. 2016;21:594–599. doi: 10.1634/theoncologist.2015-0446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Louis C, Desoteux M, Coulouarn C. Exosomal circRNAs: new players in the field of cholangiocarcinoma. Clin Sci (Lond) 2019;133:2239–2244. doi: 10.1042/CS20190940. [DOI] [PubMed] [Google Scholar]
- 28.Van Cutsem E, Sagaert X, Topal B, Haustermans K, Prenen H. Gastric cancer. Lancet. 2016;388:2654–2664. doi: 10.1016/S0140-6736(16)30354-3. [DOI] [PubMed] [Google Scholar]
- 29.Cheng J, Zhuo H, Xu M, Wang L, Xu H, Peng J, Hou J, Lin L, Cai J. Regulatory network of circRNA-miRNA-mRNA contributes to the histological classification and disease progression in gastric cancer. J Transl Med. 2018;16:216. doi: 10.1186/s12967-018-1582-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu WG, Xu Q. Upregulation of circHIPK3 promotes the progression of gastric cancer via Wnt/beta-catenin pathway and indicates a poor prognosis. Eur Rev Med Pharmacol Sci. 2019;23:7905–7912. doi: 10.26355/eurrev_201909_19004. [DOI] [PubMed] [Google Scholar]
- 31.Ghasemi S, Emadi-Baygi M, Nikpour P. Down-regulation of circular RNA ITCH and circHIPK3 in gastric cancer tissues. Turk J Med Sci. 2019;49:687–695. doi: 10.3906/sag-1806-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet. 2014;383:1490–1502. doi: 10.1016/S0140-6736(13)61649-9. [DOI] [PubMed] [Google Scholar]
- 33.Zeng K, Chen X, Xu M, Liu X, Hu X, Xu T, Sun H, Pan Y, He B, Wang S. CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell Death Dis. 2018;9:417. doi: 10.1038/s41419-018-0454-8. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.Zeng Y, Xie H, Qiao Y, Wang J, Zhu X, He G, Li Y, Ren X, Wang F, Liang L, Ding Y. Formin-like2 regulates Rho/ROCK pathway to promote actin assembly and cell invasion of colorectal cancer. Cancer Sci. 2015;106:1385–1393. doi: 10.1111/cas.12768. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35.Yan Y, Su M, Qin B. CircHIPK3 promotes colorectal cancer cells proliferation and metastasis via modulating of miR-1207-5p/FMNL2 signal. Biochem Biophys Res Commun. 2020;524:839–846. doi: 10.1016/j.bbrc.2020.01.055. [DOI] [PubMed] [Google Scholar]
- 36.Barbagallo C, Brex D, Caponnetto A, Cirnigliaro M, Scalia M, Magnano A, Caltabiano R, Barbagallo D, Biondi A, Cappellani A, Basile F, Di Pietro C, Purrello M, Ragusa M. LncRNA UCA1, upregulated in CRC biopsies and downregulated in serum exosomes, controls mRNA expression by RNA-RNA interactions. Mol Ther Nucleic Acids. 2018;12:229–241. doi: 10.1016/j.omtn.2018.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Y, Li C, Liu X, Wang Y, Zhao R, Yang Y, Zheng X, Zhang Y, Zhang X. circHIPK3 promotes oxaliplatin-resistance in colorectal cancer through autophagy by sponging miR-637. EBioMedicine. 2019;48:277–288. doi: 10.1016/j.ebiom.2019.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Znaor A, Lortet-Tieulent J, Laversanne M, Jemal A, Bray F. International variations and trends in renal cell carcinoma incidence and mortality. Eur Urol. 2015;67:519–530. doi: 10.1016/j.eururo.2014.10.002. [DOI] [PubMed] [Google Scholar]
- 39.Lai J, Xin J, Fu C, Zhang W. CircHIPK3 promotes proliferation and metastasis and inhibits apoptosis of renal cancer cells by inhibiting MiR-485-3p. Cancer Cell Int. 2020;20:248. doi: 10.1186/s12935-020-01319-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Antoni S, Ferlay J, Soerjomataram I, Znaor A, Jemal A, Bray F. Bladder cancer incidence and mortality: a global overview and recent trends. Eur Urol. 2017;71:96–108. doi: 10.1016/j.eururo.2016.06.010. [DOI] [PubMed] [Google Scholar]
- 41.Li Y, Zheng F, Xiao X, Xie F, Tao D, Huang C, Liu D, Wang M, Wang L, Zeng F, Jiang G. CircHIPK3 sponges miR-558 to suppress heparanase expression in bladder cancer cells. EMBO Rep. 2017;18:1646–1659. doi: 10.15252/embr.201643581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Okholm TLH, Nielsen MM, Hamilton MP, Christensen LL, Vang S, Hedegaard J, Hansen TB, Kjems J, Dyrskjot L, Pedersen JS. Circular RNA expression is abundant and correlated to aggressiveness in early-stage bladder cancer. NPJ Genom Med. 2017;2:36. doi: 10.1038/s41525-017-0038-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Torre LA, Trabert B, DeSantis CE, Miller KD, Samimi G, Runowicz CD, Gaudet MM, Jemal A, Siegel RL. Ovarian cancer statistics, 2018. CA Cancer J Clin. 2018;68:284–296. doi: 10.3322/caac.21456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bookman MA. Optimal primary therapy of ovarian cancer. Ann Oncol. 2016;27(Suppl 1):i58–i62. doi: 10.1093/annonc/mdw088. [DOI] [PubMed] [Google Scholar]
- 45.Liu N, Zhang J, Zhang LY, Wang L. CircHIPK3 is upregulated and predicts a poor prognosis in epithelial ovarian cancer. Eur Rev Med Pharmacol Sci. 2018;22:3713–3718. doi: 10.26355/eurrev_201806_15250. [DOI] [PubMed] [Google Scholar]
- 46.Teng F, Xu J, Zhang M, Liu S, Gu Y, Zhang M, Wang X, Ni J, Qian B, Shen R, Jia X. Comprehensive circular RNA expression profiles and the tumor-suppressive function of circHIPK3 in ovarian cancer. Int J Biochem Cell Biol. 2019;112:8–17. doi: 10.1016/j.biocel.2019.04.011. [DOI] [PubMed] [Google Scholar]
- 47.Allemani C, Weir HK, Carreira H, Harewood R, Spika D, Wang XS, Bannon F, Ahn JV, Johnson CJ, Bonaventure A, Marcos-Gragera R, Stiller C, Azevedo e Silva G, Chen WQ, Ogunbiyi OJ, Rachet B, Soeberg MJ, You H, Matsuda T, Bielska-Lasota M, Storm H, Tucker TC, Coleman MP CONCORD Working Group. Global surveillance of cancer survival 1995-2009: analysis of individual data for 25,676,887 patients from 279 population-based registries in 67 countries (CONCORD-2) Lancet. 2015;385:977–1010. doi: 10.1016/S0140-6736(14)62038-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang W, Ma J, Zhou W, Cao B, Zhou X, Zhang H, Zhao Q, Hong L, Fan D. Reciprocal regulations between miRNAs and HIF-1alpha in human cancers. Cell Mol Life Sci. 2019;76:453–471. doi: 10.1007/s00018-018-2941-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Qian W, Huang T, Feng W. Circular RNA HIPK3 promotes EMT of cervical cancer through sponging miR-338-3p to up-regulate HIF-1alpha. Cancer Manag Res. 2020;12:177–187. doi: 10.2147/CMAR.S232235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen D, Lu X, Yang F, Xing N. Circular RNA circHIPK3 promotes cell proliferation and invasion of prostate cancer by sponging miR-193a-3p and regulating MCL1 expression. Cancer Manag Res. 2019;11:1415–1423. doi: 10.2147/CMAR.S190669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cai C, Zhi Y, Wang K, Zhang P, Ji Z, Xie C, Sun F. CircHIPK3 overexpression accelerates the proliferation and invasion of prostate cancer cells through regulating miRNA-338-3p. Onco Targets Ther. 2019;12:3363–3372. doi: 10.2147/OTT.S196931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu F, Fan Y, Ou L, Li T, Fan J, Duan L, Yang J, Luo C, Wu X. CircHIPK3 facilitates the G2/M transition in prostate cancer cells by sponging miR-338-3p. Onco Targets Ther. 2020;13:4545–4558. doi: 10.2147/OTT.S242482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Van Meir EG, Hadjipanayis CG, Norden AD, Shu HK, Wen PY, Olson JJ. Exciting new advances in neuro-oncology: the avenue to a cure for malignant glioma. CA Cancer J Clin. 2010;60:166–193. doi: 10.3322/caac.20069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jin P, Huang Y, Zhu P, Zou Y, Shao T, Wang O. CircRNA circHIPK3 serves as a prognostic marker to promote glioma progression by regulating miR-654/IGF2BP3 signaling. Biochem Biophys Res Commun. 2018;503:1570–1574. doi: 10.1016/j.bbrc.2018.07.081. [DOI] [PubMed] [Google Scholar]
- 55.Hu D, Zhang Y. Circular RNA HIPK3 promotes glioma progression by binding to miR-124-3p. Gene. 2019;690:81–89. doi: 10.1016/j.gene.2018.11.073. [DOI] [PubMed] [Google Scholar]
- 56.Han C, Wang S, Wang H, Zhang J. Exosomal Circ-HIPK3 facilitates tumor progression and temozolomide resistance by regulating miR-421/ZIC5 axis in glioma. Cancer Biother Radiopharm. 2020 doi: 10.1089/cbr.2019.3492. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 57.Loscocco F, Visani G, Galimberti S, Curti A, Isidori A. BCR-ABL independent mechanisms of resistance in chronic myeloid leukemia. Front Oncol. 2019;9:939. doi: 10.3389/fonc.2019.00939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Feng XQ, Nie SM, Huang JX, Li TL, Zhou JJ, Wang W, Zhuang LK, Meng FJ. Circular RNA circHIPK3 serves as a prognostic marker to promote chronic myeloid leukemia progression. Neoplasma. 2019;67:171–177. doi: 10.4149/neo_2018_181129N908. [DOI] [PubMed] [Google Scholar]
- 59.Lee L, Fei L, Pope J, Wagner LM. Early lymphocyte recovery and outcome in osteosarcoma. J Pediatr Hematol Oncol. 2017;39:179–183. doi: 10.1097/MPH.0000000000000717. [DOI] [PubMed] [Google Scholar]
- 60.Xiao-Long M, Kun-Peng Z, Chun-Lin Z. Circular RNA circ_HIPK3 is down-regulated and suppresses cell proliferation, migration and invasion in osteosarcoma. J Cancer. 2018;9:1856–1862. doi: 10.7150/jca.24619. [DOI] [PMC free article] [PubMed] [Google Scholar]


