Abstract
Circular RNAs (circRNAs), which are considered to be important functional regulators in cancer, have provided a new perspective regarding our understanding of tumor biology, including that of breast cancer. To investigate the regulatory effect of circRPPH1 on cellular behaviors of breast cancer and the potential mechanism, the expression of circRPPH1 and miR-556-5p in breast cancer tissues and cell lines were examined by quantitative RT-PCR. The regulatory effects of the circRPPH1/miR-556-5p/YAP1 axis on cellular behaviors of breast cancer cells were evaluated through functional experiments in vitro and tumor growth in vivo. The relationship between circRPPH1 and miR-556-5p/YAP1 was assessed using dual-luciferase reporter and RNA immunoprecipitation assays. PCR results showed that circRPPH1 levels were significantly upregulated in tumor tissues and breast cancer cells. Functionally, circRPPH1 promoted the proliferation, migration, invasion, and angiogenesis of breast cancer cell lines and tumor growth in vivo. Regarding the mechanism, dual-luciferase reporter and RNA immunoprecipitation assays showed that circRPPH1 was capable of sponging miR-556-5p to increase expression of the oncogene YAP1. Our data reveal that circRPPH1 plays a vital regulatory role in breast cancer via the miR-556-5p/YAP1 axis and may serve as a promising therapeutic target for breast cancer treatment.
Keywords: Breast cancer, miRNA sponge, circRPPH1, miR-556-5p, YAP1
Introduction
Over the last few years, breast cancer has become the second-leading cause of cancer-related death in women. Significant progress has been made in the systemic therapy and earlier diagnosis areas of breast cancer [1]. However, distant metastasis is still the main obstacle to successful treatment, and the mechanism of metastasis is not fully understood [2].
Circular RNAs (circRNAs) are a novel class of endogenous noncoding RNAs, which may function as gene regulators in eukaryotes. They are either mediated by a typical splice or formed between the upstream splice receptor and the downstream splice donor, which is characterized by a covalent closed-loop structure with neither 5’ to 3’ polarity nor polyadenylated tails [3,4]. CircRNAs have strong cell- or tissuespecific expression characteristics. Moreover, circRNAs are conserved across species due to resistance to RNase R. Recent researches have demonstrated that circRNAs may play an important role in many physiological and pathophysiological processes, such as microRNA (miRNA) sponges, protein decoys, and splicing and transcription regulators [5]. Recently, deregulation of certain circRNAs has been approved for the treatment of different types of human cancers, including breast cancer, where circRNA function is related to tumor proliferation and progression [6,7]. However, the function of circRNAs in breast cancer metastasis remains largely unknown.
CircRNAs are largely found in the cytoplasm [8] and are involved in post-transcriptional regulation through their function as miRNA “sponges”. Recent evidence has shown that circRNAs regulate mRNA translation indirectly by competing for miRNAs [9,10]. In breast cancer, several circRNAs play pivotal roles as miRNA sponges. For example, circAGFG1 functioned as a sponge of miR-195-5p to aggravate triple-negative breast cancer (TNBC) progression [11], while circIRAK3 facilitates breast cancer metastasis via the sponging of miR-3607 [12]. In this study, we found that circRPPH1 was capable of sponging miR-556-5p to increase the expression of YAP1, resulting in tumorigenesis and metastasis during TNBC progression.
Yes-associated protein (YAP) acts as a key regulatory protein of the Hippo signaling transduction pathway, which controls organ size by regulating cell proliferation and survival [13]. YAP functions as a transcriptional co-activator and controls cellular responses by interacting with nuclear transcription factors. When YAP is phosphorylated, it translocates from the nucleus to the cytoplasm, and transcription of its target genes decreases [14]. Multiple studies have reported that the chromosome region 11q22 that contains the YAP1 gene is frequently amplified during multiple cancers progression [15]. Nevertheless, the role of YAP in tumor growth is controversial in breast cancer [16,17]. However, emerging studies suggests that increased activity of YAP significantly augments the metastatic potential of breast cancer cells [18,19]. Thus, we set out to elucidate the potential mechanism of YAP in the promotion of metastasis during breast cancer progression.
Collectively, in the present study, we identified a novel circRNA derived from the RPPH1 gene, termed circRPPH1 (circ-001846), as a tumor promoter in breast cancer. Expression of circRPPH1 was significantly upregulated in breast cancer and positively associated with breast cancer progression through regulation of YAP1 expression and Hippo pathway signaling. Our findings suggest that circRPPH1 may exert regulatory functions in TNBC and may be a potential target for breast cancer therapy.
Materials and methods
Clinical specimens
We collected 20 matched TNBC tissues and adjacent normal tissues from patients who underwent breast cancer surgery in the First Affiliated Hospital of Soochow University. All patients undergoing chemotherapy or radiotherapy before surgery were excluded. After extraction, clinical breast cancer tissue specimens were immediately frozen in liquid nitrogen and then stored at -80°C fridges until total RNA extraction. The Institute Research Ethics Committee of the First Affiliated Hospital of Soochow University approved all participants. All procedures were in accordance with the Helsinki Declaration and institutional guidelines.
Cell culture and transfection
Human breast cancer cells (MCF-7 and MDA-MB-231) were purchased from KeyGEN Company (Nanjing, China). Cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS; Gibco, Grand Island, NY, USA), 100 U/mL penicillin, and 100 μg/mL streptomycin (Beyotime Institute of Biotechnology, Shanghai, China). All cells were incubated at 37°C in a 5% CO2-humidified atmosphere.
The full-length cDNA of circRPPH1 was amplified in MCF-7 cells and cloned into the specific vector between two frames, while the mock plasmid lacking circRPPH1 cDNA served as the control. MCF-7 cells and MDA-MB-231 cells were seeded in a 6-well plate in DMEM supplemented with 10% FBS and cultured until 50-70% confluent, then transfected with circRPPH1 vectors or RNA oligoribonucleotides using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. Forty-eight hours after transfection, the effects of circRPPH1 knockdown or overexpression were examined by RT-qPCR.
Quantitative real-time PCR (qRT-PCR)
In brief, total endogenous RNA from breast cancer tissues and cells was extracted by using TRIzol reagent (Invitrogen). Then cDNA was synthesized using the First Strand cDNA Synthesis Kit (Poly A Tailing) (Sangon Biotech, Shanghai, China). Real-time qRT-PCR was performed using the SYBR-Green-quantitative real-time PCR Master Mix kit (Toyobo Co., Osaka, Japan). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) or U6 snRNA was used as an internal reference. The data were analyzed using the 2-ΔΔCt method. Experiments were conducted in triplicate and the mean value is reported.
Western blot analysis
First, total protein was separated using 10% or 12% SDS-PAGE and transferred onto polyvinylidene fluoride membranes (Millipore, Billerica, MA, USA). Then membranes were incubated with 5% nonfat milk and incubated with anti-YAP1 (1:500; Abcam, Cambridge, MA, USA) and anti-GAPDH (1:2000; Abcam) primary antibodies at 4°C overnight. The next day, membranes were washed with PBS for three times and then incubated with horseradish peroxidase-conjugated goat anti-rabbit IgG secondary antibody (Jackson Immunoresearch, West Grove, PA, USA). After washing with PBS three times, protein bands were visualized using enhanced chemiluminescent substrate (ECL; Pierce, USA). GAPDH was used as an internal reference.
Luciferase reporter assay
Briefly, the 3’-UTR fragments of circRPPH1 and YAP1 (containing the miR-556-5p binding site) were amplified by Shanghai GenePharma Co., Ltd. and cloned into the pmirGLO vector (Promega, Madison, WI, USA). Then, wild-type circRPPH1 or mutant circRPPH1 fragments (WT circRPPH1 or Mut circRPPH1, respectively), as well as wild-type YAP1 or mutant YAP1 3’-UTRs (WT YAP1 or Mut YAP1) were constructed. Next, the constructed plasmids were co-transfected with miR-556-5p mimetic or mimetic negative control (mimetic-NC) into MCF-7 cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. After 24 hours of transfection, luciferase activity was measured using the Dual-Luciferase Reporter System (Promega). Experiments were performed in triplicate.
RNA immunoprecipitation (RIP) assay
In brief, the Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, Billerica, MA, USA) was used to perform RIP experiments. For anti-Argonaute 2 (AGO2) RIP, cells were lysed and incubated with anti-AGO2 antibody or rabbit lgG at 4°C for 2 h. After washing, co-precipitated RNAs were detected by reverse transcription PCR.
CCK-8 assays
CCK-8 (cell counting kit 8) assays were used to evaluate breast cancer cell proliferation after transfection. In brief, MCF-7 or MDA-MB-231 cells were seeded in a 96-well plate at a density of 1×103 cells per well. After incubation, 10 μl of CCK-8 solution was added to the wells. Four hours later, absorbance was measured at 450 nm using a microplate reader (Bio-Rad, Hercules, CA, USA).
Colony formation assays
Colony formation assays were used to measure the effect of circRPPH1 on breast cancer cell proliferation. Briefly, 1.5×103 MCF-7 or MDA-MB-231 cells per well were seeded in a 6-well plate after transfection and cultured for 10 days. Colonies were then fixed with 10% formaldehyde for 40 min and stained with 0.5% crystal violet for 25 min. Pictures of the colonies were then obtained using a NIKON digital camera and analyzed using IPP Image-Pro Plus 6.0 software.
Transwell assay
In brief, Matrigel (30 μl) was diluted with culture medium and then added into each well in the upper chamber of transwell filters with 3×104 cells per well. The lower chamber of each well was supplemented with DMEM (0.5 ml) with 10% FBS. 24 hours later, cells were fixed with paraformaldehyde for 30 min and stained with 0.1% crystal violet for 0.5 h. Subsequently, cells were washed with 0.1 M PBS. Finally, cells were then counted in randomly selected fields under a light microscope. The experiment was repeated in triplicate.
In vivo tumorigenicity assay
Male nude mice (6 weeks of age) were obtained from the Animal Facility of Soochow University of Medicine. All animal experiments were approved by the responsible governmental animal ethics committee. A suspension of MDA-MB-231 cells (2×106 in 100 µl) was injected into the right flank of the nude mice. When bearing palpable tumors, mice were randomly divided into two groups: si-SCR or si-circRPPH1. Four weeks later, the mice were killed and tumor tissues were harvested for use in further experiments.
Immunohistochemistry staining
Xenograft tumors specimens were processed into paraffin-embedded sections. After deparaffinized and rehydrated, slides were immersed in 3% hydrogen peroxide for 1 h and then incubated with anti-YAP1 (1:200) and anti-Ki67 (1:200) (Abcam) primary antibodies at 4°C overnight. The next day, the slides were stained with a secondary streptavidin-horseradish peroxidase-conjugated antibody (Solarbio, Beijing, China) for 1 h. Finally, the slides were then counterstained with hematoxylin for 30 s and cover slipped.
Statistical analysis
Data were analyzed using GraphPad Prism (GraphPad Software Inc, La Jolla, CA, USA) and SPSS 17.0 software (SPSS, Chicago, IL, USA). Normal distributional data are expressed as the mean ± standard deviation. The significance of differences was assessed using a Student’s t-test or one-way ANOVA assays. P < 0.05 indicates a significant difference.
Results
CircRPPH1 is overexpressed in breast cancer tissues
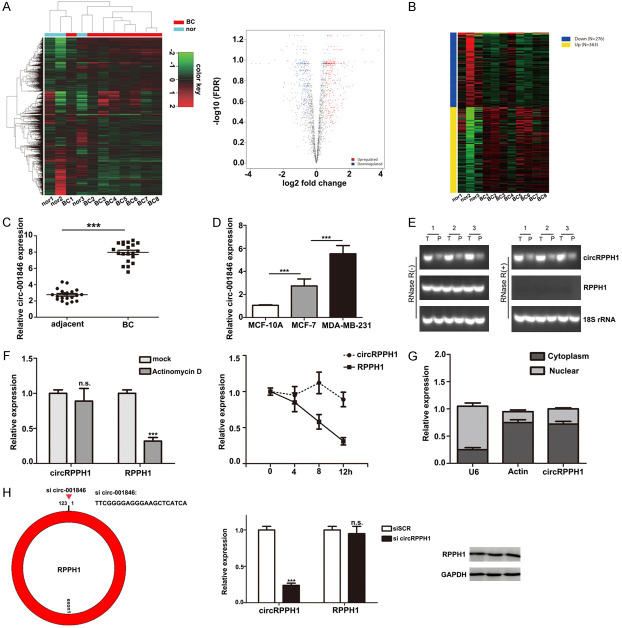
To identify essential circRNAs expressed in breast cancer tissues, we analyzed available online datasets in the GEO database that contained circRNA profiles in breast cancer tissues (GEO accession GSE101123). Our results demonstrated that multiple circRNAs are upregulated in breast cancer tissues compared to peritumor tissues (Figure 1A, 1B). Among them, circRPPH1 (circ-001846, circ-0000520) was one of the most upregulated and conserved circRNAs (Table 1). Next, we evaluated the levels of circRPPH1 in 20 paired TNBC tissues and peritumor tissues. The clinical features of the patients from whom these tissues were obtained are summarized in Table 2. Our results confirmed that circRPPH1 was significantly upregulated in tumor tissues (Figure 1C). Similarly, in comparison with circRPPH1 levels in normal breast epithelial cells (MCF-10A), increased levels of circRPPH1 were also observed in the MCF-7 and MDA-MB-231 cells. Moreover, expression of circRPPH1 in MCF-7 cells (low metastatic potential) was lower compared with that in MDA-MB-231 cells, which have a greater metastatic ability (Figure 1D). Next, we chose 3 pairs of TNBC sample tissues to subject to RT-PCR and validated that circRPPH1 was markedly overexpressed in tumor tissue. Notably, the linear RPPH1 mRNA level was only slightly elevated (Figure 1E). Furthermore, we investigated the stability of circRPPH1 in breast cancer cells using Actinomycin D (Figure 1F). Our results showed that the half-life of the circRPPH1 transcript was much more stable than the linear mRNA. Using subcellular fractionation, we detected that circRPPH1 resided predominantly in the cytoplasm of breast cancer cells (Figure 1G). Taken together, these data suggest that circRPPH1 is significantly overexpressed in breast cancer tissues.
Figure 1.
Identification of expression of different circRNAs in breast cancer. A and B. Differentially expressed genes were detected using the GEO database (GEO accession GSE101123). C. Relative expression of circRPPH1 in breast cancer tissues compared with corresponding non-tumor tissues. D. Relative circRPPH1 expression is shown in breast cancer cell lines (human normal breast cells: MCF-10A cells; breast cancer cells: MCF-7 and MDA-MB-231). E. The expression of circRPPH1 was analyzed by real-time qRT-PCR using 3 paired breast cancer samples and detected using a back-splice junction primer. P, peri-tumor; T, tumor. 18S was used as a loading control, and RNA samples for 18S detection were not treated with RNase R. F. The expression of circRPPH1 and RPPH1 was measured by real-time qRT-PCR after Actinomycin D treatment in breast cancer cells. G. The expression of circRPPH1 in the nucleus and cytoplasm was detected by subcellular fractionation. H. Schematic illustration showing the target sequences of the siRNA specific to the back-splicing junction of circRPPH1. The efficacy of circRPPH1-targeting siRNA on RPPH1 mRNA and protein was measured by qRT-PCR and western blot.
Table 1.
Top 10 up-regulated circRNAs in GSE101123
| ID | P.Value | t | B | logFC | SPOT_ID |
|---|---|---|---|---|---|
| ASCRP000343 | 0.000435 | 4.862733 | 0.28964 | 3.800033 | hsa_circRNA_001846 |
| ASCRP000018 | 0.001651 | 4.07878 | -0.93285 | 3.742085 | hsa_circRNA_000167 |
| ASCRP000389 | 0.004253 | 3.546875 | -1.8066 | 3.365067 | hsa_circRNA_002172 |
| ASCRP000385 | 0.004405 | 3.527425 | -1.83902 | 2.995608 | hsa_circRNA_002144 |
| ASCRP000017 | 0.004429 | 3.524408 | -1.84405 | 2.883005 | hsa_circRNA_000166 |
| ASCRP000071 | 0.001279 | 4.225459 | -0.69746 | 2.788805 | hsa_circRNA_000585 |
| ASCRP000315 | 0.000163 | 5.471777 | 1.17221 | 2.6775 | hsa_circRNA_001678 |
| ASCRP002307 | 0.002966 | 3.747702 | -1.4735 | 2.335477 | hsa_circRNA_101967 |
| ASCRP004099 | 0.014867 | 2.859217 | -2.95967 | 2.178211 | hsa_circRNA_103809 |
| ASCRP003437 | 0.039419 | 2.321682 | -3.84301 | 2.17781 | hsa_circRNA_103134 |
Table 2.
Association between circRPPH1 and the clinicopathological characteristics of breast cancer
| N% | circRPPH1 level | P value | ||
|---|---|---|---|---|
|
| ||||
| High (%) | Low (%) | |||
| Age (yeas) | P > 0.05 | |||
| < 55 | 8 | 4 (50.0) | 4 (50.0) | |
| ≥ 55 | 12 | 7 (58.3) | 5 (41.7) | |
| Menopause | P > 0.05 | |||
| Yes | 11 | 6 (54.5) | 5 (45.5) | |
| No | 9 | 5 (55.6) | 4 (44.4) | |
| TNM stage | P < 0.05 | |||
| I-II | 12 | 4 (33.3) | 8 (66.7) | |
| III-IV | 8 | 6 (75.0) | 2 (25.0) | |
| Lymph node metastasis | P < 0.05 | |||
| Yes | 10 | 8 (80.0) | 2 (20.0) | |
| No | 10 | 3 (30.0) | 7 (70.0) | |
| Tumor size | P < 0.05 | |||
| < 2 cm | 8 | 3 (37.5) | 5 (62.5) | |
| ≥ 2 cm | 12 | 8 (66.7) | 4 (33.3) | |
CircRPPH1 promotes breast cancer cell proliferation in vitro
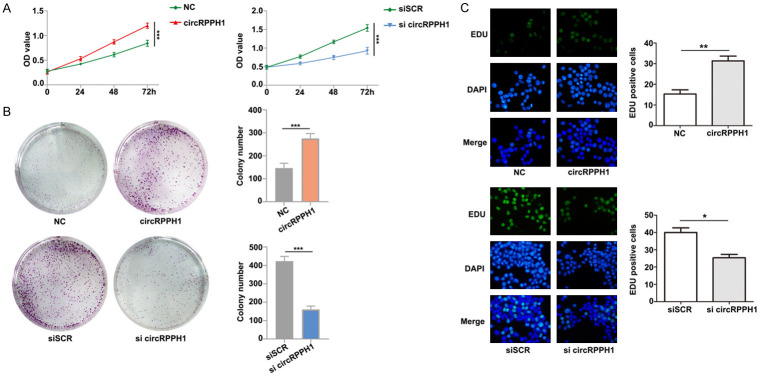
To inhibit endogenous circRPPH1 expression, we designed interference sequences targeting the junction sites of circRPPH1 using siRNAs that significantly decreased expression of circRPPH1 (Figure 1H). Of note, circRPPH1 inhibition did not affect expression of the RPPH1 mRNA and protein (Figure 1G). To explore the biological function of circRPPH1 in breast cancer cells, we constructed a circRPPH1 overexpression vector. Growth curves performed using CCK-8 assays demonstrated that upregulation of circRPPH1 significantly enhanced proliferation of MCF-7 cells, whereas downregulation of circRPPH1 inhibited cell growth (Figure 2A). The results of colony formation assays and EdU assays further elucidated that the proliferative capabilities of MCF-7 were significantly enhanced by upregulation of circRPPH1 and markedly impaired by downregulation of circRPPH1 (Figure 2B, 2C). These findings suggest that circRPPH1 enhances proliferation of breast cancer cells.
Figure 2.
CircRPPH1 promotes proliferation of breast cancer cells. A and B. CCK-8 proliferation assays and colony formation assays were performed in MCF-7 cells overexpressing circRPPH1 and in MDA-MB-231 cells with depleted circRPPH1. C. The proliferation ability of MCF-7 and MDA-MB-231 cells was measured using EdU assays.
CircRPPH1 increases breast cancer cell metastasis and facilitates angiogenesis in vitro
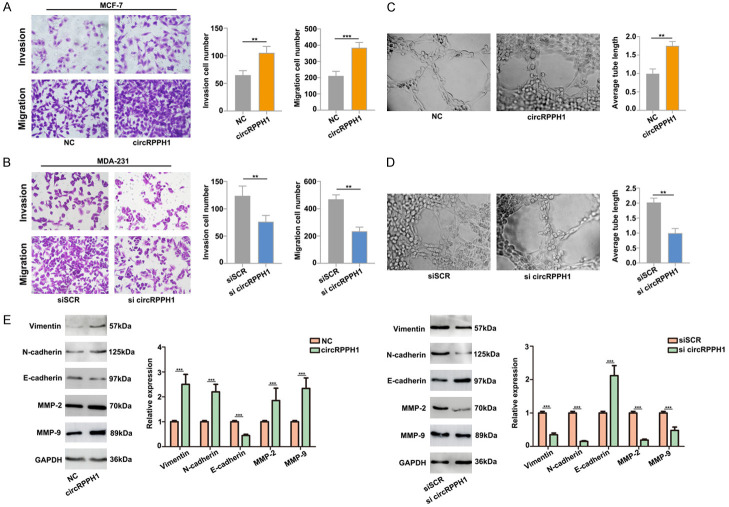
Next, we performed transwell assays to detect the effects of circRPPH1 on breast cancer cell migration and invasion. Our results show that the migration and invasion abilities of MCF-7 and MDA-MB-231 cells significantly increased after circRPPH1 overexpression, but markedly suppressed by downregulation of circRPPH1 (Figure 3A, 3B). These data indicated that circRPPH1 promotes breast cancer cell migration and invasion.
Figure 3.
CircRPPH1 promotes cell migration and invasion in vitro. A. Results of transwell and cell migration assays following circRPPH1 overexpression in MCF-7 cells. B. circRPPH1 depletion inhibited the migration and invasion of MDA-MB-231 cells. C and D. circRPPH1 regulates breast cell vasculogenic mimicry. Transfection with circRPPH1 significantly enhanced cell neovascularization rates, while circRPPH1 depletion inhibited the neovascularization rates. E. Western blot analysis for EMT-related proteins.
Furthermore, to assess the influence of circRPPH1 in tumor development, we utilized an endothelial tube formation assay. We found that tube length was significantly increased by proangiogenic stimuli from the supernatant of the circRPPH1 group (Figure 3C), and the opposing result was found using supernatant from the si-circRPPH1 group (Figure 3D). In view of the important role of epithelial mesenchymal transition (EMT) in the invasion and migration of cancer cells, we further evaluated the effect of circRPPH1 on EMT. The result of our western blot analyses demonstrated that circRPPH1 overexpression decreased expression of epithelial markers and increased expression of mesenchymal markers (Figure 3E), implicating circRPPH1 in breast cancer cell EMT.
To evaluate the influence of circRPPH1 on cell proliferation in vivo, we performed xenograft experiments. Compared with the control group, introduction of si-circRPPH1 significantly decreased mean tumor volume and tumor weight (Figure 4A). Additionally, immunostaining analysis of xenograft tissues demonstrated that Ki67 and YAP1 expression was also decreased in the si-circRPPH1 knockdown group (Figure 4B). Taken together, these data suggest that circRPPH1 promotes tumorigenesis.
Figure 4.
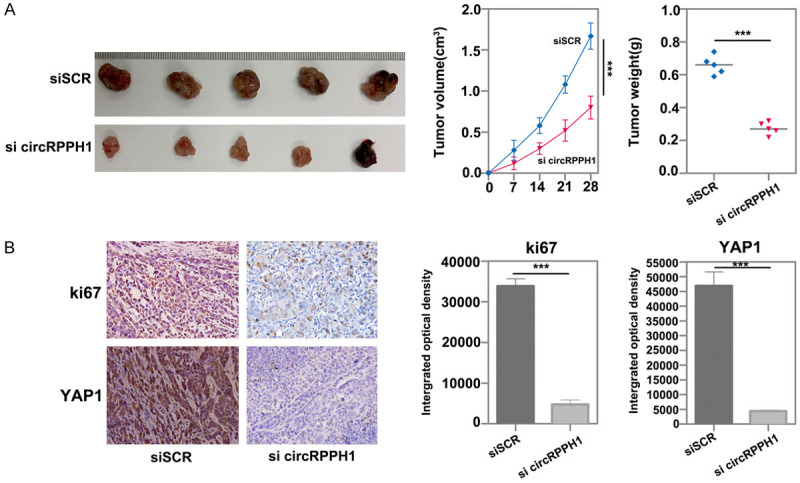
CircRPPH1 promotes breast cancer cell proliferation in vivo. A. A subcutaneous tumor formation experiment was used to measure cell proliferation ability in vivo. B. The expression of Ki67 and YAP1 in subcutaneous tumor tissues was measured by immunostaining analysis.
CircRPPH1 acts as a ceRNA to regulate YAP1
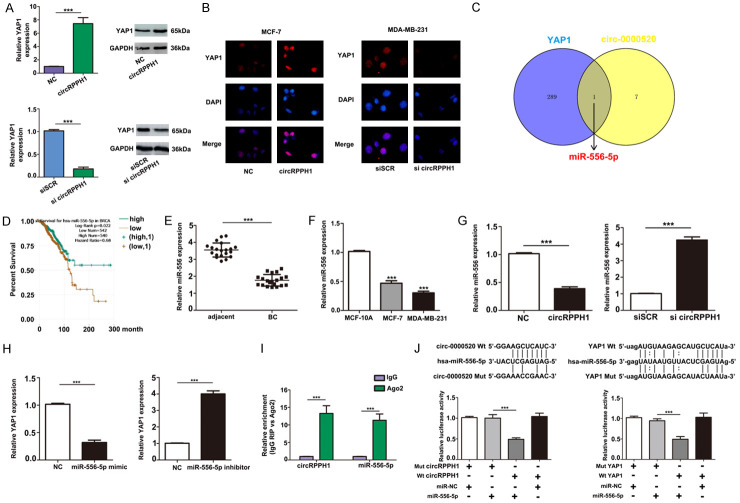
Hippo signaling plays a main role in many aspects of tumor biology [20,21]. YAP, as the main executor of the Hippo pathway, induces target gene transcription and triggers downstream biological effects through interaction with related transcription factors, particularly TEA domain family members (TEADs) [22]. YAP is considered as an oncogene during breast cancer progression, and its dysregulation often leads to increased tumor aggressiveness and metastasis [23,24]. To validate whether there was a reinforcing relationship between circRPPH1 and YAP1, RT-PCR and western blot assays were utilized to measure the level of YAP1 in breast cancer cells with modulated levels of circRPPH1. Our results showed that in MCF-7 cells, YAP1 expression increased after transfection with circRPPH1. Moreover, suppression of circRPPH1 attenuated the level of YAP1 in MDA-MB-231 cells (Figure 5A); these results were confirmed using immunofluorescence staining (Figure 5B).
Figure 5.
CircRPPH1 functions as a ceRNA by sponging miR-556-5p and indirectly regulating YAP1 expression. A. qRT-PCR and western blot assays were employed to verify the relative expression of YAP1 after circRPPH1 transfection. B. Expression of YAP1 protein was detected by immunofluorescence after circRPPH1 transfection. C. Results of miR-556-5p target analysis using the starBase database. D. Kaplan-Meier overall survival curves according to circRPPH1 expression levels using the starBase database. E and F. qRT-PCR analysis to detect expression of miR-556-5p in breast cancer tissues and cells. G and H. Expression of miR-556-5p and YAP1 was measured after transfection in breast cancer cells. I. RIP assay showing that circRPPH1 was enriched in AGO2-containing miRNAs compared with IgG; miR-556-5p was also detected in the same precipitate. J. Dual-luciferase reporter assay results to test whether circRPPH1 and YAP1 are targets of miR-556-5p.
Next, we used the starBase and circBase databases to explore the relationship between circRPPH1 and YAP1. By overlapping the results of miRNA target predictions from starBase and circBase, we found that miR-556-5p is considered a putative target of both circRPPH1 and YAP1 (Figure 5C). Using the starBase pancancer analysis (based on TCGA data), we found that miR-556-5p was associated with overall survival of patients with mutations in the breast cancer genes (BRCA) (Figure 5D). Moreover, real-time qRT-PCR confirmed that miR-556-5p was significantly downregulated in tumor tissues and breast cancer cell lines (Figure 5E, 5F). Further results showed that knockdown of circRPPH1 in breast cancer cells triggered a significant increase in miR-556-5p expression (Figure 5G). Additionally, expression of YAP1 was similarly regulated by miR-556-5p (Figure 5H). The relationship of circRPPH1 and miR-556-5p was further validated using an anti-AGO2 RIP assay (Figure 5I) in MCF-7 cells. Decreased luciferase activity was observed in MCF-7 cells transfected with miR-556-5p mimetics and wild type circRPPH1, as well as in MCF-7 cells transfected with miR-556-5p mimetics and wild type YAP1 3’-UTR (Figure 5J). These results suggest that circRPPH1 functions as a competing endogenous RNA (ceRNA) by sponging miR-556-5p and indirectly regulated YAP1 expression.
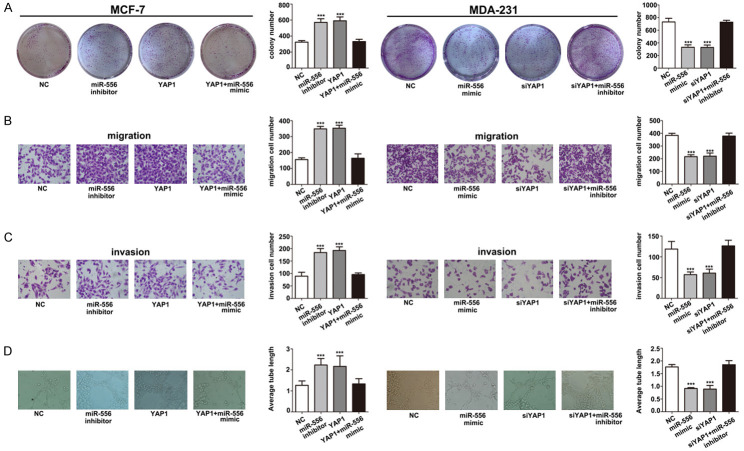
Activity of miR-556-5p is partially activated through negative regulation of YAP1
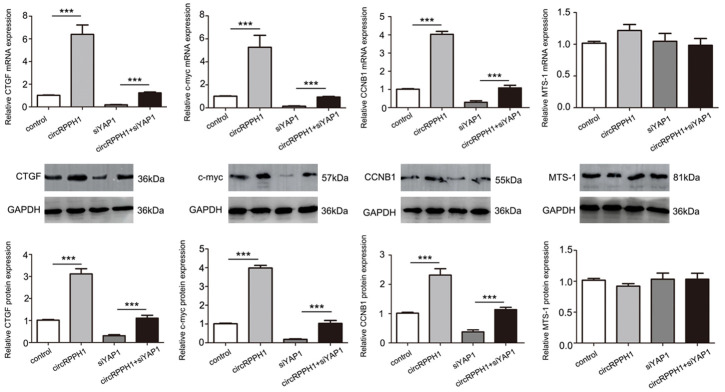
To further demonstrate the function of miR-556-5p in breast cancer progression, we performed functional experiments using MCF-7 and MDA-231 cells. Our results showed that proliferation (Figure 6A), migration, invasiveness (Figure 6B, 6C), and angiogenic ability (Figure 6D) of MCF-7 cells were all upregulated in miR-556-5p inhibitor and YAP1 groups. These MCF-7 cell properties were attenuated by the miR-556-5p mimetic. Therefore, these data indicate that the function of miR-556-5p is partially dependent on YAP1. Furthermore, we also detected YAP downstream signals such as connective tissue growth factor (CTGF), c-myc, and cyclin-B1 (CCNB1). These results show that the regulatory effects of circRPPH1 on downstream YAP signaling are dependent on YAP1, while circRPPH1 does not affect expression of proteins upstream of YAP like macrophage-stimulating protein (Mst1) (Figure 7).
Figure 6.
The function of YAP1 is partially dependent on miR-556-5p inhibition. The effects of YAP1 overexpression on (A) cell proliferation, (B) migration, (C) invasiveness, and (D) angiogenesis were attenuated by a miR-556-5p mimetic. (***P < 0.001).
Figure 7.
YAP downstream signaling is regulated by circRPPH1. Western blot analysis for expression of YAP downstream and upstream proteins after circRPPH1 transfection.
Discussion
Breast cancer is a highly heterogeneous disease. Early diagnosis, radical mastectomy, and adjuvant treatment have improved survival rates and prognosis; however, mortality rates remain high [25]. To improve the efficacy of treatments, a better understanding of the biology of breast cancer is needed. In this study, we explored the possible mechanism of circRPPH1, as a ceRNA, in the modulation of YAP1 expression through sponging of miR-556-5p. We found that circRPPH1 mediated the miR-556-5p/YAP1 axis, which regulated the proliferation, invasion, and angiogenic ability of breast cancer cell lines. We analyzed circRNA profiles in breast cancer tissues (GEO accession GSE101123) and found that many circRNAs are overexpressed in breast cancer tissues compared to peritumor tissues. Among them, circRPPH1 is one of the most upregulated and conserved circRNAs. Our real-time qRT-PCR results showed that circRPPH1 levels were significantly upregulated in tumor tissues and breast cancer cells, suggesting that circRPPH1 is highly expressed in breast cancer. Additionally, circRPPH1 enhanced the proliferation, migration, invasion, and angiogenic capacity of breast cancer cells in vitro and tumor growth in vivo. Finally, we showed that circRPPH1 functions as a ceRNA by sponging miR-556-5p and indirectly regulating YAP1 expression. In summary, our work suggests that circRPPH1 is a potent oncogene in breast cancer; however, further investigation is needed to elucidate the target of circRPPH1 in breast cancer.
CircRNA is a special type of non-coding RNA, which is the latest research hotspot in RNA field. More and more evidence indicates that circRNA is stable and conserved in eukaryotic cells, which makes it a promising tumor biomarker [26]. In particular, circPVT1 [27], circGAS8 [28], and circPAPPA [29] have been identified as potential prognostic biomarkers for gastric cancer, lung adenocarcinoma, and osteosarcoma, respectively. In this study, we identified that circRPPH1 was upregulated in breast cancer and overexpressed in 20 breast cancer tissues compared with normal tissues, and it may be a promising candidate for a tumor biomarker in breast cancer.
Over the past decade, YAP has been considered as an oncogene during breast cancer progression, and its dysregulation often leads to tumor aggressiveness and metastasis [30]. In this study, we showed that the expression of YAP1 is positively correlated with circRPPH1 levels. Furthermore, our gain- and loss-of-function analyses showed that overexpression of YAP1 promoted proliferation, migration, and invasion of breast cancer cells, consistent with its role as an oncogene [31-33]. Therefore, we hypothesized that circRPPH1 plays a role in carcinogenesis by regulating YAP1 expression.
Compelling evidence indicates that several types of RNAs, including pseudogenes, long non-coding RNAs (lncRNAs), and circRNAs, could serve as ceRNAs by competitively binding to miRNAs and regulating gene expression post-transcriptionally [34]. For instance, the lncRNA ROR promotes radioresistance in hepatocellular carcinoma cells by acting as a ceRNA for miRNA-145 to regulate RAD18 expression [35]. Additionally, several circRNAs have been identified as ceRNAs that regulate tumor progression. Of note, circGFRA1 and GFRA1 act as ceRNAs in TNBC by regulating miR-34a [36]. In our study, the starBase and circBase databases were used to explore the relationship between circRPPH1 and miR-556-5p. Real-time qRT-PCR confirmed that miR-556-5p was significantly downregulated in tumor tissues and breast cancer cells, and RIP and luciferase reporter assays confirmed that they directly interacted. In conclusion, we revealed that circRPPH1 could sponge miR-556-5p to contribute to malignant behaviors in breast cancer. Therefore, it is reasonable that circRPPH1 could promote tumor progression via the miR-556-5p/YAP1 axis. However, we are aware of the limitation of the present study that larger patient cohorts are required to validate the association between circRPPH1 expression and patient survival.
Conclusion
In summary, our study investigated for the first time, the role of circRPPH1 and identified it as a potential oncogene in breast cancer. Furthermore, mechanistic investigations showed that circRPPH1 contributes to breast cancer progression by enhancing YAP1 expression through sponging of miR-556-5p. Our results provide new insight into the mechanism that links circRPPH1 with breast cancer and suggests that circRPPH1 may serve as a promising therapeutic target for breast cancer treatment.
Disclosure of conflict of interest
None.
References
- 1.Berry DA, Cronin KA, Plevritis SK, Fryback DG, Clarke L, Zelen M, Mandelblatt JS, Yakovlev AY, Habbema JD, Feuer EJ. Effect of screening and adjuvant therapy on mortality from breast cancer. N Engl J Med. 2005;353:1784–1792. doi: 10.1056/NEJMoa050518. [DOI] [PubMed] [Google Scholar]
- 2.Eccles SA, Welch DR. Metastasis: recent discoveries and novel treatment strategies. Lancet. 2007;369:1742–1757. doi: 10.1016/S0140-6736(07)60781-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guo JU, Agarwal V, Guo H, Bartel DP. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15:409. doi: 10.1186/s13059-014-0409-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barrett SP, Wang PL, Salzman J. Circular RNA biogenesis can proceed through an exon-containing lariat precursor. Elife. 2015;4:e07540. doi: 10.7554/eLife.07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barrett SP, Salzman J. Circular RNAs: analysis, expression and potential functions. Development. 2016;143:1838–1847. doi: 10.1242/dev.128074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kristensen LS, Hansen TB, Venø MT, Kjems J. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37:555–565. doi: 10.1038/onc.2017.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu WY. Roles of the circular RNA circ-Foxo3 in breast cancer progression. Cell Cycle. 2017;16:589–590. doi: 10.1080/15384101.2017.1278935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, Chen D, Gu J, He X, Huang S. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25:981–984. doi: 10.1038/cr.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 10.Hansen TB, Kjems J, Damgaard CK. Circular RNA and miR-7 in cancer. Cancer Res. 2013;73:5609–5612. doi: 10.1158/0008-5472.CAN-13-1568. [DOI] [PubMed] [Google Scholar]
- 11.Yang R, Xing L, Zheng X, Sun Y, Wang X, Chen J. The circRNA circAGFG1 acts as a sponge of miR-195-5p to promote triple-negative breast cancer progression through regulating CCNE1 expression. Mol Cancer. 2019;18:4. doi: 10.1186/s12943-018-0933-7. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12.Wu J, Jiang Z, Chen C, Hu Q, Fu Z, Chen J, Wang Z, Wang Q, Li A, Marks JR, Guo C, Chen Y, Zhou J, Yang L, Lin C, Wang S. CircIRAK3 sponges miR-3607 to facilitate breast cancer metastasis. Cancer Lett. 2018;430:179–192. doi: 10.1016/j.canlet.2018.05.033. [DOI] [PubMed] [Google Scholar]
- 13.Zhao B, Tumaneng K, Guan KL. The Hippo pathway in organ size control, tissue regeneration and stem cell self-renewal. Nat Cell Biol. 2011;13:877–883. doi: 10.1038/ncb2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Basu S, Totty NF, Irwin MS, Sudol M, Downward J. Akt phosphorylates the Yes-associated protein, YAP, to induce interaction with 14-3-3 and attenuation of p73-mediated apoptosis. Mol Cell. 2003;11:11–23. doi: 10.1016/s1097-2765(02)00776-1. [DOI] [PubMed] [Google Scholar]
- 15.Overholtzer M, Zhang J, Smolen GA, Muir B, Li W, Sgroi DC, Deng CX, Brugge JS, Haber DA. Transforming properties of YAP, a candidate oncogene on the chromosome 11q22 amplicon. Proc Natl Acad Sci U S A. 2006;103:12405–12410. doi: 10.1073/pnas.0605579103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yuan M, Tomlinson V, Lara R, Holliday D, Chelala C, Harada T, Gangeswaran R, Manson-Bishop C, Smith P, Danovi SA, Pardo O, Crook T, Mein CA, Lemoine NR, Jones LJ, Basu S. Yes-associated protein (YAP) functions as a tumor suppressor in breast. Cell Death Differ. 2008;15:1752–1759. doi: 10.1038/cdd.2008.108. [DOI] [PubMed] [Google Scholar]
- 17.Tufail R, Jorda M, Zhao W, Reis I, Nawaz Z. Loss of Yes-associated protein (YAP) expression is associated with estrogen and progesterone receptors negativity in invasive breast carcinomas. Breast Cancer Res Treat. 2012;131:743–750. doi: 10.1007/s10549-011-1435-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lamar JM, Stern P, Liu H, Schindler JW, Jiang ZG, Hynes RO. The Hippo pathway target, YAP, promotes metastasis through its TEAD-interaction domain. Proc Natl Acad Sci U S A. 2012;109:E2441–2450. doi: 10.1073/pnas.1212021109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen D, Sun Y, Wei Y, Zhang P, Rezaeian AH, Teruya-Feldstein J, Gupta S, Liang H, Lin HK, Hung MC, Ma L. LIFR is a breast cancer metastasis suppressor upstream of the Hippo-YAP pathway and a prognostic marker. Nat Med. 2012;18:1511–1517. doi: 10.1038/nm.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pfleger CM. The hippo pathway: a master regulatory network important in development and dysregulated in disease. Curr Top Dev Biol. 2017;123:181–228. doi: 10.1016/bs.ctdb.2016.12.001. [DOI] [PubMed] [Google Scholar]
- 21.Zanconato F, Cordenonsi M, Piccolo S. YAP/TAZ at the roots of cancer. Cancer Cell. 2016;29:783–803. doi: 10.1016/j.ccell.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ji XY, Zhong G, Zhao B. Molecular mechanisms of the mammalian hippo signaling pathway. Yi Chuan. 2017;39:546–567. doi: 10.16288/j.yczz.17-094. [DOI] [PubMed] [Google Scholar]
- 23.Pegoraro S, Ros G, Ciani Y, Sgarra R, Piazza S, Manfioletti G. A novel HMGA1-CCNE2-YAP axis regulates breast cancer aggressiveness. Oncotarget. 2015;6:19087–19101. doi: 10.18632/oncotarget.4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim HM, Jung WH, Koo JS. Expression of Yes-associated protein (YAP) in metastatic breast cancer. Int J Clin Exp Pathol. 2015;8:11248–11257. [PMC free article] [PubMed] [Google Scholar]
- 25.Krell J, Stebbing J, Frampton AE, Carissimi C, Harding V, De Giorgio A, Fulci V, Macino G, Colombo T, Castellano L. The role of TP53 in miRNA loading onto AGO2 and in remodelling the miRNA-mRNA interaction network. Lancet. 2015;385(Suppl 1):S15. doi: 10.1016/S0140-6736(15)60330-0. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Z, Yang T, Xiao J. Circular RNAs: promising biomarkers for human diseases. EBioMedicine. 2018;34:267–274. doi: 10.1016/j.ebiom.2018.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen J, Li Y, Zheng Q, Bao C, He J, Chen B, Lyu D, Zheng B, Xu Y, Long Z, Zhou Y, Zhu H, Wang Y, He X, Shi Y, Huang S. Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer. Cancer Lett. 2017;388:208–219. doi: 10.1016/j.canlet.2016.12.006. [DOI] [PubMed] [Google Scholar]
- 28.Li S, Sun X, Miao S, Lu T, Wang Y, Liu J, Jiao W. hsa_circ_0000729, a potential prognostic biomarker in lung adenocarcinoma. Thorac Cancer. 2018;9:924–930. doi: 10.1111/1759-7714.12761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu Z, Shi W, Jiang C. Overexpressing circular RNA hsa_circ_0002052 impairs osteosarcoma progression via inhibiting Wnt/β-catenin pathway by regulating miR-1205/APC2 axis. Biochem Biophys Res Commun. 2018;502:465–471. doi: 10.1016/j.bbrc.2018.05.184. [DOI] [PubMed] [Google Scholar]
- 30.Shen J, Cao B, Wang Y, Ma C, Zeng Z, Liu L, Li X, Tao D, Gong J, Xie D. Hippo component YAP promotes focal adhesion and tumour aggressiveness via transcriptionally activating THBS1/FAK signalling in breast cancer. J Exp Clin Cancer Res. 2018;37:175. doi: 10.1186/s13046-018-0850-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Song Q, Mao B, Cheng J, Gao Y, Jiang K, Chen J, Yuan Z, Meng S. YAP enhances autophagic flux to promote breast cancer cell survival in response to nutrient deprivation. PLoS One. 2015;10:e0120790. doi: 10.1371/journal.pone.0120790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang X, Su L, Ou Q. Yes-associated protein promotes tumour development in luminal epithelial derived breast cancer. Eur J Cancer. 2012;48:1227–1234. doi: 10.1016/j.ejca.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 33.Wang T, Mao B, Cheng C, Zou Z, Gao J, Yang Y, Lei T, Qi X, Yuan Z, Xu W, Lu Z. YAP promotes breast cancer metastasis by repressing growth differentiation factor-15. Biochim Biophys Acta Mol Basis Dis. 2018;1864:1744–1753. doi: 10.1016/j.bbadis.2018.02.020. [DOI] [PubMed] [Google Scholar]
- 34.Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen Y, Shen Z, Zhi Y, Zhou H, Zhang K, Wang T, Feng B, Chen Y, Song H, Wang R, Chu X. Long non-coding RNA ROR promotes radioresistance in hepatocelluar carcinoma cells by acting as a ceRNA for microRNA-145 to regulate RAD18 expression. Arch Biochem Biophys. 2018;645:117–125. doi: 10.1016/j.abb.2018.03.018. [DOI] [PubMed] [Google Scholar]
- 36.He R, Liu P, Xie X, Zhou Y, Liao Q, Xiong W, Li X, Li G, Zeng Z, Tang H. circGFRA1 and GFRA1 act as ceRNAs in triple negative breast cancer by regulating miR-34a. J Exp Clin Cancer Res. 2017;36:145. doi: 10.1186/s13046-017-0614-1. [DOI] [PMC free article] [PubMed] [Google Scholar]