Abstract
Objective: The dysregulation of deubiquitinating enzymes is important in the development of many cancers, including colorectal cancer (CRC). However, the precise function and potential mode of action of the deubiquitinating enzyme UCHL3 in CRC progression are poorly elucidated. Methods: The expression levels of UCHL3 in patient samples were analyzed by western blotting, real-time PCR and immunohistochemistry and its association with overall survival was analyzed using Kaplan-Meier method. Colony formation, CCK-8 and Transwell were used to examine the effects of UCHL3 knockdown or over-expression on CRC cells growth, invasion and migration. The functional effects of UCHL3 and SOX12 on tumor growth were further examined using xenograft tumor mouse models in vivo. Results: Here, we found high expression of UCHL3 in CRC tissues which showed an association with the development of tumor and CRC patient survival. Studies conducted in vitro showed that UCHL3 overexpression facilitates proliferation, invasion, migration, and EMT (epithelial-mesenchymal transition) in cells of CRC, and a knockdown of UCHL3 had a reverse effect. Likewise, experiments conducted in vivo also showed enhanced tumor growth due to UCHL3 overexpression. In addition, UCHL3 was found regulates SOX12 expression in CRC cells. PI3K/AKT/mTOR pathway is required for UCHL3-mediated SOX12 expression. Mechanically, UCHL3 regulates SOX12 via AKT/mTOR signaling pathway and facilitated tumor progression. Conclusion: UCHL3 plays an oncogenic role through the AKT/mTOR/SOX12 axis and can be considered as a potential target for therapy and CRC prognostic biomarker.
Keywords: UCHL3, CRC, SOX12, AKT/mTOR, tumor growth
Introduction
The worldwide annual incidence of CRC (colorectal cancer) is nearly 1.4 million cases, a common cause of malignant tumors, and ranks the second in deaths due to cancer [1,2]. While there have been significant improvements made in treatment of CRC, its prognosis in patients is poor because of a dearth of effective targets for treatment [3]. While several CRC signaling pathways and aberrant gene expression have been discovered, its process of occurrence and development is not yet completely elucidated [4,5]. Therefore, it is important to look for novel molecular markers for enhanced prognosis, risk assessment and treatment of CRC patients.
Ubiquitination is a crucial posttranslational modification that has pivotal protein-regulatory functions [6,7]. This biological process is critically regulated by deubiquitinating enzymes (DUBs) [8]. UCHL3 (Ubiquitin carboxyl-terminal hydrolase L3; EC 3.4.19.12), a member of UCL subfamily, is an important DUB [9,10]. UCHL3 was found upregulated in breast cancer, besides its recent identification as a novel mediator of DNA repair [11]. The precise biological roles and potential molecular mechanism underlying UCHL3’s involvement in human malignancies, including CRC, remains unknown.
The transcription factors belonging to the sex-determining region Y-box (SOX) family are confirmed to regulate decisions of cell fate and possess the high mobility group (HMG) box, a characteristic homologous sequence [12]. Twenty SOX genes are encoded by the human genome, based on the presence of other structural motifs and homology within the HMG domain, they are categorized into subgroups A to H [13]. SOX4, SOX11 and SOX12 are proteins of the SOXC group, and play a vital part in processes of neuronal, mesenchymal, and cardiac development [14]. Interestingly, the upregulation of SOX4, SOX11, and SOX12 was observed in various cancers in humans and participate in the initiation and development of cancer [14]. SOX11 was found to promote interactions in the tumor-protective microenvironment and stalls the differentiation of terminal B cells in aggressive lymphoma of the mantle cell [15]. Recently, SOX12 was identified as a possible target in AML (acute myeloid leukemia) and as a HCC (hepatocellular carcinoma) stem-like cell marker [16-18]. Thus, while SOXC proteins are implicated as master cancer occurrence and metastasis regulators, the functional role and expression of SOX12 in human CRC is still elusive. A previously study indicated that UCHL3 as a potential oncogene, showing that UCHL3 maintains cancer stem-cells properties [19]. All SOX genes display specific expression patterns and have key roles in stem cell maintenance and cell-fate determination during development [20,21]. Therefore, in the current study, we tested the hypothesis that SOX12 regulates the oncogene function of UCHL3 in CRC cells.
In the current study, CRC and normal tissues were examined for UCHL3 expression, the relationship between UCHL3 and the patient prognosis was evaluated, as well as the direct role of UCHL3 in CRC growth was assessed, followed by the exploration of the inherent molecular mechanism.
Materials and methods
Tissue samples
Between 2014 and 2016, CRC tissues along with adjacent paired normal tissues were acquired from The First People’s Hospital of Shenyang. The Biomedical Ethics Committee of The First People’s Hospital of Shenyang consented to this study and the patients informed about the procedures to obtain their consent.
Cell lines and culture
The ATCC (VA, USA) provided the cell lines of human CRC (Lim1215, DLD1, SW48, HCT116, SW620, and SW480), which were maintained in DMEM with fetal bovine serum (10%) at 37°C in an atmosphere with CO2 (5%).
Transfection
The procurement of UCHL3 (LV-Flag UCHL3-Puromycin), UCHL3-shRNA (LV-UCHL3-shRNA-Puromycin, sh UCHL3-#1: CAGGGACAAGAUGUUACAUCA and sh UCHL3-#2: UAGAAGUUUGCAAGAAGUUUA), and viruses to be used as negative control were acquired from Genechem Co. Ltd. (China). Viruses were produced by transfecting the HEK293T cells with the UCHL3 or UCHL3-shRNA plasmids and three helper plasmids (pVSVg, RRE and REV). To create stable cell lines, cells were seeded into medium containing serial dilutions of LV and cells were incubated for 48 hours. For the selection of transduced cells, 2 mg/mL puromycin (Sigma) was added to the growth medium, and cells were maintained under this condition.
Immunohistochemistry (IHC)
IHC was carried out as described previously [22,23]. In brief, after the previous steps, incubation of slides was done along with primary antibodies, and then with HRP labeled secondary antibody. The images of three representative fields were taken at × 100 (high-power) magnification using ZEN 2012 software (blue edition). For each image captured, identical settings were used. Image J software (NIH, USA) was used for densitometric determinations.
Cell proliferation
The cell proliferation assay was carried out using the CCK-8 kit (Sigma) according to the instructions. Seeding of constructed cell lines that were stable were done in a 96-well plate and then cultured for 5 days at 24-hour intervals. Then the viability of cells was determined through CCK-8 assay. The cell proliferation was expressed in terms of absorbance at 450 nm. Each experiment was replicated at least thrice, independently.
Colony formation
Colony formation assay was performed as previously study [24,25]. In 6-well plates, 1 × 103 cells were incubated per well for the colony formation assay. The colonies that were visible were fixed after 12 days using formaldehyde (4%) and crystal violet stained. Then counting of those colonies was performed in which had more than 50 cells.
Migration and invasion
To assess the invasive and migratory ability of cells, Transwell plates (8-μm pore size; Corning) were used. Briefly, in the top chamber with serum-free medium, 3 × 105 cells were suspended again per well of the transwell plates which were layered with or without Matrigel (BD), and to the bottom chamber, medium with FBS (10%) was added. After 24 hours, the migrated cells across the membrane were fixed in paraformaldehyde (4%), and crystal violet stained. Three randomly selected fields were used for microscopic (Olympus Corporation, Japan) counting of invaded or migrated cells at 200 × magnification.
Western blotting
Western blotting was performed as previously study [26-28]. Extraction of total cellular proteins was done in a lysis buffer with PMSF phosphatase (phenylmethylsulfonyl fluoride) inhibitor. Total protein of the samples was loaded in equal amounts (50 μg) and resolved by SDS-PAGE, transferred onto PVDF membranes, and blocked with skimmed milk (5%, prepared in TBST) for 1 hour. Then, primary antibodies were added and kept throughout the night at 4°C and incubated for 1 hour with secondary antibody (CST, MA, USA) at room temperature. A chemiluminescence kit from Beyotime Biotechnology (China) was used to visualize the proteins.
Xenograft mouse model
The xenograft experiments were carried out on male mice (BALB/c nude, 6-week-old). All animal handling procedures and care were done as per the regulations of the National Institutes of Health for the use and care of laboratory animals which was then sanctioned by the Committee of Animal Care and Use, The First People’s Hospital of Shenyang. To establish the tumor growth model, the right flanks of the animal were injected with 5 × 105 cells subcutaneously. Every two days, the volume of tumors was determined as per formula: Volume (mm3) = length × width2/2. Ethical endpoint was defined as a time point when a tumor reached 1.5 cm or more in any dimension. Mice were sacrificed by CO2. Three weeks after injection, all mice were sacrificed to assess for apoptosis and tumor growth through IHC staining.
Statistical analysis
The GraphPad Prism 7 software was used for analyses and generating graphs. The survival of patients was assessed by the Kaplan-Meier method. The comparisons among the two groups were assessed by Student’s t-test. A significant value had P < 0.05.
Results
Increased expression of UCHL3 in CRC patients
To investigate the expression patterns of UCHL3 in CRC tissues, the UCHL3 expression level in 8 CRC tissues was assessed through western blotting and real-time PCR. As shown in Figure 1A and 1B, UCHL3 expression increased markedly in tumor tissues in comparison to the normal matched tissues (P < 0.05). The expression of UCHL3 and its clinical importance in CRC was established by observing increased levels of UCHL3 in tumor tissue pairs in comparison to normal matched tissues (Figure 1C). Further, as per Kaplan-Meier analysis, increased UCHL3 expression correlated with a remarkably poorer OS than those exhibiting low UCHL3 expression (Figure 1D). Thus, UCHL3 may act as a potential prognostic marker in CRC.
Figure 1.
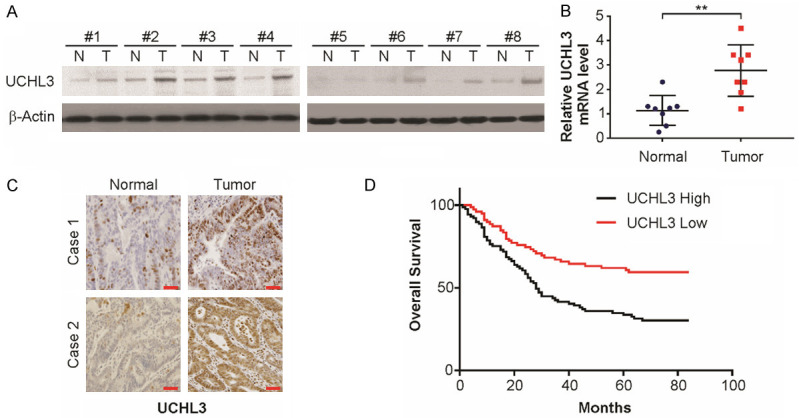
UCHL3 was up-regulated in CRC tissues. A. UCHL3 expression in 8 paired CRC samples as detected by Western blotting. B. UCHL3 expression in 8 paired CRC samples as detected by real-time PCR. C. Immunohistochemical staining of UCHL3 in tumor tissues and the matched normal tissues. D. Kaplan-Meier survival analysis of overall survival and disease-free survival according to the UCHL3 expression in CRC patients. Scale bar: 25 μm. Results represent means ± SD from three independent experiments. **P < 0.01.
UCHL3 facilitates proliferation in CRC cells
To UCHL3 level in CRC cells was regulated by carrying out lentiviral transfection and evaluated the effect of UCHL3 on CRC. For this, the level of UCHL3 was quantified in 6 CRC cell lines and found significantly high UCHL3, especially in HCT116 cells, and low in Lim1215 cells (Figure 2A). Hence, UCHL3 was knocked down in HCT116 cells by transducing with UCHL3-shRNA, and was overexpressed in Lim1215 cells by transducing with UCHL3 construct. The negative control viruses were transduced into the control cells. All these transfected cells were confirmed for stability and effcient down-regulation or up-regulation by western blotting. As shown in the Figure 2B and 2C, most robust effciency of knockdown was observed through shRNA#1 and was utilized for future studies. The role of UCHL3 on ability to form colonies and viability was evaluated respectively by colony formation assay and CCK-8 assay. The HCT116 cell viability was remarkably reduced due to UCHL3 knockdown and conversely, overexpression of UCHL3 enhanced Lim1215 cell viability (Figure 2D). The assay for colony formation and CCK-8 assay showed concurrent results (Figure 2E). Therefore, in CRC cells, UCHL3 aids in colony-formation and promoting proliferative ability.
Figure 2.
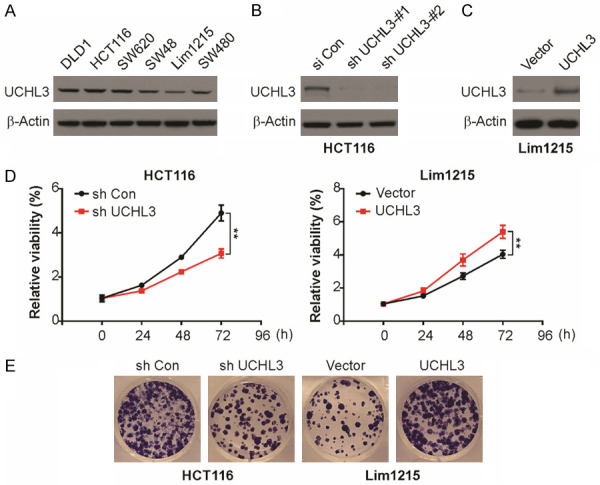
UCHL3 promoted cell proliferation and colony formation in CRC cells. A. UCHL3 expression in six CRC cell lines as detected by Western blotting. B. UCHL3 knockdown in HCT116 cells as detected by Western blotting. C. UCHL3 overexpression in Lim1215 cells as detected by Western blotting. D. Viability of UCHL3-silenced or UCHL3-overexpressed CRC cells as detected by the CCK-8 assay. E. Colony-formation ability of UCHL3-silenced or UCHL3-overexpressed CRC cells as detected by the colony formation assay. Results represent means ± SD from three independent experiments. **P < 0.01.
UCHL3 promotes invasion, migration, and EMT (epithelial-mesenchymal transition) in CRC cells
Matrigel invasion and Transwell migration assays were carried out to assess the effect of UCHL3 on invasion and migration in CRC cells, respectively. UCHL3 suppression led to significant decline in invasive and migratory potential of HCT116 cells (Figure 3A). In contrast, there was a significant enhancement in cell invasion and migration in Lim1215 cells with increased expression in UCHL3 (Figure 3B). Thus, CRC cell migration and invasion were facilitated by UCHL3. EMT has an important function in tumor metastasis, hence, we assessed the level of markers for EMT including N-cadherin, E-cadherin, and Vimentin, along with a group of transcription factors associated with EMT including Slug, Snail, and ZEB1 in Lim1215 and HCT116 cells. In HCT116 cells, UCHL3 knockdown resulted in remarkably high E-cadherin levels and declined Vimentin, N-cadherin, Slug, Snail, and ZEB1 levels (Figure 3C). Contrastingly, increased UCHL3 expression in Lim1215 cells led to a decline in E-cadherin and enhanced Vimentin, N-cadherin, Snail, Slug, and ZEB1 levels (Figure 3D). Therefore, our findings demonstrate that UCHL3 potentially induces EMT in CRC cells.
Figure 3.
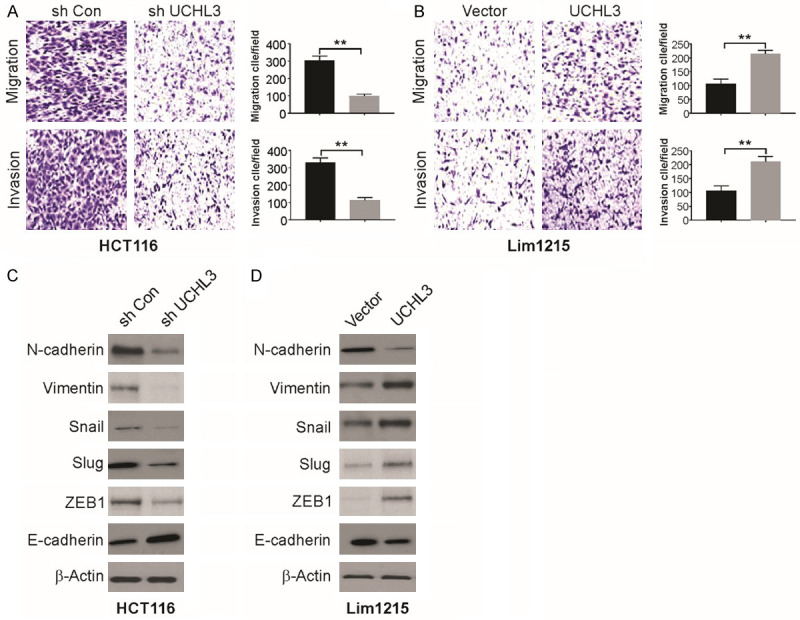
UCHL3 contributed to cell migration, invasion, and EMT in CRC cells. A. Transwell Matrigel invasion and migration assay for UCHL3-silenced HCT116 cells. B. Transwell Matrigel invasion and migration assay for UCHL3-overexpressed Lim1215 cells. C. EMT marker expression in UCHL3-silenced HCT116 cells as detected by Western blotting. D. EMT marker expression in UCHL3-overexpressed Lim1215 cells as detected by Western blotting. Results represent means ± SD from three independent experiments. **P < 0.01.
UCHL3 regulates SOX12 protein in CRC cells
To examine the association of UCHL3 with SOX12 expression in cells of CRC, protein expression was evaluated. SOX12 protein was detectable by western blotting, and the protein level of SOX12 is consistent with the level of UCHL3 (Figure 4A). To determine whether UCHL3 has a causal role in SOX12 expression, RNA interference-mediated UCHL3 silencing in HCT116 cells led to markedly reduced level of SOX12 (Figure 4B). Similarly, the ectopic expression of UCHL3 in Lim1215 resulted in increased levels of SOX12 (Figure 4C). The above data indicate that UCHL3 regulates SOX12 expression.
Figure 4.
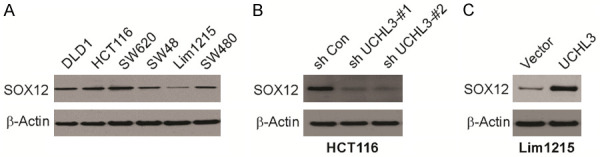
UCHL3 regulates SOX12 expression. A. SOX12 expression in six CRC cell lines as detected by Western blotting. B. SOX12 expression in HCT116 cells with UCHL3 knockdown as detected by Western blotting. C. SOX12 expression in Lim1215 cells with UCHL3 overexpression as detected by Western blotting.
PI3K/AKT/mTOR is involved UCHL3-mediated SOX12 upregulation
In a previous study, UCHL3 has been shown could affect several signaling pathways [29]. Here, Western blotting was carried out to assess the possible functional pathways in CRC. The level of UCHL3 after UCHL3 knockdown is shown, along with the consequent decline in the phosphorylation levels of mTOR and AKT (Figure 5A). Conversely, exogenous expression of UCHL3 increased the phosphorylation levels of AKT and mTOR (Figure 5B). This reveals that the effect of UCHL3 in CRC possibly works through AKT/PI3K/mTOR pathway and the protein expression downstream. On treating CRC cells, which overexpressed UCHL3, with perifosine, AKT/PI3K/mTOR is a signaling pathway inhibitor. Perifosine treatment attenuated UCHL3-induced SOX12 upregulation (Figure 5C). Furthermore, perifosine treatment UCHL3-mediated proliferation, migration and invasion in CRC cells (Figure 5D and 5F).
Figure 5.
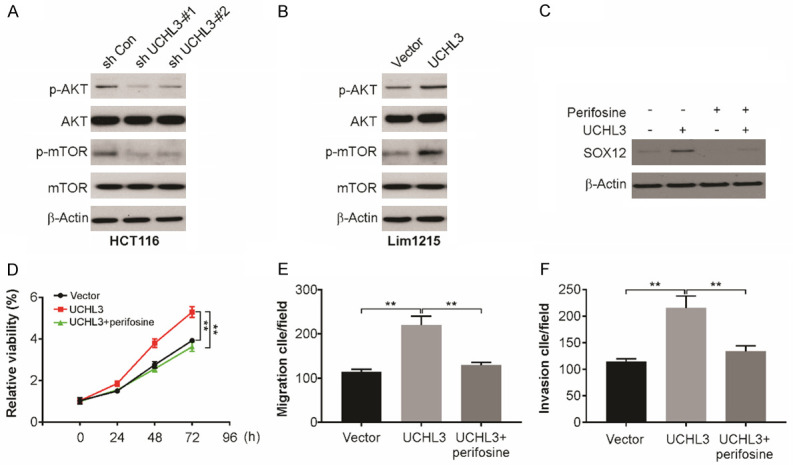
UCHL3 regulates AKT/mTOR signaling pathway in CRC cells. A. Indicated proteins expression in UCHL3-silenced HCT116 cells as detected by Western blotting. B. Indicated proteins expression in UCHL3-overexpression Lim1215 cells as detected by Western blotting. C. SOX12 expression in UCHL3-overexpression Lim1215 cells with/without AKT inhibitor treatment analyzed by Western blotting. D. Cell viability of UCHL3-overexpression Lim1215 cells with/without AKT inhibitor treatment analyzed by CCK-8 assay. E. Migration of UCHL3-overexpression Lim1215 cells with/without AKT inhibitor treatment analyzed. F. Invasion of UCHL3-overexpression Lim1215 cells with/without AKT inhibitor treatment analyzed. Results represent means ± SD from three independent experiments. **P < 0.01.
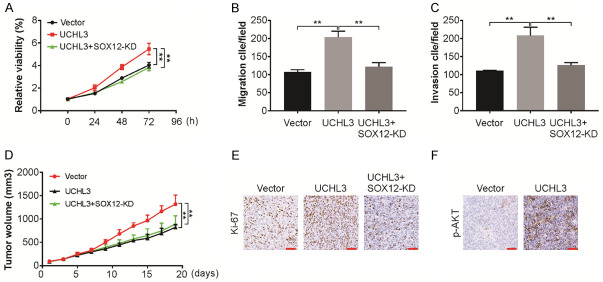
SOX12 is vital for the growth of CRC tumors induced by UCHL3
To assess the function of SOX12 in tumor growth mediated by UCHL3, SOX12 expression was stably downregulated in Lim1215 cells that overexpress UCHL3 (Figure 6A). Knockdown of SOX12 significantly decreased the in vitro proliferative, migratory and invasive capacity of Lim1215 cells (Figure 6B and 6C), indicating the important role of SOX12 in CRC cell invasion, migration and proliferation mediated by UCHL3. To further confirm this, Lim1215 and Lim1215 UCHL3 overexpressing cells with or without SOX12 knockdown were injected in nude mice. On tracking the tumor growth, mice with Lim1215-sh Con cells exhibited significantly increased tumor volume than the Lim1215-sh SOX12 cells-injected mice (Figure 6D). Moreover, enhanced Ki-67 level was observed in the UCHL3 group when compared with the NC group, with a concurrent decline in the apoptotic cell number (Figure 6E). Furthermore, there was an increase in p-AKT level in UCHL3 tumors (Figure 6F). Therefore, these observations indicate significance of SOX12 in CRC tumor growth mediated by UBHL3 in vivo.
Figure 6.
SOX12 is required for UCHL3 promoted tumor growth in vivo. A. Cell viability of UCHL3-overexpression Lim1215 cells with/without SOX12 knockdown analyzed by CCK-8 assay. B. Migration of UCHL3-overexpression Lim1215 cells with/without SOX12 knockdown analyzed. C. Invasion of UCHL3-overexpression Lim1215 cells with/without SOX12 knockdown analyzed. D. Effects of UCHL3 overexpression with or without SOX12 knockdown on tumor growth in vivo. E. Representative images of Ki-67 expression in xenograft tested by immunohistochemistry. Scale bar: 25 μm. F. Representative images of p-AKT expression in xenograft tested by immunohistochemistry. Scale bar: 25 μm. Results represent means ± SD from three independent experiments. **P < 0.01.
Discussion
CRC is among the most malignant tumors [30]. It is in the top three among all tumors and is associated with malaise and cancer-related deaths worldwide [31]. The primary reasons for death in CRC are recurrence and metastasis, and micro-metastases before radical surgery is observed in more than half of CRC patients, causing a remarkably poorer prognosis and increased mortality of patients suffering from CRC [32]. Here, we observed overexpression of UCHL3 in CRC tissues compared to the normal matched tissues. Enhanced UCHL3 expression related independently to worse DFS and OS of CRC patients. Reportedly, this is the first study to determine and relate UCHL3 expression with the symptoms and the prognosis of patients with CRC. The results obtained here indicate the role of UCHL3 as a promoter in CRC advancement, with a propensity to act as a valuable prognostic CRC biomarker. The outcomes of the present study indicate that the knockdown of UCHL3 lowered the HCT116 cell aggressiveness and malignancy decline in cell proliferation in vitro, reduced in vitro cell migration and invasion and the ability to form colonies. Concurrently, opposite effects were observed in Lim1215 cells overexpressing UCHL3 in addition to enhanced tumor growth in vivo. Thus, UCHL3 facilitated growth of tumors by enhancing CRC cell aggressiveness and malignancy and can be considered a CRC oncogene.
The role of UCHL3 has been shown in several human cancers, such as breast and prostate cancers [33,34]. Accordingly, in this study, we observed that high levels of UCHL3 correlated appreciably with a poorer prognosis for CRC patients, indicating the role of UCHL3 as a potential CRC oncogene. Considering several similarities between tumorigenesis and biological behaviors during embryonic development, tumor is currently analyzed from the perspective of developmental biology. Signaling pathways involved in the development of an embryo are aberrantly activated during the growth and advancement of several malignancies, including CRC [35]. During embryonic development, UCHL3 is one of the essential regulators of angiogenesis, therefore we hypothesized the additional role of UCHL3 in angiogenesis during CRC progression [36].
In CRC and in other cancers, EMT is an important factor causing metastasis [36,37]. During this process, the epithelial characteristics of the cells are lost, including reduced expression of mesenchymal features and E-cadherin such as enhanced vimentin and N-cadherin expressions [36]. The upstream transcription factors in EMT include proteins Slug, Snail, and ZEB1 [38]. In this study, enhanced E-cadherin expression was observed due to depletion of UCHL3, accompanied with a decline in levels of vimentin and N-cadherin, and the transcription factors Slug, Snail, and ZEB1, while a reverse effect was observed as a result of UCHL3 overexpression. Therefore, UCHL3 was found to facilitate EMT through Snail, Slug, and ZEB1 up-regulation.
Aberrant PI3K/AKT/mTOR signaling has been observed in a number of malignancies in humans, including CRC and is found to be closely associated with oncogenic effects on different cancers [39]. In CRC cells, PI3K/AKT/mTOR signaling activity is associated with chemoresistance, aggressive invasion and poor histological differentiation, and targeting this signaling pathway is a potentially promising treatment option for human cancers such as CRC [40]. Here, we observed that UCHL3 activates the PI3K/AKT/mTOR pathway. Likewise, UCHL3 knockdown led to significantly decline in p-AKT level, and an opposite effect was observed as a result of UCHL3 overexpression.
Among various transcription factors vital of development, the SOX gene family members act as human cancer modulators [41]. SOX12, a SOXC group member, is crucial for the development of the embryo and determination of cell fate during organogenesis as well as in carcinogenesis [18]. In HCC, a significant correlation of overexpressed SOX12 was observed with the progress of disease and poor prognosis [18]. SOX12 also participates in the progression of leukemia through the regulation of beta-catenin expression and ultimately disturbing the Wnt/TCF pathway, which may be targeted in AML [42]. Further, SOX12 knockdown impedes the proliferative, migratory, invasive, and metastatic capacity of breast cancer and lung cancer cells [43,44]. While these studies showed the key function of SOX12 in progress and metastasis of cancer, the status of SOX12 in human CRC still remains elusive. Accordingly, in this study, we found that UCHL3-medicated tumor growth was significantly attenuated by SOX12 knockdown, further suggesting the potential of SOX12 as a prognostic biomarker and a therapeutic target in CRC.
In conclusion, our findings are the first report on the clinical significance of UCHL3 in CRC by the evaluation of the association between UCHL3 and CRC patient prognosis and the underlying process. The outcomes of our study strongly indicate the growth-promoting effect of UCHL3 on CRC by modulating AKT-dependent SOX12 upregulation. Therefore, UCHL3 is a potential biomarker for prognostic and a target for the treatment of CRC.
Disclosure of conflict of interest
None.
References
- 1.Tan X, Tong J, Wang YJ, Fletcher R, Schoen RE, Yu J, Shen L, Zhang L. BET inhibitors potentiate chemotherapy and killing of SPOP-mutant colon cancer cells via induction of DR5. Cancer Res. 2019;79:1191–1203. doi: 10.1158/0008-5472.CAN-18-3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xie YH, Chen YX, Fang JY. Comprehensive review of targeted therapy for colorectal cancer. Signal Transduct Target Ther. 2020;5:22. doi: 10.1038/s41392-020-0116-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu IS, Cheung WY. Metastatic colorectal cancer in the era of personalized medicine: a more tailored approach to systemic therapy. Can J Gastroenterol Hepatol. 2018;2018:9450754. doi: 10.1155/2018/9450754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ciombor KK, Bekaii-Saab T. A comprehensive review of sequencing and combination strategies of targeted agents in metastatic colorectal cancer. Oncologist. 2018;23:25–34. doi: 10.1634/theoncologist.2017-0203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martini G, Troiani T, Cardone C, Vitiello P, Sforza V, Ciardiello D, Napolitano S, Della Corte CM, Morgillo F, Raucci A, Cuomo A, Selvaggi F, Ciardiello F, Martinelli E. Present and future of metastatic colorectal cancer treatment: a review of new candidate targets. World J Gastroenterol. 2017;23:4675–4688. doi: 10.3748/wjg.v23.i26.4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morrow JK, Lin HK, Sun SC, Zhang S. Targeting ubiquitination for cancer therapies. Future Med Chem. 2015;7:2333–2350. doi: 10.4155/fmc.15.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gallo LH, Ko J, Donoghue DJ. The importance of regulatory ubiquitination in cancer and metastasis. Cell Cycle. 2017;16:634–648. doi: 10.1080/15384101.2017.1288326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Senft D, Qi J, Ronai ZA. Ubiquitin ligases in oncogenic transformation and cancer therapy. Nat Rev Cancer. 2018;18:69–88. doi: 10.1038/nrc.2017.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang M, Yu T, Hu L, Cheng Z, Li M. Ubiquitin carboxy-terminal hydrolasel3 correlates with human sperm count, motility and fertilization. PLoS One. 2016;11:e0165198. doi: 10.1371/journal.pone.0165198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suzuki M, Setsuie R, Wada K. Ubiquitin carboxyl-terminal hydrolase l3 promotes insulin signaling and adipogenesis. Endocrinology. 2009;150:5230–5239. doi: 10.1210/en.2009-0332. [DOI] [PubMed] [Google Scholar]
- 11.Li G, Jin X, Zheng J, Jiang N, Shi W. UCHL3 promotes non-small cell lung cancer proliferation via accelerating cell cycle and inhibiting cell apoptosis. Biotechnol Appl Biochem. 2020;10:1002. doi: 10.1002/bab.1909. [DOI] [PubMed] [Google Scholar]
- 12.Kumar P, Mistri TK. Transcription factors in SOX family: potent regulators for cancer initiation and development in the human body. Semin Cancer Biol. 2019 doi: 10.1016/j.semcancer.2019.06.016. S1044-579X(18)30184-6. [DOI] [PubMed] [Google Scholar]
- 13.Grimm D, Bauer J, Wise P, Kruger M, Simonsen U, Wehland M, Infanger M, Corydon TJ. The role of SOX family members in solid tumours and metastasis. Semin Cancer Biol. 2019 doi: 10.1016/j.semcancer.2019.03.004. S1044-579X(18)30141-X. [DOI] [PubMed] [Google Scholar]
- 14.Castillo SD, Sanchez-Cespedes M. The SOX family of genes in cancer development: biological relevance and opportunities for therapy. Expert Opin Ther Targets. 2012;16:903–919. doi: 10.1517/14728222.2012.709239. [DOI] [PubMed] [Google Scholar]
- 15.Huang J, Ji EH, Zhao X, Cui L, Misuno K, Guo M, Huang Z, Chen X, Hu S. Sox11 promotes head and neck cancer progression via the regulation of SDCCAG8. J Exp Clin Cancer Res. 2019;38:138. doi: 10.1186/s13046-019-1146-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Du F, Chen J, Liu H, Cai Y, Cao T, Han W, Yi X, Qian M, Tian D, Nie Y, Wu K, Fan D, Xia L. SOX12 promotes colorectal cancer cell proliferation and metastasis by regulating asparagine synthesis. Cell Death Dis. 2019;10:239. doi: 10.1038/s41419-019-1481-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wan H, Cai J, Chen F, Zhu J, Zhong J, Zhong H. SOX12: a novel potential target for acute myeloid leukaemia. Br J Haematol. 2017;176:421–430. doi: 10.1111/bjh.14425. [DOI] [PubMed] [Google Scholar]
- 18.Huang W, Chen Z, Shang X, Tian D, Wang D, Wu K, Fan D, Xia L. Sox12, a direct target of FoxQ1, promotes hepatocellular carcinoma metastasis through up-regulating Twist1 and FGFBP1. Hepatology. 2015;61:1920–1933. doi: 10.1002/hep.27756. [DOI] [PubMed] [Google Scholar]
- 19.Ouyang L, Yan B, Liu Y, Mao C, Wang M, Liu N, Wang Z, Liu S, Shi Y, Chen L, Wang X, Cheng Y, Cao Y, Xiao D, Zhang L, Liu S, Tao Y. The deubiquitylase UCHL3 maintains cancer stem-like properties by stabilizing the aryl hydrocarbon receptor. Signal Transduct Target Ther. 2020;5:78. doi: 10.1038/s41392-020-0181-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sarkar A, Hochedlinger K. The sox family of transcription factors: versatile regulators of stem and progenitor cell fate. Cell Stem Cell. 2013;12:15–30. doi: 10.1016/j.stem.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dy P, Penzo-Mendez A, Wang H, Pedraza CE, Macklin WB, Lefebvre V. The three SoxC proteins--Sox4, Sox11 and Sox12--exhibit overlapping expression patterns and molecular properties. Nucleic Acids Res. 2008;36:3101–3117. doi: 10.1093/nar/gkn162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tong J, Wang P, Tan S, Chen D, Nikolovska-Coleska Z, Zou F, Yu J, Zhang L. Mcl-1 degradation is required for targeted therapeutics to eradicate colon cancer cells. Cancer Res. 2017;77:2512–2521. doi: 10.1158/0008-5472.CAN-16-3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tong J, Zheng X, Tan X, Fletcher R, Nikolovska-Coleska Z, Yu J, Zhang L. Mcl-1 phosphorylation without degradation mediates sensitivity to HDAC inhibitors by liberating BH3-only proteins. Cancer Res. 2018;78:4704–4715. doi: 10.1158/0008-5472.CAN-18-0399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He K, Chen D, Ruan H, Li X, Tong J, Xu X, Zhang L, Yu J. BRAFV600E-dependent Mcl-1 stabilization leads to everolimus resistance in colon cancer cells. Oncotarget. 2016;7:47699–47710. doi: 10.18632/oncotarget.10277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fu R, Tong JS. miR-126 reduces trastuzumab resistance by targeting PIK3R2 and regulating AKT/mTOR pathway in breast cancer cells. J Cell Mol Med. 2020;24:7600–7608. doi: 10.1111/jcmm.15396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tong J, Tan S, Zou F, Yu J, Zhang L. FBW7 mutations mediate resistance of colorectal cancer to targeted therapies by blocking Mcl-1 degradation. Oncogene. 2017;36:787–796. doi: 10.1038/onc.2016.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tong J, Tan S, Nikolovska-Coleska Z, Yu J, Zou F, Zhang L. FBW7-dependent Mcl-1 degradation mediates the anticancer effect of Hsp90 inhibitors. Mol Cancer Ther. 2017;16:1979–1988. doi: 10.1158/1535-7163.MCT-17-0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang QH, Wei L, Tong JS, Qi ST, Li S, Ou XH, Ouyang YC, Hou Y, An LG, Schatten H, Schatten H, Sun QY. Localization and function of mSpindly during mouse oocyte meiotic maturation. Cell Cycle. 2010;9:2230–2236. doi: 10.4161/cc.9.11.11895. [DOI] [PubMed] [Google Scholar]
- 29.Fang Y, Shen X. Ubiquitin carboxyl-terminal hydrolases: involvement in cancer progression and clinical implications. Cancer Metastasis Rev. 2017;36:669–682. doi: 10.1007/s10555-017-9702-0. [DOI] [PubMed] [Google Scholar]
- 30.Knickelbein K, Tong J, Chen D, Wang YJ, Misale S, Bardelli A, Yu J, Zhang L. Restoring PUMA induction overcomes KRAS-mediated resistance to anti-EGFR antibodies in colorectal cancer. Oncogene. 2018;37:4599–4610. doi: 10.1038/s41388-018-0289-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen D, Tong J, Yang L, Wei L, Stolz DB, Yu J, Zhang J, Zhang L. PUMA amplifies necroptosis signaling by activating cytosolic DNA sensors. Proc Natl Acad Sci U S A. 2018;115:3930–3935. doi: 10.1073/pnas.1717190115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xie YH, Chen YX, Fang JY. Comprehensive review of targeted therapy for colorectal cancer. Signal Transduct Target Ther. 2020;5:22. doi: 10.1038/s41392-020-0116-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luo K, Li L, Li Y, Wu C, Yin Y, Chen Y, Deng M, Nowsheen S, Yuan J, Lou Z. A phosphorylation-deubiquitination cascade regulates the BRCA2-RAD51 axis in homologous recombination. Genes Dev. 2016;30:2581–2595. doi: 10.1101/gad.289439.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Song HM, Lee JE, Kim JH. Ubiquitin C-terminal hydrolase-L3 regulates EMT process and cancer metastasis in prostate cell lines. Biochem Biophys Res Commun. 2014;452:722–727. doi: 10.1016/j.bbrc.2014.08.144. [DOI] [PubMed] [Google Scholar]
- 35.Nguyen HT, Duong HQ. The molecular characteristics of colorectal cancer: implications for diagnosis and therapy. Oncol Lett. 2018;16:9–18. doi: 10.3892/ol.2018.8679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yi YJ, Sutovsky M, Song WH, Sutovsky P. Protein deubiquitination during oocyte maturation influences sperm function during fertilisation, antipolyspermy defense and embryo development. Reprod Fertil Dev. 2015;27:1154–1167. doi: 10.1071/RD14012. [DOI] [PubMed] [Google Scholar]
- 37.Yang C, Shi S, Su Y, Tong JS, Li L. P2X7R promotes angiogenesis and tumour-associated macrophage recruitment by regulating the NF-kappaB signalling pathway in colorectal cancer cells. J Cell Mol Med. 2020;24:10830–10841. doi: 10.1111/jcmm.15708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pastushenko I, Blanpain C. EMT transition states during tumor progression and metastasis. Trends Cell Biol. 2019;29:212–226. doi: 10.1016/j.tcb.2018.12.001. [DOI] [PubMed] [Google Scholar]
- 39.Nitulescu GM, Van De Venter M, Nitulescu G, Ungurianu A, Juzenas P, Peng Q, Olaru OT, Gradinaru D, Tsatsakis A, Tsoukalas D, Spandidos DA, Margina D. The Akt pathway in oncology therapy and beyond (Review) Int J Oncol. 2018;53:2319–2331. doi: 10.3892/ijo.2018.4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shi X, Wang J, Lei Y, Cong C, Tan D, Zhou X. Research progress on the PI3K/AKT signaling pathway in gynecological cancer (Review) Mol Med Rep. 2019;19:4529–4535. doi: 10.3892/mmr.2019.10121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lou J, Zhang K, Chen J, Gao Y, Wang R, Chen LB. Prognostic significance of SOX-1 expression in human hepatocelluar cancer. Int J Clin Exp Pathol. 2015;8:5411–5418. [PMC free article] [PubMed] [Google Scholar]
- 42.Leung RKC, Leung HC, Leung AYH. Diverse pathogenetic roles of SOX genes in acute myeloid leukaemia and their therapeutic implications. Semin Cancer Biol. 2019 doi: 10.1016/j.semcancer.2019.11.001. S1044-579X(18)30139-1. [DOI] [PubMed] [Google Scholar]
- 43.Ding H, Quan H, Yan W, Han J. Silencing of SOX12 by shRNA suppresses migration, invasion and proliferation of breast cancer cells. Biosci Rep. 2016;36:e00389. doi: 10.1042/BSR20160053. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 44.Wang L, Hu F, Shen S, Xiao H, Li G, Wang M, Mei J. Knockdown of SOX12 expression inhibits the proliferation and metastasis of lung cancer cells. Am J Transl Res. 2017;9:4003–4014. [PMC free article] [PubMed] [Google Scholar]