Abstract
Long noncoding RNAs (lncRNAs) have been shown to play important roles in carcinogenesis and progression. In this study, we mainly investigate the potential influence of lncRNA NCK1 antisense RNA 1 (NCK1-AS1) on the progression of non-small cell lung cancer (NSCLC). RT-PCR was performed to determine the expression of NCK1-AS1 and miR-137 in NSCLC specimens and cell lines. The clinical significance of NCK1-AS1 in 148 patients was analyzed statistically. The receiver operating characteristic (ROC) curve was performed to estimate the diagnostic value of NCK1-AS1 and miR-137. Regulatory effects of NCK1-AS1 on proliferative, colony formation abilities, metastasis and apoptosis of SK-MES-1 and H1299 cells were assessed through a series of functional experiments. RNA-pull down and Dual-Luciferase reporter assay was performed to verify the sponge effect of NCK1-AS1 on miR-137. We observed that NCK1-AS1 expression was upregulated, while miR-137 expression was down-regulated in NSCLC specimens and cell lines. Increased NCK1-AS1 expression was positively correlated with TNM stage and lymph node metastasis and poor clinical outcome. The diagnostic value of NCK1-AS1 and miR-137 expression was also confirmed. Functionally, knockdown of NCK1-AS1 suppressed the proliferation, migration and invasion of NSCLC cells, and promoted apoptosis. Moreover, NCK1-AS1 was able to adsorb miR-137 via a sponge effect. Overall, our findings suggested that NCK1-AS1 may be a candidate biomarker and a target for new therapies in NSCLC patients.
Keywords: LncRNA NCK1-AS1, miR-137, NSCLC, biomarker, metastasis
Introduction
Lung cancer acts as a common reason for the mortality resulted from cancer all over the world, with the highest mortality rate (less than 20% 5-year survival rate) [1]. Non-small cell lung cancer (NSCLC) occupies about 85% of all cases [2]. Although experimental and clinical oncology has achieved some progressions regarding the diagnosis and treatment, NSCLC patients still exhibit gloomy prognosis mainly due to its high recurrent and distant metastasis [3,4]. Up to date, many reports have emphasized the investigation of genes and proteins underlying the progress of NSCLC; however, their sensitivity and specificity are imperfect [5]. On that account, the useful biomarkers for the diagnosis and prognosis shall be urgently identified for enhancing the clinical outcomes as well as developing effective and appropriate treatment strategies specific to NSCLC patients.
High-throughput gene sequencing analysis has advanced considerably in recent years, demonstrating the pervasive transcription of human genome, with most (about 98%) incapable of fully or obviously coding protein, that is the so-called noncoding RNAs (ncRNAs) [6]. Long non-coding RNA (lncRNAs) are long RNA transcripts (> 200 nucleotides) that do not possess protein-coding capabilities [7]. They are capable of regulating many significant physiological and biological activities, thereby having been widely concerned in recent years [8,9]. As demonstrated by emerging reports, the deregulation of lncRNAs remarkably affects many disease states, inclusive of the malignancy [10,11]. For instance, lncRNA SNHG11 was shown to exhibit a high level in NSCLC and promote the metastasis of NSCLC cells via miR-485-5p/BSG axis [12]. LncRNA ZFAS1 was reported to be a tumor promoter in NSCLC as its overexpression can target miR-1271-5p/FRS2, so as to suppress NSCLC cell proliferation and invasion [13]. In addition, more and more studies have indicated that some dysregulated and tumor-related lncRNAs can contribute to the diagnosis and prognosis of cancers, and be potential treatment targets [14,15].
LncRNA NCK1 antisense RNA 1 (NCK1-AS1), a recently identified lncRNA, is located on chromosome 3q22.3, encoding for a 1.4 kb noncoding RNA rather than protein. In recent years, the distinct overexpression of NCK1-AS1 was frequently reported in many tumors, like nasopharyngeal carcinoma, cervical cancer and prostate cancer, and in functional assays, this lncRNA was shown to exhibit a tumor-promotive role in the ability of the proliferation and metastasis via various complex mechanisms [16-18]. However, the expression and effects of NCK1-AS1 in NSCLC have not yet been characterized.
Patients and methods
Patients and tissue samples
We collected 148 primary NSCLC and corresponding normal specimens from the the Jiangjin District Central Hospital for RT-qPCR assays between June 2012 and August 2015. All the cancer tissues were confirmed by pathological diagnosis after surgery. All these patients have not underwent chemotherapy, radiotherapy or immunotherapy prior to surgery. Among these 148 NSCLC cases, there were 98 males and 50 females, and their age ranged from 26 to 75 years. We followed up all NSCLC patients each one-three months before August 2015. The median follow-up period lasted for 60 months. Follow-up studies focused on physical examination, laboratory analysis, as well as computed tomography if necessary. This study had obtained the approval of the Research Ethics Committee of the Weifang People’s Hospital, and obtained all participants’ written informed consent.
Cell culture and cell transfection
The human NSCLC lines (A549, SPC-A-1, SK-MES-1, H1299 and 95D) as well as normal bronchial epithelial cell line 16HBE came from the Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences, Shanghai, China. We cultured all cells in complete Dulbecco’s modified Eagle medium (DMEM, Qiangyao, Techinology, Guangzhou, Guangdong, China) that contained fetal bovine serum (10%, FBS, Gibco, Pudong, Shanghai, China) with 100 units/mL penicillin (Hayao Company, Haerbin, Heilongjiang, China) and 100 μg/mL streptomycin (Ruiyang Technology Company, Haidian, Biejing, China) in a humidified incubator with 5% CO2 at 37°C.
MiR-137 mimics (#HmiR0244-MR09) and scrambled miRNA oligonucleotides (miR-NC) were provided by Genecopoeia (Guangdong, Guangzhou, China). SiRNAs targeting NCK1-AS1 (si-NCK1-AS1-1, si-NCK1-AS1-2) and si-NC were provided by Shanghai Shenggong Biological corporation (Songjiang, Shanghai, China). The duplicate short hairpin RNAs for NCK1-AS1 (sh-NCK1-AS1) and their control (sh-control) were designed by Genepharm (Shanghai, China). Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) assisted in transfecting cells in line with the protocol of provider. The cells received 48 h of transfection by using a final concentration of 50 nM, followed by being harvested for later assays.
Real-time PCR assays
The Trizol Total RNA Reagent (Invitrogen, Kunshan, Jiangsu, China) was employed to isolate the total RNA from NSCLC and adjacent normal specimens. UV spectrophotometer helped to detect the purity and concentration exhibited by the total RNA at 260 nm. The SuperScript III first-strand synthesis system (Invitrogen, Carlsbad, CA, USA) was applied to the synthesis of first-strand cDNA. ABI 7500 system (Applied Biosystems, Foster City, CA, USA) was applied to the qRT-PCR assays to determine NCK1-AS1 and miR-137 expression, based on the instruction of the manufacturer. The GAPDH expression was detected as the endogenous control. The 2-ΔΔCT methods helped normalize the relative gene expression to GAPDH or U6. The primer sequences used for the studies were shown in Table 1.
Table 1.
The primers used in this study for RT-PCR
| Names | Sequences (5’-3’) |
|---|---|
| NCK1-AS1: F | GCCGCAGGAGAGACTTAACA |
| NCK1-AS1: R | CCTTCGGCTGGGATGACATT |
| miR-137: F | GAAATCCGACAGCTTAAGGAGGTTTGA |
| miR-137: R | CATTGCACAGATAGGATTTGATTTACT |
| GAPDH: F | CAATGACCCCTTCATTGACC |
| GAPDH: R | GACAAGCTTCCCGTTCTCAG |
| U6: F | GCGCGTCGTGAAGCGTTC |
| U6: R | GTGCAG GGTCCGAGGT |
Cell proliferation assays
Cell proliferation was examined by the use of cell counting assay Kit-8 (CCK-8) (DOJINDO, Pudong, Shanghai, China) as required by the manufacture. We seeded cells into 96-well plates (2 × 103 cells/well) for 24, 48, 72 and 96 hours, respectively. The Microplate Reader (Bio-Rad, Haidian, Beijing, China) was employed to measure the absorbance of cells at 450 nm, thereby detecting the proliferation rate.
Colony formation assays
si-NCK1-AS1-1 and si-NCK1-AS1-2 were used to transfect SK-MES-1 and H1299 cells. After 48 h, 3 × 103 cells were first seeded in six-well plates, followed by seven days of culturing. Methanol served for fixing cells for half an hour at room temperature. An optical microscope helped to count the colony number (with > 50 cells).
The EdU (5-ethynyl-2’-deoxyuridine) assay
The EdU labeling/detection kit (Ribobio, Pudong, Shanghai, China) was applied to assess the cell proliferation based on the previous description [19].
Wound healing assays
Cells in each group were implanted into 6-well culture plates with the density of 5.0 × 105 cells/well. We used a 200 μl pipette tip to scrape the cells, thereby manually damaging the confluent cell monolayer. The distance from one side of the injury to the other side was taken into account to measure the wound healing ability. At last, we photographed the scratched lines at 0 h and 24 h.
Transwell invasion assays
For invasion assay, we seeded 3.0 × 105 cells in DMEM free of FBS in the upper chamber of a 24-well Matrigel transwell invasion insert (BD bioscience, Xuhui, Shanghai, China), and used culture medium that contained FBS (10%) to fill the lower chamber. After cells underwent incubation for 24 hours, we removed the ono-invaded cells on membrane’s upper surface, and used crystal violet (0.1%) to stain those invaded through the membrane. We selected 5 vision fields each chamber in a random manner, followed by using an inverted microscope for counting the invasion cell number.
Flow cytometric analysis
FITC Annexin V Apoptosis Detection Kit I (BD Biosciences, Kunming, Yunnan, China) was applied for the apoptosis assay. SK-MES-1 and H1299 cells first received 48 h of si-NCK1-AS1-1, si-NCK1-AS1-2 or si-NC transfection, followed by being collected as well as resuspended in fixation fluid. Cell suspension (500 μl) was added with Annexin V-FIFC (5 μl) together with propidium iodide (100 μl). Flow cytometry was carried out to determine the cellular apoptosis.
Subcellular fractionation
The RNeasy Midie Kit (Qiagen, Xuhui, Shanghai, China) was used for detaching as well as harvesting the nuclear and cytoplasmic fractions. RT-PCR assays were performed for further determination of NCK1-AS1 cellular localization, taking GAPDH and U6 as the cytoplasm control and the nucleus control, respectively.
RNA pull-down assays
The performance of RNA pulldown assays followed previous description [20].
Luciferase reporter assays
Starbase 3.0 assisted in obtaining the predicted binding sites between miR-137 and NCK1-AS1-3’UTR. Corresponding instrument was used to clone the binding sequences as well as mutant sequences into the pmirGLO Dual-luciferase vectors respectively (Gene Pharma, Shanghai, China). SK-MES-1 and H1299 cells received pre-culturing in 96-well plates, and then underwent the co-transfection of wild-type pmirGLO-NCK1-AS1 reporter plasmid or the mutated type and mimics-miR-137 or NC using Lipofectamine 3000. Luciferase reporter assays adopted the Dual-Luciferase Reporter Assay System (Promega, Hangzhou, Zhejiang, China). After 48 h, Renilla luciferase activity was used to examine as well as normalize firefly luciferase activity.
Animal study
BALB/c female nude mice from Shanghai SIPPR-BK Laboratory Animal (Shanghai, China) were used for the functional exploration of NCK1-AS1. All mice wer maintained under SPF-condition lab. The Review Board of Weifang People’s Hospital reviewed and approved the use of animals. H1299 cells stably transfected with sh-Ctrl and sh-NCK1-AS1 were injected into nude mice at a density of 6 × 106. The size of tumour was determined every 7 days. Tumour volume was calculated as the following formula: (length × width2)/2.
Statistical analysis
The SPSS statistical software package (standard version 18.0, SPSS Inc., Chicago, IL, USA) was applied to the statistical analysis. An independent t-test assisted in comparing the two groups regarding continuous data, and the chi-square test assisted in analyzing the categorical data. Receiver-operating characteristic (ROC) curves were used for assessing the NCK1-AS1 and miR-137 levels to discriminate NSCLC specimens from non-tumor lung tissues. The Kaplan-Meier analysis was carried out to estimate the overall survival (OS) and disease-free survival (DFS) of patients who had different NCK1-AS1 expressions. The multivariate analysis adopted a Cox regression model for analyzing the prognostic parameters. p-values < 0.05 exhibited a statistical significance.
Results
Up-regulated expression of NCK1-AS1 in human NSCLC
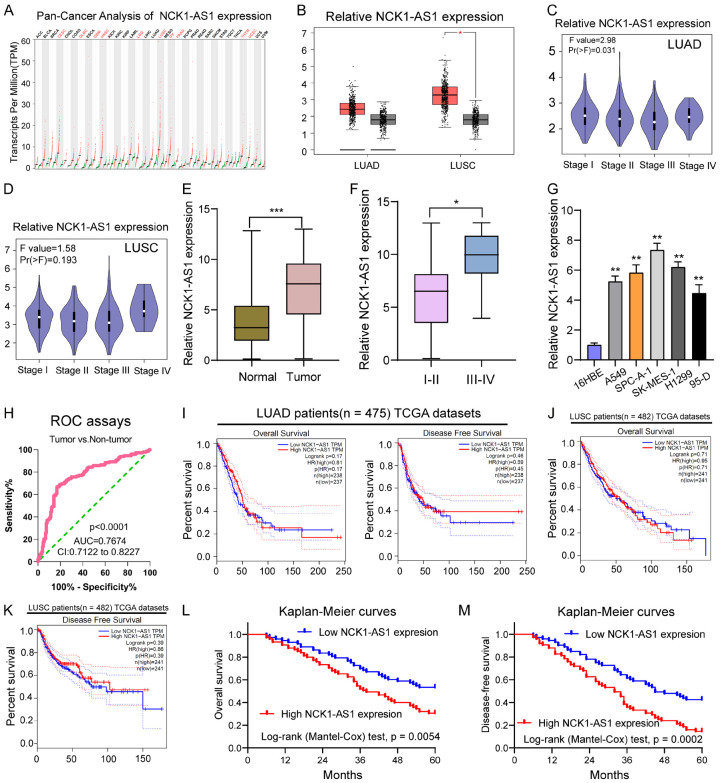
For figuring out the expression pattern exhibited by NCK1-AS1 in tumors, we performed Pan-cancer Analysis using “GEPIA” which indicated that overexpression of NCK1-AS1 was a frequent event in many tumor types, like the lung adenocarcinoma (LUAD) and the lung squamous cell carcinoma (LUSC) (Figure 1A, 1B). In addition, we observed that NSCLC samples with advanced stages showed a higher level of NCK1-AS1 in both LUAD and LUSC (Figure 1C, 1D). Then, we examined the levels of NCK1-AS1 in 148 NSCLC patients and found that NSCLC specimens exhibited a distinctly increased NCK1-AS1 expression relative to matched normal specimens (P < 0.01, Figure 1E). Moreover, NSCLC specimens with stage (III-IV) showed a higher NCK1-AS1 expression relative to NSCLC tissues with stage (I-II) (P < 0.01, Figure 1F). Besides, NCK1-AS1 expression also existed in cell lines, and NSCLC cell lines showed a higher NCK1-AS1 expression than normal 16HBE cell (Figure 1G). To determine whether NCK1-AS1 expression had a diagnostic value for NSCLC, we performed ROC assays, confirming that high NCK1-AS1 expression had an AUC value of 0.7674 (95% CI: 0.7122 to 0.8227) for NSCLC (Figure 1H). The sensitivity and specificity of NCK1-AS1 expressions for distinguishing NSCLC samples from normal samples was 69.55%/81.37%, indicating NCK1-AS1 as an early-diagnosis indicator for NSCLC patients. Overall, our findings suggested that NCK1-AS1 may be involved in the progression of NSCLC.
Figure 1.
The distinct upregulation of NCK1-AS1 expression in NSCLC patients and its clinical value. A. The Pan-Cancer Analysis of NCK1-AS1 expression based on the TCGA datasets. B. NCK1-AS1 expression was increased in LUAD and LUSC analyzed using GEPIA tool. C, D. Violin Plot for the expression of NCK1-AS1 in LUAD and LUSC tissues with different stages analyzed using GEPIA tool. E. Relative expression of NCK1-AS1 expression in NSCLC specimens (n = 148) and in paired adjacent normal specimens (n = 148). NCK1-AS1 expression was examined by qPCR and normalized to GAPDH expression (shown as 2-ΔCT). F. A high level of NCK1-AS1 was observed in NSCLC specimens with advanced stages in our cohort. G. qPCR detected the expression levels of NCK1-AS1 in 16HBE and five NSCLC cell lines. H. Receiver-operating characteristic (ROC) curves were used to evaluate the performance of NCK1-AS1 to discriminate NSCLC specimens from non-tumor specimens. I. The survival assays of the overall survival and disease-free survival of 475 LUAD patients analyzed using GEPIA tool. J, K. The survival assays of the overall survival and disease-free survival of 482 LUSC patients analyzed using GEPIA tool. L, M. Kaplan-Meier curves of the overall survival and disease-free survival of 148 NSCLC patients. ***P < 0.011, *P < 0.05.
Increased expression of NCK1-AS1 is associated with poor prognosis of NSCLC
To better understand the clinical relevance exhibited by NCK1-AS1 expression in NSCLC, the median expression level of NCK1-AS1 (6.64) in all NSCLC samples was taken into account to divide 148 NSCLC patients into a group with high expression (n = 73) and group with low expression (n = 75). The results of Chi-square test indicated that high NCK1-AS1 expression could lead to TNM stage (P = 0.021) and lymph node metastasis (P = 0.017) (Table 2). Then, we wondered whether NCK1-AS1 may impact NSCLC patients’ clinical outcomes. Using “GEPIA”, we observed that NCK1-AS1 expression did not obviously affect the OS or DFS of patients with LUAD and LUSC based on the TCGA datasets (Figure 1I-K). However, interestingly, in our cohort, the NSCLC patients with high NCK1-AS1 expression had significantly lower 5-year OS (P = 0.0054, Figure 1L) and DFS (P = 0.0002, Figure 1M) rate than those in the low NCK1-AS1 expression group. Moreover, we performed multivariate analyses for further confirmation of the prognostic value of NCK1-AS1 expression in NSCLC patients. As shown in Table 3, we showed that NCK1-AS1 expression played a significant role of independent prognostic markers in OS (HR = 2.986, 95% CI: 1.328-4.473, P = 0.015) and DFS (HR = 3.128, 95% CI: 1.328-5.137, P = 0.002) rates of NSCLC patients (Table 3).
Table 2.
Correlation between NCK1-AS1 expression and clinicopathological characteristics of NSCLC patients
| Clinicopathological features | No. of cases | NCK1-AS1 expression | p value | |
|---|---|---|---|---|
|
| ||||
| Low | High | |||
| Age (years) | 0.413 | |||
| < 58 | 76 | 41 | 35 | |
| ≥ 58 | 72 | 34 | 38 | |
| Sex | 0.907 | |||
| Male | 98 | 50 | 48 | |
| Female | 50 | 25 | 25 | |
| History of smoking | 0.611 | |||
| Ever | 80 | 39 | 41 | |
| Never | 68 | 36 | 32 | |
| Tumor size | 0.121 | |||
| ≤ 3 cm | 90 | 41 | 49 | |
| > 3 cm | 58 | 34 | 24 | |
| TNM stage | 0.021 | |||
| I/II | 98 | 43 | 55 | |
| III/IV | 50 | 32 | 18 | |
| Histological grade | 0.168 | |||
| Well and moderately | 89 | 41 | 48 | |
| Poorly | 59 | 34 | 25 | |
| Lymph node metastasis | 0.017 | |||
| Negative | 102 | 45 | 57 | |
| Positive | 46 | 30 | 16 | |
Table 3.
Multivariate analyses for overall survival and disease-free survival by Cox regression model
| Parameters | Overall survival | Disease-free survival | ||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| HR | 95% CI | p | HR | 95% CI | p | |
| Age | 0.892 | 0.472-1.763 | 0.218 | 1.137 | 0.673-1.993 | 0.378 |
| Sex | 1.217 | 0.673-1.993 | 0.137 | 1.346 | 0.773-2.138 | 0.236 |
| History of smoking | 0.985 | 0.472-1.752 | 0.228 | 1.137 | 0.632-1.993 | 0.138 |
| Tumor size | 1.327 | 0.673-1.875 | 0.138 | 1.429 | 0.773-2.138 | 0.187 |
| TNM stage | 2.895 | 1.385-4.565 | 0.013 | 3.011 | 1.428-4.884 | 0.018 |
| Histological grade | 0.896 | 0.485-1.776 | 0.118 | 0.996 | 0.582-2.328 | 0.089 |
| Lymph node metastasis | 3.018 | 1.428-4.893 | 0.006 | 3.238 | 1.594-5.329 | 0.001 |
| NCK1-AS1 expression | 2.986 | 1.328-4.473 | 0.015 | 3.128 | 1.328-5.137 | 0.002 |
NCK1-AS1 knockdown suppresses NSCLC cell proliferation, migration and invasion
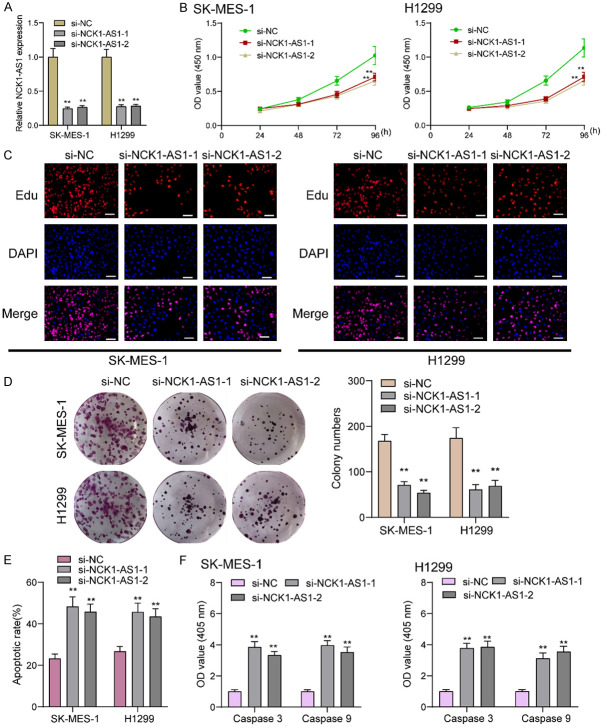
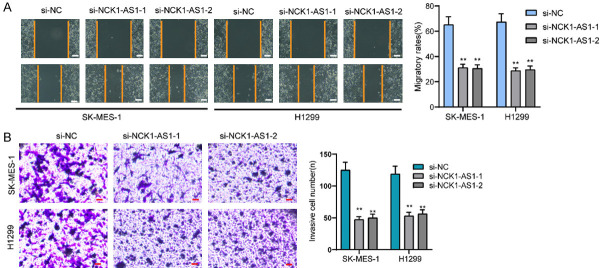
Considering the distinct overexpression of NCK1-AS1 in NSCLC patients, it is necessary to explore its biological roles in NSCLC cells. Then, we performed loss-of-function experiments. RT-PCR assisted in conforming the successful knockdown regarding NCK1-AS1 in SK-MES-1 and H1299 cells (Figure 2A). Then, the proliferation was analyzed using CCK-8 which revealed that there was a significant decrease in the absorbance in the NCK1-AS1-stably-decreasing SK-MES-1 and H1299 cells relative to the NC group (Figure 2B). EdU assays revealed that compared to si-NC group, EdU positive SK-MES-1 and H1299 cells in si-SK-MES-1 groups were reduced (Figure 2C). As showed by colony formation assays, there were less clonies in si-NCK1-AS1-transfected cells relative to si-NC-transfected cells (Figure 2D). Moreover, we performed Flow cytometric analysis for determining how NCK1-AS1 knockdown affected the apoptosis, finding that NCK1-AS1 knockdown resulted in more SK-MES-1 and H1299 cell deaths (Figure 2E). From the mechanical perspective, we confirmed the caspase 3/9 activities in NSCLC cells following NCK1-AS1 depletion, finding that NCK1-AS1 down-regulation led to a distinct increase in the caspase 3/9 activities (Figure 2F). We also performed in vivo experiments to explore the effects of NCK1-AS1 knockdown on tumor growth. Figure 3A demonstreated the transfection efficiency of sh-NCK1-AS1. In vivo assays suggested that depletion of NCK1-AS1 disticntly decreased tumor growth rate (Figure 3B). Also, we observed that the tumor volume and weight were apparently lessened in sh-NCK1-AS1 group compared with control group (Figure 3C, 3D). On the other hand, by means of scratch-wound assay, we observed the weakened migration ability possessed by SK-MES-1 and H1299 cells due to NCK1-AS1 knockdown (Figure 4A). Next, the results of transwell assays manifested that the invasion ability possessed by SK-MES-1 and H1299 cells under the transfection of si-NCK1-AS1 was markedly decreased (Figure 4B). Overall, NCK1-AS1 served as a tumor promotor in NSCLC cells.
Figure 2.
NCK1-AS1 promotes NSCLC cell proliferation in vitro. A. NCK1-AS1 expression level in SK-MES-1 and H1299 cells 48 h after NCK1-AS1-specific siRNA transfection was analyzed by qRT-PCR. B. Quantitative analysis of cell viability detected by CCK-8 assay. C. EDU staining assays were applied to detect the proliferation of si-NCK1-AS1-transfected NSCLC cells. Scale bar = 50 μm. D. Colony formation assays were used to detect the proliferation of NSCLC cells after transfection. E. Flow cytometry assay was used to detect cell apoptosis in SK-MES-1 and H1299 cells. F. The activities of caspase 3/9 were determined. **P < 0.01.
Figure 3.
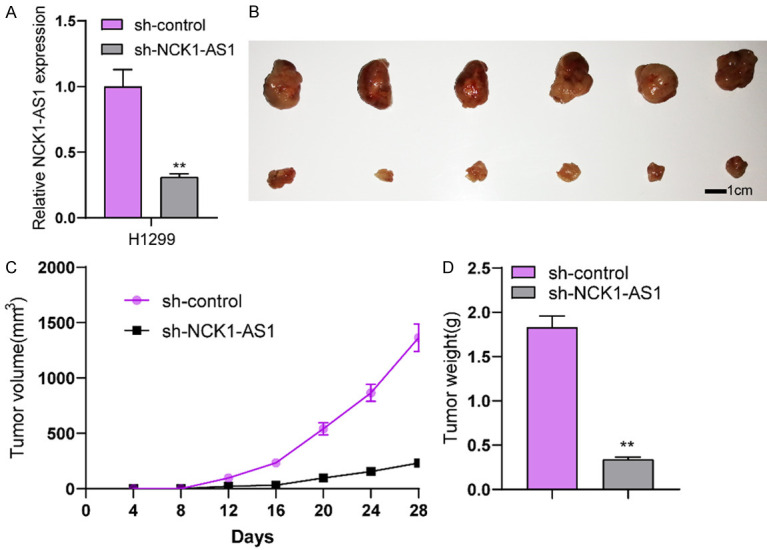
Knockdown of NCK1-AS1 suppressed tumor growth in vivo. A. RT-PCR demonstrated the distinct down-regualtion of NCK1-AS1 expression in H1299 cells after transfection with sh-NCK1-AS1. B. Tumors derived from mice in two different groups were presented. C. Tumor growth curves were shown every 7 days. D. The subcutaneous tumor weights were detected at the 28th day after injection. **P < 0.01.
Figure 4.
NCK1-AS1 promotes the migration and invasion in NSCLC cells. A. Migratory capacities in SK-MES-1 and H1299 cells increased after NCK1-AS1 knockdown, as determined by performing wound healing assays. Scale bar = 50 μm. B. The invasion ability of SK-MES-1 and H1299 cells treated with si-NCK1-AS1-1 and si-NCK1-AS1-2 was evaluated by the transwell assays. Scale bar = 50 μm. **P < 0.01.
NCK1-AS1 binds to miR-137 and represses its expression
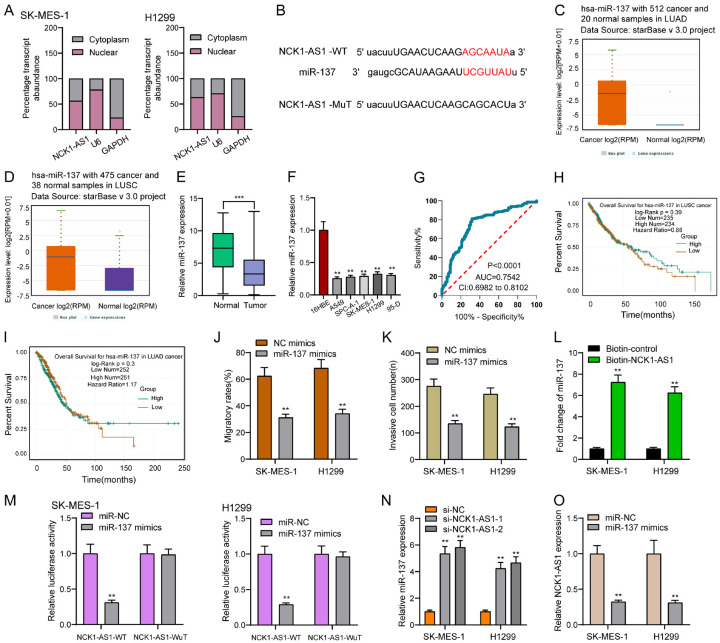
An increasing number of evidences proved that lncRNAs regulated the biological activities related to miRNAs, thereby being a molecular sponge or a competing endogenous RNA [21]. The subcellular fractionation helped to find that both the nucleus and cytoplasm saw the expression of NCK1-AS1, with the former harboring a larger part (Figure 5A). We then searched the bio-informatic tools for identifying the potential miRNAs possibly subjected to NCK1-AS1, interestingly, miR-137 could bind to NCK1-AS1. The binding sites between NCK1-AS1 and miR-137 were displayed in Figure 5B. Then, we analyzed whether miR-137 was dysregulated in NSCLC and found that miR-137 was distinctly lowly expressed in NSCLC based on the TCGA datasets (Figure 5C, 5D). In addition, we also observed the downtrend of miR-136 expression in both NSCLC tissues and cell lines (Figure 5E, 5F). Moreover, in ROC assays, we showed that high NCK1-AS1 expression had an AUC value of 0.7542 (95% CI: 0.6982 to 0.8102) for NSCLC (Figure 5G). However, there were no effects of high NCK1-AS1 expression on the OS and DFS rate of NSCLC patients (Figure 5H, 5I). Then, we explored how miR-137 overexpression affected NSCLC cells regarding the migration and invasion. As shown in Figure 5J, 5K, we observed that miR-137 overexpression exhibited a suppressing impact on SK-MES-1 and H1299 cells in terms of the migration and invasion (Figure 5J, 5K). To further verify whether miR-137 was enrichment in NCK1-AS1, we performed a pull-down assay using a biotin-labeled specific LINC00858 probe. The result of RT-PCR revealed that miR-137 was much richer in precipitate of NCK1-AS1 probe than that of in NC probe (Figure 5L). Based on Luciferase reporter assays, miR-137 mimics led to an obvious decline in the luciferase activity possessed by NCK1-AS1-WT, which, however, was incapable of changing the relative luciferase activity possessed by NCK1-AS1-MUT (Figure 5M). Besides, qRT-PCR assays confirmed that NCK1-AS1 suppression led to a distinct decrease in miR-137 expression in SK-MES-1 and H1299 cells, while miR-137 overexpression hindered the NCK1-AS1 expression. Overall, NCK1-AS1 can be a molecular sponge for miR-137in NSCLC.
Figure 5.
NCK1-AS1 sponged miR-137 in NSCLC cells. A. Relative NCK1-AS1 expression levels in nuclear and cytosolic fractions of SK-MES-1 and H1299 cells. Nuclear controls: U6; Cytosolic controls: GAPDH. B. Bioinformatic analysis indicated the predicted binding sites between miR-137 and NCK1-AS1. C, D. The expression of miR-137 in LUAD and LUSC patients based on TCGA datasets. E. Relative expression of miR-137 expression in NSCLC specimens (n = 148) and in paired adjacent normal specimens (n = 148). F. qPCR detected the expression levels of NCK1-AS1 in 16HBE and five NSCLC cell lines. G. ROC assays for the determination of the diagnostic value of miR-137 in NSCLC patients. H, I. The prognostic value of miR-137 expression analyzed using StarBase 3.0. J, K. Cell migration and invasion was measured after SK-MES-1 and H1299 cells were transfected with NC mimics or miR-137 mimics. L. RNA-pull down assays were used for the determination of the association between NCK1-AS1 and miR-137. M. Luciferase reporter assay showed the activity within miR-137 and NCK1-AS1 wild type or mutant. N. SK-MES-1 and H1299 cells were transfected with si-NCK1-AS1 or si-NC. RT-PCR measured miR-137 expression. O. Overexpression of miR-137 suppressed the expression of NCK1-AS1. **P < 0.01.
Discussion
In some of the developing countries, especially China, NSCLC is a common cancer type and the main reason for death resulted from cancer [22]. By now, many NSCLC patients still exhibit poor clinical outcomes, in spite of the clinical treatments such as radiotherapy, chemotherapy and operative treatments [23]. With the development of targeted therapy, especially targeting some important tumor-related molecules, the long-term survivals for some patients with metastasis have achieved an improvement [3,24]. On the other hand, the sensitive biomarkers were still limited, which needed to be improved. In recent years, more and more studies have paid attention to the potential possessed by lncRNAs being new biomarkers for diagnosis and prognosis of NSCLC patients [25,26]. Clinical experiments have identified some functional lncRNAs as potential biomarkers for NSCLC, such as LncRNA CCHE1, lncRNA LUCAT1, and lncRNA ADAMTS9-AS2 [27-29].
In order to screen functional lncRNAs in NSCLC, we searched TCGA datasets and identifed a series of dysregulated lncRNAs in NSCLC specimens. Many of them have been functionally explored in NSCLC based on the assays of Pubmed. However, the potential effects of NCK1-AS1 which was distinctly upregualted in NSCLC on NSCLC progression have not been investiaged. Then, we forcused on NCK1-AS1. In our cohort and cell lines, we further confirmed NCK1-AS1 as an overexpressed lncRNA in NSCLC. Then, we also determined its diagnostic value using ROC assays. Moreover, clinical assays indicated that high NCK1-AS1 expression could lead to TNM stage, lymph node metastasis as well as shorter OS and DFS, which suggested that NCK1-AS1 might facilitate NSCLC clinical progression. Above all, multivariate analyses further confirmed that NCK1-AS1 expression could be used to independently predict the gloomy prognosis in terms of the 5-year OS and DFS. Overall, findings in the study firstly proved that NCK1-AS1 was a metastasis-lncRNA in NSCLC and may be used as a novel cancer biomarker.
In recent years, as reported by several studies, NCK1-AS1 dysregulation has been involved in the progression of several tumors. For instance, NCK1-AS1 exhibited a high expression in urinary bladder cancer and promoted the proliferation and increase cell stemness of tumor cells via sponging miRNA-143 [30]. Chen et al. reported the distinct increase in NCK1-AS1 expression in glioma, and its forced expression could increase drug resistance of glioma Cells to Temozolomide via regulating miRNA-137/TRIM24 [31]. In cervical cancer, NCK1-AS1 was confirmed to be an overexpressed lncRNA and its knockdown suppressed the proliferation and metastasis via crosstalk NCK1-AS1/miRNA-6857/CDK1 pathway [32]. Interestingly, all previous findings suggested NCK1-AS1 as an oncogene in different types of tumors. Up to date, no studies have reported whether NCK1-AS1 served as a tumor suppressor in any tumors. In this study, like previous studies, we also found that NCK1-AS1 expression was distinctly increased in NSCLC. Then, we performed loss-of-function assays to determine the function of NCK1-AS1 in two NSCLC cells, finding that knockdown of NCK1-AS1 exerted a suppressing effect on SK-MES-1 and H1299 cells with regard to proliferation, migration and invasion, but facilitated the apoptosis. Further in vivo assays also demonstrated that knockdown of NCK1-AS1 subcutaneous xenograft growth in vivo. Thus, our findings suggested that NCK1-AS1 could promote tumor in NSCLC.
As demonstrated in previous studies, lncRNAs were capable of regulating miRNAs as a kind of ceRNAs or molecular sponges [33,34]. It has been confirmed that cytosolic lncRNAs have the function of modulating the stability exhibited by mRNA and the localization of protein as well as serving as microRNA sponges [35]. Then, we performed subcellular fractionation, finding that both the nucleus and cytoplasm saw NCK1-AS1 expression, with the former harboring a larger part. Then, we searched StarBase v3.0 software and observed that miR-137 shared complementary binding with the 3’-untranslated regions (UTR) of NCK1-AS1. Previously, several studies have reported the obvious decrease in miR-137 expression in several tumors, including NSCLC, and its overexpression was functionally confirmed to suppress the proliferation and metastasis of NSCLC via targeting several oncogenic genes [36-38]. In this study, we also provided evidence that miR-137 was highly expressed in NSCLC and suppressed SK-MES-1 and H1299 cell migration and invasion, which was consistent with previous findings. To further explore whether miR-137 was enrichment in NCK1-AS1, we performed pull-down assays that adopted a biotin-labeled specific NCK1-AS1 probe, which indicated that miR-137 was much richer in precipitate of NCK1-AS1 probe than that of in NC probe. Moreover, based on Dual-luciferase assay, miR-137 mimics decreased the luciferase activity exhibited by NCK1-AS1-Wt, suggesting that NCK1-AS1 directly targets miR-137 in NSCLC cells. Finally, NCK1-AS1 knockdown hindered the miR-137 expression, while miR-137 overexpression suppressed the NCK1-AS1 expression, which supported that NCK1-AS1 directly targets miR-137 in NSCLC cells. Overall, our findings, together with previous findings, suggested that NCK1-AS1 may promote the proliferation and metastasis via sponging miR-137.
Conclusions
The study focused on identifying a kind of lncRNA related to NSCLC, i.e. the NCK1-AS1, and the increased expression can affect NSCLC patients’ prognosis. NCK1-AS1 knockdown sponged miR-137, thereby hindering NSCLC progression. Taken together, NCK1-AS1 could act as a new biomarker as well as a therapeutic target of NSCLC.
Disclosure of conflict of interest
None.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Tandberg DJ, Tong BC, Ackerson BG, Kelsey CR. Surgery versus stereotactic body radiation therapy for stage I non-small cell lung cancer: a comprehensive review. Cancer. 2018;124:667–678. doi: 10.1002/cncr.31196. [DOI] [PubMed] [Google Scholar]
- 3.Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature. 2018;553:446–454. doi: 10.1038/nature25183. [DOI] [PubMed] [Google Scholar]
- 4.Osmani L, Askin F, Gabrielson E, Li QK. Current WHO guidelines and the critical role of immunohistochemical markers in the subclassification of non-small cell lung carcinoma (NSCLC): moving from targeted therapy to immunotherapy. Semin Cancer Biol. 2018;52:103–109. doi: 10.1016/j.semcancer.2017.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wood K, Hensing T, Malik R, Salgia R. Prognostic and predictive value in KRAS in non-small-cell lung cancer: a review. JAMA Oncol. 2016;2:805–812. doi: 10.1001/jamaoncol.2016.0405. [DOI] [PubMed] [Google Scholar]
- 6.Chew CL, Conos SA, Unal B, Tergaonkar V. Noncoding RNAs: master regulators of inflammatory signaling. Trends Mol Med. 2018;24:66–84. doi: 10.1016/j.molmed.2017.11.003. [DOI] [PubMed] [Google Scholar]
- 7.Romero-Barrios N, Legascue MF, Benhamed M, Ariel F, Crespi M. Splicing regulation by long noncoding RNAs. Nucleic Acids Res. 2018;46:2169–2184. doi: 10.1093/nar/gky095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang SW, Fan XN. Computational methods for predicting ncRNA-protein interactions. Med Chem. 2017;13:515–525. doi: 10.2174/1573406413666170510102405. [DOI] [PubMed] [Google Scholar]
- 9.Khanduja JS, Calvo IA, Joh RI, Hill IT, Motamedi M. Nuclear noncoding rnas and genome stability. Mol Cell. 2016;63:7–20. doi: 10.1016/j.molcel.2016.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng Y, Liu L, Shukla GC. A comprehensive review of web-based non-coding RNA resources for cancer research. Cancer Lett. 2017;407:1–8. doi: 10.1016/j.canlet.2017.08.015. [DOI] [PubMed] [Google Scholar]
- 11.Wang J, Zhu S, Meng N, He Y, Lu R, Yan GR. ncRNA-encoded peptides or proteins and cancer. Mol Ther. 2019;27:1718–1725. doi: 10.1016/j.ymthe.2019.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng R, Lu X, Xu C, Zhang F, Zhang G. SNHG11 contributes to NSCLC cell growth and migration by targeting miR-485-5p/BSG axis. Biomed Pharmacother. 2020;128:110324. doi: 10.1016/j.biopha.2020.110324. [DOI] [PubMed] [Google Scholar]
- 13.Fan G, Jiao J, Shen F, Chu F. Upregulation of lncRNA ZFAS1 promotes lung adenocarcinoma progression by sponging miR-1271-5p and upregulating FRS2. Thorac Cancer. 2020;11:2178–2187. doi: 10.1111/1759-7714.13525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29:452–463. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou DD, Liu XF, Lu CW, Pant OP, Liu XD. Long non-coding RNA PVT1: emerging biomarker in digestive system cancer. Cell Prolif. 2017;50:e12398. doi: 10.1111/cpr.12398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guan Z, Song Y, Ma J, Li F, Zhao X, Liang G, An H, Pu J. Altered expression of lncRNA NCK1-AS1 distinguished patients with prostate cancer from those with benign prostatic hyperplasia. Oncol Lett. 2019;18:6379–6384. doi: 10.3892/ol.2019.11039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu H, Li H, Feng X. Downregulation of lncRNA NCK1-AS1 inhibits cancer cell migration and invasion in nasopharyngeal carcinoma by upregulating miR-135a. Cancer Manag Res. 2019;11:10531–10537. doi: 10.2147/CMAR.S221326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang WY, Liu YJ, He Y, Chen P. Suppression of long noncoding RNA NCK1-AS1 increases chemosensitivity to cisplatin in cervical cancer. J Cell Physiol. 2019;234:4302–4313. doi: 10.1002/jcp.27198. [DOI] [PubMed] [Google Scholar]
- 19.Zhuang X, Tong H, Ding Y, Wu L, Cai J, Si Y, Zhang H, Shen M. Long noncoding RNA ABHD11-AS1 functions as a competing endogenous RNA to regulate papillary thyroid cancer progression by miR-199a-5p/SLC1A5 axis. Cell Death Dis. 2019;10:620. doi: 10.1038/s41419-019-1850-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng B, Rong A, Zhou Q, Li W. LncRNA LINC00662 promotes colon cancer tumor growth and metastasis by competitively binding with miR-340-5p to regulate CLDN8/IL22 co-expression and activating ERK signaling pathway. J Exp Clin Cancer Res. 2020;39:5. doi: 10.1186/s13046-019-1510-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the rosetta stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liang W, Zhang L, Jiang G, Wang Q, Liu L, Liu D, Wang Z, Zhu Z, Deng Q, Xiong X, Shao W, Shi X, He J. Development and validation of a nomogram for predicting survival in patients with resected non-small-cell lung cancer. J. Clin. Oncol. 2015;33:861–869. doi: 10.1200/JCO.2014.56.6661. [DOI] [PubMed] [Google Scholar]
- 23.Ko EC, Raben D, Formenti SC. The integration of radiotherapy with immunotherapy for the treatment of non-small cell lung cancer. Clin Cancer Res. 2018;24:5792–5806. doi: 10.1158/1078-0432.CCR-17-3620. [DOI] [PubMed] [Google Scholar]
- 24.Jeremić B. Standard treatment option in stage III non-small-cell lung cancer: case against trimodal therapy and consolidation drug therapy. Clin Lung Cancer. 2015;16:80–85. doi: 10.1016/j.cllc.2014.08.003. [DOI] [PubMed] [Google Scholar]
- 25.Jiang N, Meng X, Mi H, Chi Y, Li S, Jin Z, Tian H, He J, Shen W, Tian H, Pan J, Fang S, Jin X, Zhou C, Gong Z. Circulating lncRNA XLOC_009167 serves as a diagnostic biomarker to predict lung cancer. Clin Chim Acta. 2018;486:26–33. doi: 10.1016/j.cca.2018.07.026. [DOI] [PubMed] [Google Scholar]
- 26.Jing H, Qu X, Liu L, Xia H. A novel long noncoding RNA (lncRNA), LL22NC03-N64E9.1, promotes the proliferation of lung cancer cells and is a potential prognostic molecular biomarker for lung cancer. Med Sci Monit. 2018;24:4317–4323. doi: 10.12659/MSM.908359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiao Y, Li Y, Ji B, Cai H, Liu Y. Clinical value of lncRNA LUCAT1 expression in liver cancer and its potential pathways. J Gastrointestin Liver Dis. 2019;28:439–447. doi: 10.15403/jgld-356. [DOI] [PubMed] [Google Scholar]
- 28.Liao Y, Cheng S, Xiang J, Luo C. lncRNA CCHE1 increased proliferation, metastasis and invasion of non-small lung cancer cells and predicted poor survival in non-small lung cancer patients. Eur Rev Med Pharmacol Sci. 2018;22:1686–1692. doi: 10.26355/eurrev_201803_14581. [DOI] [PubMed] [Google Scholar]
- 29.Liu C, Yang Z, Deng Z, Zhou Y, Gong Q, Zhao R, Chen T. Upregulated lncRNA ADAMTS9-AS2 suppresses progression of lung cancer through inhibition of miR-223-3p and promotion of TGFBR3. IUBMB Life. 2018;70:536–546. doi: 10.1002/iub.1752. [DOI] [PubMed] [Google Scholar]
- 30.Qiao Z, Dai H, Zhang Y, Li Q, Zhao M, Yue T. LncRNA NCK1-AS1 promotes cancer cell proliferation and increase cell stemness in urinary bladder cancer patients by downregulating miR-143. Cancer Manag Res. 2020;12:1661–1668. doi: 10.2147/CMAR.S223172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen M, Cheng Y, Yuan Z, Wang F, Yang L, Zhao H. NCK1-AS1 increases drug resistance of glioma cells to temozolomide by modulating miR-137/TRIM24. Cancer Biother Radiopharm. 2020;35:101–108. doi: 10.1089/cbr.2019.3054. [DOI] [PubMed] [Google Scholar]
- 32.Li H, Jia Y, Cheng J, Liu G, Song F. LncRNA NCK1-AS1 promotes proliferation and induces cell cycle progression by crosstalk NCK1-AS1/miR-6857/CDK1 pathway. Cell Death Dis. 2018;9:198. doi: 10.1038/s41419-017-0249-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.An Y, Furber KL, Ji S. Pseudogenes regulate parental gene expression via ceRNA network. J Cell Mol Med. 2017;21:185–192. doi: 10.1111/jcmm.12952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qi X, Zhang DH, Wu N, Xiao JH, Wang X, Ma W. ceRNA in cancer: possible functions and clinical implications. J Med Genet. 2015;52:710–718. doi: 10.1136/jmedgenet-2015-103334. [DOI] [PubMed] [Google Scholar]
- 35.Smillie CL, Sirey T, Ponting CP. Complexities of post-transcriptional regulation and the modeling of ceRNA crosstalk. Crit Rev Biochem Mol Biol. 2018;53:231–245. doi: 10.1080/10409238.2018.1447542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen F, Luo N, Hu Y, Li X, Zhang K. MiR-137 suppresses triple-negative breast cancer stemness and tumorigenesis by perturbing bcl11a-dnmt1 interaction. Cell Physiol Biochem. 2018;47:2147–2158. doi: 10.1159/000491526. [DOI] [PubMed] [Google Scholar]
- 37.Jiang H, Zhang H, Hu X, Li W. Knockdown of long non-coding RNA XIST inhibits cell viability and invasion by regulating miR-137/PXN axis in non-small cell lung cancer. Int J Biol Macromol. 2018;111:623–631. doi: 10.1016/j.ijbiomac.2018.01.022. [DOI] [PubMed] [Google Scholar]
- 38.Zhang W, Chen JH, Shan T, Aguilera-Barrantes I, Wang LS, Huang TH, Rader JS, Sheng X, Huang YW. miR-137 is a tumor suppressor in endometrial cancer and is repressed by DNA hypermethylation. Lab Invest. 2018;98:1397–1407. doi: 10.1038/s41374-018-0092-x. [DOI] [PMC free article] [PubMed] [Google Scholar]