Abstract
Long noncoding RNAs (lncRNAs) play the critical biological role in many malignant tumours. MIR4435-2HG has been proven to be a novel oncogenic lncRNA. However, the exact role and mechanism of MIR4435-2HG in hepatocellular carcinoma (HCC) remain unclear. Here, we found that MIR4435-2HG is overexpressed in HCC tissue compared to normal controls and that high level of MIR4435-2HG indicates a poorer prognosis in HCC patients. MIR4435-2HG enhances the growth and metastasis ability of HCC cells. MIR4435-2HG promotes the expression of YWHAZ by sponging miR-22-3p to liberate YWHAZ mRNA transcripts. MIR4435-2HG facilitates the proliferation and metastasis of HCC by modulating the miR-22-3p/YWHAZ axis. These results demonstrated the role and mechanism of MIR4435-2HG in malignant progression of HCC. MIR4435-2HG may be used as the prognostic marker and treatment target for the patient with HCC.
Keywords: HCC, proliferation and metastasis, MIR4435-2HG, miR-22-3p, YWHAZ
Introduction
Hepatocellular carcinoma (HCC), the most common kind of primary hepatic carcinoma, is the third reason of tumour-associated death worldwide [1,2]. The incidence of HCC has gradually increased in recent years [3]. The mortality of HCC is also rising year by year, which is due to the lack of specific biomarkers [4]. Occurrence and development of HCC involve changes in many genes and signaling pathways, and its pathogenesis is still unclear [5,6]. Therefore, an in-depth study on the molecular mechanism underlying HCC carcinogenesis would help in the diagnosis and treatment of HCC patients.
Many studies have proven that noncoding RNAs (ncRNAs) directly involved in many pathophysiological processes [7]. Long noncoding RNAs (lncRNAs), a major subgroup of ncRNAs, play critical roles in cancer biology [8,9]. The affect of some specific lncRNAs in the progression of HCC has been identified [10,11]. The lncRNA MIR4435-2HG, encoded on chromosome 2q13, has been proven to be a novel oncogenic lncRNA in colorectal cancer, lung cancer, prostate carcinoma, etc [12-14]. It has also been reported that MIR4435-2HG is upregulated in HCC [15]. Nevertheless, the exact role and mechanism of MIR4435-2HG in HCC remain unclear.
Here, we found that MIR4435-2HG was overexpressed in HCC, and high level of MIR4435-2HG acted as the risk factor for the overall survival of patients with HCC. The most widely recognized mechanism of lncRNAs is that some lncRNAs affect tumour progression by acting as the competitive endogenous RNAs (ceRNAs) [16,17], and MIR4435-2HG has also been proven to have the same effect in some tumours [18]. We demonstrated that MIR4435-2HG can remove the effect of miR-22-3p on YWHAZ by sponging miR-22-3p, thus promoting the expression of YWHAZ. YWHAZ, a central hub protein for many signaling pathways, acts as an oncogene in many malignant tumours, including HCC [19,20]. We showed that MIR4435-2HG exerts its proproliferative and prometastatic effects in HCC by modulating the miR-22-3p/YWHAZ pathway. Therefore, MIR4435-2HG can be used as a new special prognostic marker and treatment target for HCC patients.
Materials and methods
Tissue samples
HCC and paracancerous normal tissues were obtained from HCC patients at Changshu No. 2 People’s Hospital. The two pathologists made the pathological diagnosis on the collected tissues independently. All HCC patients did not receive radiotherapy or chemotherapy before surgery. The ethics of this study was approved by the ethics committee of Changshu Second People’s Hospital. The HCC dataset from The Cancer Genome Atlas (TCGA) was also included in this study.
Cell lines and cell culture
HCC cell lines Huh7, SMMC7721 and BEL-7402, HepG2 were purchased from the American Type Culture Collection (ATCC, Manassas, USA). The normal hepatic cell line LO2 was purchased from Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). These cells were maintained in Dulbecco’s modified Eagle’s medium (Gibco, USA) with 10% fetal bovine serum (Invitrogen, USA) and penicillin streptomycin and incubated in an incubator with humid conditions of 37°C with 5% CO2.
Plasmids, oligonucleotides and cell transfection
The mimic and inhibitor of miR-22-3p were synthesized by GenePharma (Shanghai, China), and negative control was used as control. Small interfering RNA (siRNA) of MIR4435-2HG and YWHAZ were also synthesized by GenePharma. We inserted YWHAZ full length into the vector of pcDNA3.1 (Invitrogen, USA) to construct the YWHAZ plasmid. The related oligonucleotides and plasmids were transfected into HCC cells by using Lipofectamine 3000 (Invitrogen, USA) following the instructions.
Quantitative RT-PCR
Reverse transcription was carried out with Fermentas and special microRNA reverse transcription kits according to the experimental steps (Applied Biosystems, CA) after RNA extraction. We then set up the reaction conditions and used the ABI StepOnePlus System (Applied Biosystems, CA) to carry out the amplification reaction. The following primers were used: MIR4435-2HG forward 5’-CGGAGCATGGAACTCGACA-3’ and MIR4435-2HG reverse 5’-CAAGTCTCACACATCCGGG-3’; YWHAZ forward 5’-AGGAGCCCGTAGGTCATCTT-3’ and YWHAZ reverse 5’-TGCTTGTGAAGCATTGGGGA-3’. Specific primers for miR-22-3p were synthesized by RiboBio (Guangzhou, China), and U6 was used as normalization for the qRT-PCR of miRNA.
Western blotting
A BCA Protein Assay Kit (Beyotime, China) was used to detect the protein concentration after extracting the protein using RIPA buffer (KenGEN, China). 10% SDS-PAGE was used to separate the proteins. The protein was transferred to polyvinylidene fluoride (PVDF) membrane, and sealed with 5% skimmed milk. Next, the membranes were incubated with primary antibodies at 4°C overnight, and then with the secondary antibody at room temperature for 1 hour at the next day. Chemiluminescence reagent was used to detect the signal on the membrane, and data were analyzed by using ImageJ software. Antibodies against YWHAZ (1:1000, Abcam, Cambridgeshire, UK) were used to detect the protein expression level of YWHAZ, and GAPDH was used for standardization.
Cell proliferation assay
The proliferation ability of HCC cells were evaluated by using Cell Counting Kit-8 (CCK-8, Beyotime, China) assays. Transfected HCC cells were inoculated into a 96-well plate. At different times (12, 24, 48 and 72 h), CCK-8 was added to the cultured cells and maintained at 37°C for an additional 2 h. Microplate Reader Multiscan FC (Thermo Scientific, Waltham, USA) was used to measure the absorbance at 450 nm optical density.
Cell invasion and migration assays
Transwell assays were used to detect the invasive ability of HCC Serum-free DMEM was used to resuspend the HCC cells after transfection. We then put these cells on the upper layer of Matrigel coated chambers (BD Biosciences, Franklin Lakes, USA). DMEM containing 10% serum was placed in the lower layer of Matrigel coated chambers. The non-invasive HCC cells were removed with cotton swab and invasive HCC cells were stained with crystal violet after 24 hours. Scratch wound assay was serve to measure the migration capability of HCC cells. HCC cells were inoculated into the 6-well plate after transfection. Tip of 200 μl pipette was used to from the bruise. We measured the bruise width at 0 and 48 hours.
Luciferase reporter assay
The fragments of the YWHAZ 3’-UTR and MIR4435-2HG which matched the miR-22-3p binding site was cloned into pMIR reporter plasmid. The luciferase activity of different treatment groups was measured by using Dual Luciferase Reporter Assay System (Promega, Madison, USA) after transfection with the corresponding reporter plasmid and oligonucleotide. Mutant plasmid was used as a control.
MS2-RIP assay
miRNAs combined with MIR4435-2HG were detected by using Maltose-binding protein (MBP)-affinity purification. Three binding site of bacteriophage MS2 coat protein were added in the downstream of MIR4435-2HG. The MS2-tagged MIR4435-2HG plasmid was transfected into HCC cells to identity miRNAs that bind to MIR4435-2HG. After 48 hours, RIP assay was carried out on HCC cells [21], and miR-22-3p expression was detected.
RNA pull-down assay
Biotinylated-miR-22-3p was acquired from GenePharma (Shanghai, China), mutant and NC were used as the control. Cell lysates were cultured with M-280 streptavidin magnetic beads (Invitrogen, Carlsbad, USA) after transfection with above-mentioned oligonucleotides [22]. qRT-PCR was used to measure the expression of MIR4435-2HG in the bound RNA.
Xenograft tumour and immunohistochemistry staining assay
Nude mice were purchased from the Beijing Laboratory Animal Center (Beijing, China). Mice were subcutaneously injected with logarithmically growing HepG2 cells to form the xenograft tumour, and the NC and MIR4435-2HG siRNA was locally injected into the xenograft tumour every 4 days. The volume of xenograft tumour was calculated every four days based on the formula of 0.5 × length × width2, the mice were killed and the tumour was removed 28 days later. The ethics of this study was approved by the ethics committee of Changshu Second People’s Hospital. Immunohistochemistry was carried out as described previously [23] using the antibody YWHAZ (Abcam, Cambridgeshire, UK).
Statistical analysis
SPSS 13.0 was used to analyze the data (mean ± SD). The related data were statistically evaluated by T-test, spearman correlation analysis was performed with MATLAB, and survival curve was drawn by Kaplan Meier analysis. It is considered to be statistically significant when the P value is less than 0.05.
Results
MIR4435-2HG was considerably upregulated in HCC, and a high level of MIR4435-2HG was the risk factor for the overall survival of HCC patients
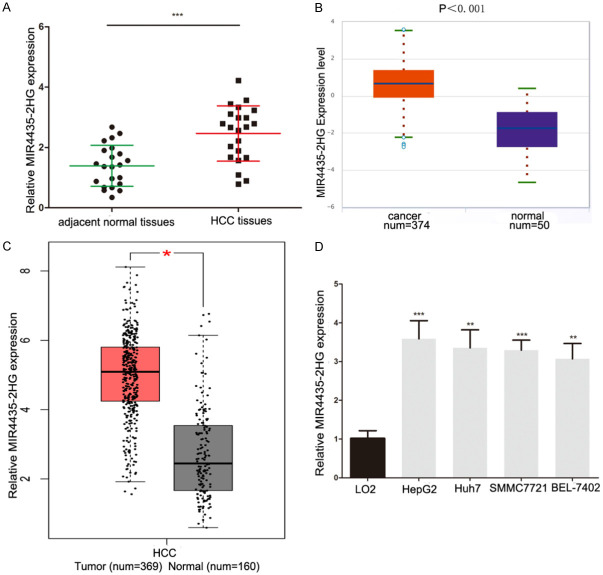
We first analyzed MIR4435-2HG expression in 22 HCC tissues and adjacent normal tissue (ANT) and discovered that the level of MIR4435-2HG in HCC tissues was raised (Figure 1A). We used GEPIA (http://gepia.cancer-pku.cn/) and StarBase (http://starbase.sysu.edu.cn/) to analyze the open HCC datasets of The Cancer Genome Atlas (TCGA), showing the same consequences (Figure 1B and 1C). Meanwhile, the level of MIR4435-2HG in HCC cell lines Huh7, SMMC7721 and BEL-7402, HepG2 was significantly higher than that in normal hepatic cell line LO2 (Figure 1D). We explored MIR4435-2HG clinical significance in HCC patients. The high MIR4435-2HG expression (≥ median ratio) was very associated with TMN stage and metastasis, not with age, sex, HBsAg infection and cirrhosis (Table 1). The HCC patients overall survival rate with high MIR4435-2HG expression was shorter by analyzing our samples (Figure 2A). We analyzed prognostic data from the TCGA-HCC database through GEPIA and Starbase and found that high MIR4435-2HG levels confer a poor prognosis in HCC patients (Figure 2B and 2C). These results suggested that MIR4435-2HG may participate in the progression of HCC.
Figure 1.
MIR4435-2HG is significantly upregulated in HCC. A. The level of MIR4435-2HG was analyzed in 22 HCC tissues and adjacent normal tissues. Statistical significance is evaluated by one-tailed t-test. B. StarBase (http://starbase.sysu.edu.cn/) was used to detect the expression of MIR4435-2HG in HCC and matched normal tissue of TCGA dataset. C. GEPIA (http://gepia.cancer-pku.cn/) was used to detect the expression of MIR4435-2HG in HCC and matched normal tissue of TCGA dataset. D. The expression profile of MIR4435-2HG in HCC cell lines (HepG2, Huh7, SMMC7721 and BEL-7402) and normal human hepatic epithelial cells (LO2). Statistical significance is evaluated by two-tailed Student’s t-test. *P < 0.05, **P < 0.01, ***P < 0.001.
Table 1.
Correlation between MIR4435-2HG expression and clinical pathological characteristic (n=22)
| Clinical characteristics | Number | High MIR4435-2HG expression | Low MIR4435-2HG expression | P-value |
|---|---|---|---|---|
| Age | 0.360 | |||
| < 50 | 7 | 2 | 5 | |
| ≥ 50 | 15 | 9 | 6 | |
| Gender | 0.580 | |||
| Male | 18 | 10 | 8 | |
| Female | 4 | 1 | 3 | |
| HBsAg infection | 0.632 | |||
| Yes | 16 | 9 | 7 | |
| No | 6 | 2 | 4 | |
| Cirrhosis | 0.386 | |||
| Yes | 13 | 5 | 8 | |
| No | 9 | 6 | 3 | |
| Metastasis | 0.032 | |||
| Yes | 12 | 9 | 3 | |
| No | 10 | 2 | 8 | |
| TMN stage | 0.042 | |||
| I-II | 5 | 0 | 5 | |
| III-IV | 17 | 10 | 6 |
Figure 2.
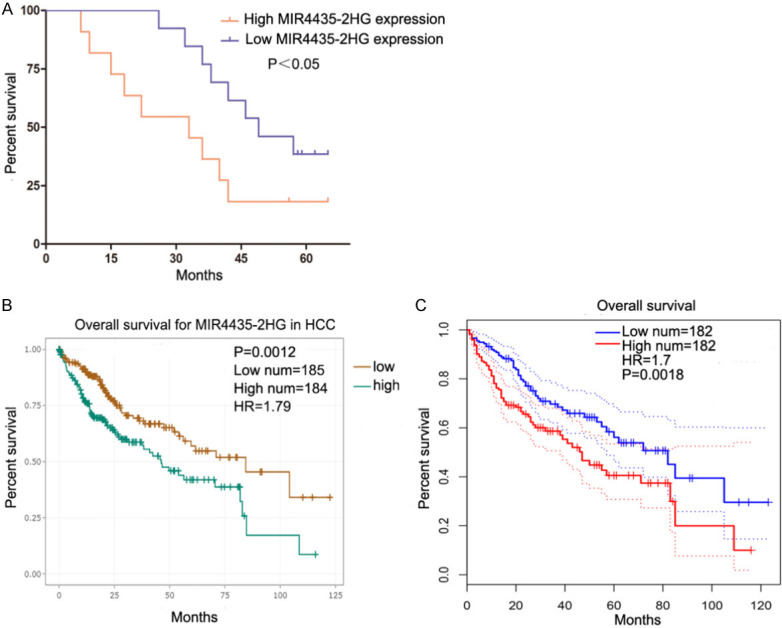
High MIR4435-2HG levels are a risk factor for the prognosis of HCC. A. The survival curves of 22 HCC patients with high and low MIR4435-2HG levels. High MIR4435-2HG expression group (n=11, MIR4435-2HG expression ratio ≥ median ratio), low MIR4435-2HG expression group (n=11, MIR4435-2HG expression ratio < median ratio). Kaplan Meier survival curve was used. B. The prognostic data of HCC patients with high and low MIR4435-2HG levels in TCGA were analyzed by using Starbase (http://http://www.sysu.edu.cn). Kaplan Meier survival curve was used. C. We analyzed the prognostic data of HCC patients in TCGA using GEPIA (http://gepia.cancer-pku.cn/). Kaplan Meier survival curve was used. *P < 0.05, **P < 0.01, ***P < 0.001.
CeRNA prediction of MIR4435-2HG
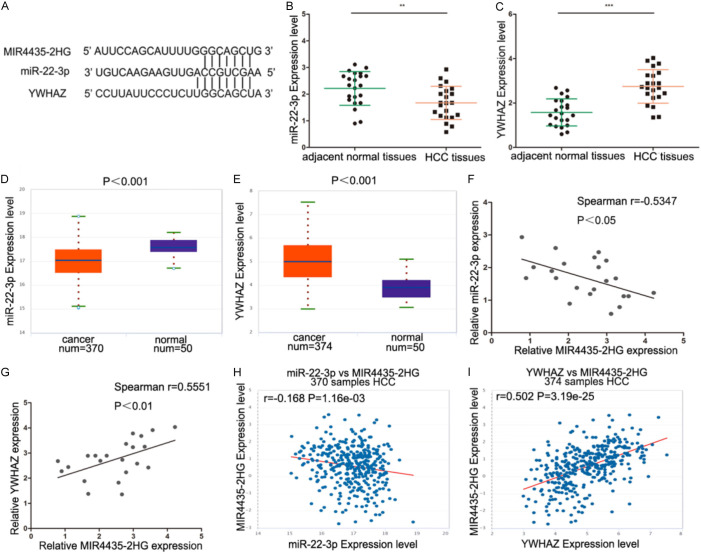
We next discussed the mechanism of MIR4435-2HG in HCC. Some lncRNAs have been proved to have pivotal influence in tumours through the ceRNA mechanism, and MIR4435-2HG has the same effect [17,18]. The interplay between MIR4435-2HG and miRNAs was identified using Starbase 3.0, and miR-22-3p may bind to MIR4435-2HG. The target genes of miR-22-3p were searched through TargetScan, miRDB, miRDIP and miRPathDB, and 3’-UTR of YWHAZ has the identical binding sites with which MIR4435-2HG binds to miR-22-3p (Figure 3A). The level of YWHAZ and miR-22-3p were detected in 22 HCC tissues. miR-22-3p expression was reduced and YWHAZ expression was raised in HCC tissues (Figure 3B and 3C), and the similar consequences were got by analyzing TCGA-HCC dataset (Figure 3D and 3E). In our HCC samples, MIR4435-2HG and miR-22-3p expression were negatively correlated, and MIR4435-2HG and YWHAZ expression were positively correlated (Figure 3F and 3G). The same correlation analysis results were also confirmed in the TCGA database (Figure 3H and 3I).
Figure 3.
ceRNA prediction of MIR4435-2HG. A. The 3’-UTR of YWHAZ has the same binding sites that MIR4435-2HG binds to miR-22-3p. B. miR-22-3p expression was detected in 22 HCC tissues and adjacent normal tissues. Statistical significance is evaluated by one-tailed t-test. C. The expression of YWHAZ was analyzed in 22 HCC tissues and adjacent normal tissues. Statistical significance is evaluated by one-tailed t-test. D. The expression of miR-22-3p was detected in HCC and matched normal tissue of TCGA dataset. E. The expression of YWHAZ was analyzed in HCC and matched normal tissue of TCGA dataset. F. The correlation of MIR4435-2HG and miR-22-3p in 22 HCC tissues was negative. Spearman correlation analysis was carried out. G. The correlation of MIR4435-2HG and YWHAZ in 22 HCC tissues was positive. Spearman correlation analysis was carried out. H. A negative correlation between MIR4435-2HG and miR-22-3p was found in the TCGA HCC dataset. Spearman correlation analysis was carried out. I. The positive correlation between MIR4435-2HG and YWHAZ mRNA levels in the TCGA HCC dataset. Spearman correlation analysis was carried out. *P < 0.05, **P < 0.01, ***P < 0.001.
MIR4435-2HG sponges miR-22-3p in HCC cells
We confirmed whether MIR4435-2HG can directly combine with miR-22-3p. MIR4435-2HG was mainly expressed in the cytoplasm of HCC cells by using analysis of nuclear and cytoplasmic fragments (Figure 4A). The miR-22-3p expression was increased in MIR4435-2HG siRNA transfected HCC cells (Figure 4B). Wild type and mutant MIR4435-2HG luciferase reporter vectors were constructed (Figure 4C). Luciferase activity of wild type plasmid was suppressed by the miR-22-3p in HepG2 and SMMC7721 cells, but the mutant plasmid was not affected (Figure 4D). MS2-RIP assay was used to confirm the direct binding combination of MIR4435-2HG and miR-22-3p. Compared with empty and mutated plasmids, MS2-tagged wild-type MIR4435-2HG plasmid enriched a large amount of miR-22-3p (Figure 4E). RNA pull-down showed that MIR4435-2HG was pulled down by biotin-labeled miR-22-3p, and the result further verified the relationship between MIR4435-2HG and miR-22-3p (Figure 4F). All results demonstrated that MIR4435-2HG can directly bind to miR-22-3p in HCC.
Figure 4.
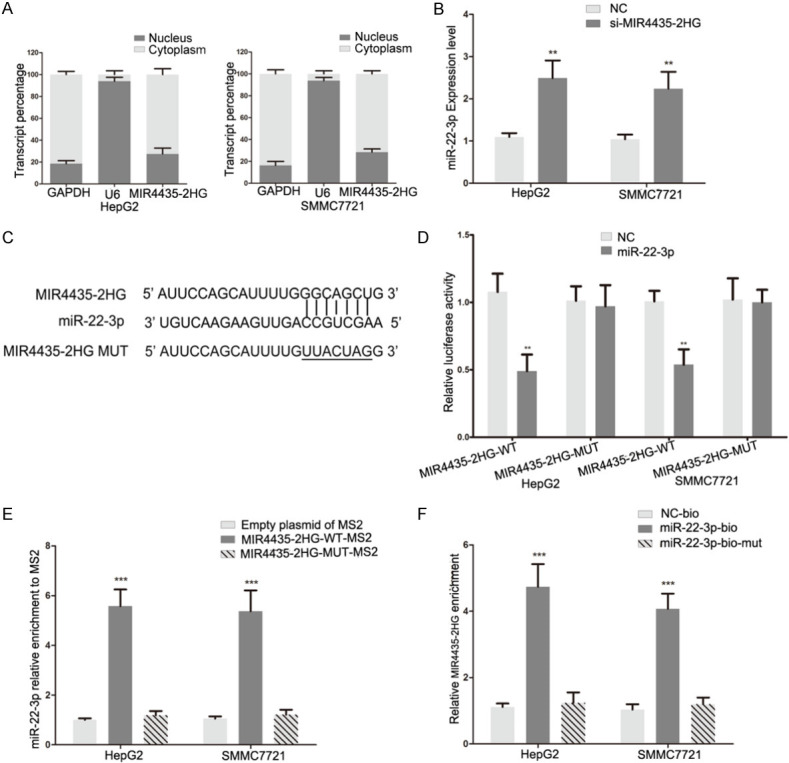
MIR4435-2HG sponges miR-22-3p in HCC cells. A. qRT-PCR of nuclear and cytoplasmic RNA fractions were used to detect MIR4435-2HG expression in the cytoplasm and nucleus. B. The levels of miR-22-3p in HCC cells following transfection with MIR4435-2HG siRNA. Statistical significance is evaluated by two-tailed Student’s t-test. C. The binding sites of miR-22-3p on MIR4435-2HG and target sequences were mutated. D. Luciferase assay of HCC cells transfected with MIR4435-2HG-WT or MIR4435-2HG-MUT plasmid together with miR-22-3p mimic. Statistical significance is evaluated by two-tailed Student’s t-test. E. MS2-RIP was used to detect endogenous miR-22-3p associated with MS2-tagged MIR4435-2HG. Empty and mutated plasmids were used as the control. Statistical significance is evaluated by two-tailed Student’s t-test. F. HCC cells transfected with biotin-labeled miR-22-3p, mutated or NC, and subjected to an RNA pull-down assay. The expression of MIR4435-2HG was detected by qRT-PCR. Statistical significance is evaluated by two-tailed Student’s t-test. *P < 0.05, **P < 0.01, ***P < 0.001.
MIR4435-2HG promote YWHAZ expression by sponging miR-22-3p in HCC cells
We further explored the effect of MIR4435-2HG on YWHAZ levels. Wild type and mutant YWHAZ 3’-UTR luciferase reporter vectors were constructed (Figure 5A), and wild type plasmid luciferase activity was repressed by miR-22-3p in HCC cells (Figure 5B). The miR-22-3p mimic inhibited YWHAZ expression both in the protein and mRNA levels in HCC cells (Figure 5D and 5E). These consequences manifested that YWHAZ is the target of miR-22-3p. Furthermore, the MIR4435-2HG siRNA inhibited the luciferase activity in wild type YWHAZ 3’-UTR luciferase reporter vectors, and the miR-22-3p inhibitor reversed the influence of the MIR4435-2HG siRNA (Figure 5C). YWHAZ expression was decreased in MIR4435-2HG siRNA-transfected HCC cells, and this inhibitory effect was attenuated when cells were cotransfected with miR-22-3p inhibitor (Figure 5D and 5E). These results indicated that MIR4435-2HG promotes YWHAZ expression by decoying miR-22-3p in HCC.
Figure 5.
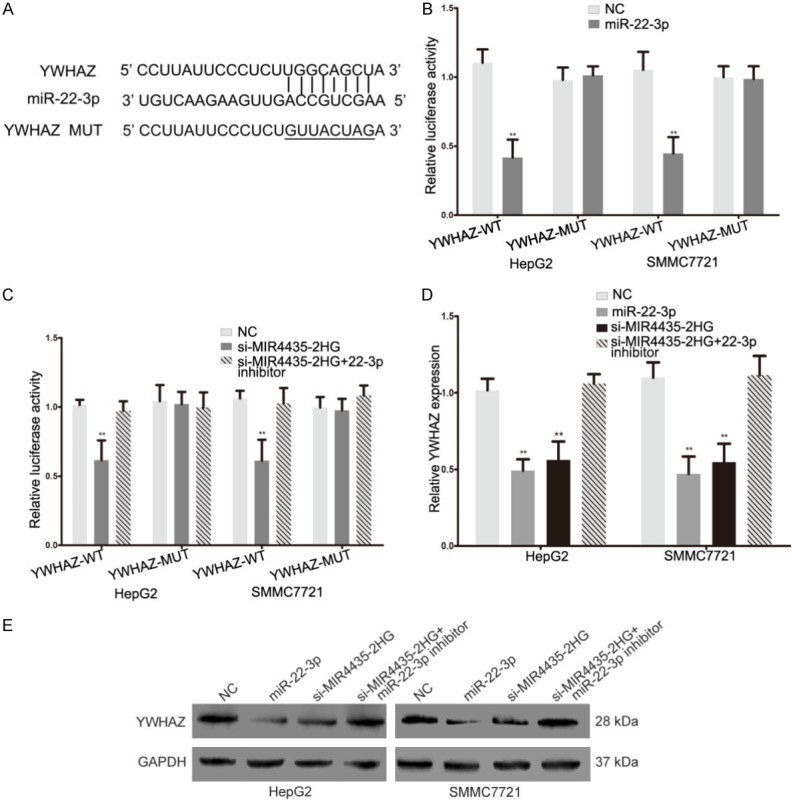
MIR4435-2HG promotes YWHAZ expression by sponging miR-22-3p in HCC cells. A. The binding sites of miR-22-3p on the 3pg sites YWHAZ, and target sequences were mutated. B. Luciferase assay of HCC cells transfected with YWHAZ-WT or YWHAZ-MUT reporter together with miR-22-3p mimic. Statistical significance is evaluated by two-tailed Student’s t-test. C. Luciferase assay of HCC cells transfected with YWHAZ-WT or YWHAZ-MUT plasmid together with MIR4435-2HG siRNA or MIR4435-2HG siRNA plus miR-22-3p inhibitor. Statistical significance is evaluated by two-tailed Student’s t-test. D. The expression of YWHAZ mRNA in HCC cells transfected with miR-22-3p mimic, MIR4435-2HG siRNA or MIR4435-2HG siRNA plus miR-22-3p inhibitor. Statistical significance is evaluated by two-tailed Student’s t-test. E. Western blotting detected YWHAZ protein expression changes in NC, miR-22-3p mimic, si-MIR4435-2HG or si-MIR4435-2HG plus miR-22-3p inhibitor transfected HCC cells, GAPDH was used as a control. *P < 0.05, **P < 0.01, ***P < 0.001.
MIR4435-2HG promotes the proliferation and metastasis of HCC via miR-22-3p/YWHAZ axis
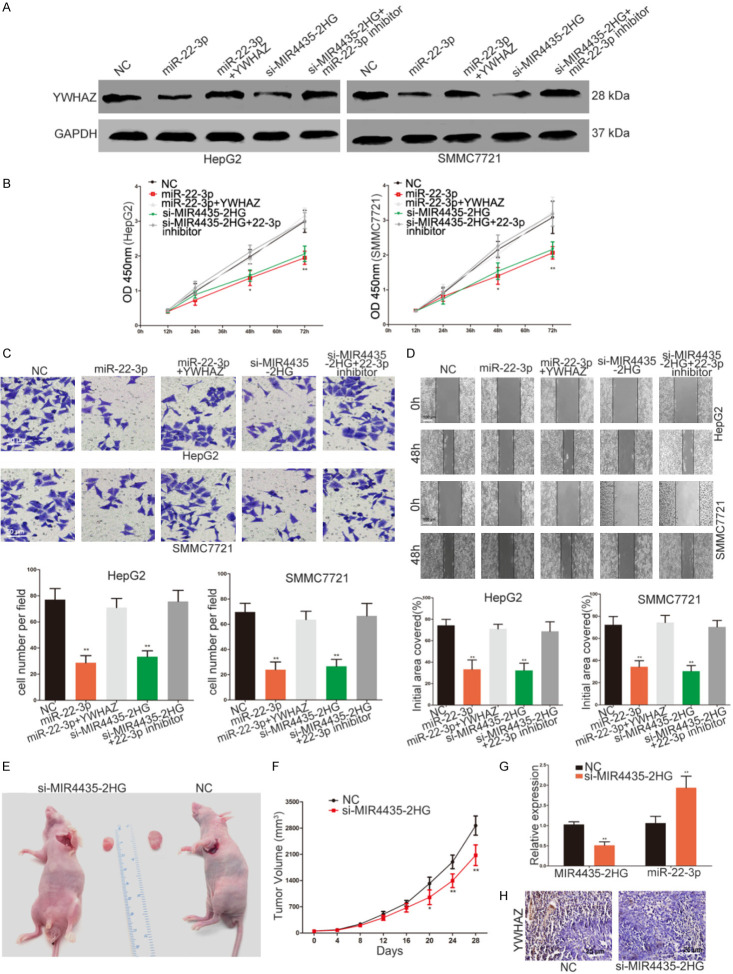
We next explored the effect of MIR4435-2HG on biological function of HCC cells. MIR4435-2HG knockdown HCC cells showed a significant decrease in proliferation (Figure 6B). Transwell and scratch wound assays showed that MIR4435-2HG siRNA suppressed the invasive and migrative capacity of HepG2 and SMMC7721 cells (Figure 6C and 6D). YWHAZ has been proven to act as an oncogene in HCC [20]. We confirmed the oncogenic effect of YWHAZ in HCC (Supplementary Figure 1A-C). We further explored whether MIR4435-2HG exerts its role in HCC by regulating YWHAZ. We subsequently transfected miR-22-3p mimic, miR-22-3p mimic with YWHAZ plasmid, MIR4435-2HG siRNA with miR-22-3p inhibitor into HCC cells. The changes in YWHAZ expression were detected by western blotting (Figure 6A). The miR-22-3p mimic significantly repressed growth and metastasis of HCC cells (Figure 6A-D). The YWHAZ plasmid reversed the influence of miR-22-3p mimic (Figure 6A-D). These results manifested that miR-22-3p plays a role in HCC by targeting YWHAZ. More importantly, the suppression effect of MIR4435-2HG siRNA on YWHAZ expression, growth and metastasis in HCC cells were attenuated by miR-22-3p inhibitor (Figure 6A-D). The xenograft model was used to further discuss the effect of MIR4435-2HG in vivo. MIR4435-2HG knockdown group showed obvious inhibitory effect on tumour growth between 20 and 28 days (Figure 6E and 6F). After 28 days, the mice were sacrificed and the tumours were stripped. The miR-22-3p level was increased, while YWHAZ expression was decreased in MIR4435-2HG knockdown group (Figure 6G and 6H). In conclusion, these results indicated that MIR4435-2HG advances proliferation and metastasis of HCC by modulating the miR-22-3p/YWHAZ axis.
Figure 6.
MIR4435-2HG promotes the proliferation and metastasis of HCC via the miR-22-3p/YWHAZ axis. A. Western blots identified YWHAZ protein expression changes in NC, miR-22-3p mimic, miR-22-3p mimic plus YWHAZ, si-MIR4435-2HG or si-MIR4435-2HG plus miR-22-3p inhibitor transfected HCC cells, GAPDH was used as a control. B. The effect of miR-22-3p mimic and si-MIR4435-2HG on the proliferative ability of HCC cells was determined by CCK8 assay. The results were further verified by cotransfection with miR-22-3p inhibitor and YWHAZ plasmid. Statistical significance is evaluated by two-tailed Student’s t-test. C. Effect of si-MIR4435-2HG and miR-22-3p mimic on the invasive capacity of HCC cells was assessed by transwell assay. The results were confirmed by the recovery experiment of cotransfection with miR-22-3p inhibitor and YWHAZ plasmid respectively. Statistical significance is evaluated by two-tailed Student’s t-test. Scale bar, 50 μm. D. The migration ability of HCC cells in different transfection groups was detected by scratch wound assay. YWHAZ plasmid and miR-22-3p inhibitor reversed the effect of miR-22-3p mimic and si-MIR4435-2HG on the migration capability of HCC cells. Statistical significance is evaluated by two-tailed Student’s t-test. Scale bar, 100 μm. E. The excision tumour of HepG2 xenografts in NC and si-MIR4435-2HG treated groups. F. The tumour volume of HepG2 xenografts in NC and si-MIR4435-2HG treated groups. Statistical significance is evaluated by two-tailed Student’s t-test. G. PCR identified miR-22-3p and MIR4435-2HG expression changes in NC and si-MIR4435-2HG treated groups. Statistical significance is evaluated by two-tailed Student’s t-test. H. The level of YWHAZ was detected by immunohistochemical staining of sections from the xenograft tumour tissues of NC and si-MIR4435-2HG treated groups. Scale bar, 25 μm. *P < 0.05, **P < 0.01, ***P < 0.001.
Discussion
Many specific lncRNAs have been proven to be involved in the malignant phenotypes of many human tumours [17,24]. MIR4435-2HG, a novel lncRNA, is encoded on chromosome 2q13. MIR4435-2HG is characterized as an oncogenic lncRNA in some malignant tumours [25,26]. MIR4435-2HG promotes the of cells by activated TGF-β pathway [12]. MIR4435-2HG also contributes to the proliferation and metastasis of gastric cancer through targeting desmoplakin to activate Wnt/β-catenin signaling [27]. MIR4435-2HG was also confirmed to be overexpressed in HCC [15]. However, the specific role and mechanism of MIR4435-2HG in HCC have not been clarified. Here, we first analyzed the MIR4435-2HG expression data of HCC using our samples and public databases. MIR4435-2HG was overexpressed in HCC compared with adjacent normal tissue, and a high MIR4435-2HG level was a risk factor for the overall survival of patients with HCC. We demonstrated that MIR4435-2HG advances the proliferation and metastasis of HCC cell.
In many human tumours, some special lncRNAs can act as ceRNAs to perform their biological functions [16,28]. ceRNAs decoy special miRNAs to inhibit miRNAs binding to their target genes, therefrom regulating the specific oncogenes and tumour suppressor genes expression [29]. The latest research showed that MIR4435-2HG participates in proliferation and metastasis of colorectal cancer by regulating miR-206/YAP1 axis [18]. MIR4435-2HG has also been proven to promote cell growth, migration and invasion in melanoma by sponging miR-802 to modulate FLOT2 [30]. In this study, we searched the potential ceRNA correlation of MIR4435-2HG in HCC based on our samples and public databases and bioinformatics prediction software. We found that MIR4435-2HG, miR-22-3p and YWHAZ maybe have a potential ceRNA relationship in HCC. MIR4435-2HG was further proven to directly bind to miR-22-3p through RNA pull-down, MS2-RIP and luciferase reporter assays. We demonstrated that YWHAZ is target gene of miR-22-3p in HCC. The miR-22-3p inhibitor abolished the suppression effect of MIR4435-2HG siRNA on expression of YWHAZ and luciferase activity of wild type YWHAZ 3’-UTR luciferase reporter vectors. Thus, we proved that MIR4435-2HG promotes YWHAZ expression by sponging miR-22-3p in HCC.
YWHAZ, a central protein involved in many important signal transduction pathways, participates in extensive cell activities in many types of tumours, including growth, cell cycle, apoptosis, and migration [19,31,32]. YWHAZ could induce the epithelial mesenchymal transition (EMT) and promote the cell autophagy through interacting with phosphorylated Atg9 in various tumours [33,34]. In addition, some scholars have confirmed that YWHAZ is also involved in the proliferation, clone formation, migration, invasion and apoptosis of HCC cells [32], and the oncogenic effect of YWHAZ was confirmed in this study. Many studies confirmed that miR-22-3p plays a role as a antioncogene in many human tumours [35,36]. Here, we demonstrated that miR-22-3p inhibited the growth and metastasis of HCC cells by targeting YWHAZ. Furthermore, the miR-22-3p inhibitor attenuated the suppression effect of MIR4435-2HG siRNA on growth, invasion and migration of HCC cells. MIR4435-2HG also advances HCC progression in vivo through miR-22-3p/YWHAZ pathway. Taken together, this study demonstrates the role of the ceRNA network of MIR4435-2HG/miR-22-3p/YWHAZ in the malignant progression of HCC.
In conclusion, these results showed that MIR4435-2HG acts as an oncogene in the malignant progression of HCC. MIR4435-2HG promotes YWHAZ expression by binding to miR-22-3p to liberate YWHAZ mRNA transcripts. MIR4435-2HG advances the proliferation and metastasis of HCC by regulating the miR-22-3p/YWHAZ axis. Understanding the mechanism of MIR4435-2HG in HCC can improve our understanding of the pathogenesis of HCC. In-depth study of the ceRNA network of MIR4435-2HG/miR-22-3p/YWHAZ in HCC will help us to identify the prognostic marker and treatment target in HCC patients.
Acknowledgements
This study was funded by the National Natural Science Foundation of China (81802726).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Wallace MC, Preen D, Jeffrey GP, Adams LA. The evolving epidemiology of hepatocellular carcinoma: a global perspective. Expert Rev Gastroenterol Hepatol. 2015;9:765–779. doi: 10.1586/17474124.2015.1028363. [DOI] [PubMed] [Google Scholar]
- 2.Shetty S, Sharma N, Ghosh K. Epidemiology of hepatocellular carcinoma (HCC) in hemophilia. Crit Rev Oncol Hematol. 2016;99:129–133. doi: 10.1016/j.critrevonc.2015.12.009. [DOI] [PubMed] [Google Scholar]
- 3.Njei B, Rotman Y, Ditah I, Lim JK. Emerging trends in hepatocellular carcinoma incidence and mortality. Hepatology. 2015;61:191–199. doi: 10.1002/hep.27388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altekruse SF, Henley SJ, Cucinelli JE, McGlynn KA. Changing hepatocellular carcinoma incidence and liver cancer mortality rates in the United States. Am J Gastroenterol. 2014;109:542–553. doi: 10.1038/ajg.2014.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dufour JF, Johnson P. Liver cancer: from molecular pathogenesis to new therapies: summary of the EASL single topic conference. J Hepatol. 2010;52:296–304. doi: 10.1016/j.jhep.2009.11.010. [DOI] [PubMed] [Google Scholar]
- 6.Dimri M, Satyanarayana A. Molecular signaling pathways and therapeutic targets in hepatocellular carcinoma. Cancers (Basel) 2020;12:491. doi: 10.3390/cancers12020491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 8.Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012;9:703–719. doi: 10.4161/rna.20481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lingadahalli S, Jadhao S, Sung YY, Chen M, Hu L, Chen X, Cheung E. Novel lncRNA LINC00844 regulates prostate cancer cell migration and invasion through AR signaling. Mol Cancer Res. 2018;16:1865–1878. doi: 10.1158/1541-7786.MCR-18-0087. [DOI] [PubMed] [Google Scholar]
- 10.Yang J, Li C, Li H, E C. LncRNA CACNA1G-AS1 facilitates hepatocellular carcinoma progression through the miR-2392/C1orf61 pathway. J Cell Physiol. 2019;234:18415–18422. doi: 10.1002/jcp.28477. [DOI] [PubMed] [Google Scholar]
- 11.Zheng YL, Li L, Jia YX, Zhang BZ, Li JC, Zhu YH, Li MQ, He JZ, Zeng TT, Ban XJ, Yuan YF, Li Y, Guan XY. LINC01554-mediated glucose metabolism reprogramming suppresses tumorigenicity in hepatocellular carcinoma via downregulating PKM2 expression and inhibiting Akt/mTOR signaling pathway. Theranostics. 2019;9:796–810. doi: 10.7150/thno.28992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang H, Meng H, Huang X, Tong W, Liang X, Li J, Zhang C, Chen M. lncRNA MIR4435-2HG promotes cancer cell migration and invasion in prostate carcinoma by upregulating TGF-beta1. Oncol Lett. 2019;18:4016–4021. doi: 10.3892/ol.2019.10757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ouyang W, Ren L, Liu G, Chi X, Wei H. LncRNA MIR4435-2HG predicts poor prognosis in patients with colorectal cancer. PeerJ. 2019;7:e6683. doi: 10.7717/peerj.6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qian H, Chen L, Huang J, Wang X, Ma S, Cui F, Luo L, Ling L, Luo K, Zheng G. The lncRNA MIR4435-2HG promotes lung cancer progression by activating beta-catenin signalling. J Mol Med (Berl) 2018;96:753–764. doi: 10.1007/s00109-018-1654-5. [DOI] [PubMed] [Google Scholar]
- 15.Kong Q, Liang C, Jin Y, Pan Y, Tong D, Kong Q, Zhou J. The lncRNA MIR4435-2HG is upregulated in hepatocellular carcinoma and promotes cancer cell proliferation by upregulating miRNA-487a. Cell Mol Biol Lett. 2019;24:26. doi: 10.1186/s11658-019-0148-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luan W, Ding Y, Ma S, Ruan H, Wang J, Lu F. Long noncoding RNA LINC00518 acts as a competing endogenous RNA to promote the metastasis of malignant melanoma via miR-204-5p/AP1S2 axis. Cell Death Dis. 2019;10:855. doi: 10.1038/s41419-019-2090-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ni X, Ding Y, Yuan H, Shao J, Yan Y, Guo R, Luan W, Xu M. Long non-coding RNA ZEB1-AS1 promotes colon adenocarcinoma malignant progression via miR-455-3p/PAK2 axis. Cell Prolif. 2020;53:e12723. doi: 10.1111/cpr.12723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dong X, Yang Z, Yang H, Li D, Qiu X. Long non-coding RNA MIR4435-2HG promotes colorectal cancer proliferation and metastasis through miR-206/YAP1 axis. Front Oncol. 2020;10:160. doi: 10.3389/fonc.2020.00160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gan Y, Ye F, He XX. The role of YWHAZ in cancer: a maze of opportunities and challenges. J Cancer. 2020;11:2252–2264. doi: 10.7150/jca.41316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wei GY, Hu M, Zhao L, Guo WS. MiR-451a suppresses cell proliferation, metastasis and EMT via targeting YWHAZ in hepatocellular carcinoma. Eur Rev Med Pharmacol Sci. 2019;23:5158–5167. doi: 10.26355/eurrev_201906_18180. [DOI] [PubMed] [Google Scholar]
- 21.Luan W, Li L, Shi Y, Bu X, Xia Y, Wang J, Djangmah HS, Liu X, You Y, Xu B. Long non-coding RNA MALAT1 acts as a competing endogenous RNA to promote malignant melanoma growth and metastasis by sponging miR-22. Oncotarget. 2016;7:63901–63912. doi: 10.18632/oncotarget.11564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Subramanian M, Li XL, Hara T, Lal A. A biochemical approach to identify direct microRNA targets. Methods Mol Biol. 2015;1206:29–37. doi: 10.1007/978-1-4939-1369-5_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luan W, Wang Y, Chen X, Shi Y, Wang J, Zhang J, Qian J, Li R, Tao T, Wei W, Hu Q, Liu N, You Y. PKM2 promotes glucose metabolism and cell growth in gliomas through a mechanism involving a let-7a/c-Myc/hnRNPA1 feedback loop. Oncotarget. 2015;6:13006–13018. doi: 10.18632/oncotarget.3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang H, Chen Z, Wang X, Huang Z, He Z, Chen Y. Long non-coding RNA: a new player in cancer. J Hematol Oncol. 2013;6:37. doi: 10.1186/1756-8722-6-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen MY, Zhou GR, Zhang ZY. LncRNA MIR4435-2HG contributes into colorectal cancer development and predicts poor prognosis. Eur Rev Med Pharmacol Sci. 2020;24:1771–1777. doi: 10.26355/eurrev_202002_20354. [DOI] [PubMed] [Google Scholar]
- 26.Shen H, Sun B, Yang Y, Cai X, Bi L, Deng L, Zhang L. MIR4435-2HG regulates cancer cell behaviors in oral squamous cell carcinoma cell growth by upregulating TGF-beta1. Odontology. 2020;108:553–559. doi: 10.1007/s10266-020-00488-x. [DOI] [PubMed] [Google Scholar]
- 27.Wang H, Wu M, Lu Y, He K, Cai X, Yu X, Lu J, Teng L. LncRNA MIR4435-2HG targets desmoplakin and promotes growth and metastasis of gastric cancer by activating Wnt/beta-catenin signaling. Aging (Albany NY) 2019;11:6657–6673. doi: 10.18632/aging.102164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu G, Li Y, Ma Y, Lu J, Chen Y, Jiang Q, Qin Q, Zhao L, Huang Q, Luo Z, Huang S, Wei Z. Long noncoding RNA LINC00511 contributes to breast cancer tumourigenesis and stemness by inducing the miR-185-3p/E2F1/Nanog axis. J Exp Clin Cancer Res. 2018;37:289. doi: 10.1186/s13046-018-0945-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma DM, Sun D, Wang J, Jin DH, Li Y, Han YE. Long non-coding RNA MIR4435-2HG recruits miR-802 from FLOT2 to promote melanoma progression. Eur Rev Med Pharmacol Sci. 2020;24:2616–2624. doi: 10.26355/eurrev_202003_20530. [DOI] [PubMed] [Google Scholar]
- 31.Qi W, Liu X, Qiao D, Martinez JD. Isoform-specific expression of 14-3-3 proteins in human lung cancer tissues. Int J Cancer. 2005;113:359–363. doi: 10.1002/ijc.20492. [DOI] [PubMed] [Google Scholar]
- 32.Zhao JF, Zhao Q, Hu H, Liao JZ, Lin JS, Xia C, Chang Y, Liu J, Guo AY, He XX. The ASH1-miR-375-YWHAZ signaling axis regulates tumor properties in hepatocellular carcinoma. Mol Ther Nucleic Acids. 2018;11:538–553. doi: 10.1016/j.omtn.2018.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen CH, Chuang SM, Yang MF, Liao JW, Yu SL, Chen JJ. A novel function of YWHAZ/beta-catenin axis in promoting epithelial-mesenchymal transition and lung cancer metastasis. Mol Cancer Res. 2012;10:1319–1331. doi: 10.1158/1541-7786.MCR-12-0189. [DOI] [PubMed] [Google Scholar]
- 34.Weerasekara VK, Panek DJ, Broadbent DG, Mortenson JB, Mathis AD, Logan GN, Prince JT, Thomson DM, Thompson JW, Andersen JL. Metabolic-stress-induced rearrangement of the 14-3-3zeta interactome promotes autophagy via a ULK1- and AMPK-regulated 14-3-3zeta interaction with phosphorylated Atg9. Mol Cell Biol. 2014;34:4379–4388. doi: 10.1128/MCB.00740-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao L, Hu K, Cao J, Wang P, Li J, Zeng K, He X, Tu PF, Tong T, Han L. lncRNA miat functions as a ceRNA to upregulate sirt1 by sponging miR-22-3p in HCC cellular senescence. Aging (Albany NY) 2019;11:7098–7122. doi: 10.18632/aging.102240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zong W, Feng W, Jiang Y, Cao Y, Ke Y, Shi X, Ju S, Cong H, Wang X, Cui M, Jing R. LncRNA CTC-497E21.4 promotes the progression of gastric cancer via modulating miR-22/NET1 axis through RhoA signaling pathway. Gastric Cancer. 2020;23:228–240. doi: 10.1007/s10120-019-00998-w. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.