Abstract
Purpose: The 5-year survival rate of patients with lung cancer in China is < 20%, and predicting their prognosis is difficult. Here, we investigated the association between two common non-synonymous single-nucleotide polymorphisms (SNPs) in the excision repair cross-complementing 2 (ERCC2) genes (rs13181 and rs1799793) and the prognosis of patients with lung cancer. Methods: Genomic DNA was extracted from the blood samples of 839 patients with lung cancer and genotyped using the SNPscan technique. The association between patient prognosis and the ERCC2 genotype was analyzed using a multivariate Cox proportional hazards model adjusted for multiple potential confounders. Results: The presence of ERCC2 rs13181 T>G significantly increased the risk of death (adjust hazard ratio (HR) = 1.29, 95% CI: 1.06-1.56, P = 0.009). Patients with the rs13181 TG genotype (adjust HR = 1.34, 95% CI: 1.08-1.65, P = 0.007) and rs13181 dominant mode TG+GG (adjust HR = 1.33, 95% CI: 1.08-1.63, P = 0.007) had significantly worse overall survival. Moreover, stratified analyses showed that patients with the TG and TG+GG rs13181 genotypes who were male, elderly (≥60 years), had a history of smoking, or without family history of malignant tumors had a significantly increased risk of death. In patients with adenocarcinoma lung cancer (ADC), the rs1799793 genotype CT (adjust HR = 1.49, 95% CI: 1.06-2.09, P = 0.023) and dominant model CT+TT (adjust HR = 1.45, 95% CI = 1.04-2.02, P = 0.027) were associated with an increased risk of death. Conclusion: ERCC2 rs13181 and rs1799793 SNPs may be significant prognostic factors for the risk of death among patients with lung cancer.
Keywords: ERCC2, rs13181, rs1799793, single-nucleotide polymorphism, lung cancer, prognosis
Introduction
Despite advances in the diagnosis and treatment of lung cancer in recent decades, reliably predicting prognosis remains challenging, particularly in China, where the five-year survival rate is lower than 20% [1,2]. Establishing accurate prognostic indicators is crucial for improving survival among patients with lung cancer, whose prognosis depends on several factors [3]. Single-nucleotide polymorphisms (SNPs), the most common form of genetic variation in humans, can affect gene expression and predict prognosis in patients with lung cancer [4-8]. In addition, smoking is an important factor associated with the risk and prognosis of lung cancer. Approximately 80-90% of lung cancer cases are caused by smoking; however, only 15% of smokers develop lung cancer. Both heredity and the living environment affect the prognosis of patients with lung cancer [3]. In a study of 104 patients with early-stage lung cancer who were treated by surgery and 107 patients with advanced lung cancer who were administered chemotherapy, Jiang [9] suggested that smoking and the rs776746 SNP of CYP3A5 were risk factors for poor prognosis among patients with lung cancer administered chemotherapy and for a lower survival rate among those undergoing surgery. These results indicated that both the risk and prognosis of lung cancer is influenced by the interaction between environmental risk factors and individual genetic factors [10-12].
Nucleotide excision repair (NER) is an important multifunctional DNA repair system that can remove exogenous or endogenous factors, induce DNA damage or interchain addition, and maintain cell function and genomic integrity [13-16]. The excision repair cross-complementing 2 (ERCC2) genes encodes an ATP-dependent DNA helicase that mediates DNA spin unfolding to initiate NER, and is a key factor in DNA transcription and the NER pathway [17-19]. Lys751Gln (rs13181) and Asp312Asn (rs1799793) are non-synonymous SNPs with a 0.5% allele frequency each in the coding region of ERCC2, which may regulate the DNA repair ability by altering the protein amino acid sequence [20,21]. ERCC2 rs13181 and rs1799793 SNPs have been shown to affect the prognosis and survival of patients with esophageal [22], gastric [23], and colorectal cancers [24] and are associated with poor curative efficacy in patients with lung cancer being administered platinum-based chemotherapy [25].
Previous studies of ERCC2 rs13181 and rs1799793 SNPs have focused on evaluating their predictive role in the curative efficacy of patients with advanced lung cancer being administered platinum-based combination chemotherapy. The association between the above loci and prognosis of lung cancer in different stages has not been reported. We collected peripheral blood samples from patients with a confirmed diagnosis of lung cancer before treatment initiation and performed genotyping and patient follow-up. The relationship between ERCC2 rs13181 and rs1799793 SNPs and the prognosis and survival of patients with lung cancer was evaluated.
Patients and methods
Study group
From January to November 2009, 888 patients with primary lung cancer were enrolled, of whom 536 were from Changhai Hospital affiliated with the Naval Military Medical University (Second Military Medical University), and the other 352 were from the Taizhou Institute of Health Sciences, Fudan University. Inclusion criteria were as follows: patients diagnosed with primary lung cancer by histopathological examination and without a history of malignancy in other organs; there were no age or gender limitations. The clinical data of the patients were obtained from medical records, and follow-up data were collected through telephone interviews. This study was approved by the ethics committee of the School of Life Sciences, Fudan University. Informed consent was obtained from each participant.
SNP selection and genotyping
All enrolled patients donated 5 mL of blood before their treatment was initiated. The genomic DNA was extracted using the Qiagen Blood DNA Extraction Kit (Qiagen, Hilden, Germany). Genotyping was performed using a 2 × 48-plex SNPscan TM kit (cat. no. G0104; Genesky Biotechnologies, Shanghai, China) [26-28]. SNPscan is a proprietary multiplex SNP genotyping system that allows simultaneous genotyping of 48/96/144/192 SNPs per sample in a single tube/sample. SNPscan uses a highly specific ligation reaction to discriminate alleles. Genotyping quality was determined using a detailed procedure consisting of an over 95% successful call rate, duplicate calling of genotypes, internal positive control samples, and Hardy-Weinberg equilibrium (HWE) testing. Laboratory personnel who performed the genotyping assays were blinded to the patients’ clinical information.
Statistical analyses
HWE was tested using Pearson’s chi-square test. Overall survival (OS) was calculated from the date of sample collection to the date of either death from any cause or last follow-up visit. The median survival time was estimated using the Kaplan-Meier method, and group differences were tested using the log-rank test. Univariate and multivariate Cox regression analyses were used to estimate the hazard ratio (HR) and its 95% confidence interval (CI) with and without adjustment for age and sex. Four genetic models (allele, genotype, dominant, and recessive) of the SNPs were applied in Cox regression analyses. Stratified analyses were performed by age, sex, smoking status, family history of malignant cancer, TNM stage, and histologic type of lung cancer. All tests were two sided and P < 0.05 was considered as statistically significant. All statistical analyses were performed using R version 3.6.2 (Vienna, Austria).
Results
Demographic and clinical characteristics and analysis of prognosis
The follow-up period was from the start of enrollment until the end of 2019. After excluding 49 patients because of incomplete clinical information, data from 839 patients were analyzed. The study sample was an ethnically homogenous group of Han Chinese, of whom 668 died, 103 survived (12.3%) for longer than five years, 68 (8.1%) were lost to follow-up, 610 (72.7%) were male, 524 (62.5%) were aged ≥60 years, 582 (69.4%) had a history of smoking, and 302 (36%) had a family history of malignant cancer. Further, 367 (43.7%) patients were diagnosed with adenocarcinoma, 282 (33.6%) with squamous cell carcinoma, 72 (8.6%) with small-cell lung cancer, and 118 (14.1%) with other types of cancer. There were 154 (18.4%) patients diagnosed with stage I and stage II disease, and 625 (74.5%) patients had stage III and stage IV disease (Figure 1).
Figure 1.
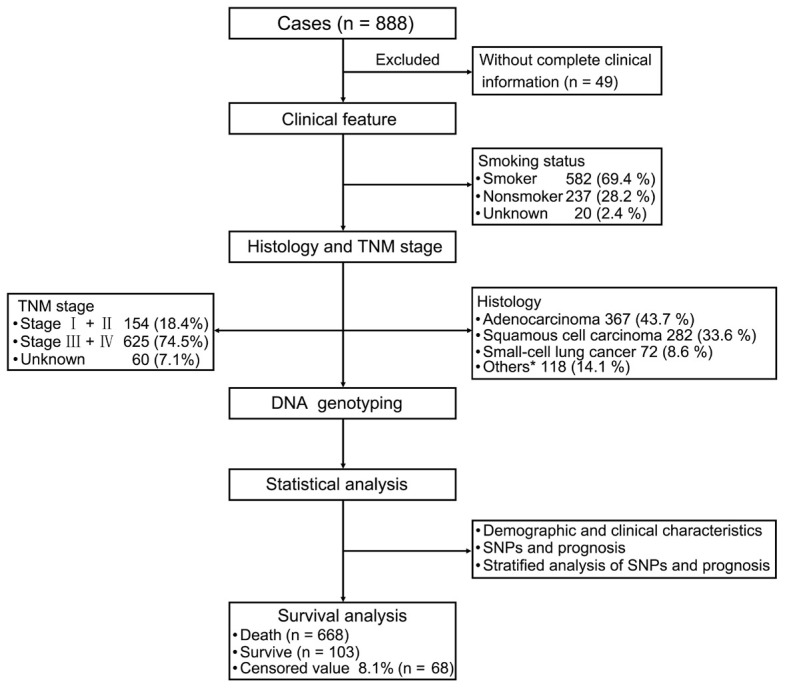
Patient’s demographic and clinical characteristics. We evaluated patients with lung cancer, clinical features, and analysis strategies. For the 839 patients with primary lung cancer (excluding 49 patients because of insufficient clinical information), we evaluated their clinical features including smoking status, TNM stage, and histology. Statistical analysis of demographic and clinical characteristics, SNPs, and prognosis. Stratified analysis of SNPs and prognosis. *Other carcinomas include adenosquamous carcinoma, large cell carcinoma, carcinosarcoma, and mucoepidermoid carcinoma.
Association between patients’ characteristics and lung cancer outcomes
We performed multivariate Cox regression analysis to assess the associations between clinical characteristics and prognosis among patients with lung cancer. As shown in Table 1, the median survival time was significantly lower among males compared to females (34.27 vs. 40.17 months; P = 0.01); among patients aged ≥60 years compared with those aged <60 years (33.2 vs. 40.87 months; P = 0.003); and among smokers compared with non-smokers (33.9 vs. 41.03 months; P < 0.001). Patients with advanced tumor stage had a significantly shorter median survival time compared with patients with early-stage tumors (29.4 vs. 113.93 months; P < 0.001).
Table 1.
Distribution of characteristics in Chinese patients with lung cancer (n = 839) and prognosis analysis
| Variables | n (%) | MST | log rank p |
|---|---|---|---|
| All | 839 | 36.73 | |
| Gender | 0.01 | ||
| Female | 229 (27.3%) | 40.17 | |
| Male | 610 (72.7%) | 34.27 | |
| Age | 0.003 | ||
| Age <60 | 315 (37.5%) | 40.87 | |
| Age ≥60 | 524 (62.5%) | 33.20 | |
| Smoking status | <0.001 | ||
| Nonsmoker | 237 (28.2%) | 41.03 | |
| Smoker | 582 (69.4%) | 33.90 | |
| Unknown | 20 (2.4%) | 67 | |
| Family history of malignant cancer | 0.462 | ||
| Yes | 302 (36%) | 33.63 | |
| No | 537 (64%) | 38.03 | |
| Histology | 0.211 | ||
| ADC | 367 (43.7%) | 38.80 | |
| SCC | 282 (33.6%) | 33.63 | |
| SCLC | 72 (8.6%) | 33.90 | |
| Others* | 118 (14.1%) | 36.20 | |
| TNM Stage | <0.001 | ||
| Stage I+II | 154 (18.4%) | 113.93 | |
| Stage III+IV | 625 (74.5%) | 29.40 | |
| Unknown | 60 (7.1%) | 66.43 |
Other carcinomas include adenosquamous carcinoma, large cell carcinoma, carcinosarcoma, and mucoepidermoid carcinoma.
MST, median survival time; CI, confidence interval.
Association between ERCC2 polymorphisms and prognosis of lung cancer
There were 677 TT, 122 TG, and 7 GG genotypes at rs13181 of ERCC2, with a genotype detection rate of 96.07% (Table 2). The genotype frequency at rs13181 T/G of ERCC2 was consistent with HWE (P = 0.426). There were 736 CC, 99 TC, and 3 TT genotypes at rs1799793 of ERCC2, with a genotype detection rate of 99.9%. The genotype frequency at rs1799793 C/T of ERCC2 was consistent with HWE (P = 0.883), indicating that the investigated population was in a genetic balance, i.e., the population survey data were credible. 1476 T genes and 136 G genes were detected at the rs13181 locus of ERCC2. The frequency of alleles T and G were 90.97% (1159/1274) and 9.03% (115/1274) in the death group respectively, and the allele frequency difference between the two groups was significant (P = 0.009). Compared with TT, the risk of death was higher among individuals with genotypes TG and the dominant model TG+GG (adjust HR for TG = 1.34; 95% CI: 1.08-1.65, P = 0.007; adjust HR for TG+GG = 1.33; 95% CI: 1.08-1.63, P = 0.007) (Table 2).
Table 2.
Association between ERCC2 polymorphisms and prognosis in Chinese patients with lung cancer
| SNP | Model | Death/Survive | HR (95% CI) | P | HRa (95% CI) | Pa |
|---|---|---|---|---|---|---|
| rs13181 | Allele | |||||
| T (ref) | 1159/317 | 1 | 1 | |||
| G | 115/21 | 1.27 (1.05-1.54) | 0.015 | 1.29 (1.06-1.56) | 0.009 | |
| Genotype | ||||||
| T/T (ref) | 528/149 | 1 | 1 | |||
| T/G | 103/19 | 1.32 (1.07-1.63) | 0.010 | 1.34 (1.08-1.65) | 0.007 | |
| G/G | 6/1 | 1.11 (0.50-2.50) | 0.793 | 1.17 (0.52-2.64) | 0.698 | |
| Dominate | ||||||
| T/T (ref) | 528/149 | 1 | 1 | |||
| T/G+G/G | 109/20 | 1.31 (1.06-1.61) | 0.011 | 1.33 (1.08-1.63) | 0.007 | |
| Recessive | ||||||
| T/T+T/G (ref) | 631/168 | 1 | 1 | |||
| G/G | 6/1 | 1.07 (0.48-2.40) | 0.868 | 1.12 (0.50-2.53) | 0.776 | |
| rs1799793 | Allele | |||||
| C (ref) | 1244/327 | 1 | 1 | |||
| T | 90/15 | 1.06 (0.86-1.32) | 0.583 | 1.03 (0.83-1.28) | 0.758 | |
| genotype | ||||||
| C/C (ref) | 580/156 | 1 | 1 | |||
| C/T | 84/15 | 1.06 (0.84-1.34) | 0.603 | 1.04 (0.82-1.31) | 0.759 | |
| T/T | 3/0 | 1.12 (0.36-3.51) | 0.846 | 1.04 (0.33-3.30) | 0.942 | |
| Dominate | ||||||
| C/C (ref) | 580/156 | 1 | 1 | |||
| C/T+T/T | 87/15 | 1.06 (0.85-1.34) | 0.587 | 1.04 (0.83-1.30) | 0.754 | |
| Recessive | ||||||
| C/C+C/T (ref) | 664/171 | 1 | 1 | |||
| T/T | 3/0 | 1.11 (0.36-3.48) | 0.856 | 1.04 (0.33-3.28) | 0.948 |
Adjusted by age, gender.
ref, reference.
Association between ERCC2 polymorphisms and prognosis of lung cancer stratified by patient characteristics
We observed that among male patients, the rs13181 G allele increased the prognostic risk of death relative to the T allele (P = 0.008) (Table 3). Compared with genotype TT, genotype TG increased the risk of death (P = 0.015) (Table 4), as did the dominant genotype TG+GG (P = 0.010) (Table 6). Among older patients (age ≥60 years), the rs13181 G allele increased the prognostic risk of death relative to the T allele (P = 0.002) (Table 3). Compared with genotype TT, genotype TG increased the risk of death (P = 0.003) (Table 4), as did the dominant genotype TG+GG (P = 0.002) (Table 6). Among patients who were smokers, the rs13181 G allele increased the prognostic risk of death relative to T (P = 0.003) (Table 3). Compared with genotype TT, genotype TG increased the risk of death (P = 0.012) (Table 4), as did the dominant genotype TG+GG (P = 0.005) (Table 6). Compared with genotype T/T+T/G, recessive genotype G/G increased the risk of death (P = 0.009) (Table 7). In patients without a family history of malignant cancer, rs13181 genotype TG (P = 0.020) (Table 4) and the dominant genotype TG+GG (P = 0.038) (Table 6) increased the prognostic risk of death compared with genotype TT. For patients with ADC, rs1799793 T allele increased the prognostic risk of death relative to C (P = 0.036) (Table 3). Compared with genotype CC, genotype CT increased the risk of death (P = 0.023) (Table 5), as did the dominant genotype CT+TT (P = 0.027) (Table 6). Among patients who were smokers, rs1799793 recessive genotype T/T increased the risk of death (P = 0.014), compared with C/C+C/T (Table 7).
Table 3.
Association between ERCC2 polymorphisms in allele model and prognosis in Chinese patients with lung cancer
| Variables | rs13181 | rs1799793 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||||||
| Death/Survive | HR (95% CI) | P | HRa (95% CI) | Pa | Death/Survive | HR (95% CI) | P | HRa (95% CI) | Pa | |||
|
|
|
|||||||||||
| T (ref) | G | C (ref) | T | |||||||||
| Gender | ||||||||||||
| Male | 874/197 | 82/15 | 1.33 (1.06-1.67) | 0.014 | 1.36 (1.08-1.70) | 0.008 | 936/205 | 66/11 | 1.05 (0.82-1.35) | 0.693 | 1.04 (0.81-1.34) | 0.737 |
| Female | 285/120 | 33/6 | 1.19 (0.83-1.71) | 0.341 | 1.14 (0.79-1.64) | 0.483 | 308/122 | 24/4 | 1.08 (0.71-1.64) | 0.712 | 1.00 (0.65-1.52) | 0.990 |
| Age (year) | ||||||||||||
| ≥60 | 754/164 | 78/8 | 1.44 (1.14-1.82) | 0.002 | 1.44 (1.14-1.82) | 0.002 | 809/166 | 63/8 | 0.99 (0.77-1.29) | 0.964 | 0.99 (0.76-1.28) | 0.929 |
| <60 | 405/153 | 37/13 | 1.05 (0.75-1.47) | 0.781 | 1.05 (0.75-1.47) | 0.772 | 435/161 | 27/7 | 1.08 (0.73-1.60) | 0.711 | 1.06 (0.72-1.58) | 0.761 |
| Smoking status | ||||||||||||
| Yes | 850/173 | 80/13 | 1.41 (1.12-1.77) | 0.004 | 1.42 (1.13-1.79) | 0.003 | 908/180 | 64/10 | 1.04 (0.81-1.35) | 0.738 | 1.03 (0.80-1.33) | 0.820 |
| No | 288/129 | 32/7 | 1.20 (0.83-1.73) | 0.332 | 1.16 (0.80-1.67) | 0.432 | 314/131 | 24/5 | 1.08 (0.71-1.63) | 0.729 | 1.03 (0.68-1.57) | 0.887 |
| Family history of malignant cancer | ||||||||||||
| Yes | 425/99 | 45/7 | 1.28 (0.94-1.74) | 0.121 | 1.30 (0.95-1.77) | 0.100 | 450/105 | 42/5 | 1.10 (0.80-1.51) | 0.553 | 1.12 (0.81-1.53) | 0.496 |
| No | 734/218 | 70/14 | 1.24 (0.97-1.59) | 0.083 | 1.25 (0.97-1.59) | 0.081 | 794/222 | 48/10 | 1.01 (0.75-1.36) | 0.946 | 0.95 (0.70-1.27) | 0.709 |
| Histology | ||||||||||||
| ADC | 491/171 | 45/9 | 1.23 (0.90-1.67) | 0.195 | 1.27 (0.93-1.73) | 0.135 | 507/176 | 45/6 | 1.45 (1.06-1.96) | 0.018 | 1.39 (1.02-1.89) | 0.036 |
| SCC | 414/77 | 36/7 | 1.24 (0.88-1.74) | 0.225 | 1.21 (0.86-1.71) | 0.267 | 452/81 | 24/5 | 0.76 (0.50-1.15) | 0.198 | 0.76 (0.50-1.15) | 0.195 |
| TNM stage | ||||||||||||
| Stage I+II | 138/143 | 12/9 | 1.24 (0.68-2.24) | 0.482 | 1.43 (0.79-2.62) | 0.241 | 142/149 | 10/7 | 1.10 (0.58-2.09) | 0.773 | 1.14 (0.59-2.17) | 0.702 |
| Stage III+IV | 923/158 | 99/12 | 1.20 (0.98-1.48) | 0.085 | 1.20 (0.98-1.48) | 0.084 | 1005/162 | 73/8 | 1.02 (0.80-1.29) | 0.879 | 1.00 (0.79-1.27) | 0.990 |
Adjusted by age, gender.
CI, confidence interval; HR, hazard ratio; ref, reference.
Table 4.
Association between ERCC2 rs13181 SNPs in genotype model and prognosis in Chinese patients with lung cancer
| Variables | Death /survive | T/G VS. T/T | G/G VS. T/T | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|||||||||
| T/T (ref) | T/G | G/G | HR (95% CI) | P | HRa (95% CI) | Pa | HR (95% CI) | P | HRa (95% CI) | Pa | |
| Gender | |||||||||||
| male | 399/92 | 76/13 | 3/1 | 1.34 (1.05-1.71) | 0.020 | 1.36 (1.06-1.74) | 0.015 | 1.70 (0.55-5.31) | 0.359 | 2.06 (0.66-6.44) | 0.216 |
| female | 129/57 | 27/6 | 3/0 | 1.32 (0.87-1.99) | 0.197 | 1.30 (0.86-1.97) | 0.216 | 0.91 (0.28-2.93) | 0.872 | 0.77 (0.24-2.51) | 0.667 |
| Age (year) | |||||||||||
| ≥60 | 344/78 | 66/8 | 6/0 | 1.53 (1.17-1.99) | 0.002 | 1.49 (1.14-1.94) | 0.003 | 1.32 (0.59-2.96) | 0.505 | 1.48 (0.65-3.38) | 0.349 |
| <60 | 184/71 | 37/11 | 0/1 | 1.12 (0.79-1.60) | 0.527 | 1.12 (0.79-1.60) | 0.531 | - | - | - | - |
| Smoking status | |||||||||||
| yes | 388/80 | 74/13 | 3/0 | 1.36 (1.06-1.75) | 0.015 | 1.38 (1.07-1.77) | 0.012 | 4.98 (1.59-15.63) | 0.006 | 4.89 (1.56-15.35) | 0.007 |
| no | 131/62 | 26/5 | 3/1 | 1.41 (0.93-2.16) | 0.109 | 1.38 (0.90-2.11) | 0.141 | 0.79 (0.25-2.49) | 0.685 | 0.75 (0.24-2.37) | 0.623 |
| Family history of malignant cancer | |||||||||||
| yes | 194/46 | 37/7 | 4/0 | 1.20 (0.84-1.71) | 0.308 | 1.23 (0.86-1.75) | 0.253 | 2.03 (0.75-5.48) | 0.161 | 2.02 (0.74-5.51) | 0.171 |
| no | 334/103 | 66/12 | 2/1 | 1.38 (1.06-1.81) | 0.016 | 1.37 (1.05-1.78) | 0.020 | 0.55 (0.14-2.22) | 0.401 | 0.60 (0.15-2.43) | 0.473 |
| Histology | |||||||||||
| ADC | 225/81 | 41/9 | 2/0 | 1.30 (0.93-1.82) | 0.127 | 1.38 (0.98-1.93) | 0.064 | 0.91 (0.22-3.76) | 0.901 | 0.82 (0.20-3.42) | 0.787 |
| SCC | 191/36 | 32/5 | 2/1 | 1.27 (0.87-1.85) | 0.214 | 1.22 (0.84-1.79) | 0.299 | 1.17 (0.29-4.73) | 0.824 | 1.32 (0.32-5.40) | 0.703 |
| Stage | |||||||||||
| Stage I+II | 64/67 | 10/9 | 1/0 | 1.42 (0.73-2.79) | 0.303 | 1.79 (0.90-3.58) | 0.097 | - | - | - | - |
| Stage III+IV | 417/74 | 89/10 | 5/1 | 1.22 (0.97-1.54) | 0.089 | 1.21 (0.96-1.52) | 0.103 | 1.24 (0.51-2.99) | 0.635 | 1.33 (0.55-3.22) | 0.528 |
Adjusted by age, gender.
CI, confidence interval; HR, hazard ratio; ref, reference.
Table 6.
Association between ERCC2 polymorphisms in dominant model and prognosis in Chinese patients with lung cancer
| Variables | rs13181 | rs1799793 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||||||
| Death/Survive | HR (95% CI) | P | HRa (95% CI) | Pa | Death/Survive | HR (95% CI) | P | HRa (95% CI) | Pa | |||
|
|
|
|||||||||||
| T/T (ref) | T/G+G/G | C/C (ref) | C/T+T/T | |||||||||
| Gender | ||||||||||||
| male | 399/92 | 79/14 | 1.35 (1.06-1.72) | 0.015 | 1.37 (1.08-1.75) | 0.010 | 437/97 | 64/11 | 1.02 (0.79-1.34) | 0.855 | 1.02 (0.78-1.33) | 0.895 |
| female | 129/57 | 30/6 | 1.26 (0.85-1.88) | 0.255 | 1.22 (0.82-1.82) | 0.326 | 143/59 | 23/4 | 1.16 (0.75-1.81) | 0.500 | 1.08 (0.69-1.69) | 0.726 |
| Age (year) | ||||||||||||
| ≥60 | 344/78 | 72/8 | 1.51 (1.17-1.94) | 0.002 | 1.49 (1.15-1.92) | 0.002 | 376/79 | 60/8 | 0.99 (0.75-1.30) | 0.947 | 0.98 (0.75-1.30) | 0.905 |
| <60 | 184/71 | 37/12 | 1.09 (0.76-1.55) | 0.635 | 1.09 (0.76-1.55) | 0.635 | 204/77 | 27/7 | 1.08 (0.72-1.64) | 0.699 | 1.07 (0.71-1.61) | 0.751 |
| Smoking status | ||||||||||||
| yes | 388/80 | 77/13 | 1.40 (1.10-1.79) | 0.007 | 1.42 (1.11-1.81) | 0.005 | 424/85 | 62/10 | 1.02 (0.78-1.33) | 0.909 | 1.00 (0.77-1.31) | 0.991 |
| no | 131/62 | 29/6 | 1.31 (0.87-1.95) | 0.195 | 1.27 (0.84-1.90) | 0.254 | 146/63 | 23/5 | 1.14 (0.73-1.77) | 0.563 | 1.10 (0.71-1.71) | 0.677 |
| Family history of malignant cancer | ||||||||||||
| yes | 194/46 | 41/7 | 1.25 (0.89-1.75) | 0.194 | 1.28 (0.91-1.80) | 0.158 | 206/50 | 40/5 | 1.06 (0.75-1.48) | 0.752 | 1.07 (0.76-1.51) | 0.680 |
| no | 334/103 | 68/13 | 1.33 (1.02-1.72) | 0.035 | 1.32 (1.02-1.72) | 0.038 | 374/106 | 47/10 | 1.05 (0.77-1.43) | 0.749 | 0.98 (0.71-1.33) | 0.875 |
| Histology | ||||||||||||
| ADC | 225/81 | 43/9 | 1.28 (0.92-1.78) | 0.149 | 1.34 (0.96-1.86) | 0.088 | 234/85 | 42/6 | 1.50 (1.08-2.09) | 0.016 | 1.45 (1.04-2.02) | 0.027 |
| SCC | 191/36 | 34/6 | 1.26 (0.88-1.82) | 0.212 | 1.23 (0.85-1.78) | 0.273 | 214/38 | 24/5 | 0.74 (0.48-1.14) | 0.177 | 0.74 (0.48-1.14) | 0.174 |
| TNM stage | ||||||||||||
| Stage I+II | 64/67 | 11/9 | 1.33 (0.70-2.53) | 0.385 | 1.60 (0.83-3.09) | 0.159 | 67/71 | 9/7 | 1.16 (0.58-2.33) | 0.676 | 1.21 (0.60-2.44) | 0.594 |
| Stage III+IV | 417/74 | 94/11 | 1.22 (0.98-1.53) | 0.081 | 1.22 (0.97-1.52) | 0.087 | 468/77 | 71/8 | 0.99 (0.77-1.28) | 0.958 | 0.97 (0.76-1.25) | 0.828 |
Adjusted by age, gender.
CI, confidence interval; HR, hazard ratio; ref, reference.
Table 7.
Association between ERCC2 polymorphisms in recessive model and prognosis in Chinese patients with lung cancer
| Variables | rs13181 | rs1799793 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||||||
| Death/Survive | HR (95% CI) | P | HRa (95% CI) | Pa | Death/Survive | HR (95% CI) | P | HRa (95% CI) | Pa | |||
|
|
|
|||||||||||
| T/T+T/G (ref) | G/G | C/C+C/T (ref) | T/T | |||||||||
| Gender | ||||||||||||
| male | 475/105 | 3/1 | 1.63 (0.52-5.08) | 0.399 | 1.96 (0.62-6.12) | 0.249 | 499/108 | 2/0 | 6.31 (1.56-25.57) | 0.010 | 5.17 (1.27-21.06) | 0.022 |
| female | 156/63 | 3/0 | 0.88 (0.27-2.82) | 0.824 | 0.74 (0.23-2.41) | 0.623 | 165/63 | 1/0 | 0.44 (0.058-3.27) | 0.420 | 0.36 (0.048-2.70) | 0.321 |
| Age (year) | ||||||||||||
| ≥60 | 410/86 | 6/0 | 1.25 (0.56-2.80) | 0.593 | 1.40 (0.61-3.19) | 0.423 | 433/87 | 3/0 | 1.05 (0.33-3.27) | 0.939 | 1.06 (0.34-3.37) | 0.916 |
| <60 | 211/82 | 0/1 | - | - | - | - | 231/84 | - | - | - | - | - |
| Smoking status | ||||||||||||
| yes | 462/93 | 3/0 | 4.73 (1.51-14.82) | 0.008 | 4.63 (1.47-14.52) | 0.009 | 484/95 | 2/0 | 6.73 (1.66-27.28) | 0.008 | 5.82 (1.43-23.73) | 0.014 |
| no | 157/67 | 3/1 | 0.75 (0.24-2.38) | 0.629 | 0.71 (0.23-2.26) | 0.566 | 168/68 | 1/0 | 0.52 (0.071-3.80) | 0.519 | 0.45 (0.062-3.32) | 0.436 |
| Family history of malignant cancer | ||||||||||||
| yes | 231/53 | 4/0 | 1.98 (0.73-5.32) | 0.178 | 1.95 (0.72-5.31) | 0.191 | 244/55 | 2/0 | 7.84 (1.90-32.34) | 0.004 | 7.17 (1.72-29.94) | 0.007 |
| no | 400/115 | 2/1 | 0.52 (0.13-2.12) | 0.364 | 0.57 (0.14-2.32) | 0.431 | 420/116 | 1/0 | 0.38 (0.053-2.76) | 0.341 | 0.38 (0.052-2.76) | 0.338 |
| Histology | ||||||||||||
| ADC | 266/90 | 2/0 | 0.88 (0.21-3.62) | 0.860 | 0.79 (0.19-3.28) | 0.745 | 273/91 | 3/0 | 1.24 (0.39-3.95) | 0.714 | 1.08 (0.33-3.49) | 0.897 |
| SCC | 223/41 | 2/1 | 1.13 (0.28-4.57) | 0.860 | 1.30 (0.32-5.32) | 0.716 | 238/43 | - | - | - | - | - |
| TNM stage | ||||||||||||
| Stage I+II | 74/76 | 1/0 | - | - | - | - | 75/78 | 1/0 | 0.78 (0.10-5.81) | 0.808 | 0.75 (0.094-5.94) | 0.783 |
| Stage III+IV | 506/84 | 5/1 | 1.20 (0.50-2.90) | 0.686 | 1.29 (0.53-3.12) | 0.572 | 537/8 | 2/0 | 5.88 (1.45-23.77) | 0.013 | 5.06 (1.24-20.56) | 0.024 |
Adjusted by age, gender.
CI, confidence interval; HR, hazard ratio; ref, reference.
Table 5.
Association between ERCC2 rs1799793 SNPs in genotype model and prognosis in Chinese patients with lung cancer
| Variables | Death/Survive | C/C VS. C/T | C/C VS. T/T | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|||||||||
| C/C (ref) | C/T | T/T | HR (95% CI) | P | HRa (95% CI) | Pa | HR (95% CI) | P | HRa (95% CI) | Pa | |
| Gender | |||||||||||
| male | 437/97 | 62/11 | 2/0 | 1.00 (0.76-1.30) | 0.986 | 0.99 (0.76-1.30) | 0.954 | 6.31 (1.56-25.58) | 0.010 | 5.17 (1.27-21.05) | 0.022 |
| female | 143/59 | 22/4 | 1/0 | 1.25 (0.80-1.96) | 0.331 | 1.18 (0.75-1.85) | 0.482 | - | - | - | - |
| Age (year) | |||||||||||
| ≥60 | 376/79 | 57/8 | 3/0 | 0.99 (0.75-1.31) | 0.933 | 0.98 (0.74-1.30) | 0.887 | 1.04 (0.33-3.27) | 0.941 | 1.06 (0.33-3.37) | 0.920 |
| <60 | 204/77 | 27/7 | - | 1.08 (0.72-1.64) | 0.699 | 1.07 (0.71-1.61) | 0.751 | - | - | - | - |
| Smoking status | |||||||||||
| yes | 424/85 | 60/10 | 2/0 | 0.99 (0.75-1.30) | 0.928 | 0.97 (0.74-1.28) | 0.855 | 6.72 (1.66-27.25) | 0.008 | 5.80 (1.42-23.65) | 0.014 |
| no | 146/63 | 22/5 | 1/0 | 1.20 (0.77-1.88) | 0.424 | 1.17 (0.74-1.83) | 0.499 | - | - | - | - |
| Family history of malignant cancer | |||||||||||
| yes | 206/50 | 38/5 | 2/0 | - | - | - | - | - | - | - | - |
| no | 374/106 | 46/10 | 1/0 | 1.09 (0.80-1.49) | 0.580 | 1.01 (0.74-1.38) | 0.964 | - | - | - | - |
| Histology | |||||||||||
| ADC | 234/85 | 39/6 | 3/0 | 1.52 (1.08-2.14) | 0.016 | 1.49 (1.06-2.09) | 0.023 | 1.29 (0.40-4.11) | 0.668 | 1.13 (0.35-3.64) | 0.844 |
| SCC | 214/38 | 24/5 | - | 0.74 (0.48-1.14) | 0.177 | 0.74 (0.48-1.14) | 0.174 | - | - | - | - |
| TNM stage | |||||||||||
| Stage I+II | 67/71 | 8/7 | 1/0 | 1.23 (0.59-2.57) | 0.581 | 1.30 (0.62-2.73) | 0.486 | - | - | - | - |
| Stage III+IV | 468/77 | 69/8 | 2/0 | 0.97 (0.75-1.25) | 0.812 | 0.95 (0.74-1.23) | 0.697 | 5.85 (1.45-23.69) | 0.013 | 5.02 (1.23-20.40) | 0.024 |
Adjusted by age, gender.
CI, confidence interval; HR, hazard ratio; ref, reference.
Discussion
In this study, we evaluated the relationship between the rs13181 and rs1799793 SNPs in ERCC2 and prognostic risk of death using blood samples from 839 Chinese patients with lung cancer. We found that ERCC2 rs13181 T>G increased the prognostic risk of death in patients with lung cancer. Further stratified analyses showed that the risk of death increased significantly in men, the elderly (60 years and older), those with a history of smoking, and those without a family history of malignant cancer.
This study also stratified the above analyses by epidemiological factors such as pathological type, disease stage, and smoking status and explored the impact of the above two loci on the prognosis of patients with lung cancer. However, previous studies of the correlation between the rs13181 and rs1799793 SNPs and prognosis of patients with lung cancer are relatively limited and mainly focused on the pharmacogenomics of platinum-based combination chemotherapy [29-36]. A study by Wu [25] included 353 patients with advanced lung cancer who were initially treated with platinum-based combination chemotherapy and whose blood-derived SNPs were genotyped before the initial treatment. The response to platinum-based chemotherapy was evaluated after first-line cycles of chemotherapy based on Response Evaluation Criteria in Solid Tumors guidelines (Version 1.1) [37]. However, Perez-Ramirez [38] found that ERCC2 rs13181 and rs1799793 SNPs were not correlated with the clinical efficacy of platinum chemotherapy in 141 Spanish patients with lung cancer.
NER plays an extremely important role in maintaining genome integrity and preventing mutation. Its effect is regulated by the transcription initiation factor TFIIH, which has ATP-dependent helicase activity. ERCC2 is an integral member of the core transcription factor IIH via p44, which, if mutated, prevents the interaction between its protein product and p44, thereby reducing the activity of helicase and affecting the repair capacity of DNA [19,39]. ERCC2 rs13181 and rs1799793 are common non-synonymous SNPs whose specific functions have been identified in the ERCC2 coding sequence and affect the function of the NER pathway by regulating mRNA expression. ERCC2 rs13181 T>G and rs1799793 C>T are missense mutations, which may alter the amino acid sequence of proteins and regulate their capacity for DNA damage repair [20,25,30].
Among smokers in this study, rs13181 mutation carriers had a higher risk of death. Smoking is generally recognized as a risk factor related to the occurrence and poor prognosis of lung cancer. More than 20 compounds in smoke are classified as lung carcinogens, as they can cause permanent mutations by forming DNA adducts and causing DNA double-strand breaks, as well as interfere with DNA repair function, resulting in a higher risk of lung cancer and worse cancer prognosis [40-43]. Among smokers, the extensive DNA damage caused by tobacco can even completely mask the slight differences in the DNA repair capacity caused by genetic polymorphisms [44,45]. Smoking is considered the most important environmental factor influencing lung cancer. Other known environmental factors include occupational exposure to carcinogens and air pollution [46,47]. In this study, the risk of death differed by gender among patients with lung cancer, and being male was a prognostic factor for poor lung cancer survival. Smoking and the SNPs may have jointly affected the prognosis of patients with lung cancer. In addition, the influence of other environmental factors on the prognosis of patients with lung cancer should also be considered in future studies.
In a study of 2724 male and 1894 female patients with lung cancer in Minnesota, USA conducted from 1997 to 2002, Visbal [48] found that men had a significantly higher risk of death compared to women following a diagnosis of lung cancer (adjusted relative risk: 1.20, 95% CI: 1.11-1.30). Smoking is a critical factor, as more men smoke and are likely to be heavier smokers than women [46,49]. In this study, the prognostic risk of death among elderly rs13181 G carriers was significantly higher, possibly because of the long-term accumulation of DNA damage over time, which in turn is due to the loss of NER function caused by rs13181 T>G [50].
Our study showed that the rs1799793 SNP of ERCC2 was not associated with the prognosis of patients with lung cancer overall, but the risk of death was significantly increased in patients with ADC with the mutant T allele. Previous studies revealed that the rs1799793 T allele might cause a decrease in NER function, thereby reducing the survival time of patients with lung cancer. However, this study did not stratify the different histologic type of lung cancer, and thus the relationship between rs1799793 SNPs and ADC was unknown [25]. Notably, a large number of studies have observed no association between the rs1799793 SNP and clinical outcomes among patients with lung cancer being administered platinum-based chemotherapy [34,38]. rs1799793 is located in the Arch transcription domain of ERCC2, and its SNP mainly affects its transcriptional activity rather than its DNA repair capacity, which may explain why rs1799793 affects patient survival but not chemotherapy efficacy [38]. A study of 360 patients with gastric cancer in North America showed that the rs1799793 mutant genotype TT was associated with a worse prognosis in patients with poorly differentiated tumors [51]; similar results were obtained in our study.
Previous studies used genome-wide association studies, which can detect millions of SNPs across the whole genome and identify associations between SNPs and complex diseases. However, the size and complexity of the data were difficult to manage and some important genes in this pathway may have been missed [52]. The candidate-gene associations explored in this study had the advantages of a limited number of candidate genes and low number of false-positive cases, enhancing the accuracy of the analyses, providing reliable estimates, and solving the problem of the insufficient association between single genes and cancer in previous studies [53]. However, this study also had some limitations. First, the relatively small sample size restricted the analysis of recessive models and may have led to unstable estimates in stratified analyses. The subjects were of Han Chinese ethnicity, limiting the ability to generalize the results to other populations. Future studies of a larger number of patients are required to evaluate the association between ERCC2 rs13181 and rs1799793 SNPs and lung cancer survival. Second, the number of SNPs analyzed in the present study was limited, although the correlation between rs13181 and the prognosis of lung cancer was clearly determined. DNA repair is a process of protein synergy, which requires multiple DNA repair pathways, and further studies of SNPs in candidate genes should be performed. Finally, although we obtained significant results, additional clinical studies are needed to verify our results; moreover, the exact biological functions and mechanisms of rs13181 and rs1799793 SNPs of ERCC2 on prognostic survival of patients with lung cancer should be further analyzed.
In summary, our results demonstrate that ERCC2 rs13181 T>G and rs1799793 C>T increase the prognostic risk of death among patients with lung cancer, with the former playing a more significant role depending on gender, age, and smoking status. These findings have potential clinical significance for predicting the prognosis of patients with lung cancer and formulating new therapeutic strategies.
Acknowledgements
The authors acknowledge all the volunteer donors involved in this study and the ChangHai Hospital and TaiZhou Institute of Health Sciences staff for supporting during sample collection. The work was supported by National Natural Science Foundation of China (81372236). Key Project of the National Science and Technology Pillar Program (2011BAI09B00). Innovation and Practice Ability Incubator for Undergraduate Students of Naval Medical University (FH2019210).
Disclosure of conflict of interest
None.
References
- 1.Cho J. The international association for the study of lung cancer--the lung cancer staging project: better data, better decisions, better outcomes. Hawaii Med J. 2008;67:220–222. [PubMed] [Google Scholar]
- 2.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 3.Woodard GA, Jones KD, Jablons DM. Lung cancer staging and prognosis. Cancer Treat Res. 2016;170:47–75. doi: 10.1007/978-3-319-40389-2_3. [DOI] [PubMed] [Google Scholar]
- 4.Sang Y, Bi X, Liu Y, Zhang W, Wang D. Adverse prognostic impact of TGFB1 T869C polymorphism in non-small-cell lung cancer. Onco Targets Ther. 2017;10:1513–1518. doi: 10.2147/OTT.S123685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shen XY, Lu FZ, Wu Y, Zhao LT, Lin ZF. XRCC3 Thr241Met polymorphism and clinical outcomes of NSCLC patients receiving platinum-based chemotherapy: a systematic review and meta-analysis. PLoS One. 2013;8:e69553. doi: 10.1371/journal.pone.0069553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qian J, Liu H, Gu S, Wu Q, Zhao X, Wu W, Wang H, Wang J, Chen H, Zhang W, Wei Q, Jin L, Lu D. Genetic variants of the MDM2 gene are predictive of treatment-related toxicities and overall survival in patients with advanced NSCLC. Clin Lung Cancer. 2015;16:e37–53. doi: 10.1016/j.cllc.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 7.Liu Y, Qing H, Su X, Wang C, Li Z, Liu S. Association of CD44 gene polymorphism with survival of NSCLC and risk of bone metastasis. Med Sci Monit. 2015;21:2694–2700. doi: 10.12659/MSM.894357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim MJ, Kang HG, Lee SY, Jeon HS, Lee WK, Park JY, Lee EB, Lee JH, Cha SI, Kim DS, Kim CH, Kam S, Jung TH, Park JY. AKT1 polymorphisms and survival of early stage non-small cell lung cancer. J Surg Oncol. 2012;105:167–174. doi: 10.1002/jso.22071. [DOI] [PubMed] [Google Scholar]
- 9.Jiang LP, Zhu ZT, He CY. Effects of CYP3A5 genetic polymorphism and smoking on the prognosis of non-small-cell lung cancer. Onco Targets Ther. 2016;9:1461–1469. doi: 10.2147/OTT.S94144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cancer Genome Atlas Research Network. Author correction: comprehensive molecular profiling of lung adenocarcinoma. Nature. 2018;559:E12. doi: 10.1038/s41586-018-0228-6. [DOI] [PubMed] [Google Scholar]
- 11.Spitz MR, Wei Q, Dong Q, Amos CI, Wu X. Genetic susceptibility to lung cancer: the role of DNA damage and repair. Cancer Epidemiol Biomarkers Prev. 2003;12:689–698. [PubMed] [Google Scholar]
- 12.Langevin SM, Kratzke RA, Kelsey KT. Epigenetics of lung cancer. Transl Res. 2015;165:74–90. doi: 10.1016/j.trsl.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spivak G. Nucleotide excision repair in humans. DNA Repair. 2015;36:13–18. doi: 10.1016/j.dnarep.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu L, Wu J, Zhong R, Wu C, Zou L, Yang B, Chen W, Zhu B, Duan S, Yu D, Tan W, Nie S, Lin D, Miao X. Multi-loci analysis reveals the importance of genetic variations in sensitivity of platinum-based chemotherapy in non-small-cell lung cancer. Mol Carcinog. 2013;52:923–931. doi: 10.1002/mc.21942. [DOI] [PubMed] [Google Scholar]
- 15.Kim SH, Lee GW, Lee MJ, Cho YJ, Jeong YY, Kim HC, Lee JD, Hwang YS, Kim IS, Lee S, Oh SY. Clinical significance of ERCC2 haplotype-tagging single nucleotide polymorphisms in patients with unresectable non-small cell lung cancer treated with first-line platinum-based chemotherapy. Lung Cancer. 2012;77:578–584. doi: 10.1016/j.lungcan.2012.04.016. [DOI] [PubMed] [Google Scholar]
- 16.Azuma K, Komohara Y, Sasada T, Terazaki Y, Ikeda J, Hoshino T, Itoh K, Yamada A, Aizawa H. Excision repair cross-complementation group 1 predicts progression-free and overall survival in non-small cell lung cancer patients treated with platinum-based chemotherapy. Cancer Sci. 2007;98:1336–1343. doi: 10.1111/j.1349-7006.2007.00557.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qixing M, Gaochao D, Wenjie X, Rong Y, Feng J, Lin X, Mantang Q, Qiang C. Predictive value of Ercc1 and Xpd polymorphisms for clinical outcomes of patients receiving neoadjuvant therapy: a prisma-compliant meta-analysis. Medicine (Baltimore) 2015;94:e1593–e1593. doi: 10.1097/MD.0000000000001593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spitz MR, Wu X, Wang Y, Wang LE, Shete S, Amos CI, Guo Z, Lei L, Mohrenweiser H, Wei Q. Modulation of nucleotide excision repair capacity by XPD polymorphisms in lung cancer patients. Cancer Res. 2001;61:1354–1357. [PubMed] [Google Scholar]
- 19.Coin F, Marinoni JC, Rodolfo C, Fribourg S, Pedrini AM, Egly JM. Mutations in the XPD helicase gene result in XP and TTD phenotypes, preventing interaction between XPD and the p44 subunit of TFIIH. Nat Genet. 1998;20:184–188. doi: 10.1038/2491. [DOI] [PubMed] [Google Scholar]
- 20.Lunn RM, Helzlsouer KJ, Parshad R, Umbach DM, Harris EL, Sanford KK, Bell DA. XPD polymorphisms: effects on DNA repair proficiency. Carcinogenesis. 2000;21:551–555. doi: 10.1093/carcin/21.4.551. [DOI] [PubMed] [Google Scholar]
- 21.Yang Y, Jin X, Yan C, Tian Y, Tang J, Shen X. Case-only study of interactions between DNA repair genes (hMLH1, APEX1, MGMT, XRCC1 and XPD) and low-frequency electromagnetic fields in childhood acute leukemia. Leuk Lymphoma. 2008;49:2344–2350. doi: 10.1080/10428190802441347. [DOI] [PubMed] [Google Scholar]
- 22.Boldrin E, Malacrida S, Rumiato E, Battaglia G, Ruol A, Amadori A, Saggioro D. Association between ERCC1 rs3212986 and ERCC2/XPD rs1799793 and OS in patients with advanced esophageal cancer. Front Oncol. 2019;9:85. doi: 10.3389/fonc.2019.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li M, Zhao Y, Zhao E, Wang K, Lu W, Yuan L. Predictive value of two polymorphisms of ERCC2, rs13181 and rs1799793, in clinical outcomes of chemotherapy in gastric cancer patients: a meta-analysis. Dis Markers. 2018;2018:3947626. doi: 10.1155/2018/3947626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gan Y, Li XR, Chen DJ, Wu JH. Association between polymorphisms of XRCC1 Arg399Gln and XPD Lys751Gln genes and prognosis of colorectal cancer in a Chinese population. Asian Pac J Cancer Prev. 2012;13:5721–5724. doi: 10.7314/apjcp.2012.13.11.5721. [DOI] [PubMed] [Google Scholar]
- 25.Wu W, Li H, Wang H, Zhao X, Gao Z, Qiao R, Zhang W, Qian J, Wang J, Chen H, Wei Q, Han B, Lu D. Effect of polymorphisms in XPD on clinical outcomes of platinum-based chemotherapy for Chinese non-small cell lung cancer patients. PLoS One. 2012;7:e33200. doi: 10.1371/journal.pone.0033200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang YH, Xu Q, Zhao Z, Wu J, Liu WX, Wang H, Jin L, Wang JC. Polymorphism rs7214723 in CAMKK1 and lung cancer risk in Chinese population. Tumour Biol. 2013;34:3147–3152. doi: 10.1007/s13277-013-0883-z. [DOI] [PubMed] [Google Scholar]
- 27.Tang LL, Chen FY, Wang H, Hu XL, Dai X, Mao J, Shen ZT, Wu YH, Wang SM, Hai J, Yan GJ, Li H, Huang J. Haplotype analysis of eight genes of the monoubiquitinated FANCD2-DNA damage-repair pathway in breast cancer patients. Cancer Epidemiol. 2013;37:311–317. doi: 10.1016/j.canep.2012.12.010. [DOI] [PubMed] [Google Scholar]
- 28.Yin J, Wang L, Shi Y, Shao A, Tang W, Wang X, Ding G, Liu C, Chen S, Gu H. Interleukin 17A rs4711998 A>G polymorphism was associated with a decreased risk of esophageal cancer in a Chinese population. Dis Esophagus. 2014;27:87–92. doi: 10.1111/dote.12045. [DOI] [PubMed] [Google Scholar]
- 29.van de Vaart PJ, Belderbos J, de Jong D, Sneeuw KC, Majoor D, Bartelink H, Begg AC. DNA-adduct levels as a predictor of outcome for NSCLC patients receiving daily cisplatin and radiotherapy. Int J Cancer. 2000;89:160–166. [PubMed] [Google Scholar]
- 30.Furuta T, Ueda T, Aune G, Sarasin A, Kraemer KH, Pommier Y. Transcription-coupled nucleotide excision repair as a determinant of cisplatin sensitivity of human cells. Cancer Res. 2002;62:4899–4902. [PubMed] [Google Scholar]
- 31.de las Penas R, Sanchez-Ronco M, Alberola V, Taron M, Camps C, Garcia-Carbonero R, Massuti B, Queralt C, Botia M, Garcia-Gomez R, Isla D, Cobo M, Santarpia M, Cecere F, Mendez P, Sanchez JJ, Rosell R. Polymorphisms in DNA repair genes modulate survival in cisplatin/gemcitabine-treated non-small-cell lung cancer patients. Ann Oncol. 2006;17:668–675. doi: 10.1093/annonc/mdj135. [DOI] [PubMed] [Google Scholar]
- 32.Li P, Wang YD, Cheng J, Chen JC, Ha MW. Association between polymorphisms of BAG-1 and XPD and chemotherapy sensitivity in advanced non-small-cell lung cancer patients treated with vinorelbine combined cisplatin regimen. Tumour Biol. 2015;36:9465–9473. doi: 10.1007/s13277-015-3672-z. [DOI] [PubMed] [Google Scholar]
- 33.Mathiaux J, Le Morvan V, Pulido M, Jougon J, Begueret H, Robert J. Role of DNA repair gene polymorphisms in the efficiency of platinum-based adjuvant chemotherapy for non-small cell lung cancer. Mol Diagn Ther. 2011;15:159–166. doi: 10.1007/BF03256406. [DOI] [PubMed] [Google Scholar]
- 34.Yang Y, Xian L. The association between the ERCC1/2 polymorphisms and the clinical outcomes of the platinum-based chemotherapy in non-small cell lung cancer (NSCLC): a systematic review and meta-analysis. Tumour Biol. 2014;35:2905–2921. doi: 10.1007/s13277-013-1493-5. [DOI] [PubMed] [Google Scholar]
- 35.Qin Q, Zhang C, Yang X, Zhu H, Yang B, Cai J, Cheng H, Ma J, Lu J, Zhan L, Liu J, Liu Z, Xu L, Sun X. Polymorphisms in XPD gene could predict clinical outcome of platinum-based chemotherapy for non-small cell lung cancer patients: a meta-analysis of 24 studies. PLoS One. 2013;8:e79864. doi: 10.1371/journal.pone.0079864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yin M, Yan J, Voutsina A, Tibaldi C, Christiani DC, Heist RS, Rosell R, Booton R, Wei Q. No evidence of an association of ERCC1 and ERCC2 polymorphisms with clinical outcomes of platinum-based chemotherapies in non-small cell lung cancer: a meta-analysis. Lung Cancer. 2011;72:370–377. doi: 10.1016/j.lungcan.2010.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S, Mooney M, Rubinstein L, Shankar L, Dodd L, Kaplan R, Lacombe D, Verweij J. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1) Eur J Cancer. 2009;45:228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 38.Perez-Ramirez C, Canadas-Garre M, Alnatsha A, Villar E, Valdivia-Bautista J, Faus-Dader MJ, Calleja-Hernandez MA. Pharmacogenetics of platinum-based chemotherapy: impact of DNA repair and folate metabolism gene polymorphisms on prognosis of non-small cell lung cancer patients. Pharmacogenomics J. 2019;19:164–177. doi: 10.1038/s41397-018-0014-8. [DOI] [PubMed] [Google Scholar]
- 39.Qiao Y, Spitz MR, Shen H, Guo Z, Shete S, Hedayati M, Grossman L, Mohrenweiser H, Wei Q. Modulation of repair of ultraviolet damage in the host-cell reactivation assay by polymorphic XPC and XPD/ERCC2 genotypes. Carcinogenesis. 2002;23:295–299. doi: 10.1093/carcin/23.2.295. [DOI] [PubMed] [Google Scholar]
- 40.Phillips DH, Venitt S. DNA and protein adducts in human tissues resulting from exposure to tobacco smoke. Int J Cancer. 2012;131:2733–2753. doi: 10.1002/ijc.27827. [DOI] [PubMed] [Google Scholar]
- 41.Lopez-Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153:1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Du Y, Su T, Zhao L, Tan X, Chang W, Zhang H, Cao G. Associations of polymorphisms in DNA repair genes and MDR1 gene with chemotherapy response and survival of non-small cell lung cancer. PLoS One. 2014;9:e99843. doi: 10.1371/journal.pone.0099843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Neumann AS, Sturgis EM, Wei Q. Nucleotide excision repair as a marker for susceptibility to tobacco-related cancers: a review of molecular epidemiological studies. Mol Carcinog. 2005;42:65–92. doi: 10.1002/mc.20069. [DOI] [PubMed] [Google Scholar]
- 44.Qian B, Zhang H, Zhang L, Zhou X, Yu H, Chen K. Association of genetic polymorphisms in DNA repair pathway genes with non-small cell lung cancer risk. Lung Cancer. 2011;73:138–146. doi: 10.1016/j.lungcan.2010.11.018. [DOI] [PubMed] [Google Scholar]
- 45.De Ruyck K, Szaumkessel M, De Rudder I, Dehoorne A, Vral A, Claes K, Velghe A, Van Meerbeeck J, Thierens H. Polymorphisms in base-excision repair and nucleotide-excision repair genes in relation to lung cancer risk. Mutat Res. 2007;631:101–110. doi: 10.1016/j.mrgentox.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 46.Dalton SO, Frederiksen BL, Jacobsen E, Steding-Jessen M, Osterlind K, Schuz J, Osler M, Johansen C. Socioeconomic position, stage of lung cancer and time between referral and diagnosis in Denmark, 2001-2008. Br J Cancer. 2011;105:1042–1048. doi: 10.1038/bjc.2011.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sekido Y, Fong KM, Minna JD. Molecular genetics of lung cancer. Annu Rev Med. 2003;54:73–87. doi: 10.1146/annurev.med.54.101601.152202. [DOI] [PubMed] [Google Scholar]
- 48.Visbal AL, Williams BA, Nichols FC 3rd, Marks RS, Jett JR, Aubry MC, Edell ES, Wampfler JA, Molina JR, Yang P. Gender differences in non-small-cell lung cancer survival: an analysis of 4,618 patients diagnosed between 1997 and 2002. Ann Thorac Surg. 2004;78:209–215. doi: 10.1016/j.athoracsur.2003.11.021. [DOI] [PubMed] [Google Scholar]
- 49.Hsu LH, Chu NM, Liu CC, Tsai SY, You DL, Ko JS, Lu MC, Feng AC. Sex-associated differences in non-small cell lung cancer in the new era: is gender an independent prognostic factor? Lung Cancer. 2009;66:262–267. doi: 10.1016/j.lungcan.2009.01.020. [DOI] [PubMed] [Google Scholar]
- 50.Moskalev AA, Shaposhnikov MV, Plyusnina EN, Zhavoronkov A, Budovsky A, Yanai H, Fraifeld VE. The role of DNA damage and repair in aging through the prism of Koch-like criteria. Ageing Res Rev. 2013;12:661–684. doi: 10.1016/j.arr.2012.02.001. [DOI] [PubMed] [Google Scholar]
- 51.Li Y, Liu Z, Liu H, Wang LE, Tan D, Ajani JA, Wei QY. ERCC1 and ERCC2 variants predict survival in gastric cancer patients. PLoS One. 2013;8:e71994. doi: 10.1371/journal.pone.0071994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hardy J, Singleton A. Genomewide association studies and human disease. N Engl J Med. 2009;360:1759–1768. doi: 10.1056/NEJMra0808700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gorlov IP, Meyer P, Liloglou T, Myles J, Boettger MB, Cassidy A, Girard L, Minna JD, Fischer R, Duffy S, Spitz MR, Haeussinger K, Kammerer S, Cantor C, Dierkesmann R, Field JK, Amos CI. Seizure 6-like (SEZ6L) gene and risk for lung cancer. Cancer Res. 2007;67:8406–8411. doi: 10.1158/0008-5472.CAN-06-4784. [DOI] [PubMed] [Google Scholar]


