Abstract
Epilepsy is a common neurological disorder characterized by recurrent and unprovoked seizures due to neuronal hyperactivity. A large proportion of epilepsy cases begin during childhood. Causes of epilepsy include stroke, infections, brain injury, genetic factors, or other factors that alter brain structure and development, but in up to 50% of cases the cause is unknown. Approximately 35% of patients have refractory seizures that do not respond to medication. Animal models and in vitro cultures have contributed to our understanding of epilepsy, but there is a clear need for better models to explore the human brain in normal and pathological conditions. Human pluripotent stem cell (PSC) technologies opened the door for new models for analyzing brain development and disease, especially conditions with a genetic component. Initially, PSCs were differentiated into 2-dimensional cultures of a homogenous population of neural cells, such as glutamatergic excitatory or γ-aminobutyric acidergic inhibitory neurons, as well as glial cells. Nevertheless, these cultures lacked the structure and complexity of a human brain. In the last decade, PSC technology has advanced to the next level through the development of 3-dimensional culture, called organoids. These organoids recapitulate features of the human brain that are missing in animal models, enabling a deeper study of the human brain. In this review, we will summarize the current status of organoid research and its application to epilepsy.
Keywords: induced pluripotent stem cells (iPSCs), embryonic stem cells (ESCs), organoids, gene editing, epilepsy
Introduction
Epilepsy is a chronic neurological condition of recurrent and unprovoked seizures due to neuronal hyperactivity.1-3 It is a common disorder, affecting 65 million people around the world.4 Most cases manifest in childhood, reflecting the vulnerability of the developing brain, with an increasing proportion identified with genetic etiologies. More than 10 monogenic epilepsies have been modeled in vitro,5,6 using pluripotent stem cells (PSCs)—both embryonic stem cells (ESCs)7 and induced PSCs (iPSCs)8—that have been differentiated into neural cells.9-11 These methods have allowed mutations to be studied: iPSCs can be reprogrammed from a skin or blood sample from patients having epilepsy with a specific mutation and then differentiated in 2-dimensional (2D) cultures of glutamatergic excitatory or γ-aminobutyric acidergic inhibitory neurons, as well as glial cells (for a review of epilepsy 2D models see reference12,13). Likewise, specific mutations in patients with epilepsy can be introduced into ESCs or iPSCs derived from healthy controls by CRISPR-Cas9 gene editing and differentiated into neurons and glial cells using the same protocols.14 Moreover, gene editing can be used to correct mutations in patient iPSCs to obtain an isogenic control line with the same genetic background. However, these 2D cultures have limitations as human cell models of development and disease, because they lack the human brain’s cell–cell interaction, structure, complexity, and heterogeneity.15 More recently, the development of 3-dimensional (3D) culture, also called organoids, may have overcome some of these limitations, at least in part.16,17 In this review, we will summarize current brain-organoid protocols, highlight several examples of neurodevelopmental disorders that these models mimic, and discuss the benefits and limitations of these models in uncovering epilepsy mechanisms.
Human Brain Development and Organoid Models
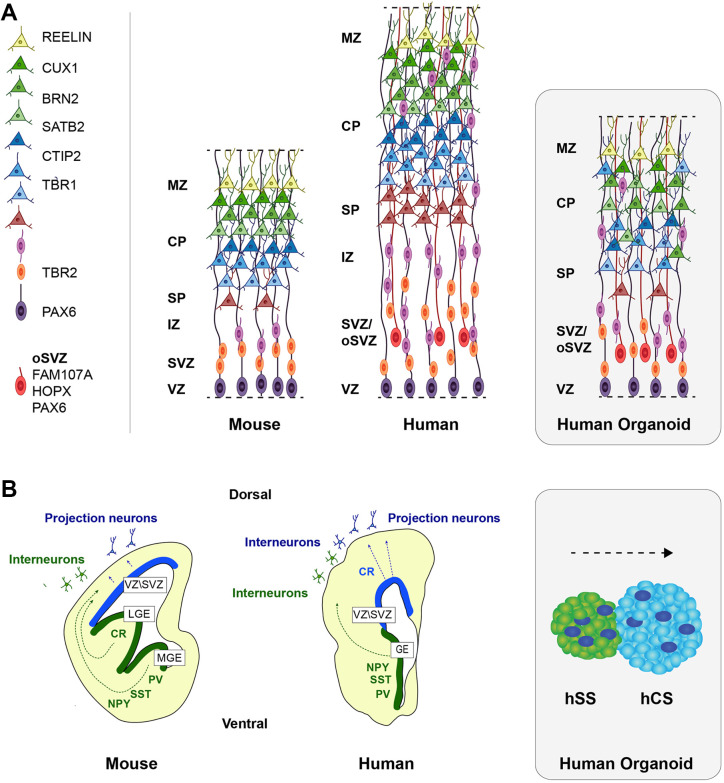
Compared to other primates, the human brain is both larger and more complex and develops differently.18 Cortical neurons derive from radial glial cells (RGCs, also known as ventral or apical RGCs), which directly contact the lateral ventricle and have long processes that extend to the pial surface.19 These RGCs give rise to cortical neurons directly or through intermediate progenitor cells (IPCs).20 In human, RGCs also differentiate into outer radial glial cells (oRGCs, also known as basal RGCs) which lack apical contacts with the lateral ventricle, as their cell bodies translocate into the outer subventricular zone but retain basal contacts with the pial surface.21 These oRGCs are considered key factors for the human cortical surface’s increased size and complexity. Daughter cells derived from RGCs migrate away from the ventricle toward the cortical plate, where they give rise to differentiated neurons. Deep-layer neurons form first, and upper-layer neurons emerge later during development, in an inside-out manner (Figure 1A).20 Once in their final destination, cortical excitatory neurons mature and are classified based on their laminar position, gene expression profile, and projection type.22 Later during development, RGCs also produce astrocytes and oligodendrocytes.20 In addition, inhibitory interneurons integrate into the cortical circuit.23 These interneurons are generated in the ganglionic eminences (ventral forebrain) and migrate tangentially toward the cortex (dorsal forebrain; Figure 1B).24 It has been proposed that a small proportion of interneurons can also be produced locally in the cortex in humans.25-28 Different types of inhibitory neurons have been described in the cortex according to their morphology, origin, location, gene expression, and electrophysiological properties.29 Moreover, specific interneuron subtypes can be observed only in humans.30 Many of these aspects of brain development can be recapitulated in organoids.16,17,31 To date, 2 general types of 3D organoids have been described: whole-brain organoids, also called cerebral organoids, or directed regional brain organoids. Formation of cerebral organoids relies on the intrinsic capacity of PSCs to self-pattern and self-organize.32 Cerebral organoids have the potential to manifest different brain regions in a single organoid (ie, cortex, hippocampus, retina), allowing the study of cell–cell interactions within these brain regions but they can be heterogeneous from batch to batch.33 On the other hand, formation of directed regional brain organoids employs small molecules and growth factors to promote neural induction (typically through inhibition of the bone morphogenetic protein [BMP]/transforming growth factor-beta [TGF-β] signaling pathways) and to specify region identity such as human cortical organoids or human cortical spheroids (hCSs),34-36 human ventral organoids or human subpallium spheroids (hSSs),37-39 among others. In addition, cellular interactions between these brain regions can be analyzed by merging 2 types of region-specified organoids, for example, fusing hCS and hSS37-39 to observe tangential migration of interneurons from the ganglionic eminences to the cortex. Some of the organoid protocols require the use of Matrigel, a gelatinous protein mixture resembling the extracellular matrix, to promote cell differentiation. Both types of organoids form similar ventricular zone-like structures surrounded by RGCs and IPCs, which give rise to cortical neurons in a time- and space-dependent pattern (Figure 1), and they resemble human brain tissue at the molecular, cellular, and electrophysiological levels.17,36,40,41 However, their organization is rudimentary. Although neurons of the 6 cortical layers are present, deep-layer neurons are formed first and are located around ventricular zone-like structures, while upper-layer neurons appear later more externally without a clear layer definition. Interestingly, they contain human-specific features, such as increased cortical thickness and the presence of oRGCs. Despite their limitations, organoids are an ideal platform to study human diseases that affect brain development.
Figure 1.
Comparative representation of mouse and human brains and organoids. A, The pictures represent cortical layers in mouse and human developing brains and in brain organoids. Human brain has an expansion of cortical layers and contains oRGCs. Brain organoids recapitulate human brain structure to a certain extent. B, The drawings show origin and migration pathways of projection neurons and interneurons in mouse and human brains, and organoid models for neuronal migration. CP indicates cortical plate; IZ, intermediate zone; MZ, marginal zone; oSVZ, outer subventricular zone; oRGCs, outer radial glial cells; SP, subplate; SVZ, subventricular zone; VZ, ventricular zone.
Organoid Models for Developmental Epilepsies
Tuberous Sclerosis
Tuberous sclerosis complex (TSC) is a developmental disorder characterized by the presence of benign tumors in multiple organs, including the brain.42,43 These brain tumors, called cortical tubers, manifest as an accumulation of enlarged and dysplastic neurons and glia in the cortex. A high percentage of patients with TSC have epilepsy, autism, and intellectual disability.44 Tuberous sclerosis complex is caused by loss-of-function mutations in the TSC1 or TSC2 genes, which encode the proteins hamartin (TSC1) and tuberin (TSC2).45 These proteins, joined with TBC1D7, form a complex which represses mammalian target of rapamycin complex 1 (mTORC1) signaling.45 Blair et al used CRISPR-Cas9 gene editing to insert loss-of function mutations in TSC1 and TSC2 human embryonic stem cells (hESCs) to model TSC in vitro.46 They showed that homozygous hESCs, but not heterozygous, exhibited an increase in mTORC1 signaling and a reduction in AKT phosphorylation. Further, they differentiated hESCs to hCSs to study the role of TSC during brain development. While they did not find significant differences in the number of progenitor or proliferative cells, they observed a reduction or delay of neuronal markers and an increase of glial cells in TSC1 −/− and TSC2 −/− hCS. Neurons and glia were enlarged and dysmorphic in hCS derived from homozygous cells, resembling cells found in cortical tubers in patients. The model also provided support for the hypothesis that in heterozygous patients, cytomegalic cells appear after a somatic second mutation (known as second-hit model) that results in the biallelic inactivation of TSC1 or TSC2. To test this hypothesis, the authors developed an hESC line with a constitutive loss-of-function mutation in 1 allele of the TSC2 gene and a Cre-inducible conditional mutation in the second allele (TSC2 −/c). Conditional hESCs were differentiated into hCS, and Cre was added after 12 days in vitro during the progenitor expansion phase. Cells with the second-hit mutation became dysmorphic and were bigger, similar to the cells found in homozygous TSC1 −/− and TSC2 −/− hCS. Finally, they showed that chronic rapamycin treatment prevented the cellular hypertrophy. A tuber-like phenotype was found in another preliminary study by Eichmüller and colleagues using cerebral organoids derived from TSC patient iPSCs with heterozygous mutations in TSC2. 47 In this study, the researchers also identified a new type of cell, the caudal late interneuron progenitors or CLIP cells, in TSC organoids by single-cell RNA sequencing. The CLIP cells differentiated into interneurons and expressed high level of epidermal growth factor receptor (EGFR). The first study provided evidence supporting that a second mutation is needed for the formation of cortical tubers. However, in the second study, the authors observed tuber-like structures even with only 1 allele mutated in TSC2 gene. Further analysis would be critical to corroborate the second-hit model. For this reason, it is essential to use isogenic controls and compare multiple iPSC lines to obtain consistent data. Although organoids contain cells that resemble those found in patients with TSC, they do not recapitulate focal tuber-like abnormalities. These studies also point to the mTORC1 signaling pathway as a potential therapeutic target for patients with TSC to prevent the formation of cortical tubers.
Rett Syndrome
Rett syndrome (RTT) is a neurodevelopmental disorder produced by mutations in the X-linked methyl-CpG-binding protein 2 (MeCP2) gene.48 Most patients with RTT are female, with intellectual disability, motor coordination problems, seizures, autism, and respiratory and cardiac abnormalities.49 Mellios et al analyzed cerebral organoids from a patient having RTT with a frameshift mutation in the MeCP2 gene resulting in a premature stop codon.50 These organoids showed an increase in ventricular area and a thinning of the ventricular wall. Moreover, patient organoids showed an increase in RGCs and neuronal cells, but a reduction in IPCs. The researchers also found that mutations in MeCP2 produced an upregulation of miR-199 and miR-214 and an increase in BMP levels. They also analyzed radial migration of cortical progenitor cells by electroporating MeCP2 short hairpin RNA expressing green fluorescent protein (GFP) in control organoids or GFP control vector in mutant organoids and found a migration reduction in both cases. This study identified novel microRNAs which regulate the MeCP2 pathway and affect early cortical development. However, it is unknown how these earlier effects on the RGCs and IPCs contribute to the epileptogenesis. A complete understanding of the MeCP2 downstream pathway would be necessary to develop better treatments for patients with RTT.
Periventricular Heterotopia
Cerebral organoids have also been used to model periventricular heterotopia (PH), a group of heterogeneous disorders characterized by intellectual disability and epilepsy.51 One study used iPSCs from 2 patients, 1 with a heterozygous mutation in the FAT atypical cadherin 4 (FAT4) gene and the other with homozygous mutations in the dachsous cadherin-related 1 (DCHS1) gene, and performed gene editing with CRISPR-Cas9 to created KO iPSC lines for both genes.52 They found accumulation of neurons in ventricular zones in organoids derived from all lines, which could be due to an aberrant morphology of RGCs and defective radial migration. The researchers also identified changes in the expression of genes involved in the formation of the cytoskeleton and cell junction and axon guidance. In another study,53 the same group examined the role of endothelin-converting enzyme 2 (ECE2) gene where 2 biallelic variants have been identified in patients with PH. They found that chronic enzymatic inhibition of ECE2 using PHOS and CRISPR/Cas9-mediated KO of ECE2 in cerebral organoids promoted an increase of neural progenitor cells and a reduction of neurons. They also described changes in the microtubule, actin cytoskeleton, and polarity of RGCs, as well as in the production of extracellular matrix proteins and their receptors. Similar ectopic neuronal positioning in ventricular zones described in the first study was found in another research in control cerebral organoids after overexpression of the pleckstrin homology domain containing family G member 6 gene, which encodes the guanine nucleotide exchange factor and was found mutated in a PH patient.54 These studies have been able to recapitulate PH phenotypes which were only partially observed in mouse models and highlight the use of PSC-derived organoids to study PH disorders. They also identified microtubule and cytoskeleton proteins as key players to maintain RGC morphology and promote correct neuronal radial migration. Targeting cytoskeleton components might be useful to prevent migration defects in patients with PH.
Angelman Syndrome
Angelman syndrome (AS) is a neurodevelopmental disorder characterized by delayed development, intellectual disability, and seizures.55 Most AS cases are caused by a loss-of function mutation of the ubiquitin protein ligase E3A (UBE3A) gene, which encodes a HECTE3 ubiquitin ligase.56 Using gene-edited KO ESCs and iPSCs derived from 1 patient with AS, Sun et al57 found that pyramidal-shaped neurons in AS hCS exhibited augmented excitability and fast components of after-hyperpolarization due to elevated levels of calcium- and voltage-dependent big potassium (BK) channels. Moreover, using Ca2+ imaging and perforated patch recordings, they observed that UBE3A loss promoted neuronal hyperactivity and synchronization. These results suggest that the inhibition of BK channels might reduce the hyperactivity in patients with AS and control seizure development. This study is also the first to observe hyperactivity in organoids derived from patients with epilepsy as a proof of concept that organoids could be used to study disruption in neuronal networks. However, it is unclear whether this hyperactivity leads to the development of seizures.
Miller-Dieker syndrome
Miller-Dieker syndrome (MDS) is a severe form of lissencephaly associated with mental retardation and epilepsy, and microcephaly in some patients.58 Miller-Dieker syndrome is caused by large heterozygous deletions in chromosome 17.58 This region contains dozens of genes, including PAFAH1B1 (LIS1 protein) and YWHAE (14-3-3ε protein).58 Using cortical organoids derived from MDS iPSCs, Bershteyn et al59 showed an increase of apoptosis in the VZ-like region and horizontal division in RGCs, as well as a decrease in the size of organoids. They also found migration defects of doublecortin+ cells after organoids were embedded in Matrigel. Miller-Dieker syndrome organoids showed an increase in deep-layer neurons, without a major change in the laminar distribution. The researchers also labeled proliferative cells (RGCs, IPCs, and oRGCs) using a cytomegalovirus (CMV)-GFP adenovirus, and they observed a delay in cell division in the oRGCs in organoids from patients with MDS. Similar results were found in an independent study performed by Lefremova et al.60 In this second study, the authors also reported smaller organoids and a switch from symmetric to asymmetric cell division of RGCs. However, they did not see significant differences in apoptotic cell death. In addition, RGCs in MDS organoids showed an impaired activation of N-cadherin/β-catenin/Wnt signaling and external activation of the Wnt pathway rescued the switch in the division mode observed in MDS organoids. The differences found between both studies could be due to variations in the organoid protocols or the organoid-to-organoid heterogeneity. These studies highlight the role of RGCs during brain development and the contribution of oRGCs to human specific phenotypes. These earlier defects could explain the microcephaly observed in some of the patients with MDS, but it is still unclear how the change of RGC division mode affects the formation of the neuronal network and promotes epilepsy.
Timothy Syndrome
Timothy syndrome (TS) is a neurodevelopmental disease characterized by autism and epilepsy.61 Birey et al analyzed interneuron migration using hCS and hSS derived from patients with TS carrying a gain-of-function mutation in the CACNA1C gene that encodes the L-type calcium channel (LTCC) subunit.38 They labelled interneuron progenitors in hSS using a Dlxi1/2b-GFP reporter and fused them to hCS. The GFP+ cells from control and patient spheroids migrated from hCS to hSS in a saltatory pattern, characterized by movement for a period of time followed by extensive pauses, as described in vivo. Patient hCS–hSS showed an increase in saltation frequency, as well as a decrease in saltation length and speed. Interestingly, this effect was cell autonomous, as the migration defect was also found when patient hSSs were fused to control hCS. Moreover, this migration phenotype was rescued after adding nimodipine, an LTCC blocker, suggesting that LTCC blockers might be used to prevent migration defects in patients with TS. It would be interesting to study whether these migration defects lead to a reduction of interneurons in hCS leading to an imbalance between excitatory and inhibitory neurons. This imbalance might be responsible for the development of seizures in patients with TS.
Developmental Epileptic Encephalopathies
Developmental epileptic encephalopathies are severe disorders characterized by intractable epileptic seizures and developmental delay. In 1 study,62 researchers analyzed 36 cases from 25 families with epileptic encephalopathies and found germline recessive mutations in the UDP-Glucose 6-Dehydrogenase (UGDH) gene. The UGDH encodes an oxidoreductase that converts UDP-glucose to UDP-glucuronic acid, a key component of proteoglycans and glycolipids. The authors generated cerebral organoids from several patients with 2 different heterozygous mutations and 1 homozygous mutation in the UGDH gene. After 10 weeks in vitro, mutant organoids were significantly smaller and showed a decrease in neuronal progenitor markers and in the number of proliferative cells. However, the authors did not find these defects in UGDH mutant zebrafish. This study brings evidence that extracellular matrix components are critical during brain development and opens up a line of investigation using dietary supplements or modulators of UGDH activity to regulate seizure formation in patients with UGDH mutations.
Progressive Myoclonus Epilepsy
Myoclonic epilepsy of Unverricht and Lundborg or epilepsy, progressive myoclonus 1 (EPM1) is an autosomal recessive disorder and the most common form of progressive myoclonus epilepsy (PME).63 Patients with PME often present with a combination of myoclonus, epilepsy, and progressive neurological deterioration, and many have mutations in the cystatin B (CSTB) gene and its promoter.64 To study the role of CSTB during human brain development, Di Matteo et al used cerebral organoids derived from patients with EPM1and healthy controls.65 They showed that overexpression of CSTB in control organoids promoted cell proliferation, while overexpression of a mutant form of CSTB inhibited it. Similarly, organoids derived from EPM1 patients were smaller and showed a decrease in cell proliferation, but an increase in the number of neurons. In line with a previous study that reported CSTB is synthesized and secreted from synaptosomes,66 CSTB was detected in the media of organoid cultures. Interestingly, control organoids showed a decrease in cell proliferation after being cultured with media from EPM1 mutant organoids, whereas media from control organoids rescued the proliferation deficit in EMP1 organoids. The researchers observed a disruption in interneuron migration when ventral organoids were fused to dorsal organoids derived from EPM1 patients. Taken together, these studies indicate that the maintenance of CSTB levels is critical for proper cell proliferation and interneuron migration during brain development. Future studies are needed to explore the use of CTSB supplements to prevent or partially correct these defects in patients with PME.
Other Cortical Malformations
Microcephaly due to loss-of-function mutations in the CDK5RAP2 gene,32 or to ZIKA virus infection,35,67-69 has also been modeled using organoids. These studies reported smaller organoids, due to a decrease in cell proliferation and/or increase in cell death. On the other hand, macrocephaly has been studied in organoids by CRISPR/Cas9-mediated phosphatase and tensin homolog (PTEN) deletion in hESCs.69 The PTEN heterozygous loss-of-function mutations have been associated with human macrocephaly. The PTEN mutant organoids were larger and exhibited more folding due to an increase in progenitor and oRGC proliferation and in transient neuronal differentiation. Interestingly, mouse PTEN mutant organoids did not show increased folding, suggesting a human-specific phenotype. These studies provide new insights into the mechanisms that control the structure of human brain and highlight the use of human organoids to analyze human characteristics like increased cortical lamination and folding which cannot be studied using animal models with smooth brains like mice.
Limitations, Challenges, and Future Directions
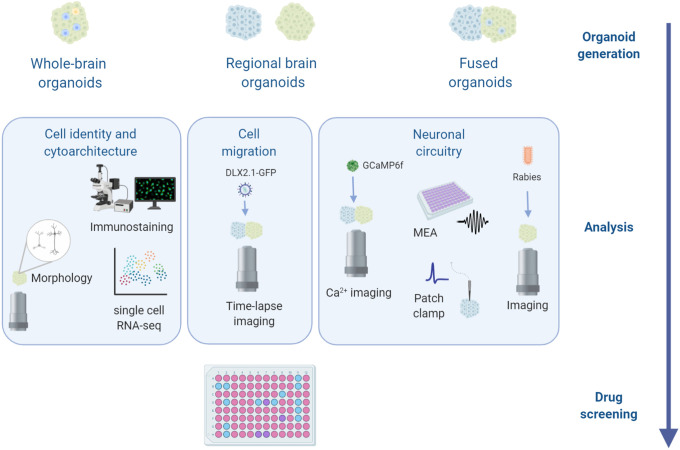
Since 2013, when brain organoid technology was first described, several studies have demonstrated its ability to recapitulate certain features of the developing human brain. By revealing phenotypes at the level of molecules, cells, and networks (Figure 2), these organoid models may be useful for studying disease mechanisms underlying neurodevelopmental disorders such as epilepsy. Nevertheless, these models also have their limitations. One of the challenges of organoid systems is the variability between organoids from independent experiments or from different cell lines, especially in whole-brain organoids.33,70 Moreover, organoids are frequently immature and resemble the developing brain only up to early-to-mid gestation. It has been difficult to recapitulate postnatal or adult stages because of the relative immaturity of the neurons and circuitry in the organoids. In addition, organoids lack specific neuronal subtypes which appear later during development, that is, parvalbumin and somatostatin interneurons; these cell types are relevant to accurately model disease phenotypes. Deep- and upper-layer neurons are present in organoids and are segregated spatially, but they do not show the clear 6-layer lamination found in vivo. Moreover, the formation of gyrus and sulcus has been observed after overexpression of PTEN, but the folding capacity in control organoids is limited. It is also important to note that organoids contain ventricular-like structures. Although these ventricular regions mimic some features of the human brain, organoids are not organized in specialized areas like the brain, making the overall organoid structure different from the human brain. In addition, organoids lack the vasculature, and the limited oxygenation in their core causes stress and cell death inside the organoids.71 Organoids also lack cells of non-neural origin, such as microglia, particularly in patterned organoids. Organoids have been very useful to study cellular phenotypes related to epilepsy. However, a key aspect of epilepsy, the development of seizures, and how electrophysiological properties of the brain can be recapitulated in organoids, is still an ongoing area of research. Overcoming these limitations will allow more accurate models of disease pathology. For example, the recent creation of blood–brain barrier organoids has opened up new avenues for characterizing phenotypes relevant to Alzheimer disease.72 In coming years, better organoid protocols, based on a better understanding of human brain development, will be critical for opening new avenues of research. We have no doubt that although the organoid field is still young, these exciting models will accelerate our knowledge of human brain development and lead to new strategies for treating epilepsy.
Figure 2.
Brain organoid applications. The use of different techniques to analyze brain organoids from molecular, cellular, and network levels will lead to a better understanding of brain development and how disruptions in this process may cause neurodevelopmental disorders such as epilepsy. The knowledge gain from these studies could be used as the basis for drug discovery with the ultimate goal of finding better treatment for patients with epilepsy.
Acknowledgments
Figure illustrations were constructed using BioRender and Adobe Illustrator. We are grateful for support from the Robert J. Kleberg Jr and Helen C. Kleberg Foundation and the Semmes Foundation, Inc.
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by grants from the National Institute of Health (R01NS093992, R01NS113516, R01NS089770, and R21AG066496 to J.H.) and from postdoctoral fellowships from the Lennox-Gastaut Syndrome Foundation and the American Epilepsy Society to V. N.-E.
ORCID iD: Jenny Hsieh  https://orcid.org/0000-0003-0214-0771
https://orcid.org/0000-0003-0214-0771
References
- 1. Stafstrom CE, Carmant L. Seizures and epilepsy: an overview for neuroscientists. Cold Spring Harb Perspect Med. 2015;5(6):a022426 doi:10.1101/cshperspect.a022426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Manford M. Recent advances in epilepsy. J Neurol. 2017;264(4):1811–1824. doi:10.1007/s00415-017-8394-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fisher RS, van Emde Boas W, Blume W, et al. Epileptic seizures and epilepsy: definitions proposed by the international league against epilepsy (ILAE) and the international bureau for epilepsy (IBE). Epilepsia. 2005;46(4):470–472. doi:10.1111/j.0013-9580.2005.66104.x [DOI] [PubMed] [Google Scholar]
- 4. Zack MM, Kobau R. National and state estimates of the numbers of adults and children with active epilepsy - United States, 2015. MMWR Morb Mortal Wkly Rep. 2017;66(31):821–825. doi:10.15585/mmwr.mm6631a1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fink JJ, Robinson TM, Germain ND, et al. Disrupted neuronal maturation in Angelman syndrome-derived induced pluripotent stem cells. Nat Commun. 2017;8:15038 doi:10.1038/ncomms15038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Higurashi N, Uchida T, Lossin C, et al. A human Dravet syndrome model from patient induced pluripotent stem cells. Mol Brain. 2013;6:19 doi:10.1186/1756-6606-6-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Thomson JA, Itskovitz-Eldor J, Shapiro SS, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282(5391):1145–1147. doi:10.1126/science.282.5391.1145 [DOI] [PubMed] [Google Scholar]
- 8. Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–676. doi:S0092-8674(06)00976-7 [pii]10.1016/j.cell.2006.07.024 [DOI] [PubMed] [Google Scholar]
- 9. Maroof AM, Keros S, Tyson JA, et al. Directed differentiation and functional maturation of cortical interneurons from human embryonic stem cells. Cell Stem Cell. 2013;12(5):559–572. doi:10.1016/j.stem.2013.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chailangkarn T, Trujillo CA, Freitas BC, et al. A human neurodevelopmental model for Williams syndrome. Nature. 2016;536(7616):338–343. doi:10.1038/nature19067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shi Y, Kirwan P, Livesey FJ. Directed differentiation of human pluripotent stem cells to cerebral cortex neurons and neural networks. Nat Protoc. 2012;7(10):1836–1846. doi:10.1038/nprot.2012.116 [DOI] [PubMed] [Google Scholar]
- 12. Lybrand ZR, Goswami S, Hsieh J. Stem cells: a path towards improved epilepsy therapies. Neuropharmacology. 2020;168:107781 doi:10.1016/j.neuropharm.2019.107781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Niu W, Parent JM. Modeling genetic epilepsies in a dish. Dev Dyn. 2020;249(1):56–75. doi:10.1002/dvdy.79 [DOI] [PubMed] [Google Scholar]
- 14. Ben Jehuda R, Shemer Y, Binah O. Genome editing in induced pluripotent stem cells using CRISPR/Cas9. Stem Cell Rev Rep. 2018;14(3):323–336. doi:10.1007/s12015-018-9811-3 [DOI] [PubMed] [Google Scholar]
- 15. Kelava I, Lancaster MA. Stem cell models of human brain development. Cell Stem Cell. 2016;18(6):736–748. doi:10.1016/j.stem.2016.05.022 [DOI] [PubMed] [Google Scholar]
- 16. Brown J, Quadrato G, Arlotta P. Studying the brain in a dish: 3d cell culture models of human brain development and disease. Curr Top Dev Biol. 2018;129:99–122. doi:10.1016/bs.ctdb.2018.03.002 [DOI] [PubMed] [Google Scholar]
- 17. Di Lullo E, Kriegstein AR. The use of brain organoids to investigate neural development and disease. Nat Rev Neurosci. 2017;18(10):573–584. doi:10.1038/nrn.2017.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Defelipe J. The evolution of the brain, the human nature of cortical circuits, and intellectual creativity. Front Neuroanat. 2011;5:29.doi:10.3389/fnana.2011.00029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Noctor SC, Flint AC, Weissman TA, Dammerman RS, Kriegstein AR. Neurons derived from radial glial cells establish radial units in neocortex. Nature. 2001;409(6821):714–720. doi:10.1038/35055553 [DOI] [PubMed] [Google Scholar]
- 20. Kwan KY, Sestan N, Anton ES. Transcriptional co-regulation of neuronal migration and laminar identity in the neocortex. Development. 2012;139(9):1535–1546. doi:10.1242/dev.069963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hansen DV, Lui JH, Parker PR, Kriegstein AR. Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature. 2010;464:554–561, doi:10.1038/nature08845 [DOI] [PubMed] [Google Scholar]
- 22. Cadwell CR, Bhaduri A, Mostajo-Radji MA, Keefe MG, Nowakowski TJ. Development and arealization of the cerebral cortex. Neuron. 2019;103(6):980–1004. doi:10.1016/j.neuron.2019.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wong FK, Bercsenyi K, Sreenivasan V, Portales A, Fernandez-Otero M, Marin O. Pyramidal cell regulation of interneuron survival sculpts cortical networks. Nature. 2018;557(3):668–673, doi:10.1038/s41586-018-0139-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mi D, Li Z, Lim L, et al. Early emergence of cortical interneuron diversity in the mouse embryo. Science. 2018;360(6384):81–85. doi:10.1126/science.aar6821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Al-Jaberi N, Lindsay S, Sarma S, Bayatti N, Clowry GJ. The early fetal development of human neocortical GABAergic interneurons. Cereb Cortex. 2015;25(3):631–645. doi:10.1093/cercor/bht254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hansen DV, Lui JH, Flandin P, et al. Non-epithelial stem cells and cortical interneuron production in the human ganglionic eminences. Nat Neurosci. 2013;16(11):1576–1587. doi:10.1038/nn.3541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Letinic K, Zoncu R, Rakic P. Origin of GABAergic neurons in the human neocortex. Nature. 2002;417(6889):645–649. doi:10.1038/nature00779 [DOI] [PubMed] [Google Scholar]
- 28. Zhong S, Zhang S, Fan X, et al. A single-cell RNA-seq survey of the developmental landscape of the human prefrontal cortex. Nature. 2018;555:524–528. doi:10.1038/nature25980 [DOI] [PubMed] [Google Scholar]
- 29. Lim L, Mi D, Llorca A, Marín O. Development and functional diversification of cortical interneurons. Neuron. 2018;100(2):294–313. doi:10.1016/j.neuron.2018.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Boldog E, Bakken TE, Hodge RD, et al. Transcriptomic and morphophysiological evidence for a specialized human cortical GABAergic cell type. Nat Neurosci. 2018;21(9):1185–1195. doi:10.1038/s41593-018-0205-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Andrews MG, Nowakowski TJ. Human brain development through the lens of cerebral organoid models. Brain Res. 2019;1725:146470 doi:10.1016/j.brainres.2019.146470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lancaster MA, Renner M, Martin CA, et al. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501(7467):373–379. doi:10.1038/nature12517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Quadrato G, Nguyen T, Macosko EZ, et al. Cell diversity and network dynamics in photosensitive human brain organoids. Nature. 2017;545(7652):48–53. doi:10.1038/nature22047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kadoshima T, Sakaguchi H, Nakano T, et al. Self-organization of axial polarity, inside-out layer pattern, and species-specific progenitor dynamics in human ES cell-derived neocortex. Proc Natl Acad Sci U S A. 2013;110(50):20284–20289. doi:10.1073/pnas.1315710110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Qian X, Nguyen HN, Song MM, et al. Brain-region-specific organoids using mini-bioreactors for modeling ZIKV exposure. Cell. 2016;165(5):1238–1254. doi:10.1016/j.cell.2016.04.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pasca AM, Sloan SA, Clarke LE, et al. Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat Methods. 2015;12(7):671–678. doi:10.1038/nmeth.3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bagley JA, Reumann D, Bian S, Levi-Strauss J, Knoblich JA. Fused cerebral organoids model interactions between brain regions. Nat Methods. 2017;14(7):743–751. doi:10.1038/nmeth.4304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Birey F, Andersen J, Makinson CD, et al. Assembly of functionally integrated human forebrain spheroids. Nature. 2017;545(7652):54–59. doi:10.1038/nature22330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Xiang Y, Tanaka Y, Patterson B, et al. Fusion of regionally specified HPSC-derived organoids models human brain development and interneuron migration. Cell Stem Cell. 2017;21(3):383–398.e7. doi:10.1016/j.stem.2017.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Trujillo CA, Gao R, Negraes PD, et al. Complex oscillatory waves emerging from cortical organoids model early human brain network development. Cell Stem Cell. 2019;25(4):558–569.e7. doi:10.1016/j.stem.2019.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Camp JG, Badsha F, Florio M, et al. Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. Proc Natl Acad Sci U S A. 2015;112(51):15672–15677. doi:10.1073/pnas.1520760112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. European Chromosome 16 Tuberous Sclerosis Consortium. Identification and characterization of the tuberous sclerosis gene on chromosome 16. Cell. 1993;75(7):1305–1315. doi:10.1016/0092-8674(93)90618-z [DOI] [PubMed] [Google Scholar]
- 43. van Slegtenhorst M, De Hoogt R, Hermans C, et al. Identification of the tuberous sclerosis gene TSC1 on chromosome 9q34. Science. 1997;277(5327):805–808. doi:10.1126/science.277.5327.805 [DOI] [PubMed] [Google Scholar]
- 44. Curatolo P, Moavero R, De Vries PJ. Neurological and neuropsychiatric aspects of tuberous sclerosis complex. Lancet Neurol. 2015;14(7):733–745. doi:10.1016/S1474-4422(15)00069 -1 [DOI] [PubMed] [Google Scholar]
- 45. Blair JD, Bateup HS. New frontiers in modeling tuberous sclerosis with human stem cell-derived neurons and brain organoids. Dev Dyn. 2020;249(1):46–55. doi:10.1002/dvdy.60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Blair JD, Hockemeyer D, Bateup HS. Genetically engineered human cortical spheroid models of tuberous sclerosis. Nat Med. 2018;24(10):1568–1578. doi:10.1038/s41591-018-0139-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Eichmüller OL, Corsini NS, Vértesy Á, et al. Cerebral organoid model reveals excessive proliferation of human caudal late interneuron progenitors in tuberous sclerosis complex. Biorxiv. 2020. doi:10.1101/2020.02.27.967802 [Google Scholar]
- 48. Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23(2):185–188. doi:10.1038/13810 [DOI] [PubMed] [Google Scholar]
- 49. Neul JL, Zoghbi HY. Rett syndrome: a prototypical neurodevelopmental disorder. Neuroscientist. 2004;10(2):118–128. doi:10.1177/1073858403260995 [DOI] [PubMed] [Google Scholar]
- 50. Mellios N, Feldman DA, Sheridan SD, et al. MeCP2-regulated miRNAs control early human neurogenesis through differential effects on ERK and AKT signaling. Mol Psychiatry. 2018;23(4):1051–1065. doi:10.1038/mp.2017.86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Cardoso C, Boys A, Parrini E, et al. Periventricular heterotopia, mental retardation, and epilepsy associated with 5q14.3-q15 deletion. Neurology. 2009;72(9):784–792. doi:10.1212/01.wnl.0000336339.08878.2d [DOI] [PubMed] [Google Scholar]
- 52. Klaus J, Kanton S, Kyrousi C, et al. Altered neuronal migratory trajectories in human cerebral organoids derived from individuals with neuronal heterotopia. Nat Med. 2019;25(4):561–568. doi:10.1038/s41591-019-0371-0 [DOI] [PubMed] [Google Scholar]
- 53. Buchsbaum IY, Kielkowski P, Giorgio G, et al. ECE2 regulates neurogenesis and neuronal migration during human cortical development. EMBO Rep. 2020;21(5):e48204 doi:10.15252/embr.201948204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. O’Neill AC, Kyrousi C, Klaus J, et al. A primate-specific isoform of PLEKHG6 regulates neurogenesis and neuronal migration. Cell Rep. 2018;25(10):2729–2741. e6. doi:10.1016/j.celrep.2018.11.029 [DOI] [PubMed] [Google Scholar]
- 55. Buiting K, Williams C, Horsthemke B. Angelman syndrome - insights into a rare neurogenetic disorder. Nat Rev Neurol. 2016;12(10):584–593. doi:10.1038/nrneurol.2016.133 [DOI] [PubMed] [Google Scholar]
- 56. Mabb AM, Judson MC, Zylka MJ, Philpot BD. Angelman syndrome: insights into genomic imprinting and neurodevelopmental phenotypes. Trends Neurosci. 2011;34(6):293–303. doi:10.1016/j.tins.2011.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sun AX, Yuan Q, Fukuda M, et al. Potassium channel dysfunction in human neuronal models of Angelman syndrome. Science. 2019;366(6472):1486–1492. doi:10.1126/science.aav5386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Dobyns WB, Stratton RF, Parke JT, Greenberg F, Nussbaum RL, Ledbetter DH. Miller-Dieker syndrome: lissencephaly and monosomy 17p. J Pediatr. 1983;102(4):552–558. doi:10.1016/s0022-3476(83)80183-8 [DOI] [PubMed] [Google Scholar]
- 59. Bershteyn M, Nowakowski TJ, Pollen AA, et al. Human iPSC-derived cerebral organoids model cellular features of lissencephaly and reveal prolonged mitosis of outer radial glia. Cell Stem Cell. 2017;20(4):435–449.e4. doi:10.1016/j.stem.2016.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lefremova V, Manikakis G, Krefft O, et al. An organoid-based model of cortical development identifies non-cell-autonomous defects in WNT signaling contributing to Miller-Dieker syndrome. Cell Rep. 2017;19(1):50–59. doi:10.1016/j.celrep.2017.03.047 [DOI] [PubMed] [Google Scholar]
- 61. Splawski I, Timothy KW, Sharpe LM, et al. Ca(V)1.2 calcium channel dysfunction causes a multisystem disorder including arrhythmia and autism. Cell. 2004;119(1):19–31. doi:10.1016/j.cell.2004.09.011 [DOI] [PubMed] [Google Scholar]
- 62. Hengel H, Bosso-Lefevre C, Grady G, et al. Loss-of-function mutations in UDP-Glucose 6-Dehydrogenase cause recessive developmental epileptic encephalopathy. Nat Commun. 2020;11(1):595 doi:10.1038/s41467-020-14360-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Berkovic SF, Andermann F, Carpenter S, Wolfe LS. Progressive myoclonus epilepsies: specific causes and diagnosis. N Engl J Med. 1986;315(5):296–305. doi:10.1056/NEJM198607313150506 [DOI] [PubMed] [Google Scholar]
- 64. Canafoglia L, Gennaro E, Capovilla G, et al. Electroclinical presentation and genotype-phenotype relationships in patients with Unverricht-Lundborg disease carrying compound heterozygous CSTB point and indel mutations. Epilepsia. 2012;53(1):2120–2127. doi:10.1111/j.1528-1167.2012.03718.x [DOI] [PubMed] [Google Scholar]
- 65. Di Matteo F, Pipicelli F, Kyrousi C, et al. Cystatin B is essential for proliferation and interneuron migration in individuals with EPM1 epilepsy. EMBO Mol Med. 2020;12(6):e11419 doi:10.15252/emmm.201911419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Penna E, Cerciello A, Chambery A, et al. Cystatin B involvement in synapse physiology of rodent brains and human cerebral organoids. Front Mol Neurosci. 2019;12:195 doi:10.3389/fnmol.2019.00195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Garcez PP, Loiola EC, Madeiro da Costa R, et al. Zika virus impairs growth in human neurospheres and brain organoids. Science. 2016;352(6287):816–818. doi:10.1126/science.aaf6116 [DOI] [PubMed] [Google Scholar]
- 68. Watanabe M, Buth JE, Vishlaghi N, et al. Self-organized cerebral organoids with human-specific features predict effective drugs to combat zika virus infection. Cell Rep 2017;21(2):517–532. doi:10.1016/j.celrep.2017.09.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Li Y, Muffat J, Omer A, et al. Induction of expansion and folding in human cerebral organoids. Cell Stem Cell. 2017;20(3):385–396.e3. doi:10.1016/j.stem.2016.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Velasco S, Kedaigle AJ, Simmons SK, et al. Individual brain organoids reproducibly form cell diversity of the human cerebral cortex. Nature. 2019;570(7762):523–527. doi:10.1038/s41586-019-1289-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Bhaduri A, Andrews MG, Mancia Leon W, et al. Cell stress in cortical organoids impairs molecular subtype specification. Nature. 2020;578(7793):142–148. doi:10.1038/s41586-020-1962-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Blanchard JW, Bula M, Davila-Velderrain J, et al. Reconstruction of the human blood-brain barrier in vitro reveals a pathogenic mechanism of APOE4 in pericytes. Nat Med. 2020;26:952–963. doi:10.1038/s41591-020-0886-4 [DOI] [PMC free article] [PubMed] [Google Scholar]