SUMMARY
Animal embryogenesis requires a precise coordination between morphogenesis and cell fate specification. During mesoderm induction, mesodermal fate acquisition is tightly coordinated with the morphogenetic process of epithelial-to-mesenchymal transition (EMT). In zebrafish, cells exist transiently in a partial EMT state during mesoderm induction. Here, we show that cells expressing the transcription factor Sox2 are held in the partial EMT state, stopping them from completing the EMT and joining the mesoderm. This is critical for preventing the formation of ectopic neural tissue. The mechanism involves synergy between Sox2 and the mesoderm-inducing canonical Wnt signaling pathway. When Wnt signaling is inhibited in Sox2-expressing cells trapped in the partial EMT, cells exit into the mesodermal territory but form an ectopic spinal cord instead of mesoderm. Our work identifies a critical developmental checkpoint that ensures that morphogenetic movements establishing the mesodermal germ layer are accompanied by robust mesodermal cell fate acquisition.
Graphical Abstract
In Brief
During embryonic development, the right tissue types must form in the proper location. Kinney et al. show that a developmental checkpoint functions during mesoderm induction, ensuring that Sox2-expressing cells do not migrate into the mesoderm. This checkpoint is critical for preventing ectopic spinal cord from forming in place of mesoderm.
INTRODUCTION
Epithelial-to-mesenchymal transition (EMT) is the process in which epithelial cells lose their adhesion to neighboring cells and adopt a mesenchymal migratory phenotype. This process was first described by observing chick mesoderm formation (Hay, 1995), and was later found to occur in many other normal processes, as well as disease states such as cancer metastasis (Nakaya and Sheng, 2013; Nieto, 2013). More recently, metastable partial (also referred to as intermediate) EMT states have been observed, in which cells maintain a transitional state that shares characteristics of both epithelial and mesenchymal cells (Ye and Weinberg, 2015; Li and Kang, 2016; Nieto et al., 2016). Partial EMT states are thought to be particularly important in the process of solid tumor metastasis, in which metastable partial EMT states exhibit increased migratory and invasive capacity, as well as more stem-cell like characteristics (Campbell, 2018; Aiello and Kang, 2019). Despite this, it is unclear what purpose, if any, metastable partial EMT states play during normal development.
Vertebrate embryos contain neuromesodermal progenitors (NMPs) (Kimelman, 2016; Martin, 2016), which make a binary decision to become spinal cord cells, or mesoderm that will primarily form the somites, with some contribution to the endothelium (Tzouanacou et al., 2009; Martin and Kimelman, 2012; Row et al., 2018). During mesoderm induction, NMPs undergo an EMT, which is tightly associated with the acquisition of mesodermal fate (Goto et al., 2017). This occurs in a two-step process, in which Wnt signaling initiates the EMT, and fibroblast growth factor (FGF) signaling promotes EMT completion by activating the expression of the transcription factors tbx16 and msgn1 (Goto et al., 2017). Zebrafish embryos deficient in the t-box transcription factor tbx16 (originally called spadetail) have a large accumulation of cells at the posterior-most structure of the embryo called the tailbud (Kimmel et al., 1989; Griffin et al., 1998), a phenotype caused by the inability of the NMPs to complete their EMT and join the developing paraxial mesoderm (Row et al., 2011; Manning and Kimelman, 2015). This phenotype is very similar to mouse embryos lacking function of the related t-box transcription factor Tbx6, which also have an enlarged tailbud and deficit in paraxial mesoderm (Chapman and Papaioannou, 1998). Cells in the partial EMT state exhibit increased adhesiveness compared to the fully mesenchymal state, and cells lacking tbx16 maintain a metastable partial EMT state until tbx16 is activated, after which they complete the EMT (Row et al., 2011).
NMPs are characterized by co-expression of the two transcription factors, sox2, which promotes spinal cord fate, and brachyury (tbxta and tbxtb in zebrafish), which specifies mesoderm in part through the activation of canonical Wnt signaling (Martin and Kimelman, 2008, 2010, 2012; Takemoto et al., 2011; Bouldin et al., 2015; Koch et al., 2017). Here, we show that a critical role of Tbx16 is to repress sox2 transcription in the partial EMT state as cells become mesoderm. Sox2 activation alone is sufficient to recapitulate the Tbx16 loss-of-function phenotype, where cells are prevented from exiting the tailbud and remain trapped in an undifferentiated partial EMT state. This acts as a developmental checkpoint, since cells with sox2 expression in mesodermal territories outside the tailbud will become neurons. Thus, the checkpoint ensures coordination of morphogenesis with proper cell fate acquisition to prevent ectopic neural formation. Our work highlights an essential normal function of a partial EMT state during development and provides insight into how the partial EMT state in cancer can be targeted by inhibiting developmental checkpoints.
RESULTS
Sox2 Activation Is Sufficient to Induce Neural Differentiation in a Context-Dependent Manner
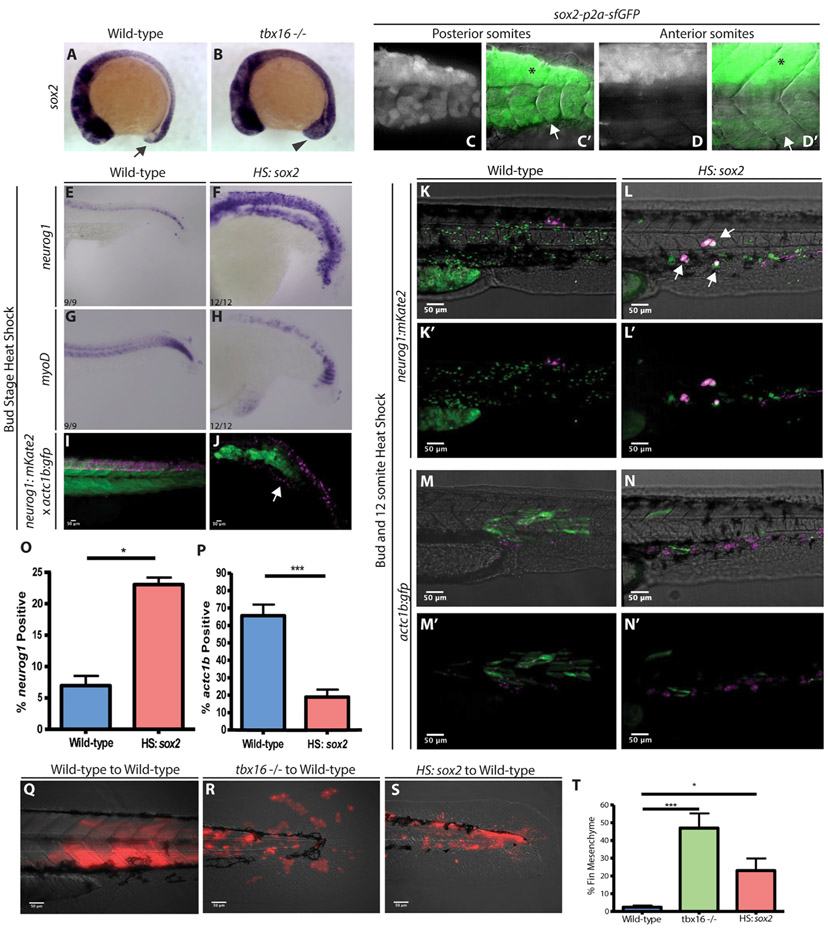
NMPs express the neural-inducing transcription factor sox2, which is downregulated as NMPs become mesoderm (Figures 1A, arrow, and 1C-1D’) (Delfino-Machín et al., 2005; Takemoto et al., 2011; Martin and Kimelman, 2012; Bouldin et al., 2015). In tbx16 mutants, sox2 expression is maintained and expanded in tailbud NMPs (Figure 1B, arrowhead). A superfolder GFP (sfGFP) sox2 transcriptional reporter shows perdurance of sfGFP in recently formed somites (Figures 1C and 1C’), which is eventually lost in older, earlier formed somites, but continues to be expressed strongly in the spinal cord (Figures 1D and 1D’) (Shin et al., 2014). This indicates that somites originate from a sox2+ source of cells. To determine the role that Sox2 plays in NMPs, we used a heat-shock-inducible sox2 transgenic line (HS:sox2) (Row et al., 2016). When sox2 was activated throughout the embryo at the end of gastrulation (bud stage) and analyzed at 24 h post-fertilization (hpf), ectopic expression of the neural marker neurog1 was observed in mesodermal territories (Figures 1E and 1F), and there was a corresponding decrease in the skeletal muscle marker myod (Figures 1G and 1H). Activation of sox2 at the bud stage in the background of reporter transgenes for skeletal muscle (actc1b:gfp) (Higashijima et al., 1997) and neurons (neurog1:mkate2) resulted in a loss of differentiated muscle and the presence of ectopic neurons in mesodermal territories (Figures 1I and 1J). To test whether the effect of Sox2 is cell autonomous, we transplanted cells transgenic for HS:sox2 and neurog1:mkate2 (Figures 1K-1L’) or HS:sox2 and actc1b:gfp (Figures 1M-1N’) into the ventral margin of shield stage wild-type embryos. The activation of sox2 by heat shock in transplanted cells caused a significant cell-autonomous induction of more neural cells, including in ectopic locations, at the expense of skeletal muscle cells (Figures 1K-1P). Intriguingly, however, the induction of ectopic neural fate by sox2 in both the whole embryo and transplant conditions was localized to more anterior regions of the embryo, in areas where somites normally form. In addition, while 72.54% of control transplanted cells differentiated into either muscle or neurons, only 42% of sox2-expressing cells differentiated into these cell types (Figures 1O and 1P). Upon further examination, a large proportion of sox2-expressing cells gave rise to fin mesenchyme (easily identified by their stellate morphology and by position between the epithelial fin folds), a phenotype that is also observed in transplanted tbx16 mutant cells, which are trapped in the partial EMT state (Figures 1Q-1T) (Ho and Kane, 1990; Row et al., 2011).
Figure 1. sox2 Activation Causes an Increase of Neural Progenitors and a Decrease in Presomitic Mesoderm.
(A and B)12-somite-stage embryos express sox2 in NMPs (A, arrow) and sox2 expression is expanded in tbx16 mutants (B, arrowhead).
(C and D) A sox2 reporter line shows perdurance of sfGFP in the most recently formed somites at 24 hpf (C, C’, arrow, spinal cord expression in dorsal region labeled with asterisk), which is absent in more anterior somites (D, D’, arrow, spinal cord expression labeled with asterisk).
(E–H) Whole-mount in situ hybridization visualizing neurog1 (neural) (E and F) or myod (skeletal muscle) (G and H) in wild type (E and G) and HS:sox2 embryos (F and H). All of the embryos for in situ hybridization were heat shocked at the bud stage at 40°C for 30 min and fixed at 24 hpf.
(I and J) Transgenic embryos with the neurog1:mKate2 and actc1b:gfp reporters show ectopic neural expansion (arrow) and muscle loss in HS:sox2 (J) embryos compared to wild type (I). Live-imaged transgenic embryos were heat shocked at the bud stage at 40°C for 30 min and imaged at 36 hpf.
(K–P) Embryos with the neurog1:mkate (K–L’) or the actc1b:gfp (M–N’) reporter were injected with NLS-KikGR mRNA, and cells from these embryos were transplanted into the ventral margin of wild-type host embryos. Donor cells with the HS:sox2 transgene exhibited an increase in the percentage of neurog1:mkate+ cells that also appeared in ectopic locations outside the spinal cord domain (arrows, L, L’, compared to K, K’ and quantified in O; 1,177 wild-type donor cells were counted in 8 host embryos, and 2,051 HS:sox2 donor cells were counted from 10 host embryos; statistics were performed using an unpaired t test, *p = 0.0105) and a decrease in the percentage of actc1b:gfp+ cells (N, N’ compared to M, M’ and quantified in P; 1,307 wild-type donor cells were counted in 8 host embryos, and 971 HS:sox2 donor cells were counted from 5 host embryos; statistics were performed using an unpaired t test, ***p = 0.0003). The NLS-KikGR protein was photoconverted to red fluorescence in (M)–(N’).
(Q–T) Wild-type, tbx16 mutant, and HS:sox2 embryos were injected with rhodamine dextran and transplanted into the ventral margin of shield stage wild-type host embryos. The percentage of transplanted cell contribution to fin mesenchyme is quantified in (T) (2,129 wild-type donor cells were counted in 6 host embryos; 2,714 tbx6−/− donor cells were counted in 6 host embryos, ***p = 0.0006; and 2,347 HS:sox2 donor cells were counted from 9 host embryos, *p = 0.0322). All transplants were heat shocked at the bud stage and 12-somites at 39°C for 30 min.
Sustained sox2 Expression in Mesoderm-Fated NMPs Traps Them in a Partial EMT State
Tbx16 is necessary and sufficient for sox2 repression (Bouldin et al., 2015), and tbx16 loss-of-function and sox2 gain-of-function both bias transplanted cells located in the tailbud of host embryos toward a fin mesenchyme fate (Figures 1Q-1T). We hypothesize, based on these observations, that the maintenance of sox2 expression in tbx16 mutant cells is responsible for the cell migration defect that prevents cells from exiting the tailbud. We transplanted HS:sox2 cells into the ventral margin of wild-type host embryos. The activation of sox2 expression at bud and 12-somite stages prevented transplanted cells from exiting the tailbud into the mesodermal territory, and the majority of cells are found at the posterior end of the host embryo (Figures 2A-2C). To better understand the migratory dynamics of the sox2-expressing cells in the tailbud, we labeled embryos with a nuclear localized kikume (NLS-kikRG), which can be photoconverted from green to red, by injecting in vitro transcribed mRNA. Small groups of cells in the NMP region were photoconverted and time-lapse imaged for 300 min. Wild-type cells move ventrally in a directed fashion (Figures 2D, 2F, and 2H-2J). While sox2-expressing cells move faster than wild-type cells, their overall displacement is reduced, due to significantly reduced migratory track straightness (Figures 2E and 2G-2J). The migratory activity but lack of directed migration has been observed as cells transition to mesoderm and defines the partial EMT state during zebrafish mesoderm induction (Row et al., 2011; Lawton et al., 2013; Manning and Kimelman, 2015; McMillen and Holley, 2015), suggesting that sox2-expressing cells are trapped in a partial EMT. The phenocopy of tbx16 loss of function that is caused by sox2 gain of function is not due to a loss of tbx16 expression. The expression of tbx16 was monitored by in situ hybridization chain reaction (HCR) in both HS:sox2 transgenic and sox2 mutant embryos (Figures 2K-2L’ and 2M-2N’) (Choi et al., 2018; Gou et al., 2018a, 2018b). Quantification of expression revealed that gain of sox2 function exerted no change on the level of tbx16 expression, while sox2 loss of function resulted in a small but significant increase in tbx16 expression (Figures 2O, 2P, and S1). However, gain of sox2 function caused a statistically significant increase on the spatial domain of tbx16 expression, which is consistent with tbx16+ cells being trapped in the tailbud and unable to exit and shut off tbx16 expression, whereas no change in spatial domain was observed in sox2 mutants (Figures 2O, 2P, and S1).
Figure 2. sox2 Levels Control the Rate of NMP Exit into the Mesoderm.
(A and B) Wild-type and HS:sox2 embryos were injected with fluorescein dextran, and the cells from these embryos were transplanted into the ventral margin of shield stage wild-type host embryos (A and B, respectively). Transplants were heat shocked at bud stage and 12-somites at 39°C for 30 min and imaged at 36 hpf.
(C) Quantification of tailbud exit was measured as a line scan of compound fluorescence from anterior to posterior, comparing wild-type transplanted cells (blue, N = 10) with HS:sox2 transplanted cells (red, N = 6). Dotted lines indicate 90% confidence.
(D–J) Wild-type (D–D”’) or HS:sox2 (E–E”’) embryos with ubiquitous NLS-KikGR expression were photoconverted in the NMP region and time-lapse imaged for 300 min. Migratory tracks of photoconverted wild-type and HS:sox2 nuclei were quantified (F, 281 cells were tracked in 5 embryos; G, 200 cells were tracked in 3 embryos, ***p < 0.0001), revealing that displacement (H) and track straightness (J) were reduced in HS:sox2 embryos, whereas average track speed was increased (I).
(K–P) HCR analysis of tbx16 expression in HS:sox2 embryos heat shocked at the 5-somite stage and fixed at the 20-somite stage shows that exogenous Sox2 does not affect the level of tbx16 expression (p = 0.122, wild type N = 6, HS:sox2 N = 6), but increases the area of tbx16 expression (p = 0.0044, wild type N = 6, HS:sox2 N = 6). The expression of tbx16 is upregulated in sox2 mutants but the expression area is unaffected (M–P, area p = 0.199, wild type N = 6, sox2 mutant N = 7, mean gray value [MGV] p = 0.017, wild type N = 6, sox2 mutant N = 7).
(Q–CC) MF-20 antibody labeling of wild-type (Q and S) and sox2 homozygous mutant (R and T) embryos showed that posterior somites are smaller in sox2 mutants. Somitic nuclei were quantified (green dots in U and V are spots generated by Imaris representing nuclei), revealing that posteriorsomites in sox2 mutants have significantly fewer cells than wild-type somites (U–Y, and W p = 0.4721, X p = 0.3208, Y *p = 0.0145). HCR analysis of tbxta expression in sox2 mutants shows that the levels of tbxta throughout the entire embryo are unchanged (p = 0.697, wild type N = 6, sox2 mutant N = 9), but expression specifically in the NMPs is significantly downregulated (Z–CC, ***p ≤ 0.001, wild type N = 6, sox2 mutant N = 9). n.s., not significant.
See Figure S1 for additional information about HCR analysis.
Our results indicate that Sox2 functions in mesoderm-fated NMPs to maintain them in an undifferentiated state within the tailbud. We next tested whether loss of sox2 function would prevent the maintenance of the undifferentiated mesoderm-fated NMPs, resulting in somite defects. To determine whether sox2 function affects the normal formation of somites from NMPs, we analyzed somite development in sox2 mutants. Somites were visualized with a muscle-specific antibody (MF20, anti-myosin heavy chain), which indicated that the posterior somites appeared smaller (Figures 2Q-2T). Total nuclei counts in somites 25–28 revealed that posterior somites contain significantly fewer cells than wild-type siblings (Figures 2U-2Y), suggesting a failure to maintain the progenitor cells. In zebrafish, the t-box transcription factor tbxta (previously called ntl, ntla, and ta) is expressed in the NMPs and is required for the maintenance of mesoderm progenitors in the tailbud, and the loss of tbxta causes a loss of posterior somite tissues (Halpern et al., 1993; Schulte-Merker et al., 1994; Martin and Kimelman, 2008). We examined tbxta expression in sox2 mutant tailbuds and found a loss of expression specifically in the NMPs but not the tailbud as a whole, which includes the notochord and notochord progenitor tbxta expression domains (Figure 2Z-2CC). These results show that Sox2 maintains mesoderm-fated NMPs at least in part through the transcriptional activation of tbxta.
sox2 Loss of Function Rescues tbx16 Loss of Function
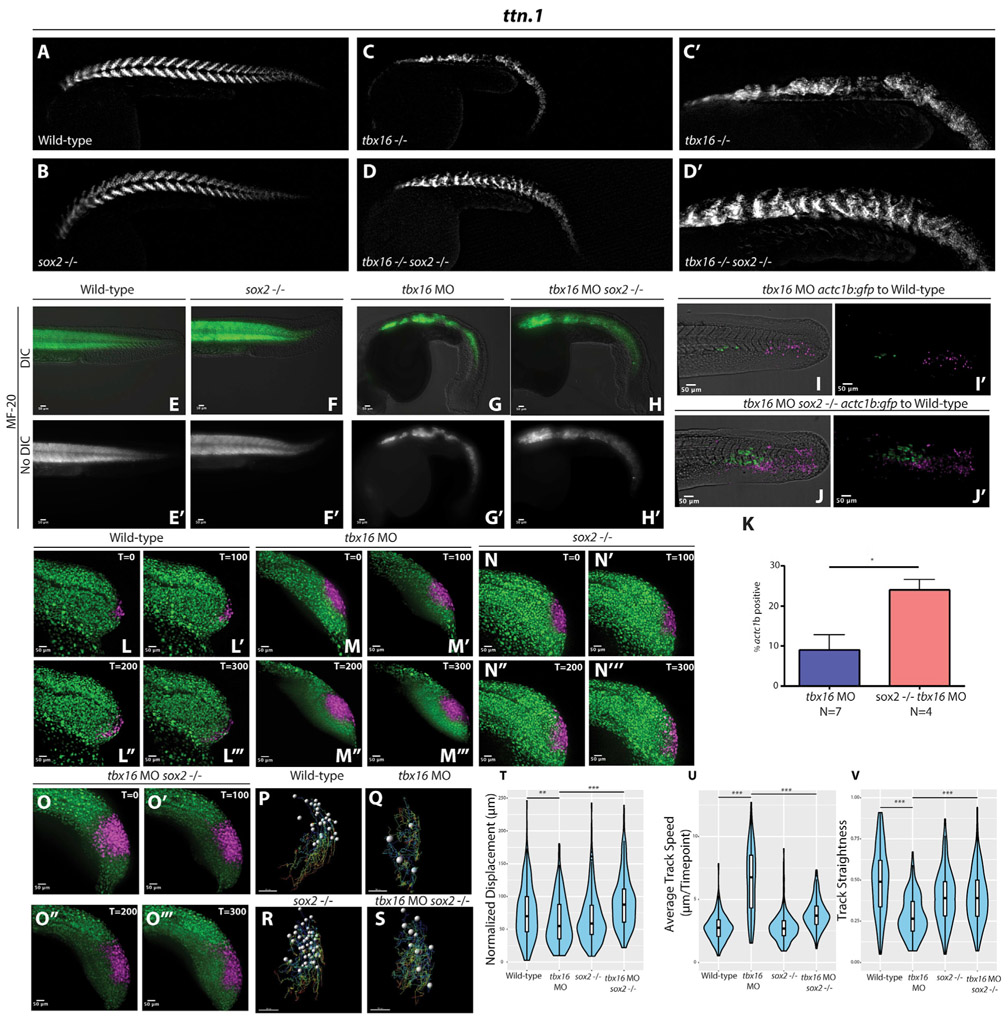
Sox2 gain of function or tbx16 loss of function in mesoderm-fated NMPs causes them to be trapped in a partial EMT state, and Tbx16 normally acts to repress sox2 expression (Figures 1A and 1B) (Bouldin et al., 2015). To determine whether sox2 is a critical target of Tbx16 accounting for the tbx16 mutant phenotype, we generated tbx16 and sox2 double mutants (Figures 3A-3D’). HCR was used to stain these embryos with a muscle marker (ttn.1), revealing that muscle formation is rescued in double mutants compared to the tbx16 single mutant (Figures 3D and 3D’ compared to 3C and 3C’). We also found that the sox2 mutation is able to rescue tbx16 morphant muscle formation in both whole embryo (Figures 3E-3H’) and transplant conditions (Figures 3I-3K), where tbx16 morpholinos were injected into embryos from a sox2+/− in-cross, and cells from these embryos were transplanted into wild-type host embryos. In addition, we performed cell tracking experiments in embryos injected with tbx16 MO, sox2−/− mutants, and sox2−/− mutants injected with tbx16 MO (Figures 3L-3S). Cells lacking tbx16 behave similarly to cells with a gain of sox2 function, including decreased track straightness and overall displacement, with an increase in track speed relative to wild-type cells (Figures 3T-3V). Cells lacking both tbx16 and sox2 function regain wild-type like behavior, including a significant rescue of displacement, track speed, and track straightness (Figures 3T-3V; the wild-type cell tracking data are the same used in Figure 2). These results indicate that sox2 is a critical target gene repressed by Tbx16. In the absence of tbx16, increased levels of sox2 cause cells to become trapped in a partial EMT state and prevent their exit into the mesodermal territory.
Figure 3. Loss of sox2 Function Rescues tbx16 Loss of Function.
(A–D) Wild type (A), sox2 mutant (B), tbx16 mutant (C and C’), or sox2 and tbx16 double mutants (D and D’) were stained for ttn.1 (skeletal muscle) by HCR, revealing a rescue of skeletal muscle in double mutants compared to tbx16 mutants.
(E–K) MF-20 labeling of wild-type, sox2−/−, tbx16 morphant, and dual sox2−/− tbx16 morphant embryos shows an increase in skeletal muscle in tbx16 morphant embryos when sox2 function is eliminated (E–H’, H, and H’ compared to G and G’), phenocopying the double mutants. Transplant experiments were performed by injecting rhodamine dextran and tbx16 MOs into embryos from a actc1b:gfp sox2+/− in-cross and transplanting cells into the ventral margin of wild-type host embryos. Donor cells with sox2 function and tbx16 loss of function showed a significantly smaller percentage of the total number of transplanted cells contributing to muscle compared to donor cells without sox2 or tbx16 function (I–K, 1,030 tbx16 morphant donor cells were counted from 7 host embryos, and 609 sox2−/− tbx16 morphant donor cells were counted from 4 host embryos, *p = 0.0082). Statistics were performed using an unpaired t test. N indicates the number of host embryos.
(L–O) Wild-type (L–L”’), tbx16 morphant (M–M”’), sox2−/− (N–N”’), or sox2−/− and tbx16 morphant (O–O”’) embryos with ubiquitous NLS-KikGR expression were photoconverted in the NMP region and time-lapse imaged for 300 min (the wild-type data presented here are the same wild-type data presented in Figure 2F).
(P–V) Migratory tracks of photoconverted nuclei were quantified (P–S, 281 wild-type cells were tracked from 5 embryos, 183 tbx16 morphant cells were tracked from 3 embryos, 210 sox2−/− cells were tracked from 3 embryos, and 218 sox2−/− tbx16 morphant cells were tracked from 3 embryos, **p = 0.0029, ***p < 0.0001), revealing that displacement (T), track speed (U), and track straightness (V) were significantly rescued toward wild-type levels in dual sox2 and tbx16 loss-of-function embryos compared to tbx16 morphants alone.
Checkpoint Activation Occurs through a Synergistic Interaction of Sox2 and Canonical Wnt Signaling
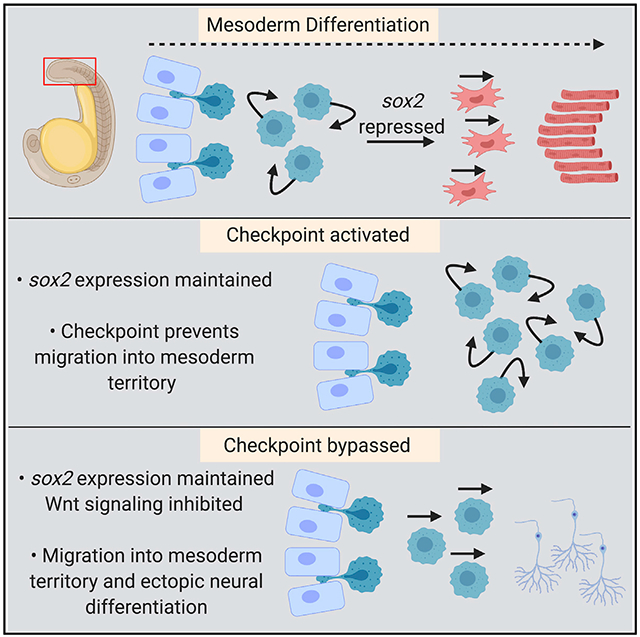
The expression of sox2 prevents mesoderm-fated NMPs from exiting the tailbud into the mesodermal territory. However, sox2 expression does not prevent the exit of NMPs from the tailbud into the spinal cord territory, suggesting that there is a local difference in the niche context of the tailbud that accounts for this differential activity of Sox2. We previously showed that in the absence of canonical Wnt signaling, NMPs sustain sox2 expression and join the spinal cord and not the mesoderm, whereas the activation of Wnt signaling using a constitutively active β-catenin transgene causes NMPs to join the mesoderm and not the spinal cord (Martin and Kimelman, 2012). These results suggest that the presence or absence of the canonical Wnt signaling pathway accounts for the context-dependent activity of sox2. To test this model, we performed transplant experiments with tbx16 morphant cells or HS:sox2 transgenic cells in the presence or absence of the HS:TCFΔC transgene, which cell autonomously inhibits canonical Wnt signaling (Martin and Kimelman, 2012). Transplanted wild-type cells contribute to various tissues throughout the body (Figure 4A). Cells lacking tbx16 fail to join the paraxial mesoderm and instead contribute predominantly to fin mesenchyme, as previously reported (Figures 4D-4D”) (Ho and Kane, 1990; Row et al., 2011). When sox2 or TCFΔC expression are activated in transplanted cells at the bud stage, fewer cells contribute to the paraxial mesoderm (Figures 4B and 4C). When Wnt signaling is inhibited in tbx16 morphant cells, cells can enter into the paraxial mesodermal territory, but rather than give rise to mesoderm, they form an ectopic spinal cord (Figures 4E-4E”). The ectopic spinal cords have the proper anatomical structure of a neural canal with motile cilia projecting into the canal (Videos S1, S2, and S3), as well as differentiated neurons sending axonal projections through the ectopic spinal cord (Figures 4G and 4H’). To determine whether this phenotype is due to sustained sox2 expression in tbx16 morphant cells, we performed transplants with cells with both the HS:sox2 and HS:TCFΔC transgenes. Combined heat-shock activation of sox2 and inhibition of Wnt signaling causes the same, yet more severe phenotype of an ectopic spinal cord in the mesodermal territory along the body axis (Figure 4F-4F”). The synergistic neural-inducing activity of sox2 activation and canonical Wnt signaling inhibition is also observed in whole embryos, where combined sox2 activation and Wnt inhibition induces the spinal cord broadly throughout the normal paraxial mesoderm domain (Figure S2). These results show that the checkpoint holding sox2-expressing cells in the partial EMT state is activated by the combined presence of sox2 and canonical Wnt signaling, and that the checkpoint can be bypassed by eliminating both Tbx16 and Sox2 function, causing the cells to exit and form mesoderm, or by eliminating Wnt signaling in sox2-expressing cells, which allows them to exit the tailbud to form an ectopic spinal cord (Figure 4I).
Figure 4. sox2 Activation in the Absence of Wnt Signaling Results in Ectopic Spinal Cords in Transplanted Cells.
(A) Wild-type-to-wild-type transplant (N = 16).
(B) HS:TCFΔC-to-wild-type transplant (N = 18).
(C) HS:sox2-to-wild-type transplant (N = 4).
(D–F) tbx16 MO-to-wild-type transplant (N = 35) (D–D”). (E–E”) HS.TCFΔC tbx16 MO-to-wild-type transplant (N = 43, 35 with ectopic spinal cords). (F–F”) HS: sox2 x HS.TCFΔC transplant (N = 17, all with ectopic spinal cords). All of the transplants were performed by injecting donor embryos with 2% fluorescein dextran (false colored magenta) and transferring donor cells to the margin of 30% epiboly wild-type host embryos. All of the transplants were heat shocked at 40°C for 30 min. The loss of tbx16 function causes donor cells that would normally form paraxial mesoderm to become fin mesenchyme (D’, blue arrowheads indicate the spinal cord; see also Video S1). Donor tbx16 morphant cells in which Wnt signaling has been inhibited can exit the tailbud into the paraxial mesoderm territory (E’, arrows), where they form an ectopic spinal cord with a neural canal (E”, arrowheads; see also Video S2). The same phenomenon occurs when sox2 is activated and Wnt signaling is inhibited, where transplanted cells leave the tailbud to form an ectopic spinal cord (F’, arrows) with a neural canal (F”, arrowheads; see also Video S3). (G–I) Ectopic spinal cords formed from the combined loss of tbx16 function and Wnt signaling have differentiated neurons (green) that form long axonal projections as revealed by the neurog1:mKate2 transgene (H, H’, arrowhead, compared to control G). See also Figure S2 for analysis of neurog1:mKate2 in whole embryos with loss of Wnt signaling and gain of Sox2 function. A model shows the normal progression of events as NMPs transition to paraxial mesoderm, as well as the conditions causing activation of the checkpoint (tbx16 loss of function) or checkpoint inhibited in which ectopic spinal cords form or when mesoderm formation is rescued (I). The genetic pathway shown in (I) is based on Figure 2 (Sox2 activation of tbxta and inhibition of tbx16), as well as previously published work showing a Tbxta/Wnt signaling autoregulatory loop (Martin and Kimelman, 2008), and the inhibition of tbxta and sox2 expression by Tbx16 (Bouldin et al., 2015). The solid lines indicate the known direct regulatory interactions.
DISCUSSION
The mesodermal EMT during development is associated with progression toward differentiation, whereas cancer EMTs are generally thought to lead to increased stem cell characteristics and a lack of differentiation. Recent evidence suggests that metastasizing cancer cells are predominantly in a partial EMT state, and that the partial state is more stem cell-like than the fully mesenchymal state (Campbell, 2018; Aiello and Kang, 2019). Here, we show that the partial EMT state during mesoderm induction is a developmental checkpoint that prevents differentiation into either neural or mesodermal fates. In addition to preventing differentiation, activation of the checkpoint alters the normal migratory properties of these cells. Thus, the initiation of metastasis in solid tumors through a partial EMT may be recapitulating a developmental state in which cells with aberrant gene expression patterns are activating a developmental checkpoint. More important, our results show that the initiation of the EMT leading to the partial EMT state is uncoupled from mesodermal fate, and is fully reversible back to the epithelial state and eventual neural differentiation by withdrawing the checkpoint activating the Wnt signal. The uncoupling of EMT initiation and mesodermal fate acquisition underscores the importance of having a developmental checkpoint.
Loss of tbx16 function in zebrafish activates the developmental checkpoint because cells maintain sox2 expression in a high canonical Wnt signaling environment. Mouse NMPs were recently described to exist in a partial EMT state within the tailbud, and the same checkpoint is likely to function in mouse embryos, as loss of function of the closely related t-box transcription factor Tbx6 causes a large accumulation of cells in the tailbud that are unable to exit into the mesodermal territory (Chapman and Papaioannou, 1998; Dias et al., 2020). In this context, sox2 also fails to be repressed and is maintained in a high Wnt environment (Takemoto et al., 2011). One key difference of the mouse Tbx6 mutant compared to the zebrafish tbx16 mutant is that in the mouse, a subset of cells exit the tailbud to form ectopic spinal cords where somites should normally form (Chapman and Papaioannou, 1998). Ectopic neural tissue is never observed in the zebrafish tbx16 mutant or in the tbx16/msgn1 or tbx16/tbx16l double mutants, which have a more severe phenotype than the tbx16 single mutant (Fior et al., 2012; Yabe and Takada, 2012; Morrow et al., 2017). Here, we show that lowering the level of Wnt signaling can recapitulate the mouse Tbx6 ectopic spinal cord phenotype in zebrafish tbx16 mutants. One implication of this result is that the relative amount of Wnt signaling is higher in the zebrafish tailbud compared to the mouse, which may help explain the species-specific differences between NMP development (Martin and Kimelman, 2009; Steventon et al., 2016; Attardi et al., 2018; Mallo, 2020), including the phenotypic differences of the Tbx6 mouse mutant and the tbx16 single or tbx16/tbx6l and tbx16/msgn1 double zebrafish mutants (Chapman and Papaioannou, 1998; Fior et al., 2012; Yabe and Takada, 2012; Morrow et al., 2017).
The developmental checkpoint preventing sox2+ cells from exiting into the mesodermal territory is activated by canonical Wnt signaling, and together these factors both prevent differentiation and delay morphogenesis of mesoderm-fated NMPs. This is in stark contrast to the roles of these factors in the absence of the other, where each promotes differentiation along the neural (sox2) or mesodermal (Wnt) lineages (Takemoto et al., 2011; Martin and Kimelman, 2012; Gouti et al., 2014, 2017; Garriock et al., 2015; Row et al., 2016; Koch et al., 2017). This type of interaction, in which two lineage-promoting factors can together prevent the differentiation down either lineage, is a well-known feature of hematopoietic stem cells (Cross and Enver, 1997; Nimmo et al., 2015). These cells are said to be in a lineage-primed state, where they are held in an undifferentiated state but are poised to rapidly differentiate into either lineage as soon as one factor becomes enriched relative to the other. Our work shows that NMPs, which express sox2 and have canonical Wnt signaling activity, are similarly in a poised state, ready to rapidly differentiate into either neural tissue or mesoderm when Wnt signaling or sox2 expression is repressed. These results help explain the dual paradoxical functions of Wnt signaling during NMP maintenance and differentiation. While Wnt signaling is required for mesoderm induction from NMPs, it is also required for the maintenance, and possible expansion, of the undifferentiated NMP population (Takada et al., 1994; Garriock et al., 2015; Wymeersch et al., 2016). How the combination of Sox2 and Wnt signaling promotes differential cell biology than either factor alone remains to be determined, but there are several instances reported of Sox transcription factors binding to β-catenin, which in some cases can affect a unique transcriptional program (Kormish et al., 2010; Ye et al., 2014). Our results suggest the difference in Wnt function may be due to whether β-catenin is interacting predominantly with Sox2 to promote NMP maintenance or with Lef1/TCF family proteins to promote mesodermal differentiation.
STAR★METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Benjamin L. Martin (benjamin.martin@stonybrook.edu).
Materials Availability
The fish line generated in this study is available upon request until availability is made at the Zebrafish International Resource Center.
Data and Code Availability
Data generated in this study is available upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Zebrafish Care and Lines
All zebrafish methods were approved by the Stony Brook University Institutional Animal Care and Use Committee. Transgenic and mutant lines used include Tg(hsp70l:sox2-2A-NLS-KikGR)sbu100 (referred to here as HS:sox2) (Row et al., 2016), Tg(hsp70l:Xla.Tcf.-EGFP)w74 (referred to here as HS:TCFΔC) (Martin and Kimelman, 2012), Tg(sox2-2A-sfGFP)stl84 (Shin et al., 2014), Tg(actc1b:gfp)zf13 (Higashijima et al., 1997), Tg(neurog1:mKate2-CAAX)sk100 (this paper), tbx16b104 (Kimmel et al., 1989), and sox2x50 (Gou et al., 2018a, 2018b). The sox2x50 line was genotyped by amplifying genomic DNA with the forward 5′- CCAGCAAAGTTACCTCCAACTG −3′ and reverse 5′- GCAGGGTGTACTTGTCCTTCTT −3′ primers, followed by digesting the PCR product with NarI (the NarI site is absent in the 8 bp sox2x50 indel mutation). Heat shock inductions were performed by immersing embryos in an elevated temperature water bath (39°C to 40°C) for 30 minutes.
Generation of a zebrafish neurogenin1 transgenic reporter line
For the neurog1:mKate2-CAAX transgene, a genomic fragment spanning 8.4 kb up-stream of the neurog1 start codon (Blader et al., 2003) was cloned into the p5E plasmid (p5E-neurog1, Invitrogen, USA) and the coding sequence of the fluorescent protein mKate2 (Evrogen, Russia) followed in-frame by a CAAX box from HRAS, was cloned into the pME plasmid (pME-mKate2-CAAX, Invitrogen, USA). Using gateway recombination (Invitrogen, USA), we fused the neurog1 genomic fragment from the p5E-neurog1 plasmid to mKate2-CAAX from pME-mKate2-CAAX plasmid followed by a SV40pA signal from the p3E-polyA plasmid into the pDestTol2pA2 plasmid (Kwan et al., 2007). The resultant plasmid is called pDest-neurog1:mKate2-CAAX-SV40pA. For transgenesis, 25 ng/μl of the pDest-neurog1:mKate2-CAAX-SV40pA plasmid was co-injected with in vitro transcribed tol2 transposase mRNA (Thermo Fisher, USA) into one-cell-stage embryos (Kawakami et al., 2000). Transgenic fish were identified by mKate2 fluorescence at 1 dpf using a Leica M165 fluorescent stereo microscope (Leica Microsystems Inc., Germany). The full name of this transgenic line is Tg(−8.4neurog1:mKate2-CAAX)sk100.
METHOD DETAILS
Imaging
For tailbud exit transplantation experiments, cells were mounted in 2% methylcellulose with tricaine. Imaging was done on a Leica DMI6000B inverted microscope. For cell tracking and transplant quantification, embryos were mounted in 1% low melt agarose with tricaine and imaged on a custom built spinning disk confocal microscope with a Zeiss Imager A.2 frame, a Borealis modified Yokogawa CSU-10 spinning disc, ASI 150uM piezo stage controlled by an MS2000, an ASI filter wheel, a Hamamatsu ImageEM x2 EMCCD camera (Hamamatsu C9100-23B), and a 63x 1.0NA water immersion lens. This microscope is controlled with Metamorph microscope control software (V7.10.2.240 Molecular Devices), with laser illumination via a Vortran laser merge controlled by a custom Measurement Computing Microcontroller integrated by Nobska Imaging. Laser power levels were set in Vortran’s Stradus VersaLase 8 software.
In Situ Hybridization and Immunohistochemistry
Whole-mount in situ hybridization was performed as previously described (Griffin et al., 1995). For skeletal muscle antibody labeling, embryos were treated with a 1:50 dilution of the MF-20 antibody (Developmental Studies Hybridoma Bank – a myosin heavy chain antibody labeling skeletal and cardiac muscle) followed by an Alexa Fluor 561-conjugated anti-mouse secondary antibody. For somite quantification embryos were injected with 100pg kikume mRNA and fixed at 36 hpf and treated with MF-20 antibody. MF-20-labeled somites were imaged on a spinning disk confocal microscope using a 40x/1.0 dip objective in embryo media. Somitic nuclei were counted using spots on Imaris software (Bitplane, Oxford Instruments).
In situ HCR analysis and quantification
For in situ HCR v3.0, embryos were heat-shocked at the 5 somite stage and fixed at the 20 somite stage. HCR was performed as previously described (Choi et al., 2018). Embryos were mounted using 50% glycerol in Phosphate-Buffered saline/Tween (PBST) on a clear glass slide, covered with a thin glass coverslip, and sealed with valap. Embryos were imaged on a spinning disk confocal microscope using a 20x air objective, with an Hamamatsu Orca EM-CCD camera controlled using Metamamorph. Images were analyzed using Fiji/ImageJ (v.1.52q) (Schindelin et al., 2012). For quantifying tbxta expression, a region of interest was drawn using the free hand selection tool (as shown in Figure S1) on the maximum intensity z projection and the MGV of the region was measured. Background subtraction was performed by subtracting the MGVs of the background for each image. For quantifying tbx16 expression, default threshold of the maximum intensity z projection was used to measure the mean gray value (MGV) and the area using the analyze particle function in Fiji. Images were also analyzed by using the plot profile function to measure the MGV of a 10 pixel wide line from posterior to anterior “PA line” and from dorsal to ventral “DV line” (as shown in Figure S1). The lines from different embryos were aligned to the central point of each individual line. Plotting and statistics were performed using Graph Pad Prism 8.
Whole Embryo Reporter Expression
Reporter lines for neural (neurog1:mKate2sk100) or muscle (actc1b:gfpzf13) were crossed to the HS:hsp70l:sox2-2A-NLS-KikGRsbu100 and imaged live on a spinning disk confocal microscope using the 10x/0.3 air objective at 36 hpf.
sox2 Overexpression Transplants
Cell transplantation experiments were performed from sphere stage to shield stage targeting the ventral margin (Martin and Kimelman, 2012). Transplanted embryos were heat shocked at 39°C for 30 minutes at bud and 12 somite stages. Embryos were imaged on the spinning disk confocal using the 10x/0.3 air objective at 36 hpf.
sox2 Mutant and sox2:sfGFP Transplants
Donor embryos were injected with 100 pg of kikume mRNA and a mix of two tbx16 morpholinos (MO1: AGCCTGCATTATTTA GCCTTCTCTA (1.5ng) MO2: GATGTCCTCTAAAAGAAAATGTCAG (0.75ng)) as previously described (Lewis and Eisen, 2004). Donor cells were transplanted from sphere stage donors to shield stage hosts targeted to the ventral margin as previously described (Martin and Kimelman, 2012). Donor embryos were screened for the sox2 genotype and presence of actc1b:gfpzf13 reporter. Embryos were imaged on the spinning disk confocal using the 10x/0.3 air objective at 36 hpf.
QUANTIFICATION AND STATISTICAL ANALYSIS
Transplant Tissue Contribution Quantification
For neural quantification embryos were imaged live on a spinning disk confocal microscope using the 10x/0.3 air objective. For muscle quantification the NLS-Kikume in the transplanted cells were photoconverted on an inverted microscope using 405 nm light for 30 s and embryos were imaged live on a spinning disk confocal microscope using the 10x/0.3 air objective. Transplanted nuclei within reporter lines were quantified using Imaris (Bitplane, Oxford Instruments). When necessary, images were stitched using Fiji (Preibisch et al., 2009). Statistics were performed using an unpaired t test.
Transplant Cell Exit Quantification
Donor embryos were in injected with 2% fluorescein dextran and cells were transplanted from sphere to shield stage targeting the ventral margin as described previously. Host embryos were imaged on an inverted Leica DMI6000B microscope using the 10x/0.4 dry objective. Compound fluorescence from transplanted cells was measured from anterior to posterior starting from somite 12 to the end of the tail using Fiji.
Tailbud Cell Tracking
Embryos were injected with 25 pg of in vitro transcribed NLS-kikume mRNA at the 1-cell stage. A small region containing NMPs was photoconverted on an inverted Leica DMI6000B microscope using 405 nm filter set for 30 s and imaged on a spinning disk confocal using the 20x/0.8 air objective and tracked on Imaris as previously described (Goto et al., 2017).
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Myosin heavy chain (chicken) | Developmental Studies Hybridoma Bank | RRID: AB_2147781 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| tbx16 antisense morpholino oligonucleotide 1: AGCCTGCATTATTTAGCCTTCTCTA | Gene tools, LLC | N/A |
| tbx16 antisense morpholino oligonucleotide 2: GATGTCCTCTAAAAGAAAATGTCAG | Gene tools, LLC | N/A |
| Fluorescein Dextran, 10,000 MW, Anionic, Lysine Fixable (Fluoro-Emerald) | Thermo Fisher | D1820 |
| Tetramethylrhodamine Dextran, 10,000 MW, Lysine Fixable (fluoro-Ruby) | Thermo Fisher | D1817 |
| Critical Commercial Assays | ||
| HCR v3.0 | Molecular Instruments | N/A |
| mMessage mMachine SP6 transcription kit | Thermo Fisher | Cat#AM1340 |
| Experimental Models: Organisms/Strains | ||
| Tg(hsp70l:sox2-2A-NLS-KikGR) | Row et al., 2016 | Allele sbu100 |
| Tg(hsp70l:Xla.Tcf.-EGFP) | Martin and Kimelman, 2012 | Allele w74 |
| sox2-2A-sfGFP | Shin et al., 2014 | Allele stl84 |
| Tg(actc1b:gfp) | Higashijima et al., 1997 | Allele zf13 |
| tbx16 | Kimmel et al., 1989 | Allele b104 |
| sox2 | Gou et al., 2018a, 2018b | Allele x50 |
| Tg(neurog1:mKate2-CAAX) | This study | Allele sk100 |
| Oligonucleotides | ||
| tbxta HCR probe pair 1: cctcaacctacctccaacaaGT ATTTCCACCGATTATTATCGGCC | This study | tbxta_B4_odd_1 |
| tbxta HCR probe pair 1: CCCACCGGGCACCCATT CACCGTTCattctcaccatattcgcttc | This study | tbxta_B4_even_1 |
| tbxta HCR probe pair 2: cctcaacctacctccaacaaTTGG CATCGAGGAAAGCTTTGGCAA | This study | tbxta_B4_odd_2 |
| tbxta HCR probe pair 2: GGGACTTCCTTGTGGTCACT TCTCTattctcaccatattcgcttc | This study | tbxta_B4_even_2 |
| tbxta HCR probe pair 3: cctcaacctacctccaacaaCGGA AGAGTTGTCCATGTAGTTATT | This study | tbxta_B4_odd_3 |
| tbxta HCR probe pair 3: ACCAGCTGTC ATGAGACGCAAGACTattctcaccatattcgcttc | This study | tbxta_B4_even_3 |
| tbxta HCR probe pair 4: cctcaacctacct ccaacaaAAGTCCATAACTGCAGCATCAGTCC | This study | tbxta_B4_odd_4 |
| tbxta HCR probe pair 4: TCTAGATTTCCTC CTGAAGCCAAGAattctcaccatattcgcttc | This study | tbxta_B4_even_4 |
| tbxta HCR probe pair 5: cctcaacctacctccaacaaGTT CTACAGAAAGCACATGTAAGAC | This study | tbxta_B4_odd_5 |
| tbxta HCR probe pair 5: CGAAACAGCAAAGTCTGTCT TTCTCattctcaccatattcgcttc | This study | tbxta_B4_even_5 |
| tbx16 HCR probe pair 1: gtccctgcctctatatcttt TGTTAAGTCCAATGCTCTGGTGTTT | This study | tbx16_B3_odd_1 |
| tbx16 HCR probe pair 1: TCTCCATCAGA ACTGGATTAATTCCttccactcaactttaacccg | This study | tbx16_B3_even_1 |
| tbx16 HCR probe pair 2: gtccctgcctctat atctttTTCTGGTACCATGTCCACCAGTAAG | This study | tbx16_B3_odd_2 |
| tbx16 HCR probe pair 2: CTTGTTCCACT TATAT CTCAGACCGttccactcaactttaacccg | This study | tbx16_B3_even_2 |
| tbx16 HCR probe pair 3: gtccctgcctctata tctttCTTTTAGAGTTTGTTCCCTCATCTC | This study | tbx16_B3_odd_3 |
| tbx16 HCR probe pair 3: GGATCAGGCAG ATTTCTGTTTGCCCttccactcaactttaacccg | This study | tbx16_B3_even_3 |
| tbx16 HCR probe pair 4: gtccctgcctctata tctttAAGACTCGGGACTCAAAGCTGGGAC | This study | tbx16_B3_odd_4 |
| tbx16 HCR probe pair 4: TCTGTGTCGGGCACGGTGG CCACGTttccactcaactttaacccg | This study | tbx16_B3_even_4 |
| tbx16 HCR probe pair 5: gtccctgcctctatat ctttCTCACAGTTTGTGTTCACTTTTGGT | This study | tbx16_B3_odd_5 |
| tbx16 HCR probe pair 5: TTCTTGCAAGTGTA GTTAGTGCGTCttccactcaactttaacccg | This study | tbx16_B3_even_5 |
| ttn.1 HCR probe pair 1: cctcaacctacctcca acaaCCTTCCTGACATGTGTCACACGTTC | This study | ttn.1_B4_odd_1 |
| ttn.1 HCR probe pair 1: TCTTCTTCTTTTC CTCCTCACTCTCattctcaccatattcgcttc | This study | ttn.1_B4_even_1 |
| ttn.1 HCR probe pair 2: cctcaacctacctcca acaaAACAGGAATATGCCACTTTGGCTTG | This study | ttn.1_B4_odd_2 |
| ttn.1 HCR probe pair 2: TCTTACAGTCTCG AGGTCATCCTGTattctcaccatattcgcttc | This study | ttn.1_B4_even_2 |
| ttn.1 HCR probe pair 3: cctcaacctacctccaa caaCTTAGGAACAGTTACTGGAGCATAG | This study | ttn.1_B4_odd_3 |
| ttn.1 HCR probe pair 3: TTTTTCTGCCACAGT TGCAGAAGGTattctcaccatattcgcttc | This study | ttn.1_B4_even_3 |
| ttn.1 HCR probe pair 4: cctcaacctacctccaacaaAGTGG CATAATCAGAGGCTTCTCCC | This study | ttn.1_B4_odd_4 |
| ttn.1 HCR probe pair 4: AGAAAATCCACCCCCAGCAA CATCAattctcaccatattcgcttc | This study | ttn.1_B4_even_4 |
| ttn.1 HCR probe pair 5: cctcaacctacctccaa caaTACGGGGAGACTCTGGAGACTTTAC | This study | ttn.1_B4_odd_5 |
| ttn.1 HCR probe pair 5: GAGACCTAATT CCAAATGGTGACTTattctcaccatattcgcttc | This study | ttn.1_B4_even_5 |
| sox2x50 forward genotyping primer: CCAGCAAAGTTACCTCCAACTG | Gou et al., 2018a, 2018b | sox2x50 forward |
| sox2x50 reverse genotyping primer: GCAGGGTGTACTTGTCCTTCTT | Gou et al., 2018a, 2018b | sox2x50 reverse |
| Recombinant DNA | ||
| pCS2+ NLS-kikume | Goto et al., 2017 | N/A |
| pCMV tol2 | Addgene #31823 | |
| Software and Algorithms | ||
| Fiji/ImageJ | Schindelin et al., 2012 | https://fiji.sc |
| Imaris | Bitplane | N/A |
| Metamorph | Molecular Devices | N/A |
| LAS (Leica Application Suite) | Leica Microsystems | N/A |
Highlights.
A checkpoint stops mesoderm-fated NMPs from exiting the tailbud when expressing Sox2
The checkpoint is activated by canonical Wnt and Sox2 interactions
The checkpoint prevents ectopic neural tissue from forming in mesodermal territories
Sox2 expression and Wnt inhibition is sufficient to induce spinal cord from NMPs
ACKNOWLEDGMENTS
We thank David Matus for use of the spinning disc confocal microscope and comments on the manuscript, Taylor Kinney for advice, Neal Bhattacharji and Stephanie Flanagan for excellent zebrafish care, and Bruce Riley for providing the sox2x50 mutant before publication. We thank Thom Geer and Nobska Imaging for microscopy support. The MF20 monoclonal antibody, developed by D.A. Fischman, was obtained from the Developmental Studies Hybridoma Bank, created by the NICHD of the NIH and maintained at the University of Iowa, Department of Biology, Iowa City, IA 52242. The graphical abstract was created with BioRender.com. This work was supported by NIH NINDS (R01NS102322), to H.K., and by NSF (IOS 1452928) and NIH NIGMS (R01GM124282) grants to B.L.M.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.108311.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Aiello NM, and Kang Y (2019). Context-dependent EMT programs in cancer metastasis. J. Exp. Med 216, 1016–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attardi A, Fulton T, Florescu M, Shah G, Muresan L, Lenz MO, Lancaster C, Huisken J, van Oudenaarden A, and Steventon B (2018). Neuromesodermal progenitors are a conserved source of spinal cord with divergent growth dynamics. Development 145, dev166728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blader P, Plessy C, and Strähle U (2003). Multiple regulatory elements with spatially and temporally distinct activities control neurogenin1 expression in primary neurons of the zebrafish embryo. Mech. Dev 120, 211–218. [DOI] [PubMed] [Google Scholar]
- Bouldin CM, Manning AJ, Peng YH, Farr GH 3rd, Hung KL, Dong A, and Kimelman D (2015). Wnt signaling and tbx16 form a bistable switch to commit bipotential progenitors to mesoderm. Development 142, 2499–2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell K (2018). Contribution of epithelial-mesenchymal transitions to organogenesis and cancer metastasis. Curr. Opin. Cell Biol 55, 30–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman DL, and Papaioannou VE (1998). Three neural tubes in mouse embryos with mutations in the T-box gene Tbx6. Nature 391, 695–697. [DOI] [PubMed] [Google Scholar]
- Choi HMT, Schwarzkopf M, Fornace ME, Acharya A, Artavanis G, Stegmaier J, Cunha A, and Pierce NA (2018). Third-generation in situ hybridization chain reaction: multiplexed, quantitative, sensitive, versatile, robust. Development 145, dev165753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross MA, and Enver T (1997). The lineage commitment of haemopoietic progenitor cells. Curr. Opin. Genet. Dev 7, 609–613. [DOI] [PubMed] [Google Scholar]
- Delfino-Machín M, Lunn JS, Breitkreuz DN, Akai J, and Storey KG (2005). Specification and maintenance of the spinal cord stem zone. Development 132, 4273–283. [DOI] [PubMed] [Google Scholar]
- Dias A, Lozovska A, Wymeersch FJ, Nóvoa A, Binagui-Casas A, Sobral D, Martins GG, Wilson V, and Mallo M (2020).A Tgfbr1/Snai1-dependent developmental module at the core of vertebrate axial elongation. eLife 9, e56615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fior R, Maxwell AA, Ma TP, Vezzaro A, Moens CB, Amacher SL, Lewis J, and Saúde L (2012). The differentiation and movement of presomitic mesoderm progenitor cells are controlled by Mesogenin 1. Development 139, 4656–4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garriock RJ, Chalamalasetty RB, Kennedy MW, Canizales LC, Lewandoski M, and Yamaguchi TP (2015). Lineage tracing of neuromesodermal progenitors reveals novel Wnt-dependent roles in trunk progenitor cell maintenance and differentiation. Development 142, 1628–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto H, Kimmey SC, Row RH, Matus DQ, and Martin BL (2017). FGF and canonical Wnt signaling cooperate to induce paraxial mesoderm from tailbud neuromesodermal progenitors through regulation of a two-step epithelial to mesenchymal transition. Development 144, 1412–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou Y, Guo J, Maulding K, and Riley BB (2018a). sox2 and sox3 cooperate to regulate otic/epibranchial placode induction in zebrafish. Dev. Biol 435, 84–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou Y, Vemaraju S, Sweet EM, Kwon HJ, and Riley BB (2018b). sox2 and sox3 Play unique roles in development of hair cells and neurons in the zebrafish inner ear. Dev. Biol 435, 73–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouti M, Tsakiridis A, Wymeersch FJ, Huang Y, Kleinjung J, Wilson V, and Briscoe J (2014). In vitro generation of neuromesodermal progenitors reveals distinct roles for wnt signalling in the specification of spinal cord and paraxial mesoderm identity. PLOS Biol. 12, e1001937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouti M, Delile J, Stamataki D, Wymeersch FJ, Huang Y, Kleinjung J, Wilson V, and Briscoe J (2017). A Gene Regulatory Network Balances Neural and Mesoderm Specification during Vertebrate Trunk Development. Dev. Cell 41, 243–261.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin K, Patient R, and Holder N (1995). Analysis of FGF function in normal and no tail zebrafish embryos reveals separate mechanisms for formation of the trunk and the tail. Development 121, 2983–2994. [DOI] [PubMed] [Google Scholar]
- Griffin KJ, Amacher SL, Kimmel CB, and Kimelman D (1998). Molecular identification of spadetail: regulation of zebrafish trunk and tail mesoderm formation by T-box genes. Development 125, 3379–3388. [DOI] [PubMed] [Google Scholar]
- Halpern ME, Ho RK, Walker C, and Kimmel CB (1993). Induction of muscle pioneers and floor plate is distinguished by the zebrafish no tail mutation. Cell 75, 99–111. [PubMed] [Google Scholar]
- Hay ED (1995). An overview of epithelio-mesenchymal transformation. Acta Anat. (Basel) 154, 8–20. [DOI] [PubMed] [Google Scholar]
- Higashijima S, Okamoto H, Ueno N, Hotta Y, and Eguchi G (1997). High-frequency generation of transgenic zebrafish which reliably express GFP in whole muscles or the whole body by using promoters of zebrafish origin. Dev. Biol 192, 289–299. [DOI] [PubMed] [Google Scholar]
- Ho RK, and Kane DA (1990). Cell-autonomous action of zebrafish spt-1 mutation in specific mesodermal precursors. Nature 348, 728–730. [DOI] [PubMed] [Google Scholar]
- Kawakami K, Shima A, and Kawakami N (2000). Identification of a functional transposase of the Tol2 element, an Ac-like element from the Japanese medaka fish, and its transposition in the zebrafish germ lineage. Proc. Natl. Acad. Sci. USA 97, 11403–11408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimelman D (2016). Tales of Tails (and Trunks): Forming the Posterior Body in Vertebrate Embryos. Curr. Top. Dev. Biol 116, 517–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Kane DA, Walker C, Warga RM, and Rothman MB (1989). A mutation that changes cell movement and cell fate in the zebrafish embryo. Nature 337, 358–362. [DOI] [PubMed] [Google Scholar]
- Koch F, Scholze M, Wittler L, Schifferl D, Sudheer S, Grote P, Timmermann B, Macura K, and Herrmann BG (2017). Antagonistic Activities of Sox2 and Brachyury Control the Fate Choice of Neuro-Mesodermal Progenitors. Dev. Cell 42, 514–526.e7. [DOI] [PubMed] [Google Scholar]
- Kormish JD, Sinner D, and Zorn AM (2010). Interactions between SOX factors and Wnt/beta-catenin signaling in development and disease. Dev. Dyn 239, 56–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan KM, Fujimoto E, Grabher C, Mangum BD, Hardy ME, Campbell DS, Parant JM, Yost HJ, Kanki JP, and Chien CB (2007). The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev. Dyn 236, 3088–3099. [DOI] [PubMed] [Google Scholar]
- Lawton AK, Nandi A, Stulberg MJ, Dray N, Sneddon MW, Pontius W, Emonet T, and Holley SA (2013). Regulated tissue fluidity steers zebrafish body elongation. Development 140, 573–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis KE, and Eisen JS (2004). Paraxial mesoderm specifies zebrafish primary motoneuron subtype identity. Development 131, 891–902. [DOI] [PubMed] [Google Scholar]
- Li W, and Kang Y (2016). Probing the Fifty Shades of EMT in Metastasis. Trends Cancer 2, 65–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallo M (2020). The vertebrate tail: a gene playground for evolution. Cell. Mol. Life Sci 77, 1021–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning AJ, and Kimelman D (2015). Tbx16 and Msgn1 are required to establish directional cell migration of zebrafish mesodermal progenitors. Dev. Biol 406, 172–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin BL (2016). Factors that coordinate mesoderm specification from neuromesodermal progenitors with segmentation during vertebrate axial extension. Semin. Cell Dev. Biol 49, 59–67. [DOI] [PubMed] [Google Scholar]
- Martin BL, and Kimelman D (2008). Regulation of canonical Wnt signaling by Brachyury is essential for posterior mesoderm formation. Dev. Cell 15, 121–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin BL, and Kimelman D (2009). Wnt signaling and the evolution of embryonic posterior development. Curr. Biol 19, R215–R219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin BL, and Kimelman D (2010). Brachyury establishes the embryonic mesodermal progenitor niche. Genes Dev. 24, 2778–2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin BL, and Kimelman D (2012). Canonical Wnt signaling dynamically controls multiple stem cell fate decisions during vertebrate body formation. Dev. Cell 22, 223–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillen P, and Holley SA (2015). The tissue mechanics of vertebrate body elongation and segmentation. Curr. Opin. Genet. Dev 32, 106–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow ZT, Maxwell AM, Hoshijima K, Talbot JC, Grunwald DJ, and Amacher SL (2017). tbx6l and tbx16 are redundantly required for posterior paraxial mesoderm formation during zebrafish embryogenesis. Dev. Dyn 246, 759–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaya Y, and Sheng G (2013). EMT in developmental morphogenesis. Cancer Lett. 341, 9–15. [DOI] [PubMed] [Google Scholar]
- Nieto MA (2013). Epithelial plasticity: a common theme in embryonic and cancer cells. Science 342, 1234850. [DOI] [PubMed] [Google Scholar]
- Nieto MA, Huang RY, Jackson RA, and Thiery JP (2016). Emt: 2016. Cell 166, 21–45. [DOI] [PubMed] [Google Scholar]
- Nimmo RA, May GE, and Enver T (2015). Primed and ready: understanding lineage commitment through single cell analysis. Trends Cell Biol. 25, 459–467. [DOI] [PubMed] [Google Scholar]
- Preibisch S, Saalfeld S, and Tomancak P (2009). Globally optimal stitching of tiled 3D microscopic image acquisitions. Bioinformatics 25, 1463–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Row RH, Maître JL, Martin BL, Stockinger P, Heisenberg CP, and Kimelman D (2011). Completion of the epithelial to mesenchymal transition in zebrafish mesoderm requires Spadetail. Dev. Biol 354, 102–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Row RH, Tsotras SR, Goto H, and Martin BL (2016). The zebrafish tailbud contains two independent populations of midline progenitor cells that maintain long-term germ layer plasticity and differentiate in response to local signaling cues. Development 143, 244–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Row RH, Pegg A, Kinney BA, Farr GH 3rd, Maves L, Lowell S, Wilson V, and Martin BL (2018). BMP and FGF signaling interact to pattern mesoderm by controlling basic helix-loop-helix transcription factor activity. eLife 7, e31018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J,Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte-Merker S, van Eeden FJ, Halpern ME, Kimmel CB, and Nüsslein-Volhard C (1994). no tail (ntl) is the zebrafish homologue of the mouse T (Brachyury) gene. Development 120, 1009–1015. [DOI] [PubMed] [Google Scholar]
- Shin J, Chen J, and Solnica-Krezel L (2014). Efficient homologous recombination-mediated genome engineering in zebrafish using TALE nucleases. Development 141, 3807–3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steventon B, Duarte F, Lagadec R, Mazan S, Nicolas JF, and Hirsinger E (2016). Species-specific contribution of volumetric growth and tissue convergence to posterior body elongation in vertebrates. Development 143, 1732–1741. [DOI] [PubMed] [Google Scholar]
- Takada S, Stark KL, Shea MJ, Vassileva G, McMahon JA, and McMahon AP (1994). Wnt-3a regulates somite and tailbud formation in the mouse embryo. Genes Dev. 8, 174–189. [DOI] [PubMed] [Google Scholar]
- Takemoto T, Uchikawa M, Yoshida M, Bell DM, Lovell-Badge R, Papaioannou VE, and Kondoh H (2011). Tbx6-dependent Sox2 regulation determines neural or mesodermal fate in axial stem cells. Nature 470, 394–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzouanacou E, Wegener A, Wymeersch FJ, Wilson V, and Nicolas J-F (2009). Redefining the progression of lineage segregations during mammalian embryogenesis by clonal analysis. Dev. Cell 17, 365–376. [DOI] [PubMed] [Google Scholar]
- Wymeersch FJ, Huang Y, Blin G, Cambray N, Wilkie R, Wong FC, and Wilson V (2016). Position-dependent plasticity of distinct progenitor types in the primitive streak. eLife 5, e10042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabe T, and Takada S (2012). Mesogenin causes embryonic mesoderm progenitors to differentiate during development of zebrafish tail somites. Dev. Biol 370, 213–222. [DOI] [PubMed] [Google Scholar]
- Ye X, and Weinberg RA (2015). Epithelial-Mesenchymal Plasticity: A Central Regulator of Cancer Progression. Trends Cell Biol. 25, 675–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X, Wu F, Wu C, Wang P, Jung K, Gopal K, Ma Y, Li L, and Lai R (2014). β-Catenin, a Sox2 binding partner, regulates the DNA binding and transcriptional activity of Sox2 in breast cancer cells. Cell. Signal 26, 492–501. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data generated in this study is available upon request.