Abstract
Background
The Mediterranean mussel Mytilus galloprovincialis is an ecologically and economically relevant edible marine bivalve, highly invasive and resilient to biotic and abiotic stressors causing recurrent massive mortalities in other bivalves. Although these traits have been recently linked with the maintenance of a high genetic variation within natural populations, the factors underlying the evolutionary success of this species remain unclear.
Results
Here, after the assembly of a 1.28-Gb reference genome and the resequencing of 14 individuals from two independent populations, we reveal a complex pan-genomic architecture in M. galloprovincialis, with a core set of 45,000 genes plus a strikingly high number of dispensable genes (20,000) subject to presence-absence variation, which may be entirely missing in several individuals. We show that dispensable genes are associated with hemizygous genomic regions affected by structural variants, which overall account for nearly 580 Mb of DNA sequence not included in the reference genome assembly. As such, this is the first study to report the widespread occurrence of gene presence-absence variation at a whole-genome scale in the animal kingdom.
Conclusions
Dispensable genes usually belong to young and recently expanded gene families enriched in survival functions, which might be the key to explain the resilience and invasiveness of this species. This unique pan-genome architecture is characterized by dispensable genes in accessory genomic regions that exceed by orders of magnitude those observed in other metazoans, including humans, and closely mirror the open pan-genomes found in prokaryotes and in a few non-metazoan eukaryotes.
Keywords: Mussel, Bivalve, Pan-genome, Presence-absence variation, Structural variants, Hemizygosity, Dispensable gene, Phylome, Innate immunity, Antimicrobial peptides
Background
The Mediterranean mussel Mytilus galloprovincialis Lamarck, 1819 (Bivalvia, Mytilida), a member of the M. edulis species complex, is an edible cosmopolitan bivalve mollusk and an important seafood product in Europe and China. This shellfish has been consumed by humans since 6000 BC, and its global production currently exceeds 400 thousand tons per year [1]. Due to its invasive nature, this species has spread far beyond its native range, and it is considered a worldwide threat for autochthonous bivalve populations [2]. Like many other marine invertebrates, mussels have separate sexes and reproduce by broadcast spawning. Upon the release of gametes into the open water, and after fertilization, larvae can travel long distances carried by the oceanic currents [3]. Metamorphosis takes place during planktonic life, which ends after 1 to 2 months—depending on water temperature and food availability—with the settlement of juveniles and the start of the sessile adult life. Mussel beds are therefore usually composed by genetically heterogeneous individuals derived from large, randomly mating populations of different geographical origin. While genetic introgression in mussels has been broadly documented [4], a number of natural and genetic barriers concur in maintaining the genetic discontinuities observed both between different species belonging to the M. edulis species complex and between different lineages belonging to the same species [5].
Due to their filter-feeding habits, mussels are constantly exposed to a wide range of potentially pathogenic microorganisms, biotoxins, and anthropogenic pollutants. However, they display a remarkable resilience to stress and infections, can evolve novel traits in response to predation within a few generations [6], and have the ability to rapidly adapt to environmental changes, such as ocean acidification [7]. Moreover, mussels are capable of significant bioaccumulation [8], without experiencing the massive mortalities often seen in other farmed bivalves [9, 10].
Although M. galloprovincialis displays a morphologically conserved karyotype compared with other mussels and has not undergone known whole-genome duplication or allopolyploidization events [11], it shares with other bivalves a relatively large and complex genome, characterized by high heterozygosity and numerous mobile elements [12–17]. These factors posed a serious challenge to previous assembly efforts, which resulted in extremely fragmented genome sequences for this species [18, 19] which, unlike the congeneric Mytilus coruscus [20] and a few other mussel species [15], still lacks a highly contiguous reference genome assembly.
The remarkable level of intraspecific sequence diversity which characterizes several bivalve immune gene families [21], together with the recent implication of high standing genetic variation within natural populations in the extraordinary capability of environmental adaptation of M. galloprovincialis [7], stimulate further investigation about the role played by the genomic complexity of this species in explaining its invasiveness and resilience [18]. While a small but growing number of studies connected gene presence-absence variation (PAV) to the generation of this molecular diversity in a few gene families encoding antimicrobial peptides (AMPs) [22–25], it is presently unclear whether and to what extent this phenomenon is widespread in bivalve genomes.
Gene PAV is intimately linked to the pan-genome concept, which can be defined as a genome that includes a set of core genes found in all individuals and dispensable genes that are entirely missing in some individuals within a population [26]. Pan-genomes have been extensively studied in prokaryotes and viruses, where a simple genome architecture and frequent lateral gene transfer events facilitate the acquisition of accessory genomic sequence that may provide an evolutionary advantage in the colonization of new ecological niches or in the interaction with the host [27–29]. In Eukaryotes, pan-genomes have been occasionally reported in plants, fungi, and microalgae, where they have been often associated with the development of phenotypic traits linked with environmental adaptation, resistance to diseases, and intraspecific differentiation [30–36]. Although a few studies have recently extended the pan-genome concept to the animal kingdom, to the best of our knowledge, these have so far mostly linked the dispensable fraction of animal genomes with intergenic regions, bringing little evidence in support of the association between accessory genomic regions and gene PAV with adaptation [37–39].
We here report an improved, highly contiguous reference genome assembly for M. galloprovincialis, obtained from the sequencing of a single female individual (nicknamed Lola) and provide evidence in support of massive gene PAV through the analysis of whole-genome resequencing (WGR) data from 14 additional individuals. The widespread observation of the gene PAV phenomenon, which involves 20,000 protein-coding genes significantly enriched in functions related with survival, provides strong evidence in support of the presence of an open pan-genome in the Mediterranean mussel.
Results
An overview of the mussel reference genome
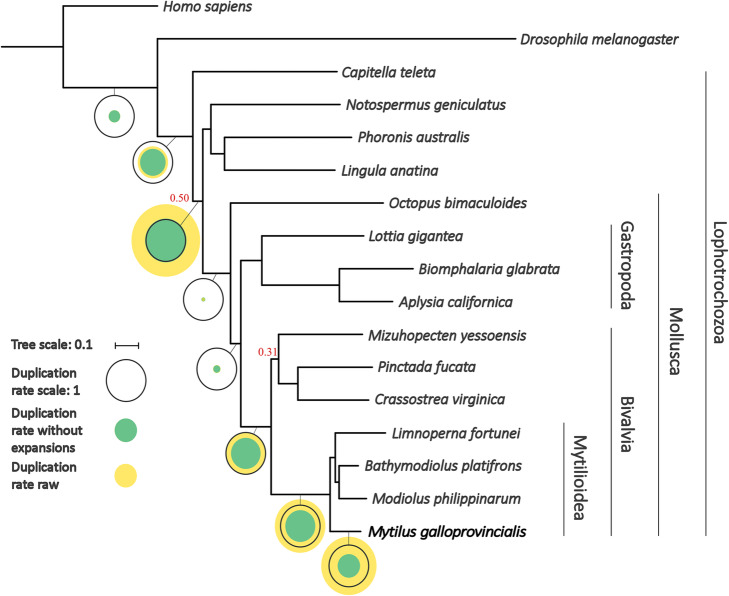
Our multi-step hierarchical de novo assembly strategy (Additional file 1: Data Note 1) resulted in a 1.28-Gb genome, of slightly smaller size compared to cytogenetic estimates [40], but of higher quality and contiguity compared to previous attempts [18, 19] (10,577 scaffolds; contig N50 = 71.42 kb; scaffold N50 = 207.64 kb). This genome shared some typical features of other bivalves, such as a low GC content (32%) and a widespread presence of repeats (43% of the assembly), but it was particularly rich in both protein-coding (60,338) and non-coding (75,973) genes. While these figures largely exceed those observed in most sequenced bivalve species [12, 14], they closely matched the numbers recently reported in the king scallop [17] and in the zebra mussel [41] (Additional file 1: Data Note 3). The reconstruction of the evolutionary relationships among M. galloprovincialis and 15 selected lophotrochozoan species [42–45], followed by an analysis of gene family trees [46], revealed that this large gene repertoire is the result of multiple lineage-specific duplication events that took place after the split between Mytilus and the rest of Mytilida (Fig. 1, Additional file 1: Data Note 5). Most mussel protein-coding genes were functionally annotated based on sequence similarity (56.17%) and were supported by transcriptomic evidence (78.70%). However, more than 5000 genes belong to recently acquired gene families specific of the Mytilus lineage, with uncharacterized function (Additional file 1: Data Note 20) [47].
Fig. 1.
Species tree obtained from the concatenation of 177 widespread single-gene families. In bold, we highlight the genome sequenced in this study. All branches were very well supported (aLRT > 0.99) except those with a number in red. In the tree, Bivalvia is a sister lineage to Gastropoda, with both molluscan classes forming a clade sister to Cephalopoda. Mollusca appears as a sister branch of a clade containing Phoronida (Phoronis), Nemertea (Notospermus), and Brachiopoda (Lingula) with low support (0.50). Open circles represent duplication rates (genome-wide average number of duplications per gene). Yellow circles represent the estimated duplication rates before removing large expansions consisting of more than 20 paralogs appearing in a single node. Green circles represent the duplication rates obtained after the removal of such events. Black circles serve as a scale and correspond to a duplication rate of 1
A genome characterized by widespread heterozygosity and hemizygosity
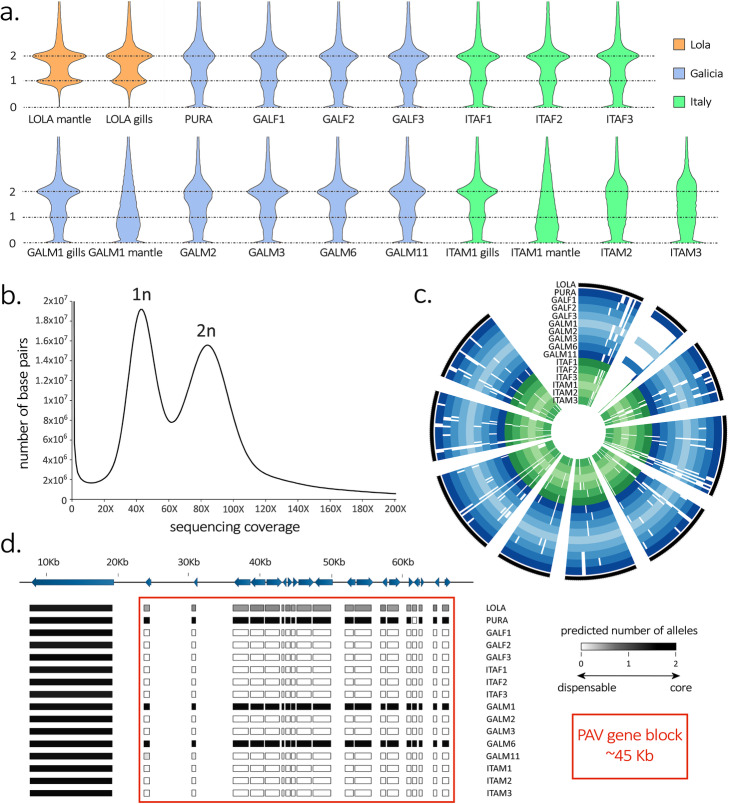
The contribution of heterozygosity to the overall intraspecific genomic variation was estimated in Lola, Pura (a female individual subject of a previous assembly effort [18]), and in the 14 resequenced genomes by analyzing only those regions shared by all individuals. The average heterozygosity rate observed across individuals was 1.73 ± 0.24%, indicating that the mussel genome harbors a very high density of single-nucleotide polymorphisms (SNPs), 12–22-fold higher than the human genome [48–50] (Additional file 1: Data Note 6). This value is in line with previous reports [4, 7] and with genomic evidence collected in other mytilids [15, 16]. However, while this high heterozygosity rate may seem rather large when compared to other animal species, it does not appear to be the main source of intraspecific genomic diversity in M. galloprovincialis. Indeed, structural variation, and large insertion/deletion polymorphisms in particular, appear to be a key aspect in the genome of this species. In spite of the assembly strategy we adopted, aimed at the removal of sequence derived from alternative haplotypes, the haploid reference genome assembly still contained a high fraction (36.78%) of sequence with low coverage. The bimodal distribution of the read mapping coverage in Lola (Fig. 2b) clearly shows that such regions are found in a hemizygous state, i.e., they are present in only one of the two homologous chromosomes.
Fig. 2.
a Violin plots displaying the “per gene” coverage of the protein-coding genes annotated in the mussel genomes, normalized by the expected haploid mussel genome size, calculated based on the mapping of Illumina PE libraries, and obtained either from the mantle or gills tissue. “0”, “1” and “2” indicate the number of alleles per individual. b Summary of the “per base” coverage of the Lola assembly (evaluated by the mapping of Illumina PE reads obtained from the mantle tissue). Two main peaks of coverage are clearly visible, corresponding to the hemizygous (41.5X) and homozygous (83X) peaks of coverage based on genome size estimates. The peak located at 0 indicates approximately 70 Mb of genome assembly which did not achieve any mapping based on Q ≥ 60. c Circus plot summarizing the presence-absence variation phenomenon on the nine longest genome scaffolds, plus the scaffold 02822, which contains a large fraction of dispensable genes. Italian and Galician mussel genomes are indicated with green and blue shades of color, respectively. Full and empty circles indicate present and absent genes, respectively. Scaffold names, reported in clockwise order, are: s00009, s029833, s00073, s00003, s00002, s00060, s00247, s00011, s00241 and s00029. d Detail of presence-absence variation in the genomic scaffold 02822; note that this scaffold contains a single core gene and a large (about 45 kb) block of dispensable genes, which are only found in four out of the analyzed genomes (including Lola, where the gene block is present in a single copy only). Gene IDs are, from left to right, as follows: MGAL10A041721, MGAL10A025495, MGAL10A021236, MGAL10A038341, MGAL10A006169, MGAL10A040493, MGAL10A046398, MGAL10A002899, MGAL10A052538, MGAL10A016640, MGAL10A080632, MGAL10A080632, MGAL10A050728, MGAL10A069289, MGAL10A086703, MGAL10A046961, MGAL10A008710, MGAL10A090544, MGAL10A069343, MGAL10A077743 and MGAL10A011823
Massive gene presence-absence variation
The hemizygous fraction of the mussel genome does not only include intergenic regions, but also contains nearly one-third of the protein-coding genes annotated in the reference genome, as well as a significant fraction of non-coding genes (Additional file 1: Data Note 8 and 9). Even more surprisingly, our analyses revealed that 24.25% of the protein-coding genes and 16.71% of the non-coding genes were entirely missing in at least one of the resequenced genomes, i.e., they were subject to gene PAV [51, 52] (Fig. 2a–c). On average, each individual lacked 4829 (8.01%) protein-coding genes and 3744 (5.12%) non-coding genes found in Lola.
Unlike the 45,518 core protein-coding genes found in homozygous genomic regions in Lola and in all the resequenced genomes, the 14,820 genes subject to PAV are dispensable and often associated with hemizygous genomic regions, i.e., they can be present in either one, two, or in neither of the two homologous chromosomes of the different mussels analyzed. Indeed, most of the genes encoded by hemizygous genomic regions in Lola either displayed a sequencing coverage consistent with hemizygosity or were entirely absent in the resequenced genomes (58.50% and 23.23% on average, respectively, Additional file 1: Fig. S56). On the other hand, the overwhelming majority (98.05%) of the genes present in homozygous genomic regions in Lola were present in all the resequenced genomes, in 85.46% of cases with a sequencing coverage also consistent with homozygosity (Additional file 1: Fig. S57).
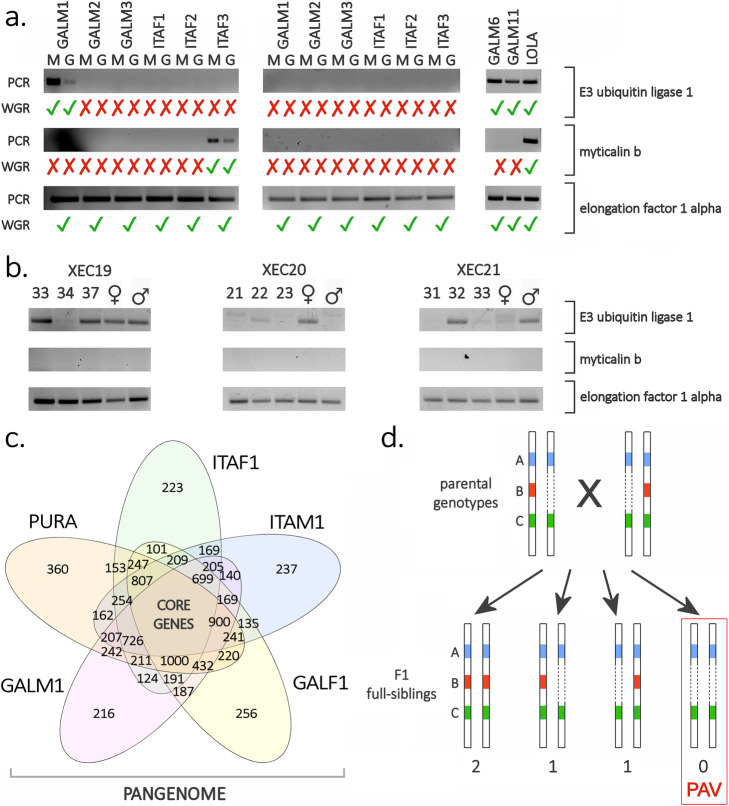
We ruled out the possibility that our observations were hampered by biases or confounding factors linked with the library preparation, sequencing, or bioinformatics analyses through a series of additional tests. First and foremost, the visual inspection on agarose gel of PCR amplification bands resulting from the analysis of twelve dispensable genes plus five core genes revealed a complete overlap between in silico predicted and experimentally observed PAV patterns (Fig. 3a, Additional file 1: Data Note 12). A similar PCR-based approach was extended to three independent families of full-sib mussels comprising the two parents and three first-generation offspring each. This allowed us to bring further support to the hypothesis that the dispensable genes are encoded by hemizygous genomic regions and that they follow a Mendelian mode of inheritance (Fig. 3d). Moreover, the presence of dispensable genes in hemizygous genomic regions was confirmed by the analysis of mapping data derived from a second round of sequencing of Lola obtained from a different tissue (gills, see Additional file 1: Fig. S33-S34), and the possible effect of mapping artifacts was excluded through computational simulations (Additional file 1: Data Note 10).
Fig. 3.
a Validation of the presence-absence variation phenomenon by PCR, carried out on the genomic DNA extracted from the mantle (M) or gills (G) of the 14 mussel individuals subjected to whole-genome resequencing. In Lola, GALM6 and GALM11, genomic DNA was extracted from the mantle tissue only. One core gene (elongation factor 1 alpha) and two dispensable genes (E3 ubiquitin ligase 1 and myticalin B1) were tested. Tick and cross symbols indicate expectations based on the in silico analysis of whole-genome resequencing (WGR) data. b Observation of the presence-absence variation phenomenon by PCR carried out in 3 full-sib mussels obtained from a controlled cross (parents were also tested and are indicated by ♂ and ♀, respectively). One core gene (elongation factor 1 alpha) and two dispensable genes (E3 ubiquitin ligase 1 and myticalin B1) were tested. XEC19, XEC20, and XEC21 indicate the three mussel families subjected to this investigation. The original photographs of the agarose gels used for the preparation of this figure and technical details about these experiments are available in Additional file 1: Data Note 12. c Structure of the mussel pangenome, as exemplified by a Venn diagram representing the overlap between the gene sets found in Pura (a female individual previously sequenced by Murgarella and colleagues [18]) and in four resequenced genomes, i.e., ITAF1, ITAM1, GALF1, and GALM1 (note that all these genes are present in Lola). Genes shared by all individuals define the core genome, whereas genes shared by some, but not all, individuals are dispensable. The entire complement of core and dispensable genes defines the mussel pangenome. d Schematic overview of the possible origin of a gene presence-absence variation phenomenon. A cross between two parents carrying two core genes (A and C) and one dispensable gene (B) is depicted. In this case, both parents carry a single copy of the dispensable gene (i.e., the dispensable gene is present in a hemizygous genomic region). Based on Mendelian inheritance, PAV should be observed in 25% of the offspring produced by this cross. “0”, “1” and “2” indicate the expected number of copies of the dispensable gene
The mussel pan-genome
Our recursive pan-genome reassembly strategy (summarized in Additional file 1: Fig. S66), paired with a strict decontamination pipeline (Additional file 1: Fig. S67), led to the generation of 267,538 contigs unambiguously taxonomically assigned to M. galloprovincialis, accounting for 578.74 Mb sequence data not present in Lola. Consistently with the high contiguity observed for some dispensable genes contained in large (up to 30 kb) hemizygous genomic regions in Lola (Fig. 2d, Additional file 1: Data Note 17), several large assembled pan-genomic contigs had protein-coding potential. This process brought the cumulative size of the mussel pan-genome assembly to 1.86 Gb, and the total number of annotated protein-coding genes to 65,625 (20,106 of which are dispensable). On average, each resequenced genome included 1974 out of the 5286 newly annotated protein-coding genes. Overall, we estimate that each resequenced genome lacked, on average, 8141 dispensable genes found in the mussel pan-genome (Additional file 1: Data Note 15).
Characterization and functional enrichment of dispensable genes
Although mussel dispensable genes generally display a shorter ORF length and a lower gene architecture complexity than core genes, they mostly retain signatures of functionality, which include the presence of conserved regulatory elements and the lack of significant GC or codon usage bias (Additional file 1: Data Note 17). While dispensable genes display, on average, expression levels 3× lower than core genes, nearly 60% of them are supported by mild or strong transcriptional evidence, accounting for 3–10% of the global transcriptional activity, depending on the tissue considered (Additional file 1: Data Note 16). They are also on average evolutionarily younger, subject to an increased lineage-specific duplication rate and four times more likely to be taxonomically restricted than core genes (Additional file 1: Data Note 19–20).
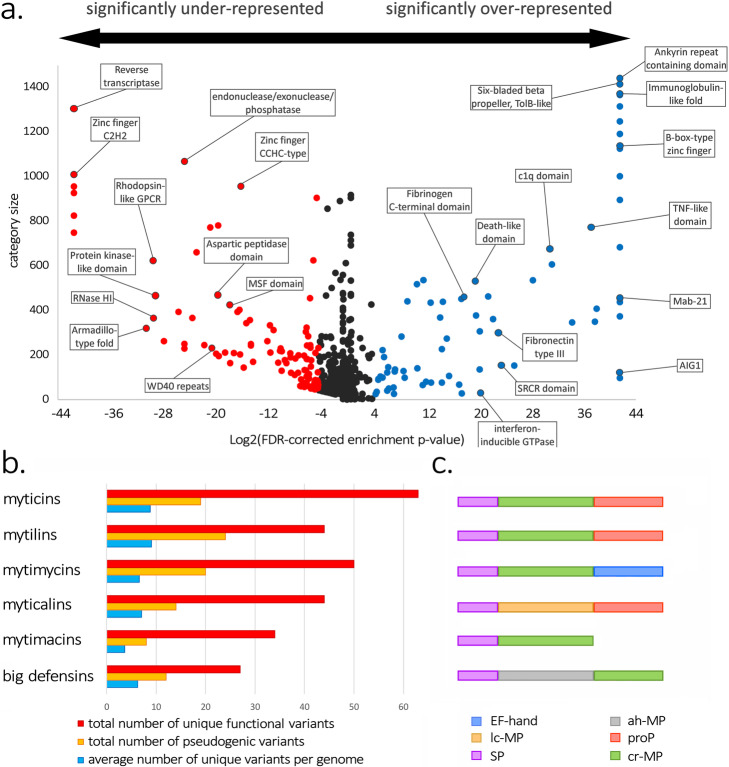
We identified several mussel gene families significantly more prone to PAV than expected by chance (Fig. 4a). The functional annotation of the dispensable genes found both in the reference genome and in the recursively re-assembled pan-genomic contigs revealed an enrichment in functions related to survival. These may be provided by proteins with marked protein- or carbohydrate-binding properties (e.g., pattern recognition receptors like C1qDC proteins, FReDs, and Ig domain-containing proteins), involved in apoptosis pathways (e.g., DEATH and BIR), or playing a role in immune signaling (e.g., interferon-inducible and IMAP GTPases) (Additional file 1: Data Note 18).
Fig. 4.
a Correlation between the abundance of conserved protein domains (Y axis) and their level of enrichment in the dispensable mussel genes (X axis). Each dot represents a conserved protein domain, with specific examples highlighted within boxes. Domains shown in red and blue are under- and over-represented, respectively, in the dispensable gene set compared to the core genome. b Total number of unique potentially functional sequence variants and pseudogenes, and average number of unique functional variants per genome for the six families of antimicrobial peptides analyzed (myticins, mytilins, mytimycins, myticalins, mytimacins, and big defensins). c Schematic organization of the precursor proteins of the six AMP families displayed in panel b. SP: signal peptide; ProP: propeptide region; EF-hand: EF-hand domain; lc-MP: linear cationic mature peptide; ah-MP: alpha helical mature peptide; cr-MP: cysteine-rich mature peptide
Mussel gene families encoding antimicrobial peptides (AMPs) were also subject to massive gene PAV. Although several dozen different sequence variants were identified for each AMP family in the resequenced genomes, each individual mussel possessed a unique combination of a small number of variants, with very little overlap with other mussels from the same population (Fig. 4b, Additional file 1: Data Note 21). On the other hand, other gene families were significantly under-represented among dispensable genes. Notably, these included genes encoding transposable elements (and therefore found in multiple copies in the genome), such as reverse transcriptase and RNase H-like proteins, or with housekeeping functions (e.g., protein kinases and G protein-coupled receptors).
Discussion
A pan-genome contains a set of core genes present in all individuals of the same species, which are fundamental for survival, and dispensable genes, which are only found in a subset of the individuals, and usually have accessory functions [26]. The expansion of pan-genomic studies to Eukaryotes has later extended this concept to intergenic genomic regions and to the activity of mobile elements [37, 39, 50, 53]. However, we will refer here to the original pan-genome definition, associated with the accessory functions of dispensable genes and with the acquisition of the ability to quickly respond to selective pressure [27] and to colonize new ecological niches [28]. The widespread occurrence of PAV in mussels is most certainly consistent with the gene-centric definition provided by Medini and colleagues [26] and reveals an “open” pan-genomic architecture with a high rate of dispensable to core genes, i.e., 1:3 (Fig. 3c).
The small size, simple organization and fast gene gain, loss, and horizontal transfer rates of bacterial and viral genomes [54–56] can explain the presence of a large number of dispensable genes in these organisms. However, pan-genomes have been also occasionally reported in eukaryotes, such as plants, fungi, and microalgae, where they may represent an unearthed source of intraspecific genomic diversity [57]. For example, in some cultivated crops, dispensable genes contribute significantly to the development of agronomic traits [30–32], such as improved resistance to disease [36]. Another key example of the adaptive importance of the pan-genome architecture is provided by some fungi, whose pathogenic potential, antimicrobial resistance, and host immune system avoidance are fueled by dispensable genes [33, 34, 58]. In the context of ecological adaptation, the cosmopolitan oceanic distribution and ability to thrive in different habitats of the coccolithophore Emiliania huxley can be explained by the acquisition of accessory metabolic activities provided by dispensable genes [35].
In spite of the growing number of reports of pan-genomes in eukaryotes, large-scale studies have been restricted to a few vertebrate species [39], targeting in particular human populations [37, 38, 50]. Moreover, the presence of accessory genomic regions not included in metazoan reference assemblies has never been associated with the massive occurrence of gene PAV, and the general impact of this phenomenon on animal intraspecific diversity has always been presumed to be minimal and in some cases deleterious [59].
Our observations, strongly supported by both experimental (Additional file 1: Data Note 12) and computational evidence (Additional file 1: Data Note 13), revealed that the dispensable fraction of the mussel pan-genome exceeds by approximately 5 and 15 times that of Sus scrofa and Homo sapiens, respectively [37, 39]. Moreover, while just 240 genes (i.e., 1.17% of the total) are presumably dispensable in humans [38], we show that 25% of mussel genes are subject to PAV and that each of the individual mussels resequenced in this study lacked on average 8141 dispensable genes identified in the pan-genome, pointing out that the fraction of protein-coding genes affected by this phenomenon in mussel is 20 times higher than in humans (Additional file 1: Data Note 22). To the best of our knowledge, PAV has been only marginally explored in bivalves as a phenomenon linked to a few gene families involved in immune functions, such as big defensins in Crassostrea gigas [22, 24] and myticins and myticalins in M. galloprovincialis [23, 25]. Therefore, this is the first study to report the widespread occurrence of gene PAV at a whole-genome scale in the animal kingdom.
Besides the 60,338 protein-coding genes present in Lola and the 5286 protein-coding genes associated with the recursively re-assembled contigs (Additional file 1: Data Note 15), the mussel pan-genome might include several thousand additional dispensable genes in natural populations which still remain unexplored (Additional file 1: Data Note 22). This open pan-genomic architecture is strongly supported by the recursive reassembly of 580 Mb DNA sequence not present in the reference assembly, by the observation that several dispensable genes were only observed in single individuals, and by the fact that the pan-genome assembly growth curve was far from reaching saturation (Additional file 1: Data Note 14).
The Mytilus genus has a complex evolutionary history, characterized by extensive gene flow among congeneric species, a process which is still ongoing in mosaic hybrid zones [60–62]. However, the analysis of nuclear and mitochondrial genetic markers ruled out the possibility that our resequenced individuals were hybrids between M. galloprovincialis and other Mytilus species (Additional file 1: Data Note 7), which suggest that dispensable genes are unlikely to be recently introgressed allelic variants that cannot be mapped to the reference genome due to their sequence divergence (Additional file 1: Data Note 10). While genetic admixture among contemporary mussel species cannot explain the mussel pan-genome architecture, the role of ancient hybridization and homologous recombination between ancestral Mytilus species remains to be investigated, as similar processes have been identified as the key drivers of PAV in plants [63]. Similarly, the possible role of transposable elements in the origin and spread of the PAV phenomenon [53] will be fully elucidated only with the availability of a chromosome-scale assembly (Additional file 1: Data Note 17.4).
Regardless of the origin of mussel dispensable genes, their absence or presence in a hemizygous or homozygous state in the mussel genome suggests that the PAV phenomenon might be strictly dependent on the matching between paternal and maternal chromosomes during the fertilization process and that dispensable genes might have Mendelian inheritance (Fig. 3d). This hypothesis was confirmed by the observation of F1 proportions fully compatible with a Mendelian pattern in full-sibs resulting from a controlled cross between individuals showing PAV at the E3 ubiquitin ligase 1 gene (Fig. 3b).
Our finding that a large fraction of the mussel genome is in a single-allele state is congruent with the presence of chromosome structural variation [64] and with the significant intra-individual and inter-population variation in nuclear DNA content reported in previous cytogenetic studies [65]. This also mirrors the situation previously described in other metazoans with high intraspecific genome diversity, such as the roundworm Caenorhabditis brenneri and the ascidian Ciona savignyi, which have genomes characterized by significant structural variations and frequent polymorphic indels [66, 67]. The very high amount of intraspecific genomic diversity revealed in our study may come at the cost of interfering with conventional homologous chromosome pairing, recombination, and segregation during meiosis.
The observation of highly skewed coverage profiles in the sequencing libraries from the gonadal tissue of some (but not all) male mussels, regardless of the stage of sexual maturation, may support this hypothesis (Additional file 1: Data Note 23). These results, confirmed by a second independent round of resequencing, were not obtained in non-reproductive tissues (i.e., gills) or in female individuals (Fig. 2a). We suspect that this observation may be the result of a significant presence of aneuploid gametes, potentially generated by an aberrant meiotic process linked with the high structural divergence between homologous chromosomes. Several studies have reported the presence of strong genetic barriers in Mytilus, acting both between and within species. Although intrinsic post-zygotic selection has been invoked as one of the most likely mechanisms underpinning the preservation of mosaic hybrid zones [62, 68], the nature of this process still remains to be elucidated. Here, we postulate that the reduced fertility of the offspring produced by individuals carrying “structurally incompatible” chromosomes may be key for explaining post-zygotic selection and the maintenance of the pan-genome architecture in mussels.
Whether the pan-genomic architecture of the mussel genome provides a selective advantage at the population level is a fundamental question. In our opinion, the large over-representation of genes involved in the response to stress and survival in the variable fraction of the mussel pan-genome (Additional file 1: Data Note 18) and the impact of PAV on the molecular diversification of AMPs (Fig. 4b, Additional file 1: Data Note 21) may suggest an adaptive role for the pan-genomic architecture. This would be consistent with the benefits provided by the development of a complex arsenal of immune molecules in sessile species characterized by high population densities such as mussels, where the spread of pathogens can be very efficient [69, 70]. We can speculate that the accessory functions provided by the 20,000 dispensable mussel genes might underpin an improved ability to adapt to challenging and varying environmental conditions, resulting in the cosmopolitan distribution and high invasiveness potential of this species [2, 6, 7] and explaining a high standing genetic variation in mussel populations [7]. Since a large number of genes subject to PAV seem to belong to recently expanded, taxonomically restricted gene families with unknown function (Additional file 1: Data Note 19–20), the putative adaptive benefits of PAV might extend well beyond immunity and survival, with a potential impact on multiple aspects of mussel biology. At the present stage, in the absence of experimental data linking phenotypic variation and fitness to PAV in different ecological contexts, this remains a working hypothesis that needs to be formally tested.
Curiously, the genome of the congeneric non-invasive mussel M. coruscus [20], whose geographical distribution is limited to the Yellow Sea, displays a significantly lower number of protein-coding genes and a much lower level of heterozygosity compared with M. galloprovincialis (Additional file 1: Data Note 3 and 6), which suggests that the prevalence of PAV may vary from species to species. Future investigations, which will hopefully benefit from the release of additional chromosome-scale genome assemblies, should be aimed at investigating whether the pan-genomic architecture we described in the Mediterranean mussel is shared by other mollusks.
Conclusions
We provide, for the first time, significant evidence in support of the existence of widespread gene PAV in a metazoan pan-genome. The unusual structure of the mussel genome is the result of the massive presence of hemizygous genomic regions, which contain several thousand dispensable protein-coding genes. The enrichment of these genes in functions associated to resilience to stress and immune response warrants further investigation on the possible links between massive PAV and the evolutionary success of mussels, exemplified by the cosmopolitan distribution of this species in temperate marine coastal waters. Most likely, extensive PAV might be found in other cosmopolitan marine invertebrates characterized by broadcast spawning, very large effective population size and subject to similar environmental pressures, including other bivalve species where similarly high heterozygosity rates have been reported.
Methods
Reference genome sequencing and assembly
The genomic DNA extracted from the mantle tissue of a single female mussel individual nicknamed Lola, collected at Ría de Vigo (Spain), was processed to generate different sequencing libraries for sequencing on an Illumina HiSeq2000 platform. A short-insert (800 bp) paired-end (PE) library, whose output accounted for an expected 110X genome coverage [18], was complemented with two long-insert mate-pair (MP) libraries, with a fragment size of 3 and 5 kb, respectively. Moreover, a fosmid library with 150,000 clones was used to generate 150 pools containing 1000 clones each, and two additional independent fosmid-end (FE) libraries were also constructed and sequenced. Overall, 330 Gb of raw sequence data were produced by Illumina sequencing, and 15.63 Gb additional raw sequence data (10.5X coverage) was obtained from the sequencing of a SMRT library on a PacBio Sequel platform.
We followed a hybrid multi-step de novo assembly strategy, which combined algorithmic strategies from the de Bruijn graph and Overlap-Layout-Consensus methods (Additional file 1: Fig. S2), with the aim to produce a highly contiguous non-redundant haploid reference assembly of the mussel genome which, based on preliminary k-mer count analyses [71], was expected to display a considerable proportion of duplicated sequence and high heterozygosity.
Briefly, the non-redundant unitigs, built with ABySS [72] and merged with ASM [73] to remove large artefactual duplicated haplotype blocks, served as anchors for the hybrid assembly of PacBio reads with DBG2OLC [74]. This noisy preliminary assembly was polished with Raccoon (https://github.com/lukud/raccoon-), using the sequencing data derived from the Illumina PE800 library. SSPACEv3.0 [75] was then used to perform a first round of scaffolding using all the available PE, MP, and FE libraries available, and a second round of scaffolding with PacBio reads was performed with SSPACE-LongRead [76]. The scaffolding procedure was re-iterated a second time, with both Illumina and PacBio reads, and was followed by a MP and FE libraries-derived gap-closing step performed with PBJelly [77].
The resulting assembly was subjected to an additional round of polishing with Proovread [78], and the coding portion of the genome was further refined with GATK [79], based on the alignment between the genome sequence and available transcriptome data generated with STAR [80]. RNA-seq data was also used for an additional round of scaffolding with AGOUTI v0.2.4 [81]. Finally, a local region of assembly, which included the myticin gene cluster, was improved by the combination of Platanus [82] and DBG2OLC [74].
All the aforementioned steps of the assembly were paired with strict decontamination procedures, which employed KRAKEN 2 [83] and BLASTN [84, 85]. These were aimed at removing exogenous DNA sequence which may have resulted from accidental environmental or laboratory contamination, a common issue in NGS approaches [86]. A final analysis of our final assembly (mg10) using Blobtools v1.1.1 [87, 88] confirmed the absence of known contaminants in the genome sequence. Detailed information concerning the DNA extraction, library preparation, sequencing, genome assembly, and decontamination processes are provided in Additional file 1: Data Note 1.2.
Quality evaluation, gene model construction, and functional annotation
The completeness of the genome assembly was estimated with BUSCO v.3 [89], using a set of 843 conserved metazoan single-copy orthologs as a reference, and the resulting data about the present, fragmented, duplicated, and missing gene models were compared with previous genome assembly efforts carried out in M. galloprovincialis [18, 19] (Additional file 1: Data Note 1.3.3). The residual presence of artefactual duplications was assessed with the Kmer Analysis Toolkit [90]. Consensus gene models were obtained by combining transcript alignments generated with PASA v 2.0.2 [91], bivalve protein alignments created with SPALN v2.2.2 [92], and ab initio gene predictions obtained with GeneID [93], GeneMark-ES [94], GlimmerHMM [94], and Augustus [95]. Evidences derived from these methods were assigned different weights and combined into consensus CDS predictions with EvidenceModeler-1.1.1. Gene models were subjected to an additional round of quality control to refine the annotation of UTRs and alternatively spliced exons (Additional file 1: Data Note 2.1 and 2.2). Gene models were functionally annotated with InterPro [96], KEGG [97], Blast2GO [98], SignalP [99], and NCBI CDsearch [100] (Additional file 1: Data Note 2.3). The gene models supported by PASA, but lacking a CDS, were considered as non-coding genes and included in a separate annotation track (Additional file 1: Data Note 2.5).
The completeness and integrity of gene models, as well as the genome assembly size and the number and density of gene models, were compared with several other recently sequenced molluscan genomes (Additional file 1: Data Note 3). Each gene model was assigned a support level (high, mild, or low) based on evidence obtained from Lola gills and digestive gland transcriptomes, as well as from several publicly available M. galloprovincialis RNA-seq datasets (Additional file 1: Data Note 4).
Whole-genome resequencing of 14 additional individuals and pan-genome recursive assembly
The genome of 14 additional adult M. galloprovincialis specimens, belonging to two independent European populations (Ría de Vigo, Spain, 9 individuals, and Goro lagoon, Italy, 6 individuals, Additional file 1: Data Note 6.1), was resequenced on an Illumina HiSeq 2500 platform, aiming at achieving a 35X genome sequencing coverage. Raw sequencing data from the previous assembly of Pura was also included in this analysis [18]. In total, besides Lola, whole-genome resequencing (WGR) data of comparable quality was obtained for six female and eight male individuals. Trimmed sequencing reads were mapped on the mussel reference genome, and unmapped reads were collected and de novo assembled with the CLC Genomics Workbench v.20 (Qiagen, Hilden, Germany). Newly assembled contigs were added to the reference assembly, building a mussel pan-genome. This process, inspired by a similar approach previously carried out by other authors [32], was performed recursively for the 14 individuals (+ Pura), mapping the reads obtained from each genome against the growing pan-genome (Additional file 1: Fig. S66). All the de novo assembled contigs underwent a strict filtering process, aimed at removing exogenous contaminants, based on strict coverage and GC content criteria, and the detection of BLAST matches against known contaminants (Additional file 1: Fig. S67). Assembled contigs satisfying threshold quality criteria (detailed in Additional file 1: Data Note 14) were annotated following the same procedure described above for the reference genome.
Presence-absence variation analysis
Quality-trimmed sequencing reads obtained from all individuals were independently mapped to the reference assembly and to the accessory pan-genomic contigs, with BWA mem (v0.7.15) [101]. As detailed in Additional file 1: Data Note 8, the mapping strategy we used aimed at tolerating multi-mappings (i.e., the alignment of reads with similar scores on different genomic positions). Exon mapping data, extracted with BEDtools [102], were used to calculate the average read coverage per base within the coding region of each gene. These values, normalized by the expected haploid genome size, allowed us to classify genes either as “present” or “absent,” depending on whether their normalized coverage exceeded 0.25 (i.e., less than 25% of expectations for a gene found in a hemizygous genomic region). This strict threshold was set to put a major focus on the high-confidence identification of putatively absent genes, at the cost of the detection of some false positives in the set of “present” genes. The same procedure was extended to non-coding genes annotated in the reference genome (Additional file 1: Data Note 9). The expected hemizygous and homozygous peaks of coverage for each genome were estimated with an accurate calibration pipeline, based on a set of over 4000 genes displaying high coverage stability across individuals, as explained in Additional file 1: Data Note 23.
The comparative analysis of gene PAV among individuals led to their categorization either as core (i.e., present on all individuals) or dispensable (i.e., absent in one or more individuals) genes. Please note that all the genes annotated in the accessory pan-genomic contigs were, by definition, dispensable, as they were not found in Lola, as verified by an accurate re-mapping of genomic reads obtained from both gills and mantle (Additional file 1: Data Note 14).
Validation of PAV patterns and further characterization of dispensable genes
We explored whether the PAV patterns observed could be explained by technical artifacts linked with mapping stringency criteria or by high sequence diversity between allelic variants, computationally simulating the effect of decreasing mapping stringency on mapping rates, and of increasing diversity between allelic variants on the drop of observed sequencing coverage (Additional file 1: Data Note 10). We further confirmed the widespread nature of PAV in mussels though the analysis of the distribution of the genes encoded by the accessory pan-genomic contigs in the resequenced individuals (Additional file 1: Data Note 15) and identified further cases of PAV through the analysis of several publicly available M. galloprovincialis transcriptomes (Additional file 1: Data Note 13).
The PAV phenomenon was further confirmed by PCR on 13 mussel genomes, through the evaluation of the presence-absence of specific amplification bands on agarose for 12 selected dispensable gene targets, expected to produce discordant PCR results across individuals due to PAV, and 5 core genes. These experimental observations were compared with in silico predictions (see the details in Additional file 1: Data Note 12.1). Moreover, similar PCR analyses were extended to three different families of full-sib mussels, produced after induced spawning of a single male and a single female individual, to test whether the presence-absence of dispensable genes could be explained by Mendelian inheritance (see the details in Additional file 1: Data Note 12.2.).
We assessed to what extent dispensable genes were associated with hemizygous genomic regions by evaluating whether their coverage was consistent with the presence of zero, one, or two alleles in each individual (Additional file 1: Data Note 11). Particular attention was focused on the analysis of a few selected large genomic regions characterized by the presence of several contiguous dispensable genes (see Additional file 1: Data Note 17.1). We characterized the transcriptional activity of dispensable genes in different tissues through the mapping of several RNA-seq datasets (as detailed in Additional file 1: Data Note 16) and evaluated whether they were associated with significant codon usage bias, functional promoters, architectural alterations, and flanking transposable elements (Additional file 1: Data Note 17).
Dispensable genes were subjected to functional enrichment analyses with hypergeometric tests [103], which identified over- or under-represented associated Gene Ontology terms and conserved domain annotations, based on a FDR-corrected p value threshold of 0.05 (Additional file 1: Data Note 18). The phylome data (see below, and Additional file 1: Data Note 5) allowed us to investigate the association of dispensable genes with recent linage-specific gene cluster expansions, through the comparison between the rates of gene duplication with the background rate of the genome (Additional file 1: Data Note 19), and to evaluate their overlap with taxonomically restricted gene (TRG) families (Additional file 1: Data Note 20).
Phylome reconstruction
The mussel phylome was reconstructed using the PhylomeDB pipeline [46], as detailed in Additional file 1: Data Note 5.1. This approach, which involved 16 target species, enabled the detection of orthology and paralogy relationships (Additional file 1: Data Note 5.2), lineage-specific gene duplications (Additional file 1: Data Note 5.4), and associated significantly enriched annotations, based on the genes annotated in Lola (Additional file 1: Data Note 5.5). Moreover, species trees were built using three different approaches: (i) a maximum likelihood analysis, carried out with PhyML v3.0 on a concatenated gene alignment dataset [42]; (ii) a gene-tree parsimony analysis, carried with the dup-tree algorithm [43]; and (iii) a coalescent-based analysis, performed with ASTRAL-III [44, 104] (Additional file 1: Data Note 5.3).
Assessment of genetic introgression from congeneric species
Exploiting previously published data and experimentally validated haplotypes, we inspected whether Lola, Pura, and the resequenced mussel genomes displayed genetic signatures consistent with their identification as part of a “pure” M. galloprovincialis lineage, or any evidence of hybridization with congeneric species M. edulis and M. trossulus existed. For this purpose, we recovered in each individual the two alleles for three target nuclear loci Glu-5′ [105, 106], mac-1 [107–109], and EFbis [110, 111], and the sequence of the mitochondrial markers 16S rRNA and COI [60, 112, 113]. As detailed in Additional file 1: Data Note 7, amplicon size was predicted by in silico PCR, and the nucleotide sequences, aligned with MUSCLE [114] with sequences of known taxonomic origin retrieved from GenBank, were used to build neighbor joining (NJ) phylogenetic trees [115].
Analysis of target PAV-associated gene families
We collected the nucleotide sequences of the core gene EEF1A1 and its dispensable paralogous gene EEF1A1-bis from Lola, Pura, and the 14 resequenced individuals. Similarly, all sequence variants available for the myticin, mytilin, big defensins, mytimycin, mytimacin, and myticalin gene families were recovered, with particular focus on the exons encoding the mature peptide region of these AMPs. We studied their association with the PAV phenomenon and investigated their molecular phylogeny and the ongoing process of pseudogenization of several variants with a NJ phylogenetic reconstruction approach (Additional file 1: Data Note 21).
Supplementary information
Additional file 1. Manuscript data notes. This file includes additional data notes 1–24, along with all the supplementary figures and most supplementary tables referenced in the main text.
Additional file 2. Large supplementary tables. This file includes Table S1, Table S3, Table S4, Table S6, Table S16, Table S17, Table S18, Table S19, Table S20, Table S21, Table S22, Table S23, Table S24, Table S25, Table S26, Table S27, Table S28, Table S29, Table S30, Table S31, Table S32, Table S33, Table S34, Table S40, Table S52, Table S53, Table S54, Table S55, Table S56 and Table S57.
Acknowledgements
We would like to thank the following members of the CNAG Data Analysis Team for their help evaluating intermediate assemblies and their useful comments on Variant Calling: Sophia Derdak, Marcos Fernández, Steve Laurie, Jordi Morata, and Raúl Tonda. We are grateful to Edoardo Turolla from Istituto Delta (Goro, Italy) for the support provided in mussel sampling. We would like to thank María Gasset for critically reading the manuscript and providing suggestions for its improvement.
Peer review information
Kevin Pang was the primary editor of this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Review history
The review history is available as Additional file 3.
Authors’ contributions
BN, CC, DP, and AF planned and granted the funding to start the project. CC, DP, AF, TA, and MGu designed the sequencing strategy. RM extracted the genomic DNA from Lola and all resequenced individuals. MGu performed the Illumina sequencing of Lola. FC, LF, AC, and TA performed the genome assembly. JGG, AV, and TA performed the genome annotation. MM and CC carried out preliminary analyses of the genome of Lola and provided their inputs for the integration of genomic data from Pura in the present manuscript. MGe, AP, and AF planned the whole-genome resequencing approach from additional individuals. PV and UR provided mussels from the Adriatic Sea. AF, BN, AP, and MGe managed and analyzed the sequencing data. MGe and SG performed the recursive pan-genome reassembly. FC and TA performed the single-nucleotide variation analysis. PB generated the full-sib mussel families. RM validated PAV from the sequenced individuals and full-sib mussel families. TG, MNO, and DP carried out the phylogenetic analyses and studied the evolutionary origin of dispensable genes. MGe and UR analyzed RNA-sequencing data. MGe, AP, PV, UR, RM, AF, and BN performed the analysis of genes encoding antimicrobial peptides. MGe, AF, and DP wrote the manuscript with inputs from the other authors. All authors contributed to the writing of the supplementary data notes and to the preparation of supplementary tables and figures. All the authors read and approved the manuscript. DP and AF supervised the whole study.
Authors’ information
Twitter handles: @MGerdol (Marco Gerdol); @Mol_Biol_Marine (Pablo Balseiro); @toni_gabaldon (Toni Gabaldón); @aukicha (Carlos Canchaya); @dposada_ (David Posada); @antoniofigueras (Antonio Figueras).
Funding
This work was conducted with the support of the projects AGL2011-14507-E, AGL2015-65705-R, RTI2018-095997-B-I00 (Ministerio de Ciencia, Innovación y Universidades, Spain) and INCITE 10PXIB402096PR, IN607B 2016/12 (Consellería de Economía, Emprego e Industria - GAIN, Xunta de Galicia). Antonio Figueras, Beatriz Novoa, Rebeca Moreira, Alberto Pallavicini, Marco Gerdol, Paola Venier, and Umberto Rosani are supported by the European Union’s Horizon 2020 research and innovation programme under grant agreement no. 678589. David Posada is supported by the European Research Council, the Spanish Ministry of Economy and Competitiveness, and Xunta de Galicia. We acknowledge the support of the Spanish Ministry of Science and Innovation to the EMBL partnership, the Centro de Excelencia Severo Ochoa, the CERCA Programme/Generalitat de Catalunya, the Spanish Ministry of Science and Innovation through the Instituto de Salud Carlos III, the Generalitat de Catalunya through Departament de Salut and Departament d’Empresa i Coneixement, and the Co-financing by the Spanish Ministry of Science and Innovation with funds from the European Regional Development Fund (ERDF) corresponding to the 2014-2020 Smart Growth Operating Program.
Availability of data and materials
All the genome sequencing data obtained in this work, as well as the genome assembly and the annotation, are available in the European Nucleotide Archive (ENA) under the project IDs PRJEB24883 (Lola) [116] and PRJNA230138 (resequencing) [117]. A genome browser and a blast server for this genome can be accessed in our local server (https://denovo.cnag.cat/mussel/). The mussel phylome is available for download or browsing at PhylomeDB [118].
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Marco Gerdol, Email: mgerdol@units.it.
Rebeca Moreira, Email: rebecamoreira@iim.csic.es.
Fernando Cruz, Email: fernando.cruz@cnag.crg.eu.
Jessica Gómez-Garrido, Email: jessica.gomez@cnag.crg.eu.
Anna Vlasova, Email: vlasova.av@gmail.com.
Umberto Rosani, Email: umberto.rosani@unipd.it.
Paola Venier, Email: paola.venier@unipd.it.
Miguel A. Naranjo-Ortiz, Email: miguelangel.naranjo@crg.eu
Maria Murgarella, Email: mmurgarella@uvigo.es.
Samuele Greco, Email: samuele.greco@phd.units.it.
Pablo Balseiro, Email: pbal@norceresearch.no.
André Corvelo, Email: acorvelo@nygenome.org.
Leonor Frias, Email: leonor.frias@gmail.com.
Marta Gut, Email: marta.gut@cnag.crg.eu.
Toni Gabaldón, Email: toni.gabaldon@crg.eu.
Alberto Pallavicini, Email: pallavic@units.it.
Carlos Canchaya, Email: canchaya@uvigo.es.
Beatriz Novoa, Email: beatriznovoa@iim.csic.es.
Tyler S. Alioto, Email: tyler.alioto@cnag.crg.eu
David Posada, Email: dposada@uvigo.es.
Antonio Figueras, Email: antoniofigueras@iim.csic.es.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s13059-020-02180-3.
References
- 1.FAO Fisheries and Aquaculture Department. Cultured aquatic species information programme. Mytilus galloprovincialis. Cultured aquatic species information programme. Rome: FAO Fisheries and Aquaculture Department; 2020. http://www.fao.org/fishery/culturedspecies/Mytilus_galloprovincialis/en.
- 2.Bonham V. Mytilus galloprovincialis. Invasive species compendium. Wallingford: CAB; 2017.
- 3.Gosling E. Bivalve molluscs: biology, ecology and culture. Hoboken: Blackwell Publishing Ltd; 2003.
- 4.Fraïsse C, Belkhir K, Welch JJ, Bierne N. Local interspecies introgression is the main cause of extreme levels of intraspecific differentiation in mussels. Mol Ecol. 2016;25:269–286. doi: 10.1111/mec.13299. [DOI] [PubMed] [Google Scholar]
- 5.El Ayari T, Trigui El Menif N, Hamer B, Cahill AE, Bierne N. The hidden side of a major marine biogeographic boundary: a wide mosaic hybrid zone at the Atlantic–Mediterranean divide reveals the complex interaction between natural and genetic barriers in mussels. Heredity. Nat Publ Group; 2019;122:770–784. [DOI] [PMC free article] [PubMed]
- 6.Freeman AS, Byers JE. Divergent induced responses to an invasive predator in marine mussel populations. Science. 2006;313:831–833. doi: 10.1126/science.1125485. [DOI] [PubMed] [Google Scholar]
- 7.Bitter MC, Kapsenberg L, Gattuso J-P, Pfister CA. Standing genetic variation fuels rapid adaptation to ocean acidification. Nat Commun. 2019;10:5821. doi: 10.1038/s41467-019-13767-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goldberg ED. The mussel watch — a first step in global marine monitoring. Mar Pollut Bull. 1975;6:111. doi: 10.1016/0025-326X(75)90271-4. [DOI] [Google Scholar]
- 9.Barbosa Solomieu V, Renault T, Travers M-A. Mass mortality in bivalves and the intricate case of the Pacific oyster, Crassostrea gigas. J Invert Pathol. 2015;131:2–10. doi: 10.1016/j.jip.2015.07.011. [DOI] [PubMed] [Google Scholar]
- 10.Xiao J, Ford SE, Yang H, Zhang G, Zhang F, Guo X. Studies on mass summer mortality of cultured zhikong scallops (Chlamys farreri Jones et Preston) in China. Aquaculture. 2005;250:602–615. doi: 10.1016/j.aquaculture.2005.05.002. [DOI] [Google Scholar]
- 11.Pérez-García C, Morán P, Pasantes JJ. Karyotypic diversification in Mytilus mussels (Bivalvia: Mytilidae) inferred from chromosomal mapping of rRNA and histone gene clusters. BMC Genet. 2014;15:84. doi: 10.1186/1471-2156-15-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang G, Fang X, Guo X, Li L, Luo R, Xu F, et al. The oyster genome reveals stress adaptation and complexity of shell formation. Nature. 2012;490:49–54. doi: 10.1038/nature11413. [DOI] [PubMed] [Google Scholar]
- 13.Du X, Fan G, Jiao Y, Zhang H, Guo X, Huang R, et al. The pearl oyster Pinctada fucata martensii genome and multi-omic analyses provide insights into biomineralization. Gigascience. 2017;6:1–12. doi: 10.1093/gigascience/gix059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang S, Zhang J, Jiao W, Li J, Xun X, Sun Y, et al. Scallop genome provides insights into evolution of bilaterian karyotype and development. Nat Ecol Evol. 2017;1:s41559–017–0120–017. doi: 10.1038/s41559-017-0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun J, Zhang Y, Xu T, Zhang Y, Mu H, Zhang Y, et al. Adaptation to deep-sea chemosynthetic environments as revealed by mussel genomes. Nat Ecol Evol. 2017;1:0121. doi: 10.1038/s41559-017-0121. [DOI] [PubMed] [Google Scholar]
- 16.Uliano-Silva M, Dondero F, Dan Otto T, Costa I, Lima NCB, Americo JA, et al. A hybrid-hierarchical genome assembly strategy to sequence the invasive golden mussel, Limnoperna fortunei. Gigascience. 2018;7:1–10. doi: 10.1093/gigascience/gix128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kenny NJ, McCarthy SA, Dudchenko O, James K, Betteridge E, Corton C, et al. The gene-rich genome of the scallop Pecten maximus. Gigascience. 2020;9 Oxford Academic:giaa037. [DOI] [PMC free article] [PubMed]
- 18.Murgarella M, Puiu D, Novoa B, Figueras A, Posada D, Canchaya C. A first insight into the genome of the filter-feeder mussel Mytilus galloprovincialis. PLoS One. 2016;11:e0151561. doi: 10.1371/journal.pone.0151561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nguyen TTT, Hayes BJ, Ingram BA. Genetic parameters and response to selection in blue mussel (Mytilus galloprovincialis) using a SNP-based pedigree. Aquaculture. 2014;420–421:295–301. doi: 10.1016/j.aquaculture.2013.11.021. [DOI] [Google Scholar]
- 20.Li R, Zhang W, Lu J, Zhang Z, Mu C, Song W, et al. The whole-genome sequencing and hybrid assembly of Mytilus coruscus. Front Genet. 2020;11 Frontiers:440. [DOI] [PMC free article] [PubMed]
- 21.Gerdol M, Gomez-Chiarri M, Castillo MG, Figueras A, Fiorito G, Moreira R, et al. Immunity in molluscs: recognition and effector mechanisms, with a focus on bivalvia. In: Cooper EL, et al., editors. Advances in comparative immunology. Cham: Springer International Publishing; 2018. pp. 225–341. [Google Scholar]
- 22.Rosa RD, Alonso P, Santini A, Vergnes A, Bachère E. High polymorphism in big defensin gene expression reveals presence–absence gene variability (PAV) in the oyster Crassostrea gigas. Dev Comp Immunol. 2015;49:231–238. doi: 10.1016/j.dci.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 23.Leoni G, De Poli A, Mardirossian M, Gambato S, Florian F, Venier P, et al. Myticalins: a novel multigenic family of linear, cationic antimicrobial peptides from marine mussels (Mytilus spp.) Marine Drugs. 2017;15:261. doi: 10.3390/md15080261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gerdol M, Schmitt P, Venier P, Rocha G, Rosa RD, Destoumieux-Garzón D. Functional insights from the evolutionary diversification of big defensins. Front Immunol. 2020;11:758. [DOI] [PMC free article] [PubMed]
- 25.Rey-Campos M, Novoa B, Pallavicini A, Gerdol M, Figueras A. Comparative genomics reveals a significant sequence variability of myticin genes in Mytilus galloprovincialis. Biomolecules. 2020;10:943. doi: 10.3390/biom10060943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Medini D, Donati C, Tettelin H, Masignani V, Rappuoli R. The microbial pan-genome. Curr Opin Genet Dev. 2005;15:589–594. doi: 10.1016/j.gde.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 27.de Brito AF, Braconi CT, Weidmann M, Dilcher M, Alves JMP, Gruber A, et al. The pangenome of the Anticarsia gemmatalis multiple nucleopolyhedrovirus (AgMNPV) Genome Biol Evol. 2015;8:94–108. doi: 10.1093/gbe/evv231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McInerney JO, McNally A, O’Connell MJ. Why prokaryotes have pangenomes. Nat Microbiol. 2017;2:17040. doi: 10.1038/nmicrobiol.2017.40. [DOI] [PubMed] [Google Scholar]
- 29.Choudoir MJ, Panke-Buisse K, Andam CP, Buckley DH. Genome surfing as driver of microbial genomic diversity. Trends Microbiol. 2017;25:624–636. doi: 10.1016/j.tim.2017.02.006. [DOI] [PubMed] [Google Scholar]
- 30.Hirsch CN, Foerster JM, Johnson JM, Sekhon RS, Muttoni G, Vaillancourt B, et al. Insights into the maize pan-genome and pan-transcriptome. Plant Cell. 2014;26:121–35. [DOI] [PMC free article] [PubMed]
- 31.Marroni F, Pinosio S, Morgante M. Structural variation and genome complexity: is dispensable really dispensable? Curr Opin Plant Biol. 2014;18:31–36. doi: 10.1016/j.pbi.2014.01.003. [DOI] [PubMed] [Google Scholar]
- 32.Golicz AA, Batley J, Edwards D. Towards plant pangenomics. Plant Biotechnol J. 2016;14:1099–1105. doi: 10.1111/pbi.12499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Plissonneau C, Hartmann FE, Croll D. Pangenome analyses of the wheat pathogen Zymoseptoria tritici reveal the structural basis of a highly plastic eukaryotic genome. BMC Biol. 2018;16:5. doi: 10.1186/s12915-017-0457-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCarthy CGP, Fitzpatrick DA. Pan-genome analyses of model fungal species. Microb Genom. 2019;5:e000243. [DOI] [PMC free article] [PubMed]
- 35.Read BA, Kegel J, Klute MJ, Kuo A, Lefebvre SC, Maumus F, et al. Pan genome of the phytoplankton Emiliania underpins its global distribution. Nature. 2013;499:209–213. doi: 10.1038/nature12221. [DOI] [PubMed] [Google Scholar]
- 36.Hübner S, Bercovich N, Todesco M, Mandel JR, Odenheimer J, Ziegler E, et al. Sunflower pan-genome analysis shows that hybridization altered gene content and disease resistance. Nat Plants. 2019;5:54–62. doi: 10.1038/s41477-018-0329-0. [DOI] [PubMed] [Google Scholar]
- 37.Li R, Li Y, Zheng H, Luo R, Zhu H, Li Q, et al. Building the sequence map of the human pan-genome. Nature Biotechnol. 2010;28:57–63. doi: 10.1038/nbt.1596. [DOI] [PubMed] [Google Scholar]
- 38.Sudmant PH, Rausch T, Gardner EJ, Handsaker RE, Abyzov A, Huddleston J, et al. An integrated map of structural variation in 2,504 human genomes. Nature. 2015;526:75–81. doi: 10.1038/nature15394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tian X, Li R, Fu W, Li Y, Wang X, Li M, et al. Building a sequence map of the pig pan-genome from multiple de novo assemblies and Hi-C data. Sci China Life Sci. 2020;63:750–763. doi: 10.1007/s11427-019-9551-7. [DOI] [PubMed] [Google Scholar]
- 40.Ieyama H, Kameoka O, Tan T, Yamasaki J. Chromosomes and nuclear DNA contents of some species of Mytilidae. Venus. 1994;53:327–331. [Google Scholar]
- 41.McCartney MA, Auch B, Kono T, Mallez S, Zhang Y, Obille A, et al. The genome of the zebra mussel, Dreissena polymorpha: a resource for invasive species research. bioRxiv. 2019. https://www.biorxiv.org/content/10.1101/696732v1. [DOI] [PMC free article] [PubMed]
- 42.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 43.Wehe A, Bansal MS, Burleigh JG, Eulenstein O. DupTree: a program for large-scale phylogenetic analyses using gene tree parsimony. Bioinformatics. 2008;24:1540–1541. doi: 10.1093/bioinformatics/btn230. [DOI] [PubMed] [Google Scholar]
- 44.Zhang C, Rabiee M, Sayyari E, Mirarab S. ASTRAL-III: polynomial time species tree reconstruction from partially resolved gene trees. BMC Bioinformatics. 2018;19:153. doi: 10.1186/s12859-018-2129-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gabaldón T. Large-scale assignment of orthology: back to phylogenetics? Genome Biol. 2008;9:235. doi: 10.1186/gb-2008-9-10-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huerta-Cepas J, Gabaldón T. Assigning duplication events to relative temporal scales in genome-wide studies. Bioinformatics. 2011;27:38–45. doi: 10.1093/bioinformatics/btq609. [DOI] [PubMed] [Google Scholar]
- 47.Khalturin K, Hemmrich G, Fraune S, Augustin R, Bosch TCG. More than just orphans: are taxonomically-restricted genes important in evolution? Trends Genet. 2009;25:404–413. doi: 10.1016/j.tig.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 48.Leffler EM, Bullaughey K, Matute DR, Meyer WK, Ségurel L, Venkat A, et al. Revisiting an old riddle: what determines genetic diversity levels within species? PLoS Biol. 2012;10:e1001388. doi: 10.1371/journal.pbio.1001388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.The International SNP Map Working Group A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature. 2001;409:928–933. doi: 10.1038/35057149. [DOI] [PubMed] [Google Scholar]
- 50.Sherman RM, Forman J, Antonescu V, Puiu D, Daya M, Rafaels N, et al. Assembly of a pan-genome from deep sequencing of 910 humans of African descent. Nat Genet. 2019;51:30–35. doi: 10.1038/s41588-018-0273-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Springer NM, Ying K, Fu Y, Ji T, Yeh C-T, Jia Y, et al. Maize inbreds exhibit high levels of copy number variation (CNV) and presence/absence variation (PAV) in genome content. PLoS Genet. 2009;5:e1000734. doi: 10.1371/journal.pgen.1000734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Trowsdale J, Barten R, Haude A, Andrew Stewart C, Beck S, Wilson MJ. The genomic context of natural killer receptor extended gene families. Immunol Rev. 2001;181:20–38. doi: 10.1034/j.1600-065X.2001.1810102.x. [DOI] [PubMed] [Google Scholar]
- 53.Morgante M, De Paoli E, Radovic S. Transposable elements and the plant pan-genomes. Curr Opin Plant Biol. 2007;10:149–155. doi: 10.1016/j.pbi.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 54.Tettelin H, Riley D, Cattuto C, Medini D. Comparative genomics: the bacterial pan-genome. Curr Opin Microbiol. 2008;11:472–477. doi: 10.1016/j.mib.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 55.Kuo C-H, Ochman H. The fate of new bacterial genes. FEMS Microbiol Rev. 2009;33:38–43. doi: 10.1111/j.1574-6976.2008.00140.x. [DOI] [PubMed] [Google Scholar]
- 56.Aherfi S, Andreani J, Baptiste E, Oumessoum A, Dornas FP, Andrade AC dos SP, et al. A large open pangenome and a small core genome for giant pandoraviruses. Front Microbiol. 2018;9 Frontiers:1486. [DOI] [PMC free article] [PubMed]
- 57.Khan AW, Garg V, Roorkiwal M, Golicz AA, Edwards D, Varshney RK. Super-pangenome by integrating the wild side of a species for accelerated crop improvement. Trends in Plant Science. 2020;25:148–158. doi: 10.1016/j.tplants.2019.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Badet T, Oggenfuss U, Abraham L, McDonald BA, Croll D. A 19-isolate reference-quality global pangenome for the fungal wheat pathogen Zymoseptoria tritici. BMC Biol. 2020;18:12. doi: 10.1186/s12915-020-0744-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stammnitz MR, Coorens THH, Gori KC, Hayes D, Fu B, Wang J, et al. The origins and vulnerabilities of two transmissible cancers in Tasmanian devils. Cancer Cell. 2018;33:607–619.e15. doi: 10.1016/j.ccell.2018.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Śmietanka B, Burzyński A, Hummel H, Wenne R. Glacial history of the European marine mussels Mytilus, inferred from distribution of mitochondrial DNA lineages. Heredity. 2014;113:hdy201423. doi: 10.1038/hdy.2014.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bierne N, Borsa P, Daguin C, Jollivet D, Viard F, Bonhomme F, et al. Introgression patterns in the mosaic hybrid zone between Mytilus edulis and M. galloprovincialis. Mol Ecol. 2003;12:447–461. doi: 10.1046/j.1365-294X.2003.01730.x. [DOI] [PubMed] [Google Scholar]
- 62.Ayari TE, Menif NTE, Hamer B, Cahill AE, Bierne N. The hidden side of a major marine biogeographic boundary: a wide mosaic hybrid zone at the Atlantic–Mediterranean divide reveals the complex interaction between natural and genetic barriers in mussels. Heredity. 2019;122:770–84. [DOI] [PMC free article] [PubMed]
- 63.Hurgobin B, Golicz AA, Bayer PE, Chan CK, Tirnaz S, Dolatabadian A, et al. Homoeologous exchange is a major cause of gene presence/absence variation in the amphidiploid Brassica napus. Plant Biotechnol J. 2018;16:1265–1274. doi: 10.1111/pbi.12867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Martínez-Lage A, González-Tizón A, Méndez J. Chromosome differences between European mussel populations (genus Mytilus) Caryologia. 1996;49:343–355. doi: 10.1080/00087114.1996.10797379. [DOI] [Google Scholar]
- 65.Bihari N, Mičić M, Batel R, Zahn RK. Flow cytometric detection of DNA cell cycle alterations in hemocytes of mussels (Mytilus galloprovincialis) off the Adriatic coast, Croatia. Aquat Toxicol. 2003;64:121–129. doi: 10.1016/S0166-445X(03)00040-7. [DOI] [PubMed] [Google Scholar]
- 66.Small KS, Brudno M, Hill MM, Sidow A. Extreme genomic variation in a natural population. Proc Natl Acad Sci U S A. 2007;104:5698–5703. doi: 10.1073/pnas.0700890104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dey A, Chan CKW, Thomas CG, Cutter AD. Molecular hyperdiversity defines populations of the nematode Caenorhabditis brenneri. Proc Natl Acad Sci U S A. 2013;110:11056–11060. doi: 10.1073/pnas.1303057110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bierne N, Bonhomme F, Boudry P, Szulkin M, David P. Fitness landscapes support the dominance theory of post-zygotic isolation in the mussels Mytilus edulis and M. galloprovincialis. Proc R Soc London B Biol Sci. 2006;273:1253–1260. doi: 10.1098/rspb.2005.3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rolff J, Siva-Jothy MT. Invertebrate ecological immunology. Science. 2003;301:472–475. doi: 10.1126/science.1080623. [DOI] [PubMed] [Google Scholar]
- 70.Cremer S, Pull CD, Fürst MA. Social immunity: emergence and evolution of colony-level disease protection. Annu Rev Entomol. 2018;63:105–123. doi: 10.1146/annurev-ento-020117-043110. [DOI] [PubMed] [Google Scholar]
- 71.Marçais G, Kingsford C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics. 2011;27:764–770. doi: 10.1093/bioinformatics/btr011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJM, Birol İ. ABySS: a parallel assembler for short read sequence data. Genome Res. 2009;19:1117–1123. doi: 10.1101/gr.089532.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cruz F, Julca I, Gómez-Garrido J, Loska D, Marcet-Houben M, Cano E, et al. Genome sequence of the olive tree, Olea europaea. GigaScience. 2016;5:29. doi: 10.1186/s13742-016-0134-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ye C, Hill CM, Wu S, Ruan J, Ma Z(S) DBG2OLC: efficient assembly of large genomes using long erroneous reads of the third generation sequencing technologies. Sci Rep. 2016;6:31900. doi: 10.1038/srep31900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics. 2011;27:578–579. doi: 10.1093/bioinformatics/btq683. [DOI] [PubMed] [Google Scholar]
- 76.Boetzer M, Pirovano W. SSPACE-LongRead: scaffolding bacterial draft genomes using long read sequence information. BMC Bioinformatics. 2014;15:211. doi: 10.1186/1471-2105-15-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.English AC, Richards S, Han Y, Wang M, Vee V, Qu J, et al. Mind the gap: upgrading genomes with Pacific Biosciences RS long-read sequencing technology. PLoS One. 2012;7:e47768. doi: 10.1371/journal.pone.0047768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hackl T, Hedrich R, Schultz J, Förster F. proovread: large-scale high-accuracy PacBio correction through iterative short read consensus. Bioinformatics. 2014;30:3004–3011. doi: 10.1093/bioinformatics/btu392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang SV, Zhuo L, Hahn MW. AGOUTI: improving genome assembly and annotation using transcriptome data. GigaScience. 2016;5:31. doi: 10.1186/s13742-016-0136-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kajitani R, Toshimoto K, Noguchi H, Toyoda A, Ogura Y, Okuno M, et al. Efficient de novo assembly of highly heterozygous genomes from whole-genome shotgun short reads. Genome Res. 2014;24:1384–1395. doi: 10.1101/gr.170720.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wood DE, Lu J, Langmead B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019;20:257. doi: 10.1186/s13059-019-1891-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, et al. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lusk RW. Diverse and widespread contamination evident in the unmapped depths of high throughput sequencing data. PLoS One. 2014;9:e110808. doi: 10.1371/journal.pone.0110808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Laetsch DR, Blaxter ML. BlobTools: interrogation of genome assemblies. F1000Res. 2017;6:1287. doi: 10.12688/f1000research.12232.1. [DOI] [Google Scholar]
- 88.Challis R, Richards E, Rajan J, Cochrane G, Blaxter M. BlobToolKit – interactive quality assessment of genome assemblies. G3 (Bethesda) 2020;10:1361–1374. doi: 10.1534/g3.119.400908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 90.Mapleson D, Garcia Accinelli G, Kettleborough G, Wright J, Clavijo BJ. KAT: a K-mer analysis toolkit to quality control NGS datasets and genome assemblies. Bioinformatics. 2017;33:574–576. doi: 10.1093/bioinformatics/btw663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Haas BJ, Salzberg SL, Zhu W, Pertea M, Allen JE, Orvis J, et al. Automated eukaryotic gene structure annotation using EVidenceModeler and the Program to Assemble Spliced Alignments. Genome Biol. 2008;9:R7. doi: 10.1186/gb-2008-9-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Iwata H, Gotoh O. Benchmarking spliced alignment programs including Spaln2, an extended version of Spaln that incorporates additional species-specific features. Nucleic Acids Res. 2012;40:e161. doi: 10.1093/nar/gks708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Parra G, Blanco E, Guigó R. GeneID in drosophila. Genome Res. 2000;10:511–515. doi: 10.1101/gr.10.4.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lomsadze A, Ter-Hovhannisyan V, Chernoff YO, Borodovsky M. Gene identification in novel eukaryotic genomes by self-training algorithm. Nucleic Acids Res. 2005;33:6494–6506. doi: 10.1093/nar/gki937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Stanke M, Waack S. Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics. 2003;19(Suppl 2):ii215–ii225. doi: 10.1093/bioinformatics/btg1080. [DOI] [PubMed] [Google Scholar]
- 96.Hunter S, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, et al. InterPro: the integrative protein signature database. Nucleic Acids Res. 2009;37:D211–D215. doi: 10.1093/nar/gkn785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016;44:D457–D462. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Conesa A, Götz S. Blast2GO: a comprehensive suite for functional analysis in plant genomics. Int J Plant Genomics. 2008;2008:619832. doi: 10.1155/2008/619832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Meth. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 100.Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, et al. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011;39:D225–D229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Falcon S, Gentleman R. Hypergeometric testing used for gene set enrichment analysis. Bioconductor case studies. New York: Springer; 2008. pp. 207–220. [Google Scholar]
- 104.Mirarab S, Warnow T. ASTRAL-II: coalescent-based species tree estimation with many hundreds of taxa and thousands of genes. Bioinformatics. 2015;31:i44–i52. doi: 10.1093/bioinformatics/btv234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rawson PD, Joyner KL, Meetze K, Hilbish TJ. Evidence for intragenic recombination within a novel genetic marker that distinguishes mussels in the Mytilus edulis species complex. Heredity. 1996;77:599–607. doi: 10.1038/hdy.1996.187. [DOI] [PubMed] [Google Scholar]
- 106.Inoue K, Waite JH, Matsuoka M, Odo S, Harayama S. Interspecific variations in adhesive protein sequences of Mytilus edulis, M. galloprovincialis, and M. trossulus. Biol Bull. 1995;189:370–375. doi: 10.2307/1542155. [DOI] [PubMed] [Google Scholar]
- 107.Daguin C, Borsa P. Genetic characterisation of Mytilus galloprovincialis Lmk. in North West Africa using nuclear DNA markers. J Exp Mar Biol Ecol. 1999;235:55–65. doi: 10.1016/S0022-0981(98)00163-4. [DOI] [Google Scholar]
- 108.Daguin C, Bonhomme F, Borsa P. The zone of sympatry and hybridization of Mytilus edulis and M. galloprovincialis, as described by intron length polymorphism at locus mac-1. Heredity (Edinb) 2001;86:342–354. doi: 10.1046/j.1365-2540.2001.00832.x. [DOI] [PubMed] [Google Scholar]
- 109.Ohresser M, Borsa P, Delsert C. Intron-length polymorphism at the actin gene locus mac-1: a genetic marker for population studies in the marine mussels Mytilus galloprovincialis Lmk. and M. edulis L. Mol Marine Biol Biotechnol. 1997;6:123–130. [PubMed] [Google Scholar]
- 110.Bierne N, David P, Boudry P, Bonhomme F. Assortative fertilization and selection at larval stage in the mussels Mytilus edulis and M. galloprovincialis. Evolution. 2002;56:292–298. doi: 10.1111/j.0014-3820.2002.tb01339.x. [DOI] [PubMed] [Google Scholar]
- 111.Bierne N, David P, Langlade A, Bonhomme F. Can habitat specialisation maintain a mosaic hybrid zone in marine bivalves? Mar Ecol Prog Ser. 2002;245:157–170. doi: 10.3354/meps245157. [DOI] [Google Scholar]
- 112.Gérard K, Bierne N, Borsa P, Chenuil A, Féral J-P. Pleistocene separation of mitochondrial lineages of Mytilus spp. mussels from Northern and Southern Hemispheres and strong genetic differentiation among southern populations. Mol Phylogenet Evol. 2008;49:84–91. doi: 10.1016/j.ympev.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 113.Stewart DT, Sinclair-Waters M, Rice A, Bunker RA, Robicheau BM, Breton S. Distribution and frequency of mitochondrial DNA polymorphisms in blue mussel (Mytilus edulis) populations of southwestern Nova Scotia (Canada) Can J Zool. 2018;96:608–613. doi: 10.1139/cjz-2017-0212. [DOI] [Google Scholar]
- 114.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 116.Gerdol M, Moreira R, Cruz F, Gómez-Garrido J, Vlasova A, Rosani U, et al. PRJEB24883. ENA. https://www.ebi.ac.uk/ena/browser/view/PRJEB24883 (2020).
- 117.Gerdol M, Moreira R, Cruz F, Gómez-Garrido J, Vlasova A, Rosani U, et al. PRJNA230138. ENA. https://www.ebi.ac.uk/ena/browser/view/PRJNA230138 (2020).
- 118.Huerta-Cepas J, Capella-Gutiérrez S, Pryszcz LP, Marcet-Houben M, Gabaldón T. Phylome ID: 599. PhylomeDB. http://phylomedb.org/phylome_599?q=phylome_browser&phyid=599 (2020).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Manuscript data notes. This file includes additional data notes 1–24, along with all the supplementary figures and most supplementary tables referenced in the main text.
Additional file 2. Large supplementary tables. This file includes Table S1, Table S3, Table S4, Table S6, Table S16, Table S17, Table S18, Table S19, Table S20, Table S21, Table S22, Table S23, Table S24, Table S25, Table S26, Table S27, Table S28, Table S29, Table S30, Table S31, Table S32, Table S33, Table S34, Table S40, Table S52, Table S53, Table S54, Table S55, Table S56 and Table S57.
Data Availability Statement
All the genome sequencing data obtained in this work, as well as the genome assembly and the annotation, are available in the European Nucleotide Archive (ENA) under the project IDs PRJEB24883 (Lola) [116] and PRJNA230138 (resequencing) [117]. A genome browser and a blast server for this genome can be accessed in our local server (https://denovo.cnag.cat/mussel/). The mussel phylome is available for download or browsing at PhylomeDB [118].