Abstract
BACKGROUND
Colorectal cancer is a leading cause of cancer-related death. Early onset colorectal cancer (age ≤45 years) is increasing and associated with advanced disease. While distinct molecular subtypes of colorectal cancer have been characterized, it is unclear whether age-related molecular differences exist.
OBJECTIVE
We sought to identify differences in gene expression between early and late onset (age ≥65 years) colorectal cancer.
DESIGN
We performed a review of our institution’s colorectal cancer registry and identified patients with colorectal cancer with tissue specimens available for analysis. We used The Cancer Genome Atlas to initially identify differences in gene expression between early and late onset colorectal cancer. In vitro experiments were performed on two colorectal cancer cell lines.
SETTINGS
The study was conducted at a tertiary medical center.
PATIENTS
Patients with early onset (N=28) or late onset (age ≥65 years old, N=38) at time of diagnosis were included.
MAIN OUTCOME MEASURES
The primary outcome was differential gene expression in patients with early versus late onset colorectal cancer. The secondary outcome was patient mortality.
RESULTS
Seven genes had increased expression in younger patients using The Cancer Genome Atlas. Only PEG10 was sufficiently expressed with quantitative polymerase chain reaction and had increased expression in our early onset group. Multivariable linear regression analysis identified age as a significant independent predictor of increased PEG10 expression. Outcomes data from The Cancer Genome Atlas suggests that PEG10 is associated with poor overall survival. In vitro studies in HCT-116 and HT-29 cell lines showed PEG10 contributes to cellular proliferation and invasion in colorectal cancer.
LIMITATIONS
Tissue samples were from formalin-fixed paraffin-embedded slides. Many patients did not have mutational status for review.
CONCLUSIONS
PEG10 is differentially expressed in early onset colorectal cancer and may functionally contribute to tumor cell proliferation and invasion. Increase in PEG10 expression correlates with decreased overall survival. See Video Abstract at http://links.lww.com/DCR/Bxxx.
Keywords: Colon cancer, Early onset, PEG10
Abstract
ANTECEDENTES
El cáncer colorrectal es una de las principales causas de muerte relacionada con el cáncer. El cáncer colorrectal de inicio temprano (edad ≤45 años) está en aumento y asociado con enfermedad avanzada. Aunque se han caracterizado distintos subtipos moleculares del cáncer colorrectal, no está claro si existen diferencias moleculares relacionadas con la edad.
OBJETIVO
Se buscó identificar diferencias en la expresión génica entre el cáncer colorrectal de inicio temprano y tardío (edad ≥ 65 años).
DISEÑO
Realizamos una revisión del registro de cáncer colorrectal de nuestra institución e identificamos pacientes con cáncer colorrectal con muestras de tejido disponibles para su análisis. Utilizamos el Atlas del Genoma del Cáncer para identificar inicialmente las diferencias en la expresión génica entre el cáncer colorrectal de inicio temprano y de inicio tardío. Se realizaron experimentos in vitro en dos líneas celulares de cáncer colorrectal.
AJUSTES
El estudio se realizó en un centro médico de tercer nivel.
PACIENTES
Se incluyeron pacientes con inicio temprano (N = 28) e inicio tardío (edad ≥65 años, N = 38) al momento del diagnóstico.
PRINCIPALES MEDIDAS DE RESULTADO
El resultado primario fue la expresión diferencial de genes en pacientes con cáncer colorrectal de inicio temprano versus tardío. El resultado secundario fue la mortalidad de los pacientes.
RESULTADOS
Siete genes aumentaron su expresión en pacientes más jóvenes usando el Atlas del Genoma del Cáncer. Solo PEG10 se expresó suficientemente con la reacción en cadena de la polimerasa cuantitativa y tuvo una mayor expresión en nuestro grupo de inicio temprano. El análisis de regresión lineal multivariable identificó la edad como un predictor independiente significativo del aumento de la expresión de PEG10. Los datos de resultados de el Atlas del Genoma del Cáncer sugieren que PEG10 está asociado con una pobre supervivencia general. Los estudios in vitro en líneas celulares HCT-116 y HT-29 mostraron que PEG10 contribuye a la proliferación e invasión celular en el cáncer colorrectal.
LIMITACIONES
Las muestras de tejido fueron de portaobjetos embebidos en parafina fijados con formalina. Muchos pacientes no tenían el estado de mutación para su revisión.
CONCLUSIONES
El PEG10 se expresa diferencialmente en el cáncer colorrectal de inicio temprano y puede contribuir funcionalmente a la proliferación e invasión de células tumorales. El aumento en la expresión de PEG10 se correlaciona con la disminución de la supervivencia general.
Consulte Video Resumen en http://links.lww.com/DCR/Bxxx. (Traducción—Dr. Gonzalo Hagerman)
INTRODUCTION
Colorectal cancer (CRC) remains the second leading cause of cancer-related death in the United States.1 While the mortality rate from CRC has decreased significantly over the past two decades, the disease has risen alarmingly in young people under the age of recommended initial screening.1–3 The incidence of rectal cancer in people 20–39 years old has quadrupled since 1980, and based on these trends, the incidence of colon and rectal cancer are predicted to increase by 90% and 124%, respectively, for people aged 20–34 by the year 2030.3,4 Early onset CRC is more often diagnosed in non-white patients, in the rectum, at an advanced stage, and with concerning pathological features, such as mucinous differentiation and signet ring histology.5–7 The majority of early onset CRC (80%) do not appear to be related to hereditary cancer syndromes.8 While experts postulate that the rise in early onset CRC is likely multifactorial, the actual etiologies are not known.
Gaps in understanding the possible molecular mechanism of this disease led us to explore the differences in gene expression between early and late onset CRC. Identifying different genetic signatures between the two cohorts will improve our overall understanding of this unique disease process. We hypothesized that early onset CRC has a distinct molecular signature in comparison to late onset disease which would be identifiable using a bioinformatics approach.
METHODS
TCGA Analysis
RNA sequencing data was obtained from The Cancer Genome Atlas (TCGA) via the cBioPortal. Data from patients with early onset disease (age ≤ 45 years) were compared to data from patients with late onset disease (age ≥ 65 years) to assess differences in gene expression. A log2 (fold change) was calculated, and genes with increased expression in early onset disease with a Benjamini-Hochberg adjusted p-value of < 0.15 were selected for further evaluation. This false discovery rate was selected to limit missing potentially important differential gene expression in this initial exploratory step.9,10 Analysis was performed with limma VOOM, part of the R software package. This analysis yielded seven genes with increased expression in early onset disease: GDF11, EPHX3, RPL39L, CACNA1I, PEG10, DSC3, and ZNF334. The TCGA survival data was obtained from the PanCancer Atlas cohort on August 14, 2019. Characteristics of the TCGA cohort are provided in supplemental table 1.
Cohort Analysis
Patient Selection
Patients diagnosed with CRC between 2008 and 2017 were identified using the Oregon Colorectal Cancer Registry and stratified into early onset if their age was ≤ 45 years old versus late onset if ≥ 65 years old at time of diagnosis. Ten patients who underwent colectomy for benign pathology were identified to serve as a control group. Patient demographics, neoadjuvant treatment regimens, clinical stage at time of diagnosis, and outcomes were obtained from the electronic medical record (EMR). Additionally, MSI and genetic mutational status were obtained from the EMR when available.
Tissue Specimens
CRC specimens were predominantly obtained at time of surgical oncologic resection, with the remainder from pre-treatment endoscopic biopsies. The formalin-fixed paraffin-embedded (FFPE) blocks were sectioned into 5μm-thick slides of tissue with at least 70% of the sectioned tissue containing tumor. Hematoxylin and Eosin slides were prepared for each sample to identify the tumor and were used as guides to sharply dissect the tumor from the unstained slides.
RNA extraction and qPCR
RNA was isolated using the RNeasy FFPE kit (Qiagen). RNA concentration and purity was confirmed with the NanoDrop spectrophotometer (ThermoFisher Scientific). Quantitative-PCR analysis was performed to assess expression of the seven genes identified in the TCGA analysis using TaqMan Gene Expression Assays (Applied Biosystems, Cat# 4448892 (Hs00227184_m1, Hs00227328_m1, Hs00738983_m1, Hs01096207_m1, Hs00170032_m1, Hs00737840_m1)) and the ViiA7 qRT-PCR machine (Applied Biosystems). Two housekeeping genes, GAPDH and ACTB (Applied Biosystems, Cat#433182 (Hs02758991_g1, Hs001060665_g1)), were used for all samples and expression was calculated using the 2−ΔCt method. Housekeeping gene expression Ct values were consistent (Supplement 2) with FFPE sample standards and did not vary across our age groups. We analyzed expression of all seven genes for the first ten samples, though in doing so, it became clear that PEG10 was the only gene with expression levels consistently above the detection limit (Ct values >36).
Statistical Analysis
Gene expression was reported as the change in the cycle threshold (−2ΔCt, normalized to housekeeping genes). For analysis, the change in cycle threshold was log-transformed to avoid violating the assumptions behind statistical tests. Pre-operative treatment was considered a single variable and separated into four groups: no pre-op treatment, systemic chemotherapy only, chemoradiation, and both systemic chemotherapy and chemoradiation. Associations with age for tissue type (colon or rectum), pre-op treatment, and AJCC stage (I-IV) were evaluated with univariate chi-square tests. Differences in PEG10 expression between age groups were evaluated with t-tests for univariable associations, and with multivariable linear regression accounting for pre-op treatment, sex, tissue type, and cancer stage. Significance was set at alpha=0.05. Statistical analysis was conducted in JMP-14 (SAS Inc., Cary, NC).
Survival Analysis
A Kaplan-Meier curve of overall survival was created from both our cohort and the TCGA cohort using GraphPad Prism 8. Survival was calculated from date of diagnosis. Patients were separated into high and low PEG10 expression groups based on the median value of PEG10 of our patient cohort. For the TCGA cohort, we excluded missense PEG10 mutations of unclear significance and only included patients with amplification mutations and high mRNA PEG10 in our “increased PEG10” group in comparison to the unaltered PEG10 group.
In Vitro Experiments
Cell Culture
HCT-116 and HT-29 cells (American Type Culture Collection) were selected because each line has moderate intrinsic PEG10 expression allowing for knockdown and gain of function assays. Additionally, each was originally harvested from individuals < 50 years old and present two different baseline mutational profiles. HT-29 cells are MSS and BRAF mutant and originally from a 44-year-old individual. HCT-116 cells are MSI and KRAS mutant and originally from a 48-year-old individual.11,12 Cells were cultured in McCoy’s media supplemented with 10% FBS under 5% CO2 conditions. Short tandem repeat profiling of the cell lines was performed to ensure integrity of our cell lines.
Transfection
For the knockdown assays, both cell lines were transfected with siRNA targeting PEG10 (Applied Biosystems, Cat#4392420) or control siRNA (Applied Biosystems, Cat#4390847) for 18hrs and plated as required for the assay. For the gain of function assay, cells were transfected with PEG10 plasmid (OriGene, Cat#RC208683) or control plasmid for 18hrs using the XtremeGENE Transfection Reagent (SigmaAldrich, Cat#06365779001). At 48hrs post-transfection, qPCR was performed to confirm that PEG10 gene expression was decreased for the knockdown assays in both cell lines and increased for the gain of function assay in the HT-29 cell line (Supplement 3).
Colony Formation
Following knockdown, cells were plated (100, 200, 400 or 800 cells/well) in triplicate in six-well culture plates. Ten days after plating, cells were fixed and stained with crystal violet and the colonies were counted. As cells were plated in triplicate for each density, mean colony number was used for calculations. In addition, ImageJ (National Institutes of Health) was used to quantify differences in cellular area given variable colony sizes in the HCT-116 cell line at higher densities.13
Cellular Proliferation and Death
Eighteen hours following transfection of each cell line, cells were transferred to 96-well plates at a density of 1000 cells/well and assayed for proliferation and cell death. Cell Titer-Glo (proliferation) and Caspase 3/7 Glo (cell death) assays were performed at 48hrs and 96hrs post-transfection, according to manufacturer’s (Promega) instructions. The same methods were repeated for the proliferation study using the HT-29 cells transfected with PEG10 plasmid or normal control.
Cellular Invasion
Twenty-four hours after knockdown, both cell lines were plated in a 96-well plate at 2.0 × 105 cells per well. Invasion was evaluated using the QCM 96-well cell invasion assay (8 μm, EMD Millipore Corp, Cat# ECM 555). Incubation was performed for 24hrs allowing for invasion across the ECMatrix. Cells were then dissociated, lysed, and quantified using fluorescence as detailed in manufacturer instructions.
RESULTS
TCGA Analysis
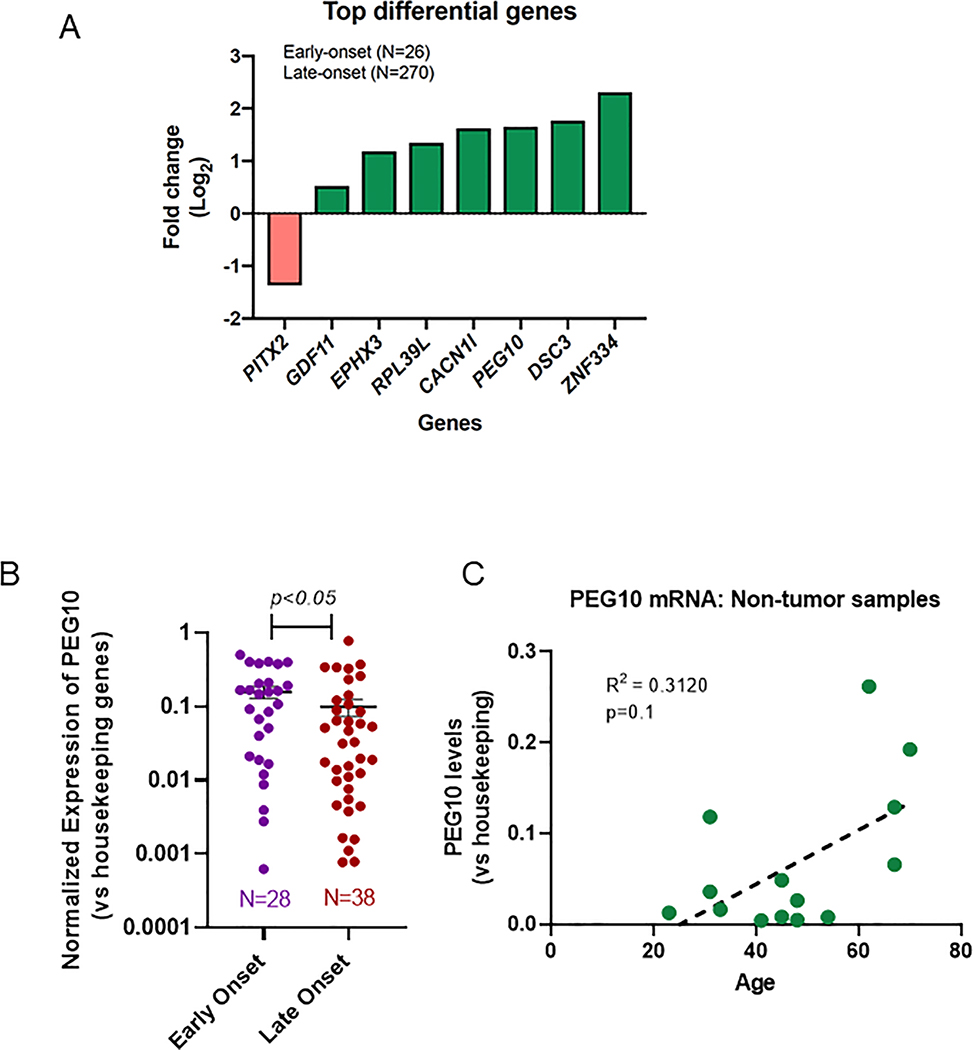
Analysis of the TCGA included 26 early onset patients and 270 late onset patients. One gene was found to have lower expression in the early onset group compared to the late onset group: PITX2 (log2(fold change) = −1.37, p<0.15). Seven genes were found to have higher expression in the early onset group compared to the late onset group: GDF11 (log2(fold change) = 0.52), EPHX3 (1.18), RPL39L (1.34), CACNA1I (1.62), PEG10 (1.65), DSC3 (1.77), and ZNF334 (2.31) (p<0.15) (Figure 1a).
Figure 1: Differential gene expression in early versus late colorectal cancer.
(A) Analysis of TCGA for molecular differences in early versus late onset CRC identified a total of 26 early onset cases and 270 late onset cases. Bars depict log2(fold change) of the most significant genes (Benjamini-Hochberg adjusted p-value<0.15) expressed in early onset tumors over late onset tumors. (B) PEG10 RNA expression (reported as −2^(ΔCt) scale) in early vs late onset samples measured via qPCR (mean + SEM 0.0158±0.029 vs. 0.099±0.026, p<0.05). (C) PEG10 expression by age for normal (non-tumor) controls with best fit line (R2 =0.3120), p=0.1.
Cohort Analysis
Patient Cohort
PEG10 expression was analyzed in a total of 66 patients with CRC, 28 with early onset (mean age 38.7 years) and 38 with late onset (mean age 72.9 years) disease. Aside from age, there were no significant demographic differences in tissue source, preoperative treatment, stage at diagnosis, or sex (Table 1). There were 11 rectal samples in the early onset group (39.3%) and 23 rectal samples in the late onset group (60.5%), but this difference was not significant. Using qPCR, PEG10 expression was higher in the early onset group compared to late onset group (Figure 1b). We were therefore able to validate the findings from the TCGA in our own patient cohort. Additionally, we analyzed PEG10 expression by age in a normal tissue (non-tumor) cohort and found that PEG10 expression trended towards an increase with age, however this was not significant (Figure 1c). This finding supports the hypothesis that PEG10 plays a role in early onset CRC and is not just a function of patient age independent of presence or absence of neoplastic disease.
Table 1:
Patient Characteristics
| Early Onset |
Late Onset |
||||
|---|---|---|---|---|---|
| Characteristic | N | % | N | % | p-value |
| Biopsy/Surg | |||||
| biopsy | 4 | 14.3% | 5 | 13.2% | 0.895 |
| surg | 24 | 85.7% | 33 | 86.8% | |
| Tissue | |||||
| Colon | 17 | 60.7% | 15 | 39.5% | 0.088 |
| Rectum | 11 | 39.3% | 23 | 60.5% | |
| Pre-op Treatment | |||||
| Chemo and chemorad | 3 | 10.7% | 4 | 10.5% | 0.771 * |
| Chemo only | 6 | 21.4% | 5 | 13.2% | |
| Chemorad only | 3 | 10.7% | 3 | 7.9% | |
| No pre-op treatment | 16 | 57.1% | 26 | 68.4% | |
| Stage at Diagnosis | |||||
| I | 2 | 7.1% | 6 | 15.8% | 0.280 * |
| II | 8 | 28.6% | 18 | 47.4% | |
| III | 9 | 32.1% | 7 | 18.4% | |
| IV | 9 | 32.1% | 7 | 18.4% | |
| Sex | |||||
| Male | 13 | 46.4% | 21 | 55.3% | 0.619 |
| Female | 15 | 53.6% | 17 | 44.7% | |
The differences in group characteristics are not statistically significant. Chi-squared test where not indicated
indicates fisher exact test. Pre-op = pre-operative, chemo = chemotherapy, chemorad = chemoradiation.
Univariable analysis demonstrated that PEG10 expression varied by age (mean 0.068, 95%CI: 0.35–0.13 vs. 0.027, 95%CI: 0.015–0.050, p=0.04) (Table 2). This remained significant with multivariable analysis and early stage was also associated with increased PEG10 expression (Table 3). The relationship of PEG10 with mutational status of the tumors was not assessed in the current study given the prevalence of missing information.
Table 2:
Mean Univariable Estimates and 95% Confidence Intervals for mRNA PEG10 Expression.
| PEG10 (N=66) | ||||||
|---|---|---|---|---|---|---|
| 95% CI |
||||||
| Term | N | % | Mean | Lower | Upper | Prob>|t| |
| Age Group | ||||||
| ≤45 | 28 | 42.4% | 0.0683 | 0.0350 | 0.1332 | 0.0429 |
| ≥65 | 38 | 57.6% | 0.0272 | 0.0147 | 0.0503 | |
| Tissue | ||||||
| Colon | 33 | 50.0% | 0.0270 | 0.0135 | 0.0537 | 0.0795 |
| Rectum | 33 | 50.0% | 0.0600 | 0.0329 | 0.1094 | |
| Pre-op Treatment | ||||||
| Chemo and chemorad | 7 | 10.6% | 0.0421 | 0.0095 | 0.1871 | 0.9895 |
| Chemo only | 11 | 16.7% | 0.0348 | 0.0091 | 0.1337 | |
| Chemorad only | 6 | 9.1% | 0.0478 | 0.0042 | 0.5388 | |
| None | 42 | 63.6% | 0.0405 | 0.0227 | 0.0720 | |
| Stage at Diagnosis | ||||||
| I | 8 | 12.1% | 0.1453 | 0.0476 | 0.4437 | 0.0858 |
| II | 26 | 39.4% | 0.0228 | 0.0099 | 0.0526 | |
| III | 16 | 24.2% | 0.0500 | 0.0226 | 0.1108 | |
| IV | 16 | 24.2% | 0.0428 | 0.0165 | 0.1111 | |
| Sex | ||||||
| Male | 32 | 48.5% | 0.1026 | 0.0421 | 0.1631 | 0.1786 |
| Female | 34 | 51.5% | 0.1439 | 0.0928 | 0.1949 | |
Age was significant in both univariable and multivariable analyses. Early stage was also significantly associated with increased PEG10, though only in the multivariable analysis. For both analyses, the outcome variable was log-transformed and results reported on −2^(ΔCt) scale. Pre-op= pre-operative, chemo= chemotherapy, chemorad= chemoradiation.
Table 3.
Multivariable Estimates and 95% Confidence Intervals for mRNA PEG10 Expression
| PEG10 (N=66) | ||||
|---|---|---|---|---|
| 95% CI | ||||
| Term | Estimate | Lower | Upper | Prob>|t| |
| Age (< 45) | 1.87 | 1.18 | 2.95 | 0.0085 |
| Pre-op Treatment | 0.82 | 0.26 | 2.62 | 0.7388 |
| Tissue (Colon) | 0.62 | 0.37 | 1.04 | 0.0712 |
| Stage at Diagnosis | ||||
| I | 3.08 | 1.11 | 8.56 | 0.0312 |
| II | 0.48 | 0.23 | 1.02 | 0.0548 |
| III | 0.69 | 0.30 | 1.58 | 0.3751 |
| Sex (Female) | 0.65 | 0.41 | 1.01 | 0.0533 |
Age was significant in both univariable and multivariable analyses. Early stage was also significantly associated with increased PEG10, though only in the multivariable analysis. For both analyses, the outcome variable was log-transformed and results reported on −2^(ΔCt) scale. Pre-op= pre-operative, chemo= chemotherapy, chemorad= chemoradiation.
PEG10 and Outcomes
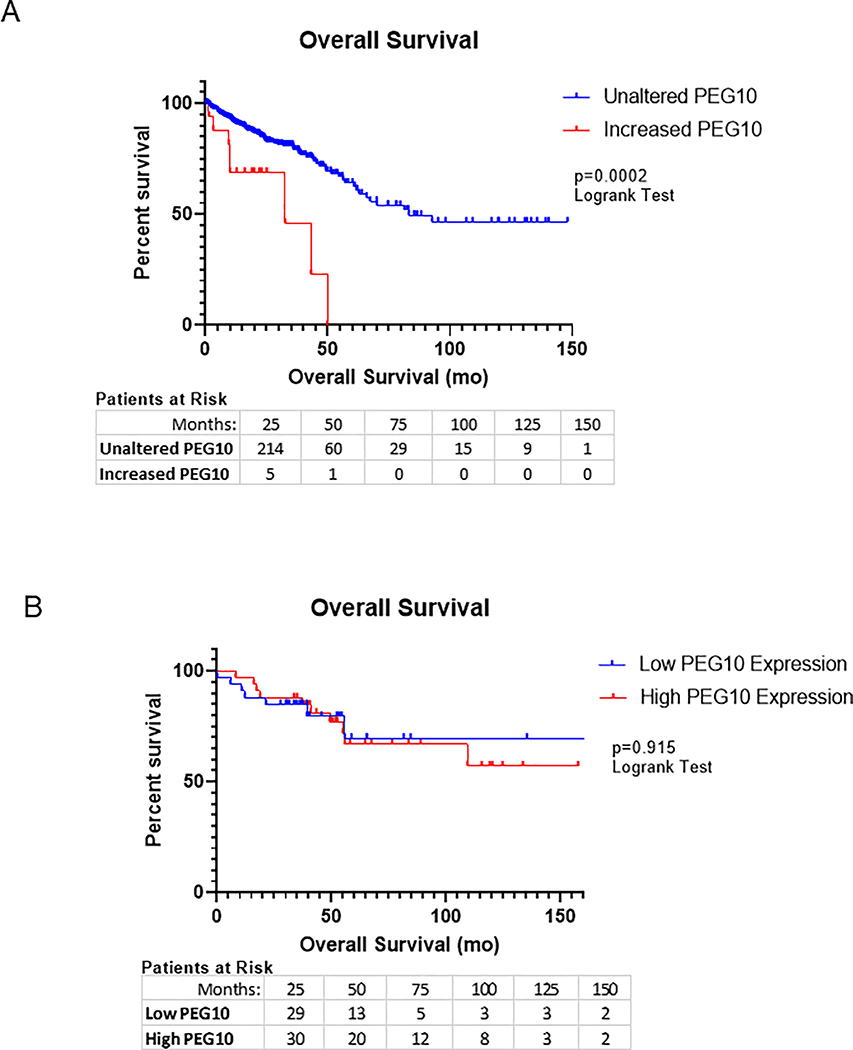
PEG10 alteration also appears to negatively impact patient outcomes. Using the TCGA PanCancer Atlas for colorectal adenocarcinoma, we identified altered PEG10 in 25 of 524 samples, approximately 5% of patients. The majority of these alterations were either upregulation in mRNA levels (4%) or mutations (1%).14,15 Patients with increased PEG10 expression had significantly worse overall survival compared to those without an alteration in PEG10 (Figure 2a), with median survival decreasing to 38 months in those with abnormal PEG10 versus 83 months in the control. We then evaluated survival in our cohort though were unable to appreciate the same trend (Figure 2b).
Figure 2: PEG10 expression correlates with overall survival in CRC.
(A) Kaplan-Meier plot of overall patient survival measured in months in the TCGA cohort. Median survival for increased PEG10 was 32mo vs. 83mo for unaltered PEG10 cases (log-rank test p-value= 0.0002). (B) Kaplan-Meier estimates generated for our own patient cohort (n=66).
In vitro Experiments
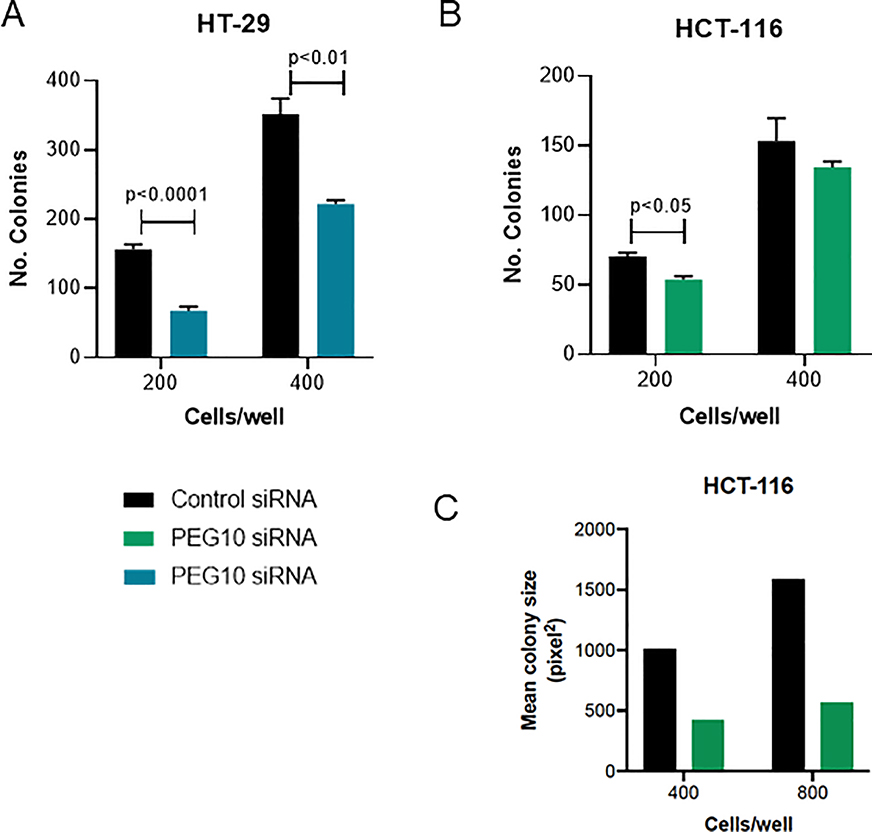
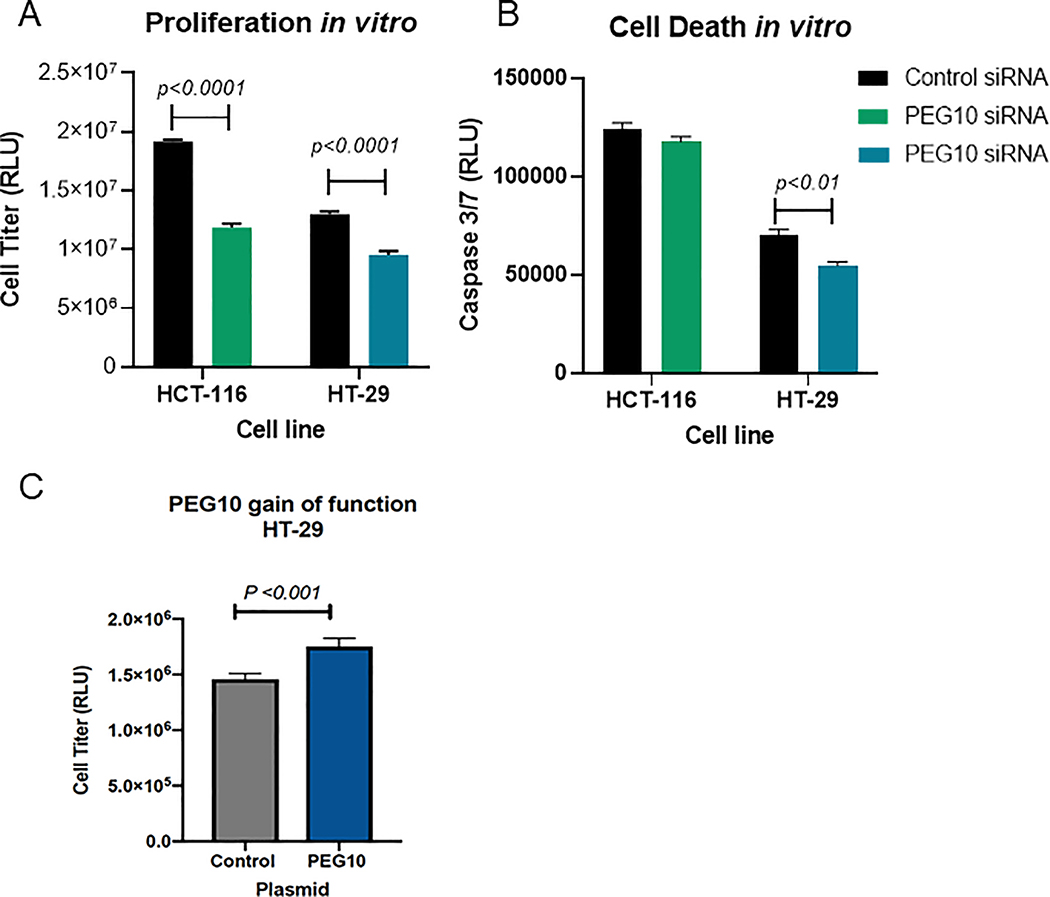
PEG10 knockdown resulted in a significant decrease in colony formation in both cell lines (Figure 3a-c). Colony numbers in the HCT-116 cells were not significantly different at higher cell numbers. This was largely due to confluence of the colonies and increased colony volume at these time points as supported by measurement of the area fraction using ImageJ (Figure 3c). PEG10 therefore appears to have an important role in colony proliferation in both colorectal cancer cell types, with PEG10 knockdown resulted in significantly fewer and less robust colonies. Knockdown cells in both lines had decreased proliferation compared to controls (Figure 4a). We observed less cell death in HT-29 PEG10 knockdown cells, but no significant difference in HCT-116 (Figure 4b). Conversely, we found that ectopic expression of PEG10 using a plasmid vector in the HT-29 cell line resulted in increased cellular proliferation (Figure 4c).
Figure 3: PEG10 silencing decreases tumor cell colony formation.
(A-B) HCT-116 and HT-29 cells were transfected with indicated siRNAs. Colony formation was assayed by crystal violet staining 10 days post-transfection. Bars show mean + SEM of three wells. P<0.05 via Student’s two-tailed t test was considered statistically significant. (C) Colony Area Fraction was calculated using ImageJ for the HCT-116 cell line in the higher density plates (400 & 800 cells/well).
Figure 4: PEG10 is associated with CRC cell proliferation in vitro.
siPEG10 knockdown studies in HCT-116 and HT-29 cell lines (A-B) demonstrated the effect of PEG10 on proliferation and cell death and gain of function study (C) demonstrating cell proliferation in HT-29 cells. Cell proliferation was measured at 96 hrs using the CellTiter Glo assay and cell death using the Caspase 3/7 assay. Bars show mean + SEM of technical replicates. P<0.05 via Student’s two-tailed t test was considered statistically significant.
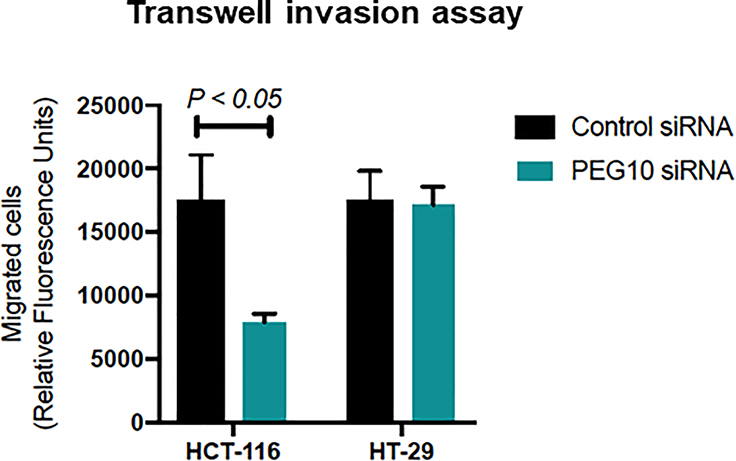
Alterations in PEG10 may also affect cellular behavior in regard to invasion and migration. Using a transwell invasion assay, we demonstrated that PEG10 may confer increased invasion capabilities (Figure 5), though this effect was only observed in the HCT-116 cell line. This may be due to silencing being more effective in the HCT-116 cell line, which is consistent with the fact that HCT-116 has higher intrinsic PEG10 expression than does HT-29 (Supplement #3). However, we did not observe a significant difference in migration in either cell line using a scratch assay between 0 and 8hrs (data not shown). There were differences at 24hrs, however this was likely secondary to proliferation instead of migration alone. Overall, our data demonstrate that PEG10 expression appears to predominantly play a role in cellular proliferation.
Figure 5: PEG10 expression in HCT-116 is associated with invasion.
A transwell invasion assay with HCT-116 and HT-29 cells show decreased invasion in HCT-116 cells following knockdown of PEG10, though not in HT-29. Bars show mean + SEM of technical replicates and a Student’s two-tailed t test was used.
DISCUSSION
Early onset colorectal cancer is increasing dramatically in the United States, a trend which is concerning yet unexplained. While physicians and scientists speculate that this is likely multifaceted, diverse approaches to understanding this phenomenon are needed.16 We have hypothesized that early onset disease may have a unique molecular signature, potentially contributing to unique disease characteristics. While there may be multiple factors or molecular signatures at play, using a combination of bioinformatics and experimental approaches, we have identified PEG10 as a differentially expressed gene in early versus late onset CRC. We found PEG10 expression in a limited number of non-tumor samples was not correlated with age, suggesting that the increased expression of PEG10 in early onset CRC specimens appears to be a result of the tumor itself and not just due to age related differences in PEG10 expression. Using the TCGA, we found that PEG10 had negative impacts on survival, though we were not able to support these findings in our own smaller patient cohort. Using two colorectal cancer cell lines with differing molecular drivers, we identified that increased levels of PEG10 resulted in increased colony formation, primarily through increased cellular proliferation. We also found that PEG10 may play a role in the ability of cellular invasion by neoplastic cells.
PEG10 is a paternally expressed imprinted gene which is located on human chromosome 7q21 and encodes for two protein isoforms which have been shown to be primarily cytosolic, though with association with the nuclear membrane.17,18 PEG10 has been implicated in multiple neoplastic processes, including lymphoma, hepatocellular carcinoma (HCC), lung cancer, breast cancer, and prostate cancer.18–26 We performed several experiments to better understand how PEG10 contributes to neoplastic behavior in colorectal cancer. Increased levels of PEG10 were associated with increased colony formation, and we found that this was largely driven by an increase in cellular proliferation in contrast to a decrease in cellular death. PEG10 proliferative effects have been identified in HCC, breast cancer and pancreatic cancer cell lines, with PEG10 knockdown resulting in G0/G1 cell cycle arrest.26–28 Additionally, our in vitro studies suggest that increased PEG10 expression is associated with increased invasion capacity. This has been attributed to a downstream increase of β-catenin and reduction of E-cadherin expression resulting in weakened cell junctions, allowing for migration and metastasis.23,26 PEG10 has also been shown to result in extracellular matrix degradation, through upregulation of MMP-1, MMP-2 and MMP-9 in HCC,23,26 and through ERK/MMP7 pathway in pancreatic cancer.28
PEG10 expression in colorectal cancer cell lines is associated with cellular proliferation and invasion which may contribute to more aggressive disease. We observed that the expression of PEG10 is upregulated in ~5% of CRC patients in the TCGA pan-cancer dataset, and these patients have significantly decreased overall survival. We did not appreciate this relationship with survival in our cohort, though we were limited by our cohort size in comparison to the TCGA. Our limited cohort suggests that increased PEG10 levels may be associated with younger stage and earlier stage, though this merits further evaluation in larger cohorts. If this trend is confirmed in larger cohorts, this may suggest that PEG10 contributes to more aggressive disease. Our results join a growing body of literature demonstrating that PEG10 is oncogenic in various types of cancer, particularly hepatocellular carcinoma, lung cancer, prostate cancer, and breast cancer, and is associated with worse disease.18,25–27 Specifically, Deng et al demonstrated that increased PEG10 correlated with advanced stage and conferred worse survival in lung cancer; Li et al reported an association with aggressive clincopathological features and worse outcomes in patients with increased PEG10 expression in patients with breast cancer.23,26 Similarly, this association was demonstrated in patients with gallbladder and hepatocellular carcinoma.21,22
In the case of CRC, we demonstrate that PEG10 expression appears to be associated with younger age, something which has not been appreciated in other cancers. With CRC incidence increasing rapidly among younger individuals, especially those under the age of routine screening, it is important that we begin to understand some of the divergent characteristics of the disease seen with age. PEG10 is one factor which may contribute to early and proliferative disease and joins several other proteins which have been identified as differentially expressed in early versus late onset CRC. Wang et al identified PRL, RBM3, and Wrap53 as proteins associated with cancer-specific survival with different expression in early and late onset CRC, but these proteins do not appear to share a common pathway with PEG10.29 There also appear to be differences in early and late onset rectal carcinogenesis, with MAPK and cell cycle signaling more commonly affected in early onset rectal cancer compared to PI3K-AKT signaling in late onset rectal cancer.30 Of note, the CTNNB1 gene was found to be over-expressed in early onset CRC by Kirzin et al, resulting in over-activation of β-catenin.31 In the CRC cell line HCT-116, PEG10 overexpression increased expression of Wnt1 and β-catenin thereby increasing cellular proliferation. This may indicate that this pathway is activated more frequently in early onset CRC through various upstream molecules such as PEG10.
Our study is limited by the use of FFPE specimens that have been archived for varying lengths of time. However, our comparison groups had similar distribution of RNA quality metrics. For example, the expression of housekeeping genes may have decreased slightly with sample age but did not differ significantly between early and late onset groups. Another limitation is that the TCGA cohort rectal cancer patients did not receive neoadjuvant chemoradiation. We attempted to mitigate this limitation with the use of own patient cohort that included rectal cancer patients that were treated with neoadjuvant chemoradiation. In order to ensure the difference in survival in the TCGA cohort was not due to this deviation from standard care, we also performed the survival analysis with colon cancer patients only, but found that this did not affect our findings. In addition, we were unable to evaluate the relationship of PEG10 to known mutations in patient specimens given the prevalence of absent genetic information. In order to overcome this limitation, we used cancer cell lines with known mutations. Next steps would include evaluation of PEG10 function in vivo and increasing our human cohort sample size.
CONCLUSION
We hypothesized there may be unique molecular signatures between early and late onset colorectal cancer and identifying these signatures may help us understand the increasing CRC rates in younger individuals. We have identified elevated PEG10 levels as a potentially unique marker in early onset CRC and have also found it to be associated with more aggressive disease. PEG10 is associated with cellular proliferation and invasion in vitro which may explain why it is associated with worse overall survival in the TCGA cohort. Additional studies addressing the mechanism of PEG10 in CRC may lead to therapeutic adjuncts for patients with early onset colorectal cancer.
Supplementary Material
ACKNOWLEDGMENTS
Authors would like to thank Abhinav Nellore, PhD, for his assistance with processing of the TCGA data and Raga Siddarthan, MD, for his multiple contributions to this project.
Funding/Support: NIH R01 (5R01 HL137779–02) and NIH R01 (5R01 HL143803–02) to SA
Footnotes
Financial Disclaimers: The authors have no conflicts of interest.
Watson and Gardner had equal contributions (first authors) and Tsikitis and Anand had equal contributions (last authors).
REFERENCES
- 1.Noone A, Howlader N, Krapcho M, et al. Cancer Statistics Review, 1975–2015 - SEER Statistics. 2018. [Google Scholar]
- 2.Zauber AG, Winawer SJ, O’Brien MJ, et al. Colonoscopic polypectomy and long-term prevention of colorectal-cancer deaths. N Engl J Med. 2012;366:687–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bailey CE, Hu CY, You YN, et al. Increasing disparities in the age-related incidences of colon and rectal cancers in the United States, 1975–2010. JAMA Surg. 2015;150:17–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tawadros PS, Paquette IM, Hanly AM, Mellgren AF, Rothenberger DA, Madoff RD. Adenocarcinoma of the rectum in patients under age 40 is increasing: impact of signet-ring cell histology. Dis Colon Rectum. 2015;58:474–478. [DOI] [PubMed] [Google Scholar]
- 5.Dozois EJ, Boardman LA, Suwanthanma W, et al. Young-onset colorectal cancer in patients with no known genetic predisposition: can we increase early recognition and improve outcome? Medicine (Baltimore). 2008;87:259–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O’Connell JB, Maggard MA, Liu JH, Etzioni DA, Ko CY. Are survival rates different for young and older patients with rectal cancer? Dis Colon Rectum. 2004;47:2064–2069. [DOI] [PubMed] [Google Scholar]
- 7.Abdelsattar ZM, Wong SL, Regenbogen SE, Jomaa DM, Hardiman KM, Hendren S. Colorectal cancer outcomes and treatment patterns in patients too young for average-risk screening. Cancer. 2016;122:929–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cavestro GM, Mannucci A, Zuppardo RA, Di Leo M, Stoffel E, Tonon G. Early onset sporadic colorectal cancer: worrisome trends and oncogenic features. Dig Liver Dis. 2018;50:521–532. [DOI] [PubMed] [Google Scholar]
- 9.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
- 10.Reiner A, Yekutieli D, Benjamini Y. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics. 2003;19:368–375. [DOI] [PubMed] [Google Scholar]
- 11.Berg KCG, Eide PW, Eilertsen IA, et al. Multi-omics of 34 colorectal cancer cell lines - a resource for biomedical studies. Mol Cancer. 2017;16:116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ahmed D, Eide PW, Eilertsen IA, et al. Epigenetic and genetic features of 24 colon cancer cell lines. Oncogenesis. 2013;2:e71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Connell LC, Mota JM, Braghiroli MI, Hoff PM. The rising incidence of younger patients with colorectal cancer: questions about screening, biology, and treatment. Curr Treat Options Oncol. 2017;18:23. [DOI] [PubMed] [Google Scholar]
- 17.Ono R, Kobayashi S, Wagatsuma H, et al. A retrotransposon-derived gene, PEG10, is a novel imprinted gene located on human chromosome 7q21. Genomics. 2001;73:232–237. [DOI] [PubMed] [Google Scholar]
- 18.Tsou AP, Chuang YC, Su JY, et al. Overexpression of a novel imprinted gene, PEG10, in human hepatocellular carcinoma and in regenerating mouse livers. J Biomed Sci. 2003;10:625–635. [DOI] [PubMed] [Google Scholar]
- 19.Kainz B, Shehata M, Bilban M, et al. Overexpression of the paternally expressed gene 10 (PEG10) from the imprinted locus on chromosome 7q21 in high-risk B-cell chronic lymphocytic leukemia. Int J Cancer. 2007;121:1984–1993. [DOI] [PubMed] [Google Scholar]
- 20.Hu C, Xiong J, Zhang L, et al. PEG10 activation by co-stimulation of CXCR5 and CCR7 essentially contributes to resistance to apoptosis in CD19+CD34+ B cells from patients with B cell lineage acute and chronic lymphocytic leukemia. Cell Mol Immunol. 2004;1:280–294. [PubMed] [Google Scholar]
- 21.Bang H, Ha SY, Hwang SH, Park CK. Expression of PEG10 is associated with poor survival and tumor recurrence in hepatocellular carcinoma. Cancer Res Treat. 2015;47:844–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Z, Yang Z, Liu D, et al. TSG101 and PEG10 are prognostic markers in squamous cell/adenosquamous carcinomas and adenocarcinoma of the gallbladder. Oncol Lett. 2014;7:1128–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deng X, Hu Y, Ding Q, et al. PEG10 plays a crucial role in human lung cancer proliferation, progression, prognosis and metastasis. Oncol Rep. 2014;32:2159–2167. [DOI] [PubMed] [Google Scholar]
- 24.Li CM, Margolin AA, Salas M, et al. PEG10 is a c-MYC target gene in cancer cells. Cancer Res. 2006;66:665–672. [DOI] [PubMed] [Google Scholar]
- 25.Akamatsu S, Wyatt AW, Lin D, et al. The placental gene PEG10 promotes progression of neuroendocrine prostate cancer. Cell Rep. 2015;12:922–936. [DOI] [PubMed] [Google Scholar]
- 26.Li X, Xiao R, Tembo K, et al. PEG10 promotes human breast cancer cell proliferation, migration and invasion. Int J Oncol. 2016;48:1933–1942. [DOI] [PubMed] [Google Scholar]
- 27.Okabe H, Satoh S, Furukawa Y, et al. Involvement of PEG10 in human hepatocellular carcinogenesis through interaction with SIAH1. Cancer Res. 2003;63:3043–3048. [PubMed] [Google Scholar]
- 28.Peng YP, Zhu Y, Yin LD, et al. PEG10 overexpression induced by E2F-1 promotes cell proliferation, migration, and invasion in pancreatic cancer. J Exp Clin Cancer Res. 2017;36:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang MJ, Ping J, Li Y, et al. The prognostic factors and multiple biomarkers in young patients with colorectal cancer. Sci Rep. 2015;5:10645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nfonsam V, Xu W, Koblinski J, Jandova J. Gene expression analysis of sporadic early-onset rectal adenocarcinoma. Gastrointest Cancer (Jersey City). 2016;1:1. [PMC free article] [PubMed] [Google Scholar]
- 31.Kirzin S, Marisa L, Guimbaud R, et al. Sporadic early-onset colorectal cancer is a specific sub-type of cancer: a morphological, molecular and genetics study. PLoS One. 2014;9:e103159. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.