Abstract
Background
Carbapenem resistant Acinetobacter species have caused great difficulties in clinical therapy in the worldwide. Here we describe an Acinetobacter johnsonii M19 with a novel blaOXA-23 containing transposon Tn6681 on the conjugative plasmid pFM-M19 and the ability to transferand carbapenem resistance.
Methods
A. johnsonii M19 was isolated under selection with 8 mg/L meropenem from hospital sewage, and the minimum inhibitory concentrations (MICs) for the representative carbapenems imipenem, meropenem and ertapenem were determined. The genome of A. johnsonii M19 was sequenced by PacBio RS II and Illumina HiSeq 4000 platforms. A homologous model of OXA-23 was generated, and molecular docking models with imipenem, meropenem and ertapenem were constructed by Discovery Studio 2.0. Type IV secretion system and conjugation elements were identified by the Pathosystems Resource Integration Center (PATRIC) server and the oriTfinder. Mating experiments were performed to evaluate transfer of OXA-23 to Escherichia coli 25DN.
Results
MICs of A. johnsonii M19 for imipenem, meropenem and ertapenem were 128 mg/L, 48 mg/L and 24 mg/L, respectively. Genome sequencing identified plasmid pFM-M19, which harbours the carbapenem resistance gene blaOXA-23 within the novel transposon Tn6681. Molecular docking analysis indicated that the elongated hydrophobic tunnel of OXA-23 provides a hydrophobic environment and that Lys-216, Thr-217, Met-221 and Arg-259 were the conserved amino acids bound to imipenem, meropenem and ertapenem. Furthermore, pFM-M19 could transfer blaOXA-23 to E. coli 25DN by conjugation, resulting in carbapenem-resistant transconjugants.
Conclusions
Our investigation showed that A. johnsonii M19 is a source and disseminator of blaOXA-23 and carbapenem resistance. The ability to transfer blaOXA-23 to other species by the conjugative plasmid pFM-M19 raises the risk of spread of carbapenem resistance.
Graphic abstract
The carbapenem resistance gene blaOXA-23 is disseminated by a conjugative plasmid containing the novel transposon Tn6681 in Acinetobacter johnsonii M19.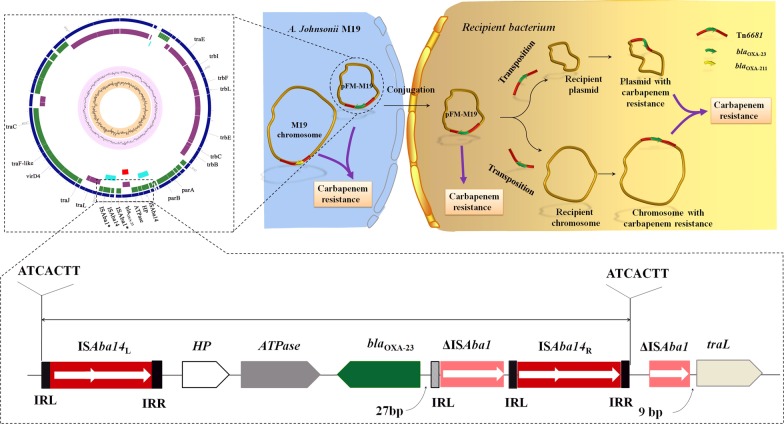
Keywords: Acinetobacter johnsonii, Carbapenem resistance, Conjugative plasmid, Novel transposon Tn6681, blaOXA-23
Introduction
Carbapenems are considered to be reliable and effective antibiotic agents against most pathogenic bacteria because of their broad antibacterial spectrum [1] and are used in the treatment of serious nosocomial infections caused by cephalosporin-resistant bacteria [1]. Species of the Acinetobacter genus are extremely well adapted to the hospital environment and can easily become resistant to available antimicrobial agents; therefore, the isolation of carbapenem-resistant Acinetobacter species has raised increasing concerns [2–6]. Acinetobacter johnsonii is an opportunistic human pathogen that colonizes humans but rarely causes clinical infections. Nevertheless, verification of a carbapenem-resistant strain of A. johnsonii encoding an extended-spectrum β-lactamase raises concern [7, 8].
β-lactamases are common mediators of β-lactam resistance and have been divided into four classes: A, B, C and D [2]. Members of class D, which are also referred to oxacillinases (OXAs), are notable contributors to carbapenem resistance and have been frequently observed in Acinetobacter species [9]. OXAs with carbapenemase activity were classified into 12 subgroups based on their amino acid sequences [10], and OXA-23 is the major source of carbapenem resistance in Acinetobacter [11].
From the bacterial perspective, conjugative plasmids are an ideal vehicle for transferring resistance genes among species. Fortunately, only a few types of plasmids in Acinetobacter species are conjugative and able to transfer resistance genes into new hosts [12]. However, numerous transposons, such as Tn2006, Tn2007, Tn2008 and Tn2009, have frequently been found to be associated with OXA genes [13]. The migration of OXA genes onto transposons has allowed them to become transmissible factors [14]. In this study, we isolated the high-level carbapenem-resistant strain A. johnsonii M19 from hospital sewage and discovered that it contained a novel transposon in a conjugative plasmid, thus allowing us to explore the potential for dissemination of carbapenem resistance by this species. These results provide new insights into the mechanisms of dissemination of carbapenem resistance.
Materials and methods
Isolation and identification of the carbapenem-resistant strain M19
Hospital sewage was obtained from the influx of the wastewater treatment facility in Shandong province, China. The sewage samples were diluted and spread onto Luria–Bertani (LB) agar plates containing 8 mg/L meropenem (Sigma Co. Shanghai, China) and then incubated at 30 °C for 24 h. A single clone, named M19, was isolated and cultured in LB medium containing meropenem at 30 °C overnight and stored in 15% glycerin at − 20 °C.
A partial fragment of the 16S rRNA gene of M19 was amplified with the universal primers 27F (5′-agagtttgatcctggctcag-3′) and 1492R (5′-ggttaccttgttacgactt-3′) and sequenced. Similarity analyses of the 16S rRNA sequences were conducted using BLASTn (https://blast.ncbi.nlm.nih.gov/Blast.cgi). A phylogenetic tree was produced by using the neighbor-joining algorithms with the Molecular Evolutionary Genetics Analysis 7 (MEGA 7) software based on BLAST results of the 16S rRNA sequence [15]. Antimicrobial susceptibility tests were performed to determine the MICs for carbapenems based on the breakpoints defined by the Clinical and Laboratory Standards Institute [4].
Whole-genome sequencing, annotation and analysis
The M19 genome was sequenced by PacBio RS II and Illumina HiSeq 4000 platforms at BGI Co., Ltd. (Wuhan, China). Gene prediction was performed on the M19 genome assembly by glimmer3 (https://www.cbcb.umd.edu/software/glimmer/) with Hidden Markov models [16]. Genome annotation was performed using the Prokaryotic Genome Annotation Pipeline on NCBI (https://ncbi.nlm.nih.gov/genome/annotation_prok/). Virulence factors and pathogenicity analysis were identified based on the core dataset in the Virulence Factors of Pathogenic Bacteria database (VFDB) [17] and the Pathogen Host Interactions (PHI) database [18].
Bioinformatics analyses of resistance genes, transposon and conjugation system
Antibiotic resistance genes (ARGs) were analyzed by RAST and BLASTp based on the core dataset in the Antibiotic resistance genes database (ARDB) [19]. Multi-sequence comparison was carried out by Clustal Omega [20] and ESPript [21]. Homologous model construction was operated by Discovery Studio 2.0 [22]. Molecular docking was performed by the CDOCKER protocol of Discovery Studio 2.0 [22]. IS transposases were detected by IS-Finder [23]. The new transposon was denominated and registered as Tn6681, according to the Transposon Registry (https://transposon.lstmed.ac.uk/). The genetic context of Tn6681 was compared with Tn2008 and Tn2008B using BLASTn. The conjugation system was identified by PATRIC server [24] and oriTfinder [25].
Mating experiments
Broth-based mating experiments were carried out using M19 as the donor and Escherichia coli 25DN as the recipient as described previously [26]. M19 and 25DN were cultivated overnight in LB medium containing 8 mg/L meropenem and 220 mg/L sodium azide. The mixture was incubated at 37 °C for 30 min, and transconjugants were selected on plates containing 8 mg/L meropenem, 220 mg/L sodium azide and 0.1 mg/L 5-bromo-4-chloro-3-indolyl-beta-D-glucuronic acid. The conjugal transfer efficiency was calculated, and six transconjugants, named MAT-1 to MAT-6, were isolated and purified. The MICs of carbapenems for these transconjugants were determined as above. To determine whether the plasmid pFM-M19 and blaOXA-23 were transferred to transconjugants, DNA fragments of the plasmid in transconjugants MAT-1 to MAT-6 were extracted and used as templates. The transconjugants were analyzed by PCR with a primer pair (Plasmid-For: 5′-tgtataggtgtgatgccttgta-3′; Plasmid-Rev: 5′-agaaacacagtgatgggagata-3′) for pFM-M19 or a primer pair (OXA23-For: 5′-ctgtcaagctcttaaataatattcagc-3′; OXA23-Rev: 5′-tattcgtcgttagaaaaacaattattg-3′) for the blaOXA-23 gene, and DNA sequencing was performed to confirm the presence of blaOXA-23 and plasmid-related genes.
Results
Acinetobacter johnsonii M19 has high carbapenem resistance
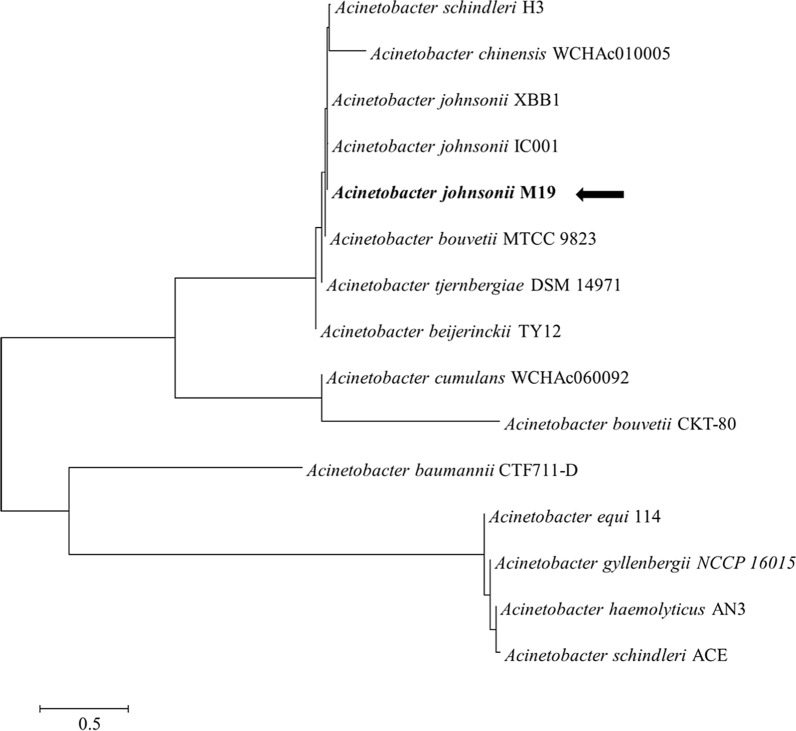
Strain M19 was isolated from hospital sewage and identified as A. johnsonii based on the 16S rDNA sequence (Fig. 1). MICs of imipenem, meropenem and ertapenem for A. johnsonii M19 were 128 mg/L, 48 mg/L and 24 mg/L, respectively, which were higher than those reported for most A. johnsonii strains (Table 1) [4, 27–32], indicating that strain M19 had striking resistance to carbapenems.
Fig. 1.
Neighbor-joining tree generated on the basis of 16S rDNA gene sequences and showing the relationship of M19 to other Acinetobacter species. Bootstrap values are shown as percentages of 1000 replicates when those values were greater than 50%. The scale bar represents 0.5% substitution per nucleotide position
Table 1.
MICs of carbapenems for the A. johnsonii strains
| Strains | Carbapenem resistance MIC (mg/L) | References | ||
|---|---|---|---|---|
| IPM | MEM | ERP | ||
| A. johnsonii M19 | ≥ 128 | 48 | 24 | This work |
| A. johnsonii XBB1 | 4 | ≥ 2 | / | [27] |
| A. johnsonii XBC1 | 4 | ≥ 2 | / | [27] |
| A. johnsonii Aj306, Aj289, Aj286, Aj205 | ≤ 1 | ≤ 0.25 | / | [28] |
| A. johnsonii CIP70.16 | 0.12 | 0.19 | 3 | [4] |
| A. johnsonii 2199 | 2 | 2 | / | [29] |
| A. johnsonii 370, 371, 372, 373 | / | ≥ 128 | / | [30] |
| A. johnsonii 363, 366, 364, 367 | / | 0.25–1 | / | [30] |
| A. johnsonii ATCC 17909 | / | 2 | / | [31] |
| A. johnsonii Z4SZ2 | 0.125 | 0.19 | / | [32] |
| A. johnsonii ST-2 | 0.75 | 0.38 | / | [32] |
| A. johnsonii J6 | 0.5 | 0.19 | / | [32] |
| A. johnsonii 6/1 | 0.5 | 0.38 | / | [32] |
IPM, imipenem; MEM, meropenem; ERP, ertapenem; /, not determined
The whole genome of M19 was sequenced, and the assembled genome contained one 3.75 Mb circular chromosome with 41.4% GC content, and one 55 kb circular plasmid, here named pFM-M19, with 35.8% GC content. The general features of the complete genome sequence are included in Additional file 1: Table S1. Overall, 197 genes (5.24% of the total genes) could be assigned to a VFDB number, and 228 genes (6.07% of the total genes) to a PHI number, indicating that M19 has a high pathogenic potential for humans or other hosts.
Dozens of ARGs were identified in the genome of M19 (Additional file 1: Table S2). Three classes of β-lactamase-encoding genes (class B, class C and class D) were identified, including genes encoding six metallo-β-lactamases (MBLs), two AmpCs and two OXAs (OXA-23 and OXA-211). Furthermore, other antibiotic resistance genes, including efflux pumps, a porin and an aminoglycoside-modifying enzyme gene, were also identified.
M19 harbours two oxacillinases genes, blaOXA-23 and blaOXA-211
Genome annotation of A. johnsonii M19 revealed the presence of two OXA-encoding genes, which are responsible for carbapenem resistance, blaOXA-211 in the chromosome and blaOXA-23 in plasmid pFM-M19. In addition, blaOXA-211 in M19 has the same genetic context conserved in other A. johnsonii strains (Additional file 1: Fig. S1) and which appears to be ubiquitous in this species [28].
OXA-23 encoded by plasmid pFM-M19 exhibited extremely high amino acid identity with OXA-23 found in A. baumannii (CAB69042.1), A. pittii (AUF80820.1), A. wuhouensis (AYO52469.1), A. indicus (ANG65640.1), A. nosocomialis (AKL90363.1), E. coli 521 (AIE13834.1), A. baylyi (AER61544.1), A. radioresistens (ABX00637.1) and Klebsiella pneumoniae (WP_063864531.1) (Additional file 1: Fig. S2). M19 OXA-23 also has the conserved active-sites (Additional file 1: Fig. S2) (for example Ser-79, Ser-126, Lys-216, Phe-110 and Met-221) that essential to carbapenemase activity [33, 34].
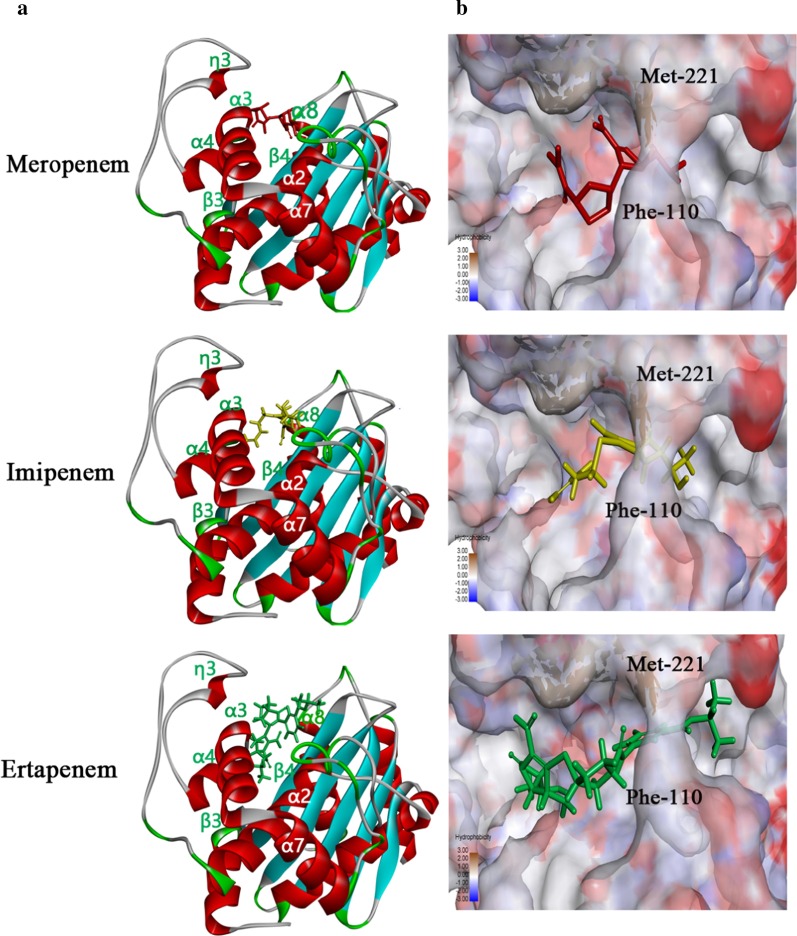
The tertiary structure of M19 OXA-23 was modelled based on the crystal structure of 4JF4, which is an OXA-23 from A. baumannii (GenBank accession number CAB69042.1) and which had the highest amino acid similarity with M19 OXA-23 in the Protein Data Bank (Fig. 2). In this model, the hydrophobic tunnel was formed by Phe-110 and Met-221, and had an elongated shape. Meropenem, imipenem and ertapenem were able to traverse the hydrophobic tunnel and bound to similar positions in the tunnel (Fig. 2). Additionally, Phe-110, Lys-216, Thr-217, Met-221 and Arg-259 were the conserved reactive amino acids (Additional file 1: Fig. S3).
Fig. 2.
Tertiary structure modelling of A. johnsonii M19 OXA-23 with carbapenems. a Molecular docking models showing embedding of carbapenems in the OXA-23 cavity. b Hydrophobicity of OXA-23 surface and hydrophobic tunnel. Meropenem, imipenem and ertapenem are represented, respectively, by red, blue and green stick models
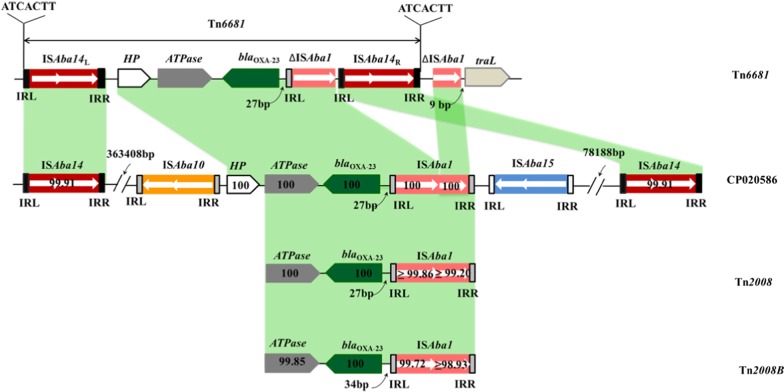
blaOXA-23 is located in the novel transposon Tn6681 in pFM-M19
To evaluate the potential for horizontal transfer of blaOXA-23, the genetic context of blaOXA-23 was investigated. Notably, sequence analysis found that the region containing blaOXA-23 formed a composite transposon with the components ISAba14-HP-ATPase-blaOXA-23-ΔISAba1-ISAba14 (Fig. 3); this novel transposon has been named Tn6681 in the Transposon Registry and GenBank (Accession number: MN081614). Further alignment analysis showed that Tn6681 was highly similar to a chromosome fragment of A. baumannii CBA7, which was isolated in Korea (Accession number: CP020586.1) [35]. However, the blaOXA-23 context region of CBA7 is ISAba10-HP-ATPase-blaOXA-23-ISAba1-ISAba15, which differs somewhat from Tn6681. In addition, two ISAba14 genes, marked as ISAba14L and ISAba14R, were found upstream (3,63,408 bp) of the ISAba10 gene and downstream (78,188 bp) of the ISAba15 gene in the CBA7 chromosome and share 99.91% identity with ISAba14 in Tn6681 (Fig. 3). Given their overall similarity, we propose that Tn6681 and this region of the CBA7 chromosome have the same ancestor.
Fig. 3.
Linear comparison of structures harbouring blaOXA-23. IRR, right inverted repeat; IRL, left inverted repeat. All block arrows indicate length and orientation. The numbers in each block arrow or box represent the nucleotide sequence identity compared with Tn6681. The distance between ISAba1 or ΔISAba1 and the blaOXA-23 start codon is indicated by the number of base pairs below a curved arrow. Direct repeat sequences are indicated
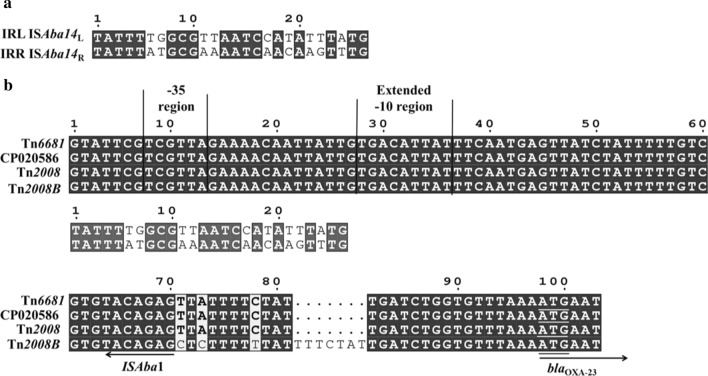
ISAba14 genes belong to the IS3 family and have been previously identified as part of the active composite transposon Tn2114 in A. baumannii RAB [36]. Analysis of the inverted repeats (IRs) of ISAba1 showed that the right inverted repeat (IRR) of ISAba1 remained only 9 bp and the direct repeat and inverted repeat sequences vanished, but the left inverted repeat (IRL) of ISAba14L shared sequence similarity with IRR of ISAba14R (17/26) in particular the motif TATTT(TG/AT)GCG in their extremities (Fig. 4a). The direct repeat sequences (ATCACTT) of 7 bp were also identified (Fig. 3). This overall structure formed a composite transposon Tn6681, which is a novel blaOXA-23 containing transposon.
Fig. 4.
Sequence comparisons of the ISAba1 junction upstream of blaOXA-23. a Sequence alignment of left inverted repeat (IRL) of ISAba14L and right inverted repeat (IRR) of ISAba14R. b Sequence comparisons of the ISAba1 junctions upstream of blaOXA-23 genes. The start codon of the blaOXA-23 gene is underlined. The − 35 and extended − 10 regions of the promoter are marked
Additionally, the arrangement ATPase-blaOXA-23-ISAba1 constitutes a classic genomic organisation found in Tn2008 of A. pittii (GenBank accession number MF078634) and Tn2008B from A. baumannii (GenBank accession number LN877214.1) [13]. In Tn2008, the promoter of blaOXA-23 was overlapped by ISAba1 upstream of the start codon of OXA-23, and both the -10 and -35 regions of this promoter are within the sequence of the ISAba1 gene [37–39]. In Tn6681, the insertion of ISAba14 into ISAba1 generated two ΔISAba1, but the complete -10 and -35 regions of the blaOXA-23 promoter were fully maintained (Fig. 4b), indicating that blaOXA-23 should be expressed normally in M19.
Conjugative plasmid pFM-M19 disseminates blaOXA-23 and carbapenem resistance
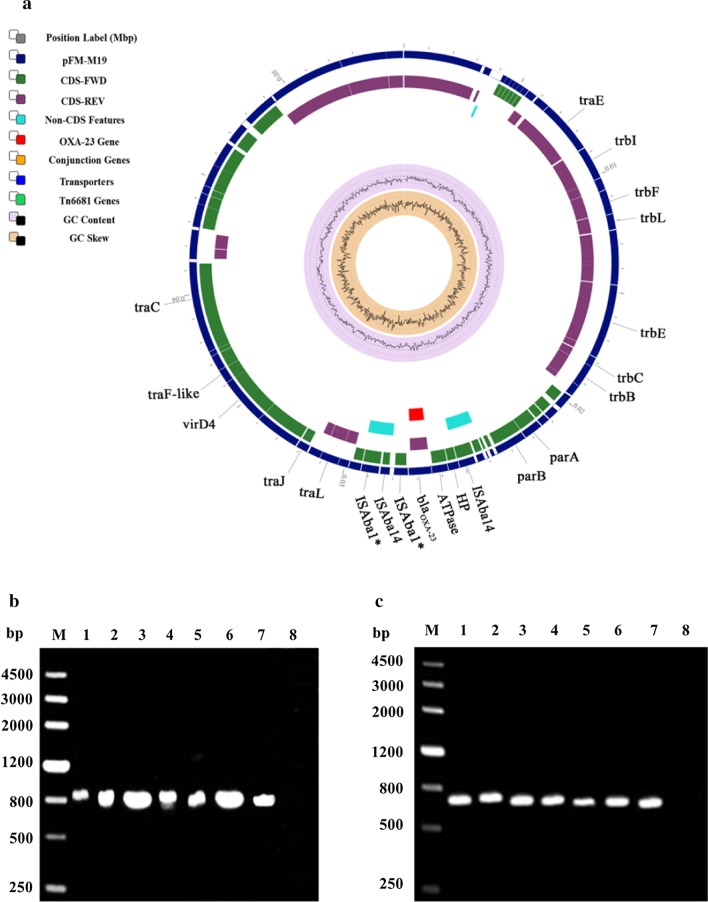
To evaluate the ability to transfer blaOXA-23 and carbapenem resistance, the conjugation systems of pFM-M19 were analyzed. Components of conjugative machinery were identified in pFM-M19, such as a relaxase; the type IV coupling protein (T4CP) gene (traG) for initiation of conjugation; type IV secretion system (T4SS)-related genes, including the translocation channel protein genes (trbD, trbL, trbF, trbG and trbI); the pilus protein genes (trbC and trbJ) and the ATPase genes (trbE, trbB and traG), indicating that pFM-M19 was a conjugative plasmid (Fig. 5a).
Fig. 5.
Schematic representation of plasmid pFM-M19 and amplification of blaOXA-23 and specific region of pFM–M19 from six transconjugant. a The asterisk on ISAba1 indicates a truncated gene. b, c M, DNA marker. The following templates were used in PCR: Lanes 1, plasmid fragments extracted from M19; Lanes 2–7, plasmid fragments extracted from the six transconjugants MAT-1 to MAT-6; Lanes 8, genome of 25DN
Mating experiments between M19 and E. coli 25DN were carried out. The results revealed that the conjugal transfer efficiency of pFM-M19 from M19 to E. coli 25DN was approximately 1.6 × 10–4 CFU/donor when 8 mg/L meropenem was used as the selective pressure. The MICs of carbapenems in six transconjugants were 20 mg/L (imipenem), 16 mg/L (meropenem) and 4 mg/L(ertapenem), which were weaker than that of the donor strain M19 but much higher than that of strain 25DN (Table 2). PCR analysis was performed to confirm the dissemination of carbapenem resistance via plasmid pFM-M19 and blaOXA-23. The results showed that both pFM-M19 and blaOXA-23 were detected in all transconjugants (Fig. 5b and c) and suggested that E. coli 25DN obtained carbapenem resistance due to the acquisition of blaOXA-23 along with pFM-M19.
Table 2.
MICs of carbapenem-resistant strains
| Strains | MICs (mg/L) | ||
|---|---|---|---|
| IPM | MEM | ERP | |
| M19 | ≥ 128 | 48 | 24 |
| 25DN | < 2 | < 2 | < 2 |
| MAT-1 | 20 | 16 | 4 |
| MAT-2 | 20 | 16 | 4 |
| MAT-3 | 20 | 16 | 4 |
| MAT-4 | 20 | 16 | 4 |
| MAT-5 | 20 | 16 | 4 |
| MAT-6 | 20 | 16 | 4 |
25DN, sodium azide-resistant E. coli strain derived from ATCC25922; MAT, transconjugants of A. johnsonii M19 and E. coli 25DN; IPM, imipenem; MEM, meropenem; ERP, ertapenem
Discussion
Acinetobacter johnsonii strain M19 presented higher carbapenem resistance than that of most A. johnsonii strains [2, 4, 28–31, 40], Pseudomonas aeruginosa [41], Proteus mirabilis [42] or A. baumannii [43]. β-lactam antibiotic resistance of Acinetobacter is mainly due to the inactivation of β-lactams catalysed by four classes (A, B, C and D) of β-lactamases [9, 13]. In this work, three classes (class B, C and D) of β-lactamase genes, including six MBL-encoding genes, two AmpC-encoding genes and two OXA-encoding genes, were identified in the genome of A. johnsonii M19, suggesting that M19 is a reservoir of β-lactam resistance genes.
Class D β-lactamases, commonly referred to as OXA, are responsible for carbapenem resistance, and their encoding genes are conserved and widespread in Acinetobacter species [44, 45]. M19 harbours blaOXA-211 in the chromosome and blaOXA-23 in plasmid pFM-M19. OXA-23 was the first reported class D β-lactamase and was originally detected in a patient isolate of A. baumannii in Scotland in 1993 [46]. Twenty years later, the structure of OXA-23 was resolved and revealed that the elongated pocket of OXA-23 provides a hydrophobic environment for high reaction efficiency with carbapenems [11]. OXA-23 is considered to be the major β-lactamase for carbapenem resistance in Acinetobacter [11], suggesting that the OXA-23 encoded by plasmid pFM-M19 plays a key role in carbapenem resistance in M19. Interestingly, the blaOXA-23 found in plasmid pFM-M19 has not been previously reported in A. johnsonii strains, indicating that M19 obtained this gene from other bacterial species and further increasing concern over the ability of blaOXA-23 to spread among species.
Mobile elements are considered to be responsible for the movement and dissemination of blaOXA-23, and all of the reported genetic structures that contain blaOXA-23 have been classified as transposons [13]. Previously, blaOXA-23 had been found in five transposons, with ISAba1 upstream of the start codon of blaOXA-23 in four of these (Tn2006 [47], Tn2008 [48], Tn2008B [49], Tn2009 [50]) and with ISAba4 preceding blaOXA-23 in Tn2007 [47]. In plasmid pFM-M19, we found that blaOXA-23 was located in the new transposon Tn6681, which has the genetic context ISAba14-HP-ATPase-blaOXA-23-ΔISAba1-ISAba14-ΔISAba1 and which was likely formed as two copies of ISAba14 were inserted into the ISAba1 of Tn2008. The structure of Tn6681 has some differences from that of the other transposons previously reported to contain blaOXA-23 [13]. In addition, in Tn2008, a high level of expression of the blaOXA-23 gene is associated with significant resistance to carbapenems in A. baumannii, and this expression is controlled by promoter elements within ISAba1 [37–39]. However, although sequence analysis revealed that ISAba14 inserted into ISAba1 in Tn6681, the promoter of blaOXA-23 appears to be intact, indicating that the blaOXA-23 gene may still be highly expressed and responsible for the striking carbapenem resistance in M19.
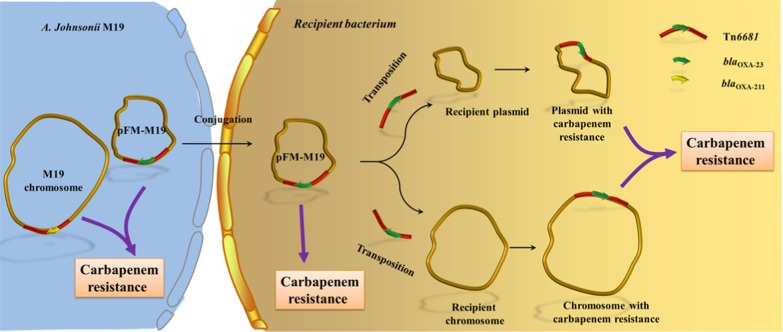
The acquisition of novel genes by plasmids, especially conjugative plasmids [51], along with mobile genetic elements such as transposons or insertion sequences, makes them perfect vehicles for the spread of antibiotic resistance [52]. The conjugative DNA transfer mechanism is well conserved and depends on a T4SS [53, 54]. Genes of T4SS and T4CP modules, including genes for a translocation channel protein, pilus protein and ATPase, were identified in pFM-M19, indicating that pFM-M19 is a conjugative plasmid, which has also been confirmed by mating experiments in this study. However, it should be pointed out that the typical origin of transfer site (oriT) was not detected on pFM-M19 by oriT finder, suggesting that pFM-M19 might initiate the transfer from a cryptic oriT. Moreover, the combination of transposon Tn6681 and the conjugative plasmid pFM-M19 may provide a robust means for blaOXA-23 transfer, with the potential for Tn6681 to shift blaOXA-23 between the chromosome and plasmid in one bacterial strain, and the conjugative plasmid pFM-M19 disseminating Tn6681 and blaOXA-23 between different bacterial species (Fig. 6).
Fig. 6.
Proposed pathway for transfer of the blaOXA-23 gene and dissemination of carbapenem resistance from A. johnsonii M19
Conclusions
In conclusion, A. johnsonii strain M19, which contains the conjugative plasmid pFM-M19 and Tn6681, is not only a reservoir of blaOXA-23 but also an effective disseminator of blaOXA-23. To our knowledge, our investigation is the first to provide evidence that blaOXA-23 was transferred into A. johnsonii. The presence of blaOXA-23 on conjugative plasmids of A. johnsonii enhances the risk of carbapenem resistance spread to the environment and needs to be monitored closely.
Supplementary information
Additional file 1. Table S1. General features of the A. johnsonnii M19 genome. Table S2. Antibiotic resistant genes of the A. johnsonnii M19 genome. Table S3. Predicted genes of plasmid pFM-M19. Fig. S1. Comparison of the genetic context of blaOXA-23 in M19 and other A. johnsonii strains. Fig. S2. Multi-sequence comparison of OXA-23 proteins from A. johnsonii M19 and various other bacteria. Fig. S3. Stick models of the active sites of OXA-23 during carbapenem binding and the carbapenem β-lactam ring.
Acknowledgements
Not applicable.
Nucleotide sequence accession number
Complete sequences of the chromosome of A. johnsonii strain M19 and of plasmid pFM-M19 were deposited in GenBank under accession numbers CP037424 and CP037425, respectively. The 16S rDNA sequence of strain M19 was deposited in GenBank under accession number MT226917.
Abbreviations
- MICs
Minimum inhibitory concentrations
- PATRIC
Pathosystems Resource Integration Center
- OXA
Oxacillinase
- LB
Luria–Bertani
- MEGA
Molecular evolutionary genetics analysis
- VFDB
Virulence factors of pathogenic bacteria database
- PHI
Pathogen host interactions
- ARGs
Antibiotic resistance genes
- ARDB
Antibiotic resistance genes database
- MBLs
Metallo-β-lactamases
- IRs
Inverted repeats
- IRR
Right inverted repeat
- IRL
Left inverted repeat
- T4CP
Type IV coupling protein
- T4SS
Type IV secretion system
- oriT
Origin of transfer site
Authors’ contributions
GZ conceived and designed and conducted experiments, analyzed and interpreted data, and wrote the manuscript. YZ, JF, PZ and CZ analyzed data and performed the calculation. WZ and YX helped to analyze data and revised the manuscript. RZ and GC led the project and revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the National Key Research and Development Program of China (No. 2018YFA0900302), the Key Research and Development Program of Shandong Province (No. 2019GSF107021) and the Academic Promotion Program of Shandong First Medical University (No. LJ001), the Postgraduate Research & Practice Innovation Program of Jiangsu Province (1012050205205969).
Availability of data and materials
All data generated or analyzed during this study are included in this published article [and its supplementary information files] Original data are available from the corresponding author upon reasonable request.
Ethics approval and consent to participate
All activities undertaken did not require Ethics approval. An informed written consent was obtained from the legal guardian of the case.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Guangxiang Cao, Email: caozhong0402@163.com.
Rongzhen Zhang, Email: rzzhang@jiangnan.edu.cn.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s13756-020-00832-4.
References
- 1.Lee YR, Baker NT. Meropenem-vaborbactam: a carbapenem and β-lactamase inhibitor with activity against carbapenem-resistant Enterobacteriaceae. Eur J Clin Microbiol Infect Dis. 2018;37(8):1411–1419. doi: 10.1007/s10096-018-3260-4. [DOI] [PubMed] [Google Scholar]
- 2.Zong Z, Zhang X. blaNDM-1-carrying Acinetobacter johnsonii detected in hospital sewage. J Antimicrob Chemother. 2013;68(5):1007–1010. doi: 10.1093/jac/dks505. [DOI] [PubMed] [Google Scholar]
- 3.Quinteira S, Grosso F, Ramos H, Peixe L. Molecular epidemiology of imipenem-resistant Acinetobacter haemolyticus and Acinetobacter baumannii isolates carrying plasmid-mediated OXA-40 from a Portuguese hospital. Antimicrob Agents Chemother. 2007;51(9):3465–3466. doi: 10.1128/AAC.00267-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Figueiredo S, Bonnin RA, Poirel L, Duranteau J, Nordmann P. Identification of the naturally occurring genes encoding carbapenem-hydrolysing oxacillinases from Acinetobacter haemolyticus, Acinetobacter johnsonii, and Acinetobacter calcoaceticus. Clin Microbiol Infect. 2012;18(9):907–913. doi: 10.1111/j.1469-0691.2011.03708.x. [DOI] [PubMed] [Google Scholar]
- 5.Feng Y, Yang P, Wang X, Zong Z. Characterization of Acinetobacter johnsonii isolate XBB1 carrying nine plasmids and encoding NDM-1, OXA-58 and PER-1 by genome sequencing. J Antimicrob Chemother. 2016;71(1):71–75. doi: 10.1093/jac/dkv324. [DOI] [PubMed] [Google Scholar]
- 6.Octavia S, Xu W, Ng OT, Marimuthu K, Venkatachalam I, Cheng B, Lin RTP, Teo JWP. Identification of AbaR4 Acinetobacter baumannii resistance island in clinical isolates of blaOXA-23-positive Proteus mirabilis. J Antimicrob Chemother. 2020;75(3):521–525. doi: 10.1093/jac/dkz472. [DOI] [PubMed] [Google Scholar]
- 7.Zong Z. The complex genetic context of blaPER-1 flanked by miniature inverted-repeat transposable elements in Acinetobacter johnsonii. PLoS ONE. 2014;9(2):e90046. doi: 10.1371/journal.pone.0090046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tian S, Ali M, Xie L, Li L. Genome-sequence analysis of Acinetobacter johnsonii MB44 reveals potential nematode-virulent factors. Springerplus. 2016;5(1):986. doi: 10.1186/s40064-016-2668-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee C-R, Lee JH, Park M, Park KS, Bae IK, Kim YB, Cha C-J, Jeong BC, Lee SH. Biology of Acinetobacter baumannii: pathogenesis, antibiotic resistance mechanisms, and prospective treatment options. Front Cell Infect Microbiol. 2017;7:55. doi: 10.3389/fcimb.2017.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jeon JH, Lee JH, Lee JJ, Park KS, Karim AM, Lee C-R, Jeong BC, Lee SH. Structural basis for carbapenem-hydrolyzing mechanisms of carbapenemases conferring antibiotic resistance. Int J Mol Sci. 2015;16(5):9654–9692. doi: 10.3390/ijms16059654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith CA, Antunes NT, Stewart N, Toth M, Kumarasiri M, Chang M, Mobashery S, Vakulenko SB. Structural basis for carbapenemase activity of the OXA-23 β-lactamase from Acinetobacter baumannii. Chem Biol. 2013;20(9):1107–1115. doi: 10.1016/j.chembiol.2013.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blackwell G, Hall RM. Mobilisation of a small Acinetobacter plasmid carrying an oriT transfer origin by conjugative RepAci6 plasmids. Plasmid. 2019;103:36–44. doi: 10.1016/j.plasmid.2019.04.002. [DOI] [PubMed] [Google Scholar]
- 13.Nigro SJ, Hall RM. Structure and context of Acinetobacter transposons carrying the oxa23 carbapenemase gene. J Antimicrob Chemother. 2016;71(5):1135–1147. doi: 10.1093/jac/dkv440. [DOI] [PubMed] [Google Scholar]
- 14.Evans BA, Amyes S. OXA β-lactamases. Clin Microbiol Rev. 2014;27(2):241–263. doi: 10.1128/CMR.00117-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeong S, Hong JS, Kim JO, Kim K-H, Lee W, Bae IK, Lee KJ, Jeong SH. Identification of Acinetobacter species using matrix-assisted laser desorption ionization-time of flight mass spectrometry. Ann Lab Med. 2016;36(4):325–34. doi: 10.3343/alm.2016.36.4.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki E, Pruitt KD, Borodovsky M, Ostell J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016;44(14):6614–6624. doi: 10.1093/nar/gkw569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen L, Dandan Z, Liu B, Yang J, Jin Q. VFDB 2016: hierarchical and refined dataset for big data analysis-10 years on. Nucleic Acids Res. 2016;44(D1):D694–D697. doi: 10.1093/nar/gkv1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Urban M, Pant R, Raghunath A, Irvine AG, Pedro H, Hammond-Kosack K. The Pathogen-Host Interactions database (PHI-base): additions and future developments. Nucleic Acids Res. 2015;41(D1):D645–D655. doi: 10.1093/nar/gku1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu B, Pop M. ARDB—antibiotic resistance genes database. Nucleic Acids Res. 2009;37(Database issue):D443–D447. doi: 10.1093/nar/gkn656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Madeira F, Park YM, Lee J, Buso N, Gur T, Madhusoodanan N, Basutka P, Tivey ARN, Potter SC, Finn RD, Lopez R. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019;47:W636–W641. doi: 10.1093/nar/gkz268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xavier R, Patrice G. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42(W1):W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.BIOVIA., Discovery Studio Modeling Environment, Release 2017. San Diego, CA: Dassault Systèmes. 2017.
- 23.Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 2006;34(Suppl 1):D32–D36. doi: 10.1093/nar/gkj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brettin T, Davis JJ, Disz T, Edwards RA, Gerdes S, Olsen GJ, Olson R, Overbeek R, Parrello B, Pusch GD, Shukla M, Stevens R, Vonstein V, Wattam AR, Xia F. RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci Rep. 2015;5:8365. doi: 10.1038/srep08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li X, Xie Y, Liu M, Tai C, Sun J, Deng Z, Ou HY. oriTfinder: a web-based tool for the identification of origin of transfers in DNA sequences of bacterial mobile genetic elements. Nucleic Acids Res. 2018;46(W1):W229–W34. doi: 10.1093/nar/gky352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang P, Liu M, Fu J, Zhong C, Zong G, Cao G. Identification of a mobilizable, multidrug-resistant genomic island in Myroides odoratimimus isolated from Tibetan pasture. Sci Total Environ. 2020;723:137970. doi: 10.1016/j.scitotenv.2020.137970. [DOI] [PubMed] [Google Scholar]
- 27.Penzak SR, Gubbins PO, Stratton SL, Anaissie EJ. Investigation of an outbreak of gram-negative bacteremia among hematology–oncology outpatients. Infect Control Hosp Epidemiol. 2000;21(9):597–599. doi: 10.1086/501810. [DOI] [PubMed] [Google Scholar]
- 28.Montaña S, Schramm STJ, Traglia GM, Chiem K, Noto GPD, Almuzara MN, Barberis CM, Vay C, Quiroga C, Tolmasky M, Iriarte A, Ramirez MS. The genetic analysis of an Acinetobacter johnsonii clinical strain evidenced the presence of horizontal genetic transfer. PLoS ONE. 2016;11(8):e0161528. doi: 10.1371/journal.pone.0161528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rodríguez CH, Nastro M, Dabos L, Barberis C, Vay C, Famiglietti A. First isolation of Acinetobacter johnsonii co-producing PER-2 and OXA-58 β-lactamases. Diagn Microbiol Infect Dis. 2014;80(4):341–342. doi: 10.1016/j.diagmicrobio.2014.09.013. [DOI] [PubMed] [Google Scholar]
- 30.Maravić A, Skočibušić M, Fredotović Ž, Šamanić I, Cvjetan S, Knezović M, Puizina J. Urban riverine environment is a source of multidrug-resistant and ESBL-producing clinically important Acinetobacter spp. Environ Sci Pollut Res. 2016;23(4):3525–3535. doi: 10.1007/s11356-015-5586-0. [DOI] [PubMed] [Google Scholar]
- 31.Ito A, Sato T, Ota M, Takemura M, Nishikawa T, Toba S, Kohira N, Miyagawa S, Ishibashi N, Matsumoto S, Nakamura R, Tsuji M, Yamano Y. In vitro antibacterial properties of cefiderocol, a novel siderophore cephalosporin, against gram-negative bacteria. Antimicrob Agents Chemother. 2018;62(1):e01454–e1517. doi: 10.1128/AAC.01454-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Radó J, Kaszab E, Benedek T, Kriszt B, Szoboszlay S. First isolation of carbapenem-resistant Acinetobacter beijerinckii from an environmental sample. Acta Microbiol Immunol Hung. 2019;66(1):113–130. doi: 10.1556/030.66.2019.004. [DOI] [PubMed] [Google Scholar]
- 33.Couture F, Lachapelle J, Levesque RC. Phylogeny of LCR-1 and OXA-5 with class A and class D β-lactamases. Mol Microbiol. 1992;6(12):1693–1705. doi: 10.1111/j.1365-2958.1992.tb00894.x. [DOI] [PubMed] [Google Scholar]
- 34.Schneider KD, Ortega CJ, Renck NA, Bonomo RA, Powers RA, Leonard DA. Structures of the Class D carbapenemase OXA-24 from Acinetobacter baumannii in complex with doripenem. J Mol Biol. 2011;406(4):583–594. doi: 10.1016/j.jmb.2010.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoon EJ, Kim JO, Yang JW, Kim HS, Lee KJ, Jeong SH, Lee H, Lee K. The blaOXA-23-associated transposons in the genome of Acinetobacter spp. represent an epidemiological situation of the species encountering carbapenems. J Antimicrob Chemother. 2017;72(10):2708–2714. doi: 10.1093/jac/dkx205. [DOI] [PubMed] [Google Scholar]
- 36.Bonnin RA, Poirel L, Nordmann P. A novel and hybrid composite transposon at the origin of acquisition of blaRTG-5 in Acinetobacter baumannii. Int J Antimicrob Agents. 2012;40(3):257–259. doi: 10.1016/j.ijantimicag.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 37.Walther-Rasmussen J, Høiby N. OXA-type carbapenemases. J Antimicrob Chemother. 2006;57(3):373–383. doi: 10.1093/jac/dki482. [DOI] [PubMed] [Google Scholar]
- 38.Choi JY, Ko G, Jheong W, Huys G, Seifert H, Dijkshoorn L, Ko KS. Acinetobacter kookii sp. nov., isolated from soil. Int J Syst Evol Microbiol. 2013;63(Pt 12):4402–4406. doi: 10.1099/ijs.0.047969-0. [DOI] [PubMed] [Google Scholar]
- 39.Héritier C, Poirel L, Lambert T, Nordmann P. Contribution of acquired carbapenem-hydrolyzing oxacillinases to carbapenem resistance in Acinetobacter baumannii. Antimicrob Agents Chemother. 2005;49(8):3198–3202. doi: 10.1128/AAC.49.8.3198-3202.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krizova L, Maixnerova M, Sedo O, Nemec A. Acinetobacter bohemicus sp. nov. widespread in natural soil and water ecosystems in the Czech Republic. Syst Appl Microbiol. 2014;37(7):467–473. doi: 10.1016/j.syapm.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 41.Rostami S, Sheikh AF, Shoja S, Farahani A, Tabatabaiefar MA, Jolodar A, Sheikhi R. Investigating of four main carbapenem-resistance mechanisms in high-level carbapenem resistant Pseudomonas aeruginosa isolated from burn patients. J Chin Med Assoc. 2018;81(2):127–132. doi: 10.1016/j.jcma.2017.08.016. [DOI] [PubMed] [Google Scholar]
- 42.Shaker MA, Shaaban MI. Formulation of carbapenems loaded gold nanoparticles to combat multi-antibiotic bacterial resistance: In vitro antibacterial study. Int J Pharm. 2017;525(1):71–84. doi: 10.1016/j.ijpharm.2017.04.019. [DOI] [PubMed] [Google Scholar]
- 43.Breilh D, Texier-Maugein J, Allaouchiche B, Saux M-C, Boselli E. Carbapenems. J Chemother. 2013;25(1):1–17. doi: 10.1179/1973947812Y.0000000032. [DOI] [PubMed] [Google Scholar]
- 44.Wong EW, Yusof MYM, Mansor M, Anbazhagan D, Ong G, Sekaran SD. Disruption of adeB gene has a greater effect on resistance to meropenems than adeA gene in Acinetobacter spp. isolated from University Malaya Medical Centre. Singapore Med J. 2009;50(8):822–26. [PubMed] [Google Scholar]
- 45.Iacono M, Villa L, Fortini D, Bordoni R, Imperi F, Bonnal RJP, Sicheritz-Ponten T, Bellis GD, Visca P, Cassone A, Carattoli A. Whole-genome pyrosequencing of an epidemic multidrug-resistant Acinetobacter baumannii strain belonging to the European clone II group. Antimicrob Agents Chemother. 2008;52(7):2616–2625. doi: 10.1128/AAC.01643-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paton R, Miles RS, Hood J, Amyes S. ARI 1: β-lactamase-mediated imipenem resistance in Acinetobacter baumannii. Int J Antimicrob Agents. 1993;2(2):81–87. doi: 10.1016/0924-8579(93)90045-7. [DOI] [PubMed] [Google Scholar]
- 47.Corvec S, Poirel L, Naas T, Drugeon H, Nordmann P. Genetics and expression of the carbapenem-hydrolyzing oxacillinase gene blaOXA-23 in Acinetobacter baumannii. Antimicrob Agents Chemother. 2007;51(4):1530–1533. doi: 10.1128/AAC.01132-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Adams-Haduch JM, Paterson DL, Sidjabat HE, Pasculle AW, Potoski BA, Muto CA, Harrison LH, Doi Y. Genetic basis of multidrug resistance in Acinetobacter baumannii clinical isolates at atertiary medical center in Pennsylvania. Antimicrob Agents Chemother. 2008;52:3837–3843. doi: 10.1128/AAC.00570-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nigro S, Hall RM. Distribution of the blaOXA-23-containing transposons Tn2006 and Tn2008 in Australian carbapenem-resistant Acinetobacter baumannii isolates. J Antimicrob Chemother. 2015;70(8):2409–2411. doi: 10.1093/jac/dkv102. [DOI] [PubMed] [Google Scholar]
- 50.Zhou H, Zhang T, Yu D, Pi B, Yang Q, Zhou J, Hu S, Yu Y. Genomic analysis of the multidrugresistant Acinetobacter baumannii strain MDR-ZJ06 widely spread in China. Antimicrob Agents Chemother. 2011;55(10):4506–4512. doi: 10.1128/AAC.01134-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.de Lagarde M, Larrieu C, Praud K, Lallier N, Trotereau A, Sallé G, Fairbrother JM, Schouler C, Doublet B. Spread of multidrug resistance IncHI1 plasmids carrying ESBL gene blaCTX-M-1 and metabolism operon of prebiotic oligosaccharides in commensal Escherichia coli from healthy horses, France: blaCTX-M-1 plasmids in healthy horses, France. Int J Antimicrob Agents. 2020;5:105936. doi: 10.1016/j.ijantimicag.2020.105936. [DOI] [PubMed] [Google Scholar]
- 52.Rozwandowicz M, Brouwer MSM, Fischer J, Wagenaar JA, Gonzalez-Zorn B, Guerra B, Mevius DJ, Hordijk J. Plasmids carrying antimicrobial resistance genes in Enterobacteriaceae. J Antimicrob Chemother. 2018;73(5):1121–1137. doi: 10.1093/jac/dkx488. [DOI] [PubMed] [Google Scholar]
- 53.Makart L, Gillis A, Hinnekens P, Mahillon J. A novel T4SS-mediated DNA transfer used by pXO16, a conjugative plasmid from Bacillus thuringiensis serovar israelensis. Environ Microbiol. 2018;20(4):1550–1561. doi: 10.1111/1462-2920.14084. [DOI] [PubMed] [Google Scholar]
- 54.Grohmann E, Christie PJ, Waksman G, Backert S. Type IV secretion in Gram-negative and Gram-positive bacteria. Mol Microbiol. 2018;107(4):455–471. doi: 10.1111/mmi.13896. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Table S1. General features of the A. johnsonnii M19 genome. Table S2. Antibiotic resistant genes of the A. johnsonnii M19 genome. Table S3. Predicted genes of plasmid pFM-M19. Fig. S1. Comparison of the genetic context of blaOXA-23 in M19 and other A. johnsonii strains. Fig. S2. Multi-sequence comparison of OXA-23 proteins from A. johnsonii M19 and various other bacteria. Fig. S3. Stick models of the active sites of OXA-23 during carbapenem binding and the carbapenem β-lactam ring.
Data Availability Statement
All data generated or analyzed during this study are included in this published article [and its supplementary information files] Original data are available from the corresponding author upon reasonable request.