In this issue of Circulation Research, Bi et al1 presents functional data linking a GWAS locus at chromosome 6p21, known as chromosome 6 open reading frame 106 (C6ORF106), to the regulation of hepatic lipid metabolism. In these studies, the mouse homologue of ILRUN (D17wsu92e) was deleted in mice resulting in a significant reduction in plasma cholesterol levels due to decreased liver lipoprotein production. Analysis of ILRUN deficient murine livers further showed that mRNA for several lipid-related transcription factors were up regulated. The greatest increase was for murine peroxisome proliferator activated receptor α (PPARα) protein. Thus, these findings establish ILRUN as a novel regulator of lipid metabolism promoting hepatic lipoprotein production and likely the casual gene underlying the observed genetic associations with plasma lipids at 6p21 in humans.
The biological function of the protein coded by C6ORF106 has been elusive. Independent groups working together investigated the locus C6ORF106 and provided the official moniker for the protein product from this site named, Inflammation and Lipid Regulator with ubiquitin associated domain or UBA-like and a next to BRACA1 gene 1 protein or NBR1-like domain, abbreviated, ILRUN2, 3. Ambrose et al2 reported details of the physical and genetic aspects of ILRUN showing that that it codes for a 298 amino acid protein in humans having an N-terminal UBA-like domain, an internal NBR1-like domain, and a disordered domain having 3 phosphorylation sites. They also showed that ILRUN is expressed in a variety of human tissues with the greatest expression in adipose, heart, ovary, and immune cells with very high expression in activated B-cells2. Three isoforms of ILRUN have been found in most tissues with ILRUNa being the most abundant. ILRUN is also thought to be an “old” protein that is highly conserved from fish and amphibians to humans2.
Bi et al1, show that plasma high density lipoprotein (HDL) levels were reduced in ILRUN KO mice, which was largely attributable to a decrease in HDL apoA-I production. Consistent with these findings, a significant decrease in apoA-I mRNA and protein secretion was seen from primary hepatocytes obtained from ILRUN KO mice. ILRUN KO hepatocytes from diet-fed animals also showed low apoA-I secretion without further lowering of apoA-I mRNA suggesting that additional post-transcriptional pathways may participate in regulating apoA-I protein levels. Additionally, plasma very low density lipoprotein (VLDL) concentrations were also reduced in ILRUN KO mice compared to wild-type controls. There was no indication that murine low density lipoprotein (LDL) levels were directly affected, and preliminary evidence suggests that the lower LDL levels were a consequence of lower plasma VLDL and/or triglyceride levels. Expression of a variety of genes involved in lipid homeostasis were significantly reduced, with lower murine liver apolipoprotein A-4 (apoA-4) levels possibly responsible for the lower VLDL levels.
Intriguingly, while basal overnight fast plasma insulin levels were nearly identical between wild-type and ILRUN KO mice, insulin levels after glucose injection were significantly lower in ILRUN KO mice. Dyslipidemia in commonly associated with obesity and type 2 diabetes. This study showed that ILRUN is expressed in murine pancreatic islet cells suggesting that ILRUN may control different metabolic pathways in different cell types. Therefore, a global mutation in a single gene that affects the “activity” or concentration of the protein in several tissues may contribute to diseases that often occur together, like dyslipidemia and type 2 diabetes.
Global transcriptome profiling of whole liver tissue followed by pathway analysis showed that there was enrichment of gene expression for several nuclear receptor pathways. The greatest increase was for murine PPARα mRNA which also showed a corresponding increase in PPARα protein. Activation of PPARα resulting from post-translational control by ILRUN reduces HDL cholesterol in chow fed ILRUN KO mice through reduced apoAI transcription. In primary hepatocytes there was no increase in PPARα mRNA, but net increase in PPARα protein, suggesting to the authors that in hepatocyte the function of ILRUN was to retard post-translational catabolism of PPARα. These findings from hepatocytes suggested that the ubiquitin associated domain may play a role in the catabolism of PPARα. In an elegant set of experiments using ILRUN mutants lacking either an UBA-like or NBDR1-like region the authors showed that ILRUN interacts with PPARα through the UBA-like region. Future experiments will yield a more complete picture of how the UBA-region of ILRUN interacts with ubiquitinated regions to modulate protein turnover. Additional studies will be needed to elucidate detailed mechanisms of how the various, related transcription pathways are modulated.
Here, a major milestone has been achieved by demonstrating a mechanism that explains the GWAS association between locus C6orf106 and lipid metabolism. The identification of the ILRUN signaling mechanism that influences lipoprotein metabolism is a significant step toward elucidating a comprehensive set of signaling processes that modulate lipid metabolism and yields insight into how dysregulation of ILRUN may lead to disease state. However, the control of lipid metabolism is only one facet of ILRUN control. Other processes that are modulated by ILRUN protein include pathways whose dysregulation can lead to cancers or other diseases. The most firmly established effect of ILRUN in humans and mice is the regulation of type-I interferon (INF) synthesis by promoting degradation of transcription coactivators p300 and cAMP-response element-binding protein and inhibiting IFN regulatory factor 3 binding to DNA reducing transcription of IFNs and TNFα,4 and, thereby, inhibiting, or turning off, the innate immune response to viral infection to prevent extensive tissue damage after the infection has been cleared. Regulation of the IFN response is essential to prevent abnormal signaling that may promote disease. GWAS have associated region C6orf106 with several diseases including, but not limited to, progressive chronic lymphocytic leukemia,5 systemic lupus erythematosus,6 breast cancer,7 and pancreatic cancer.8 Follow up studies on pancreatic cancer suggest that the ERK signaling pathway was involved. A fuller understanding of how ILRUN modulates signaling in various tissues is an important goal for the future. Only with more research on these signaling pathways will it be possible to selectively target signaling in specific tissues to stop or control disease related to dysregulation.
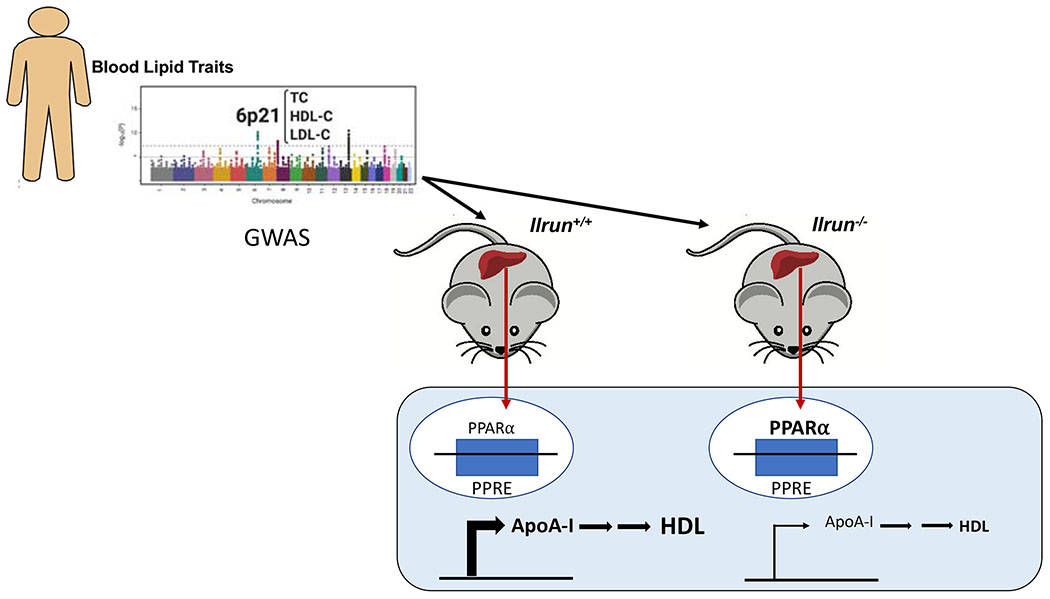
ILRUN expression attenuates HDL production. Human GWAS identified a gene locus 6p21 (C60RF106) as a robust modulator of plasma lipid concentrations. In mice lacking ILRUN, liver PPARa expression was increased and associated with a concomitant reduction in the production of plasma HDL. This study suggests that ILRUN modulates the binding of PPARa to PPRE and ultimately controls the transcription of apoA-I and thus, liver HDL production.( Illustration modified from Bi X, Kuwano T, Lee PC, Millar JS, Li L, Shen Y, Soccio RE, Hand NJ, Rader DJ. Ilrun, a human plasma lipid gwas locus, regulates lipoprotein metabolism in mice. Circ Res. 2020; 127: xx-xxx, with permission).
Acknowledgments
Sources of Funding
AHA19TPA34890023 (MJT) and RO1HL127649 and HL138907(MGS-T).
Footnotes
Disclosures
None.
Contributor Information
Michael J. Thomas, Department of Pharmacology & Toxicology, Medical College of Wisconsin
Mary G. Sorci-Thomas, Department of Pharmacology & Toxicology, Medical College of Wisconsin; Department of Medicine, Section on Endocrinology, Medical College of Wisconsin
REFERENCES
- 1.Bi X, Kuwano T, Lee PC, Millar JS, Li L, Shen Y, Soccio RE, Hand NJ, Rader DJ. Ilrun, a human plasma lipid gwas locus, regulates lipoprotein metabolism in mice. Circ Res. 2020; 127: xx–xxx. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ambrose RL, Brice AM, Caputo AT, Alexander MR, Tribolet L, Liu YC, Adams TE, Bean AGD, Stewart CR. Molecular characterisation of ilrun, a novel inhibitor of proinflammatory and antimicrobial cytokines. Heliyon. 2020;6:e04115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bi X, Kuwano T, Lee PC, Millar JS, Soccio RE, Hand NJ, Rader DJ. A novel plasma lipid and cad locus in humans, regulates lipoprotein metabolism and atherogenesis in mice. American Heart Association Scientific Sessions. 2020 [Google Scholar]
- 4.Ambrose RL, Liu YC, Adams TE, Bean AGD, Stewart CR. C6orf106 is a novel inhibitor of the interferon-regulatory factor 3-dependent innate antiviral response. J Biol Chem. 2018;293:10561–10573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allsup D, Lin WY, Fordham SF, et al. Multicentre genome wide association study identifies risk alleles for progressive chronic lymphocytic leukaemia. Blood. 2019;134:1740 [Google Scholar]
- 6.Bentham J, Morris DL, Graham DSC, et al. Genetic association analyses implicate aberrant regulation of innate and adaptive immunity genes in the pathogenesis of systemic lupus erythematosus. Nat Genet. 2015;47:1457–1464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang G, Zhang X, Zhang Y, Wang L, Fan C, Xu H, Miao Y, Wang E. A novel biomarker c6orf106 promotes the malignant progression of breast cancer. Tumour Biol. 2015;36:7881–7889 [DOI] [PubMed] [Google Scholar]
- 8.Li X, Dong M, Zhou J, Zhu D, Zhao J, Sheng W. C6orf106 accelerates pancreatic cancer cell invasion and proliferation via activating erk signaling pathway. Mol Cell Biochem. 2019;454:87–95 [DOI] [PubMed] [Google Scholar]