Abstract
Objective
Infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is now a global pandemic. Emerging results indicate a dysregulated immune response. Given the role of CCR5 in immune cell migration and inflammation, we investigated the impact of CCR5 blockade via the CCR5-specific antibody leronlimab on clinical, immunological, and virological parameters in severe COVID-19 patients.
Methods
In March 2020, 10 terminally ill, critical COVID-19 patients received two doses of leronlimab via individual emergency use indication. We analyzed changes in clinical presentation, immune cell populations, inflammation, as well as SARS-CoV-2 plasma viremia before and 14 days after treatment.
Results
Over the 14-day study period, six patients survived, two were extubated, and one discharged. We observed complete CCR5 receptor occupancy in all donors by day 7. Compared with the baseline, we observed a concomitant statistically significant reduction in plasma IL-6, restoration of the CD4/CD8 ratio, and resolution of SARS-CoV2 plasma viremia (pVL). Furthermore, the increase in the CD8 percentage was inversely correlated with the reduction in pVL (r = −0.77, p = 0.0013).
Conclusions
Our study design precludes clinical efficacy inferences but the results implicate CCR5 as a therapeutic target for COVID-19 and they form the basis for ongoing randomized clinical trials.
Keywords: COVID-19, Immunotherapy, Plasma viral load, CCR5, Leronlimab
Introduction
Since the initial cases of coronavirus disease 2019 (COVID-19) were reported from Wuhan, China in December 2019 (Huang et al., 2020), severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has emerged as a global pandemic and increasing numbers of severe and critical cases have required invasive external ventilation, which threatens to overwhelm health care systems (World Health Organization, 2020). It remains unclear why COVID-19 patients experience a spectrum of clinical outcomes ranging from asymptomatic to severe disease, where the features of critical COVID-19 include rampant inflammation and cytokine release syndrome (CRS) leading to acute respiratory distress syndrome (ARDS) (Mehta et al., 2020, Qin et al., 2020). Indeed, excessive immune cell infiltration into the lung, CRS, and ARDS have previously been described as defining features of severe disease in humans infected with the closely related betacoronaviruses SARS-CoV and MERS-CoV (Channappanavar and Perlman, 2017, Nicholls et al., 2003). SARS-CoV-infected airway epithelial cells and macrophages express high levels of CCL5 (Law et al., 2005, Yen et al., 2006), which is a chemotactic molecule that can amplify inflammatory responses toward immunopathology, so we hypothesized that disrupting the CCL5–CCR5 axis via the leronlimab-mediated CCR5 blockade might prevent pulmonary trafficking of pro-inflammatory leukocytes and dampen pathogenic immune activation in COVID-19.
Leronlimab, formerly known as PRO 140, is a CCR5-specific human IgG4 monoclonal antibody in development for HIV therapy as a once weekly, at home subcutaneous injection. In five completed and four ongoing HIV clinical trials where over 800 individuals received leronlimab, no drug-related deaths, serious injection site reactions, or drug–drug interactions were reported (Jacobson et al., 2008, Jacobson et al., 2010a, Jacobson et al., 2010b, Dhody et al., 2018). Subcutaneous, self-administration of leronlimab by patients facilitates simple, once weekly dosing. In contrast to the small molecule CCR5 inhibitors that prevent HIV Env binding to CCR5 via allosteric modulation, leronlimab binds to the CCR5 extracellular loop 2 domain and N-terminus, thereby directly blocking the binding of HIV Env to the CCR5 co-receptor via a competitive mechanism. Leronlimab does not downregulate CCR5 surface expression or deplete CCR5-expressing cells, but it does prevent CCL5-induced calcium mobilization in CCR5+ cells with an IC50 of 45 μg/mL (Olson et al., 1999). This ability to specifically prevent CCL5-induced activation and chemotaxis of inflammatory CCR5+ macrophages and T cells suggests that leronlimab might be effective in mitigating pathologies involving the CCR5-ligand pathway.
Methods
Patients
All leronlimab-treated patients were enrolled in this study under an individual patient emergency use investigation new drug (EIND) via a United States Food and Drug Administration (FDA) emergency use authorization. The Albert Einstein College of Medicine Institution Review Board reviewed and approved this study. One 8 mL ethylenediaminetetraacetic acid (EDTA) tube and one 4 mL plasma preparation tube were drawn by venipuncture at day 0 (pre-treatment) and day 3, day 7, and day 14 post-treatment. Peripheral blood mononuclear cells (PBMCs) were isolated from peripheral blood using a Lymphoprep density gradient (STEMCELL Technologies, Vancouver, Canada). Aliquots of cells were frozen in media that contained 90% fetal bovine serum (Hyclone, Logan, UT, USA) and 10% dimethyl sulfoxide (Sigma–Aldrich, St. Louis, MO, USA), and stored at −70 °C. The five COVID observational control patients were part of a prospective observational cohort of subjects with viral respiratory illness symptoms who presented to Barnes Jewish Hospital, St. Louis Children’s Hospital, or affiliated Barnes Jewish Hospital testing sites located in Saint Louis, MO, USA. Inclusion criteria required that subjects were symptomatic and had a physician-ordered SARS-CoV-2 test performed in the course of their normal clinical care. All samples were collected at the time of enrollment, which was during or immediately following evaluation in a medical facility. The study was reviewed and approved by the Washington University Saint Louis Institutional Review Board (WU-350 study approval # 202003085). The study complied with the ethical standards of the Helsinki Declaration.
Assessment of plasma cytokine and chemokine levels
Fresh plasma was employed for cytokine quantification using a customized 13-plex bead-based flow cytometric assay (LegendPlex, Biolegend, Inc., San Diego, CA, USA) conducted with a CytoFlex flow cytometer. Raw data were analyzed using LegendPlex software (Biolegend Inc., San Diego, CA, USA). Samples were run in duplicate. In addition, split sample confirmation testing was performed by enzyme-linked immunosorbent assay (MDBiosciences, Minneapolis, MN, USA). Samples falling outside the linear range of the appropriate standard curves were diluted and repeated, where the dilution factor was incorporated into the final average. The cytokines, chemokines, and growth factors analyzed comprised: sCD40L, EGF, eotaxin, fibroblast growth factor (FGF)-2, fms-like tyrosine kinase 3 (Flt-3), fractalkine, granulocyte colony-stimulating factor (G-CSF), granulocyte-macrophage colony-stimulating factor (GM-CSF), growth-regulated oncogene (GRO)-α, FNα2, interferon (IFN)γ, interleukin (IL)-1α, IL-1β, IL-1ra, IL-2, IL-3, Il-4, IL-5, IL-5, IL-6, IL-7, IL-8, IL-9, IL-10, IL-2 (p40), IL-12 (p70), IL-13, IL-15, IL-17A, IL-17E/IL-25, IL-17F, IL-18, IL-22, IL-27, interferon gamma-induced protein 10 (IP-10), monocyte chemoattractant protein (MCP)-1, monocyte-chemotactic protein (MCP)-3, macrophage colony-stimulating factor (M-CSF), macrophage-derived chemokine (MDC), monokine induced by IFN-γ (MIG), macrophage inflammatory protein (MIP)-1α, MIP-1β, platelet-derived growth factor (PDGF)-AA, PDGF-AB/BB, RANTES (regulated on activation, normal T cell expressed and secreted), transforming growth factor (TGF)-α, tumor necrosis factor (TNF)-α, TNF-β, and vascular endothelial growth factor (VEGF).
Flow cytometry
PBMCs were isolated from peripheral blood using a Lymphoprep density gradient (STEMCELL Technologies, Vancouver, Canada). Each sample received a cocktail containing 10 μL Brilliant Stain Buffer (BD Biosciences, Franklin Lakes, NJ, USA), 5 μL True-Stain Monocyte Blocker (BioLegend, San Diego, CA, USA), and the following surface marker antibodies: anti-CD19 (PE-Dazzle594), anti-CD3 (APC), anti-CD16 (Alexa700), HLA-DR (APC/Fire750), and anti-CTLA-4 (PE-Cy7). The following antibodies were then added to each tube individually: anti-CD8 (BUV496), anti-CD4 (BUV661), anti-CD45 (BUV805), anti-CD103 (BV421), anti-TIM3 (BV605), anti CD56 (BV650), anti-LAG-3 (BV711), anti-CD14 (BB785), and anti-PD-1 (BB700), before incubating at room temperature for 30 min. Cells were fixed and permeabilized in 1× incellMAX (IncellDx, San Carlos, CA, USA) for 60 min at room temperature in the dark. Cells were washed once with 2% bovine serum albumin (BSA) solution, and analyzed using a Cytoflex LX system with 355 nm (20 mW), 405 nm (80 mW), 488 nm (50 mW), 561 nm (30 mW), 638 nm (50 mW), and 808 nm (60 mW) lasers (Beckman Coulter Life Sciences, Indianapolis, IN, USA). Analysis was performed with Kaluza version 2.1 software. The panel used in this study is shown in Supplementary Table 1.
CCR5 receptor occupancy
We determined CCR5 receptor occupancy by leronlimab using phycoerythrin-labeled leronlimab (IncellDx Inc.) in a competitive flow cytometry assay. CCR5-expressing immune cells including CD4+, CD45RO+ T-lymphocytes, CD4+, FoxP3+ T-regulatory cells, and CD14+, CD16+ monocytes/macrophages were included in the panel using the appropriate immunophenotypic markers for each population in addition to phycoerythrin-labeled leronlimab. Cells were incubated for 30 min in the dark at room temperature and washed twice with 2% BSA solution. Flow acquisition was conducted with a three-laser CytoFLEX fitted with 405 nm (80 mW), 488 nm (50 mW), and 638 nm (50 mW) lasers (Beckman Coulter, Indianapolis, IN, USA). Receptor occupancy was determined based on the loss of CCR5 detection over time in the subpopulations, and calculated using the following equation: 1–A/B × 100, where A is day 0 and B is day 7.
Measurement of plasma SARS-CoV-2 viral loads
A QIAamp Viral Mini Kit (Qiagen, Catalog #52906) was used to extract nucleic acids from 300 to 400 μL of plasma sample according to the manufacturer’s instructions and eluted in 50 μL of AVE buffer (RNase-free water with 0.04% sodium azide). The purified nucleic acids were tested immediately with a Bio-Rad SARS-CoV-2 ddPCR Kit (Bio-Rad, Hercules, CA, USA). The panel was designed for specifically detecting 2019-nCoV (two primer/probe sets). An additional primer/probe set was used to detect the human RNase P gene in control samples and clinical specimens. RNA isolated and purified from the plasma samples (5.5 μL) was added to a master mix comprising 1.1 μL of 2019-nCoV triplex assay, 2.2 μL of reverse transcriptase, 5.5 μL of supermix, 1.1 μL of dithiothreitol, and 6.6 μL of nuclease-free water.
The mixtures were then fractionated into up to 20,000 nanoliter-sized droplets in the form of a water-in-oil emulsion in a QX200 Automated Droplet Generator (Bio-Rad, Hercules, CA). The 96-well real-time-digital droplet polymerase chain reaction (RT-ddPCR) ready plate containing droplets was sealed with foil using a plate sealer and thermocycled to reverse transcribe the RNA, before PCR amplification of cDNA in a C1000 Touch thermocycler (Bio-Rad, Hercules, CA, USA). After PCR, the plate was loaded into a QX200 Droplet Reader (Bio-Rad, Hercules, CA, USA) and the fluorescence intensity of each droplet was measured in two channels (FAM and HEX). The fluorescence data were then analyzed with QuantaSoft 1.7 and QuantaSoft Analysis Pro 1.0 Software (Bio-Rad, Hercules, CA, USA) to determine the presence of SARS-CoV-2 N1 and N2 in the specimen. The limit of detection (LOD)was determined as shown in Supplementary Figure 1.
Bio-Rad SARS-CoV-2 RT-ddPCR Thermal Cycling Protocol
| Cycling Step | Temperature (°C) | Time | Number of Cycles |
|---|---|---|---|
| Reverse | 50 | 60 min | 1 |
| Transcription | |||
| PCR Enzyme | 95 | 10 min | 1 |
| Activation | |||
| Template | 94 | 30 s | 40 |
| Denaturation | |||
| Annealing/Extension | 55 | 60 s | |
| Droplet Stabilization | 4 | 30 min | 1 |
| Hold (Optional) | 4 | Overnight | 1 |
10. × Genomics 5′ single-cell RNA sequencing
Cells were then diluted to a concentration of 1 million cells per mL before loading onto the 10× chip. A single-cell RNA sequencing library was prepared with Chromium Next GEM Single cell Immune Profiling (v.1.1 Chemistry) according to the manufacturer’s protocols using a Chromium Controller instrument. The library was sequenced using a High Output Flowcell and Illumina NextSeq 500 instrument. The healthy control reference data were obtained from public 10× genomics data sets (“5k_pbmc_v3” and “pbmc4k” from https://support.10xgenomics.com/single-cell-gene-expression/datasets, and “vdj_nextgem_hs_pbmc3” from https://support.10xgenomics.com/single-cell-vdj/datasets). Raw FASTQ reads for the experimental samples and reference data were processed using 10× Genomics Cellranger version 3.1.0, where the data were aligned with human genome build GRCh38.p13, including unplaced contigs. The Ensembl release 98 gene build was used for feature counting. The raw count matrix provided by Cellranger was filtered using EmptyDrops, and processed using Seurat version 3.1.4. Briefly, count matrices were imported for each sample, merged, and processed using a standard Seurat pipeline. Cells were filtered based on the following criteria: less than 200 or more than 3000 distinct genes, more than 15% unique molecular identifiers from mitochondrial genes, and a maximum library size of 20,000. Log normalization was then performed and the top variable genes were identified using the “vst” method. Differential expression (DE) analysis was performed using the Wilcoxon rank-based test based on the normalized counts. Only pertinent cells were used in the contrast for the cell-type DE level. In each comparison, the inclusion criteria for significant genes were set with the Bonferroni adjusted p-values generated with Seurat’s FindMarkers function, with an error rate less than 0.1.
Statistical analysis
We performed statistical analyses between groups using the nonparametric Kruskal–Wallis test followed by Dunn’s multiple comparison correction to control the experiment-wise error rate. We compared categorical variables using Fisher’s exact test and continuous variables with the Mann–Whitney U test We assessed data based on the repeated measure correlation (rmcorr).
Results
Ten critical COVID-19 patients at the Montefiore Medical Center received leronlimab via FDA-approved EIND requests for individual patient use (Table 1). These confirmed SARS-CoV-2 positive patients had significant pre-existing co-morbidities and were receiving intensive care treatment, including mechanical ventilation or supplemental oxygen for ARDS. Consistent with previous reports of severe COVID-19 disease (Huang et al., 2020), these patients showed evidence of lymphopenia with liver and kidney damage (Supplementary Figure 2) (Akalin et al., 2020). Four of the patients died during the 14-day study period due to a combination of disease complications and severe constraints on medical equipment culminating in medical triage. Although this EIND study lacked a placebo control group for comparison, a recent study of other critically ill COVID-19 patients in the New York City area indicated mortality rates as high as 88% (Richardson et al., 2020).
Table 1.
Leronlimab-treated critical COVID-19 patient summaries.
| Day relative to leronlimab |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient | Age/Gender | Pre-existing conditions | Renal transplant year | Dialysis in hospital | Onset Sx | PCR+ | Admit | Treatments for COVID and concomitant infections | Vasopressors used | Baseline status | Extubated (day relative to leronlimab) | Respiratory outcome | Subject status (day relative to leronlimab) |
| L1 | 74/M | AKI, HTHD, Prostate CA (s/p prostatectomy), DM, Gout | N/A | Yes | −14 | −8 | −8 | CQ, HCQ, TZP, AZT | Yes | Intubated | TBD | Remains intubated | Still admitted |
| L2 | 74/F | ESRD, HTHD, DM, HLD | 2018 | Yes | −3 | −2 | −2 | HCQ, CQ, AZM, LPV/RTV | Yes | Intubated | No | Worsened | Deceased (8) |
| L3 | 54/M | RF, HTHD, HLD | N/A | Yes | −1 | −1 | −1 | CQ, HCQ, TZP, VAN | Yes | Venture mask, same day intubated | TBD | Remains intubated | Still admitted |
| L4 | 56/M | HTHD, Skin CA, Papillary thyroid CA (s/p thyroidectomy), DM | N/A | Yes | −1 | 2 | −8 | CQ, LPV/RTV | Yes | Intubated | Yes (1) | RA by day 8 | Still admitted |
| L5 | 58/M | ESRD, CKD stage 3 in renal allograft, recurrent UTI with MDR E.coli, DM, DR, HTHD, HLD | 2016 | Yes | −8 | −5 | −6 | AZM, HCQ | Yes | Intubated | Yes (13) | Improving | Still admitted |
| L6 | 42/M | FSGS, CKD stage 3, DVT/PE, Gout | 2005, 2016 | No | −4 | −1 | −1 | HCQ, AZM, CAX, VAN, CEF | No | On 2 L NC | N/A | Stable on RA | Discharged (13) |
| L7 | 68/M | ESRD, Hydronephrosis (s/p stent placement), HTHD, HLD, DM with retinopathy and neuropathy | 2018 | Yes | −13 | −10 | −10 | AZM, HCQ, VAN, TZP | Yes | On NRB | TBD | Improving | Still admitted |
| L8 | 56/F | ESRD, lung CA (s/p bilateral upper lobectomy), COPD, Asthma, DM, HTHD, HLD, Hepatitis C | 2009 | No | −7 | −5 | −5 | CAX, AZT, HCQ, CEF, VAN | No | 3−4 L NCa | No | Worsened | Deceased (6) |
| L9 | 51/F | AKI, HTHD, OSA (on Bilevel Positive Airway Pressure) | 2006 | Yes | −6 | −5 | −6 | HCQ | Yes | Intubated | No | Worsened | Deceased (4) |
| L10 | 79/M | AKI, CAD, Prostate CA, GERD, HTHD, HLD | N/A | Yes | −13 | −6 | −6 | HCQ | Yes | Intubated | No | Remained intubated | Deceased (7) |
N/A = not applicable, s/p = status post-, AKI = acute kidney injury, HTHD = hypertensive heart disease, DM = diabetes mellitus, HLD = hyperlipidemia, ESRD = end-stage renal disease, HD = hemodialysis, CA = cancer, COPD = chronic obstructive pulmonary disease, LUL = left upper lobe, RUL = right upper lobe, MDR = multi-drug resistant, CKD = chronic kidney disease, UTI = urinary tract infection, FSGS = focal segmental glomerulosclerosis, DVT = deep vein thrombosis, PE = pulmonary embolism, OSA = obstructive sleep apnea, CAD = coronary artery disease, GERD = gastroesophageal reflux disease, RF = renal failure, DR = diabetic retinopathy, HCQ = hydroxychloroquine, CQ = chloroquine, AZM = azithromycin, VAN = vancomycin, CAX = ceftriaxone, LPV/RTV = lopinavir/ritonavir, TZP = piperacillin-tazobactam, CEF = cefepime, NC = nasal canula, NRB = non-rebreather mask, TBD = to be determined.
Patient declined intubation due to poor baseline pulmonary status.
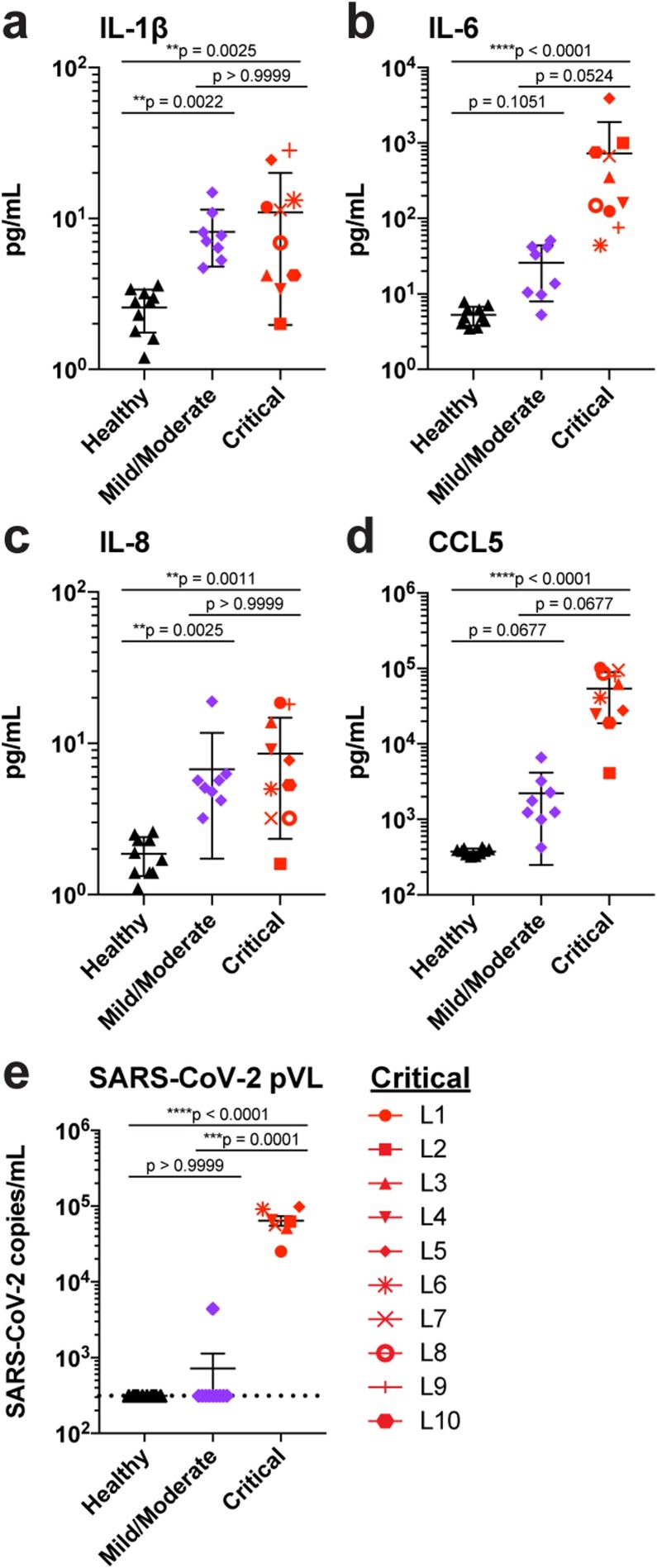
Hyper-immune activation and CRS are present in cases of severe COVID-19 (Mehta et al., 2020). Indeed, at leronlimab treatment baseline, signatures of CRS were present in the plasma of all 10 patients in the form of significantly elevated levels of the inflammatory cytokines IL-1β, IL-6, and IL-8 (Figure 1A–C) compared with the healthy controls. In contrast to patients with mild or moderate COVID-19, only IL-6 was present at significantly higher levels in the critically ill patients. It should be noted that the plasma CCL5 levels of the 10 critically ill patients were markedly elevated compared with those of both the healthy controls and mild or moderate COVID-19 patients (Figure 1D). High levels of CCL5 can cause acute renal failure and liver toxicity (Yu et al., 2016, Chen et al., 2020), which are both common findings with COVID-19 infection. Indeed, the critically ill patients presented with varying degrees of kidney and liver injury, although many had also previously received kidney transplants (Akalin et al., 2020) (Table 1 and Supplementary Figure 2). Low levels of SARS-CoV-2 have been detected but not yet quantified in the plasma of COVID-19 patients (Lescure et al., 2020). We used high sensitivity, ddPCR to quantify plasma SARS-CoV-2 viremia at baseline. SARS-CoV-2 was found in the plasma of all 10 critically ill patients, but no viremia was detected in the healthy controls and in only one patient with mild/moderate COVID-19, thereby highlighting the critical nature of COVID-19 in these patients (Figure 1E).
Figure 1.
Elevated cytokine, chemokine, and SARS-CoV-2 levels in critically ill COVID-19 patients. (A–E) Plasma levels of IL-1β (A), IL-6 (B), IL-8 (C), CCL5 (D), and SARS-CoV-2 RNA copies (E) in patients with mild/moderate (purple symbols, n = 8) and critical (red symbols, n = 10 in panels a–d, and n = 7 in panel e) COVID-19 disease compared with healthy controls (black symbols, n = 10). Dashed line indicates the LOD. Graphs show p-values calculated using Dunn’s Kruskal–Wallis test: *p ≤ 0.05, ** p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001.
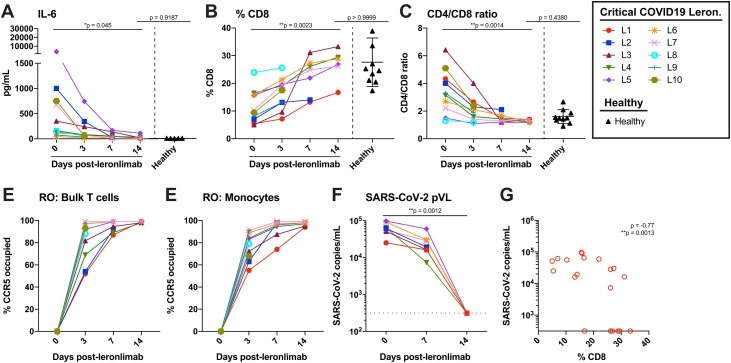
At study day 0, all 10 critically ill patients received a subcutaneous injection of 700 mg leronlimab following baseline blood collection. Subsequently, the patients received a second subcutaneous injection of 700 mg leronlimab at study day 7. The defining features of severe COVID-19 disease include the plasma IL-6 and T cell lymphopenia (Huang et al., 2020, Lescure et al., 2020), so we longitudinally monitored these parameters for 2 weeks after the first leronlimab treatment. A reduction in the plasma IL-6 was observed as early as 3 days following leronlimab with a return to healthy control levels by day 14 (Figure 2A). In contrast to IL-6, the levels of other cytokines and chemokines were more variable after leronlimab treatment (Supplementary Figure 3). Following leronlimab administration, a marked restoration of CD8+ T cells (Figure 2B) and normalization of the CD4+ and CD8+ T cell ratio were observed in the blood samples (Figure 2C). These immunological changes occurred concomitant with leronlimab CCR5 receptor occupancy on the surface of CCR5+ T cells and monocytes (Figure 2D and E). The percentage of other CCR5-expressing cell types (CD4 T-cells, NK cells, and monocyte/macrophages) did not change significantly over 14 days of leronlimab treatment. Following leronlimab administration, SARS-CoV-2 plasma viremia decreased in all patients at day 7, and all but one patient had resolved SARS-CoV-2 plasma viremia to undetectable levels by day 14 (Figure 2F left, p = 0.0012). Finally, SARS-CoV-2 plasma viremia in leronlimab-treated COVID-19 patients was inversely correlated with the frequency of CD8+ T cells in the blood, which suggested immune restoration (Figure 2G).
Figure 2.
Reversal of immune dysfunction, CCR5 receptor occupancy, and decrease in SARS-CoV-2 plasma viral loads in critically ill COVID-19 patients after leronlimab administration. (A–C) Plasma levels of IL-6 (A), and peripheral blood CD8+ T cell percentages of CD3+ cells (B) and CD4/CD8 T cell ratio (C) at days 0 (n = 10), 3 (n = 10), 7 (n = 7), and 14 (n = 6) post-leronlimab administration. Healthy controls (n = 10) are shown in black triangles. Critically ill COVID-19 patients not treated with leronlimab are shown in panel A (right, open symbols, n = 5). Graphs show p-values calculated using Dunn’s Kruskal–Wallis test: not significant p > 0.05, *p ≤ 0.05, ** p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001. (D–E) CCR5 receptor occupancy on peripheral blood bulk T cells (D), and monocytes (E). (F), SARS-CoV-2 plasma viral loads at days 0, 7, and 14 post-leronlimab (left panel, closed symbols, n = 7). Critically ill COVID-19 patients not treated with leronlimab are shown in panel F (right panel, open symbols, n = 5). Horizontal dashed line indicates the LOD. Graph show p-values calculated using the Mann–Whitney test: *p ≤ 0.05, ** p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001. (G) Plot showing CD8 percentages in blood and SARS-CoV- 2 plasma viral load in seven critically ill COVID-19 patients at days 0, 7, and 14 post-leronlimab (n = 20). Graph shows rho (ρ) and p- values calculated by repeated measures correlation: *p ≤ 0.05, ** p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001. The 95% confidence interval for the repeated measures correlation was −0.93 to 0.35.
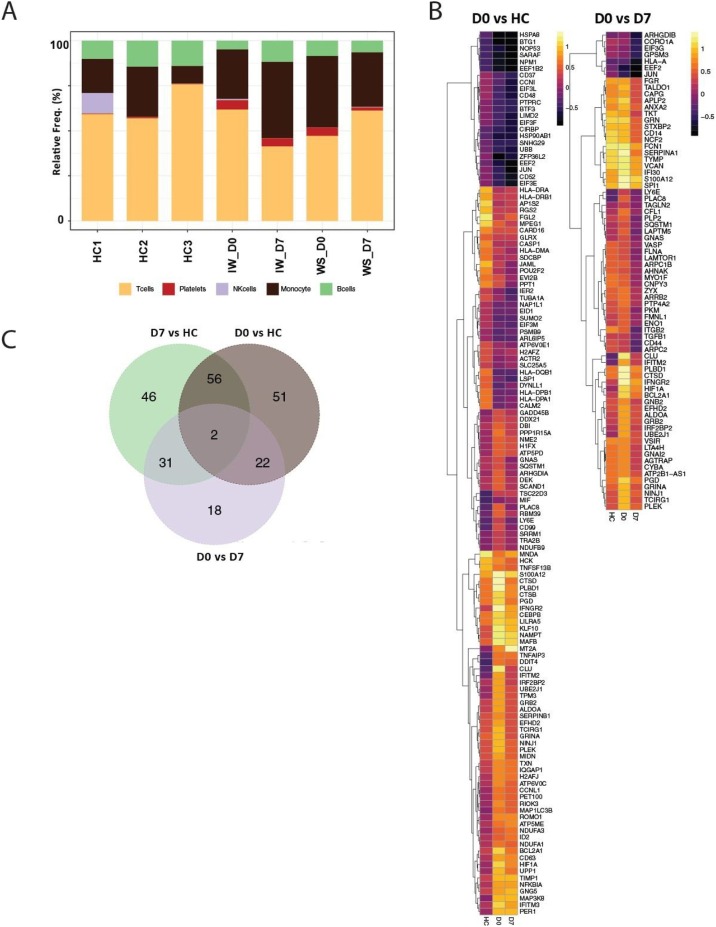
To establish an unbiased gene repertoire for these COVID-19 patients, we performed 10× Genomics 5′ single cell RNA sequencing for PBMCs from two of the severe COVID-19 patients (L2 and L4) for which sufficient baseline, pre-leronlimab treatment COVID-19 samples were available. We also performed scRNA-seq for the same patients at day 7 post-treatment, and included three reference PBMC scRNA-seq data sets from healthy controls provided by 10× Genomics. After quality control, our longitudinal COVID-19 single cell data set profiled a total of 3785 cells at baseline, 5056 cells at the 7-day post-leronlimab time point, and 18,603 cells from healthy controls. We were able to assign cell types to most cells based on a combination of the expression of known marker genes and automated cell type classification using the SingleR package (Aran et al., 2019) to identify five primary clusters that corresponded to monocytes, B, T, and NK cells, and some platelets (Figure 3). We initially compared the baseline COVID samples with the healthy controls. We detected a total of 75 DE transcripts (Wilcoxon adjusted p < 0.1) (Supplementary Table 2). The top pathways enriched in this set included multiple TCR signaling pathways, such as the genes LCK, CD3E, and NFKBIA (Figure 3). Differences in cellularity between donors will confound global gene expression analyses, so we extracted the myeloid cell cluster and performed DE analyses using only these cells. We identified 131 DE transcripts between the healthy controls and COVID-19 on day 0 (Supplementary Table 2 and Figure 3). This gene set was enriched for genes involved in interferon signaling (IFITM2, IFNGR2, and NFKBIA), and cytokine signaling (HSP8A, JUN, and TIMP1). We found that several downstream targets of IL-6 were also upregulated: ZFP36L2, CEBPB, and LY6E.
Figure 3.
Phenotyping and differential gene expression analyses based on scRNA-seq data. (A) Graph showing the proportion of each cell type in healthy control and leronlimab-treated COVID patient samples. (B) Heatmap of significantly differentially expressed genes (Wilcoxon adjusted p < 0.1) by contrasting either COVID day 0 vs. healthy control (HC) or COVID day 9 vs day 7 post-leronlimab treatment. (C) Venn diagram showing the overlap between genes identified from the contrasts shown.
To identify markers that could inform effective leronlimab treatment, we then compared gene expression within the two severe COVID-19 participants by contrasting the baseline and day 7 post-leronlimab. After performing bulk DE based on the total PBMCs, we identified 14 DE genes, although this could also have been confounded by the differences in the cell compositions. It should be noted that this set included a significant drop in CD44, a downstream target of IL-6 signaling (Vincent and Mechti, 2004), and a number of genes involved in neutrophil degranulation (ITGB2, EEF2, HSP90AA1, and ALDOA). Next, we performed DE specifically based on myeloid cells and identified 73 DE transcripts in this subset (Wilcoxon adjusted p < 0.1; Supplementary Table 2). In agreement with the decrease in the IL-6 protein levels observed in plasma, we observed significant decreases in the expression of many IL-6 responsive genes, including TGFB1, IFI30, and LY6E, which are consistent with previous reports of monocyte/macrophages repolarization following CCR5 blockade (Halama et al., 2016). We also observed myeloid cells expressing chemokine and IFN-related genes, such as IFNGR2, IFITM2, and TALDO1, which were significantly downregulated at day 7 post-leronlimab compared with baseline (Supplementary Table 2). These transcriptomic findings further highlight the potential impact of leronlimab-mediated CCR5 blockade on the inflammatory state in COVID-19.
Discussion
In this study, we investigated the involvement of the chemokine receptor CCR5 in COVID-19 and obtained data from 10 critically ill patients with severe COVID-19 that demonstrated reductions in inflammation, restoration of T cell lymphocytopenia, and reduced SARS-CoV-2 plasma viremia following leronlimab-mediated CCR5 blockade. We found statistically significant increases in IL-1β, IL-6, IL-8, and RANTES in these critically ill patients compared with healthy controls. The increases in these cytokines are the hallmark of COVID-19 and elevation of the chemokine CCL5/RANTES has been demonstrated across the COVID-19 disease spectrum from mild (Zhao et al., 2020) to severe (Li et al., 2020) patients. RANTES was elevated to a greater extent than the other CCR5 binding chemokines MIP-1α and MIP-1β, probably due to the production of RANTES in respiratory epithelial cells (in addition to immune cells) in respiratory viral infections (Schroth et al., 1999). We also found a profound reduction in the CD8 percentage with concomitant increases in the CD4/CD8 ratio. In this cohort of critical COVID-19 patients, SARS-CoV-2 RNA was detectable and quantifiable in plasma samples from all patients. For the first time, we demonstrated that restoration of the CD8 T-cell numbers was significantly correlated with decreases in the plasma viral load. Overall, our results showed that therapy with a CCR5 antagonist could reduce the cytokine storm, resolve the profound CD-8 T-cell lymphopenia, and reduce the plasma viral load to undetectable levels by day 14. These data support recent studies that suggested the potential for targeting CCR5 as a therapeutic in COVID-19 (Chua et al., 2020).
Recent studies showed that a significant number of COVID-19 patients had increased risks of strokes, blood clots, and other thromboembolic events (Grillet et al., 2020). Platelet activation leads to initiation of the coagulation cascade and it can be triggered by chemokines including CCL5/RANTES (Machlus et al., 2016), thereby suggesting that leronlimab treatment may be beneficial beyond its immunomodulatory effects on inflammation and hemostasis in COVID-19 patients. Given medical triage resulting in patient death and the lack of a placebo control group, we cannot comment on the impact of leronlimab on the clinical outcomes in these patients. Anecdotal evidence has been reported of clinical improvements in COVID-19 patients following leronlimab treatment (Mottet, 2020), but randomized controlled trials are required to determine the efficacy of leronlimab in COVID-19. Indeed, randomized, double blind, placebo controlled clinical trials are underway to assess the efficacy of leronlimab treatments in patients with mild to moderate (NCT04343651) (ClinicalTrials.gov, 2020) and severe to critical (NCT04347239) COVID-19 (ClinicalTrials.gov). In summary, our results suggest the involvement of CCR5 in the pathology of SARS-CoV-2, and that inhibiting the activity of CCL5 via CCR5/RANTES blockade represents a novel therapeutic strategy for COVID-19 with both immunological and virological implications.
Author contributions
BKP, HS, KD, KK, JL, SK, NP, and JBS conceived the study, HS and EA coordinated patient care, JAO, PAM, MP, LCN, RG, JAO, PAM, and OA led control patient recruitment, BKP, HS, AP, EBF, HR, WR, AL, LK, MH, EH, JAO, and PM acquired data, BKP, MJC, APSP, CS, BF, HLW, GMW, BSP, SK, JL, AL, LK, MH, EH, LCN, and NP analyzed data, and BKP, MJC, KD, JL, HLW, GMW, BSO, SK, NP, LCN, and JBS wrote the manuscript.
Data availability
All primary data presented in this study are available from the corresponding author upon reasonable request. Primary data exist for all figures
Funding
No funding.
Conflict of interest
Dr. Sacha has received compensation for consulting for CytoDyn Inc., a company that may have a commercial interest in the results of this research. The potential conflict of interest has been reviewed and managed by Oregon Health & Science University. Drs. Kelly and Pourhassan are employees of CytoDyn Inc., owner and developer of Leronlimab. Dr. Lalezari is a principal investigator for CytoDyn Inc. through his company Quest Clinical Research. Dr. Patterson, Brian Francisco, Amruta Pise, Matthew Ryou, and Hallison Rodrigues are employees of IncellDx, Inc., a diagnostic company that providied assays to Cytodyn Inc. Dr. Ndhlovu has received compensation for serving on a scientific advisory board for Abbvie. Lama Kdouh and Alina Lelic are employees of Beckman Coulter Life Sciences, and Monica Herrera and Eric Hall are employees of Bio-Rad, Inc. Drs. Kush Dhody and Kazem Kazempour are employees of Amarex Clinical Research, LLC, a company that manages clinical trials and regulatory matters for CytoDyn Inc.
Ethical approval
Approved.
Acknowledgments
We gratefully acknowledge the patient participants and their caregivers Kristine Stryker, Victoria Caputo, Vagish Hemminge, Scott A Scheinin, Magdalena Mamczur-Madry, Sana Ahmed, Pamela Philllippsborn, Vagish Hemmige, Reena Joseph, and Jasmine Thallipllill who made this work possible. We acknowledge Lawrence Drew, Parviz Lalezari, Shaheed Abdulhaqq, Justin Greene, Whitney Weber, Jason Reed, Cleiton Pessoa, Katherine Bateman, and Jason Reed for critical reading of the manuscript. The WU-350 cohort was funded by a grant from the Barnes Jewish Hospital Foundationand by support from the Washington University Institute of Clinical and Translational Sciences grant UL1TR002345 from the N ational Center for Advancing Translational Sciences (NCATS)of the National Institutes of Health (NIH). This study was also supported partly by NIH R01 AI129703 to JBS. The content is solely the responsibility of the authors and does not necessarily represent the official view of the NIH.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.ijid.2020.10.101.
Appendix A. Supplementary data
The following are Supplementary data to this article:
List of antibodies utilized in this study.
Differentially expressed genes. The table shows all of the genes identified as differentially expressed (Wilcoxon adjusted p < 0.1) in for the contrasts listed.
COVID-19 plasma viral load lower limit of detection (LOD) determination. A dilution series of SARS-CoV-2 RNA diluted in normal human plasma was tested in 15 extraction replicates for each dilution. The LOD was defined as the concentration where ≥ 95% of the replicates tested were positive for the N1 and N2 markers, which was 313 cp/mL. This value was used as the LOD.
Complete blood count and serum chemistry values in critically ill COVID-19 patients after leronlimab administration. a–d, Peripheral blood levels of platelets (a), hemoglobin (b), white blood cells (c), and lymphocytes (d). Serum levels of ferritin (e), C-reactive protein (f), procalcitonin (g), D-dimer, where values >20 are shown as 20 (h), aspartate aminotransferase (AST), where values <20 are shown as 0 (i), alanine aminotransferase (ALT), where values <10 are shown as 0 (j), blood urea nitrogen (BUN) (k), creatinine (CREA) (l), sodium (m), and total bilirubin (n). Gray boxes indicate the standard reference range for each parameter.
Immunological parameters in critically ill COVID19 patients untreated and treated with leronlimab. a–af, Plasma levels of cytokines and chemokines in critically ill COVID19 patients treated with leronlimab for the cytokine or chemokine listed.
References
- Akalin E., Azzi Y., Bartash R., Seethamraju H., Parides M., Hemmige V., et al. Covid-19 and kidney transplantation. N Engl J Med. 2020 doi: 10.1056/NEJMc2011117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aran D., Looney A.P., Liu L., Wu E., Fong V., Hsu A., et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat Immunol. 2019;20:163–172. doi: 10.1038/s41590-018-0276-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Channappanavar R., Perlman S. Pathogenic human coronavirus infections: causes and consequences of cytokine storm and immunopathology. Semin Immunopathol. 2017;39:529–539. doi: 10.1007/s00281-017-0629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Zhang Q., Yu C., Wang F., Kong X. Functional roles of CCL5/RANTES in liver disease. Liver Res. 2020;4:28–34. [Google Scholar]
- Chua R.L., Lukassen S., Trump S., Hennig B.P., Wendisch D., Pott F., et al. COVID-19 severity correlates with airway epithelium-immune cell interactions identified by single-cell analysis. Nat Biotechnol. 2020;8:970–979. doi: 10.1038/s41587-020-0602-4. [DOI] [PubMed] [Google Scholar]
- ClinicalTrials.gov-Study to Evaluate the Efficacy and Safety of Leronlimab for Mild to ModerateCOVID-19. https://clinicaltrials.gov/ct2/show/NCT04343651.ClinicalTrials.gov, Study to Evaluate the Efficacy and Safety of Leronlimab for Patients with Severe or Critical Coronavirus Disease 2019 (COVID-19). https://clinicaltrials.gov/ct2/show/NCT04347239.
- Dhody K., Pourhassan N., Kazempour K., Green D., Badri S., Mekonnen H., et al. PRO 140, a monoclonal antibody targeting CCR5, as a long-acting, single-agent maintenance therapy for HIV-1 infection. HIV Clin Trials. 2018;19:85–93. doi: 10.1080/15284336.2018.1452842. [DOI] [PubMed] [Google Scholar]
- Grillet F., Behr J., Calame P., Aubry S., Delabrousse E. Acute pulmonary embolism associated with COVID-19 pneumonia detected by pulmonary CT angiography. Radiology. 2020 doi: 10.1148/radiol.2020201544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halama N., Zoernig I., Berthel A., Kahlert C., Klupp F., Suarez-Carmona M., et al. Tumoral immune cell exploitation in colorectal cancer metastases can be targeted effectively by anti-CCR5 therapy in cancer patients. Cancer Cell. 2016;29:587–601. doi: 10.1016/j.ccell.2016.03.005. [DOI] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson J.M., Saag M.S., Thompson M.A., Fischl M.A., Liporace R., Reichman R.C., et al. Antiviral activity of single-dose PRO 140, a CCR5 monoclonal antibody, in HIV-infected adults. J Infect Dis. 2008;198:1345–1352. doi: 10.1086/592169. [DOI] [PubMed] [Google Scholar]
- Jacobson J.M., Thompson M.A., Lalezari J.P., Saag M.S., Zingman B.S., D’Ambrosio P., et al. Anti-HIV-1 activity of weekly or biweekly treatment with subcutaneous PRO 140, a CCR5 monoclonal antibody. J Infect Dis. 2010;201:1481–1487. doi: 10.1086/652190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson J.M., Lalezari J.P., Thompson M.A., Fichtenbaum C.J., Saag M.S., Zingman B.S., et al. Phase 2a study of the CCR5 monoclonal antibody PRO 140 administered intravenously to HIV-infected adults. Antimicrob Agents Chemother. 2010;54:4137–4142. doi: 10.1128/AAC.00086-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law H.K., Cheung C.Y., Ng H.Y., Sia S.F., Chan Y.O., Luk W., et al. Chemokine up-regulation in SARS-coronavirus-infected, monocyte-derived human dendritic cells. Blood. 2005;106:2366–2374. doi: 10.1182/blood-2004-10-4166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescure F.X., Bouadma L., Nguyen D., Parisey M., Wicky P.H., Behillil S., et al. Clinical and virological data of the first cases of COVID-19 in Europe: a case series. Lancet Infect Dis. 2020 doi: 10.1016/S1473-471.3099(20)30200-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Jiang L., Li X., Lin F., Wang Y., Li B., et al. Clinical and pathological investigation of patients with severe COVID-19. JCI Insight. 2020;5(12) doi: 10.1172/jci.insight.138070. PMID: 32427582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machlus K.R., Johnson K.E., Kulenthirarajan R., Forward J.A., Tippy M.D., Soussou T.S., et al. CCL5 derived from platelets increases megakaryocyte proplatelet formation. Blood. 2016;127:921–926. doi: 10.1182/blood-2015-05-644583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta P., McAuley D.F., Brown M., Sanchez E., Tattersall R.S., Manson J.J., et al. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395:1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottet S. Los Angeles CBS Local; 2020. Coronavirus Survivor Credits Artificial Antibody Experimental Treatment for Recovery.https://losangeles.cbslocal.com/2020/04/10/coronavirus-survivor-leronlimab/ [Google Scholar]
- Nicholls J.M., Poon L.L., Lee K.C., Ng W.F., Lai S.T., Leung C.Y., et al. Lung pathology of fatal severe acute respiratory syndrome. Lancet. 2003;361:1773–1778. doi: 10.1016/S0140-6736(03)13413-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson W.C., Rabut G.E., Nagashima K.A., Tran D.N., Anselma D.J., Monard S.P., et al. Differential inhibition of human immunodeficiency virus type 1 fusion, gp120 binding, and CC-chemokine activity by monoclonal antibodies to CCR5. J Virol. 1999;73:4145–4155. doi: 10.1128/jvi.73.5.4145-4155.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin C., Zhou L., Hu Z., Zhang S., Yang S., Tao Y., et al. Dysregulation of immune response in patients with COVID-19 in Wuhan, China. Clin Infect Dis. 2020;71:762–768. doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson S., Hirsch J.S., Narasimhan M., Crawford J.M., McGinn T., Davidson K.W., et al. Presenting characteristics, comorbidities, and outcomes among 5700 patients hospitalized with COVID-19 in the New York City Area. JAMA. 2020;26(323):2052–2059. doi: 10.1001/jama.2020.6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroth M.K., Grimm E., Frindt P., Galagan D.M., Konno S.-I., Love R., et al. Rhinovirus replication causes RANTES production in primary bronchial epithelial cells. Am J Respir Cell Mol Biol. 1999;20:1220–1228. doi: 10.1165/ajrcmb.20.6.3261. [DOI] [PubMed] [Google Scholar]
- Vincent T., Mechti N. IL-6 regulates CD44 cell surface expression on human myeloma cells. Leukemia. 2004;18:967–975. doi: 10.1038/sj.leu.2403333. [DOI] [PubMed] [Google Scholar]
- World Health Organization. Coronavirus Disease (COVID-2019) Situation Reports. Available at: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports. [Accessed 23 April 2020].
- Yen Y.T., Liao F., Hsiao C.H., Kao C.L., Chen Y.C., Wu-Hsieh B.A. Modeling the early events of severe acute respiratory syndrome coronavirus infection in vitro. J Virol. 2006;80:2684–2693. doi: 10.1128/JVI.80.6.2684-2693.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu T.M., Palanisamy K., Sun K.T., Day Y.J., Shu K.H., Wang I.K., et al. RANTES mediates kidney ischemia reperfusion injury through a possible role of HIF-1α and LncRNA PRINS. Sci Rep. 2016;6:18424. doi: 10.1038/srep18424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Qin L., Zhang P., Li K., Liang L., Sun J., et al. Longitudinal COVID-19 profiling associates IL-1RA and IL-10 with disease severity and RANTES with mild disease. JCI Insight. 2020;5 doi: 10.1172/jci.insight.139834. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of antibodies utilized in this study.
Differentially expressed genes. The table shows all of the genes identified as differentially expressed (Wilcoxon adjusted p < 0.1) in for the contrasts listed.
COVID-19 plasma viral load lower limit of detection (LOD) determination. A dilution series of SARS-CoV-2 RNA diluted in normal human plasma was tested in 15 extraction replicates for each dilution. The LOD was defined as the concentration where ≥ 95% of the replicates tested were positive for the N1 and N2 markers, which was 313 cp/mL. This value was used as the LOD.
Complete blood count and serum chemistry values in critically ill COVID-19 patients after leronlimab administration. a–d, Peripheral blood levels of platelets (a), hemoglobin (b), white blood cells (c), and lymphocytes (d). Serum levels of ferritin (e), C-reactive protein (f), procalcitonin (g), D-dimer, where values >20 are shown as 20 (h), aspartate aminotransferase (AST), where values <20 are shown as 0 (i), alanine aminotransferase (ALT), where values <10 are shown as 0 (j), blood urea nitrogen (BUN) (k), creatinine (CREA) (l), sodium (m), and total bilirubin (n). Gray boxes indicate the standard reference range for each parameter.
Immunological parameters in critically ill COVID19 patients untreated and treated with leronlimab. a–af, Plasma levels of cytokines and chemokines in critically ill COVID19 patients treated with leronlimab for the cytokine or chemokine listed.
Data Availability Statement
All primary data presented in this study are available from the corresponding author upon reasonable request. Primary data exist for all figures