Abstract
Human primary breast cancers and breast cancer cell lines are classified by microarray‐defined molecular subtypes, which reflect differentiation characteristics. Estrogen receptor (ER) expression is indicative of the luminal molecular subtype. We have previously established IPH‐926, the first well–characterized cell line from infiltrating lobular breast cancer. IPH‐926 displays an ER/PR/ErbB2 triple‐negative immunophenotype, which is due to a loss of ER expression in its in vivo clonal ancestry. Loss of ER might indicate a fundamental change of cellular differentiation and it is unclear whether a luminal subtype is preserved beyond ER conversion. Using Affymetrix microarray analysis, seven different classifier gene lists (PAM305, DISC256, TN1288, PAM50, UNC1300, LAB704, INT500) and a background population of 50 common mammary carcinoma cell lines, we have now determined the molecular subtype of IPH‐926. Strikingly, the IPH‐926 expression profile is highly consistent with a luminal subtype. It is nearest to luminal/ER‐positive breast cancer cell lines and far apart from basal breast cancer cell lines. Quantitative real–time RT–PCR confirmed enhanced expression of luminal marker genes (AGR2, CLU, CA12, EMP2, CLDN3) and low or absent expression of basal marker genes (KRT5, CD44, CAV1, VIM). Moreover, IPH‐926 lacked androgen receptor (AR) expression, a transcription factor previously associated with luminal‐like gene expression in a subset of triple‐negative or molecular apocrine breast cancers. In conclusion, IPH‐926 is triple‐negative but belongs to the luminal subtype. Luminal differentiation characteristics can be preserved beyond ER conversion and might not require a compensatory expression of AR.
Microarray expression analyses identified breast cancer molecular subtypes, such as luminal, basal‐like and ErbB2‐positive carcinomas, which share differentiation characteristics.1, 2, 3, 4 This molecular taxonomy is continually refined.5 However, for clinical diagnostics, molecular subtypes are approximated by immunohistochemical surrogate markers.6 The 2011 and 2013 St Gallen consensus conferences have confirmed immunohistochemistry for estrogen receptor (ER), progesterone receptor (PR) and ErbB2 as the clinical gold standard to assess tumor phenotypes corresponding to microarray‐defined molecular subtypes.7, 8 However, this has not been without dissenting votes, as these immunohistochemical markers are primarily directed towards finding the optimal targeted therapy and show only a limited concordance with microarray‐defined molecular subtypes.8 For instance, an ER/PR/ErbB2 triple‐negative (TN) immunophenotype is typically seen in carcinomas classified as basal‐like by microarray analysis, but only 30–70% of TN carcinomas are basal, as defined by gene expression profiling.9, 10 Accordingly, basal and TN phenotypes must not be equated.9
Human breast cancer cell lines are also classified by microarray‐defined subtypes.11, 12, 13, 14 This is relevant for pre‐clinical models.15 The PAM305 classifier gene list has been established specifically for subtype assignment in breast cancer cell lines.11 Like primary tumors, cell lines might show a luminal subtype (typically ER‐positive) or a basal subtype (typically TN).11, 12 The basal subtype is commonly stratified into groups termed basal‐A and basal‐B.11, 12 An alternative subdivision for TN breast cancer has been described by Lehmann et al.10 and includes subclasses termed basal‐1 and basal‐2 (BL1/BL2), immunomodulatory (IM), mesenchymal and mesenchymal stem–like (M/MSL) and luminal androgen receptor (AR)‐positive (LAR). Contrary to primary tumors, the luminal subtype is not further subdivided and ErbB2–positive samples do not form a class of their own in breast cancer cell lines.11, 12
Infiltrating lobular breast cancer (ILBC) is a special histological entity associated with ER expression and the luminal subtype.16, 17 IPH‐926 is the first well–characterized cell line from ILBC.18, 19, 20 We have previously shown that IPH‐926 displays a TN immunophenotype, which is due to a progression‐related loss of ER in its in vivo clonal ancestry.18 Estrogen receptor conversion is observed in approximately 10% of initially ER‐positive carcinomas and might indicate a loss of luminal differentiation.21 Hence, the molecular subtype of IPH‐926 is enigmatic. Here we report the first comprehensive microarray‐based classification of IPH‐926, which adds important new information as to the exact categorization of this unique cell line and the stability of luminal differentiation.
Materials and Methods
Cell lines
IPH‐926 cells were authenticated by short tandem repeat profiling and PCR–based detection of the unique CDH1 241ins4 mutation.18
Immunohistochemistry
Immunohistochemical stainings were performed on a Benchmark Ultra (Ventana, Tucson, AZ, USA) automated stainer using the CC1 mild protocol for antigen retrieval and the monoclonal anti‐ER (clone SP1, undiluted read‐to‐use; Ventana), anti‐PR (clone 1E2, undiluted read‐to‐use; Ventana), anti‐ErbB2 (clone 4B5, undiluted read‐to‐use; Ventana) and anti‐AR (clone AR441, 1:40; Dako, Glostrup, Denmark) antibodies.
Microarray analyses
Affymetrix U133Plus2.0 (Santa Clara, CA, USA) GeneChip raw data of IPH‐926 (GEO GSE28089)20 and 50 common breast cancer cell lines (GEO GSE12777)14 were combined and analyzed using Expression Console and BRB‐array tools software as outlined in the Data S1.
Quantitative real‐time RT‐PCR
Quantitative assessment of gene expression normalized to the housekeeping gene GUSB was performed with Platinum Taq DNA polymerase (Invitrogen, Darmstadt, Germany), Sybr Green I (Invitrogen) and QuantiTect primer assays (Qiagen, Hilden, Germany) on an ABI Prism 7700 system (Applied Biosystems, Foster City, CA, USA).
Results
The IPH‐926 ILBC cells displayed a TN immunophenotype (Fig. 1a).18 To assess the molecular subtype of IPH‐926, we analyzed the Affymetrix U133Plus2.0 microarray expression data of IPH‐926 (internal data set) on a background population of 50 common breast cancer cell lines (external data set) (Fig. 1b). Sufficient comparability of internal data and external expression data was confirmed by evaluating two cell lines (MCF‐7, BT‐474) provided in the external data and profiled at our own laboratory, which showed near perfect whole array signal log values (SLV) correlation (r s > 0.95, not shown). Hierarchical clustering of the combined data set based on the PAM305 classifier gene list reproduced distinct clusters of cell lines (basal‐A, basal‐B, luminal) as described previously.11, 12 IPH‐926 was included in such an analysis for the first time and was part of the luminal cluster (Fig. 1c).
Figure 1.
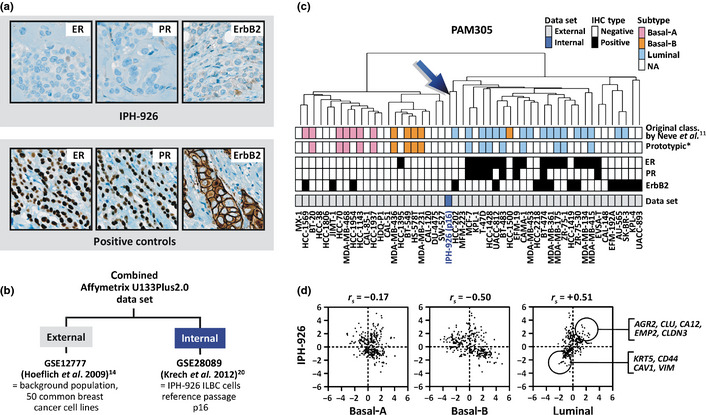
(a) Immunohistochemical staining for estrogen receptor (ER), progesterone receptor (PR) and ErbB2. (b) Overview on the external and internal microarray data sets and their Gene Expression Omnibus (GEO) accession numbers.14, 20 (c) Hierarchical clustering of 51 human breast cancer cell lines using the PAM305 classifier gene list.11 IPH‐926 (p16) refers to the reference passage p16. *Cell lines considered as prototypic based on their repeated synonymous classification in independent studies11, 12, 13, 14 are highlighted. The ER, PR and ErbB2 status was retrieved from the literature11, 12, 13, 14, 18. NA, not assessed/not classified in previous studies.11 (d) Spearman correlation of the IPH‐926 expression profile with the PAM305 basal‐A, basal‐B and luminal centroids based on prototypic cell lines computed from the external data set. Each dot represents a single probe set. Marker genes for quantitative real‐time RT–PCR validation were chosen from the numerous probe sets with high signal log values (SLV) in IPH‐926 and the luminal centroid (upper circle) or low SLV in IPH‐926 and the luminal class centroid (lower circle).
Hierarchical clustering is not reliable for molecular subtype assignment.3 A more objective approach is to test for a Spearman correlation r s > 0.1 of the test sample's expression profile with the mean expression profiles (e.g. centroids) of prototypic samples representative for each subtype.3, 17 Thus, PAM305 centroids were computed for the basal‐A, basal‐B and the luminal subtype based on prototypic cell lines highlighted in several independent studies.11, 12, 13, 14 Prototypic cell lines selected for the luminal subtype included ER‐positive/ErbB2‐negative cell lines only. Prototypic cell lines selected for the basal subtypes included TN cell lines only. Then, Spearman correlation of the IPH‐926 expression profile with the three PAM305 class centroids was determined. IPH‐926 was nearest to the PAM305 luminal centroid (r s = +0.51) and was far apart from the basal‐A (r s = −0.17) and basal‐B (r s = −0.50) centroids, indicating a bona fide luminal subtype (Fig. 1d).
Next, the IPH‐926 microarray profile was validated using quantitative real‐time RT‐PCR. For this purpose, n = 10 marker genes were chosen. AGR2, CLU, CA12, EMP2 and CLDN3 were included based on their high SLV in the IPH‐926 microarray profile (Fig. 1d, upper circle). These genes are established luminal markers.11, 12 KRT5, CD44, CAV1 and VIM were included based on their low SLV in the IPH‐926 microarray profile (Fig. 1d, lower circle). These genes are preferentially expressed in the basal‐A (KRT5, CD44) or basal‐B (CAV1, VIM) subtype.11, 12 ESR1, encoding for ER, was also included, although we have previously shown that IPH‐926 cells express little or no ESR1 mRNA.18 Quantitative real‐time RT‐PCR was carried out for IPH‐926, three prototypic basal‐A cell lines, two prototypic basal‐B cell lines and two prototypic luminal cell lines (Fig. 2). Consistent with the microarray data, IPH‐926 showed enhanced expression of all luminal marker genes, except ESR1 (Fig. 2a), and low or absent expression of basal‐A/basal‐B marker genes (Fig. 2b). Hence, quantitative real‐time RT‐PCR confirmed the validity of the IPH‐926 microarray profile.
Figure 2.
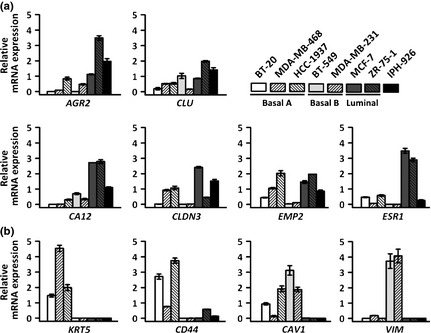
(a) Validation of the IPH‐926 microarray profile using quantitative real–time RT–PCR for luminal marker genes. (b) Quantitative real–time RT–PCR for basal‐A/basal‐B marker genes. Data are presented as relative mRNA expression with the mean of each gene's expression across the eight cell lines analyzed adjusted to a value of 1 for better visualization. Error bars indicate SEM calculated from three independent experiments.
The PAM305 classifier gene list was devised for breast cancer cell lines.11 Other classifier gene lists have been established for primary breast cancers.2, 3, 4, 5 Altered in vitro growth and the lack of tumor stroma are reasonable grounds to suppose that classifiers optimized for primary tumors should not be used for cell lines and vice versa.11 In fact, studies using laser capture microdissection have suggested that subtype assignment with some classifiers is at least partially dependent on the specific gene expression characteristics of the stroma associated with different types of tumors.22 However, as this is a matter of debate, we repeated all analyses for the INT500, LAB704, UNC1300, PAM50, TN1288 and DISC256 classifiers, which were all optimized for primary tumors. Hierarchical clustering based on these alternative classifiers reproduced essentially the same clusters of cell lines (basal‐A, basal‐B, luminal) as seen with the PAM305 classifier (Fig. S1). Spearman correlation of the IPH‐926 profile with the centroids of prototypic basal‐A, basal‐B and luminal cell lines showed that IPH‐926 was always nearest to the luminal centroid, irrespective of the classifier gene list (Table 1). This additionally corroborated that IPH‐926 cells belong to the luminal molecular subtype.
Table 1.
Spearman correlation of the IPH‐926 profile with the centroids of prototypic basal‐A, basal‐B and luminal cell lines
| Classifier gene list | Original reference | Established for classification of | Original platform | Representation on Affymetrix U133Plus2.0 | Cluster of cell lines that contains IPH‐926 | IPH‐926 [r s] | ||
|---|---|---|---|---|---|---|---|---|
| Basal‐A | Basal‐B | Luminal | ||||||
| PAM305 | Neve et al.11 | Breast cancer cell lines | Affymetrix | 305 probe sets | Luminal | −0.17 | −0.50 | +0.51 |
| DISC256 | Guedj et al.5 | Primary breast cancers | Affymetrix | 375 probe sets | Luminal | −0.16 | −0.37 | +0.27 |
| TN1288 | Lehmann et al.10 | Primary breast cancers and cell lines | Cross‐platform compilation | 5213 probe sets | Luminal | −0.03 | −0.21 | +0.19 |
| PAM50 | Parker et al.4 | Primary breast cancers | qRT‐PCR | 132 probe set | Separate | −0.16 | −0.21 | +0.03 |
| UNC1300 | Hu et al.3 | Primary breast cancers | Agilent | 2925 probe set | Luminal | −0.07 | −0.13 | +0.19 |
| LAB704 | Farmer et al.23 | Primary breast cancers | Affymetrix | 902 probe sets | Luminal | −0.18 | −0.29 | +0.16 |
| INT500 | Sorlie et al.2 | Primary breast cancers | Stanford cDNA array | 1309 probe sets | Luminal | −0.05 | −0.15 | +0.20 |
Androgen receptor (AR) induces a luminal‐like transcriptional program in some TN breast cancers.24 A new molecular subclass, termed luminal AR‐positive (LAR), has been established to cover this phenotype, which overlaps with the molecular apocrine subtype.10, 23 Based on the TN1288 classifier gene list, Lehman et al.10 have defined prototypic LAR‐type breast cancer cell lines, such as TN/AR‐positive MDA‐MB‐453 cells. To assess whether the gene expression of IPH‐926 reflects a LAR‐type cellular differentiation, the combined microarray data set was reanalyzed (Fig. 3). Similar to the approach of Lehmann et al., only TN cell lines were retained for a refined hierarchical clustering based on the TN1288 classifier gene list. As a result of this, IPH‐926 clustered together with LAR‐type cell lines (Fig. 3a). The IPH‐926 profile was positively correlated with the centroid of LAR‐type cell lines, but it was nearest to the centroid of prototypic luminal/ER‐positive cell lines, which were excluded from the clustering analysis (Fig. 3b). In line with the microarray data, IPH‐926 cells were AR‐negative, as determined using immunohistochemistry and quantitative real‐time RT‐PCR, suggesting that IPH‐926 is not a LAR‐type breast cancer cell line (Fig. 3c,d).
Figure 3.
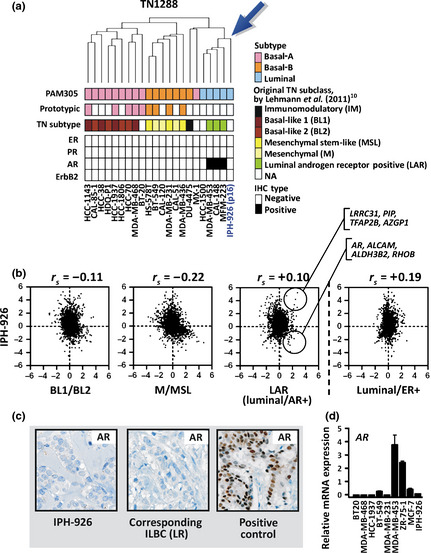
(a) Hierarchical clustering of 22 human triple‐negative (TN) breast cancer cell lines using the TN1288 classifier gene list.10 The estrogen receptor (ER), progesterone receptor (PR), ErbB2 and androgen receptor (AR) status was retrieved from the literature.10, 11, 12, 13, 14, 18 Please note that the immunophenotype of HCC‐1500 cells is contro‐versial.10, 11, 12 (b) Spearman correlation of the IPH‐926 expression profile with the TN1288 BL1/BL2, M/MSL and LAR (luminal/AR+) centroids based on prototypic TN breast cancer cell lines computed from the external data set. Spearman correlation of the IPH‐926 expression profile with the TN1288 luminal/ER‐positive centroid was also calculated (right plot). Each dot represents a single probe set. The upper circle highlights marker genes with high signal log values (SLV) in the LAR subclass and IPH‐926. The lower circle highlights marker genes with low SLV in IPH‐926 but high SLV in the LAR subclass. (c) Immunohistochemical staining for AR. The middle panel shows the original locally recurrent (LR) lobular carcinoma corresponding to IPH‐926. (d) Quantitative real–time RT–PCR for AR. Data are presented as relative mRNA expression with the mean across the nine cell lines tested adjusted to a value of 1. Error bars indicate SEM calculated from three independent experiments.
Discussion
IPH‐926 is the first well‐characterized ILBC cell line and displays a TN immunophenotype.18, 19, 20 This is due to a progression‐related loss of ER in its in vivo clonal ancestry.18 More specifically, the corresponding patient had experienced ER conversion in the locally recurrent lobular carcinoma corresponding to the IPH‐926 cell line.18 Loss of ER might indicate a fundamental change of cellular differentiation. Accordingly, the molecular subtype of IPH‐926 has remained undefined. Determining the molecular subtype of IPH‐926 cells is important for two reasons: (i) IPH‐926 is increasingly used as a pre–clinical model, urging for its categorization in relation to other breast cancer cell lines; and (ii) IPH‐926 may provide evidence that luminal differentiation can be preserved beyond ER conversion.
Here we report that the IPH‐926 microarray expression profile is consistent with a luminal subtype. It was nearest to luminal/ER‐positive breast cancer cell lines and far apart from basal breast cancer cell lines. This was independent from the classifier gene list, arguing for the robustness of this categorization. Quantitative real‐time RT‐PCR confirmed that IPH‐926 cells express luminal marker genes, such as AGR2, but lack basal marker genes, such as KRT5 or VIM. Unfortunately, a direct comparison of microarray profiles before and after loss of ER was impossible because of unavailability of cryoconserved tissue of the early, still ER‐positive ILBC corresponding to the IPH‐926 cell line.18
Recent studies have drawn attention to TN breast cancers with luminal gene expression characteristics. The AR has emerged as an important driver of luminal differentiation in TN breast cancer and the molecular apocrine and LAR breast cancer subtypes were established to cover this phenotype.10, 23, 24 Among breast cancer cell lines, TN/AR‐positive MDA–MB–453 cells were first described as luminal breast cancer cells, but were later defined more precisely as LAR‐type breast cancer cells.10 IPH‐926 cells are AR‐negative and their gene expression profile is more closely related to prototypic luminal/ER‐positive than to prototypic luminal/AR‐positive representatives, suggesting that IPH‐926 is not a LAR‐type breast cancer cell line.
In conclusion, IPH‐926 is best classified as a luminal breast cancer cell line, similar to MCF‐7, despite its TN immunophenotype. This will impact on the future use of IPH‐926 in breast cancer research. Moreover, our finding implies that a luminal expression profile is not simply erased if its prime clinical surrogate marker, ER, is lost. Maintenance of a luminal differentiation following to ER conversion might not even require compensatory expression of AR. This may help to further improve our understanding of TN mammary carcinomas with otherwise luminal gene expression.
Supporting information
Fig. S1. Hierarchical clustering of 51 human breast cancer cell lines using the INT500, LAB704, UNC1300, PAM50, TN1288 and DISC256 classifier gene list.2, 3, 4, 5
Data S1. Including: microarray data analyses; and classifier gene lists.
Disclosure Statement
The authors have no conflict of interest.
(Cancer Sci 2013; 104: 1726–1730)
References
- 1. Perou CM, Sorlie T, Eisen MB et al Molecular portraits of human breast tumours. Nature 2000; 406: 747–52. [DOI] [PubMed] [Google Scholar]
- 2. Sorlie T, Tibshirani R, Parker J et al Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci USA 2003; 100: 8418–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hu Z, Fan C, Oh DS et al The molecular portraits of breast tumors are conserved across zicroarray platforms. BMC Genomics 2006; 7: 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Parker JS, Mullins M, Cheang MC et al Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol 2009; 27: 1160–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Guedj M, Marisa L, de Reynies A et al A refined molecular taxonomy of breast cancer. Oncogene 2012; 31: 1196–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Weigelt B, Pusztai L, Ashworth A, Reis‐Filho JS. Challenges translating breast cancer gene signatures into the clinic. Nat Rev Clin Oncol 2011; 9: 58–64. [DOI] [PubMed] [Google Scholar]
- 7. Gnant M, Harbeck N, Thomssen CS. St Gallen 2011: Summary of the consensus discussion. Breast Care (Basel) 2011; 6: 136–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Harbeck N, Thomssen C, Gnant MS. St Gallen 2013: Brief preliminary summary of the consensus discussion. Breast Care (Basel) 2013; 8: 102–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bertucci F, Finetti P, Cervera N et al How basal are triple‐negative breast cancers? Int J Cancer 2008; 123: 236–40. [DOI] [PubMed] [Google Scholar]
- 10. Lehmann BD, Bauer JA, Chen X et al Identification of human triple‐negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest 2011; 121: 2750–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Neve RM, Chin K, Fridlyand J et al A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell 2006; 10: 515–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kao J, Salari K, Bocanegra M et al Molecular profiling of breast cancer cell lines defines relevant tumor models and provides a resource for cancer gene discovery. PLoS ONE 2009; 4: e6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hollestelle A, Nagel JH, Smid M et al Distinct gene mutation profiles among luminal‐type and basal‐type breast cancer cell lines. Breast Cancer Res Treat 2010; 121: 53–64. [DOI] [PubMed] [Google Scholar]
- 14. Hoeflich KP, O'Brien C, Boyd Z et al In vivo antitumor activity of MEK and phosphatidylinositol 3‐kinase inhibitors in basal‐like breast cancer models. Clin Cancer Res 2009; 15: 4649–64. [DOI] [PubMed] [Google Scholar]
- 15. Holliday DL, Speirs V. Choosing the right cell line for breast cancer research. Breast Cancer Res 2011; 13: 215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sabatier R, Finetti P, Cervera N et al A gene expression signature identifies two prognostic subgroups of basal breast cancer. Breast Cancer Res Treat 2011; 126: 407–20. [DOI] [PubMed] [Google Scholar]
- 17. Weigelt B, Geyer FC, Natrajan R et al The molecular underpinning of lobular histological growth pattern: a genome‐wide transcriptomic analysis of invasive lobular carcinomas and grade‐ and molecular subtype‐matched invasive ductal carcinomas of no special type. J Pathol 2010; 220: 45–57. [DOI] [PubMed] [Google Scholar]
- 18. Christgen M, Bruchhardt H, Hadamitzky C et al Comprehensive genetic and functional characterization of IPH‐926: a novel CDH1‐null tumour cell line from human lobular breast cancer. J Pathol 2009; 217: 620–32. [DOI] [PubMed] [Google Scholar]
- 19. Christgen M, Noskowicz M, Heil C et al IPH‐926 lobular breast cancer cells harbor a p53 mutant with temperature‐sensitive functional activity and allow for profiling of p53‐responsive genes. Lab Invest 2012; 92: 1635–47. [DOI] [PubMed] [Google Scholar]
- 20. Krech T, Scheuerer E, Geffers R, Kreipe H, Lehmann U, Christgen M. ABCB1/MDR1 contributes to the anticancer drug‐resistant phenotype of IPH‐926 human lobular breast cancer cells. Cancer Lett 2012; 315: 153–60. [DOI] [PubMed] [Google Scholar]
- 21. Hoefnagel LD, Moelans CB, Meijer SL et al Prognostic value of estrogen receptor alpha and progesterone receptor conversion in distant breast cancer metastases. Cancer 2012; 118: 4929–35. [DOI] [PubMed] [Google Scholar]
- 22. Boersma BJ, Reimers M, Yi M et al A stromal gene signature associated with inflammatory breast cancer. Int J Cancer 2008; 122: 1324–32. [DOI] [PubMed] [Google Scholar]
- 23. Farmer P, Bonnefoi H, Becette V et al Identification of molecular apocrine breast tumours by microarray analysis. Oncogene 2005; 24: 4660–71. [DOI] [PubMed] [Google Scholar]
- 24. Robinson JL, Macarthur S, Ross‐Innes CS et al Androgen receptor driven transcription in molecular apocrine breast cancer is mediated by FoxA1. EMBO J 2011; 30: 3019–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Hierarchical clustering of 51 human breast cancer cell lines using the INT500, LAB704, UNC1300, PAM50, TN1288 and DISC256 classifier gene list.2, 3, 4, 5
Data S1. Including: microarray data analyses; and classifier gene lists.


