New classes of antiviral drugs are needed to treat the ever-changing viral disease landscape. Current antiviral drugs treat only a small number of viral diseases, leaving many patients with established or emerging infections to be treated solely with supportive care. Recent antiviral peptide research has produced numerous membrane-interacting peptides that inhibit diverse enveloped viruses in vitro and in vivo. Peptide therapeutics are becoming more common, with over 60 FDA-approved peptides for clinical use. Included in this class of therapeutics is enfuvirtide, a 36-residue peptide drug that inhibits HIV entry/fusion. Due to their broad-spectrum mechanism of action and enormous potential sequence diversity, peptides that inhibit virus entry could potentially fulfill the need for new antiviral therapeutics; however, a better understanding of their mechanism is needed for the optimization or evolution of sequence design to combat the wide landscape of viral disease.
KEYWORDS: entry inhibitor, interfacial activity, membrane, peptide
ABSTRACT
Numerous peptides inhibit the entry of enveloped viruses into cells. Some of these peptides have been shown to inhibit multiple unrelated viruses. We have suggested that such broad-spectrum antiviral peptides share a property called interfacial activity; they are somewhat hydrophobic and amphipathic, with a propensity to interact with the interfacial zones of lipid bilayer membranes. In this study, we further tested the hypothesis that such interfacial activity is a correlate of broad-spectrum antiviral activity. In this study, several families of peptides, selected for the ability to partition into and disrupt membrane integrity but with no known antiviral activity, were tested for the ability to inhibit multiple diverse enveloped viruses. These include Lassa pseudovirus, influenza virus, dengue virus type 2, herpes simplex virus 1, and nonenveloped human adenovirus 5. Various families of interfacially active peptides caused potent inhibition of all enveloped viruses tested at low and submicromolar concentrations, well below the range in which they are toxic to mammalian cells. These membrane-active peptides block uptake and fusion with the host cell by rapidly and directly interacting with virions, destabilizing the viral envelope, and driving virus aggregation and/or intervirion envelope fusion. We speculate that the molecular characteristics shared by these peptides can be exploited to enable the design, optimization, or molecular evolution of novel broad-spectrum antiviral therapeutics.
IMPORTANCE New classes of antiviral drugs are needed to treat the ever-changing viral disease landscape. Current antiviral drugs treat only a small number of viral diseases, leaving many patients with established or emerging infections to be treated solely with supportive care. Recent antiviral peptide research has produced numerous membrane-interacting peptides that inhibit diverse enveloped viruses in vitro and in vivo. Peptide therapeutics are becoming more common, with over 60 FDA-approved peptides for clinical use. Included in this class of therapeutics is enfuvirtide, a 36-residue peptide drug that inhibits HIV entry/fusion. Due to their broad-spectrum mechanism of action and enormous potential sequence diversity, peptides that inhibit virus entry could potentially fulfill the need for new antiviral therapeutics; however, a better understanding of their mechanism is needed for the optimization or evolution of sequence design to combat the wide landscape of viral disease.
INTRODUCTION
Viral diseases are among the leading global causes of morbidity and mortality (1–3). Yet for most viruses, there are few, if any, effective therapeutics (4, 5). New antiviral therapies are needed, especially broad-spectrum antivirals that could be useful in outbreaks of new viruses or in regions where diagnostics are limited. For example, an effective broad-spectrum antiviral therapeutic could have dramatically decreased global morbidity and mortality during the SARS-CoV-2 pandemic that began in 2019 (6).
In this study, we tested the hypothesis that peptides or peptide-like entry inhibitors could fill this void. We note that thousands of naturally occurring and synthetic peptides with potentially useful bioactivities have been described, including many with antibacterial, antifungal, or antiviral activity in vitro (7). There are over 60 FDA-approved peptide drugs and hundreds of candidates in preclinical testing (8, 9). Fuseon (enfuvirtide), developed in 1994 and approved by the FDA in 2003, is a peptide entry inhibitor used to treat individuals infected with HIV (10), proving that peptides can be effective antiviral drugs.
In recent years, many peptides with antiviral activity in vitro have been identified (11) using a variety of methods, such as structure-based identification (12–18), brute force trial and error (19–22), and bioinformatics-based techniques (23–27). For example, numerous peptides with antiviral activity have been identified by scanning the sequence of viral fusion proteins (glycoproteins) for sequences that have membrane binding potential, determined by the Wimley-White interfacial hydrophobicity scale (WWIHS) (28, 29). Using this method, peptides have been discovered that inhibit dengue virus (23, 30), West Nile virus (23), severe acute respiratory syndrome-associated coronavirus (SARS-CoV) (24), cytomegalovirus (25), Rift Valley fever virus (26), Pichinde virus (27), and hepatitis C virus (11). Efficacies among these peptides vary, but some have concentrations of 50% inhibition (EC50) in the single-digit micromolar range. Interestingly, some of these virus-derived antiviral peptides inhibited not only the virus from which their sequences were derived but also other diverse viruses, indicating a mechanism that is not dependent on sequence-specific peptide-protein interactions (23, 24, 26). We have noted previously that many antiviral peptides share the common physical characteristics of moderate to high interfacial hydrophobicity and amphipathicity (11), giving them a propensity to partition into and disrupt membranes. This combination of properties has been termed “interfacial activity” (31, 32). We hypothesize that these molecular characteristics drive antiviral activity through direct peptide-virus interaction.
In this study, we tested this hypothesis by measuring the antiviral activity of multiple families of peptides that were selected in high-throughput screens for interfacial activity. These peptides were not selected for antiviral activity or for low toxicity. Instead, they were selected for other manifestations of interfacial activity: the ability to permeabilize synthetic lipid vesicles or the ability to sterilize bacteria by membrane disruption. We show here that these interfacially active peptide families potently inhibit a variety of enveloped viruses with class I, II, and III fusion proteins. Virus entry inhibition, the underlying mechanism, occurs at concentrations much lower than the cytotoxic concentrations. These results support the hypothesis that interfacially active peptides can have broad-spectrum entry inhibition against enveloped viruses. Exploitation of this knowledge could lead to the discovery of potent, broad-spectrum antiviral molecules with a unique mechanism of action and should ultimately help guide the development of clinically useful antiviral peptides.
RESULTS
Potential antiviral peptides.
The peptides tested in this study were previously selected in three separate screens of two libraries for interfacially active peptides that cause distinct types of membrane disruption (Table 1). The vesicle-selected (VS) membrane-permeabilizing peptides were selected from a de novo-designed library for permeabilization of anionic, bacterium-like synthetic lipid vesicles (32, 33). The library contained 9- to 15-residue peptides which had RRG- and -GRR cassettes randomly absent or present at the termini. The core sequence was a mixture of alternating hydrophobic and combinatorically varied residues, designed to favor β-sheet secondary structure. Combinatorial sites could contain hydrophobic, polar, anionic, or cationic residues. The selected positive peptides had 0, 1, or 2 terminal cassettes and were generally hydrophobic in the core, with a single polar residue, often arginine. These peptides permeabilized lipid bilayer vesicles with moderate potency. They were subsequently found to have broad-spectrum antibacterial activity at low micromolar concentrations that was due to bacterial membrane disruption (32, 34). Their toxicity against red blood cells (RBCs) and nucleated mammalian cells was moderate despite the fact that they were not down selected for toxicity in any way (32, 34). The biologically selected (BS) peptides were identified from the same library. In this case, the library was screened either for simultaneous sterilization of Escherichia coli, a Gram-negative bacterium, Staphylococcus aureus, a Gram-positive bacterium, and Cryptococcus neoformans, a fungus, or for sterilization of E. coli at a very high cell count (34). The selected positive peptides always have 2 basic terminal cassettes and are 15 residues long. The residues in the core frequently contain two polar or charged residues that include arginine, asparagine, or threonine. While apparently distinct from the VS peptides, the BS peptides have antimicrobial activities that are very similar, but have somewhat higher toxicity (34).
TABLE 1.
Names, sequences, and some properties of interfacially active and control peptidesa
N, number of amino acids. Charge, the net charge of the peptides at neutral pH. All peptides are C-terminal amides. WWIHS, Wimley-White interfacial hydrophobicity scale (28, 29) values in kilocalories per mole. Greater negative values indicate stronger interactions with membranes. SS, secondary structure. All of the interfacially active peptides studied have β-sheet secondary structure in membranes (β). Control peptides have random-coil (rc) or α-helical (α) secondary structures.
The synthetically evolved second-generation pore-forming peptides, which we call the iteration 1 (It1) peptides, were selected as part of a series of studies on the bioengineering of pore-forming peptides. The goal was to evolve potent equilibrium pore-forming peptides with β-sheet secondary structure from a 26-residue template sequence that was a moderately active, transient pore former in synthetic vesicles. An iterative, second-generation library of 24- to 26-residue peptides was screened for very potent, equilibrium pore formation in synthetic lipid vesicles (35). The selected positives are highly potent against synthetic vesicles, with activity at peptide-to-lipid ratios as low as 1:1,000, and have β-sheet secondary structure. This combination of properties is unique among thousands of known membrane-permeabilizing peptides. Against biological membranes, the It1 peptides are less potent. They have moderate antimicrobial activity and are moderately toxic to some eukaryotic cells (36).
While none of these families were selected specifically for antiviral activity, and none were down selected for lack of toxicity, they all possess interfacial activity and are not highly toxic to mammalian cells. We have previously noted that such interfacial activity is found in many known antimicrobial (31) and antiviral (11) peptides from diverse sources (37). In this study, we tested these families of interfacially active peptides specifically for antiviral activity.
Various peptides were selected as controls. TP2 is a spontaneous membrane-translocating peptide with a composition that is similar to that of the interfacially active peptides, but with no known bactericidal or membrane-permeabilizing activity (38). Arg9 is a cell-penetrating peptide (CPP) with a charge of +10 that was used as a highly cationic, but not amphipathic, control. Indolicidin is a natural 13-residue antimicrobial peptide. Mel-L16G and MelN4 are decrease-of-function analogs of melittin, a potent membrane-permeabilizing peptide found in bee venom, with a decreased propensity for membrane interaction and lysis (39).
Interfacially active peptides inhibit infectivity of Lassa pseudovirus and show low cytotoxicity.
Peptides from the three families described above and control peptides (Table 1) were tested for antiviral activity in a cell culture-based Lassa pseudovirus system (LASVpv). The pseudovirus was made by cotransfecting an HIV core plasmid (pNL4-3.Luc.R-E-), containing a luciferase reporter gene, with a Lassa virus glycoprotein precursor (GPC) plasmid that was optimized in a previous study (40). This pseudovirus has an envelope containing only the Lassa glycoprotein, mimicking the envelope of Lassa virus.
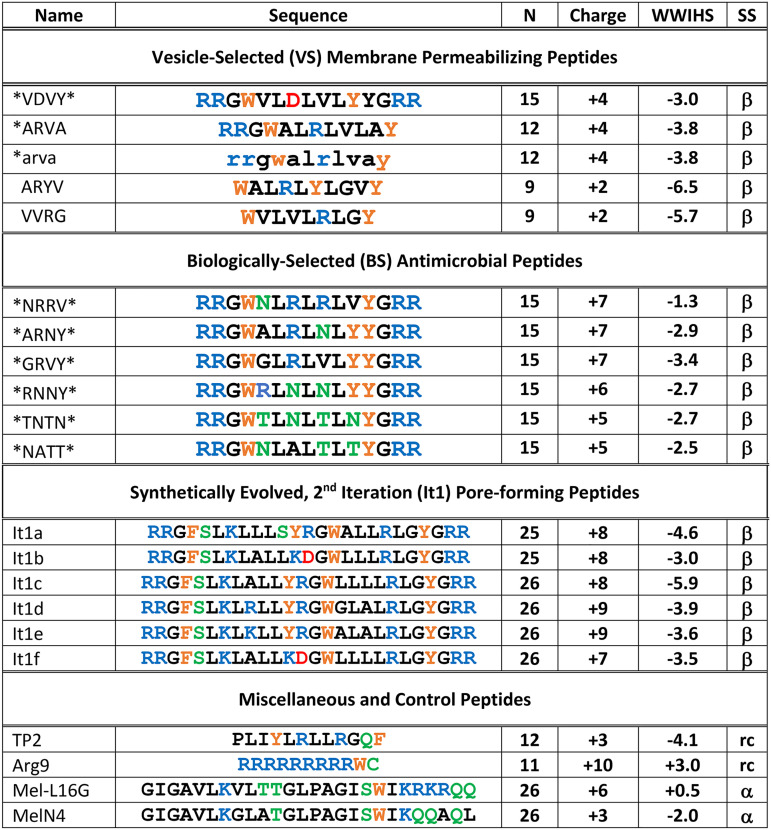
To measure inhibition, Lassa pseudovirus, at 1,000× 50% tissue culture infective dose (TCID50), was incubated with serially diluted peptide for 1 h and then added to HEK 293T/17 cells. After approximately 72 h, the infection levels were quantified by measuring total luminescence. In parallel, cells were also incubated with peptide alone to determine cytotoxicity using alamarBlue, a reagent that measures cellular aerobic respiration (41). All the selected interfacially active peptides tested inhibited Lassa pseudovirus activity (Fig. 1). Of the peptides tested, 13 of 14 had concentrations of 50% inhibition (EC50) between 1 and 10 μM. The control peptides Arg9 and MelN4 had little or no activity in this assay. The melittin analog Mel-L16G showed some activity, with an EC50 of ∼20 μM, but was significantly less active than the other peptides tested. Of the peptides that were shown to have antiviral activity, 12 of 13 had relatively low cytotoxicity (cell viability > 80%) at the antiviral EC50. EC50 values for cytotoxicity in the absence of virus are shown in Table 2. Peptide cytotoxicity against HEK 293T/17 cells was also evaluated in the presence of virus. For some of the peptides tested, toxicity was much lower with virus present, possibly because peptide was bound to virion aggregates instead of cells (see below).
FIG 1.
Peptide inhibition of LASVpv and cytotoxicity. Pseudovirus was incubated with various concentrations of peptide for 1 h and was then added to HEK 293T/17 cells. Infectivity was quantified by measuring luciferase expression translated from the LASVpv genome approximately 72 h after infection. Cells were also treated with peptide alone, and cell viability was determined by alamarBlue approximately 72 h after treatment. Infectivity is shown as solid points, and respective nonlinear curve fits are shown as solid lines. Cell viability is shown as empty points with dashed lines. Peptide libraries tested were the It1 family (A), BS family (B), and VS family (C), as well as miscellaneous and control peptides (D). Points are means ± SE; n = 4 to 8.
TABLE 2.
Therapeutic indicesa
| Name | EC50, INFV | EC50, LASVpv | EC50, Tox HEK | EC50, Tox MDCK | EC50, Tox Vero | Index (Tox HEK/LASVpv) |
|---|---|---|---|---|---|---|
| Vesicle-selected membrane-permeabilizing peptides | ||||||
| *VDVY* | 12.0 | 5.0 | >100 | ND | ND | >20 |
| *ARVA | 0.9 | 6.2 | >75 | >100 | >100 | >12 |
| *arva | 0.4 | ND | ND | ND | ND | ND |
| ARYV | 3.7 | 7.9 | ND | ND | ND | ND |
| VVRG | 6.1 | ND | ND | ND | ND | ND |
| Biologically selected antimicrobial peptides | ||||||
| *NRRV* | 8.0 | 26 | 15 | ND | ND | 0.6 |
| *ARNY* | 4.5 | 2.2 | >100 | ND | ND | >45 |
| *GRVY* | 9.1 | 2.1 | ND | ND | ND | ND |
| *RNNY* | 2.3 | 2.1 | 19 | ND | ND | 9 |
| *TNTN* | 6.1 | 1.1 | 22 | ND | ND | 20 |
| *NATT* | 0.7 | 1.2 | 43 | 60 | 41 | 36 |
| Synthetically evolved, 2nd-iteration (It1) pore-forming peptides | ||||||
| It1a | 1.6 | 1.6 | 19 | ND | ND | 12 |
| It1b | 2.3 | 1.4 | >75 | >100 | >100 | >54 |
| It1c | 1.2 | ND | ND | ND | ND | ND |
| It1d | 0.4 | 1.3 | >100 | ND | ND | >77 |
| It1e | 0.6 | 1.2 | >50 | ND | ND | >42 |
| It1f | 1.5 | 1.1 | 20 | ND | ND | 18 |
| Miscellaneous and control peptides | ||||||
| Arg9 | 6.7 | >100 | >75 | ND | ND | ND |
| Mel-L16G | ND | 28 | 21 | ND | ND | 0.8 |
| MelN4 | ND | >100 | >100 | >100 | >100 | ND |
Columns 2 and 3 contain EC50 values (micromolar) for inhibition of competent influenza virus (INFV) and inhibition of LASV pseudovirus, respectively. Column 3 contains EC50 values for cytotoxicity in HEK 293T/17 cells used in LASVpv assays. These values are from Fig. 1 and 2. Columns 4 and 5 contain EC50 values for cytotoxicity (Tox) for the four representative peptides against MDCK cells (used for influenza virus experiments) and Vero E6 cells (used for dengue virus, herpes simplex virus, and adenovirus experiments). Column 7 is a pseudo-therapeutic index containing the ratio of EC50 for cytotoxicity in HEK cells to EC50 for LASVpv inhibition. “ND” represents measurements that were not determined.
Interfacially active peptides also inhibit influenza virus.
We also tested the same peptide families against replication-competent H3N2 influenza virus (A/Hong Kong/8/68) in a cell culture-based assay of cytopathic effects (CPE) (Fig. 2). Virus at 50× TCID50 was incubated for 1 h with serially diluted peptide before being added to a confluent monolayer of MDCK cells. After 48 h, the number of viable cells remaining on the plate was quantified by staining with 4′,6-diamidino-2-phenylindole (DAPI) and measuring fluorescence. Infected control monolayers were completely destroyed by 48 h and contained essentially no viable cells due to the cytopathic effects of the virus. Pretreatment with a human convalescent-phase antibody (1:50,000 dilution) completely protected the cell monolayer from viral cytopathic effects. In all cases, the interfacially active peptides effectively inhibited influenza virus infection, with EC50 values ranging from 20 μM to ≤1 μM. In most cases, 100% cell viability (i.e., a lack of CPE produced by virus infection or by peptide) was observed up to a concentration of 50 μM peptide, indicating that antiviral activity occurs at lower concentrations than cytotoxicity. Interestingly, against influenza virus, the cell-penetrating peptide Arg9 had potent inhibitory activity, whereas it was inactive against the Lassa pseudovirus. The other control peptide tested, TP2, also showed some activity against influenza virus, but at higher concentrations than the interfacially active peptides. We also measured cytotoxicity against MDCK cells independently for some peptides (Table 2 and Fig. 2E). Only one of the peptides tested (*NATT*) had substantial cytotoxicity and was only toxic at high peptide concentrations.
FIG 2.
Inhibition of influenza virus. Inhibition of influenza A/Hong Kong/8/68 (H3N2) virus was measured for the It1 peptides (A), the BS peptides (B), the VS peptides (C), and the set of control peptides (D; see the text). We also used mock inhibition as a buffer-only positive control and used a 1:50,000 dilution of a human convalescent-phase antibody (BEI Resources) as an inhibition control. Virus at 50× TCID50 was incubated in 96-well plates with serially diluted peptide for 1 h and was then added to MDCK cells for 1 h, followed by washing and the addition of fresh media. After 48 h at 37°C, the supernatants were removed for further assays; the plates were then washed, fixed with 4% paraformaldehyde, and stained with DAPI. DAPI fluorescence enables the measurement of intact cells remaining in the well. Viral cytopathic effects destroy the cell monolayer under these conditions, unless inhibited; cells that have succumbed to infection are washed away before fixation. Points are means ± SE; n = 4 to 10. (E) Viability of MDCK cells after treatment with one representative peptides from each set of peptides measured by SYTOX green staining.
We verified that influenza virus infection and propagation are blocked by interfacially active peptides by assaying for virus protein expression. For this, we used Western blotting to measure viral hemagglutinin protein in the supernatants (Fig. 3A) and measured hemagglutination directly using cell culture supernatant at the 48-h endpoint (Fig. 3B). The results for two example peptides demonstrate that viral replication does not occur when the CPE assay reports complete inhibition (Fig. 2). By Western blotting, no viral hemagglutinin was detected in the cell culture supernatant at peptide concentrations that were inhibitory, measured by CPE, whereas it was abundant at peptide concentrations that were not inhibitory. Similarly, hemagglutination assays verified that virus was not produced when the peptide concentration was inhibitory. In a few cases, hemagglutination assays also showed red blood cell aggregation caused by higher concentrations of some peptides, including d-ARVA, one of the most potent antiviral peptides.
FIG 3.
Confirmation of influenza virus growth and inhibition for some example peptides. (A) Western blotting. Aliquots of the supernatant from viral inhibition experiments (Fig. 2) were subjected to SDS-PAGE, transferred, and then blotted with an anti-hemagglutinin primary antibody and a peroxidase-labeled secondary antibody. One Western blot from the biologically selected peptides (*RNNY*) and another from the It1 family (It1f) are shown. (B) Hemagglutination assay. d-ARVA (*arva) is the d-amino acid variant of *ARVA. Example experiments were performed with infection supernatant from the inhibition assay (Fig. 2). In the top row, infection was done with virus treated with a peptide concentration of 50 μM or antibody dilution of 1/1,000. Treatments were serially diluted by a factor of 2 down the plate. Culture supernatants were mixed with 1% turkey RBCs and then incubated at room temperature for 1 h. Hemagglutination prevented the RBCs from settling into a distinct pellet. (C) Virus propagation time course. MDCK cells were infected with various titers of influenza A (H1N1) virus, untreated, or treated with peptide 30 min before or 30 min after infection. Virus propagation was measured by qPCR at multiple time points postinfection (n = 3; mean ± SE).
We also conducted a growth curve assay for influenza virus to support the endpoint measurements reported above. Infection was done with 20× to 20,000× TCID50 of H1N1 influenza virus (A/Puerto Rico/8/34). Virus propagation was measured at time points up to 48 h by quantitative PCR (qPCR). Treatment with the peptide It1b was done at 30 min before or 30 min after infection. At 2,000× or lower TCID50, pretreatment with 25 μM It1b completely inhibited viral propagation at all time points from 0 to 48 h. Postinfection treatment caused less inhibition, presumably due to the relatively rapid entry of influenza virus under these conditions. The added peptides were expected to be metabolized by the cells within the first 24 h. This observation suggests that direct peptide-virus interactions are responsible for entry inhibition; peptide-cell interactions beginning 30 min after infection has only a small effect on virus propagation. When we infected with a very high virus titer, 20,000× TCID50 (multiplicity of infection [MOI] = 1), some virus propagation occurred with time even in the peptide pretreatment samples. However, virus production was delayed compared to that in untreated samples, and the total amount of virus produced was reduced.
Interfacially active peptides inhibit diverse viruses.
Both Lassa pseudovirus and influenza virus are single-stranded RNA (ssRNA) enveloped viruses that have class I fusion proteins. To determine if these peptides possess broad-spectrum antiviral activity, viruses with other classes of fusion proteins were also tested. We tested dengue virus serotype 2, which has a class II fusion protein, and herpes simplex virus 1 (HSV-1), which has a class III fusion protein and has a large double-stranded DNA (dsDNA) genome. We also tested adenovirus as a representative nonenveloped virus.
For these experiments, a representative set of peptides were used, one from each peptide family and one from the control peptide group: ARVA from the VS peptides, NATT from the BS peptides, It1b from the iteration peptides, and MelN4 as the control peptide. Plaque assays were used to determine the antiviral activity of each peptide for each virus. When dengue virus was challenged with the representative peptides, all interfacially active peptides showed very high antiviral activity, with It1b showing the greatest inhibition of any challenge performed (Fig. 4A). MelN4, the representative control peptide, did not show significant anti-dengue virus activity. When HSV-1 was challenged, all selected peptides inhibited the virus, with activity similar to that observed against influenza virus (Fig. 4B). When adenovirus was challenged, all peptides inhibited the virus except for the control, MelN4 (Fig. 4C), although the activity for the interfacially active peptides was much lower against adenovirus than that measured for enveloped viruses.
FIG 4.
Peptides inhibit multiple diverse viruses. Peptides were tested against dengue virus type 2 (A), herpes simplex virus 1 (B), and human adenovirus 5 (C) (n = 6, mean ± SE). Plaque assays in Vero E6 cells were used to quantify infectivity. Virus and peptide were incubated for 1 h. The inoculum was transferred to cells for 1 h and then washed from the cells. Avicel overlay was added to cells and then incubated until viral plaques were visible by crystal violet staining. (D) Peptide cytotoxicity against Vero E6 cells was measured by SYTOX green staining.
We tested the cytotoxicity of the representative peptides on the Vero E6 cells used for these plaque assays and found that there was little cytotoxicity in this cell type (Fig. 4D and Table 2). The highest toxicity was observed for the peptide NATT, which is one of the more toxic peptides against HEK 293T/17 and MDCK cells (Table 2). Similar to the case with Lassa pseudovirus assays in HEK 293T/17 cells, these representative peptides inhibit multiple viruses at concentrations well below the concentrations at which they have toxic effects on cells.
Mechanism of action is fast and inhibits nearly all virions in the supernatant.
To help identify the mechanism of action of these peptides, we performed a time-of-addition assay (Fig. 5A to D). The four representative peptides at various concentrations were added to HEK 293T/17 cells at specific time points before or after infection with Lassa pseudovirus. Virus inhibition was similar when peptide was added at any time (−60, −45, −30, and −15 min) before infection, which is done at 0 min. Peptides added after infection inhibited virus for as long as 60 min, although inhibition steadily decreased. We attribute these results to rapid inhibition of virus by peptides in the extracellular space, coupled with the relatively slow binding and entry of the pseudovirus into cells. To prove that entry is slow enough to account for these observations, we used anti-Lassa virus antibody 8.9F (42) added at the same time intervals (Fig. 5D). We observed a similar time dependence of inhibition; antibody inhibited pseudovirus entry into the cells and displayed a steady decrease in inhibition after pseudovirus infection.
FIG 5.
Time-of-addition assay. Peptides, at 10 μM, were added to cells at various time points before and after LASVpv infection (0 min). Representatives of each interfacially active peptide family (A to C) were tested, as well as two controls (D). Controls include the negative peptide MelN4 and an anti-Lassa virus antibody. Infectivity was quantified approximately 72 h after infection by measuring luciferase expression translated from the LASVpv genome (n = 4; mean ± SE). Significance relative to −60 min was determined by one-way analysis of variance (ANOVA) with Dunnett’s posttest. *, P < 0.05; **, P < 0.01; ***, P < 0.001; n.s., no statistical difference. (E) Effect of preincubation with cells instead of virus. Standard series (s) was treated in the same manner as that for Fig. 1; various concentrations of It1b were first incubated with LASVpv for 1 h and then added to HEK 239T/17 cells. For the wash series (w), It1b was first incubated on cells for 1 h. Cells were then washed with PBS to remove excess unbound peptide, followed by the addition of LASVpv to the cells. Infectivity of both series was quantified by measuring luciferase expression translated from the LASVpv genome approximately 72 h after infection. Data are represented as the difference in inhibition between the two series (w − s). n = 8; mean ± SE.
We also used peptide It1b to measure the decrease in inhibition when peptide was preincubated with cells for 1 h, instead of virus, followed by washing off of the peptide before immediate infection with pseudovirus (Fig. 5E). Even though It1b should have bound strongly to cells because of its high cationic charge and many hydrophobic residues, the inhibition was reduced as much as 75% by preincubation with cells. This result indicates that peptide-cell interactions are not responsible for viral inhibition.
Antiviral activity is due to entry inhibition.
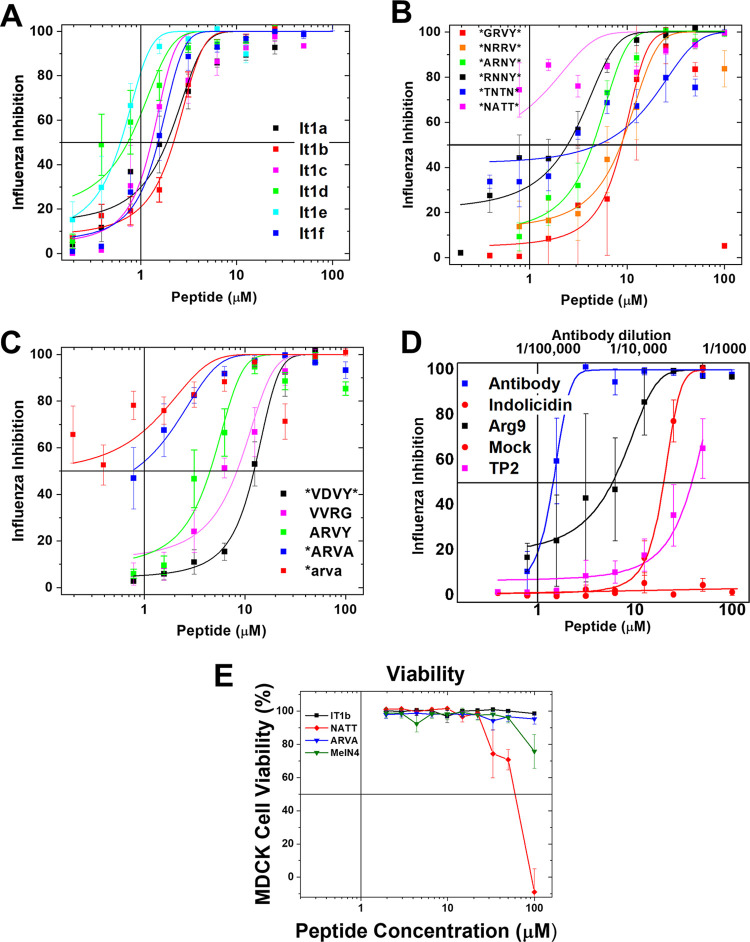
We used confocal fluorescence microscopy to examine the effect of these interfacially active peptides on the binding and entry of influenza virus in live cells (Fig. 6). Influenza virus H1N1, labeled with rhodamine 18 (R18), was not fluorescent due to self-quenching. Virus was first incubated with peptide at 37°C, and then the mixture was incubated with cells for 1 h at 4°C to enable binding but not uptake of virus. Prior to raising of the temperature, unbound virus and peptide were washed from the cells and Alexa Fluor 350-labeled dextran was added to mark cell boundaries. Cells were imaged at room temperature (time zero) and then brought to 37°C for 120 min to enable uptake and fusion of virus. All cells were then imaged again at 120 min. Example images are shown in Fig. 6A. We used ImageJ to count fluorescent intracellular particles above a threshold size and intensity (Fig. 6B). Because the R18 in the virion envelope was self-quenched, intracellular fluorescent objects indicate endosomes with which labeled virions fused and contained dequenched R18 due to lipid mixing. These results are shown in Fig. 6D.
FIG 6.
Antiviral peptides prevent viral entry into mammalian cells. (A) R18-labeled H1N1 influenza virus was preincubated with peptides and then incubated with A549 cells for 1 h at 4°C to allow binding but not uptake of virus. After the incubation, cells were washed. Confocal microscopy images were taken at 0 and 2 h at 37°C. Negative and positive controls are shown in the first three columns. Peptide-treated samples are shown in the right four columns. The blue channel is dextran-Cascade Blue to show cell borders. The red is R18, which is self-quenched in intact virions and becomes fluorescent upon endosomal fusion. (B) Confocal images with depth profiles on left and lower edge. At 0 h, viral particles were bound only to the cell surface. After 2 h at 37°C, particles were largely within the cell and were more fluorescent due to virus-endosome fusion. (C) With peptide treatment, large viral aggregates were observed in the extracellular space or on the cell surface, but they did not enter cells. Scale bar = 5 μm. (D) Quantitation of viral entry and fusion. Red particles (RP) brighter than a background-level threshold that are inside treated cells were counted with ImageJ and compared to the number in untreated cells. Large particles (>1 μm) (see panel C) were excluded, based on size and also because they were always located outside the cells. The results are expressed as percentages of virus-only control. Individual experiments are shown as points. Bars are means ± SE; n = 3 to 8.
In the absence of peptide, only a few particles are visible per cell after binding at 4°C (Fig. 6A and B). This is due to self-quenching of R18 in the intact virus. After the temperature was raised to 37°C for 2 h, virus-treated cells each had dozens of small, fluorescent intracellular objects. Their size and location (Fig. 6B) indicate that these were endosomal/lysosomal membranes that had undergone fusion with R18-labeled virus, thereby relieving the self-quenching of the dye.
Preincubation of virus with human convalescent-phase antibody blocked uptake completely at a dilution of 1:10,000. When viruses were preincubated with interfacially active peptides, the number of fluorescent intracellular particles counted at 2 h was dramatically reduced compared to that of the control, showing that the peptides were blocking productive entry at a stage before viral fusion. The control peptide MelN4 had a much smaller effect on viral entry. In confocal images of virus treated with these interfacially active peptides, a small number of very large aggregates, 20 to 100 μm, containing virus, could sometimes be observed (Fig. 6C). These particles always were found outside the cell, sometimes adhered to the cell surface, but were not endocytosed.
Peptides cause virus aggregation.
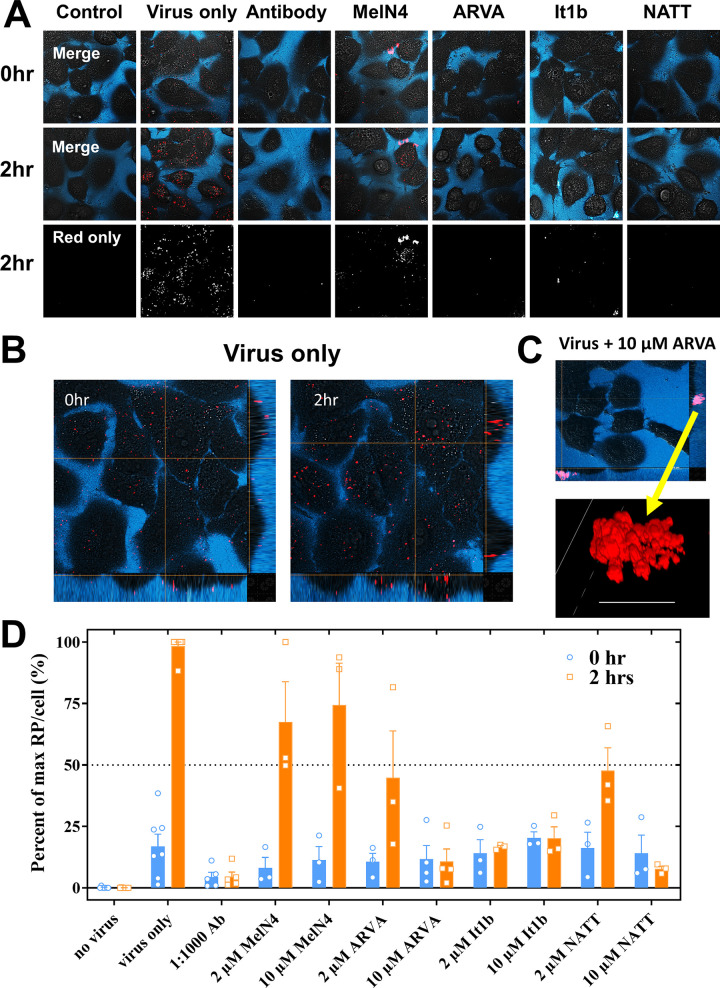
We used cryo-electron microscopy to observe the morphology of H1N1 influenza virus virions treated with various concentrations of peptide. H1N1 was incubated for 30 min with NATT peptide, followed by UV inactivation. These solutions were rapidly frozen in liquid ethane and imaged by cryo-transmission electron microscopy (Fig. 7A). At 0.5 μM peptide, a concentration below the EC50 for inhibition, virions were indistinguishable from those in control samples without peptide. Virions were distinct and easily observed. At 5 μM peptide, a concentration slightly above the EC50, virions were mostly observed in clusters, although a few individual virions could be observed outside these clusters. At 25 μM, a concentration well above the EC50 for virus inhibition but below the EC50 for cytotoxicity, virion integrity was destroyed, and no single virions were observed. Viral components were observed only in large aggregates.
FIG 7.
Virions rapidly form aggregates as peptide concentration increases. (A) H1N1 influenza virus was incubated with various concentrations of NATT peptide for 30 min and then inactivated by UV light. Samples were then visualized by cryo-electron microscopy. (B) UV-inactivated H1N1 was incubated with 25 μM NATT peptide for 1 and 5 min and then immediately frozen in liquid ethane. Samples were visualized by cryo-electron microscopy. Insets are shown from addition images of the same sample.
To determine the time frame in which peptide inclusion occurs in the virus, we added NATT peptide, at 25 μM, to UV-inactivated H1N1 for 1 and 5 min before freezing in liquid ethane and imaging by cryo-transmission electron microscopy. After 1 min of peptide exposure, we observed virion clustering and the formation of irregular structures (Fig. 7B). After only 5 min of peptide exposure, virions formed large aggregates and structural integrity was severely compromised. We conclude that peptide physically affects virions and does so rapidly, consistent with the time-of-addition data (Fig. 5).
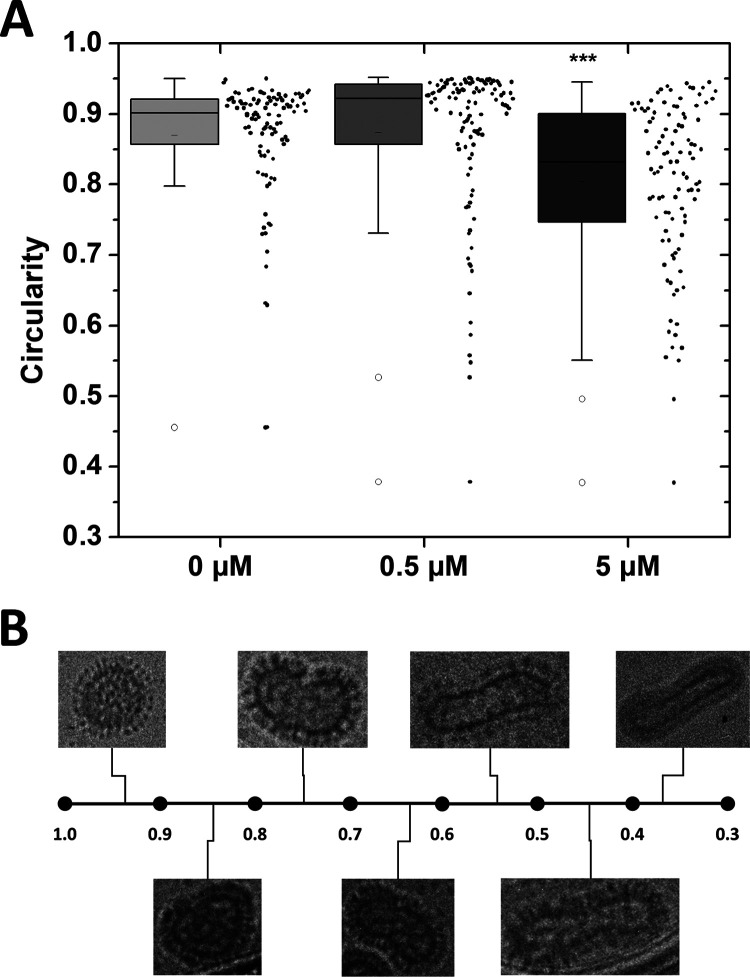
To determine if significant morphological changes were occurring with increasing peptide concentration, we measured the circularity of individual virions from the experiment shown in Fig. 7A using ImageJ (Fig. 8). We found that samples with 0.5 μM peptide were not different from untreated virions. Samples with 5 μM peptide were significantly less circular than samples with 0.5 μM peptide and untreated virions. Samples with 25 μM peptide could not be measured because no intact virions were observed. Figure 8B shows examples of virions from this study with circularities ranging from almost 1 (circular) to <0.4 (highly elliptical) for visual reference. Based on these results, we conclude that peptides were directly interacting with the virus, causing viral membrane perturbations, virion aggregation, and intervirion fusion and destroying viral structural integrity.
FIG 8.
Virion morphology changes as peptide concentration increases. (A) Viral envelopes from Fig. 7A were traced, and the circularity of each virion was measured using ImageJ. Data are represented by box plot (left of x axis tick) as well as individual measurements (right of x axis tick). Significance was determined by one-way ANOVA with Tukey’s posttest. ***, P < 0.001. n = 103 to 115. (B) Representative set of virions displaying a wide range of circularity.
DISCUSSION
Interfacially active, broad-spectrum antiviral peptides.
Numerous broad-spectrum peptide entry inhibitors have been described in the literature (11). These have been identified by a variety of techniques. Some were identified by scanning viral fusion proteins for segments with high Wimley-White interfacial hydrophobicity (28, 29), resulting in inhibitors of numerous viruses (23, 24, 27, 30). Other peptide entry inhibitors have been identified using rational structural approaches intended to inhibit particular protein-protein interactions (12–18). Others were identified by empirical testing of peptides representing entire viral proteomes (20). Interestingly, some antiviral peptides designed to target a particular virus or virus family have been shown to inhibit viruses other than the intended targets (23, 24, 26). Even these peptides almost always have interfacial hydrophobicity, and therefore are expected to interact with membranes (11). This observation supports the hypothesis that we tested in this study, that peptides with interfacial activity inhibit viral entry.
As an excellent example of this effect, Cho and colleagues tested all overlapping peptides from the proteome of hepatitis C virus in a search for inhibitory sequences. They identified one particularly potent peptide inhibitor, called AH, which is a classical interfacially active amphipathic helix. At nanomolar concentrations, the AH peptide interacts strongly with and lyses lipid vesicles and enveloped viruses that have high curvature (43, 44). In vitro, the AH peptide inhibits hepatitis C virus, as well as other unrelated viruses with extraordinary potency, including dengue (DENV), West Nile, and Zika viruses (45), and it does so without cytotoxicity. Furthermore, this peptide has potent, protective antiviral activity in animals. Systemic administration of d-amino acid AH peptide in Zika virus-infected mice reduced viral propagation and clinical symptoms, such as brain damage, and protected against death (45). This highly active, broad-spectrum antiviral peptide interacts with membranes via its interfacial activity and physically disrupts viral envelopes. In this work, we tested the hypothesis that broad-spectrum antiviral activity is a generic bioactivity of interfacially active peptides.
Broad-spectrum inhibitory peptides.
We measured the antiviral activity of three families of peptides that were selected in screens for interfacial activity in membranes (Table 1). These screens were not for virus inhibition or for lack of cytotoxicity but instead selected for permeabilization of synthetic lipid bilayers or sterilization of bacteria and fungi by membrane permeabilization. We show that these interfacially active peptides, with moderate hydrophobicity scores and β-sheet secondary structure, possess broad-spectrum antiviral activity at low micromolar concentrations and have relatively low cytotoxicity. Nearly all the peptides we tested had antiviral activity against Lassa pseudovirus, influenza virus, dengue virus, and herpes simplex virus while having relatively low toxicity (Table 2). EC50 values of library peptides ranged from 30 μM (*NRRV* inhibition of LASVpv) to 225 nM (It1b inhibition of DENV), with an average around 1 to 5 μM. Ratios of EC50 values for cytotoxicity and viral inhibition ranged from about 10 to greater than ∼80 (Table 2). Selected control peptides that did not have the characteristics of moderate hydrophobicity with β-sheet secondary structure were not able to significantly inhibit all viruses. These included the polycationic CPP Arg9 and the conformationally inhibited melittin analogs Mel-L16G and MelN4 (39).
The common features of these peptides do not give rise to an active sequence motif. However, examination of the sequences reveals common physical chemical characteristics. All of the active peptides are cationic, hydrophobic, and amphipathic. Many of them have one or two double arginine motifs at the termini, a feature found in both libraries but not always subject to selection. Aromatic residues can also be found near the termini which can stabilize insertion of the peptide into membranes. Segments of hydrophobic residues alternating with a few polar residues are present in the middle of VS and BS peptides and may be buried in the core of the envelope or interact with hydrophobic segments of the fusion protein. Segments of continuous leucine and alanine residues found in the It1 and VS peptides may serve the same purpose. Of all the library peptides, the It1 library peptides were the most consistently potent. These are the longest and most cationic peptides tested and have the most potent permeabilizing activity against synthetic vesicles. These traits are likely to increase interaction with the viral envelope, leading to structural destabilization.
Antiviral activity occurs by entry inhibition.
These families of interfacially active peptides block virus entry into cells by interacting directly with virus, destabilizing the envelope, and driving aggregation and/or intervirion envelope fusion. Cationic, interfacially active peptides can interact with viral envelopes in a variety of nonexclusive ways. Direct electrostatic and hydrophobic interaction with the envelope is likely, perhaps enhanced by the high curvature of viral membranes compared to host cells as observed with some antiviral peptides (44). Peptides may also interact with hydrophobic segments of the fusion proteins, especially during the critical conformational changes that drive fusion. This interaction could block the oligomeric formation of the fusion complex or could block the hydrophobic segment of the fusion protein necessary for virus-cell membrane fusion. In either case, these various interactions could destabilize the viral envelope, not allowing for a proper prefusion protein complex to form. This can lead to viral aggregation and fusion between virions, as suggested by cryo-electron microscopy experiments. We speculate that selectivity for viruses over host cells is driven by the high positive curvature of viral envelopes, perhaps combined with the lack of repair mechanisms and the lack membrane homeostasis systems, such as lipid flippases.
Interfacially active peptides were also able to inhibit human adenovirus, a nonenveloped virus, although with much lower potency. The EC50 values for the library peptides were up to 10 times higher for adenovirus than for enveloped viruses. We hypothesize that peptides can also bind electrostatically to viral capsid surface and/or to hydrophobic sections of capsid proteins, even in the absence of a membrane. Based on the lower EC50 values against enveloped viruses, we believe that envelope disruption is much more effective than viral protein binding.
A basis for synthetic molecular evolution.
The peptides we studied are not likely to be antiviral drugs; their activities are likely too low. Additionally, their residual toxicities, although relatively low, would nonetheless be prohibitive for a therapeutic. Furthermore, activity may decrease in the presence of concentrated host cells and tissue, as we have seen for some antimicrobial peptides (46). Yet this work shows unequivocally that interfacially active peptides can have broad-spectrum antiviral activity at peptide concentrations much lower than concentrations that are cytotoxic, an important first step toward a new class of therapeutics. We envision that the next step toward development of peptide antiviral drugs will be synthetic molecular evolution in which a peptide library based on the principles revealed in this work will be screened simultaneously for potent antiviral activity and lack of toxicity in the presence of concentrated host cells and serum. We have recently applied this approach to the discovery of host cell-compatible antibacterial peptides (47) and successfully identified antibacterial peptides with very strong selectivity for bacteria over host cells. We expect the same result to be achievable in one generation of synthetic molecular evolution of antiviral peptides.
Summary.
Interfacially active peptides demonstrate potent and broad-spectrum viral entry inhibition at concentrations 10 to 100 times lower than the concentrations at which they are toxic in cell culture. We found that these peptides rapidly inhibit virus through direct nonspecific interactions with viral surface components. Although the peptides tested in this study are unlikely to be viable therapeutics, further rational optimization or synthetic molecular evolution (35, 48, 49) of these peptides, or other interfacially active peptides, could lead to identification of broad-spectrum antiviral therapeutics to treat a wide variety of viral diseases.
MATERIALS AND METHODS
Cells.
HEK 293T/17 (ATCC CRL-11268), Vero E6 (ATCC CRL-1586), and MDCK (ATCC CCL-34) cells were cultured with Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum (FBS).
Peptides.
Peptides were purchased from Bio-synthesis Inc. (Lewisville, TX) with C-terminal amidation and purified to >95%. It1 library peptides were dissolved in water, and all other peptides were dissolved in 0.025% acetic acid.
Viruses.
In this work, we used human influenza A/Puerto Rico/8/34 (H1N1) purified virus obtained from Advanced Biotechnologies Inc. (catalog number 10-210-500 and human influenza A/Hong Kong/8/68 (H3N2) purified virus from BEI Resources (catalog number NR-346) which were propagated in cell culture. Growth medium consisted of DMEM, 0.2% bovine serum albumin, 25 mM HEPES, 2 mM l-glutamine, and 2 μg/ml of tosyl phenylalanyl chloromethyl ketone (TPCK)-trypsin. Cells were first washed with phosphate-buffered saline (PBS). Original virus stock was diluted 1:100 in a low volume of growth medium (3 ml for a T150 flask) then added to cells for 1 h. Growth medium was then added to cell at a normal culture volume (30 ml for a T150 flask). Cells were incubated at 37°C 5% CO2 until approximately 75% of cells showed CPE (about 4 to 6 days). Supernatant was then collected and clarified by centrifugation at 1,000 × g for 15 min at 4°C. When necessary to remove trypsin or concentrate virus, influenza virus was then purified by ultracentrifugation. Clarified virus was added on top of a cushion of 20% sorbitol in PBS. This was then spun down at 22,000 rpm at 4°C for 160 min, using an SW28 rotor in a Beckman XL-90 ultracentrifuge. Pelleted virus was resuspended in PBS. This solution was then aliquoted and stored at –80°C.
Dengue virus serotype 2 (New Guinea C strain) was propagated on Vero E6 cells. Cells were first washed with DMEM. Virus was added to 30 ml of DMEM with 2% FBS (for a T150 flask) and then added to cells. Cells were incubated until approximately 75% of cells showed CPE (about 16 days). Supernatant was then collected, clarified by centrifugation at 1,000 × g for 15 min at 4°C, and then aliquoted and stored at –80°C.
Herpes simplex virus 1 (ATCC VR-539) and human adenovirus 5 (ATCC VR-1516) were cultured in the same manner as dengue virus. The incubation times were 3 and 4 days, respectively.
Lassa pseudovirus propagation.
Lassa pseudovirus was produced by cotransfecting HEK 293T/17 cells with two plasmids: pNL4-3.Luc.R-E- (NIH AIDS Reagent Program; catalog number 3418) and LASV GPC plasmid (40). Ten-centimeter dishes were first coated with 100 μg/ml of poly-d-lysine hydrobromide, and then cells were seeded at 1 × 107/dish. When monolayers were ≥90% confluent, 30 μg of each plasmid was transfected with 74 μl of Lipofectamine 2000 using the manufacturer’s protocol. Supernatant was collected from the dishes 3 days posttransfection and was clarified by centrifugation at 1,000 × g for 15 min at 4°C. This was then aliquoted and stored at –80°C.
To obtain a viral titer, pseudovirus was serially diluted on HEK 293T/17 cells. Infectivity was quantified by measuring luciferase expression translated from the pseudovirus genome approximately 72 h after infection using Bright-Glo (Promega). The Reed-Muench (50) method was used to determine a TCID50, where positivity for infection was defined as follows: (signal) > (background signal) + (3 × standard deviation of background signal).
Lassa pseudovirus neutralization assay.
HEK 293T/17 cells were first seeded in 96-well plates at a density of 1 × 104/well in DMEM plus 10% FBS and were incubated overnight. Lassa pseudovirus was added at a concentration of 1 × 103 TCID50/well to various concentrations of peptide in DMEM and incubated for 1 h. The culture medium was then aspirated from the cells and replaced with a virus-peptide solution. Infectivity was quantified by measuring luciferase expression translated from the pseudovirus genome approximately 72 h after infection using Bright-Glo (Promega).
For experiments in which peptide was washed from cells, peptide was first added to plated cells at various concentrations and incubated for 1 h. Cells were then carefully washed twice with PBS, and then Lassa pseudovirus was added to cells at a titer of 1 × 103 TCID50/well. Incubation and measurement steps were identical to those described above.
Peptide cytotoxicity assay.
HEK 293T/17, Vero E6, and MDCK cells were first seeded in 96-well plates in DMEM plus 10% FBS at a density of 1 × 104/well and were incubated overnight. Peptide was first diluted in DMEM. The culture medium was then aspirated from the cells and replaced with peptide solutions at various concentrations. Viability was determined by alamarBlue or SYTOX green staining. alamarBlue was added to cells to a final concentration of 1%. After a 4-h incubation at 37°C, fluorescence was measured at excitation/emission wavelengths of 560/590 nm. SYTOX green was added at 5 μM at the same time as peptide. SYTOX fluorescence was read at 1 h using a 485/20 filter for excitation and a 530/25 filter for emission.
Influenza virus inhibition.
MDCK (ATCC) cells were seeded into 96-well plates. The assay was conducted once the monolayers became confluent. On the day of inoculation, two preliminary plates were prepared for each 96-well cell plate. All media used beyond this stage were FBS and bovine serum albumin (BSA) free. The influenza virus strain used was human influenza A/Hong Kong/8/68 (H3N2) virus propagated in cell culture from purified virus obtained from BEI Resources (catalog number NR-346). The first plate was to dilute the H3N2 influenza virus into cell overlay medium which was used to infect cells. The second plate was used to serially dilute the peptide to create a gradient of concentrations and also to act as a preincubation plate for the virus and peptide to mix before the infection occurred. Diluted virus was added to the peptide plate and incubated at 37°C for 60 min. After the 60-min incubation, the cell plate was washed with Dulbecco’s phosphate-buffered saline (DPBS) twice and 100 μl/well of inoculum was added to each well. The plate was then incubated at 37°C for 1 h. The infection medium was then aspirated, and each well was again washed twice with DPBS before fresh medium was added. After 48 h, the wells were washed and the cells were fixed with 4% paraformaldehyde (PFA) and then stained with DAPI. DAPI fluorescence, used to measure remaining viable cells, was quantified in a Biotek Synergy plate reader. The supernatant medium was also saved in a plate for further testing as described in the next section.
Influenza hemagglutination assay and Western blotting.
Supernatant from the influenza virus inhibition assay (described above) was tested with hemagglutination assays and Western blotting for the presence of viral particles. For the hemagglutination assay, we added 100 μl/well of the infection supernatant from the primary influenza virus inhibition assay to a prepared 96-well plate of turkey RBCs, to a final concentration of 1% RBCs. The plate was then allowed to incubate at room temperature for 45 min and was then photographed. Western blotting was used as a method of secondary confirmation. The supernatant medium was loaded into an SDS-PAGE gel and then blotted with anti-hemagglutinin primary antibody and a horseradish peroxidase (HRP)-labeled secondary antibody for development.
Influenza virus growth curve assay.
MDCK cells were plated at a density of 2 × 104/well in DMEM plus 10% FBS in a 96-well plate and incubated overnight. The next day, in a separate plate, various titers of human influenza A/Puerto Rico/8/34 (H1N1) virus, measured by genome copy number, were either incubated with 25 μM It1b (−30-min samples) or alone (−30-min and no-treatment samples) in a total volume of 50 μl of serum-free DMEM for 30 min at 37°C. TCID50 values were assumed to be 50 genomes/TCID50 based on the approximate average of several studies (51–53). Cells were then washed with 200 μl of PBS and the inoculum was added to cells. For +30-min samples, 25 μM It1b was added to cells 30 min after infection. All samples were then incubated for an additional 30 min. Cells were washed 3 times with 200 μl of PBS, followed by the addition of 150 μl of growth medium (DMEM, 2 μg/ml of TPCK-trypsin, 0.2% BSA, 25 mM HEPES). At designated time points, 100 μl of supernatant was removed, followed immediately by RNA extraction using the Quick-RNA viral kit (Zymo Research) and storage at –80°C. After all RNA extractions were completed, samples were analyzed by reverse transcription-quantitative PCR (qRT-PCR) (n = 3 per sample). Primers amplified the influenza virus genome, and detection was by fluorescent probe and comparison to a standard curve of gBlock. The protocol was adapted from the CDC protocol for the detection of influenza A virus using qRT-PCR (54). Influenza A virus forward primer GACCRATCCTGTCACCTCTGAC (GenBank accession number HM590431; residues 146 to 167), reverse primer AGGGCATTYTGGACAAAKCGTCTA (reverse complement of residues 228 to 251 at GenBank accession number HM590431), and probe 6-carboxyfluorescein (FAM)-TGCAGTCCTCGCTCACTGGGCACG-BHQ1 (reverse complement of residues 201 to 224 at GenBank accession number HM590431) were used to detect influenza virus RNA. An influenza virus gBlock (GenBank accession number HM641211; residues 141 to 265) was used to construct standard curves. Samples were run using the SuperScript III one-step RT-PCR system with Platinum Taq DNA polymerase (Invitrogen) supplemented with ROX dye on a QuantStudio 6 Flex real-time PCR system using the following protocol: 50°C for 30 min, 94°C for 2 min, and 50 amplification cycles of 94°C for 15 s, 55°C for 30 s, and 68°C for 60 s, with fluorescence data being gathered at each 68°C incubation step.
Plaque assay.
Vero E6 cells were plated in 12-well plates in DMEM plus 10% FBS at a density of 5 × 105/well and were incubated overnight. Totals of 100 PFU/well for dengue virus and adenovirus and 150 PFU/well for herpes simplex virus were mixed with various concentrations of peptide in a total of 150 μl of DMEM per well and were incubated for 1 h. Culture medium was aspirated from cells and was replaced with the virus-peptide solutions. This was incubated for 1 h while rocking the plate every 15 min to ensure that the cells were covered with inoculum. After the incubation, the inoculum was aspirated from cells. Minimal essential medium (MEM) at 2× plus 4% FBS was mixed with 2.4% Avicel in equal amounts to make the overlay. A total of 1 ml of overlay was added to each well and was incubated until plaques were visible. Incubation times were 4 days for dengue virus, 2 days for herpes simplex virus, and 6 days for adenovirus. Plaques were visualized by crystal violet staining.
Time-of-addition assay.
HEK 293T/17 cells were first seeded in 96-well plates at a density of 1 × 104/well in DMEM plus 10% FBS and were incubated overnight. A 10 μM concentration of peptide or 200 μg/ml of anti-Lassa virus antibody 8.9F (42) was added before, during, or after infection at various time points. Lassa pseudovirus was added to cells at a concentration of 1 × 103 TCID50/well at 0 min. Infectivity was quantified by measuring luciferase expression translated from the pseudovirus genome approximately 72 h after infection using Bright-Glo (Promega).
Influenza virus entry inhibition analysis by confocal microscopy.
A549 cells (ATCC) were seeded into an 8-well chambered confocal microscope plate and grown to ∼60% confluence. Rhodamine 18 (R18) dye was added to 20 μg of human influenza A/Puerto Rico/8/34 (H1N1) purified virus obtained from Advanced Biotechnologies Inc. (catalog number 10-210-500) in DPBS to a final concentration of 67 μM R18. The mixture was then light protected and shaken at room temperature for 1 h. Labeled virus was then passed through a 0.22-μm filter and stored on ice (light protected). Peptides and antibody, diluted in DMEM to appropriate concentrations, were mixed with 1.75 μg of labeled virus and incubated for 30 min at room temperature. Cells were then washed once with DPBS, and then the virus-peptide mixture was added to cells and incubated for 10 min on ice. The inoculum was then aspirated, and the cells were washed once with DPBS. Finally, 50 μM Cascade Blue dextran in medium with 10% FBS was added to cells. Confocal microscopy was done in a stage incubator with 5% CO2 that was prewarmed to 37°C. Imaging was done at time zero (within 5 min of the start of incubation) and again after 2 h of incubation at 37°C.
Electron microscopy.
NATT, at various concentrations, and human influenza A/Puerto Rico/8/34 (H1N1) virus were incubated in DMEM. The virus was inactivated by exposing the solution to 30 min of UV radiation in a biosafety cabinet (55) followed by treatment with 200 mJ/cm2 UV-C radiation (56). This sample was plated onto a grid and then frozen in liquid ethane. The samples were visualized on an FEI Tecnai G2 F30 TWIN.
Virion circularity was measured using ImageJ. Outlines of the viral envelop were traced freehand three times, and the circularity of each trace was averaged to minimize error due to variance in tracing. Circularity was calculated, by the software, as follows:
ACKNOWLEDGMENTS
We thank James Robinson and Ashley Smira for their training and assistance with the Lassa pseudovirus assay system. We thank Jibao He for assistance with electron microscopy. We also thank Luis Branco for providing us with the Lassa virus glycoprotein plasmid and the NIH AIDS Reagent program for providing us with the pNL4-3.Luc.R-E- plasmid.
This work was supported by NIH grants 5R01GM111824-04 (W. C. Wimley, principal investigator [PI]), 1U19AI135995-01 (K. Andersen, PI), 2U54HG007480-05 (C. Happi, PI), 5U19AI115589-05 (R. F. Garry, PI), 1U19AI142790-01 (E. Saphire, PI), and 1R44AI082778-01 (R. Wilson, PI).
REFERENCES
- 1.Heron M. 2019. National Vital Statistics Reports deaths: leading causes for 2017. Natl Vital Stat Rep 68:1–77. [PubMed] [Google Scholar]
- 2.Murphy SL, Xu J, Kochanek KD, Arias E. 2018. Mortality in the United States, 2017. NCHS Data Brief 2018(328):1–8. [PubMed] [Google Scholar]
- 3.Johnson NB, Hayes LD, Brown K, Hoo EC, Ethier KA, Centers for Disease Control and Prevention. 2014. CDC National Health Report: leading causes of morbidity and mortality and associated behavioral risk and protective factors—United States, 2005–2013. MMWR Suppl 63:3–27. [PubMed] [Google Scholar]
- 4.De Clercq E, Li G. 2016. Approved antiviral drugs over the past 50 years. Clin Microbiol Rev 29:695–747. doi: 10.1128/CMR.00102-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Littler E, Oberg B. 2005. Achievements and challenges in antiviral drug discovery. Antivir Chem Chemother 16:155–168. doi: 10.1177/095632020501600302. [DOI] [PubMed] [Google Scholar]
- 6.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M, Xiao Y, Gao H, Guo L, Xie J, Wang G, Jiang R, Gao Z, Jin Q, Wang J, Cao B. 2020. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang G, Li X, Wang Z. 2016. APD3: the antimicrobial peptide database as a tool for research and education. Nucleic Acids Res 44:D1087–D1093. doi: 10.1093/nar/gkv1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaspar AA, Reichert JM. 2013. Future directions for peptide therapeutics development. Drug Discov Today 18:807–817. doi: 10.1016/j.drudis.2013.05.011. [DOI] [PubMed] [Google Scholar]
- 9.Fosgerau K, Hoffmann T. 2015. Peptide therapeutics: current status and future directions. Drug Discov Today 20:122–128. doi: 10.1016/j.drudis.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 10.Ashkenazi A, Wexler-Cohen Y, Shai Y. 2011. Multifaceted action of Fuzeon as virus-cell membrane fusion inhibitor. Biochim Biophys Acta 1808:2352–2358. doi: 10.1016/j.bbamem.2011.06.020. [DOI] [PubMed] [Google Scholar]
- 11.Badani H, Garry RF, Wimley WC. 2014. Peptide entry inhibitors of enveloped viruses: the importance of interfacial hydrophobicity. Biochim Biophys Acta 1838:2180–2197. doi: 10.1016/j.bbamem.2014.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu S, Lu H, Niu J, Xu Y, Wu S, Jiang S. 2005. Different from the HIV fusion inhibitor C34, the anti-HIV drug fuzeon (T-20) inhibits HIV-1 entry by targeting multiple sites in gp41 and gp120. J Biol Chem 280:11259–11273. doi: 10.1074/jbc.M411141200. [DOI] [PubMed] [Google Scholar]
- 13.Chong H, Yao X, Qiu Z, Sun J, Zhang M, Waltersperger S, Wang M, Liu SL, Cui S, He Y. 2013. Short-peptide fusion inhibitors with high potency against wild-type and enfuvirtide-resistant HIV-1. FASEB J 27:1203–1213. doi: 10.1096/fj.12-222547. [DOI] [PubMed] [Google Scholar]
- 14.Rapaport D, Ovadia M, Shai Y. 1995. A synthetic peptide corresponding to a conserved heptad repeat domain is a potent inhibitor of Sendai virus-cell fusion: an emerging similarity with functional domains of other viruses. EMBO J 14:5524–5531. doi: 10.1002/j.1460-2075.1995.tb00239.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lambert DM, Barney S, Lambert AL, Guthrie K, Medinas R, Davis DE, Bucy T, Erickson J, Merutka G, Petteway SR Jr. 1996. Peptides from conserved regions of paramyxovirus fusion (F) proteins are potent inhibitors of viral fusion. Proc Natl Acad Sci U S A 93:2186–2191. doi: 10.1073/pnas.93.5.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lamb D, Schüttelkopf AW, van Aalten DM, Brighty DW. 2008. Highly specific inhibition of leukaemia virus membrane fusion by interaction of peptide antagonists with a conserved region of the coiled coil of envelope. Retrovirology 5:70. doi: 10.1186/1742-4690-5-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu L, Liu Q, Zhu Y, Chan KH, Qin L, Li Y, Wang Q, Chan JF, Du L, Yu F, Ma C, Ye S, Yuen KY, Zhang R, Jiang S. 2014. Structure-based discovery of Middle East respiratory syndrome coronavirus fusion inhibitor. Nat Commun 5:3067. doi: 10.1038/ncomms4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee KK, Pessi A, Gui L, Santoprete A, Talekar A, Moscona A, Porotto M. 2011. Capturing a fusion intermediate of influenza hemagglutinin with a cholesterol-conjugated peptide, a new antiviral strategy for influenza virus. J Biol Chem 286:42141–42149. doi: 10.1074/jbc.M111.254243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Akkarawongsa R, Pocaro NE, Case G, Kolb AW, Brandt CR. 2009. Multiple peptides homologous to herpes simplex virus type 1 glycoprotein B inhibit viral infection. Antimicrob Agents Chemother 53:987–996. doi: 10.1128/AAC.00793-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng G, Montero A, Gastaminza P, Whitten-Bauer C, Wieland SF, Isogawa M, Fredericksen B, Selvarajah S, Gallay PA, Ghadiri MR, Chisari FV. 2008. A virocidal amphipathic α-helical peptide that inhibits hepatitis C virus infection in vitro. Proc Natl Acad Sci U S A 105:3088–3093. doi: 10.1073/pnas.0712380105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Si Y, Liu S, Liu X, Jacobs JL, Cheng M, Niu Y, Jin Q, Wang T, Yang W. 2012. A human claudin-1-derived peptide inhibits hepatitis C virus entry. Hepatology 56:507–515. doi: 10.1002/hep.25685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bai F, Town T, Pradhan D, Cox J, Ashish, Ledizet M, Anderson JF, Flavell RA, Krueger JK, Koski RA, Fikrig E. 2007. Antiviral peptides targeting the West Nile virus envelope protein. J Virol 81:2047–2055. doi: 10.1128/JVI.01840-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hrobowski YM, Garry RF, Michael SF. 2005. Peptide inhibitors of dengue virus and West Nile virus infectivity. Virol J 2:49. doi: 10.1186/1743-422X-2-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sainz B Jr, Mossel EC, Gallaher WR, Wimley WC, Peters CJ, Wilson RB, Garry RF. 2006. Inhibition of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) infectivity by peptides analogous to the viral spike protein. Virus Res 120:146–155. doi: 10.1016/j.virusres.2006.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Melnik LI, Garry RF, Morris CA. 2011. Peptide inhibition of human cytomegalovirus infection. Virol J 8:76. doi: 10.1186/1743-422X-8-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koehler JW, Smith JM, Ripoll DR, Spik KW, Taylor SL, Badger CV, Grant RJ, Ogg MM, Wallqvist A, Guttieri MC, Garry RF, Schmaljohn CS. 2013. A fusion-inhibiting peptide Against Rift Valley fever virus inhibits multiple, diverse viruses. PLoS Negl Trop Dis 7:e2430. doi: 10.1371/journal.pntd.0002430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spence JS, Melnik LI, Badani H, Wimley WC, Garry RF. 2014. Inhibition of arenavirus infection by a glycoprotein-derived peptide with a novel mechanism. J Virol 88:8556–8564. doi: 10.1128/JVI.01133-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wimley WC, White SH. 1996. Experimentally determined hydrophobicity scale for proteins at membrane interfaces. Nat Struct Biol 3:842–848. doi: 10.1038/nsb1096-842. [DOI] [PubMed] [Google Scholar]
- 29.White SH, Wimley WC. 1999. Membrane protein folding and stability: physical principles. Annu Rev Biophys Biomol Struct 28:319–365. doi: 10.1146/annurev.biophys.28.1.319. [DOI] [PubMed] [Google Scholar]
- 30.Lok SM, Costin JM, Hrobowski YM, Hoffmann AR, Rowe DK, Kukkaro P, Holdaway H, Chipman P, Fontaine KA, Holbrook MR, Garry RF, Kostyuchenko V, Wimley WC, Isern S, Rossmann MG, Michael SF. 2012. Release of dengue virus genome induced by a peptide inhibitor. PLoS One 7:e50995. doi: 10.1371/journal.pone.0050995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wimley WC. 2010. Describing the mechanism of antimicrobial peptide action with the interfacial activity model. ACS Chem Biol 5:905–917. doi: 10.1021/cb1001558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rathinakumar R, Walkenhorst WF, Wimley WC. 2009. Broad-spectrum antimicrobial peptides by rational combinatorial design and high-throughput screening: the importance of interfacial activity. J Am Chem Soc 131:7609–7617. doi: 10.1021/ja8093247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rathinakumar R, Wimley WC. 2008. Biomolecular engineering by combinatorial design and high-throughput screening: small, soluble peptides that permeabilize membranes. J Am Chem Soc 130:9849–9858. doi: 10.1021/ja8017863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rathinakumar R, Wimley WC. 2010. High-throughput discovery of broad-spectrum peptide antibiotics. FASEB J 24:3232–3238. doi: 10.1096/fj.10-157040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krauson AJ, He J, Wimley AW, Hoffmann AR, Wimley WC. 2013. Synthetic molecular evolution of pore-forming peptides by iterative combinatorial library screening. ACS Chem Biol 8:823–831. doi: 10.1021/cb300598k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.He J, Krauson AJ, Wimley WC. 2014. Toward the de novo design of antimicrobial peptides: lack of correlation between peptide permeabilization of lipid vesicles and antimicrobial, cytolytic, or cytotoxic activity in living cells. Biopolymers 102:1–6. doi: 10.1002/bip.22281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guha S, Ghimire J, Wu E, Wimley WC. 2019. Mechanistic landscape of membrane-permeabilizing peptides. Chem Rev 119:6040–6085. doi: 10.1021/acs.chemrev.8b00520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.He J, Kauffman WB, Fuselier T, Naveen SK, Voss TG, Hristova K, Wimley WC. 2013. Direct cytosolic delivery of polar cargo to cells by spontaneous membrane-translocating peptides. J Biol Chem 288:29974–29986. doi: 10.1074/jbc.M113.488312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krauson AJ, Hall OM, Fuselier T, Starr CG, Kauffman WB, Wimley WC. 2015. Conformational fine-tuning of pore-forming peptide potency and selectivity. J Am Chem Soc 137:16144–16152. doi: 10.1021/jacs.5b10595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Illick MM, Branco LM, Fair JN, Illick KA, Matschiner A, Schoepp R, Garry RF, Guttieri MC. 2008. Uncoupling GP1 and GP2 expression in the Lassa virus glycoprotein complex: implications for GP1 ectodomain shedding. Virol J 5:161. doi: 10.1186/1743-422X-5-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O'Brien J, Wilson I, Orton T, Pognan F. 2000. Investigation of the Alamar Blue (resazurin) fluorescent dye for the assessment of mammalian cell cytotoxicity. Eur J Biochem 267:5421–5426. doi: 10.1046/j.1432-1327.2000.01606.x. [DOI] [PubMed] [Google Scholar]
- 42.Robinson JE, Hastie KM, Cross RW, Yenni RE, Elliott DH, Rouelle JA, Kannadka CB, Smira AA, Garry CE, Bradley BT, Yu H, Shaffer JG, Boisen ML, Hartnett JN, Zandonatti MA, Rowland MM, Heinrich ML, Martinez-Sobrido L, Cheng B, De La Torre JC, Andersen KG, Goba A, Momoh M, Fullah M, Gbakie M, Kanneh L, Koroma VJ, Fonnie R, Jalloh SC, Kargbo B, Vandi MA, Gbetuwa M, Ikponmwosa O, Asogun DA, Okokhere PO, Follarin OA, Schieffelin JS, Pitts KR, Geisbert JB, Kulakoski PC, Wilson RB, Happi CT, Sabeti PC, Gevao SM, Khan SH, Grant DS, Geisbert TW, Saphire EO, Branco LM, Garry RF. 2016. Most neutralizing human monoclonal antibodies target novel epitopes requiring both Lassa virus glycoprotein subunits. Nat Commun 7:11544. doi: 10.1038/ncomms11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cho NJ, Dvory-Sobol H, Xiong A, Cho SJ, Frank CW, Glenn JS. 2009. Mechanism of an amphipathic alpha-helical peptide’s antiviral activity involves size-dependent virus particle lysis. ACS Chem Biol 4:1061–1067. doi: 10.1021/cb900149b. [DOI] [PubMed] [Google Scholar]
- 44.Tabaei SR, Rabe M, Zhdanov VP, Cho NJ, Höök F. 2012. Single vesicle analysis reveals nanoscale membrane curvature selective pore formation in lipid membranes by an antiviral α-helical peptide. Nano Lett 12:5719–5725. doi: 10.1021/nl3029637. [DOI] [PubMed] [Google Scholar]
- 45.Jackman JA, Costa VV, Park S, Real ALCV, Park JH, Cardozo PL, Ferhan AR, Olmo IG, Moreira TP, Bambirra JL, Queiroz VF, Queiroz-Junior CM, Foureaux G, Souza DG, Ribeiro FM, Yoon BK, Wynendaele E, De Spiegeleer B, Teixeira MM, Cho NJ. 2018. Therapeutic treatment of Zika virus infection using a brain-penetrating antiviral peptide. Nat Mater 17:971–977. doi: 10.1038/s41563-018-0194-2. [DOI] [PubMed] [Google Scholar]
- 46.Starr CG, He J, Wimley WC. 2016. Host cell interactions are a significant barrier to the clinical utility of peptide antibiotics. ACS Chem Biol 11:3391–3399. doi: 10.1021/acschembio.6b00843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Starr CG, Ghimire J, Guha S, Hoffmann JP, Wang Y, Sun L, Landreneau BN, Kolansky ZD, Kilanowski-Doroh IM, Sammarco MC, Morici LA, Wimley WC. 2020. Synthetic molecular evolution of host cell-compatible, antimicrobial peptides effective against drug-resistant, biofilm-forming bacteria. Proc Natl Acad Sci U S A 117:8437–8448. doi: 10.1073/pnas.1918427117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kauffman WB, Guha S, Wimley WC. 2018. Synthetic molecular evolution of hybrid cell penetrating peptides. Nat Commun 9:2568. doi: 10.1038/s41467-018-04874-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li S, Kim SY, Pittman AE, King GM, Wimley WC, Hristova K. 2018. Potent macromolecule-sized poration of lipid bilayers by the macrolittins, a synthetically evolved family of pore-forming peptides. J Am Chem Soc 140:6441–6447. doi: 10.1021/jacs.8b03026. [DOI] [PubMed] [Google Scholar]
- 50.Reed LJ, Muench H. 1938. A simple method of estimating fifty per cent endpoints. Am J Epidemiol 27:493–497. doi: 10.1093/oxfordjournals.aje.a118408. [DOI] [Google Scholar]
- 51.Donald HB, Isaacs A. 1954. Counts of influenza virus particles. J Gen Microbiol 10:457–464. doi: 10.1099/00221287-10-3-457. [DOI] [PubMed] [Google Scholar]
- 52.Wei Z, Mcevoy M, Razinkov V, Polozova A, Li E, Casas-Finet J, Tous GI, Balu P, Pan AA, Mehta H, Schenerman MA. 2007. Biophysical characterization of influenza virus subpopulations using field flow fractionation and multiangle light scattering: correlation of particle counts, size distribution and infectivity. J Virol Methods 144:122–132. doi: 10.1016/j.jviromet.2007.04.008. [DOI] [PubMed] [Google Scholar]
- 53.Fonville JM, Marshall N, Tao H, Steel J, Lowen AC. 2015. Influenza virus reassortment is enhanced by semi-infectious particles but can be suppressed by defective interfering particles. PLoS Pathog 11:e1005204. doi: 10.1371/journal.ppat.1005204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.World Health Organization. 2009. CDC protocol of realtime RT-PCR for influenza A (H1N1). World Health Organization, Geneva, Switzerland. [Google Scholar]
- 55.Zou S, Guo J, Gao R, Dong L, Zhou J, Zhang Y, Dong J, Bo H, Qin K, Shu Y. 2013. Inactivation of the novel avian influenza A (H7N9) virus under physical conditions or chemical agents treatment. Virol J 10:289. doi: 10.1186/1743-422X-10-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang J, Mauser A, Chao SF, Remington K, Treckmann R, Kaiser K, Pifat D, Hotta J. 2004. Virus inactivation and protein recovery in a novel ultraviolet-C reactor. Vox Sang 86:230–238. doi: 10.1111/j.0042-9007.2004.00485.x. [DOI] [PubMed] [Google Scholar]