Previous reports assumed that sphingomyelin (SM) is essential for HCV replication, but the mechanism was unclear. In this study, we showed for the first time that SM and ceramide transfer protein (CERT), which is in the SM biosynthesis pathway, are essential for the biosynthesis of double-membrane vesicles (DMVs), the sites of viral replication. Low numbers of DMVs were observed in CERT-KO cells transfected with replicon RNA or with constructs that drive HCV protein production in a replication-independent system. HCV replication was rescued by ectopic expression of the CERT protein, but not by CERT mutants, that abolishes the binding of CERT to vesicle-associated membrane protein-associated protein (VAP) or phosphatidylinositol 4-phosphate (PI4P), indicating new roles for VAP and PI4P in HCV replication. The biosynthesis of DMVs has great importance to replication by a variety of plus-stranded RNA viruses. Understanding of this process is expected to facilitate the development of diagnosis and antivirus.
KEYWORDS: sphingomyelin, ceramide transfer protein, double-membrane vesicles, hepatitis C virus, phosphatidylinositol 4-phosphate, replication, vesicle-associated membrane protein-associated protein
ABSTRACT
Some plus-stranded RNA viruses generate double-membrane vesicles (DMVs), one type of the membrane replication factories, as replication sites. Little is known about the lipid components involved in the biogenesis of these vesicles. Sphingomyelin (SM) is required for hepatitis C virus (HCV) replication, but the mechanism of SM involvement remains poorly understood. SM biosynthesis starts in the endoplasmic reticulum (ER) and gives rise to ceramide, which is transported from the ER to the Golgi by the action of ceramide transfer protein (CERT), where it can be converted to SM. In this study, inhibition of SM biosynthesis, either by using small-molecule inhibitors or by knockout (KO) of CERT, suppressed HCV replication in a genotype-independent manner. This reduction in HCV replication was rescued by exogenous SM or ectopic expression of the CERT protein, but not by ectopic expression of nonfunctional CERT mutants. Observing low numbers of DMVs in stable replicon cells treated with a SM biosynthesis inhibitor or in CERT-KO cells transfected with either HCV replicon or with constructs that drive HCV protein production in a replication-independent system indicated the significant importance of SM to DMVs. The degradation of SM of the in vitro-isolated DMVs affected their morphology and increased the vulnerability of HCV RNA and proteins to RNase and protease treatment, respectively. Poliovirus, known to induce DMVs, showed decreased replication in CERT-KO cells, while dengue virus, known to induce invaginated vesicles, did not. In conclusion, these findings indicated that SM is an essential constituent of DMVs generated by some plus-stranded RNA viruses.
IMPORTANCE Previous reports assumed that sphingomyelin (SM) is essential for HCV replication, but the mechanism was unclear. In this study, we showed for the first time that SM and ceramide transfer protein (CERT), which is in the SM biosynthesis pathway, are essential for the biosynthesis of double-membrane vesicles (DMVs), the sites of viral replication. Low numbers of DMVs were observed in CERT-KO cells transfected with replicon RNA or with constructs that drive HCV protein production in a replication-independent system. HCV replication was rescued by ectopic expression of the CERT protein, but not by CERT mutants, that abolishes the binding of CERT to vesicle-associated membrane protein-associated protein (VAP) or phosphatidylinositol 4-phosphate (PI4P), indicating new roles for VAP and PI4P in HCV replication. The biosynthesis of DMVs has great importance to replication by a variety of plus-stranded RNA viruses. Understanding of this process is expected to facilitate the development of diagnosis and antivirus.
INTRODUCTION
Hepatitis C virus (HCV) is among the most important human pathogens and is capable of causing chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma (HCC) (1). Worldwide, HCV prevalence is estimated at 2.5% of the general population, given that the virus is believed to infect 177 million people around the globe (2). HCV is a plus-stranded RNA virus that is classified into 7 recognized genotypes (3). The virus belongs to the genus Hepacivirus of the Flaviviridae family, a family that includes flaviviruses (such as Zika virus, dengue virus [DENV], yellow fever virus, and tick-borne encephalitis virus), and animal pestivirus genera (such as bovine viral diarrhea virus) (4).
Plus-stranded RNA viruses depend on cellular membranes in all steps of the viral life cycle (5). These viruses, including HCV, share the characteristic of remodeling intracellular membranes in order to create membrane replication factories or replication organelles, which are vesicles where viral RNA replication occurs (6–8). These vesicles not only represent the site of viral replication but also act as one of the strategies of viral immune evasion mechanisms, shielding the viral RNA from cellular innate immune sensors (9–11). Electron microscopic investigations have defined the membrane replication factory or vesicle as a complex structure of remodeled membranes with negative and positive curvatures; these vesicles exhibit little similarity to the parental organelles from which the factories are remodeled (5, 12–14). Almost all of the intracellular membranes in eukaryotic cells, including the endoplasmic reticulum (ER), the Golgi, the outer membrane of the mitochondria, and the peroxisomal membranes, represent substrates for the biosynthesis of membrane replication factories induced by plus-stranded RNA viruses (5). These vesicles exist in two different morphological types, the invaginated vesicle/spherule type and the double-membrane vesicle (DMV) type. Infection by HCV, picornaviruses, and coronaviruses leads to the formation of DMVs, whereas infection by DENV and West Nile virus (WNV) induces the formation of invaginated vesicles (15). The differences from their parental organelles reflect changes in both structure and in protein and lipid composition (5).
Lipids represent the major macromolecule responsible for the unique physical properties of different membranes, which include permeability, fluidity, and bending characteristics (16). Sphingomyelin (SM) is among the most common sphingolipids in many mammalian cells and tissues; SM plays significant structural and functional roles in mammalian cells through the generation of lipid rafts, participation in cell signaling pathways, and membrane homeostasis and curvature (17–19). SM is also involved in many viral infections, such as those by Ebola virus (20), human immune deficiency virus (HIV) (21), Japanese encephalitis virus (22), rabies virus (23), and bovine viral diarrhea virus (24). SM plays several roles in the HCV infection cycle, including in the replication, morphogenesis, and egress steps (25, 26). As indicated in Fig. 1A, SM is biosynthesized in the ER by two different pathways, the de novo and salvage pathways. Both pathways yield ceramide. The ceramide transfer protein (CERT) is responsible for extracting ceramide from the ER and carrying this compound to the Golgi apparatus in a nonvesicular manner; the efficient CERT-mediated trafficking of ceramide occurs at membrane contact sites between the ER and the Golgi apparatus (27). In the Golgi, ceramide is converted to SM by the sphingomyelin synthase (SGMS) enzyme. Within the CERT protein, three sites are known to be important for CERT’s ability to transfer ceramide from the ER to the Golgi. The START domain is responsible for extracting ceramide from the ER membrane and transferring the compound to membranes of the Golgi. Two other CERT domains contribute to START function. The first, the FFAT motif, directs CERT to the ER by binding to the ER-resident vesicle-associated membrane protein-associated protein (VAP); the second, the PH domain, targets ceramide to the Golgi membrane by binding to phosphatidylinositol 4-phosphate (PI4P) on the surface of the Golgi apparatus (27, 28). While the function of CERT in SM synthesis, including the involvement of SM in binding and activation of RNA-dependent RNA polymerase of some HCV genotypes (29), has been detailed in multiple reports, the role of CERT and/or SM in HCV replication remains unclear.
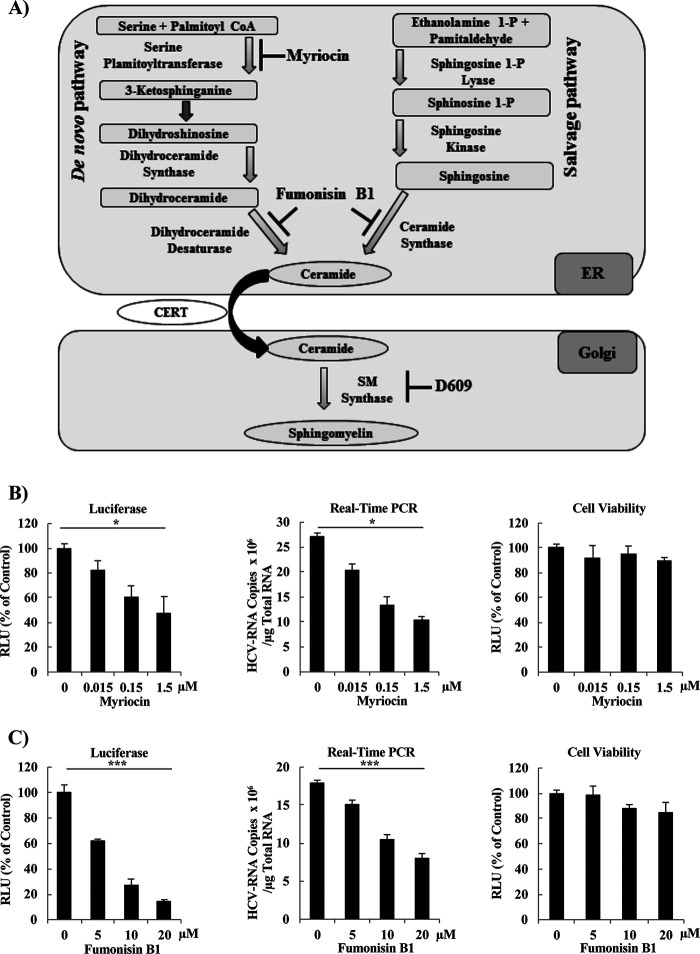
FIG 1.
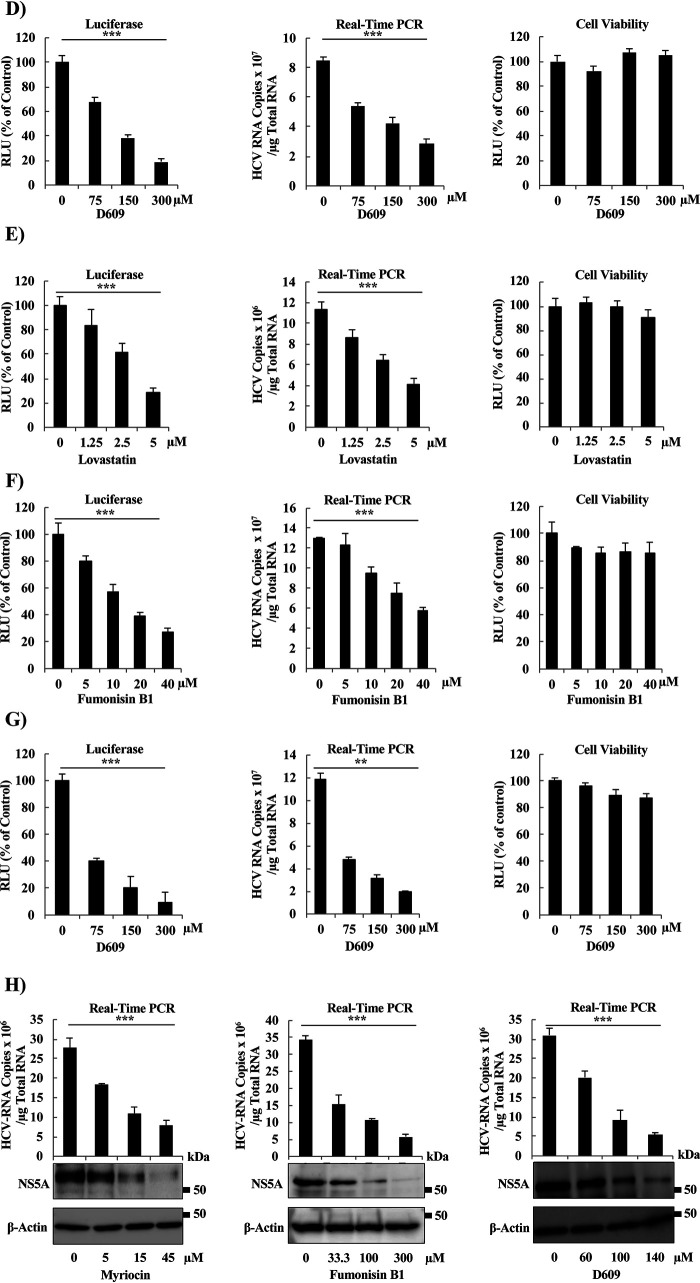
Treatment with small-molecule inhibitors of SM biosynthesis decreases HCV replication in SGR cells and HCV-infected cells. (A) Schematic representation of SM biosynthesis pathways. SM biosynthesis starts in the ER and proceeds by either of two pathways, the de novo pathway or the salvage pathway. Both pathways yield ceramide, which then is transported from the ER into the Golgi by the CERT. Within the Golgi, ceramide is converted to SM by the activity of the SM synthases. The steps blocked by each of the following three SM biosynthesis inhibitors tested in the present study (to analyze the role of SM in HCV replication) are indicated: myriocin (inhibitor of the de novo pathway), fumonisin B1 (inhibitor of both the de novo and salvage pathways), and D609 (a direct, and the sole commercially available, inhibitor of SM synthases). (B to E) LN#2 cells were treated for 72 h with vehicle (indicated as zero) or the indicated concentrations of (B) myriocin, (C) fumonisin B1, (D) D609, or (E) lovastatin (a known inhibitor of cholesterol biosynthesis; used here as a control); the effects on replication then were determined by measurement of luciferase activities (left panels) and by quantification of HCV-RNA copy number by real-time RT-PCR (middle panels), in which the HCV-RNA copy number was normalized to 1 μg total RNA. Fold changes in HCV replication were calculated by dividing relative luciferase unit (RLU) values at 72 h by those at 4 h; the results were normalized to those of the control. Cell viability was evaluated using a Cell TiterGlo luminescent cell viability assay (right panels). (F and G) JFH1/SGR were treated for 96 h with the indicated concentrations of (F) fumonisin B1 or (G) D609, and the effects on replication were then determined by measurement of luciferase activities (left panels) and by quantification of HCV-RNA copy number by real-time reverse transcription-PCR (RT-PCR) (middle panels), in which RNA copy number was normalized to 1 μg of total RNA. Cell viability was evaluated using a Cell TiterGlo luminescent cell viability assay (right panels). (H) Huh7.5.1-8 cells were infected with cell culture-derived infectious HCVcc at a multiplicity of infection (MOI) of 0.1 after pretreatment for 24 h with the indicated concentrations of the various SM biosynthesis inhibitors, including myriocin (left), fumonisin B1 (right), and D609 (right). Intracellular HCV RNA copy number then was analyzed by real-time RT-PCR and normalized to 1 μg total RNA. HCV protein levels were analyzed by Western blot (WB) using anti-NS5A rabbit polyclonal antibodies (H, lower panels). Values were obtained from quadruplicate wells in at least three independent experiments and are presented as means ± standard deviations (SDs). ***, P < 0.005; *, P < 0.05 (two-sided Student’s t test).
Despite extensive progress in the molecular characterization of HCV replication and the accumulation of an increasing list of host factors necessary for HCV replication, the structure of the membrane replication factories or vesicles and the cooperative action between HCV proteins and host factors to remodel the ER membranes, the parental organelle of DMVs, remain poorly characterized (6, 8, 12, 15, 30). Although the important requirement of SM in HCV replication has been reported previously, it remains unclear whether SM is an essential component of the functional membrane replication factories. In the present study, we sought to analyze the importance of SM in the biogenesis of the functional membrane replication factories. Specifically, we provide several lines of evidence demonstrating the essential role of SM in the biosynthesis of DMVs induced by HCV infection. We also show that SM is required for the replication of poliovirus, which is known to generate DMVs. This investigation will serve as a cornerstone for the future analysis of a lipid host factor in the biogenesis of membrane replication factories induced by various plus-stranded RNA viruses. Based on our findings, we suggest that the DMV, a structure unique to cells infected by positive-stranded RNA viruses, may serve as a target for viral diagnosis, prognosis, and therapeutics.
RESULTS
SM biosynthesis inhibitors reduce HCV replication in concentration-dependent and genotype-independent manners.
We and others have previously shown that HCV replication occurs in lipid raft-like membranes enriched for cholesterol and SM (31–34). To analyze the importance of SM in HCV replication and the biogenesis of membrane replication factories, we used two different methods to inhibit the SM biosynthesis pathways, either by using well-known small-molecule inhibitors of SM biosynthesis pathways or by establishing CERT knockout (CERT-KO) cells.
We used three different inhibitors of SM biosynthesis pathways (including myriocin, fumonisin B1, and D609) to assess the importance of SM to HCV replication, as in previous studies (25, 35). Myriocin, a well-known inhibitor of the serine palmitoyltransferase enzyme that inhibits the de novo pathway (36), reduced HCV replication, as analyzed using the luciferase assay, in Huh7.5.1-8 human hepatocellular carcinoma cells (37) harboring the genotype-1b subgenomic replicon (SGR) (LucNeo#2; here designated LN#2) (38), in a dose-dependent manner (Fig. 1B, left). The effect of myriocin on HCV replication, analyzed by luciferase assay, was confirmed by quantification of HCV RNA copy number (Fig. 1B, middle) and was further shown to have no significant effect on cell viability (Fig. 1B, right). Fumonisin B1, a potent inhibitor of (dihydro) ceramide synthases that inhibits both the de novo and salvage pathways (39), suppressed HCV replication as measured by luciferase activity (Fig. 1C, left). This reduction in HCV replication was comparable to that observed with D609 (Fig. 1D, left), the only commercially available compound that directly inhibits SGMS. As with myriocin, the effects of fumonisin B1 and D609 on replication were confirmed by quantification of HCV RNA copy number using real-time reverse transcription-PCR (RT-PCR) (Fig. 1C and D, middle panels), and the effect exerted by both compounds was not due to cellular toxicity (Fig. 1C and D, right panels). Lovastatin, a known inhibitor of both cholesterol biosynthesis (specifically of 3-hydroxy-3-methylglutaryl coenzyme A [HMG-CoA] reductase) and HCV replication, as it is required for the stability of DMVs (15, 40), was used as a positive control. As expected, lovastatin yielded a dose-dependent decrease in HCV replication when assessed by luciferase activity or real-time RT-PCR (Fig. 1E, left and middle), again with no significant effect on cell viability (Fig. 1E, right). To further confirm these results and to test if the compounds have pangenotypic suppression activity of HCV replication the effects of fumonisin B1 and D609 on replication were tested using Huh7.5.1-8 cells (37) harboring the genotype-2a SGR derived from HCV strain JFH1 (SGRlucneo; designated JFH1/SGR) (41). Fumonisin B1 and D609 yielded a decrease in luciferase activity (Fig. 1F and G, left panels) and in HCV RNA copy number (Fig. 1F and G, middle panels), without showing significant toxic effects on the cells (Fig. 1F and G, right panels). To confirm the effects of myriocin (Fig. 1H, left), fumonisin B1 (Fig. 1H, middle), and D609 (Fig. 1G, right) on HCV-infected cells, JFH1-infected Huh7.5.1-8 cells were treated with each of the inhibitors starting at 24 h before infection, and the effect on HCV replication was monitored by real-time RT-PCR (Fig. 1H, upper panels) and Western blotting (WB) (Fig. 1H, lower panels). The results of the infection experiments of infectious cell culture-produced HCV/JFH1 (HCVcc) were consistent with those seen with the replicon cell lines.
CERT-KO cells show significant decreases in both SM level and HCV replication.
To further analyze the role of SM in HCV replication, a CRISPR/Cas9 system was used to generate CERT-KO cells. Five different CERT-KO clones were obtained, either by insertion or deletion of sequences in exon 1 or 2; frameshift mutations were detected in both CERT alleles of each clone (Fig. 2A). All five CERT-KO clones showed significant reductions of SM levels as detected using 14C-serine metabolic labeling and thin-layer chromatography (TLC) (Fig. 2B) and complete disappearance of CERT protein expression as assessed by WB analysis (Fig. 2C, lower). Replication of HCV in the five different CERT-KO clones was analyzed by both measurement of luciferase activities (Fig. 2C, upper) and WB analysis (Fig. 2C, lower) after transfection of these cells with in vitro-transcribed JFH1/SGR. HCV replication was significantly reduced in all CERT-KO cell clones compared that in to Huh7.5.1-8 cells (the parental cell line from which CERT-KO cell clones were derived). To ensure that the observed effect was not due to differences in transfection efficiencies, we quantified firefly luciferase and Renilla luciferase activity at 4 h posttransfection. No significant effect on luciferase activities was observed, suggesting similar transfection efficiencies in different cells used in this experiment (Fig. 2D and E).
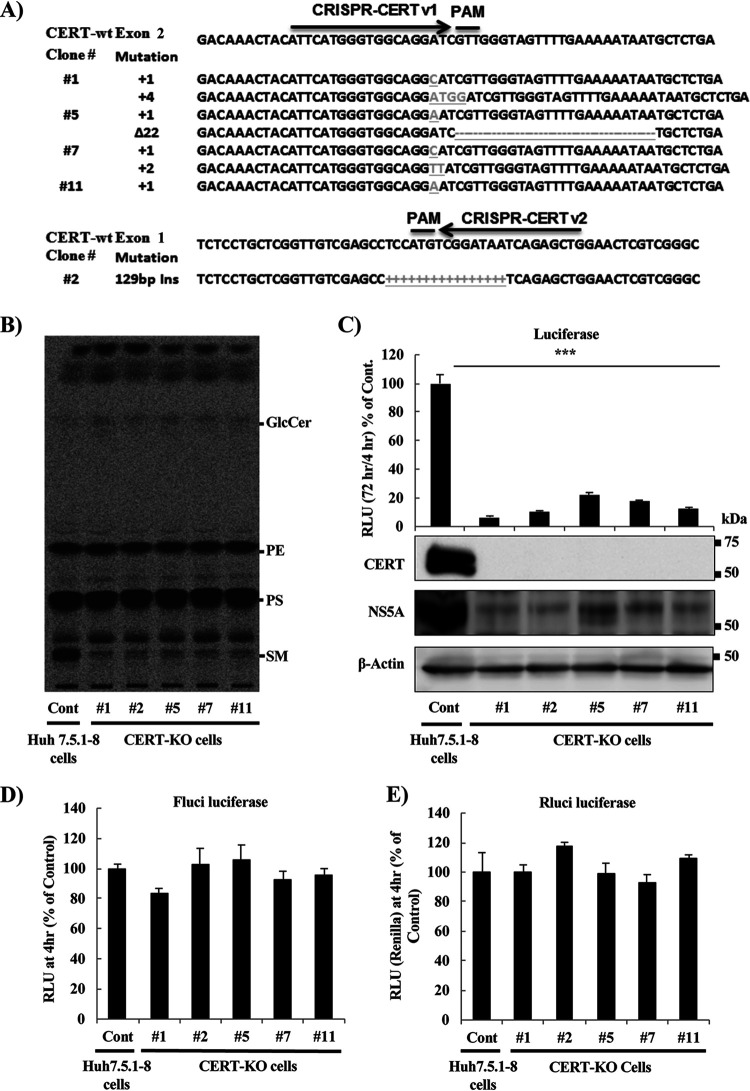
FIG 2.
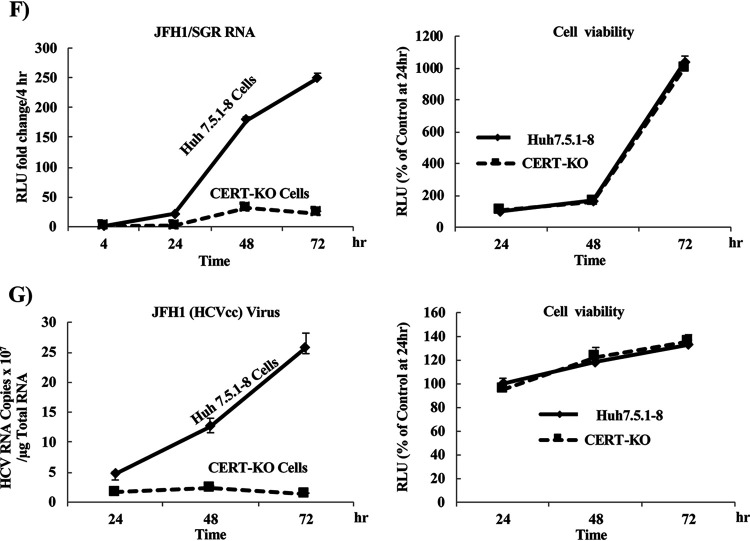
HCV replicates at significantly lower levels in CERT-KO cells. (A) Using a CRISPR/Cas9 system, two different sequences were used to generate CERT-KO cells from Huh7.5.1-8 parent cells. CRISPR-CERT v1 and its PAM sequence targeted exon 2 (upper), while CRISPR-CERT v2 and its PAM sequence targeted exon 1 (lower). The sequence alignments show exon 2 of the gene encoding the wild-type protein (CERT-wt) aligned with the gene sequences recovered from CERT-KO clones 1, 5, 7, and 11 (upper) and exon 1 of the gene encoding CERT-wt aligned with the gene sequences recovered from CERT-KO clone 2 (lower panel). The observed nucleotide insertions (gray letters or + signs) or deletions (frame shifts; gray − signs) in either both CERT alleles (clones 1, 5, and 7) or one CERT allele (clones 11 and 2) are indicated. (B) The levels of SM were analyzed by TLC after 14C-serine metabolic labeling in each of the CERT-KO clones and in the parental Huh7.5.1-8 cells (Cont). The bands corresponding to SM and other labeled lipid species, including glucosylceramide (GlcCer), phosphatidylethanolamine (PE), and phosphatidylserine (PS), are indicated. (C to E) The parental Huh7.5.1-8 cells (Cont) and the five CERT-KO clones each were electroporated with the combination of 10 μg of in vitro-transcribed JFH1/SGR RNA and 1 μg of in vitro-transcribed Renilla luciferase-encoding RNA (as an internal control); HCV replication was then analyzed by measuring luciferase activities at 4 and 72 h posttransfection. Fold changes in HCV replication were calculated by dividing RLU values at 72 h by those at 4 h; the results were normalized to those of the control (Cont; upper). In parallel, HCV replication was analyzed in transfected Huh7.5.1-8 and CERT-KO cells by measuring HCV protein expression by analysis with WB with anti-NS5A antibodies. CERT protein expression levels also were analyzed using anti-CERT antibodies (lower). Blotting with anti-β-actin antibodies was performed to normalize protein loading. At 4 h postelectroporation, (D) firefly luciferase activities and (E) Renilla luciferase activities in the cell lysates were measured to assess transfection and translation efficacies. (F) Both Huh7.5.1-8 and CERT-KO clone 7 were electroporated with 10 μg JFH1/SGR RNA, and luciferase activities were measured at 4, 24, 48, and 72 h posttransfection; fold changes (left) were calculated as in panel C. Cell viability was evaluated using a Cell TiterGlo luminescent cell viability assay (right). (G) Both Huh7.5.1-8 and CERT-KO clone 7 were infected with HCVcc at a multiplicity of infection (MOI) of 0.1; intracellular HCV RNA levels then were analyzed by real-time RT-PCR at 24, 48, and 72 h postinfection. RNA copy numbers were normalized to 1 μg total RNA (left). Cell viability was evaluated using a Cell TiterGlo luminescent cell viability assay (right). Values were obtained from quadruplicate wells in at least three independent experiments and are presented as means ± SDs. ***, P < 0.005 (two-sided Student’s t test).
Since different CERT-KO clones gave similar results and to simplify the experimental setup, clone 7 was selected for further analysis and will be referred to here as “CERT-KO cells.” To follow the kinetics of HCV replication in CERT-KO cells, we monitored HCV replication at different time points after electroporation with in vitro-transcribed JFH1/SGR RNA. Measurement of luciferase activity showed a decrease in HCV replication at all tested time points compared to that in parental Huh7.5.1-8 cells (Fig. 2F, left) with no significant effect on cell viability (Fig. 2F, right). The reduction in HCV replication was further confirmed by infection of both cell lines, CERT-KO clone 7 cells and Huh7.5.1-8 cells, with HCVcc and by subsequently following HCV-RNA over time by real-time RT-PCR (Fig. 2G, left); again, no significant effects on cell viability were observed (Fig. 2G, right).
Ectopic expression of CERT protein and exogenous SM supply rescue replication in CERT-KO cells.
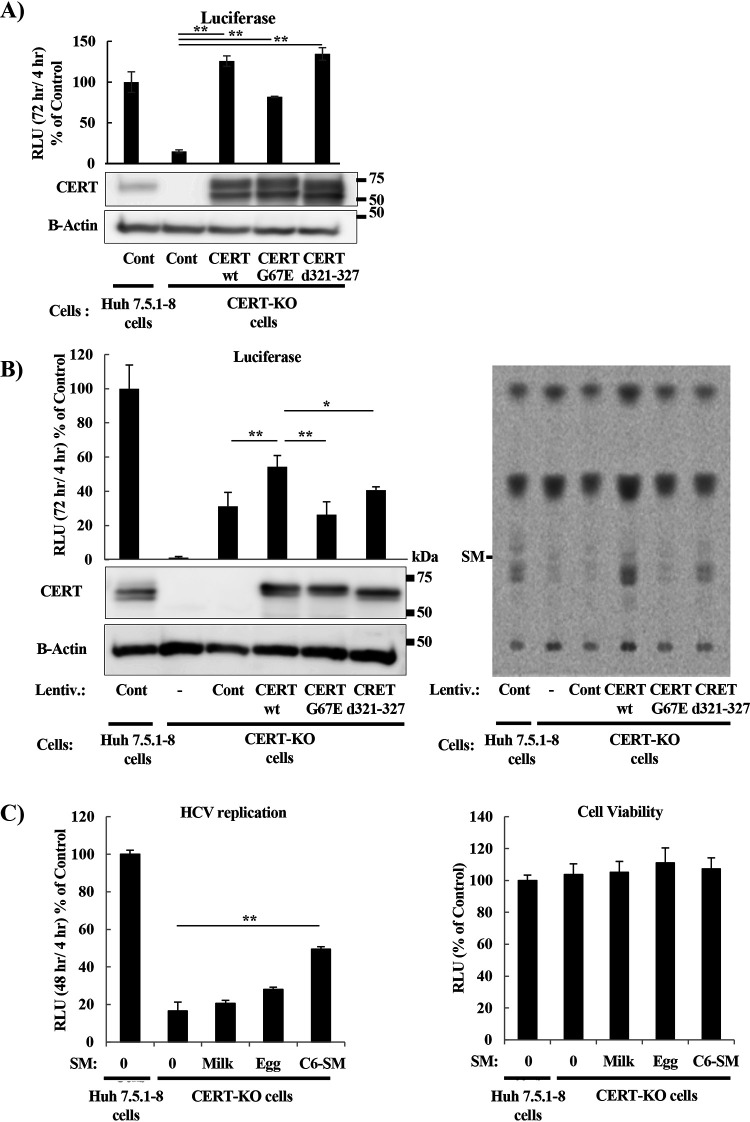
It has been reported that ectopic expression in CERT-defective cells of CERT mutant proteins (harboring mutations in either the FFAT or PH domains) at levels matching the endogenous expression results in impairment of ER-to-Golgi trafficking of ceramide (28) while enhancing the frequency of random ceramide transfer (27). To test whether impairment of ceramide trafficking to the Golgi would affect HCV replication, we generated mutant CERT genes encoding proteins harboring either of the following two mutations in CERT: ΔFFAT (d321-327), a deletion mutation that abolishes the binding of CERT to the ER-resident protein VAP, and the G67E mutation, which abolishes the PI4P-binding capacity of the PH domain, consequently impairing ER-Golgi transport of ceramide (27). First, CERT-KO cells were transfected with CERT-wt (that is, the wild-type protein), the G67E, or the ΔFFAT (d321-327) mutant with the G418 resistance gene, and were cultured in G418-containing medium to establish cell lines that stably express wild or mutant CERT (Fig. 3A). CERT expression levels in these CERT-KO cells were much higher than those of endogenous CERT expression. HCV replication was fully restored in wild-type and mutant (d321-327) CERT cells. CERT mutants, when strongly overexpressed, are known to substantially support ceramide transport from the ER to sphingomyelin production (28). Therefore, the expression levels of wild or mutant CERT had to match that of endogenous CERT. As shown in Fig. 3A, establishing such cell lines by G418 selection was difficult. Then, various concentrations of lentivirus were used to establish cell lines with different levels of wild/mutant CERT expression (data not shown). Cell lines with CERT expression levels similar to that of endogenous CERT were selected for subsequent experiments (Fig. 3B). CERT-KO cells expressing CERT-wt showed significant restoration of HCV replication compared to a CERT-KO cell line transformed with an empty vector, while expression of the G67E and ΔFFAT (d321-327) mutant genes did not rescue viral replication (Fig. 3B, left). Notably, infection with lentivirus alone restored HCV replication (Fig. 3B, left). It was confirmed that overexpression of CERT-wt rescued SM biosynthesis (Fig. 3B, right). We also analyzed the rescue of HCV replication in CERT-KO cells grown in medium supplemented with various types of SM. Supplementation of CERT-KO cells with N-[6-[(7-nitro-2-1,3-benzoxadiazol-4-yl)amino]hexanoyl]-sphingosine-1 phosphocholine (C6-SM) partially restored HCV replication, while supplementation with milk and egg SMs, supplied as mixtures of SMs that are purified from milk and egg, respectively, did not significantly restore replication (Fig. 3C, left); none of these SM sources had significant effects on cell viability at the used concentrations (Fig. 3C, right). Note that egg-SM has a mixture of SMs with fatty acid compositions of 16:0, 18:0, 22:0, and 24:1, while milk-SM has a mixture of SMs with fatty acid compositions of 16:0, 22:0, 23:0, 24:0, and 24:1. Use of N-(4,4-difluoro-5,7-dimethyl-4-bora-3a,4a-diaza-s-indacene-3 pentanoyl) sphingosyl phosphocholine (SM-BODIPY) restored the replication of the HCV replicon RNA in CERT-KO cells as assessed by luciferase levels (Fig. 3D, left), NS5A protein accumulation (Fig. 3E), and double-stranded RNA (dsRNA; a mediator and indicator of HCV replication) signal (Fig. 3F), and also permitted us to show that SM was colocalized with both NS5A and dsRNA (Fig. 3E and F, respectively) in both Huh7.5.1-8 cells and CERT-KO cells. In contrast, HCV replication was difficult to detect in untreated CERT-KO cells.
FIG 3.
Analyzing HCV replication after production of CERT protein or exogenous SM supplementation in CERT-KO cells. (A) CERT-KO cells were transfected with CERT as the intact protein (CERT-wt), as a mutant lacking PI4P binding (CERT-G67E), or as a mutant lacking VAP binding [CERT-(d321-327)], and with pEF321swxneo containing the G418 resistance gene, and were cultured in G418-containing medium. Cells were seeded in 24-well plates and transfected with JFH1/SGR RNA, and luciferase activities were measured at 4 h and 72 h. Fold changes in HCV replication were calculated as in Fig. 2C, and the results were plotted as a percentage of control. CERT expression was analyzed by WB using anti-CERT antibodies. (B) Lentiviruses were constructed in 293T cells either as empty virus (mock control) or encoding CERT-wt, CERT-G67E, or CERT-(d321-327). Spent medium from each line (i.e., containing each of the viruses) was collected and titrated for viral number; equalized titers were used to infect Huh7.5.1-8 (empty virus only) and CERT-KO cells (empty and CERT protein-encoding viruses). The infected cells were selected on G418 for approximately 2 weeks. Then, cells were seeded in 24-well plates and transfected with JFH1/SGR RNA, and luciferase activities were measured at 4 h and 72 h. Fold changes in HCV replication were calculated as in Fig. 2C, and the results were plotted as a percentage of control. CERT expression was analyzed by WB using anti-CERT antibodies (left). The levels of SM were analyzed by TLC after 14C-serine metabolic labeling in the indicated conditions (right panel). (C) Huh7.5.1-8 cells and CERT-KO cells were incubated for 48 h with ethanol (indicated as zero) or with the indicated SM types, including milk-SM, egg-SM, or C6-SM. Then either mock-treated or treated CERT-KO cells or mock-treated Huh7.5.1-8 cells were electroporated with JFH1/SGR RNA and luciferase activities were measured at 4 and 48 h; fold changes were calculated as in Fig. 2D (C, left). Cell viability was evaluated using a Cell TiterGlo luminescent cell viability assay (C, right). (D, E, and F) Huh7.5.1-8 cells and CERT-KO cells were electroporated with JFH1/SGR RNA, and then the transfected Huh7.5.1-8 cells or CERT-KO cells were treated with 0.02% ethanol [SM(−)] or 1 μM N-(4,4-difluoro-5,7-dimethyl-4-bora-3a,4a-diaza-s-indacene-3 pentanoyl) sphingosyl phosphocholine (SM-BODIPY [SM(+)]). (D) HCV replication activities were analyzed as for panel B, including measurement and analysis of luciferase activities at 4 and 48 h posttransfection (D, left) and determination of cell viability (D, right). (E) Cells were fixed, and viral replication complexes were stained with fluorescently labeled anti-NS5A antibodies (red); nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI; blue). The SM was localized by the use of SM-BODIPY (green). (F) Cells were fixed, and viral replication complexes were stained with fluorescently labeled anti-dsRNA (a mediator and indicator of HCV replication) antibodies (red); nuclei were stained with DAPI (blue). The panels on the left present low-magnification overviews; the boxed areas are enlarged in the corresponding panels on the right. Quantities for the degree of colocalization (overlapping area/red area) are given at the top of the enlarged pictures and are indicated as means ± SDs. Values were obtained from quadruplicate wells in at least three independent experiments and are presented as means ± SDs. ***, P < 0.005; **, P < 0.01; *, P < 0.05 (two-sided Student’s t test).
Inhibition of SM biosynthesis in CERT-KO cells decreases the number of replication vesicles.
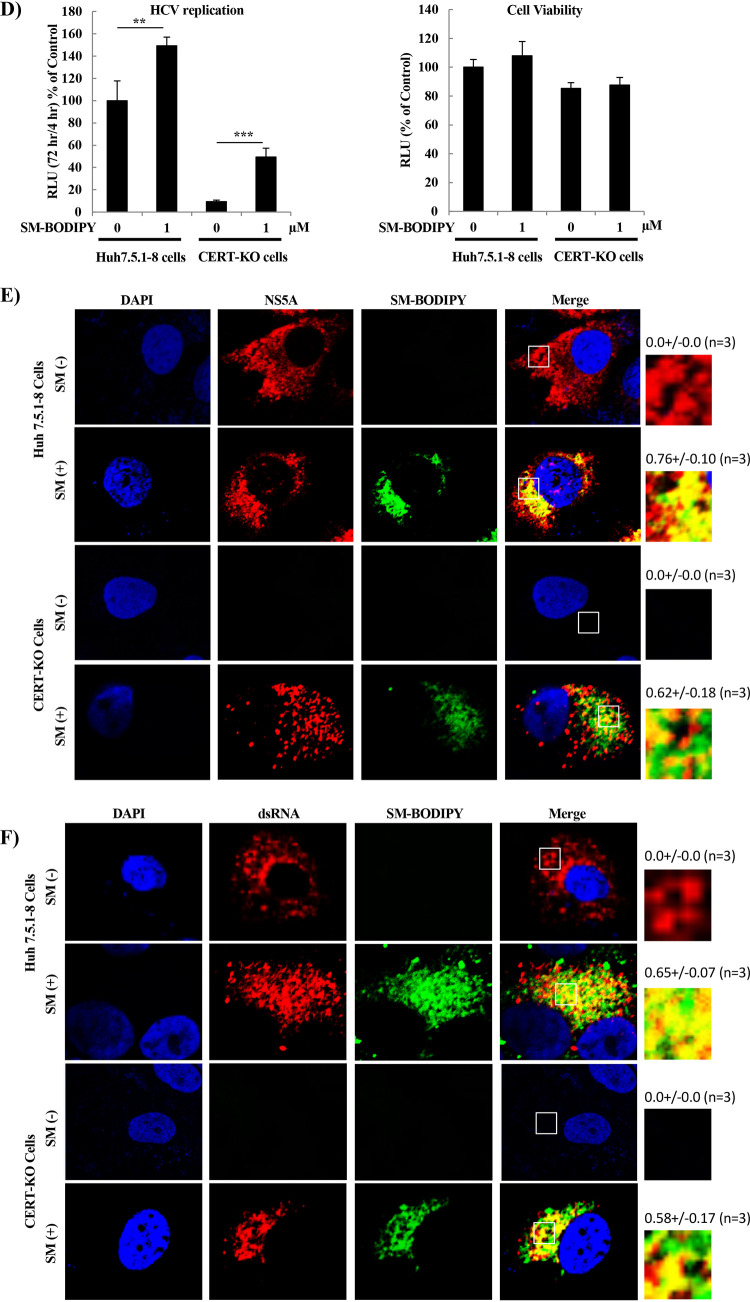
In the above-described work, we showed that SM is necessary for HCV replication. In previous work, we and others have reported that HCV replication occurs in lipid raft-associated membranes enriched for cholesterol and SM (31, 32). Several previous reports hypothesized that cholesterol, the partner of SM in lipid rafts, is required for the stability of DMVs, a major site of viral replication (15, 40). Other papers have shown that inhibition of SM biosynthesis by serine palmitoyltransferase inhibitors suppresses HCV replication (29, 35, 42, 43), but there is no evidence of the importance of SM for the biosynthesis of membrane replication factories. To investigate the importance of SM for DMV formation, we performed analysis using a transmission electron microscope (TEM). The numbers and sizes of DMVs in JFH1/SGR cells treated with D609 were determined and compared to the same parameters in untreated control cells. As shown in Fig. 4A, the numbers of DMVs were significantly decreased in D609-treated cells compared to those in cells subjected to the control treatment. Similar results were obtained in CERT-KO cells compared to control cells (Fig. 4B). As shown in Fig. 4C, the numbers of DMVs of various sizes, especially those of 100 to 200 nm in diameter, were significantly decreased in CERT-KO cells compared to control cells.
FIG 4.
Effect of inhibition of SM biosynthesis by either D609 or in CERT-KO cells on the number of induced membrane replication vesicles. (A) JFH1/SGR were treated with D609 (150 μM), and after 72 h, cells were examined using transmission electron microscopy (TEM). (B) Huh7.5.1-8 cells and CERT-KO cells were electroporated with JFH1/SGR RNA and analyzed by TEM at 48 h. (C) The numbers and sizes of the membrane vesicles in the boxed areas were quantified and plotted. Values were obtained from three samples and represent means ± SDs. (D and E) Huh7.5.1-8 cells and CERT-KO cells were infected with AdexCAT7, a recombinant adenovirus encoding the bacteriophage T7 RNA polymerase, and Huh7.5.1-8 cells were infected with AdexCAwt, a mock virus containing the promoter sequence but lacking the gene encoding the T7 RNA polymerase. All cells then were transfected with pSGRlucneoGND, an HCV construct that encodes a mutant NS5B protein harboring a GND-inactivating mutation. (D) At 3 days posttransfection, accumulation of NS5A protein was analyzed by WB using anti-NS5A antibodies (lower); luciferase activities were measured and analyzed as in Fig. 2C (upper), and (E) cells were fixed and examined by TEM.
To confirm that the reduced numbers of DMVs are a direct result of SM inhibition and not a consequence of the suppression of HCV replication, a replication-independent system for HCV protein expression was used to induce DMV formation. As shown in Fig. 4D, Huh7.5.1-8 cells and CERT-KO cells showed comparable levels of protein expression when assessed for luciferase activity (Fig. 4D, upper) or for NS5A expression by WB analysis (Fig. 4D, lower). As shown in Fig. 4E, the numbers of DMVs induced in CERT-KO cells were significantly decreased compared to those in control Hu7.5.1-8 cells.
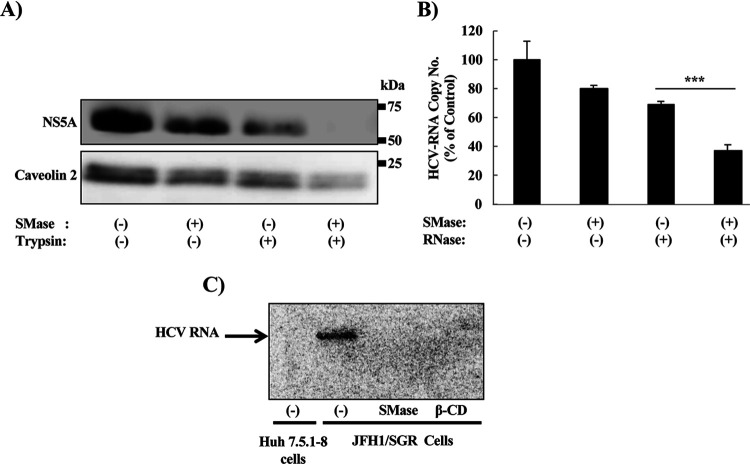
Degradation of SM into ceramide by sphingomyelinase causes deformation and shrinkage of DMVs.
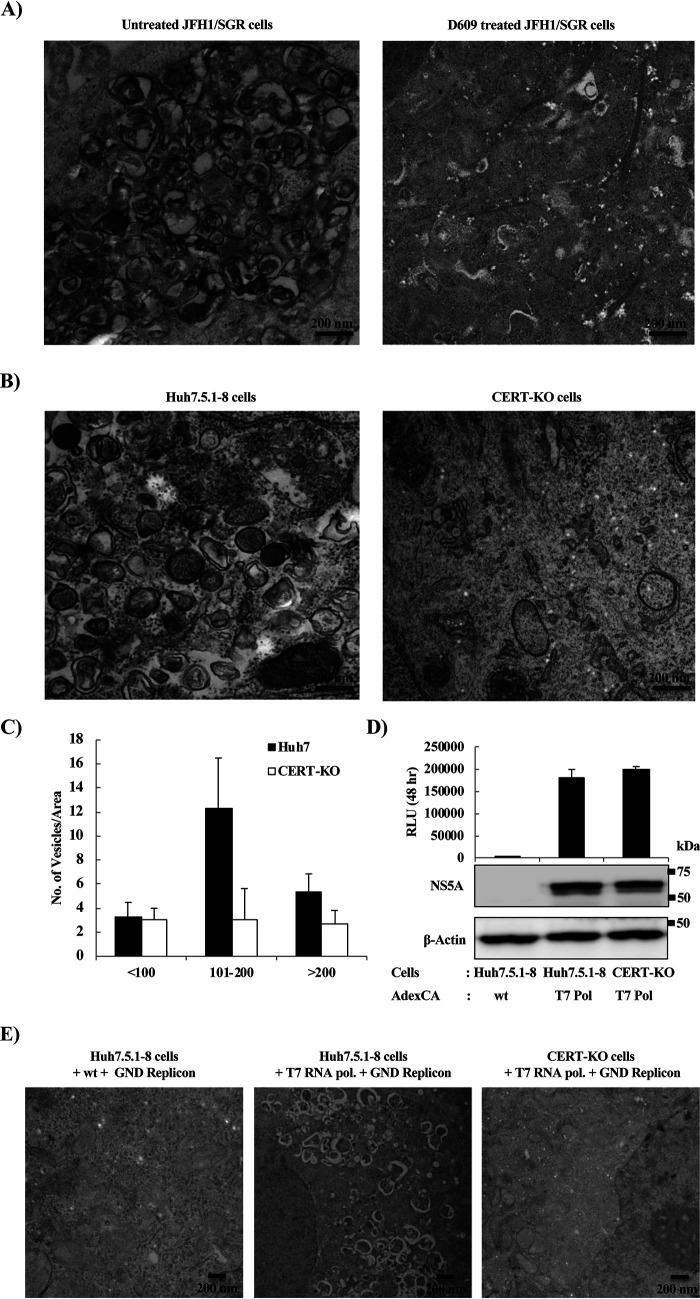
Another approach to analyzing the importance of SM in the biosynthesis of DMVs was to analyze the effect on DMV morphology and function of degradation of SM. We first isolated vesicles by detergent treatment and membrane flotation. As previously reported, the HCV RNA replication complex is associated with detergent-resistant membranes (DRMs) known as lipid rafts. As expected, immunoblotting analysis (Fig. 5A) showed that caveolin 2, a marker of the DRM fraction, cofractionated in the top fraction (fraction 1) with NS5A, a marker of the HCV replication complex; in contrast, calnexin, a marker of the detergent-soluble membrane (DSM) fraction, localized primarily in the bottom fractions (fractions 7, 8, and 9).
FIG 5.
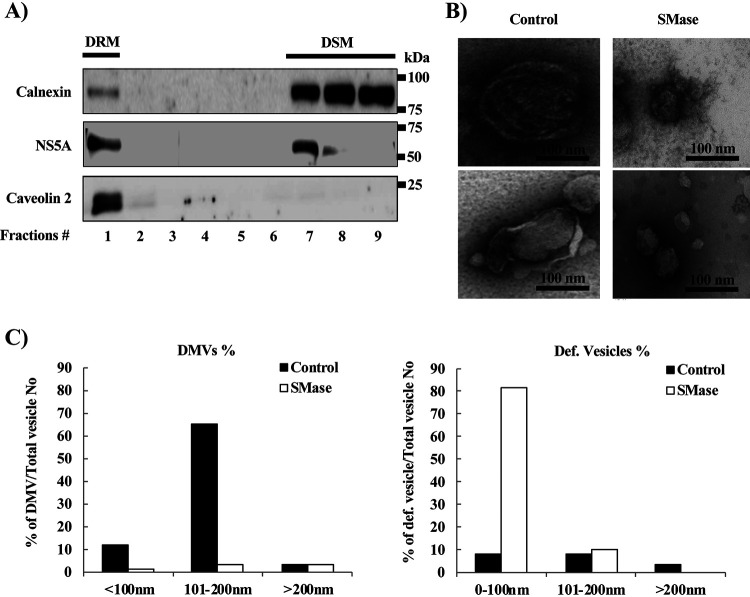
Effect of degradation of SM by sphingomyelinase (SMase) on the morphology of DMVs. JFH1/SGR cell lysates were treated with 1% Triton X-100 for 60 min on ice and fractionated by discontinuous sucrose gradient centrifugation. (A) Equal volumes of the recovered concentrated fractions were analyzed by WB using antibodies against NS5A, calnexin, or caveolin 2. Fractions are numbered from 1 to 9 in order from top to bottom. (B) The DRM fraction (fraction 1) from the experiment shown in panel A was subjected to SMase treatment (10 U/ml) and examined using TEM. Representative DMVs and deformed vesicle membrane structures are shown. (C) In the control or SMase-treated fractions, the percentages and diameters of DMVs (left) and deformed (Def.) vesicles (right) were determined for 200 membrane structures in each and plotted as a percentage of the total number of vesicles.
Observation of the DRM fraction by transmission electron microscopy (TEM) revealed vesicles with the known DMV morphology (Fig. 5B, left panels). On the other hand, degradation of SM into ceramide by treatment with sphingomyelinase (SMase) caused deformation and shrinkage of these DMVs (Fig. 5B right panels). After counting the DMVs and measuring their diameters, we observed that a significant percentage of the DMVs had a mean diameter of 100 to 200 nm (Fig. 5C, left), whereas after SMase treatment, most of the DMVs disappeared; deformed membranes with a mean diameter of less than 100 nm become the predominant structure (Fig. 5C, right). These data suggested that SMase treatment caused deformation as well as shrinkage of the DMVs.
SMase treatment of DRM fraction increases the sensitivity of HCV RNA and proteins to RNase and protease treatment, respectively, and inhibits active replication.
HCV-induced membrane replication factories have been reported to protect viral RNA and proteins involved in the replication complex from degradation by cellular nucleases and proteases, respectively (40, 44). Indeed, the degradation of SM in the DRM fraction (isolated in Fig. 5A) by SMase increased the sensitivity of NS5A to protease treatment (Fig. 6A) and the sensitivity of HCV RNA to RNase treatment (Fig. 6B). Active replication of HCV in the isolated replication complex was inhibited by SMase treatment; the degree of inhibition was comparable to that observed with the positive-control treatment, methyl-β-cyclodextrin (β-CD), a compound known to extract cholesterol from the membranes (Fig. 6C).
FIG 6.
Effect of degradation of SM content by SMase on the sensitivity of HCV RNA and proteins to RNase and protease treatment, respectively, and on the active replication of HCV. DRM fraction (fraction 1) from Fig. 5A was subjected to SMase treatment (10 U/ml). Equal amounts of proteins from the DRM fractions purified in the experiment in Fig. 5A were subjected or not to treatment with SMase (10 U/ml) at 37°C for 2 h and then analyzed as follows: (A) with or without trypsin digestion (25 μg/ml) at 37°C for 10 min, with protein levels analyzed by WB using anti-NS5A and anti-caveolin 2 antibodies, or (B) with or without RNase A treatment (250 ng/ml) at 37°C for 5 min, with HCV RNA levels quantified by real-time RT-PCR. Real-time RT-PCR values were obtained from quadruplicate wells in two independent experiments and represent means ± SD. (C) Cell lysates of Huh7.5.1-8 cells or JFH1/SGR cells were incubated with SMase, β-cyclodextrin (β-CD), or control, then incubated with [32P]-CTP for 90 min. The RNA from these lysates was purified and fractionated by agarose gel electrophoresis in Tris-acetate-EDTA buffer. The gel was dried and exposed overnight to X-ray film. Values were obtained from quadruplicate wells in at least three independent experiments and are presented as means ± SDs. ***, P < 0.005 (two-sided Student’s t test).
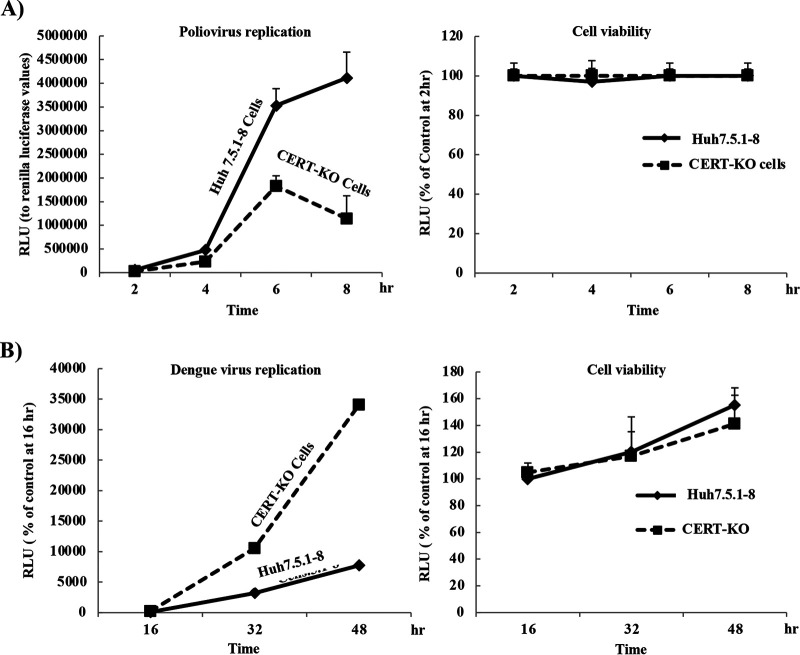
Replication of poliovirus is downregulated in CERT-KO cells, while that of DENV is upregulated.
As seen with HCV, replication of a poliovirus replicon was decreased at all tested time points (Fig. 7A, left) in CERT-KO cells compared to that in Huh7.5.1-8 cells (the parental cell line from which CERT-KO cells were derived); as noted previously, the two infected cell lines did not exhibit significant differences in cell viability (Fig. 7A, right). Interestingly, replication of a DENV replicon was increased approximately 3- to 4-fold in CERT-KO cells compared to that in the control cells at all tested time points, especially at 32 and 48 h (Fig. 7B, left); again, no significant difference in cell viability was observed in the two cell lines (Fig. 7B, right).
FIG 7.
Replication of poliovirus and dengue virus in CERT-KO cells. (A) Either Huh7.5.1-8 cells or CERT-KO cells were transfected with in vitro-transcribed poliovirus replicon RNA, and then replication was analyzed by measurement of luciferase activities at 2, 4, 6, and 8 h posttransfection; the data were normalized to Renilla luciferase values (right). Cell viability was evaluated using a Cell TiterGlo luminescent cell viability assay (left). (B) Huh7.5.1-8 cells and CERT-KO cells were transfected with a DNA-based dengue virus replicon (DGL2) or with an equivalent replicon encoding a NS5 protein harboring a mutation (DGL2-mut). GLuci activities in the medium then were measured at 16, 32, and 48 h posttransfection and the resulting luciferase values were normalized to those at 16 h (left panel). Cell viability was evaluated using a Cell TiterGlo luminescent cell viability assay (right). Values represent means ± SDs obtained from quadruplicate wells in at least three independent experiments.
DISCUSSION
Previous reports assumed that SM is essential for HCV replication, but the mechanism of SM’s involvement in the process was not clear. In this study, we showed for the first time that SM is essential for the biosynthesis of the membrane replication factories, the site of viral replication.
To assess the importance of SM to HCV replication, we used two methods to inhibit SM biosynthesis; one by using small molecule inhibitors of SM biosynthesis pathways and another by KO of CERT, a key player in SM biosynthesis pathways. We showed that suppression of HCV replication by three inhibitors of SM biosynthesis, namely myriocin, fumonisin B1, and D609, was concentration dependent and genotype independent. Notably, all three compounds inhibited SGR replication of HCV of both genotypes 1b and 2a; the sole exception was that the effect of myriocin could not be analyzed on JFH1/SGR because of cell cytotoxicity (data not shown). However, all three SM biosynthesis inhibitors, including myriocin, suppressed genotype 2a replication in an HCV infection system (Fig. 1). These data were consistent with those of previous studies showing that inhibition of SM biosynthesis using small-molecule inhibitors suppressed HCV replication in a genotype-independent manner (43). This result suggested the existence of an additional role of SM in HCV replication, separate from the previous report that SM activates RdRp in a genotype-specific manner (29).
Suppression of HCV replication in different CERT-KO cell clones that showed significant reductions of SM levels at different time points gives another line of evidence of the importance of SM to HCV replication (Fig. 2C). Expression of CERT-wt in CERT-KO cells, which rescued SM biosynthesis, efficiently restored viral replication, indicating the importance of ceramide trafficking, from the ER to the Golgi, and that of SM to HCV replication (Fig. 3A). In contrast, the expression of the two mutant CERT forms that impaired ER-Golgi transport of ceramide, namely ΔFFAT (d321-327), which abolishes the binding of CERT to the ER-resident protein VAP, and the G67E mutation, which abolishes the PI4P-binding capacity of the PH domain, failed to rescue viral replication. These data confirm the requirement of ER to Golgi ceramide trafficking and SM to HCV replication. The suppression of HCV replication in CERT-KO cells was counteracted by supplementation with various kinds of SM as well (Fig. 3B). C6-SM and C5-SM-BODIPY (i.e., derivatives with the shortest-chain fatty acids) exhibited the most efficient rescue of HCV replication in CERT-KO cells (Fig. 3B and C). These data are consistent with previous reports showing that short-chain fatty acid SMs are imported through the cell membrane and accumulate inside the cells (45). Supplementation with SM rescued the signals for both NS5A and dsRNA, while SM localized with both NS5A and dsRNA (Fig. 3D and E). These results indicated a specific requirement for SM in HCV replication, along with the accumulation of SM at the site of viral replication. Some previous reports have suggested that ceramide accumulation suppresses the replication of certain RNA viruses (46, 47), while others have indicated that both SM and ceramide accumulate in HCV-infected cells (35). Consequently, it is possible that the inhibition of HCV replication in both CERT-KO cells and cells treated with D609, a direct inhibitor of SGMS, and that in myriocin- and fumonisin B1-treated cells might be attributed to accumulation or depletion of ceramide, respectively (Fig. 1A) (35). In the present work, inhibition of HCV replication was observed using any of three inhibitors of the SM biosynthesis pathways, which included the two inhibitors (myriocin and fumonisin B1) that inhibit steps upstream of ceramide synthesis and another inhibitor (D609, a direct inhibitor of SGMS) that inhibits a step downstream of ceramide synthesis; an effect on the suppression in HCV replication also was seen in cells harboring a KO for the genes encoding CERT, a protein that transfers CERT from the ER to the Golgi, as well (Fig. 1A). Together, these results demonstrated that the observed suppression of HCV replication was not due to ceramide accumulation or depletion. Additionally, the rescue of HCV replication in CERT-KO cells by exogenous supplementation with SM confirmed that inhibition of HCV replication results from decreased SM content rather than from ceramide accumulation (Fig. 3B to E). These data were consistent with the results of Hirata et al. (35), a report that indicated that SM levels were upregulated in humanized chimeric mouse livers and human hepatocytes upon infection with HCV and that also showed that SM was concentrated in the DRM fractions in which HCV replicates. This report also showed that our data represent strong evidence that SM is required for HCV replication. It was previously reported that the exogenously supplied short-chain SM (either C6-SM or C5-SM-BODIPY) are directed more effectively to the plasma membrane and the Golgi, while the long-chain SMs (either egg-SM or milk-SM) are transported along the late endocytic pathway and recycled back (45). This may explain why C6-SM and C5-SM could restore the replication of HCV in CERT-KO cells, whereas egg-SM or milk-SM could not rescue the HCV replication in these cells.
Previous reports indicated that membrane replication vesicles are enriched in the DRM fraction of HCV-infected cells (31–35). Therefore, membrane vesicles containing replication complexes in this DRM fraction were roughly purified and analyzed. Our data showed that the degradation of the SM in these vesicles resulted in deterioration of the vesicles and shrinkage of their size (Fig. 5B and C). Previous work reported that depletion of cholesterol from purified DMVs resulted in decreased DMV diameters (40). These data suggested that both SM and cholesterol, the major components of lipid rafts, are essential structural components of HCV DMVs, consistent with previous reports showing that the membrane replication factories are composed of lipid raft-associated membranes enriched in cholesterol and SM (31–34). Inhibition of RNA replication activity by degradation of SM in isolated vesicles or by treatment of the isolated vesicles with β-CD (a compound that extracts cholesterol from the membrane) supported the same conclusion (Fig. 6C). It is believed that the membrane replication factories protect the contained viral replication complex from cellular innate immune responses by limiting the replication complex’s interaction with the pattern recognition receptors and also by protecting the replication complex from intracellular RNase and protease activities (5, 15, 48). Our data showed that the degradation of SM in purified vesicles increased the sensitivity of HCV RNA and proteins to RNase and protease treatments, respectively (Fig. 6A and B). The SM content of the isolated membranes correlated with protection of the replication complex in DMVs. Approximately 30% of HCV RNA was degraded by RNase treatment alone, and more than 60% of HCV RNA was degraded by treatment with both RNase and SMase (Fig. 6B), supporting the previously proposed localization of the HCV replication process to the outer and inner parts of the membrane replication vesicles (40). This result suggested that one of the proposed functions of the membrane replication factory is to increase the local concentration of the factors required for the replication process (5, 15). Collectively, the degradation of SM of the isolated vesicles by SMase treatment affected the morphology, size, and function of the DMVs. SM degradation of the isolated vesicles not only resulted in a change of the morphology from DMVs to deformed vesicles and the reduction of the size (Fig. 5B and C), but the protective function of these vesicles was affected as well (Fig. 6).
CERT has previously been reported to be involved in the HCV life cycle (49), but the detailed mechanism of CERT’s role in virus replication is unknown. We constructed genes encoding CERT-wt (that is, the wild-type protein) or either of two mutant versions of the CERT protein, ΔFFAT (d321-327), harboring a mutation that abolishes binding to the ER-resident VAP protein, and the G67E mutant, in which the PI4P-binding activity of PH domain is abolished, yielding impairment of Golgi trafficking of ceramide (27). Compared to control CERT-KO cells that were infected with empty lentivirus, CERT-KO cells engineered to overproduce CERT-wt showed a significant rescue of HCV replication, an effect that was not seen in CERT-KO cells overproducing ΔFFAT (d321-327) or the G67E mutant CERT (Fig. 3A). However, it has been demonstrated in an experiment with semi-intact CHO cells that overexpression of the G67E or ΔFFAT mutations can restore SM synthesis (22). In Fig. 3A, the rescue of HCV replication in CERT-KO cells infected with a control lentiviral vector may reflect the induction of autophagosomal membranes by lentiviral infection. Several reports have shown (using large-scale small RNA interference [RNAi] screens, stable knockdown of some autophagic cofactors, or inhibition of autophagy) that lentivirus infection induces autophagy and biogenesis of autophagosomal DMVs (50–54), which presumably facilitates HCV replication (33, 55, 56). Although PI4P and SM, which are essential for HCV replication, are enriched in the Golgi, several studies have reported that DMVs are derived from the ER (7, 57, 58). Therefore, the identity of the parental membrane from which DMVs are derived remains unclear. We propose two models. In the first model, CERT combines ER and Golgi membranes at a membrane contact site to form DMVs, because CERT has binding sites for both VAP (an ER protein) and PI4P (a Golgi component). In the second model, an entirely new membrane containing both ER and Golgi components is formed. The latter model is consistent with the replication of HCV occurring on both the outside and the inside of the membrane vesicles (Fig. 6B). Several recent publications have reported that HCV induces autophagy to enhance the replication process (56, 59–61) and that replication occurs in autophagosomal DMVs (33, 55, 56). Proteomic analysis of proteins associated with HCV-induced autophagosomes has identified several proteins associated with lipid rafts and confirmed the involvement of cholesterol in such vesicles (33). Further analysis is required to connect the role of SM and autophagosomes in the construction of DMVs.
The requirement for SM is not unique to HCV viral replication. It is specific for DMV-inducing viruses, such as HCV and poliovirus (Fig. 7A), since infection with viruses that induce invaginated vesicles, such as DENV, did not require SM in their replication (Fig. 7B). The inhibitory effect of SM on DENV replication shown in Fig. 7B has been reported in several previous studies. Either myriocin or fumonisin B1 treatment enhanced DENV replication (47), while genome-wide analysis showed that SGMS is a significant host factor required for HCV infection but not for DENV infection (62). The inhibitory mechanism of SM to DENV replication has not been examined yet (47). Others reported that sphingolipids, especially ceramide, may play a protective or antiviral role by the host cell against some other viral infection, such as those caused by influenza A virus and HIV (46, 63). The mechanism underlying this protective role is unidentified, but it may represent a target for discovering new therapies for some viral infections. It is noteworthy that DMV induction by HCV or poliovirus shares the requirement for PI4P and oxysterol binding protein (OSBP), suggesting an evolutionarily conserved mechanism (64). Some reports have shown that DENV replication does not require cholesterol, OSBP, or PI4P (65), a pattern that is distinct from those of both HCV and poliovirus. Further analysis will be needed to understand the differences of the morphological forms of membrane replication vesicles and cell types.
In summary, we have demonstrated that SM is an essential constituent of HCV DMVs and in their function; SM appears to participate in the biosynthesis of membrane replication factories by contributing to the integrity and function of these structures. DMVs are considered abnormal organelles, given that these structures do not exist under normal conditions. Thus, DMVs represent diagnostic and prognostic markers for infection by many positive-stranded viruses, and additionally may serve as excellent therapeutic targets.
MATERIALS AND METHODS
Chemicals and enzymes.
Lovastatin (Sigma-Aldrich, St. Louis, MO), D609 (Santa Cruz Biotechnology, Dallas, TX), myriocin (ISP-1; Santa Cruz Biotechnology), and fumonisin B1 (derived from Fusarium moniliforme; Sigma-Aldrich) were dissolved in nuclease-free water (NSFW) and dimethyl sulfoxide (DMSO) to generate stock solutions; neat water and 0.05% DMSO, respectively, were used as a negative control for treatment with these reagents. Sphingomyelin analogues, including SM-BODIPY (Thermo Fisher, Waltham, MA), C6-SM, milk-SM, and egg-SM (Avanti Polar Lipids, Alabaster, AL), were dissolved in absolute ethanol to generate stock solutions; 0.02% ethanol was used as a negative control for treatment with these reagents. After optimization by serial dilution (data not shown), the highest concentration of each SM type that caused no significant cell toxicity (10 μM C6-SM, 2 μM SM-BODIPY, 400 μM milk-SM, or 400 μM egg-SM) was used in the presented experiments. Incubations with SM-BODIPY were carried out in serum-free medium in the dark (45). Methyl-β-cyclodextrin (β-CD; Sigma-Aldrich) was dissolved in NSFW. SMase derived from Bacillus cereus was obtained from Sigma-Aldrich. RNase A was obtained from Ambion (Austin, TX); trypsin was obtained from Invitrogen (Carlsbad, CA).
Cell culture.
Huh7.5.1-8 cells (37) and 293T human embryonic kidney cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (FBS). LN#2 cells (38) or JFH1/SGR cells (41) were maintained in DMEM containing 10% FBS supplemented with 0.5 mg/ml G418 (38).
Construction of CERT-KO cell lines.
Starting on day 0, Huh7.5.1-8 cells (1.5 × 105 cells/well in 12-well plates) were cultured overnight. On day 1, the cells were incubated with CRISPR plasmid mixed with Polyethylenimine Max (Polysciences, Inc., Warrington, PA) according to the manufacturer’s instructions. On day 2, the cells were transferred to a 24-well plate and cultured at 37°C in medium supplemented with 2 μg/ml of puromycin. The medium was replaced with puromycin-free medium at day 5, and the cells were maintained in this medium for 1 week. The cells then were subjected to limiting dilution to isolate gene-disrupted clones. The resulting Huh7.5.1-8 CR-CERTv1 Clones 1, 5, 7, and 11 (obtained using the CERTver1 guide sequence) and Huh7.5.1-8 CR-CERTv2 Clone 2 (obtained using the CERTver2 guide sequence) (CERT-KO cells) were subjected to further culturing before being stored at −80°C until subsequent analysis. Insertion/deletion analysis was performed as previously described (66). All CERT-KO cells were maintained in DMEM containing 10% FBS.
Plasmid construction and lentivirus construction and transduction.
The plasmids pJFH1 (containing the full-length HCV genotype-2a JFH1 strain cDNA) (67) and pSGRlucneo (containing the JFH1/SGR), and pSGRlucneoGND (containing JFH1/SGR with an inactivating mutation within the sequences encoding NS5B) have been described previously (41), as have a poliovirus replicon encoding firefly luciferase (Fluci) in place of the capsid genes (68) and a DNA-based Gaussia luciferase (GLuci)-encoding DENV replicon (DGL2) (69). Template plasmids were used for PCR amplification of the sequences encoding CERT and mutants thereof (G67E and ΔFFAT [d321-327]) (28). The amplified fragments were cloned (separately) into the pLVSIN-EF1α Neo vector using the In-Fusion HD cloning kit (TaKaRa Bio, Mountain View, CA). All constructs were verified by DNA sequencing.
Establishment of stable cell lines expressing wild or mutant CERT in CERT-KO cells and effects on HCV replication.
To establish cell lines that stably express wild or mutant CERT, CERT-KO cells were transfected with CERT-wt (that is, the wild-type protein), the G67E mutant, or the ΔFFAT (d321-327) mutant and pEF321swxneo containing the G418 resistance gene, and were cultured in G418-containing medium as described previously (70). G418-resistent colonies were clonally isolated and screened for abilities to express CERT proteins by indirect immunofluorescence staining. On the other hand, lentiviral production of CERT-wt was carried out using TransIT1 transfection reagent (Mirus, Madison, WI) to transfect 293T cells with a mixture of pMDL (Riken BRC, Tsukuba, Japan), VSVG (Riken BRC), pREV (Riken BRC), and the effector plasmids. Two days after transfection, cell culture supernatants were collected, centrifuged to remove cell debris, and then aliquoted and stored at −80°C for further analysis. CERT-KO cells were infected with various concentrations of lentiviral vectors encoding CERT-wt or its mutant forms, and then the cells were subjected to neomycin selection by incubation with G418 for 2 weeks. Cell lines with expression levels similar to those for endogenous CERT were selected for the experiments. Huh7.5.1-8 cells, CERT-KO cells, or Neo-selected CERT-KO cells transduced with lentivirus-CERT-wt or its mutant forms were transfected with in vitro-transcribed JFH1/SGR RNAs using a TransIT-mRNA transfection kit, and luciferase activities were then measured at 4 and 72 h. CERT protein expression levels were analyzed by WB using anti-CERT antibodies.
Antibodies.
Mouse monoclonal antibodies against β-actin, caveolin 2, and dsRNA were obtained from Sigma-Aldrich, BD Transduction Laboratories (San Jose, CA), and Biocenter, Ltd. (Szirák, Hungary), respectively. Rabbit polyclonal anti-calnexin antibodies were obtained from Stressgen Bioreagents (Victoria, BC, Canada). Anti-CERT antibodies were obtained from Abcam (Cambridge, UK). Rabbit polyclonal anti-NS5A antibodies were obtained as described elsewhere (71).
Immunoblots.
Immunoblotting was performed as reported previously (44).
Indirect immunofluorescence assay.
Huh7.5.1-8 cells and CERT-KO cells were transfected with JFH1/SGR RNA using a TransIT-mRNA transfection kit according to the manufacturer’s protocol and as previously described (44), with some modifications. Transfected cells were incubated with 2 μM (final concentration) SM-BODIPY in Opti-minimal essential medium (MEM) at 37°C in the dark, as described previously (45). Next, the cells were washed 2 times with ice-cold Hanks’ balanced salt solution (HBSS), and plasma membrane-bound SM-BODIPY was removed by 6 rounds of back exchange, each consisting of incubating cells in 5% fatty acid-free bovine serum albumin in ice-cold HBSS (6 times for 10 min each, on ice). The indirect immunofluorescence assay then was performed as reported previously (44).
Metabolic labeling of sphingolipids and TLC analysis.
Cells were seeded in the regular culture medium. After overnight incubation, the medium was replaced with Opti-MEM supplemented with 1% Neutridoma-SP (Roche), and the cells were incubated with 18.5 kBq of l-[U-14C] serine or 6.1 kBq of d-[1-14C] galactose for 16 h. The cells then were lysed with 0.1% sodium dodecyl sulfate (SDS), and total lipid was extracted with chloroform-methanol (1:2, vol/vol). The extracts were spotted onto silica gel 60 plates (Merck, Darmstadt, Germany) and chromatographed with methyl acetate–1-propanol chloroform-methanol-0.25% KCl (25:25:25:10:9, vol/vol). Extraction of lipids from cells and their separation by TLC were performed as described previously (27). The radioactive lipids on TLC plates were visualized using a Typhoon FLA 7000 instrument (GE Healthcare, Little Chalfont, UK).
HCV replication assays (luciferase and real-time RT-PCR).
For analyzing HCV replication, either LN#2 cells or JFH1/SGR cells (in which HCV was replicating) were treated with each of the various inhibitors of SM biosynthesis pathways or with lovastatin under the conditions described for each experiment. Luciferase activities were measured in cell lysates as described previously (44). The intracellular HCV RNA levels were measured by real-time RT-PCR, and HCV copy numbers were normalized to the total cellular RNA of each sample, as described previously (67).
JFH1/SGR RNA and Renilla luciferase (Rluci)-encoding RNA (Promega, Madison, WI) were synthesized by in vitro transcription as described previously (67). Huh7.5.1-8 cells were electroporated with 10 μg of JFH1/SGR RNA and 1 μg of Rluci RNA, with the latter serving as an internal control. For analysis of RNA translation, the RNA-transfected cells were harvested to determine Fluci and Rluci activities at 4 h posttransfection by use of a dual-luciferase reporter assay system (Promega) following the manufacturer’s instructions. The relative luciferase unit (RLU) values were measured with a luminometer (Berthold, Wildbad, Germany), and Fluci activity was normalized to Rluci activity. For analysis of RNA replication, transfected cells were harvested to determine the JFH1/SGR RNA levels at 24, 48, and 72 h posttransfection. Plasmid pJFH1 was used to generate HCVcc in Huh7.5.1-8 cells as described previously (67).
The restoration of SM in CERT-KO cells was performed either by engineered production of CERT protein or by supplementation with exogenous SM. CERT protein production was provided by transfection of Huh7.5.1-8 cells or CERT-KO cells with in vitro-transcribed JFH1/SGR RNA using a TransIT-mRNA transfection kit. Transfection of JFH1/SGR RNA was performed according to the manufacturer’s protocol, with minor modifications. In brief, 24 h after seeding the cells in 48-well plates, a mixture (per well) of 26 μl Opti-MEM, 0.14 μg JFH1/SGR RNA, 0.14 μl mRNA Boost reagent and 0.28 μl TransIT-mRNA reagent was incubated together for 2 min before being distributed to each well and incubated for the indicated time intervals.
DENV replication assay and GLuci assay.
DENV replication was assessed using a DNA-based replicon employing internal ribosome entry site (IRES)-dependent translation and encoding a version of the GLuci protein that was secreted into the culture medium (69). The DENV-replicon construct was transfected into either Huh7.5.1-8 cells or CERT-KO cells, as described previously, and GLuci activities in the supernatants were analyzed using the Renilla luciferase assay system kit (Promega) as described previously (72).
Cell viability.
Cell viability was analyzed using a CellTiter-Glo luminescent cell viability assay (Promega) according to the manufacturer’s protocol.
Replication-independent system (expression of HCV proteins under the control of the T7-promoter).
Huh7.5.1-8 cells and CERT-KO cells were infected with AdexCAT7, a recombinant adenovirus encoding the bacteriophage T7 RNA polymerase, and AdexCAwt, a control virus containing only the promoter sequence and lacking the T7 RNA polymerase-encoding gene, as described previously (73). pSGRlucneoGND was then transfected into the infected cells using the TransIT transfection reagent.
Membrane flotation assay.
The membrane flotation assay was performed as described previously (44).
RNase and protease sensitivity assays.
DRM extracted from the cells was used to analyze the sensitivity of HCV RNA and proteins to RNase and protease treatments, respectively. For analysis of RNA sensitivity to RNase, the DRM sample was treated with RNase A (500 ng/ml) at 37°C for 5 min, and RNA then was extracted using a Qiagen extraction kit; HCV RNA levels were determined by real-time RT-PCR. For analysis of HCV protein sensitivity to protease, the DRM sample was treated with 25 μg/ml trypsin (Invitrogen) at 37°C for 10 min, and the protein levels then were determined by immunoblot analysis using antibodies specific for NS5A and caveolin 2.
Ultrastructural analysis.
Cells were fixed with 2.5% glutaraldehyde in 0.1 M phosphate buffer (pH 7.4) for 30 min at room temperature; the cells then were scraped from the plastic plate into a fixative solution in which the cells were incubated for another 90 min at 4°C. The cells then were postfixed with 1% OsO4 in phosphate buffer (pH 7.4) for 2 h, embedded in 2% agar, dehydrated in ethanol, and embedded in Epon 812. Ultrathin sections were double stained with uranyl acetate and lead citrate and examined by TEM (H-7100; Hitachi, Ltd., Tokyo, Japan).
For examination of the isolated DRM fraction, either intact or following SMase treatment as described previously (38), the samples were applied to freshly glow-discharged carbon-coated 300-mesh Cu grids, and excess solution was blotted away before immersion of the grids in 10 μl of 1% uranyl acetate for 1 min. Excess stain then was removed by blotting, and the grids were air-dried. The percentages of DMVs and deformed vesicles in 200 membranes each in either control-treated or SMase-treated fractions were determined.
Statistical analysis.
Statistical analyses were performed using two-sided Student’s t tests assuming equal variance. The P values are indicated according to the following convention: P > 0.05 (nonsignificant, ns); P < 0.05 (*); P < 0.01 (**); P < 0.005 (***).
ACKNOWLEDGMENTS
We thank Y. Hirama for technical assistance. We also thank T. Miyamura for helpful discussions.
The Ministry of Health, Labor and Welfare of Japan provided funding to Hideki Aizaki (grants 10851601, 10850101, and 10850201). The Ministry of Education, Culture, Sports, Science and Technology of Japan provided funding to Hideki Aizaki (grant 15K09034). The Advanced Research & Development Programs for Medical Innovation (AMED, AMED-CREST) provided funding to Hideki Aizaki (grants 20fk0210066j0001, 20fk0210047j1102, and 20fk0310112j704) and Kentaro Hanada (grant JP19gm0910005).
We have no conflicts of interest to report.
REFERENCES
- 1.Lauer GM, Walker BD. 2001. Hepatitis C virus infection. N Engl J Med 345:41–52. doi: 10.1056/NEJM200107053450107. [DOI] [PubMed] [Google Scholar]
- 2.Petruzziello A, Marigliano S, Loquercio G, Cozzolino A, Cacciapuoti C. 2016. Global epidemiology of hepatitis C virus infection: an up-date of the distribution and circulation of hepatitis C virus genotypes. WJG 22:7824–7840. doi: 10.3748/wjg.v22.i34.7824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith DB, Bukh J, Kuiken C, Muerhoff AS, Rice CM, Stapleton JT, Simmonds P. 2014. Expanded classification of hepatitis C virus into 7 genotypes and 67 subtypes: updated criteria and genotype assignment web resource. Hepatology 59:318–327. doi: 10.1002/hep.26744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mayo MP. 1998. Virus taxonomy—1997. J Gen Virol 79:649–657. doi: 10.1099/0022-1317-79-4-649. [DOI] [PubMed] [Google Scholar]
- 5.Altan-Bonnet N. 2017. Lipid tales of viral replication and transmission. Trends Cell Biol 27:201–213. doi: 10.1016/j.tcb.2016.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shulla A, Randall G. 2016. (+) RNA virus replication compartments: a safe home for (most) viral replication. Curr Opin Microbiol 32:82–88. doi: 10.1016/j.mib.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Egger D, Wolk B, Gosert R, Bianchi L, Blum HE, Moradpour D, Bienz K. 2002. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J Virol 76:5974–5984. doi: 10.1128/jvi.76.12.5974-5984.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang H, Tai AW. 2016. Mechanisms of cellular membrane reorganization to support hepatitis C virus replication. Viruses 8:142. doi: 10.3390/v8050142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kikkert M. 2020. Innate immune evasion by human respiratory RNA viruses. J Innate Immun 12:4–20. doi: 10.1159/000503030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kao YT, Lai MMC, Yu CY. 2018. How dengue virus circumvents innate immunity. Front Immunol 9:2860. doi: 10.3389/fimmu.2018.02860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garcia-Sastre A. 2017. Ten strategies of interferon evasion by viruses. Cell Host Microbe 22:176–184. doi: 10.1016/j.chom.2017.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Romero-Brey I, Bartenschlager R. 2014. Membranous replication factories induced by plus-strand RNA viruses. Viruses 6:2826–2857. doi: 10.3390/v6072826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Belov GA, Nair V, Hansen BT, Hoyt FH, Fischer ER, Ehrenfeld E. 2012. Complex dynamic development of poliovirus membranous replication complexes. J Virol 86:302–312. doi: 10.1128/JVI.05937-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paul D, Bartenschlager R. 2015. Flaviviridae replication organelles: oh, what a tangled web we weave. Annu Rev Virol 2:289–310. doi: 10.1146/annurev-virology-100114-055007. [DOI] [PubMed] [Google Scholar]
- 15.Paul D, Madan V, Bartenschlager R. 2014. Hepatitis C virus RNA replication and assembly: living on the fat of the land. Cell Host Microbe 16:569–579. doi: 10.1016/j.chom.2014.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neufeldt CJ, Cortese M, Acosta EG, Bartenschlager R. 2018. Rewiring cellular networks by members of the Flaviviridae family. Nat Rev Microbiol 16:125–142. doi: 10.1038/nrmicro.2017.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peter Slotte J. 2013. Molecular properties of various structurally defined sphingomyelins – correlation of structure with function. Prog Lipid Res 52:206–219. doi: 10.1016/j.plipres.2012.12.001. [DOI] [PubMed] [Google Scholar]
- 18.Krishna A, Prakash S, Sengupta D. 2020. Sphingomyelin effects in caveolin-1 mediated membrane curvature. J Phys Chem B doi: 10.1021/acs.jpcb.0c02962. [DOI] [PubMed] [Google Scholar]
- 19.Sodt AJ, Venable RM, Lyman E, Pastor RW. 2016. Nonadditive compositional curvature energetics of lipid bilayers. Phys Rev Lett 117:138104. doi: 10.1103/PhysRevLett.117.138104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller ME, Adhikary S, Kolokoltsov AA, Davey RA. 2012. Ebolavirus requires acid sphingomyelinase activity and plasma membrane sphingomyelin for infection. J Virol 86:7473–7483. doi: 10.1128/JVI.00136-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brugger B, Glass B, Haberkant P, Leibrecht I, Wieland FT, Krausslich HG. 2006. The HIV lipidome: a raft with an unusual composition. Proc Natl Acad Sci U S A 103:2641–2646. doi: 10.1073/pnas.0511136103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taniguchi M, Tasaki T, Ninomiya H, Ueda Y, Kuremoto KI, Mitsutake S, Igarashi Y, Okazaki T, Takegami T. 2016. Sphingomyelin generated by sphingomyelin synthase 1 is involved in attachment and infection with Japanese encephalitis virus. Sci Rep 6:37829. doi: 10.1038/srep37829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang HB, Lu ZL, Wei XK, Zhong TZ, Zhong YZ, Ouyang LX, Luo Y, Xing XW, Liao F, Peng KK, Deng CQ, Minamoto N, Luo TR. 2016. Viperin inhibits rabies virus replication via reduced cholesterol and sphingomyelin and is regulated upstream by TLR4. Sci Rep 6:30529. doi: 10.1038/srep30529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Callens N, Brugger B, Bonnafous P, Drobecq H, Gerl MJ, Krey T, Roman-Sosa G, Rumenapf T, Lambert O, Dubuisson J, Rouille Y. 2016. Morphology and molecular composition of purified bovine viral diarrhea virus envelope. PLoS Pathog 12:e1005476. doi: 10.1371/journal.ppat.1005476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aizaki H, Morikawa K, Fukasawa M, Hara H, Inoue Y, Tani H, Saito K, Nishijima M, Hanada K, Matsuura Y, Lai MM, Miyamura T, Wakita T, Suzuki T. 2008. Critical role of virion-associated cholesterol and sphingolipid in hepatitis C virus infection. J Virol 82:5715–5724. doi: 10.1128/JVI.02530-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Syed GH, Amako Y, Siddiqui A. 2010. Hepatitis C virus hijacks host lipid metabolism. Trends Endocrinol Metab 21:33–40. doi: 10.1016/j.tem.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hanada K, Kumagai K, Yasuda S, Miura Y, Kawano M, Fukasawa M, Nishijima M. 2003. Molecular machinery for non-vesicular trafficking of ceramide. Nature 426:803–809. doi: 10.1038/nature02188. [DOI] [PubMed] [Google Scholar]
- 28.Kawano M, Kumagai K, Nishijima M, Hanada K. 2006. Efficient trafficking of ceramide from the endoplasmic reticulum to the Golgi apparatus requires a VAMP-associated protein-interacting FFAT motif of CERT. J Biol Chem 281:30279–30288. doi: 10.1074/jbc.M605032200. [DOI] [PubMed] [Google Scholar]
- 29.Weng L, Hirata Y, Arai M, Kohara M, Wakita T, Watashi K, Shimotohno K, He Y, Zhong J, Toyoda T. 2010. Sphingomyelin activates hepatitis C virus RNA polymerase in a genotype-specific manner. J Virol 84:11761–11770. doi: 10.1128/JVI.00638-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kong L, Aoyagi H, Yang Z, Ouyang T, Matsuda M, Fujimoto A, Watashi K, Suzuki R, Arita M, Yamagoe S, Dohmae N, Suzuki T, Suzuki T, Muramatsu M, Wakita T, Aizaki H. 2019. Surfeit 4 contributes to the replication of hepatitis C virus using double-membrane vesicles. J Virol 94. doi: 10.1128/JVI.00858-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aizaki H, Lee KJ, Sung VM, Ishiko H, Lai MM. 2004. Characterization of the hepatitis C virus RNA replication complex associated with lipid rafts. Virology 324:450–461. doi: 10.1016/j.virol.2004.03.034. [DOI] [PubMed] [Google Scholar]
- 32.Shi ST, Lee KJ, Aizaki H, Hwang SB, Lai MMC. 2003. Hepatitis C virus RNA replication occurs on a detergent-resistant membrane that cofractionates with caveolin-2. J Virol 77:4160–4168. doi: 10.1128/jvi.77.7.4160-4168.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim JY, Wang L, Lee J, Ou JJ. 2017. Hepatitis C virus induces the localization of lipid rafts to autophagosomes for its RNA replication. J Virol 91. doi: 10.1128/JVI.00541-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shanmugam S, Saravanabalaji D, Yi M, Diamond MS. 2015. Detergent-resistant membrane association of NS2 and E2 during hepatitis C virus replication. J Virol 89:4562–4574. doi: 10.1128/JVI.00123-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hirata Y, Ikeda K, Sudoh M, Tokunaga Y, Suzuki A, Weng L, Ohta M, Tobita Y, Okano K, Ozeki K, Kawasaki K, Tsukuda T, Katsume A, Aoki Y, Umehara T, Sekiguchi S, Toyoda T, Shimotohno K, Soga T, Nishijima M, Taguchi R, Kohara M. 2012. Self-enhancement of hepatitis C virus replication by promotion of specific sphingolipid biosynthesis. PLoS Pathog 8:e1002860. doi: 10.1371/journal.ppat.1002860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miyake Y, Kozutsumi Y, Nakamura S, Fujita T, Kawasaki T. 1995. Serine palmitoyltransferase is the primary target of a sphingosine-like immunosuppressant, ISP-1/myriocin. Biochem Biophys Res Commun 211:396–403. doi: 10.1006/bbrc.1995.1827. [DOI] [PubMed] [Google Scholar]
- 37.Shirasago Y, Sekizuka T, Saito K, Suzuki T, Wakita T, Hanada K, Kuroda M, Abe R, Fukasawa M. 2015. Isolation and characterization of an Huh.7.5.1-derived cell clone highly permissive to hepatitis C virus. Jpn J Infect Dis 68:81–88. doi: 10.7883/yoken.JJID.2014.231. [DOI] [PubMed] [Google Scholar]
- 38.Goto K, Watashi K, Murata T, Hishiki T, Hijikata M, Shimotohno K. 2006. Evaluation of the anti-hepatitis C virus effects of cyclophilin inhibitors, cyclosporin A, and NIM811. Biochem Biophys Res Commun 343:879–884. doi: 10.1016/j.bbrc.2006.03.059. [DOI] [PubMed] [Google Scholar]
- 39.Kitatani K, Idkowiak-Baldys J, Hannun YA. 2008. The sphingolipid salvage pathway in ceramide metabolism and signaling. Cell Signal 20:1010–1018. doi: 10.1016/j.cellsig.2007.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paul D, Hoppe S, Saher G, Krijnse-Locker J, Bartenschlager R. 2013. Morphological and biochemical characterization of the membranous hepatitis C virus replication compartment. J Virol 87:10612–10627. doi: 10.1128/JVI.01370-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matsumoto Y, Matsuura T, Aoyagi H, Matsuda M, Hmwe SS, Date T, Watanabe N, Watashi K, Suzuki R, Ichinose S, Wake K, Suzuki T, Miyamura T, Wakita T, Aizaki H. 2013. Antiviral activity of glycyrrhizin against hepatitis C virus in vitro. PLoS One 8:e68992. doi: 10.1371/journal.pone.0068992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Katsume A, Tokunaga Y, Hirata Y, Munakata T, Saito M, Hayashi H, Okamoto K, Ohmori Y, Kusanagi I, Fujiwara S, Tsukuda T, Aoki Y, Klumpp K, Tsukiyama-Kohara K, El-Gohary A, Sudoh M, Kohara M. 2013. A serine palmitoyltransferase inhibitor blocks hepatitis C virus replication in human hepatocytes. Gastroenterology 145:865–873. doi: 10.1053/j.gastro.2013.06.012. [DOI] [PubMed] [Google Scholar]
- 43.Sakamoto H, Okamoto K, Aoki M, Kato H, Katsume A, Ohta A, Tsukuda T, Shimma N, Aoki Y, Arisawa M, Kohara M, Sudoh M. 2005. Host sphingolipid biosynthesis as a target for hepatitis C virus therapy. Nat Chem Biol 1:333–337. doi: 10.1038/nchembio742. [DOI] [PubMed] [Google Scholar]
- 44.Kong L, Fujimoto A, Nakamura M, Aoyagi H, Matsuda M, Watashi K, Suzuki R, Arita M, Yamagoe S, Dohmae N, Suzuki T, Sakamaki Y, Ichinose S, Suzuki T, Wakita T, Aizaki H, Ou JHJ. 2016. Prolactin regulatory element binding protein is involved in hepatitis C virus replication by interaction with NS4B. J Virol 90:3093–3111. doi: 10.1128/JVI.01540-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nusshold C, Uellen A, Bernhart E, Hammer A, Damm S, Wintersperger A, Reicher H, Hermetter A, Malle E, Sattler W. 2013. Endocytosis and intracellular processing of BODIPY-sphingomyelin by murine CATH.a neurons. Biochim Biophys Acta 1831:1665–1678. doi: 10.1016/j.bbalip.2013.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Soudani N, Hage-Sleiman R, Karam W, Dbaibo G, Zaraket H. 2019. Ceramide suppresses influenza A virus replication in vitro. J Virol 93:e00053-19. doi: 10.1128/JVI.00053-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aktepe TE, Pham H, Mackenzie JM. 2015. Differential utilisation of ceramide during replication of the flaviviruses West Nile and dengue virus. Virology 484:241–250. doi: 10.1016/j.virol.2015.06.015. [DOI] [PubMed] [Google Scholar]
- 48.Neufeldt CJ, Joyce MA, Van Buuren N, Levin A, Kirkegaard K, Gale M Jr, Tyrrell DL, Wozniak RW. 2016. The hepatitis C virus-induced membranous web and associated nuclear transport machinery limit access of pattern recognition receptors to viral replication sites. PLoS Pathog 12:e1005428. doi: 10.1371/journal.ppat.1005428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Amako Y, Syed GH, Siddiqui A. 2011. Protein kinase D negatively regulates hepatitis C virus secretion through phosphorylation of oxysterol-binding protein and ceramide transfer protein. J Biol Chem 286:11265–11274. doi: 10.1074/jbc.M110.182097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brass AL, Dykxhoorn DM, Benita Y, Yan N, Engelman A, Xavier RJ, Lieberman J, Elledge SJ. 2008. Identification of host proteins required for HIV infection through a functional genomic screen. Science 319:921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- 51.Espert L, Varbanov M, Robert-Hebmann V, Sagnier S, Robbins I, Sanchez F, Lafont V, Biard-Piechaczyk M. 2009. Differential role of autophagy in CD4 T cells and macrophages during X4 and R5 HIV-1 infection. PLoS One 4:e5787. doi: 10.1371/journal.pone.0005787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eekels JJ, Sagnier S, Geerts D, Jeeninga RE, Biard-Piechaczyk M, Berkhout B. 2012. Inhibition of HIV-1 replication with stable RNAi-mediated knockdown of autophagy factors. Virol J 9:69. doi: 10.1186/1743-422X-9-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang X, Gao Y, Tan J, Devadas K, Ragupathy V, Takeda K, Zhao J, Hewlett I. 2012. HIV-1 and HIV-2 infections induce autophagy in Jurkat and CD4+ T cells. Cell Signal 24:1414–1419. doi: 10.1016/j.cellsig.2012.02.016. [DOI] [PubMed] [Google Scholar]
- 54.Campbell GR, Bruckman RS, Herns SD, Joshi S, Durden DL, Spector SA. 2018. Induction of autophagy by PI3K/MTOR and PI3K/MTOR/BRD4 inhibitors suppresses HIV-1 replication. J Biol Chem 293:5808–5820. doi: 10.1074/jbc.RA118.002353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang L, Kim JY, Liu HM, Lai MMC, Ou JJ. 2017. HCV-induced autophagosomes are generated via homotypic fusion of phagophores that mediate HCV RNA replication. PLoS Pathog 13:e1006609. doi: 10.1371/journal.ppat.1006609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sir D, Kuo CF, Tian Y, Liu HM, Huang EJ, Jung JU, Machida K, Ou JH. 2012. Replication of hepatitis C virus RNA on autophagosomal membranes. J Biol Chem 287:18036–18043. doi: 10.1074/jbc.M111.320085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gosert R, Egger D, Lohmann V, Bartenschlager R, Blum HE, Bienz K, Moradpour D. 2003. Identification of the hepatitis C virus RNA replication complex in Huh-7 cells harboring subgenomic replicons. J Virol 77:5487–5492. doi: 10.1128/jvi.77.9.5487-5492.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Romero-Brey I, Merz A, Chiramel A, Lee JY, Chlanda P, Haselman U, Santarella-Mellwig R, Habermann A, Hoppe S, Kallis S, Walther P, Antony C, Krijnse-Locker J, Bartenschlager R. 2012. Three-dimensional architecture and biogenesis of membrane structures associated with hepatitis C virus replication. PLoS Pathog 8:e1003056. doi: 10.1371/journal.ppat.1003056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dreux M, Gastaminza P, Wieland SF, Chisari FV. 2009. The autophagy machinery is required to initiate hepatitis C virus replication. Proc Natl Acad Sci U S A 106:14046–14051. doi: 10.1073/pnas.0907344106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tanida I, Fukasawa M, Ueno T, Kominami E, Wakita T, Hanada K. 2009. Knockdown of autophagy-related gene decreases the production of infectious hepatitis C virus particles. Autophagy 5:937–945. doi: 10.4161/auto.5.7.9243. [DOI] [PubMed] [Google Scholar]
- 61.Fahmy AM, Labonte P. 2017. The autophagy elongation complex (ATG5-12/16L1) positively regulates HCV replication and is required for wild-type membranous web formation. Sci Rep 7:40351. doi: 10.1038/srep40351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Marceau CD, Puschnik AS, Majzoub K, Ooi YS, Brewer SM, Fuchs G, Swaminathan K, Mata MA, Elias JE, Sarnow P, Carette JE. 2016. Genetic dissection of Flaviviridae host factors through genome-scale CRISPR screens. Nature 535:159–163. doi: 10.1038/nature18631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Finnegan CM, Rawat SS, Puri A, Wang JM, Ruscetti FW, Blumenthal R. 2004. Ceramide, a target for antiretroviral therapy. Proc Natl Acad Sci U S A 101:15452–15457. doi: 10.1073/pnas.0402874101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Altan-Bonnet N, Balla T. 2012. Phosphatidylinositol 4-kinases: hostages harnessed to build panviral replication platforms. Trends Biochem Sci 37:293–302. doi: 10.1016/j.tibs.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang H, Perry JW, Lauring AS, Neddermann P, De Francesco R, Tai AW. 2014. Oxysterol-binding protein is a phosphatidylinositol 4-kinase effector required for HCV replication membrane integrity and cholesterol trafficking. Gastroenterology 146:1373–85e1-11. doi: 10.1053/j.gastro.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yamaji T, Hanada K. 2014. Establishment of HeLa cell mutants deficient in sphingolipid-related genes using TALENs. PLoS One 9:e88124. doi: 10.1371/journal.pone.0088124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Krausslich HG, Mizokami M, Bartenschlager R, Liang TJ. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med 11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Arita M, Philipov S, Galabov AS. 2015. Phosphatidylinositol 4-kinase III beta is the target of oxoglaucine and pachypodol (Ro 09–0179) for their anti-poliovirus activities, and is located at upstream of the target step of brefeldin A. Microbiol Immunol 59:338–347. doi: 10.1111/1348-0421.12261. [DOI] [PubMed] [Google Scholar]
- 69.Kato F, Kobayashi T, Tajima S, Takasaki T, Miura T, Igarashi T, Hishiki T. 2014. Development of a novel dengue-1 virus replicon system expressing secretory Gaussia luciferase for analysis of viral replication and discovery of antiviral drugs. Jpn J Infect Dis 67:209–212. doi: 10.7883/yoken.67.209. [DOI] [PubMed] [Google Scholar]
- 70.Aizaki H, Harada T, Otsuka M, Seki N, Matsuda M, Li YW, Kawakami H, Matsuura Y, Miyamura T, Suzuki T. 2002. Expression profiling of liver cell lines expressing entire or parts of hepatitis C virus open reading frame. Hepatology 36:1431–1438. doi: 10.1053/jhep.2002.36937. [DOI] [PubMed] [Google Scholar]
- 71.Suzuki R, Saito K, Kato T, Shirakura M, Akazawa D, Ishii K, Aizaki H, Kanegae Y, Matsuura Y, Saito I, Wakita T, Suzuki T. 2012. Trans-complemented hepatitis C virus particles as a versatile tool for study of virus assembly and infection. Virology 432:29–38. doi: 10.1016/j.virol.2012.05.033. [DOI] [PubMed] [Google Scholar]
- 72.Matsuda M, Suzuki R, Kataoka C, Watashi K, Aizaki H, Kato N, Matsuura Y, Suzuki T, Wakita T. 2014. Alternative endocytosis pathway for productive entry of hepatitis C virus. J Gen Virol 95:2658–2667. doi: 10.1099/vir.0.068528-0. [DOI] [PubMed] [Google Scholar]
- 73.Aoki Y, Aizaki H, Shimoike T, Tani H, Ishii K, Saito I, Matsuura Y, Miyamura T. 1998. A human liver cell line exhibits efficient translation of HCV RNAs produced by a recombinant adenovirus expressing T7 RNA polymerase. Virology 250:140–150. doi: 10.1006/viro.1998.9361. [DOI] [PubMed] [Google Scholar]