Oncolytic viruses kill tumors without affecting normal cells. A variety of oncolytic viruses are used as cancer therapeutics. Mammalian reovirus (MRV), which belongs to the genus Orthoreovirus, family Reoviridae, is one such natural oncolytic virus. The anticancer effects of MRV are being evaluated in clinical trials. Unlike other oncolytic viruses, MRV has not been genetically modified for use as a cancer therapeutic in clinical trials. Here, we used a reverse genetic approach to introduce an integrin-affinity peptide sequence into the MRV cell attachment protein σ1 to alter the natural tropism of the virus. The recombinant viruses were able to infect cancer cell lines expressing very low levels of the MRV entry receptor, junctional adhesion molecule A (JAM-A), and cause tumor cell death while maintaining its original tropism via JAM-A. This is a novel report of a genetically modified oncolytic MRV by introducing a peptide sequence into σ1.
KEYWORDS: mammalian reovirus, oncolytic virotherapy, reverse genetics system
ABSTRACT
Mammalian reovirus (MRV) strain type 3 Dearing (T3D) is a naturally occurring oncolytic virus that has been developed as a potential cancer therapeutic. However, MRV treatment cannot be applied to cancer cells expressing low levels of junctional adhesion molecule A (JAM-A), which is the entry receptor of MRV. In this study, we developed a reverse genetics system for MRV strain T3D-L, which showed high oncolytic potency. To modify the cell tropism of MRV, an arginine–glycine–aspartic acid (RGD) peptide with an affinity to integrin was inserted at the C terminus or loop structures of the viral cell attachment protein σ1. The recombinant RGD σ1-modified viruses induced remarkable cell lysis in human cancer cell lines with marginal JAM-A expression and in JAM-A knockout cancer cell lines generated by a CRISPR/Cas9 system. Pretreatment of cells with anti-integrin antibody decreased cell death caused by the RGD σ1-modified virus, suggesting the infection to the cells was via a specific interaction with integrin αV. By using mouse models, we assessed virulence of the RGD σ1-modified viruses in vivo. This system will open new avenues for the use of genetically modified oncolytic MRV for use as a cancer therapy.
IMPORTANCE Oncolytic viruses kill tumors without affecting normal cells. A variety of oncolytic viruses are used as cancer therapeutics. Mammalian reovirus (MRV), which belongs to the genus Orthoreovirus, family Reoviridae, is one such natural oncolytic virus. The anticancer effects of MRV are being evaluated in clinical trials. Unlike other oncolytic viruses, MRV has not been genetically modified for use as a cancer therapeutic in clinical trials. Here, we used a reverse genetic approach to introduce an integrin-affinity peptide sequence into the MRV cell attachment protein σ1 to alter the natural tropism of the virus. The recombinant viruses were able to infect cancer cell lines expressing very low levels of the MRV entry receptor, junctional adhesion molecule A (JAM-A), and cause tumor cell death while maintaining its original tropism via JAM-A. This is a novel report of a genetically modified oncolytic MRV by introducing a peptide sequence into σ1.
INTRODUCTION
Oncolytic virotherapy is an approach to treat tumors using viruses that replicate in and kill tumor cells. Viruses belonging to a number of families, including Adenoviridae, Herpesviridae, Paramyxoviridae, Parvoviridae, Picornaviridae, Poxviridae, Reoviridae, and Retroviridae have been used to develop cancer therapeutics (1–3). To improve the cancer therapeutic efficacy and reduce potential toxic effects of oncolytic viruses, reverse genetics systems have been used to manipulate the viral genome of many oncolytic viruses. For example, talimogene laherparepvec (or T-VEC, trade name Imlygic), an oncolytic herpesvirus, was generated by deleting the viral ICP34.5 and ICP47 genes, which are involved in tumor-specific replication, in addition to inserting GM-CSF (granulocyte/macrophage colony-stimulating factor) into the ICP34.5 locus (4). GM-CSF amplifies antitumor immune responses induced by the virus. Another example is recombinant adenovirus Tasadenoturev (trade name DNX-2401, formerly known as AdΔ24-RGD); in this case, a high-affinity integrin-binding (arginine–glycine–aspartic acid [RGD]) peptide was introduced into the fiber protein to alter virus tropism (5). Adenovirus binds to the coxsackievirus adenovirus receptor (CAR) via the cell attachment fiber protein (6). The recombinant adenovirus (harboring an RGD peptide within the fiber protein) was able to infect endothelial cells and smooth muscle cells, both of which express low levels of CAR (7).
Mammalian reovirus (MRV; genus Orthoreovirus, family Reoviridae) possesses a 10-segment double-stranded RNA (dsRNA) genome that is classified according to size: L, M, and S (8). Three prototypic serotypes of MRV (serotypes 1 [T1], 2 [T2], and 3 [T3]) have been identified by hemagglutination inhibition and neutralization tests (8). Three MRV strains, type 1 Lang (T1L), type 2 Jones, and type 3 Dearing (T3D), have been characterized in vitro and in vivo, providing insight into the molecular mechanisms underlying replication and pathogenesis of this family (8). T3D strains from different laboratories show different capacities for replication in cell culture, pathogenesis in vivo, and oncolysis due to nucleotide polymorphisms (9–13). Among the strains, a T3D strain with oncolysis has been studied for use as a cancer therapeutic. In humans, MRV infection is normally asymptomatic; however, the T3D strain replicates effectively in cancer cells in which the Ras signaling pathway is activated, thereby triggering tumor cell death (14–20). Thus, the effects of oncolytic virotherapy using the wild-type T3D strain (Pelareorep; trade name Reolysin) have been studied, or are currently being evaluated in clinical trials (21). Reolysin was granted orphan drug registration by the U.S. Food and Drug Administration and the European Medicines Agency for treatment of gastric cancer, ovarian cancer, and pancreatic cancer (22).
Entry of MRV into cells is initiated by the interaction between cell attachment protein σ1 and the host cell surface sialic acid (SA) and junctional adhesion molecule A (JAM-A) (23–26). The σ1 protein, encoded by the S1 gene segment, comprises an N-terminal tail, body, and C-terminal head domains (27–29). After low-affinity binding to SA via the body domain, σ1 binds to JAM-A via the head domain (25, 26). This interaction is followed by an internalization of the virion by endocytosis. Proteolytic disassembly of the outer capsid releases the infectious subvirion particle into the cytoplasm, where the virus replicates (8). Since JAM-A plays a critical role in MRV entry, very low/lack of JAM-A expression is a potential barrier to cancer therapy using MRV. To overcome this problem, previous studies identified mutations in the tail domain of σ1 that confer SA-dependent infection to cells (30, 31). In addition to altering tropism, the mutation facilitated the introduction of exogenous genes into the C-terminal head domain of σ1, which is required for its binding to JAM-A. By employing the mutation in the tail domain, replication-competent MRV-expressing iLOV, UnaG, E4orf4, or GM-CSF in place of the σ1 head domain have been generated (32–35). Infection by these MRVs relies on the expression of SA on the cell surface. Interestingly, reovirus σ1 and the adenovirus fiber protein share structural similarities even though these viruses belong to different families. Both proteins form homotrimers with a fibrous tail and a head containing triple β-spiral motifs (28, 36). The receptors for MRV and adenovirus, JAM-A and CAR, respectively, are immunoglobulin superfamily-like proteins that form homodimers (37–39). This similarity indicates that genetic modification of σ1 may allow us to modify the cellular tropism of MRV for cancer cells, as demonstrated for recombinant adenoviruses encoding genetically modified fiber proteins (5). Introduction of an RGD peptide into the σ1 protein would provide another means to modify tropism of reovirus; however, generation of reovirus harboring RGD in σ1 have not been reported.
Here, we developed an reverse genetics system for highly oncolytic MRV strain T3D (T3D-L) and then modified its tissue tropism. To expand the utility of MRV as a cancer therapeutic, we attempted to generate recombinant T3D-L that can infect cancer cells in a JAM-A-independent manner via cell surface molecules. An integrin αV-binding RGD peptide was introduced into the cell attachment protein σ1 to increase reovirus-mediated gene transduction into tumors lacking JAM-A. Analysis of several RGD σ1-modified MRVs revealed that they infected and killed reovirus-resistant cancer cells lacking JAM-A much more efficiently than the wild-type virus. In addition, these mutants may be useful oncolytic agents that can be used to treat tumors inaccessible to wild-type reoviruses. The data suggest that genetically modified oncolytic MRV systems can be used as effective cancer therapeutics.
RESULTS
Oncolytic activity of MRV strains T1L, T3D-D, and T3D-L against human cancer cell lines.
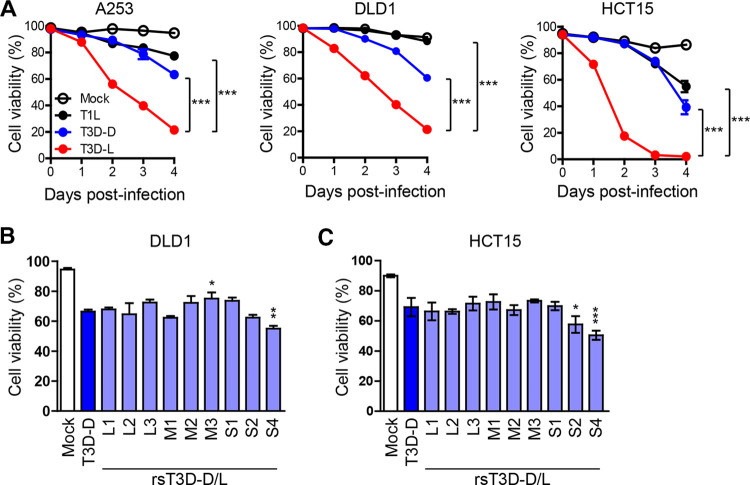
All tested reovirus serotypes show oncolytic activity in tumor cells (40, 41). Among them, a T3D strain has been evaluated in clinical trials as a potential oncolytic agent (42–48). Recent studies show that laboratory strains of T3D have different phenotypes with respect to viral replication and pathogenesis (9–13). Therefore, we assessed the oncolytic activity of T3D-D (obtained from Terence S. Dermody laboratory), T3D-L (obtained from Patrick W. K. Lee), and T1L strains. Human cancer cell lines A253, DLD1, and HCT15 were infected with T1L, T3D-D, or T3D-L at a multiplicity of infection (MOI) of 30 PFU/cell and cell viability was monitored for 4 days postinfection. Oncolysis caused by T3D-L was significantly higher than that caused by T1L or T3D-D (Fig. 1A). These results suggest that T3D-L has stronger oncolytic activity than other MRV strains, and that viral gene segment(s) derived from T3D-L play a role in increased oncolysis. Previously, we developed reverse genetics systems for the T1L and T3D-D strains (49, 50), but not for the T3D-L strain. First, we sequenced the entire viral genome of the T3D-L strain. Comparison of this sequence with that of T3D-D revealed 44 nucleotide substitutions in the genome of T3D-L. Importantly, these amino acid substitutions were observed in only eight of the gene segments (none were found in the S2 and S3 gene segments) (Table 1). These results suggest that certain amino acids play a critical role in the higher oncolytic activity of T3D-L, and that the S2 and S3 gene segments are unlikely to be involved. We also compared whole-genome sequence of T3D-L with that of Reolysin, which is a derivative of T3D-L and is being evaluated in clinical trials (51). Sequencing results revealed seven nucleotide substitutions between strain T3D-L and Reolysin; only one of these nucleotide substitutions is a nonsynonymous change (Table 2). To construct rescue plasmids for generation of recombinant strain T3D-L (rsT3D-L), each of the 10 gene segment cDNAs derived from T3D-L were flanked by the T7 promoter and hepatitis delta virus (HDV) ribozyme sequences (49). To identify the viral gene segment(s) responsible for the increased oncolysis demonstrated by strain T3D-L, we generated monoreassortant viruses by introducing one gene segment from strain T3D-L into the genetic background of strain T3D-D (rsT3D-D/L-L1, -L2, -L3, -M1, -M2, -M3, -S1, -S2, and -S4), and then infected DLD1 and HCT15 cancer cell lines with either T3D-D or monoreassortant viruses. We then examined cell viability at 3 or 4 days postinfection. Among the monoreassortant viruses tested, rsT3D-D/L-S2 showed higher oncolytic activity than rsT3D-D in HCT15 cells, and rsT3D-D/L-S4 showed increased oncolytic activity in DLD1 and HCT15 cells (Fig. 1B and C). Collectively, these data suggest that S4 gene segment plays a role in highly oncolysis by T3D-L in both cancer cell lines.
FIG 1.
Comparative analysis of the oncolytic activity of different MRV strains in cancer cell lines. (A) A253, DLD1, and HCT15 cells were infected with T1L, T3D-D, and T3D-L (MOI = 30 PFU/cell). Cells were then collected and stained with propidium iodide (PI) prior to flow cytometry analysis. Cell viability was determined by calculating the ratio of PI-negative cells to total cells. Each value represents an average from triplicate samples. The error bars indicate the standard deviation (SD). Representative results from three independent experiments are shown. Significant differences were determined using two-way ANOVA; ***, P < 0.001. (B and C) Oncolytic activity of T3D-D/T3D-L monoreassortant viruses in cancer cell lines. DLD1 (B) and HCT15 (C) cells were infected with T3D-D/T3D-L monoreassortant viruses at an MOI of 10 PFU/cell. The cells were collected 3 days after infection and viability was measured after PI staining. Each value represents an average from triplicate samples. The error bars indicate the standard deviation. Representative results from two independent experiments are shown. Significance differences were determined by one-way ANOVA; *, P < 0.05; **, P < 0.01; ***, P < 0.001.
TABLE 1.
Nucleotide and amino acid differences between T3D-L and T3D-D
| Gene segment (protein) | Nucleotide |
Amino acid |
||||
|---|---|---|---|---|---|---|
| Position (nt) | T3D-D | T3D-L | Position (aa) | T3D-D | T3D-L | |
| L1 (λ3) | 9–14 | TTCCAC | deletion | |||
| 1440 | G | A | ||||
| 2205 | A | G | ||||
| 2959 | A | C | 979 | M | L | |
| 2973 | A | G | 1045 | S | R | |
| 3159 | A | C | 1048 | N | S | |
| 3167 | A | G | ||||
| 3237 | T | G | ||||
| L2 (λ2) | 7 | A | T | |||
| 1524 | G | A | 504 | G | E | |
| 1538 | G | C | 509 | G | R | |
| 2251 | C | T | ||||
| 3079 | C | T | ||||
| L3 (λ1) | 1512 | T | G | 500 | I | S |
| 2347 | T | C | ||||
| 2569 | G | T | 852 | Q | H | |
| M1 (μ2) | 247 | A | G | |||
| 462 | A | G | 150 | Q | R | |
| 635 | T | C | 208 | S | P | |
| 985 | G | T | ||||
| 1038 | G | A | 342 | R | Q | |
| 1228 | T | C | ||||
| 1454 | C | T | ||||
| 1595 | G | T | 528 | A | S | |
| 2284 | A | G | ||||
| M2 (μ1) | 248 | A | C | 73 | E | D |
| 1173 | T | C | ||||
| 1808 | T | C | ||||
| M3 (μNS) | 357 | T | C | |||
| 556 | A | G | 180 | K | E | |
| 1908 | T | C | ||||
| 2133 | C | T | 705 | A | V | |
| 2139 | G | A | 707 | G | D | |
| S1 (σ1) | 77 | T | C | 22 | V | A |
| 438 | C | T | ||||
| 504 | A | G | ||||
| 1234 | A | G | 408 | T | A | |
| S1 (σ1s) | 77 | T | C | 3 | Y | H |
| S2 (σ2) | 702 | C | T | |||
| S3 (σNS)a | ||||||
| S4 (σ3) | 74 | G | A | |||
| 206 | C | T | ||||
| 429 | T | C | 133 | W | R | |
| 624 | G | A | 198 | G | K | |
| 625 | G | A | ||||
| 719 | G | T | 229 | E | D | |
Nucleotide substitution was not observed in the S3 gene segment.
TABLE 2.
Nucleotide and amino acid differences between T3D-L and Reolysin
| Gene segment (protein) | Nucleotide |
Amino acid |
||||
|---|---|---|---|---|---|---|
| Position (nt) | Reolysin | T3D-L | Position (aa) | Reolysin | T3D-L | |
| L1 (λ3) | 3071 | A | T | 1018 | Q | L |
| L2 (λ2)a | ||||||
| L3 (λ1)a | ||||||
| M1 (μ2) | 247 | A | G | |||
| 2284 | A | G | ||||
| M2 (μ1) | 1808 | T | C | |||
| M3 (μNS) | 357 | T | C | |||
| 1908 | T | C | ||||
| S1 (σ1)a | ||||||
| S1 (σ1s)a | ||||||
| S2 (σ2) | 1306 | A | C | |||
| S3 (σNS)a | ||||||
| S4 (σ3)a | ||||||
Nucleotide substitution was not observed in the L2, L3, S1, S3, nor S4 gene segments.
Generation of rsT3D-L from cloned viral cDNAs.
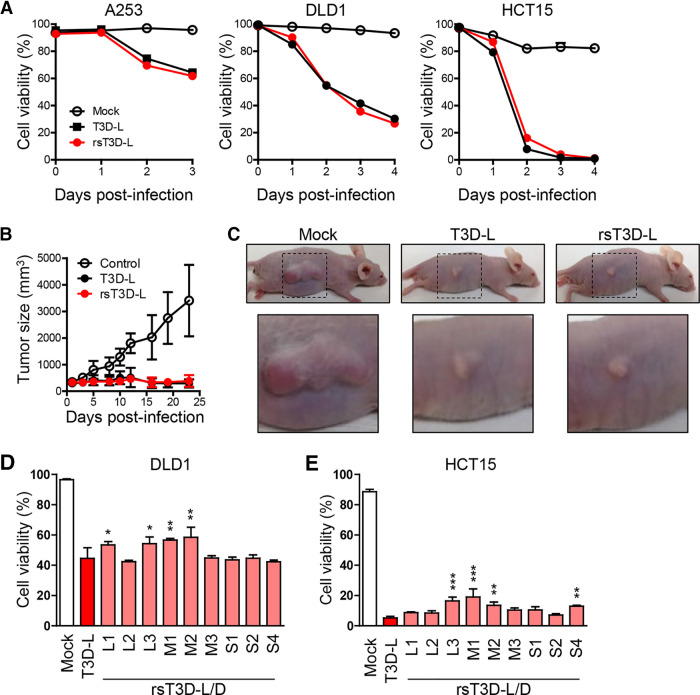
Since oncolysis by the T3D-D-based monoreassortant viruses was not as high as that by the T3D-L strain, we generated a complete rsT3D-L using the cDNAs derived from the T3D-L strain. The rsT3D-L was recovered by transfection of 10 rescue plasmids, each coding 10 gene segments of the T3D-L in L929 cells infected with vaccinia virus rDIs-T7pol expressing T7 RNA polymerase. To confirm whether rsT3D-L viruses have the same phenotype as wild-type T3D-L, we infected human cancer cell lines A253, DLD1, and HCT15 with either wild-type or recombinant virus. We then monitored cell viability for 3 or 4 days postinfection. We found that the oncolytic activity of rsT3D-L in these three tumor cell lines was similar to that of native T3D-L (Fig. 2A). These data suggest that rsT3D-L retains the oncolytic activity of the parent strain (T3D-L) in vitro. Next, we examined the oncolytic activity of native T3D-L and rsT3D-L in vivo. A253 cells were subcutaneously transplanted into BALB/cAJcl-nu nude mice, followed by intratumoral injection of either T3D-L or rsT3D-L (5 × 107 PFU). Injections were repeated every 2 to 3 days, and changes in tumor size were monitored for 3 weeks. The results showed that tumors injected with either T3D-L or rsT3D-L did not grow, although mock-injected tumor cells did (Fig. 2B and C). These results indicate that the oncolytic activity of rsT3D-L is indistinguishable from that of native T3D-L in a mouse xenograft model. To identify which viral gene segment(s) is irrelevant to increased oncolysis by T3D-L, we generated monoreassortant viruses on the genetic background of strain T3D-L. rsT3D-L/D-L1, -L3, -M1, and -M2 showed reduced oncolytic activity in DLD1 cells, and rsT3D-L/D-L3, -M1, -M2, and -S4 showed reduced oncolytic activity in HCT15 cells, indicating that the L1, L3, M1, M2, and S4 gene segments play a role in the reduced oncolytic activity of T3D-D (Fig. 2D and E). Taken together, the results suggest that replacement of a single gene does not fully explain differences between the oncolytic activity of T3D-L and T3D-D, implying that a combination of several gene segments is critical for oncolytic activity.
FIG 2.
Reverse genetics approach for T3D-L. (A) Oncolytic activity of T3D-L and rsT3D-L in A253, DLD1 and HCT15 cells. Cells were infected with viruses at an MOI of 10 PFU/cell for A253 and 30 PFU/cell for DLD1 and HCT15, then collected and stained with propidium iodide (PI), and subjected to flow cytometry analysis. Cell viability was determined by calculating the ratio of PI-negative cells to total cells. Each value represents an average from triplicate samples. The error bars indicate the standard deviation. Representative results from three independent experiments are shown. (B) Oncolytic activity of T3D-L and rsT3D-L in a mouse xenograft model. A253 cells were transplanted into 4-week-old BALB/cAJcl-nu nude mice. After tumors developed, mice received an intratumoral injection of T3D-L (n = 6), rsT3D-L (n = 6), or the same volume of DMEM (n = 6), every 2 to 3 days. Tumor growth was monitored for 23 days. Each value represents the average volume (six tumors). (C) Representative images showing tumor size at 14 days. (D and E) Oncolytic activity of T3D-L/T3D-D monoreassortant viruses in cancer cell lines. DLD1 (D) and HCT15 (E) cells were infected with T3D-L/T3D-D monoreassortant viruses at an MOI of 10 PFU/cell. The cells were collected 3 days after infection. Cell viability was determined by PI staining. Each value represents an average from triplicate samples. The error bars indicate the standard deviation. Representative results from two independent experiments are shown. The significance of differences was determined by one-way ANOVA; *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Generation of RGD σ1-modifed viruses showing altered cell tropism.
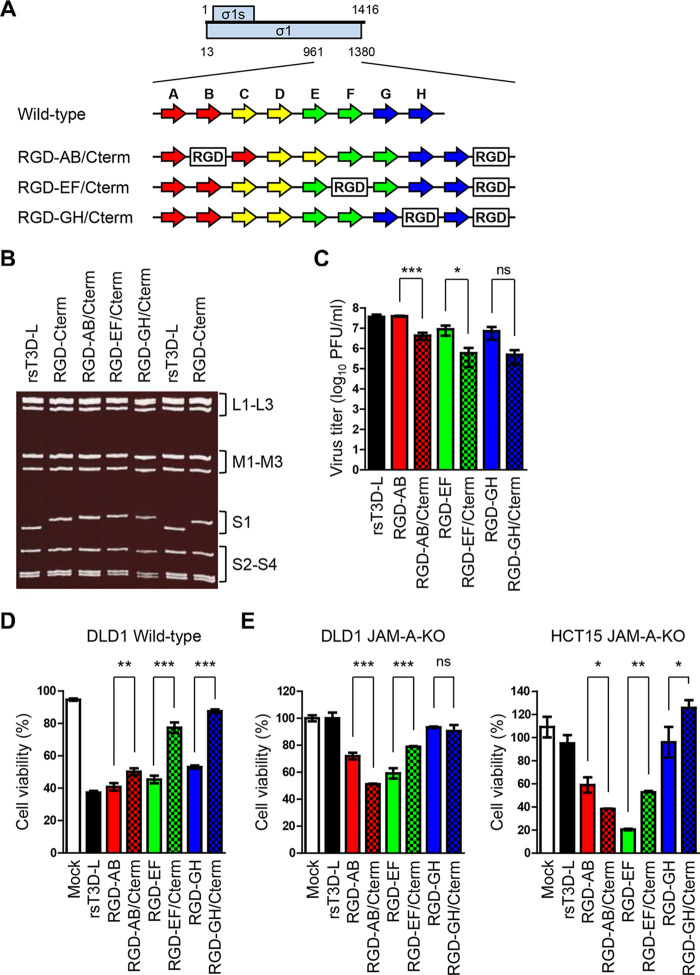
MRVs use JAM-A as an entry receptor. An interaction between cell attachment protein σ1 and JAM-A is required for MRV entry (25). While MRVs exhibit oncolytic activity in various cancer cell lines, some cell lines (including human glioblastoma U118MG) are resistant because they express low levels of JAM-A; thus, receptor-dependent MRV infection limits its clinical application as an oncolytic MRV (30, 52). Therefore, to increase the infection efficiency of MRV in tumor cell lines lacking JAM-A, we attempted to use a reverse genetics approach to generate a recombinant T3D-L reovirus with altered cell tropism and high oncolytic capacity. Reovirus σ1 protein forms a homotrimer with head-and-tail morphology. The crystal structure of the σ1 protein shows that the C-terminal globular head domain contains a receptor-binding domain comprising a Greek key motif composed of eight β-sheets and loop structures (28). To generate cell tropism-modified MRVs, we introduced an RGD motif, which binds integrins expressed on the cell surface, into the σ1 protein (53). Based on a previous study that reported the generation of recombinant viruses harboring an exogenous peptide sequence in the C terminus of σ1 (54), we introduced the RGD sequence into the C terminus of σ1 to yield RGD-Cterm virus (Fig. 3A and B). In addition, we attempted to introduce the RGD peptide into the loop regions that connect β-sheet structures to avoid disrupting the native structure of σ1. The RGD peptide was inserted within either of four candidate loops: the AB, CD, EF, or GH loops, exposed to the surface of the head domain, to yield RGD-AB, RGD-CD, RGD-EF, and RGD-GH viruses, respectively (Fig. 3A and B). Gel electrophoresis revealed that S1 gene segments purified from RGD viruses migrated more slowly than those from rsT3D-L due to addition of the RGD peptide (Fig. 3C). The nucleotide sequence of the S1 gene segment of the RGD viruses was confirmed by direct sequencing. Next, we compared the growth kinetics of RGD σ1-modified viruses with those of rsT3D-L in L929 cells. Replication of RGD viruses was comparable with that of rsT3D-L (Fig. 3D), suggesting that insertion of the RGD peptide into the loops or C terminus of σ1 does not disrupt propagation of the modified viruses. We were concerned that addition of an RGD motif might increase virulence in vivo by inducing nonspecific infection; therefore, we assessed the virulence of the RGD σ1-modified virus in newborn and adult mice. Newborn mice were orally inoculated with rsT3D-L, RGD-AB, or RGD-EF and monitored for survival (Fig. 3E). Mice infected with rsT3D-L succumbed to infection and all the mice died 15 days postinfection. In contrast, both RGD-AB (mortality 66.7%) and RGD-EF (mortality 25%) showed attenuated pathogenicity in newborn mice (Fig. 3E). Adult mice infected intranasally with rsT3D-L, RGD-AB, or RGD-EF showed no sign of illness, although they gained weight more slowly than uninfected mice (Fig. 3F). There were, however, no significant differences in body weight between mice infected with rsT3D-L and RGD-AB or RGD-EF. Both the experimental infections of RGD viruses in newborn and adult mice indicate that insertion of the RGD peptide did not increase pathogenicity, but rather caused it to be attenuated.
FIG 3.
Generation and characterization of RGD σ1-modified viruses. (A and B) Schematic presentations of the C-terminal head domain of σ1. (A) Schematics of the plasmids used to generate RGD σ1-modified viruses. The ORFs of the σ1 and σ1s proteins are shown as blue boxes (top). Each number indicates a nucleotide position. The eight β-sheet structures (A to H) are indicated by arrows. The RGD peptide sequence is represented by the white boxes. (B) Structure of the monomeric head domain of σ1 (PDB code 3EOY). The eight β-sheet structures are colored as in Fig. 3A. The RGD peptide insertion sites are represented by white circles (left panel). The amino acids contacting JAM-A (distance cutoff 4 Å) are orange (right panel). (C) Electrophoretic analysis of RGD σ1-modified viruses. Viral dsRNA was extracted from virions and separated by SDS-PAGE, followed by ethidium bromide staining. The different gene segments are indicated. (D) Growth kinetics of RGD σ1-modified viruses in L929 cells. Cells were infected with viruses at an MOI of 0.01 PFU/cell. Samples were collected every day. The virus titer in the cell lysate was determined in a plaque assay. Each value represents an average from triplicate samples. The error bars indicate the standard deviation. Representative results from two independent experiments are shown. (E) Survival curves of newborn mice after infection with RGD viruses. Two-day-old mice (n = 12 per group) were orally inoculated with 1.0 × 103 PFU of rsT3D-L, RGD-AB, or RGD-EF viruses. Mice were monitored for 21 days postinoculation. (F) Body weight changes in adult mice infected with RGD viruses. Four-week-old ICR mice were infected intranasally with rsT3D-L (n = 5), RGD-AB (n = 4), RGD-EF (n = 4), or DMEM (n = 5), and changes in body weight were monitored for 15 days. Each value represents an average score. The error bars indicate the standard deviation. Significant differences at 14 days postinoculation were determined using two-way ANOVA; ns, not significant. The experiment was repeated twice.
Generation of cancer cell lines lacking expression of JAM-A.
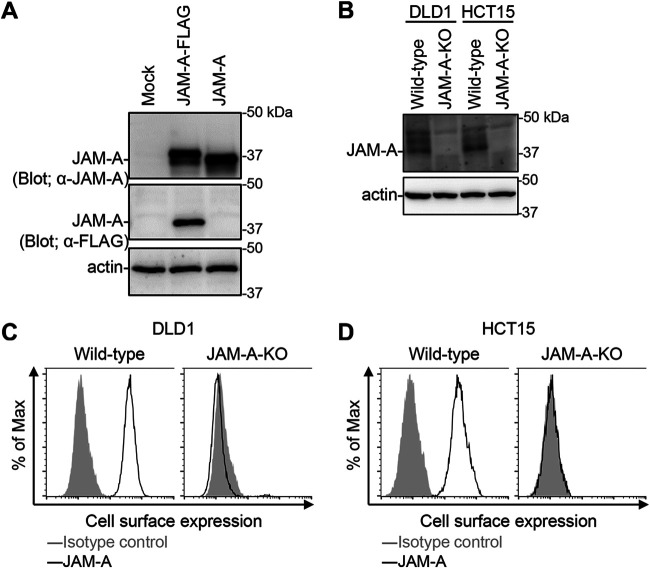
To assess oncolytic activity of RGD σ1 modified viruses in cancer cell lines lacking JAM-A, we generated JAM-A knockout (KO) cell lines (DLD1 JAM-A-KO and HCT15 JAM-A-KO) using the CRISPR/Cas9 system (55). Nucleotide deletions within the JAM-A coding region were confirmed by DNA sequencing. KO of JAM-A in established DLD1 JAM-A-KO and HCT15 JAM-A-KO cell lines was confirmed by immunoblotting with mouse antiserum against JAM-A. The antiserum recognized JAM-A expressed by 293T cells (Fig. 4A). Western blotting with anti-JAM-A antibody did not detect expression of JAM-A in lysates of DLD1 JAM-A-KO and HCT15 JAM-A-KO cells, and no cell surface JAM-A protein was detected by flow cytometry (Fig. 4B to D). These data demonstrate that both DLD1 JAM-A-KO and HCT15 JAM-A-KO cell lines lack functional expression of JAM-A on the cell surface.
FIG 4.
Generation of JAM-A knockout cell lines. (A) Detection of JAM-A protein expression using mouse antiserum. Human 293T cells were transfected with pCXN2-JAM-A-FLAG and pCXN2-JAM-A. The cells were lysed with RIPA buffer and the lysates were centrifuged to remove cell debris. Proteins were separated by SDS-PAGE, followed by immunoblotting with antiserum against JAM-A and antibodies specific for FLAG and β-actin. The molecular weights of the proteins are shown in kilodaltons (kDa). (B) Expression of JAM-A in JAM-A-KO cell lines. Cells were lysed with RIPA buffer and centrifuged to remove cell debris. Proteins were separated by SDS-PAGE, followed by immunoblotting with antiserum against JAM-A and with an anti-β-actin antibody. The molecular weights of the proteins are indicated in kDa. (C and D) Cell-surface expression of JAM-A by wild-type and JAM-A-KO cell lines. DLD1 (C) and HCT15 (D) cells were collected in a nonenzymatic solution and then stained with mouse antiserum against JAM-A and an Alexa488-conjugated secondary antibody, followed by flow cytometry analysis.
Oncolytic activity of RGD σ1-modified viruses in cancer cell lines lacking JAM-A.
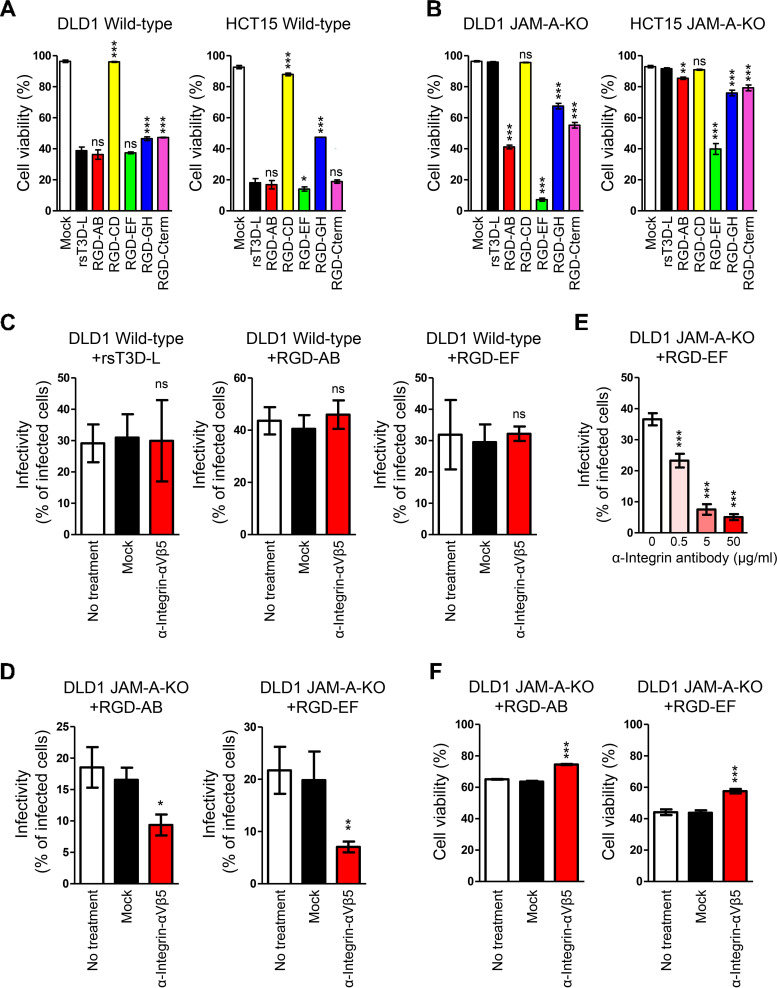
Next, we used the JAM-A-KO cell lines to examine the oncolytic activity of RGD σ1-modified viruses. We found that rsT3D-L, RGD-AB, and RGD-EF showed effective oncolytic activity in wild-type DLD1 and HCT15 cells (Fig. 5A). RGD-CD, RGD-GH, and RGD-Cterm viruses showed lower oncolytic activity than rsT3D-L in DLD1 cells, and RGD-CD and RGD-GH viruses showed impaired oncolytic activity in HCT15 cells, suggesting that insertion of the RGD motif into the CD and GH loops had negative effects on oncolytic activity (Fig. 5A). Taken together, the data showed that RGD σ1-modified viruses (except RGD-CD and RGD-GH) retain RGD sequence activity and possess a functional σ1 protein. DLD1 JAM-A-KO and HCT15 JAM-A-KO cells were completely resistant to infection by rsT3D-L, indicating that JAM-A is a critical virus entry factor for wild-type T3D (Fig. 5B). In contrast, RGD-AB, RGD-EF, RGD-GH, and RGD-Cterm showed higher oncolytic activity than rsT3D-L in DLD1 JAM-A-KO cells. Although RGD-AB, RGD-EF, RGD-GH, and RGD-Cterm killed HCT15 JAM-A-KO cells, oncolytic activity was weaker than that against DLD1 JAM-A-KO cells. Notably, the RGD-EF virus showed the highest oncolytic activity in both DLD1 JAM-A-KO and HCT15 JAM-A-KO cells (Fig. 5B). To confirm whether RGD σ1-modified viruses infect cells in an integrin-dependent manner, we infected wild-type DLD1 and DLD1 JAM-A-KO cells with rsT3D-L, RGD-AB, or RGD-EF viruses following treatment with an antibody specific for integrin αVβ5. Infectivity of rsT3D-L, RGD-AB, and RGD-EF did not decrease in wild-type DLD1 cells in the presence of the anti-integrin αVβ5 antibody (Fig. 5C). However, virus infectivity of RGD-AB and RGD-EF did fall in DLD1 JAM-A-KO cells in the presence of the anti-integrin αVβ5 antibody, and the effect was dose-dependent (Fig. 5D and E). These data show that treatment with an anti-integrin αVβ5 antibody is effective against infectivity of RGD viruses in DLD1 JAM-A-KO cells. Furthermore, we assessed whether treatment with an antibody specific for integrin αVβ5 inhibits oncolytic activity of the RGD-AB and RGD-EF viruses. Oncolysis of DLD1 JAM-A-KO cells treated with the antibody was less evident than that in mock-treated cells (Fig. 5F). These data suggest the RGD peptide sequence allows viruses to infect cells in a JAM-A-independent manner.
FIG 5.
Integrin-dependent oncolysis of JAM-A knockout cell lines by RGD σ1-modified viruses. (A and B) Oncolytic activity of RGD σ1-modified viruses in cell lines. Wild-type (A) and JAM-A-KO (B) cells were infected with wild-type and RGD σ1-modified viruses at an MOI of 10 PFU/cell. Cells were collected every 24 h and cell viability was measured by propidium iodide staining. Each value represents an average from triplicate samples. The error bars indicate the standard deviation. Representative results from three independent experiments are shown. Significant differences were determined using one-way ANOVA; *, P < 0.05; ***, P < 0.0001; ns, not significant. (C and D) Infection of cancer cells by RGD σ1-modified viruses following treatment with anti-integrin antibodies. Wild-type DLD1 (C) and DLD1 JAM-A-KO (D) cells were incubated at 37°C for 1 h with an anti-integrin αVβ5 antibody (P1F6) or an anti-FLAG antibody and then infected for 16 h with rsT3D-L, RGD-AB, or RGD-EF at an MOI of 50 PFU/cell. Infectivity was analyzed by indirect immunofluorescence analysis using an anti-T3D antibody. Infectivity was calculated as the ratio of infected cells to the total number of cells. The results are expressed as the mean score for four independent fields of view. The error bars indicate the standard deviation. Representative results from two independent experiments are shown. Significant differences were determined using Student’s t test; *, P < 0.05; **, P < 0.001; ns, not significant. (E) Infection of JAM-A-KO cells by RGD-EF following treatment with different concentrations of an anti-integrin antibody. DLD1 JAM-A-KO cells were incubated at 37°C for 1 h with an anti-integrin αVβ5 antibody (P1F6) or an anti-FLAG antibody, and then infected for 16 h with RGD-EF at an MOI of 50 PFU/cell. Infectivity was analyzed by indirect immunofluorescence analysis using an anti-T3D antibody. Infectivity was calculated as the ratio of infected cells to the total number of cells. The results are expressed as the mean score for four independent fields of view. The error bars indicate the standard deviation. Representative results from two independent experiments are shown. Significant differences were determined using one-way ANOVA; ***, P < 0.0001. (F) Oncolysis of RGD σ1-modified viruses following treatment with an anti-integrin antibody. DLD1 JAM-A-KO cells were incubated at 37°C for 1 h with an anti-integrin αVβ5 antibody (P1F6) or an anti-FLAG antibody, and then infected for 16 h with RGD-AB or RGD-EF at an MOI of 50 PFU/cell. Cell viability was measured by propidium iodide staining. Each value represents an average from triplicate samples. The error bars indicate the standard deviation. Representative results from two independent experiments are shown. Significant differences were determined using Student’s t test; ***, P < 0.0001.
Oncolytic activity of RGD σ1-modified viruses harboring multiple RGD sequences.
To test whether insertion of multiple RGD motifs into RGD σ1-modified viruses leads to a synergistic increase in JAM-A-independent infectivity, we generated RGD-Cterm viruses harboring an additional RGD motif in the AB, EF, and GH loops (RGD-AB/Cterm, RGD-EF/Cterm, and RGD-GH/Cterm viruses, respectively) (Fig. 6A). Electrophoretic analysis of viral dsRNA genomes demonstrated that the S1 gene segment of RGD-AB/Cterm, RGD-EF/Cterm, and RGD-GH/Cterm migrated more slowly than that of RGD-Cterm (Fig. 6B). Next, we assessed replication of RGD σ1-modified viruses harboring multiple RGD sequences. Interestingly, replication of RGD-AB/Cterm and RGD-EF/Cterm was lower than that of RGD-AB and RGD-EF, respectively (Fig. 6C). In addition, replication of RGD-GH/Cterm was lower than that of RGD-GH, although the difference was not statistically significant (Fig. 6C). These data suggest that insertion of multiple RGD sequences affects replication of the viruses. Next, we compared the oncolytic activity of the viruses in wild-type DLD1 cells. Oncolytic activity of recombinant viruses harboring multiple RGD sequences (RGD-AB/Cterm, RGD-EF/Cterm, and RGD-GH/Cterm) was lower than that of viruses harboring a single RGD sequence (RGD-AB, RGD-EF, and RGD-GH, respectively) (Fig. 6D). The oncolytic activity of these viruses was investigated in DLD1 JAM-A-KO and HCT15 JAM-A-KO cells. Oncolytic activity of RGD-AB/Cterm was higher than that of RGD-AB in both cell lines (Fig. 6E). In contrast, oncolytic activity of RGD-EF/Cterm was lower than that of RGD-EF in both cell lines (Fig. 6E). Although oncolytic activity of RGD-GH/Cterm was similar to that of RGD-GH in DLD1 JAM-A-KO cells, it was less than that of RGD-GH in HCT15 JAM-A-KO cells (Fig. 6E). The results demonstrate that inserting multiple RGD motifs into σ1-modified viruses affects viral replication and alters the ability of MRV to infect tumor cells lacking JAM-A; these effects appear to be dependent on the insertion sites within the exposed loops of the σ1 protein.
FIG 6.
Generation and in vitro characterization of viruses harboring multiple RGD sequences. (A) Schematic presentation of the C-terminal head domain of σ1 and the plasmids used to recover RGD σ1-modified viruses. The ORFs of the σ1 and σ1s proteins are denoted by blue boxes (top). Each number indicates a nucleotide position. The eight β-sheet structures (A to H) are indicated by arrows (colors are the same as in Fig. 3A). The RGD peptide sequence is denoted by white boxes. (B) Electrophoretic analysis of viruses harboring multiple RGD sequences. Viral dsRNA was extracted from virions, separated by SDS-PAGE, and stained with ethidium bromide. The electrophoretic patterns of rsT3D-L and RGD-Cterm are shown for reference. Gene segments are indicated. (C) Replication of viruses harboring multiple RGD sequences in L929 cells. Cells were infected with viruses at an MOI of 0.01 PFU/cell. Samples were collected 72 h postinfection. The virus titer in the cell lysate was determined in a plaque assay. Each value represents an average from triplicate samples. The error bars indicate the standard deviation. Significant differences were determined using Student’s t test; *, P < 0.05; ***, P < 0.0001; ns, not significant. (D and E) In vitro oncolytic activity of the viruses. Wild-type DLD1 (D) and DLD1 JAM-A-KO and HCT15 JAM-A-KO cells (E) were infected with each virus at an MOI of 10 PFU/cell or mock-infected with DMEM. Cell viability was determined in a WST1 colorimetric assay. Cell viability is shown as a score relative to that of mock samples. Each value represents an average from triplicate samples. The error bars indicate the standard deviation. Representative results from two independent experiments are shown. Significant differences were determined using Student’s t test; *, P < 0.05; **, P < 0.001; ***, P < 0.0001; ns, not significant.
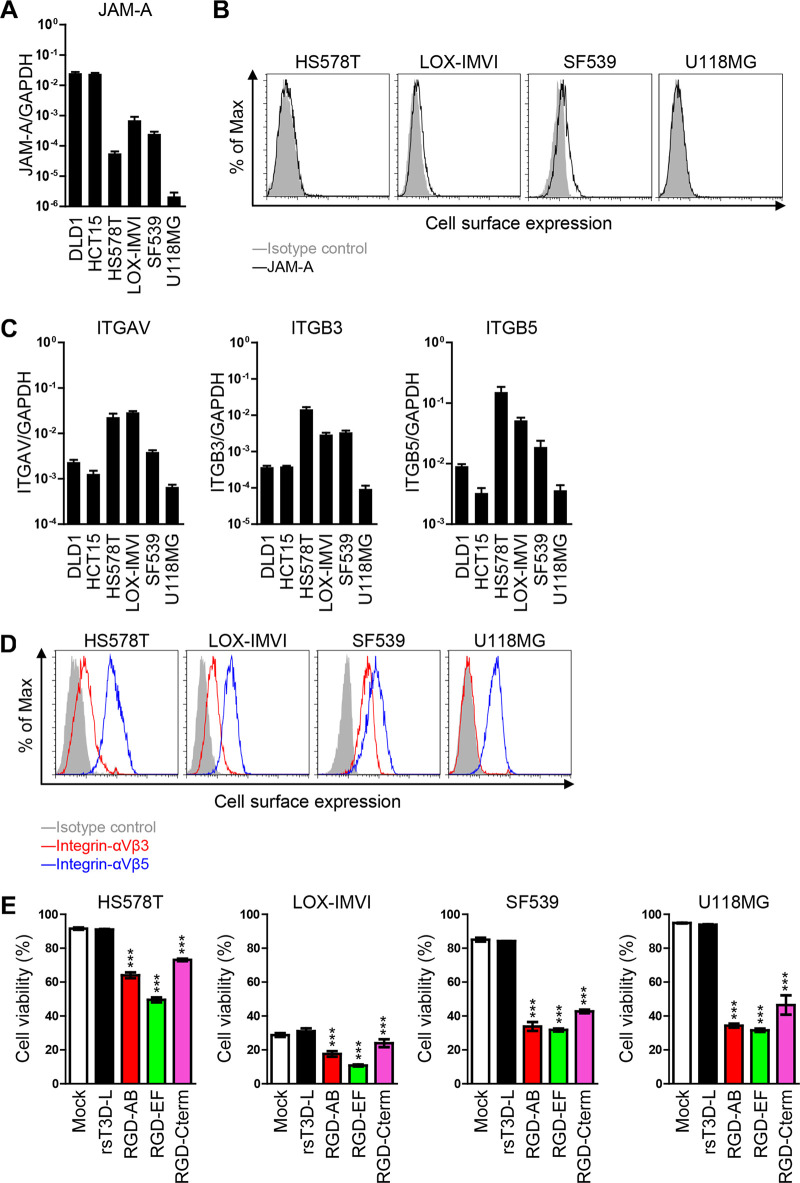
Oncolytic activity of RGD σ1-modified viruses in cancer cell lines expressing low levels of JAM-A.
To explore the ability of RGD viruses to act as oncolytic agents under natural conditions, we tested them against human cancer cells expressing low levels of JAM-A. We screened cancer cell lines to identify those with lower JAM-A transcription levels than the DLD1 and HCT15 cell lines. Four cell lines, HS578T, LOX-IMVI, SF539, and U118MG, were identified (Fig. 7A). All four cell lines express marginal levels of JAM-A on the cell surface (Fig. 7B). However, these cell lines showed similar expression of mRNA encoding integrins αV, β3, and β5, raising the possibility that RGD σ1-modified viruses would infect these cell lines more efficiently than rsT3D-L (Fig. 7C). We confirmed that these cell lines express integrins αVβ3 and αVβ5 on the cell surface (Fig. 7D). Therefore, we infected these cell lines with RGD viruses and measured cell viability. As expected, the cells were resistant to infection by rsT3D-L. However, the cells were killed by the RGD σ1-modified viruses. The RGD-EF virus showed highest oncolytic activity, which was comparable with that in JAM-A KO cells (Fig. 7E). These data suggest that RGD σ1-modified viruses are effective against human cancer cells showing low JAM-A expression.
FIG 7.
Characterization of viruses harboring RGD sequence using cancer cell lines expressing low levels of JAM-A. (A and C) Transcription of JAM-A, ITGAV, ITGB3, and ITGB5 in cancer cell lines. mRNA was extracted from cells and levels were measured by quantitative reverse transcription-PCR (RT-qPCR). Each value represents an average from triplicate samples. The error bars indicate the standard deviation. Representative results from two independent experiments are shown. (B and D) Cell surface expression of JAM-A, integrin αVβ3, and integrin αVβ5 by cancer cell lines. Cancer cell lines were collected in a nonenzymatic solution and then stained with mouse antiserum against JAM-A (B) or with an anti-integrin αVβ3 (LM609) or an anti-integrin αVβ5 antibody (P1F6) (D), followed by an Alexa 488-conjugated secondary antibody. Cells were then subjected to flow cytometry analysis. (E) Human cancer cells were infected with each virus at an MOI of 10 PFU/cell. Cell viability was determined by propidium iodide staining. Each value represents an average from triplicate samples. The error bars indicate the standard deviation. Representative results from two independent experiments are shown. Significant differences were determined using one-way ANOVA; ***, P < 0.0001.
DISCUSSION
Oncolytic MRV Reolysin has been exploited for use as a cancer therapeutic in combination with chemotherapy. Although Reolysin increased overall survival for breast cancer, it did not improve progression-free survival in phase II clinical trials for several cancers performed in the U.S and Canada (56–59). Alternative treatment strategies are required as a cancer therapeutic. Since the establishment of a plasmid-based reverse genetics system, the efforts have been taken to improve oncolytic activity of T3D by gene modification (49, 60, 61). Currently, major progress in improving the oncolytic effect of T3D as a cancer therapeutic relies on the identification of mutations in the σ1 tail domain that confer SA-dependent infection (30–35). While viruses with the mutation can infect cancer cells with marginal expression levels of JAM-A, the viruses lost their original ability to effect JAM-A-dependent infection. As a different strategy to kill cancer cells with marginal expression levels of JAM-A, we generated recombinant viruses expressing an RGD motif within the cell attachment protein σ1, which enabled the reovirus to infect cell lines in an integrin-dependent manner. We selected the C-terminal region and four loop structures as insertion sites for the RGD peptide sequence (Fig. 3). While recombinant virus expressing small peptides (histidine and hemagglutinin) in the C-terminal end of σ1 have been recovered (54), the C-terminal end of σ1 contains amino acids required for binding to JAM-A (26). Thus, we were concerned that insertion of an RGD sequence into the C terminus of σ1 would inhibit viral replication of RGD-Cterm. However, RGD-Cterm replicated efficiently and showed oncolytic activity in both wild-type and JAM-A KO cancer cells and other cancer cells with marginal expression levels of JAM-A (Fig. 3, Fig. 5, and Fig. 7). This suggests that insertion of an RGD sequence at the C terminus did not interrupt entry of virus into cells. Interestingly, only RGD-CD significantly lost oncolytic activity in tumor cells (Fig. 5). Since the CD loop structure is located between the AB and EF loops in the tertiary structure of σ1 (Fig. 3), insertion of the RGD sequence may disrupt the structure of the σ1 protein. These data suggest that the spatial arrangement between the RGD sequence and the integrin is critical for efficient infection. So far, only the C terminus position has been reported as a site of foreign peptide insertion in the head domain of σ1 protein (54). In this study, it was initially shown that the loop regions of the σ1 head domain can accommodate exogenous peptides without affecting virus infectivity. Moreover, we revealed the possibility that two different peptides could be inserted simultaneously. We successfully infected JAM-A KO cancer cells with RGD viruses. However, the entry mechanism is unclear. A previous study demonstrated that the outer capsid protein λ2, which serves as the structural base for σ1, contains an RGD sequence, and that β1 integrin is required for entry after attachment (62). Thus, the RGD sequence in σ1 may interact with integrins, bypassing the interaction between σ1 and JAM-A. Future studies should focus on the molecular mechanism underlying JAM-A-independent entry.
As it was of concern that addition of RGD peptide might increase the virulence of T3D-L due to excessive dissemination of virus, attenuated virulence of RGD-AB and RGD-EF was not expected. Interestingly, there were difference in the virulence between RGD-AB and RGD-EF, although the replication and infectivity of these two RGD viruses were similar in in vitro experiments. One of the hypotheses to explain the attenuated virulence of RGD viruses is that viruses become adhered to miscellaneous cells which do not support virus replication. Although the MRVs are low-pathogenicity viruses, vast amounts of oncolytic MRVs could be inoculated in clinical settings, such that the virulence of the virus is an important factor along with oncolytic activity. Further investigations of virus virulence with higher virus titers and various inoculation routes will be continued.
We obtained the whole-genome sequence of T3D-L (the original strain of Reolysin) and identified viral gene segments essential for effective oncolysis, noting that the T3D-L and T3D-D strains differ with respect to 21 amino acids within the L1, L2, L3, M1, M2, M3, S1, and S4 gene segments (Table 1). These amino acid differences are exactly the same as the results recently shown by Mohamed et al. (12). The T3D-L and Reolysin strains differed by one amino acid (Table 2) (51, 63). These data confirm that T3D-L is genetically similar to strain Reolysin, and that the identified amino acid differences contribute to the higher oncolytic activity of T3D-L. While we genetically modified strain T3D-L in this study, recombinant viruses with a sequence identical to that of Reolysin can be generated by introducing seven nucleotide mutations into the T3D-L strain.
In this study, we also compared oncolysis of tumor cells by monoreassortant of T3D-L and T3D-D viruses and found that segments L1 (coding λ3), L3 (λ1), M1 (μ2), M2 (μ1), and S4 (σ3) contributed to increased oncolytic activity of T3D-L (Fig. 1 and Fig. 2). Among the gene segments, L3, M1, and S4 gene segments have been reported to be involved in large, plaque-sized T3D-L (12). Mohamed et al. showed that T3D-L forms larger plaques than T3D-D in murine and human cancer cells. They demonstrated that L3, M1, and S4 gene segments independently contributed to a higher mRNA transcription of T3D-L than T3D-D, which leads to the rapid replication of T3D-L (13). While we used different cancer cells, the rapid replication of T3D-L can be raised as one of the mechanisms underlying efficient cell death caused by T3D-L. In addition to these three gene segments, we obtained L1 and M2 gene segments as determinants of T3D-L oncolysis. Since the L1 gene segment encodes the RNA-dependent RNA polymerase (RdRp) λ3 (64–66), it is tempting to speculate that differences in RdRp activity also contributed to the efficient replication of T3D-L. Regarding the M2 gene segments, previous studies have demonstrated the M2 gene segment plays an important role in the ability of MRV strains to induce apoptosis (9, 67). Since it was reported that the transformation of Ras increased reovirus-induced apoptosis in cancer cells (68), differences in the induction of apoptosis could be another mechanism for the efficient oncolysis by T3D-L.
In the context of oncolytic virotherapy, the reverse genetics system has facilitated tumor-selective virus replication, induction of antitumor immunity, and induction of immunostimulatory cytokines. The reovirus genome is smaller than that of DNA viruses such as adenovirus and herpesvirus; therefore, it is difficult to generate a replication-competent virus harboring a foreign gene. However, previous studies reported successful insertion of a foreign gene (SIV gag or NanoLuc) into the 5′ terminal region of the L1 gene segment (69, 70). Therefore, the 5′ terminus of the L1 gene segment may be an optimal region for insertion and expression of a foreign gene. In conclusion, a reverse genetics approach led to the creation of a highly oncolytic MRV strain, T3D-L. The system described herein provides a platform for improving cancer therapeutics based on MRV.
MATERIALS AND METHODS
Cells and viruses.
Human colorectal cancer DLD1 (TKG 0379) and HCT15 (TKG 0504) cells were obtained from the Cell Resource Center for Biomedical Research, Institute of Development, Aging and Cancer, Tohoku University. Murine fibroblast L929 cells were obtained from the American Type Culture Collection. DLD1, HCT15, and L929 cells were maintained in Dulbecco modified Eagle medium (DMEM) supplemented with 5% fetal bovine serum (FBS), 100 units/ml penicillin, and 100 μg/ml streptomycin (Nacalai Tesque). Human cancer cell lines HS578T, LOX-IMVI, and SF539 were obtained from Toru Okamoto (the Walter & Eliza Hall Institute), and U118MG cells were obtained from Shinichi Yokota (Sapporo Medical University). These cells were maintained in RPMI 1640 supplemented with 10% FBS, 100 units/ml penicillin, and 100 μg/ml streptomycin. Human head and neck cancer cell line A253 (HTB-41) was obtained from the ATCC and maintained in McCoy’s 5A medium supplemented with 5% FBS, 100 units/ml penicillin, and 100 μg/ml streptomycin. Mammalian reovirus (MRV) T1L and T3D-D strains were obtained from Terence S. Dermody. The T3D-L strain was obtained from Patrick W. K. Lee and Tsuyoshi Etoh. MRV strains were propagated in L929 cells. Infectious virus titers were determined in a plaque assay, as previously described (71).
Sequencing of the T3D-L strain.
The nucleotide sequence of the T3D-L strain was determined as previously described (71). Briefly, viral dsRNAs were extracted from purified virions using Sepasol-RNA I Super G (Nacalai Tesque). A self-anchoring primer was ligated to the 3′ termini of viral dsRNAs using T4 RNA ligase (Thermo Scientific) and viral cDNAs were reverse transcribed using Superscript III (Invitrogen). Viral gene segments were amplified from cDNA using KOD-plus-Neo (TOYOBO) by annealing primers to the self-anchoring primer. The PCR product was cloned into the pBluescript KS (+) vector and the sequence was determined using an ABI 3130 genetic analyzer (Life Technologies).
Plasmid construction.
To construct rescue plasmids for recombinant strain T3D-L (rsT3D-L), cDNA encoding each of the 10 viral gene segments was cloned between the T7 promoter sequence and the hepatitis delta virus (HDV) ribozyme sequence by replacing the T3D-D L1 gene in pT7-T3D-D-L1 (49). The resulting plasmids were named pT7-T3D-L-L1, pT7-T3D-L-L2, pT7-T3D-L-L3, pT7-T3D-L-M1, pT7-T3D-L-M2, pT7-T3D-L-M3, pT7-T3D-L-S1, pT7-T3D-L-S2, and pT7-T3D-L-S4. To generate rescue plasmids for arginine–glycine–aspartic acid (RGD) viruses, the RGD peptide coding sequence (CDCRGDCFC) was inserted into the S1 gene segment at nucleotide positions 999 to 1000 (AB loop), 1095 to 1096 (CD loop), 1233 to 1234 (EF loop), or 1317 to 1318 (GH loop) by PCR mutagenesis. The resulting plasmids were named pT7-T3D-L-S1-RGD-AB, pT7-T3D-L-S1-RGD-CD, pT7-T3D-L-S1-RGD-EF, and pT7-T3D-L-S1-RGD-GH, respectively. To generate T3D-L-RGD-C-terminal, the RGD peptide coding sequence (GGGGSGGGGSGGGGSCDCRGDCFC) was fused at the C terminus of the σ1 protein (1377 to 1378) by PCR mutagenesis. pX330 (a gift from Feng Zhang) (Addgene plasmid number 42230) and pCAG-EGxxFP (a gift from Masahito Ikawa) (Addgene plasmid number 50716) were used to construct the plasmid for the CRISPR/Cas9 system (72, 73). The 5′-gaagttgtcctgtgcctact-3′ sequence within the JAM-A gene was used as the single guide RNA sequence for Cas9 and was cloned into the BbsI site of the pX330 plasmid (72). The resulting plasmid was named pX330-JAM-A. The genomic DNA sequence of JAM-A (nucleotides [nt] 20008 to 20548) was cloned into the pCAG-EGxxFP plasmid between the BamHI and EcoRI sites (73). The resulting plasmid was named pCAG-EGxxFP-JAM-A. To construct pCXN2-JAM-A, the ORF of JAM-A (NM_016946) was cloned into the EcoRI site of the pCXN2 vector. To construct pCXN2-JAM-A-FLAG, a FLAG tag sequence was fused to the C terminus of JAM-A by PCR mutagenesis.
Reverse genetics system to generate MRVs.
Recombinant viruses were recovered from cells following plasmid transfection as previously described (71). Briefly, L929 cells were infected for 1 h with recombinant vaccinia virus rDIs-T7pol expressing T7 RNA polymerase (74) at an MOI of ∼3 50% tissue culture infective dose (TCID50)/cell. Plasmids harboring cDNA encoding the 10 viral gene segments were transfected into the cells using 2 μl of TransIT-LT1 (Mirus) per microgram of plasmid. The cells were collected 5 days after plasmid transfection, and recombinant virus was purified in a plaque assay. To generate RGD viruses, recombinant S1 plasmids were cotransfected along with the other nine plasmids. To generate monoreassortant viruses, the rescue plasmid for T3D-L was combined with that of T3D-D. L929 cells were transfected with 10 plasmids harboring cDNAs encoding T3D-L and T3D-D.
Genotyping of recombinant viruses.
Viral genomic dsRNA was extracted from purified virions using Sepasol-RNA I Super G (Nacalai Tesque) and separated by polyacrylamide electrophoresis, stained with 5 μg/ml ethidium bromide, and analyzed using a UV transilluminator.
Generation of JAM-A-knockout cell lines.
The JAM-A-knockout (KO) cancer cell lines were generated using the CRISPR/Cas9 system. Briefly, DLD1 and HCT15 cells were cotransfected with pX330-JAM-A and pCAG-EGxxFP-JAM-A. Two days later, GFP-positive cells were isolated using a FACSAria II cytometer (Becton, Dickinson) and single-cell clones were isolated by limiting dilution. Genomic DNA was extracted from each clone using proteinase K and deletion of JAM-A was confirmed by DNA sequencing. Single-cell clones of DLD1 JAM-A-KO cells and HCT15 JAM-A-KO cells were used for the experiments.
Death assay.
Cancer cells were seeded into a 48-well plate and incubated overnight at 37°C. The cells were infected with viruses at an MOI of 10 PFU/cell. After 1 h, cells were washed with phosphate-buffered saline (PBS) and cultured for different times in DMEM containing 2% FBS. For cell viability analysis, cells were collected and resuspended in PBS containing 2% FBS and 10 μg/ml propidium iodide solution. The percentage of living cells was analyzed using a FACSCalibur cytometer (Becton, Dickinson). Data were analyzed using FlowJo software. Alternatively, cancer cells were seeded into a 96-well plate and infected with viruses at an MOI of 10 PFU/cell. After 1 h, the inoculum was removed and cells were incubated for 3 days with DMEM containing 2% FBS. Viability was measured using Cell Proliferation Reagent WST-1 (Roche Diagnostics). Absorbance at 440 nm was determined in a microplate reader (DS Pharma Biomedical). The viability score relative to that of uninfected cells was calculated. To assess the effect of treatment with an anti-integrin antibody on oncolysis, DLD1 JAM-A-KO cells were incubated at 37°C for 1 h with an antibody specific for integrin αVβ5 (10 μg/ml) (P1F6, Merck). Cells were washed with PBS and then infected with RGD-AB or RGD-EF at an MOI of 50 PFU/cell. After 1 h, the inoculum was removed and cells were washed with PBS and incubated with DMEM for 48 h. Then, cells were collected and stained with propidium iodide solution. The percentage of cells was analyzed using a FACSCalibur flow cytometer. Data were analyzed using FlowJo software.
Growth kinetics of recombinant viruses.
A monolayer of L929 cells was infected with recombinant viruses at an MOI of 0.01 PFU/cell. After incubation for 1 h, the cells were washed twice with PBS and incubated with DMEM containing 2% FBS. The samples were collected at various intervals for use in a plaque assay.
Generation of antiserum against JAM-A.
The ORF of human JAM-A was cloned into the pTrcHisA vector (Life Technologies). Next, Escherichia coli strain BL21 was transformed with the pTrcHisA-JAM-A plasmid. The recombinant JAM-A protein harboring an N-terminal polyhistidine (6×His) tag was expressed by addition of IPTG (isopropyl-β-D-thiogalactopyranoside) to bacteria and collected by passage over cOmplete His-Tag purification resin (Roche Diagnostics). ICR mice (4 weeks old) were purchased from CLEA Japan, Inc. The recombinant protein was mixed with Alhydrogel adjuvant 2% (InvivoGen) and injected subcutaneously into mice. Antiserum was collected after three rounds of boosting.
Immunoblot analysis.
Wild-type or JAM-A-KO cancer cells were seeded into a 6-well plate and incubated at 37°C overnight. The cells were lysed with RIPA buffer (25 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, and 0.1% SDS) containing protease inhibitor, the lysate centrifuged at 17,700 × g for 5 min at 4°C, and the supernatant collected. The supernatant was mixed with 2× SDS sample buffer, boiled at 95°C, and separated on SDS-PAGE gels. Proteins were then transferred to a polyvinylidene difluoride (PVDF) membrane and reacted with a polyclonal antibody specific for JAM-A (dilution, 1:1,000), followed by a horseradish peroxidase (HRP)-conjugated anti-mouse secondary antibody (Sigma-Aldrich) (dilution, 1:10,000).
Flow cytometry.
Cells were collected using Cell Dissociation Solution Non-enzymatic (Sigma-Aldrich) and resuspended in PBS containing 2% FBS (1 × 106 cells/ml). Cells (5 × 105) were incubated with PBS containing 2% FBS, followed by incubation for 30 min at 4°C with a polyclonal antibody specific for JAM-A (dilution, 1:500), an antibody specific for integrin αVβ3 (LM609) (2.5 μg/ml), or an antibody specific for integrin αVβ5 (P1F6) (2.5 μg/ml). The cells were washed three times with PBS and incubated for 30 min at 4°C with PBS containing 2% FBS and a CF 488-conjugated anti-mouse secondary antibody (Biotium; dilution, 1:1,000) plus propidium iodide (10 μg/ml). Finally, cells were washed three times with PBS. The signal intensity of living cells was quantified using a FACSCalibur flow cytometer (Becton, Dickinson). Data were analyzed using FlowJo software.
Immunofluorescence and infectivity assays.
DLD1 JAM-A-KO cells seeded onto glass coverslips were incubated at 37°C for 1 h with DMEM containing an anti-FLAG monoclonal antibody (clone M2, Sigma-Aldrich) (50 μg/ml). The cells were washed three times with PBS and then infected for 16 h with RGD-EF at an MOI of 50 PFU/cell. The cells were fixed with 4% paraformaldehyde for 15 min and permeabilized for 15 min with PBS containing 0.5% Triton X. After 1 h of incubation with PBS containing 2% FBS, the cells were incubated with a polyclonal antibody specific for T3D (dilution, 1:1,000). After washing three times with PBS, the cells were incubated with CF 488 goat anti-rabbit IgG (Biotium). After washing three times with PBS, the cells were incubated with 4’,6-diamidino-2-phenylindole (DAPI) to stain nuclei and then washed with PBS. Images were acquired with a Fluo View FV1000 laser scanning confocal microscope (Olympus). The number of nuclei in the image was counted using ImageJ software (75). To compare infectivity of viruses following treatment with antibodies specific for integrins, wild-type DLD1 or DLD1 JAM-A-KO cells were incubated at 37°C for 1 h with DMEM containing an anti-integrin αVβ5 antibody (clone P1F6) (10 μg/ml) or an anti-FLAG monoclonal antibody (clone M2) (10 μg/ml). The cells were washed three times with PBS and then infected for 16 h with rsT3D-L, RGD-AB, or RGD-EF at an MOI of 50 PFU/cell. Finally, infected cells were detected using a polyclonal antibody specific for anti-T3D (dilution, 1:1,000), followed by a CF 488 goat anti-rabbit IgG secondary antibody (Biotium). Infectivity was calculated as the ratio of infected cells to the total number of cells.
Quantitative reverse transcription-PCR.
Total RNA was extracted from cell pellets using Sepasol-RNA I Super G (Nacalai Tesque) and cDNA was reverse transcribed using ReverTra Ace (TOYOBO). The amount of mRNA was determined using Fast SYBR green Master Mix (Applied Biosystems). The following primers were used: F11R, 5′-GAGACACCACCAGACTCGTTTG-3′ and 5′-CAAGTGTATGTCCCAGTGTCT-3′; ITGAV, 5′-TGCAGATGTGTTTATTGGAGCACC-3′ and 5′-CAAAGGAGCTATGGCACTGCC-3′; ITGB3, 5′-CTGCTATGATATGAAGACCACCTGC-3′ and 5′-GACTGTAGCCTGCATGATGG-3′; ITGB5, 5′-GGACATCTCTCCTTTCTCCTACAC-3′ and 5′-CATTGAAGCTGTCCACTCTGTCT-3′; GAPDH, 5′-ACATCGCTCAGACACCATG-3′ and 5′-TGTAGTTGAGGTCAATGAAGGG-3′. Expression of each gene was expressed relative to that of GAPDH (internal control).
Mouse experiments.
To analyze tumor cell death in vivo, BALB/cAJcl-nu nude mice (4 weeks old; CLEA Japan, Inc.,) were injected subcutaneously in the lower back with A253. Mice were inoculated intratumorally with recombinant viruses (5 × 107 PFU) and the tumor volume was calculated as follows:
For analyses of viral pathogenicity in newborn mice, pregnant C57BL/6J mice were purchased from CLEA Japan. Two days after delivery, newborn mice were orally inoculated with rsT3D-L, RGD-AB, or RGD-EF (1 × 103 PFU). Mice were monitored for 21 days postinoculation. To examine the viral pathogenicity in adult mice, ICR mice (4 weeks old) were purchased from CLEA Japan, Inc. Mice were infected intranasally with 3 × 106 PFU/ml of virus and body weight was monitored.
Statistical analysis.
Statistical analysis was performed using Prism Software (GraphPad Software, Inc.). P values in Fig. 1A and Fig. 3F were calculated using two-way ANOVA. Those in Fig. 1B, 1C, 2D, 2E, 5A, 5B, 5E, 6D, 6E, and 7E were calculated using one-way ANOVA. Those in Fig. 5C, D, F, and 6C were calculated using Student’s t test. A P value of <0.05 was considered significant.
Ethical statement.
All animal experiments were approved by the Animal Research Committee of the Research Institute for Microbial Diseases, Osaka University, and conducted under the guidelines for the Care and Use of Laboratory Animals of the Ministry of Education, Culture, Sports, Science and Technology, Japan.
ACKNOWLEDGMENTS
We thank K. Yukawa and M. Yoshida for secretarial work, M. Onishi for technical assistance, and F. Sakurai and T. Okamoto for technical advice.
This work was supported in part by a grant from AMED (number 19fk0108080) and grants from JSPS KAKENHI (numbers JP18K15167, JP19H04835, and JP16K19138).
Author contributions are as follows: conception and design, Takahiro Kawagishi, Yuta Kanai, and Takeshi Kobayashi; acquisition of data, Takahiro Kawagishi, Yuta Kanai, Ryotaro Nouda, Ichika Fukui, and Jeffery A. Nurdin; data analysis, Takahiro Kawagishi, Yuta Kanai, and Takeshi Kobayashi; manuscript, Takahiro Kawagishi, Yuta Kanai, Yoshiharu Matsuura, and Takeshi Kobayashi.
REFERENCES
- 1.Bell J, McFadden G. 2014. Viruses for tumor therapy. Cell Host Microbe 15:260–265. doi: 10.1016/j.chom.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miest TS, Cattaneo R. 2014. New viruses for cancer therapy: meeting clinical needs. Nat Rev Microbiol 12:23–34. doi: 10.1038/nrmicro3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaufman HL, Kohlhapp FJ, Zloza A. 2015. Oncolytic viruses: a new class of immunotherapy drugs. Nat Rev Drug Discov 14:642–662. doi: 10.1038/nrd4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu BL, Robinson M, Han ZQ, Branston RH, English C, Reay P, McGrath Y, Thomas SK, Thornton M, Bullock P, Love CA, Coffin RS. 2003. ICP34.5 deleted herpes simplex virus with enhanced oncolytic, immune stimulating, and anti-tumour properties. Gene Ther 10:292–303. doi: 10.1038/sj.gt.3301885. [DOI] [PubMed] [Google Scholar]
- 5.Suzuki K, Fueyo J, Krasnykh V, Reynolds PN, Curiel DT, Alemany R. 2001. A conditionally replicative adenovirus with enhanced infectivity shows improved oncolytic potency. Clin Cancer Res 7:120–126. [PubMed] [Google Scholar]
- 6.Bergelson JM, Cunningham JA, Droguett G, Kurt JE, Krithivas A, Hong JS, Horwitz MS, Crowell RL, Finberg RW. 1997. Isolation of a common receptor for coxsackie B viruses and adenoviruses 2 and 5. Science 275:1320–1323. doi: 10.1126/science.275.5304.1320. [DOI] [PubMed] [Google Scholar]
- 7.Wickham TJ, Tzeng E, Shears LL 2nd, Roelvink PW, Li Y, Lee GM, Brough DE, Lizonova A, Kovesdi I. 1997. Increased in vitro and in vivo gene transfer by adenovirus vectors containing chimeric fiber proteins. J Virol 71:8221–8229. doi: 10.1128/JVI.71.11.8221-8229.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dermody T, Parker J, Sherry B. 2013. Orthoreoviruses, p 1304–1346. In Fields virology. Wolters Kluwer/Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 9.Coffey CM, Sheh A, Kim IS, Chandran K, Nibert ML, Parker JS. 2006. Reovirus outer capsid protein micro1 induces apoptosis and associates with lipid droplets, endoplasmic reticulum, and mitochondria. J Virol 80:8422–8438. doi: 10.1128/JVI.02601-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nygaard RM, Lahti L, Boehme KW, Ikizler M, Doyle JD, Dermody TS, Schiff LA. 2013. Genetic determinants of reovirus pathogenesis in a murine model of respiratory infection. J Virol 87:9279–9289. doi: 10.1128/JVI.00182-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lanoie D, Lemay G. 2018. Multiple proteins differing between laboratory stocks of mammalian orthoreoviruses affect both virus sensitivity to interferon and induction of interferon production during infection. Virus Res 247:40–46. doi: 10.1016/j.virusres.2018.01.009. [DOI] [PubMed] [Google Scholar]
- 12.Mohamed A, Clements DR, Gujar SA, Lee PW, Smiley JR, Shmulevitz M. 2019. Single amino acid differences between closely related reovirus T3D lab strains alter oncolytic potency in vitro and in vivo. J Virol 94:e01688-19. doi: 10.1128/JVI.01688-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mohamed A, Smiley JR, Shmulevitz M. 2019. Polymorphisms in the most oncolytic reovirus strain confer enhanced cell attachment, transcription, and single-step replication kinetics. J Virol 94:e01937-19. doi: 10.1128/JVI.01937-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coffey MC, Strong JE, Forsyth PA, Lee PW. 1998. Reovirus therapy of tumors with activated Ras pathway. Science 282:1332–1334. doi: 10.1126/science.282.5392.1332. [DOI] [PubMed] [Google Scholar]
- 15.Strong JE, Coffey MC, Tang D, Sabinin P, Lee PW. 1998. The molecular basis of viral oncolysis: usurpation of the Ras signaling pathway by reovirus. EMBO J 17:3351–3362. doi: 10.1093/emboj/17.12.3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alain T, Hirasawa K, Pon KJ, Nishikawa SG, Urbanski SJ, Auer Y, Luider J, Martin A, Johnston RN, Janowska-Wieczorek A, Lee PW, Kossakowska AE. 2002. Reovirus therapy of lymphoid malignancies. Blood 100:4146–4153. doi: 10.1182/blood-2002-02-0503. [DOI] [PubMed] [Google Scholar]
- 17.Norman KL, Coffey MC, Hirasawa K, Demetrick DJ, Nishikawa SG, DiFrancesco LM, Strong JE, Lee PW. 2002. Reovirus oncolysis of human breast cancer. Hum Gene Ther 13:641–652. doi: 10.1089/10430340252837233. [DOI] [PubMed] [Google Scholar]
- 18.Hirasawa K, Nishikawa SG, Norman KL, Alain T, Kossakowska A, Lee PW. 2002. Oncolytic reovirus against ovarian and colon cancer. Cancer Res 62:1696–1701. [PubMed] [Google Scholar]
- 19.Hirasawa K, Nishikawa SG, Norman KL, Coffey MC, Thompson BG, Yoon CS, Waisman DM, Lee PW. 2003. Systemic reovirus therapy of metastatic cancer in immune-competent mice. Cancer Res 63:348–353. [PubMed] [Google Scholar]
- 20.Thirukkumaran CM, Nodwell MJ, Hirasawa K, Shi ZQ, Diaz R, Luider J, Johnston RN, Forsyth PA, Magliocco AM, Lee P, Nishikawa S, Donnelly B, Coffey M, Trpkov K, Fonseca K, Spurrell J, Morris DG. 2010. Oncolytic viral therapy for prostate cancer: efficacy of reovirus as a biological therapeutic. Cancer Res 70:2435–2444. doi: 10.1158/0008-5472.CAN-09-2408. [DOI] [PubMed] [Google Scholar]
- 21.Gong J, Sachdev E, Mita AC, Mita MM. 2016. Clinical development of reovirus for cancer therapy: an oncolytic virus with immune-mediated antitumor activity. World J Methodol 6:25–42. doi: 10.5662/wjm.v6.i1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bourhill T, Mori Y, Rancourt DE, Shmulevitz M, Johnston RN. 2018. Going (reo)viral: factors promoting successful reoviral oncolytic infection. Viruses 10:421. doi: 10.3390/v10080421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chappell JD, Duong JL, Wright BW, Dermody TS. 2000. Identification of carbohydrate-binding domains in the attachment proteins of type 1 and type 3 reoviruses. J Virol 74:8472–8479. doi: 10.1128/JVI.74.18.8472-8479.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barton ES, Connolly JL, Forrest JC, Chappell JD, Dermody TS. 2001. Utilization of sialic acid as a coreceptor enhances reovirus attachment by multistep adhesion strengthening. J Biol Chem 276:2200–2211. doi: 10.1074/jbc.M004680200. [DOI] [PubMed] [Google Scholar]
- 25.Barton ES, Forrest JC, Connolly JL, Chappell JD, Liu Y, Schnell FJ, Nusrat A, Parkos CA, Dermody TS. 2001. Junction adhesion molecule is a receptor for reovirus. Cell 104:441–451. doi: 10.1016/s0092-8674(01)00231-8. [DOI] [PubMed] [Google Scholar]
- 26.Kirchner E, Guglielmi KM, Strauss HM, Dermody TS, Stehle T. 2008. Structure of reovirus sigma1 in complex with its receptor junctional adhesion molecule-A. PLoS Pathog 4:e1000235. doi: 10.1371/journal.ppat.1000235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fraser RD, Furlong DB, Trus BL, Nibert ML, Fields BN, Steven AC. 1990. Molecular structure of the cell-attachment protein of reovirus: correlation of computer-processed electron micrographs with sequence-based predictions. J Virol 64:2990–3000. doi: 10.1128/JVI.64.6.2990-3000.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chappell JD, Prota AE, Dermody TS, Stehle T. 2002. Crystal structure of reovirus attachment protein sigma1 reveals evolutionary relationship to adenovirus fiber. EMBO J 21:1–11. doi: 10.1093/emboj/21.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dietrich MH, Ogden KM, Long JM, Ebenhoch R, Thor A, Dermody TS, Stehle T. 2018. Structural and functional features of the reovirus σ1 tail. J Virol 92:e00336-18. doi: 10.1128/JVI.00336-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van den Wollenberg DJ, van den Hengel SK, Dautzenberg IJ, Cramer SJ, Kranenburg O, Hoeben RC. 2008. A strategy for genetic modification of the spike-encoding segment of human reovirus T3D for reovirus targeting. Gene Ther 15:1567–1578. doi: 10.1038/gt.2008.118. [DOI] [PubMed] [Google Scholar]
- 31.van den Wollenberg DJ, Dautzenberg IJ, van den Hengel SK, Cramer SJ, de Groot RJ, Hoeben RC. 2012. Isolation of reovirus T3D mutants capable of infecting human tumor cells independent of junction adhesion molecule-A. PLoS One 7:e48064. doi: 10.1371/journal.pone.0048064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van den Wollenberg DJ, Dautzenberg IJ, Ros W, Lipińska AD, van den Hengel SK, Hoeben RC. 2015. Replicating reoviruses with a transgene replacing the codons for the head domain of the viral spike. Gene Ther 22:267–279. doi: 10.1038/gt.2014.126. [DOI] [PubMed] [Google Scholar]
- 33.Eaton HE, Kobayashi T, Dermody TS, Johnston RN, Jais PH, Shmulevitz M. 2017. African swine fever virus NP868R capping enzyme promotes reovirus rescue during reverse genetics by promoting reovirus protein expression, virion assembly, and RNA incorporation into infectious virions. J Virol 91:e02416-16. doi: 10.1128/JVI.02416-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kemp V, Dautzenberg IJC, Cramer SJ, Hoeben RC, van den Wollenberg DJM. 2018. Characterization of a replicating expanded tropism oncolytic reovirus carrying the adenovirus E4orf4 gene. Gene Ther 25:331–344. doi: 10.1038/s41434-018-0032-9. [DOI] [PubMed] [Google Scholar]
- 35.Kemp V, van den Wollenberg DJM, Camps MGM, van Hall T, Kinderman P, Pronk-van Montfoort N, Hoeben RC. 2019. Arming oncolytic reovirus with GM-CSF gene to enhance immunity. Cancer Gene Ther 26:268–281. doi: 10.1038/s41417-018-0063-9. [DOI] [PubMed] [Google Scholar]
- 36.van Raaij MJ, Mitraki A, Lavigne G, Cusack S. 1999. A triple beta-spiral in the adenovirus fibre shaft reveals a new structural motif for a fibrous protein. Nature 401:935–938. doi: 10.1038/44880. [DOI] [PubMed] [Google Scholar]
- 37.van Raaij MJ, Chouin E, van der Zandt H, Bergelson JM, Cusack S. 2000. Dimeric structure of the coxsackievirus and adenovirus receptor D1 domain at 1.7 A resolution. Structure 8:1147–1155. doi: 10.1016/S0969-2126(00)00528-1. [DOI] [PubMed] [Google Scholar]
- 38.Prota AE, Campbell JA, Schelling P, Forrest JC, Watson MJ, Peters TR, Aurrand-Lions M, Imhof BA, Dermody TS, Stehle T. 2003. Crystal structure of human junctional adhesion molecule 1: implications for reovirus binding. Proc Natl Acad Sci U S A 100:5366–5371. doi: 10.1073/pnas.0937718100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mandell KJ, McCall IC, Parkos CA. 2004. Involvement of the junctional adhesion molecule-1 (JAM1) homodimer interface in regulation of epithelial barrier function. J Biol Chem 279:16254–16262. doi: 10.1074/jbc.M309483200. [DOI] [PubMed] [Google Scholar]
- 40.Alloussi SH, Alkassar M, Urbschat S, Graf N, Gartner B. 2011. All reovirus subtypes show oncolytic potential in primary cells of human high-grade glioma. Oncol Rep 26:645–649. doi: 10.3892/or.2011.1331. [DOI] [PubMed] [Google Scholar]
- 41.Simon EJ, Howells MA, Stuart JD, Boehme KW. 2017. Serotype-specific killing of large cell carcinoma cells by reovirus. Viruses 9:140. doi: 10.3390/v9060140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vidal L, Pandha HS, Yap TA, White CL, Twigger K, Vile RG, Melcher A, Coffey M, Harrington KJ, DeBono JS. 2008. A phase I study of intravenous oncolytic reovirus type 3 Dearing in patients with advanced cancer. Clin Cancer Res 14:7127–7137. doi: 10.1158/1078-0432.CCR-08-0524. [DOI] [PubMed] [Google Scholar]
- 43.Karapanagiotou EM, Roulstone V, Twigger K, Ball M, Tanay M, Nutting C, Newbold K, Gore ME, Larkin J, Syrigos KN, Coffey M, Thompson B, Mettinger K, Vile RG, Pandha HS, Hall GD, Melcher AA, Chester J, Harrington KJ. 2012. Phase I/II trial of carboplatin and paclitaxel chemotherapy in combination with intravenous oncolytic reovirus in patients with advanced malignancies. Clin Cancer Res 18:2080–2089. doi: 10.1158/1078-0432.CCR-11-2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Galanis E, Markovic SN, Suman VJ, Nuovo GJ, Vile RG, Kottke TJ, Nevala WK, Thompson MA, Lewis JE, Rumilla KM, Roulstone V, Harrington K, Linette GP, Maples WJ, Coffey M, Zwiebel J, Kendra K. 2012. Phase II trial of intravenous administration of Reolysin((R)) (Reovirus serotype-3-Dearing strain) in patients with metastatic melanoma. Mol Ther 20:1998–2003. doi: 10.1038/mt.2012.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morris DG, Feng X, DiFrancesco LM, Fonseca K, Forsyth PA, Paterson AH, Coffey MC, Thompson B. 2013. REO-001: a phase I trial of percutaneous intralesional administration of reovirus type 3 dearing (Reolysin(R)) in patients with advanced solid tumors. Invest New Drugs 31:696–706. doi: 10.1007/s10637-012-9865-z. [DOI] [PubMed] [Google Scholar]
- 46.Sborov DW, Nuovo GJ, Stiff A, Mace T, Lesinski GB, Benson DM Jr, Efebera YA, Rosko AE, Pichiorri F, Grever MR, Hofmeister CC. 2014. A phase I trial of single-agent reolysin in patients with relapsed multiple myeloma. Clin Cancer Res 20:5946–5955. doi: 10.1158/1078-0432.CCR-14-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Noonan AM, Farren MR, Geyer SM, Huang Y, Tahiri S, Ahn D, Mikhail S, Ciombor KK, Pant S, Aparo S, Sexton J, Marshall JL, Mace TA, Wu CS, El-Rayes B, Timmers CD, Zwiebel J, Lesinski GB, Villalona-Calero MA, Bekaii-Saab TS. 2016. Randomized phase 2 trial of the oncolytic virus pelareorep (Reolysin) in upfront treatment of metastatic pancreatic adenocarcinoma. Mol Ther 24:1150–1158. doi: 10.1038/mt.2016.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mahalingam D, Fountzilas C, Moseley J, Noronha N, Tran H, Chakrabarty R, Selvaggi G, Coffey M, Thompson B, Sarantopoulos J. 2017. A phase II study of REOLYSIN((R)) (pelareorep) in combination with carboplatin and paclitaxel for patients with advanced malignant melanoma. Cancer Chemother Pharmacol 79:697–703. doi: 10.1007/s00280-017-3260-6. [DOI] [PubMed] [Google Scholar]
- 49.Kobayashi T, Antar AA, Boehme KW, Danthi P, Eby EA, Guglielmi KM, Holm GH, Johnson EM, Maginnis MS, Naik S, Skelton WB, Wetzel JD, Wilson GJ, Chappell JD, Dermody TS. 2007. A plasmid-based reverse genetics system for animal double-stranded RNA viruses. Cell Host Microbe 1:147–157. doi: 10.1016/j.chom.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kobayashi T, Ooms LS, Ikizler M, Chappell JD, Dermody TS. 2010. An improved reverse genetics system for mammalian orthoreoviruses. Virology 398:194–200. doi: 10.1016/j.virol.2009.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chakrabarty R, Tran H, Fortin Y, Yu Z, Shen SH, Kolman J, Onions D, Voyer R, Hagerman A, Serl S, Kamen A, Thompson B, Coffey M. 2014. Evaluation of homogeneity and genetic stability of REOLYSIN (pelareorep) by complete genome sequencing of reovirus after large scale production. Appl Microbiol Biotechnol 98:1763–1770. doi: 10.1007/s00253-013-5499-0. [DOI] [PubMed] [Google Scholar]
- 52.Wilcox ME, Yang W, Senger D, Rewcastle NB, Morris DG, Brasher PM, Shi ZQ, Johnston RN, Nishikawa S, Lee PW, Forsyth PA. 2001. Reovirus as an oncolytic agent against experimental human malignant gliomas. J Natl Cancer Inst 93:903–912. doi: 10.1093/jnci/93.12.903. [DOI] [PubMed] [Google Scholar]
- 53.Ruoslahti E. 1996. RGD and other recognition sequences for integrins. Annu Rev Cell Dev Biol 12:697–715. doi: 10.1146/annurev.cellbio.12.1.697. [DOI] [PubMed] [Google Scholar]
- 54.Brochu-Lafontaine V, Lemay G. 2012. Addition of exogenous polypeptides on the mammalian reovirus outer capsid using reverse genetics. J Virol Methods 179:342–350. doi: 10.1016/j.jviromet.2011.11.021. [DOI] [PubMed] [Google Scholar]
- 55.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. 2013. RNA-guided human genome engineering via Cas9. Science 339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cohn DE, Sill MW, Walker JL, O'Malley D, Nagel CI, Rutledge TL, Bradley W, Richardson DL, Moxley KM, Aghajanian C. 2017. Randomized phase IIB evaluation of weekly paclitaxel versus weekly paclitaxel with oncolytic reovirus (Reolysin) in recurrent ovarian, tubal, or peritoneal cancer: an NRG Oncology/Gynecologic Oncology Group study. Gynecol Oncol 146:477–483. doi: 10.1016/j.ygyno.2017.07.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bernstein V, Ellard SL, Dent SF, Tu D, Mates M, Dhesy-Thind SK, Panasci L, Gelmon KA, Salim M, Song X, Clemons M, Ksienski D, Verma S, Simmons C, Lui H, Chi K, Feilotter H, Hagerman LJ, Seymour L. 2018. A randomized phase II study of weekly paclitaxel with or without pelareorep in patients with metastatic breast cancer: final analysis of Canadian Cancer Trials Group IND.213. Breast Cancer Res Treat 167:485–493. doi: 10.1007/s10549-017-4538-4. [DOI] [PubMed] [Google Scholar]
- 58.Bradbury PA, Morris DG, Nicholas G, Tu D, Tehfe M, Goffin JR, Shepherd FA, Gregg RW, Rothenstein J, Lee C, Kuruvilla S, Keith BD, Torri V, Blais N, Hao D, Korpanty GJ, Goss G, Melosky BL, Mates M, Leighl N, Ayoub JP, Sederias J, Feilotter H, Seymour L, Laurie SA. 2018. Canadian Cancer Trials Group (CCTG) IND211: a randomized trial of pelareorep (Reolysin) in patients with previously treated advanced or metastatic non-small cell lung cancer receiving standard salvage therapy. Lung Cancer 120:142–148. doi: 10.1016/j.lungcan.2018.03.005. [DOI] [PubMed] [Google Scholar]
- 59.Jonker DJ, Tang PA, Kennecke H, Welch SA, Cripps MC, Asmis T, Chalchal H, Tomiak A, Lim H, Ko YJ, Chen EX, Alcindor T, Goffin JR, Korpanty GJ, Feilotter H, Tsao MS, Theis A, Tu D, Seymour L. 2018. A randomized phase II study of FOLFOX6/bevacizumab with or without pelareorep in patients with metastatic colorectal cancer: IND.210, a Canadian Cancer Trials Group Trial. Clin Colorectal Cancer 17:231–239. doi: 10.1016/j.clcc.2018.03.001. [DOI] [PubMed] [Google Scholar]
- 60.Kemp V, Hoeben RC, van den Wollenberg DJ. 2015. Exploring reovirus plasticity for improving its use as oncolytic virus. Viruses 8:4. doi: 10.3390/v8010004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mohamed A, Johnston RN, Shmulevitz M. 2015. Potential for improving potency and specificity of reovirus oncolysis with next-generation reovirus variants. Viruses 7:6251–6278. doi: 10.3390/v7122936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Maginnis MS, Forrest JC, Kopecky-Bromberg SA, Dickeson SK, Santoro SA, Zutter MM, Nemerow GR, Bergelson JM, Dermody TS. 2006. Beta1 integrin mediates internalization of mammalian reovirus. J Virol 80:2760–2770. doi: 10.1128/JVI.80.6.2760-2770.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sandekian V, Lemay G. 2015. A single amino acid substitution in the mRNA capping enzyme lambda2 of a mammalian orthoreovirus mutant increases interferon sensitivity. Virology 483:229–235. doi: 10.1016/j.virol.2015.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Drayna D, Fields BN. 1982. Activation and characterization of the reovirus transcriptase: genetic analysis. J Virol 41:110–118. doi: 10.1128/JVI.41.1.110-118.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Starnes MC, Joklik WK. 1993. Reovirus protein lambda 3 is a poly(C)-dependent poly(G) polymerase. Virology 193:356–366. doi: 10.1006/viro.1993.1132. [DOI] [PubMed] [Google Scholar]
- 66.Tao Y, Farsetta DL, Nibert ML, Harrison SC. 2002. RNA synthesis in a cage—structural studies of reovirus polymerase lambda3. Cell 111:733–745. doi: 10.1016/S0092-8674(02)01110-8. [DOI] [PubMed] [Google Scholar]
- 67.Tyler KL, Squier MK, Brown AL, Pike B, Willis D, Oberhaus SM, Dermody TS, Cohen JJ. 1996. Linkage between reovirus-induced apoptosis and inhibition of cellular DNA synthesis: role of the S1 and M2 genes. J Virol 70:7984–7991. doi: 10.1128/JVI.70.11.7984-7991.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Marcato P, Shmulevitz M, Pan D, Stoltz D, Lee PW. 2007. Ras transformation mediates reovirus oncolysis by enhancing virus uncoating, particle infectivity, and apoptosis-dependent release. Mol Ther 15:1522–1530. doi: 10.1038/sj.mt.6300179. [DOI] [PubMed] [Google Scholar]
- 69.Demidenko AA, Blattman JN, Blattman NN, Greenberg PD, Nibert ML. 2013. Engineering recombinant reoviruses with tandem repeats and a tetravirus 2A-like element for exogenous polypeptide expression. Proc Natl Acad Sci U S A 110:E1867–76. doi: 10.1073/pnas.1220107110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kanai Y, Kawagishi T, Matsuura Y, Kobayashi T. 2019. In vivo live imaging of oncolytic mammalian orthoreovirus expressing NanoLuc luciferase in tumor xenograft mice. J Virol 93:e00401-19. doi: 10.1128/JVI.00401-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kawagishi T, Kanai Y, Tani H, Shimojima M, Saijo M, Matsuura Y, Kobayashi T. 2016. Reverse genetics for fusogenic bat-borne orthoreovirus associated with acute respiratory tract infections in humans: role of outer capsid protein σC in viral replication and pathogenesis. PLoS Pathog 12:e1005455. doi: 10.1371/journal.ppat.1005455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mashiko D, Fujihara Y, Satouh Y, Miyata H, Isotani A, Ikawa M. 2013. Generation of mutant mice by pronuclear injection of circular plasmid expressing Cas9 and single guided RNA. Sci Rep 3:3355. doi: 10.1038/srep03355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ishii K, Ueda Y, Matsuo K, Matsuura Y, Kitamura T, Kato K, Izumi Y, Someya K, Ohsu T, Honda M, Miyamura T. 2002. Structural analysis of vaccinia virus DIs strain: application as a new replication-deficient viral vector. Virology 302:433–444. doi: 10.1006/viro.2002.1622. [DOI] [PubMed] [Google Scholar]
- 75.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]