Zika virus (ZIKV) has been linked to serious neurologic disorders and causes widespread concern in the field of global public health. Inositol requiring enzyme 1α (IRE1α) is an ER-related transmembrane protein that mediates unfolded protein response (UPR) pathway. Here, we revealed that IRE1α is a proviral factor for ZIKV replication both in culture cells and mice model, which relies on its kinase and RNase activities. Importantly, we further provided evidence that upon ZIKV infection, IRE1α is activated and splices XBP1 mRNA which enhances the expression of monounsaturated fatty acids rate-limiting enzyme stearoyl coenzyme A (stearoyl-CoA) desaturase 1 (SCD1) and subsequent lipid droplet production. Our data uncover a novel mechanism of IRE1α proviral effect by modulating lipid metabolism, providing the first evidence of a close relationship between IRE1α-mediated UPR, lipid metabolism, and ZIKV replication and indicating IRE1α inhibitors as potentially effective anti-ZIKV agents.
KEYWORDS: ZIKV, IRE1α, UPR, SCD1, lipid metabolism
ABSTRACT
Zika virus (ZIKV) is an emerging mosquito-borne flavivirus which has become a global epidemic threat due to its rapid spread and association with serious consequences of infection, including neonatal microcephaly. Inositol-requiring enzyme 1α (IRE1α) is an endoplasmic reticulum (ER)-related transmembrane protein that mediates unfolded protein response (UPR) pathway and has been indicated to play an important role in flavivirus replication. However, the mechanism of how IRE1α affects ZIKV replication remains unknown. In this study, we explored the role of IRE1α in ZIKV infection in vitro and in vivo by using CRISPR/Cas9-based gene knockout and RNA interference-based gene knockdown techniques. Both knockout and knockdown of IRE1α dramatically reduced ZIKV replication levels, including viral RNA levels, protein expression, and titers in different human cell lines. Trans-complementation with IRE1α restored viral replication levels decreased by IRE1α depletion. Furthermore, the proviral effect of IRE1α was dependent on its kinase and RNase activities. Importantly, we found that IRE1α promoted the replication of ZIKV through upregulating the accumulation of monounsaturated fatty acid (MUFA) rate-limiting enzyme stearoyl coenzyme A (stearoyl-CoA) desaturase 1 (SCD1), which further affected the production of oleic acid (OA) and lipid droplet. Finally, our data demonstrated that in the brain tissues of ZIKV-infected mice, the replication levels of ZIKV and virus-related lesions were significantly suppressed by both the kinase and RNase inhibitors of IRE1α. Taken together, our results identified IRE1α as a ZIKV dependency factor which promotes viral replication through affecting SCD1-mediated lipid metabolism, potentially providing a novel molecular target for the development of anti-ZIKV agents.
IMPORTANCE Zika virus (ZIKV) has been linked to serious neurologic disorders and causes widespread concern in the field of global public health. Inositol requiring enzyme 1α (IRE1α) is an ER-related transmembrane protein that mediates unfolded protein response (UPR) pathway. Here, we revealed that IRE1α is a proviral factor for ZIKV replication both in culture cells and mice model, which relies on its kinase and RNase activities. Importantly, we further provided evidence that upon ZIKV infection, IRE1α is activated and splices XBP1 mRNA which enhances the expression of monounsaturated fatty acids rate-limiting enzyme stearoyl coenzyme A (stearoyl-CoA) desaturase 1 (SCD1) and subsequent lipid droplet production. Our data uncover a novel mechanism of IRE1α proviral effect by modulating lipid metabolism, providing the first evidence of a close relationship between IRE1α-mediated UPR, lipid metabolism, and ZIKV replication and indicating IRE1α inhibitors as potentially effective anti-ZIKV agents.
INTRODUCTION
Zika virus (ZIKV) belongs to the Flavivirus genus of the Flaviviridae family, together with dengue virus (DENV), yellow fever virus (YFV), West Nile virus (WNV), tick-borne encephalitis virus (TBEV), and Japanese encephalitis virus (JEV) (1–3). ZIKV was first discovered in Uganda in 1947. In the wake of its emergence in French Polynesia and the Americas, ZIKV has been linked to serious neurologic disorders, such as Guillain-Barré syndrome and neonatal microcephaly, and causes widespread concern in the field of global public health (2–4). Currently, specific treatment and vaccine for ZIKV infection are not available in clinics.
ZIKV genome is a positive-sense single-stranded RNA which encodes three structural proteins and seven nonstructural proteins. Following virion endocytosis and uncoating, viral proteins are translated and endoplasmic reticulum (ER) membrane can be induced to form viral replication complex in the perinuclear region, where ZIKV replication takes place (4–6).
The ER is an important organelle which is involved in multiple cell functions, including protein folding and secretion, lipid synthesis, and calcium homeostasis. Several quality control mechanisms ensure that those properly folded proteins exit the ER, while improperly folded proteins are retained and degraded (7). The life cycle of viruses within the Flaviviridae family, including ZIKV, DENV, WNV, YFV, JEV, and hepatitis C virus (HCV), relies on the extensive membrane network of the ER for translation, replication, and packaging (7–11). Infection of these ER-tropic viruses disrupts normal ER functions and triggers ER stress. In order to alleviate the ER stress, the unfolded protein response (UPR) pathway is activated for translation attenuation, properly protein folding, and protein degradation (12). The sensors of the UPR pathway are inositol-requiring enzyme 1α (IRE1α), protein kinase R (PKR)-like ER kinase (PERK), and transcription factor 6 (ATF6) (12).
IRE1α is the most evolutionarily conserved sensor of the UPR. In response to ER stress, IRE1α is activated through oligomerization and autophosphorylation. In mammals, the cytosolic domain of activated IRE1α induces the activation of the c-Jun N-terminal kinase (JNK) pathway (13, 14). IRE1α has been reported to modulate autophagy upon ER stress by activating the JNK pathway (15). Moreover, the cytosolic RNase activity of IRE1α can be activated to initiate the splicing of XBP1 mRNA (16). The spliced XBP1 mRNA encodes transcription factor XBP1, which regulates the expression of a broad spectrum of UPR-related genes involved in protein folding, ER-associated degradation (ERAD), and protein quality control (17). In addition, IRE1α also targets other specific mRNAs, leading to a process known as regulated IRE1-dependent decay (RIDD) (18, 19).
Previously, an association between flavivirus and the UPR has been established by several groups. The IRE1α-mediated UPR pathway is activated by virus infection and plays a pivotal role in DENV, JEV, TBEV, and ZIKV infection (1, 7, 20). Importantly, the UPR contributes to cortical neurogenesis in mammalian brain development. Triggering of the URP in mouse brain impairs the generation of intermediate progenitors and ultimately leads to microcephaly (21). In 2017, Gladwyn-Ng et al. further showed that the UPR pathway is dramatically activated in ZIKV-infected mice and associates with ZIKV-related microcephaly, which was reversed by treatment of IRE1α and PERK inhibitors (22). In a later study, the IRE1α-XBP1-mediated UPR pathway was reported to promote ZIKV infection (23). However, Carletti’s group showed that priming with an inducer of the UPR during flavivirus infection (DENV, ZIKV, WNV, and TBEV) caused a decrease of viral titers and depletion of IRE1α-enhanced viral replication (1). Therefore, whether IRE1α-mediated URP pathway plays a proviral or antiviral role in ZIKV replication remains controversial.
To clarify the role of IRE1α in ZIKV replication, our study utilized several strategies: depletion of IRE1α through CRISPR/Cas9-based gene editing technology in human lung carcinoma epithelial cell line (A549), knockdown of IRE1α in several cell lines, and employment of IRE1α inhibitors in in vitro-cultured cells and an in vivo mouse model. We found that IRE1α is a proviral factor for ZIKV replication both in vitro and in vivo. The proviral function of IRE1α relies on its kinase and RNase activities via the downstream XBP1 pathway. Our data further showed that IRE1α-XBP1 pathway activation by ZIKV infection upregulated the accumulation of stearoyl coenzyme A (stearoyl-CoA) desaturase 1 (SCD1), a rate-limiting enzyme catalyzing the synthesis of monounsaturated fatty acids (MUFA), which affected oleic acid (OA) and lipid droplet production. These findings demonstrated that the UPR sensor IRE1α promotes ZIKV infection through affecting SCD1-mediated lipid metabolism, suggesting that inhibition of IRE1α is a potential antiviral strategy, and revealed a close relationship among ZIKV, the IRE1α-mediated UPR, and lipid metabolism.
RESULTS
ZIKV replication is reduced in IRE1α-deficient cells.
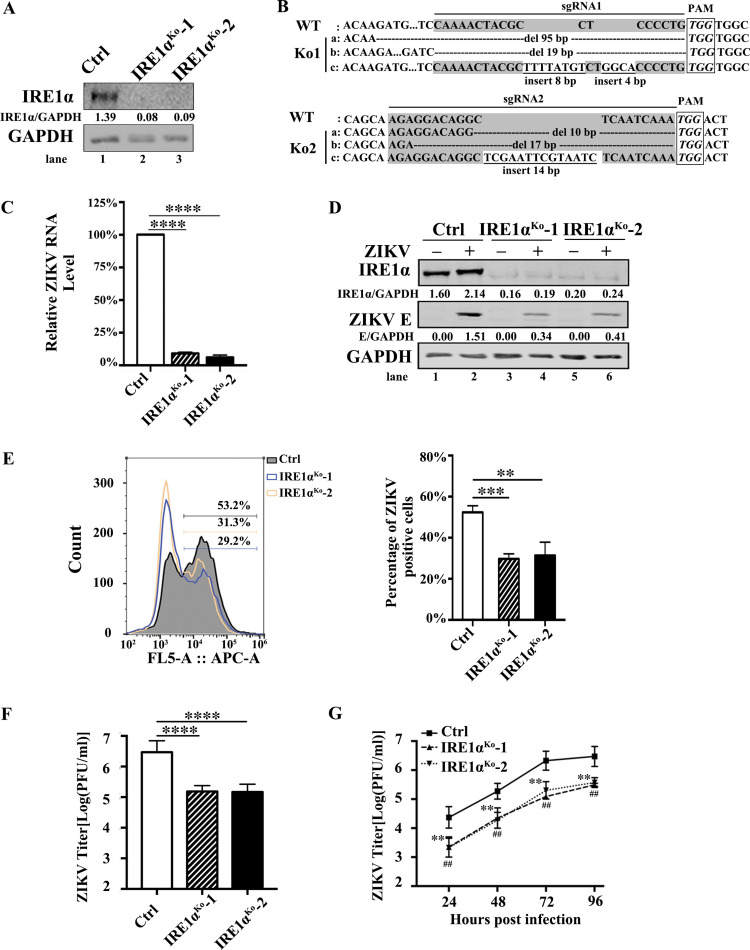
To test the role of IRE1α in ZIKV (H/PF/2013 strain) replication, IRE1α knockout (IRE1αKo) A549 cells were generated by a CRISPR/Cas9 gene-editing technique and two monoclonal cell lines were isolated. To monitor the knockout efficiency, the expression of IRE1α in IRE1αKo cells was examined by Western blotting. IRE1α protein was not detected in IRE1αKo-1 or IRE1αKo-2 cells (Fig. 1A), indicating that the IRE1α gene was successfully edited in IRE1αKo cells (Fig. 1B). To examine whether IRE1α affects ZIKV replication, control cells and IRE1αKo cells were infected with ZIKV at a multiplicity of infection (MOI) of 3 and harvested at 24 h postinfection (p.i.). Then, the viral RNA levels, protein expression, infection rate, and titers were detected in these cells through quantitative real-time PCR (qRT-PCR), Western blotting, flow cytometry, and plaque assay, respectively. Compared to those in the control cells, the knockout of IRE1α led to a significant reduction of viral RNA levels (∼90% [Fig. 1C]). Similarly, the Western blotting result showed that ZIKV E protein levels in IRE1αKo cells were dramatically lower than in the control group (Fig. 1D). Moreover, the flow cytometry data showed that the percentages of E-positive-staining cells in IRE1α-deficient cells were markedly downregulated (Fig. 1E). Likewise, the plaque assay data showed that the viral titers of IRE1α knockout cells were ∼90% lower than in the control cells (Fig. 1F). We then performed a multistep virus growth assay to explore whether IRE1α affects the replication and virus multiplication cycle of ZIKV. The control and IRE1αKo cells were infected with ZIKV at an MOI of 0.01. The supernatants were collected at different time points (24, 48, 72, and 96 h p.i.) and then titers were determined. In the absence of IRE1α, the viral yields were significantly decreased (∼12-fold at 24 h p.i., ∼10-fold at 48 h p.i., ∼11-fold at 72 h p.i., and ∼10-fold at 96 h p.i., respectively) (Fig. 1G), suggesting that IRE1α enhances the ZIKV replication and virus multiplication cycle.
FIG 1.
ZIKV replication is reduced in IRE1α knockout cells. (A) The IRE1α knockout A549 cells were generated by the CRISPR/Cas9 technique. Two different knockout cell lines (IRE1αKo-1 and IRE1αKo-2) were confirmed by Western blotting. (B) Confirmation of IRE1α knockout efficiency by DNA sequencing. (C to F) Viral replication levels in IRE1αKo A549 cells. Control cells and IRE1αKo cells were infected with ZIKV at an MOI of 3. Then, the cells were harvested at 24 h p.i. for qRT-PCR (C), Western blotting (D), flow cytometry (E), and plaque assay (F). (G) Multistep virus growth assay. Control cells and IRE1αKo cells were infected with ZIKV at an MOI of 0.01. The supernatants were collected at 24, 48, 72, and 96 h p.i. for the measurement of viral titers by standard plaque assay. Human β-ACTIN mRNA level was measured as an internal control for qRT-PCR. Data are shown as means ± SD from at least three independent experiments. ##, P < 0.01; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 (unpaired, two-tailed Student t test).
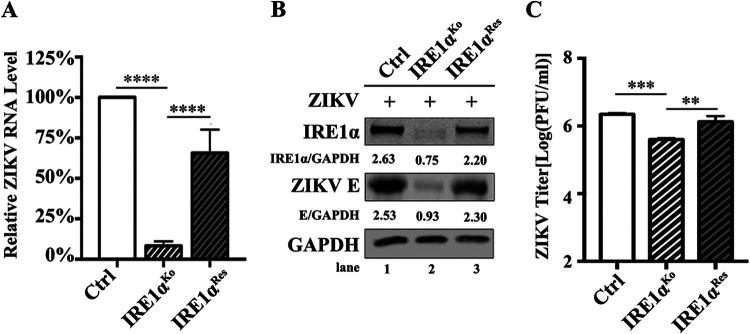
To rule out the possibility that potential off-target effects of IRE1α-specific sgRNAs lead to the reduction of virus infection, we introduced the wild-type (wt) IRE1α gene into IRE1αKo cells by lentivirus-mediated transduction. The IRE1αRes cells were sorted by flow cytometry. The control, IRE1αKo, and IRE1αRes cells were infected with ZIKV at an MOI of 3 and harvested at 24 h p.i. for viral replication detection. The viral RNA levels (Fig. 2A), ZIKV E protein levels (Fig. 2B), and viral yields (Fig. 2C) were significantly rescued in IRE1αRes cells. Together, these observations demonstrated that IRE1α promotes the replication of ZIKV.
FIG 2.
ZIKV replication is restored in IRE1α-rescuing cells. (A to C) Viral replication levels in IRE1αRes A549 cells. Control cells, IRE1αKo cells, and IRE1αRes cells were infected with ZIKV at an MOI of 3 and harvested at 24 h p.i. for qRT-PCR (A) or Western blotting (B). The supernatants were collected at 24 h p.i. for plaque assay (C). Human β-ACTIN mRNA level was measured as an internal control for qRT-PCR. Data are shown as means ± SD from at least three independent experiments. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 (unpaired, two-tailed Student t test).
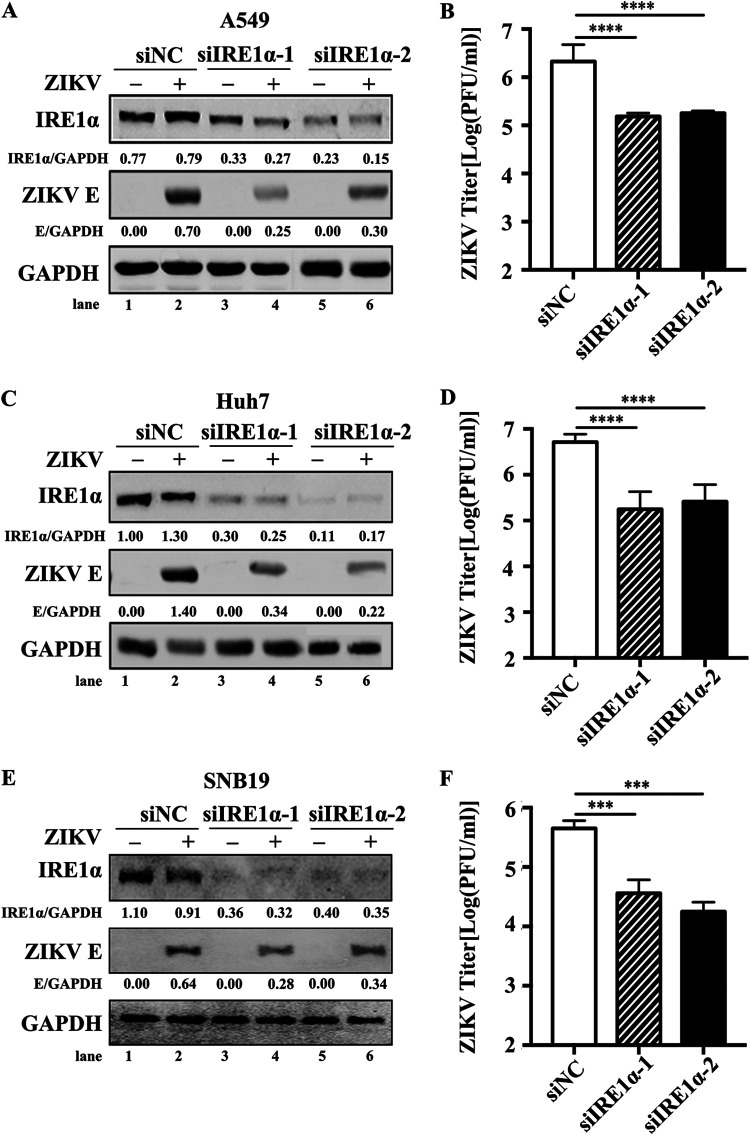
We further validated the proviral effect of IRE1α in different cell lines (A549, Huh7, and SNB19 cells) using an RNA interference (RNAi) strategy. Cells were transfected with a negative-control (NC) small interfering RNA (siRNA) (siNC) or IRE1α-specific siRNAs (siIRE1α-1 or siIRE1α-2). The protein levels of IRE1α in A549 cells transfected with siIRE1αs were dramatically lower than those in the control group (Fig. 3A, lanes 1, 3, and 5), indicating that the knockdown of IRE1α was effective. To test whether IRE1α has an impact on ZIKV replication, A549 cells were transfected with siNC or siIRE1αs for 48 h and then infected with ZIKV at an MOI of 3. Cells and supernatants were harvested at 24 h p.i. for Western blotting and plaque assay. The levels of ZIKV E protein were markedly reduced by the knockdown of IRE1α (Fig. 3A). The viral yields in IRE1α knockdown (IRE1αKd) cells were decreased by ∼90% (Fig. 3B). Similarly, a significant reduction of ZIKV E protein levels and viral titers was also observed in Huh7 IRE1α knockdown cells, suggesting that IRE1α also plays a vital role in Huh7 cells during ZIKV infection (Fig. 3C and D). As ZIKV infection is associated with serious nervous system diseases, we also examined the role of IRE1α in ZIKV replication in human glioma cells (SNB19) (24). Compared to those in the control cells, siIRE1α led to a dramatic reduction of viral E protein level and titers in SNB19 cells (Fig. 3E and F), suggesting that the proviral effect of IRE1α is not cell specific.
FIG 3.
ZIKV replication is reduced in IRE1α knockdown cells. (A and B) Viral replication levels in IRE1α knockdown A549 cells. A549 cells were transfected with siNC, siIRE1α-1, or siIRE1α-2. At 48 h posttransfection, the cells were infected with ZIKV at an MOI of 3. Then, the cells were harvested at 24 h p.i. for the measurement of IRE1α and ZIKV E protein levels (A). The supernatants were collected at 24 h p.i. for plaque assay (B). (C and D) Viral replication levels in IRE1α knockdown Huh7 cells. Huh7 cells were transfected with siRNAs, followed by ZIKV infection at an MOI of 3. The cells and supernatants were harvested at 24 h p.i. for Western blotting (C) and plaque assay (D). (E and F) Viral replication levels in IRE1α knockdown SNB19 cells. SNB19 cells were transfected with siRNAs, followed by ZIKV infection at an MOI of 3. The cells and supernatants were then harvested at 24 h p.i. to detect viral E protein levels (E) and viral titers (F) as described above. GAPDH was probed as an internal control (A, C, and E). Data are shown as means ± SD from at least three independent experiments. ***, P < 0.001; ****, P < 0.0001 (unpaired, two-tailed Student t test).
Both kinase and RNase activities of IRE1α are required for its proviral effect.
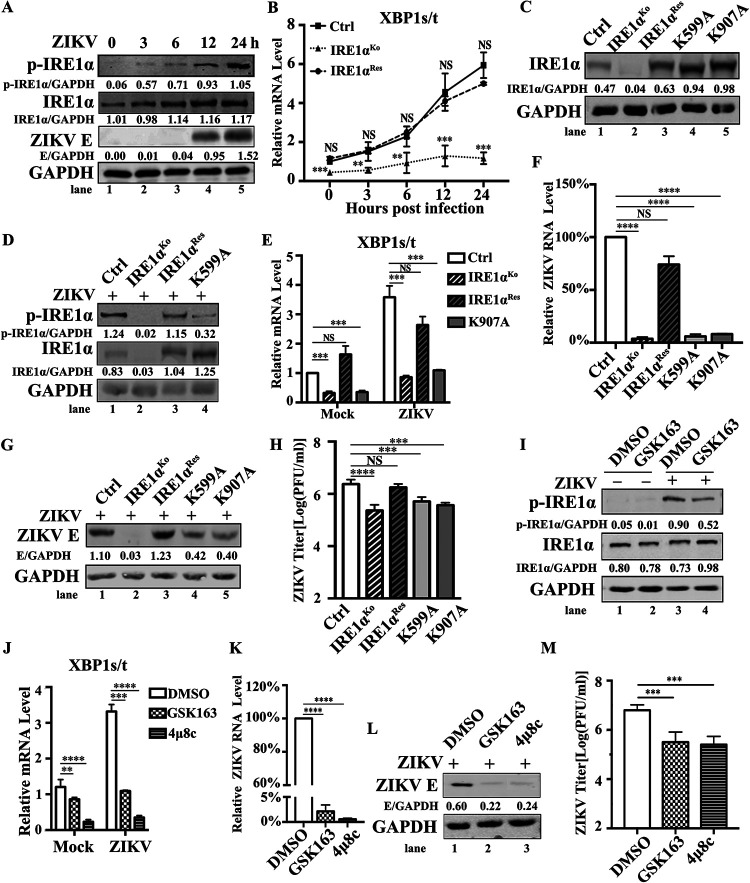
As the IRE1α-mediated UPR pathway can be activated in neural cells upon ZIKV infection, we explored whether ZIKV infection induces the activation of IRE1α in A549 cells (20). A549 cells were infected with ZIKV at an MOI of 3 and collected at different time points (0, 3, 6, 12, and 24 h p.i.). The level of phosphorylated IRE1α was analyzed by Western blotting. Compared with that at 0 h p.i., ZIKV-infected A549 cells exhibited an upregulation of phosphorylated IRE1α at 12 and 24 h p.i. (Fig. 4A). Moreover, because ZIKV-mediated activation of IRE1α further splices XBP1u to XPB1s, we also compared the mRNA levels of XBP1t (total XBP1, including both XBP1u and XBP1s) and XBP1s in the control, IRE1αKo, and IRE1αRes cells via qRT-PCR (20, 25–27). As shown in Fig. 4B, ZIKV infection dramatically promoted the splicing of XBP1 in the control cells (Fig. 4B, full line). As expected, IRE1αKo cells exhibited downregulation of splicing XBP1 (Fig. 4B, dotted line) upon ZIKV infection, and IRE1αRes cells rescued this phenomenon (Fig. 4B, dashed line). Collectively, the findings demonstrated that ZIKV infection leads to the phosphorylation of IRE1α and subsequent cleavage of XBP1 in A549 cells.
FIG 4.
The kinase and RNase activities of IRE1α are required for ZIKV replication. (A) IRE1α phosphorylation levels upon ZIKV infection in A549 cells. A549 cells were infected with ZIKV at an MOI of 3. The cells were collected at the indicated time points for Western blotting using antibodies against IRE1α, p-IRE1α, ZIKV E, or GAPDH. (B) XBP1 splicing levels upon ZIKV infection in IRE1αKo and IRE1αRes cells. Control cells, IRE1αKo cells, and IRE1αRes cells were infected with ZIKV at an MOI of 3 and harvested at 24 h p.i. for qRT-PCR to measure XBP1 splicing levels. (C) IRE1α mutant restoration levels. Control cells, IRE1αKo cells, IRE1αRes cells, IRE1α-K599A cells, and IRE1α-K907A cells were harvested for Western blotting. Primary antibodies against IRE1α or GAPDH were used. (D) Phosphorylation levels of IRE1α in IRE1α-K599A cells. Control cells, IRE1αKo cells, IRE1αRes cells, and IRE1α-K599A cells were infected with ZIKV at an MOI of of 3 and collected at 24 h p.i. for Western blotting to examine the phosphorylation levels of IRE1α. (E) XBP1 splicing levels in IRE1α-K907A cells. Control cells, IRE1αKo cells, IRE1αRes cells, and IRE1α-K907A cells were infected with ZIKV at an MOI of 3 and harvested for qRT-PCR to measure the splicing of XBP1. (F to H) Viral replication levels in IRE1α-K599A and IRE1α-K907A cells. Control cells, IRE1αKo cells, IRE1αRes cells, IRE1α-K599A cells, and IRE1α-K907A cells were infected with ZIKV at an MOI of 3. The cells and supernatants were collected at 24 h p.i. for the measurement of viral RNA levels (F), viral E protein levels (G), and viral yields (H). (I) Impact of GSK163 on phosphorylation levels of IRE1α. A549 cells were treated with 30 μM GSK163 and infected with ZIKV at an MOI of 3 at 1 h posttreatment. The cells were then collected at 24 h p.i. for Western blotting to detect the phosphorylation levels of IRE1α. (J) Effect of GSK163 or 4μ8c on XBP1 splicing levels. A549 cells were treated with 30 μM GSK163 or 4μ8c and infected with ZIKV at an MOI of 3 at 1 h posttreatment. The cells were harvested at 24 h p.i. for qRT-PCR to measure XBP1 splicing levels. (K to M) Impact of IRE1α inhibitors on ZIKV replication levels. A549 cells were treated with 30 μM GSK163 or 4μ8c and infected with ZIKV (MOI, 3) at 1 h posttreatment. The cell samples were collected at 24 h p.i. for the examination of viral RNA levels (K), viral E protein levels (L), and viral titers (M). Human β-ACTIN mRNA level was measured as an internal control for qRT-PCR. Data are representative of those from three independent experiments. NS, no statistical significance. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 (two-tailed Student t test).
Considering that the kinase and RNase activities of IRE1α are essential for its sensor function, we further investigated whether the kinase and RNase activities of IRE1α are required for its proviral effect. Two lentivirus vectors, CSII-EF-MCS-IRES2-Venus-IRE1αK599A-FLAG (K599, a reserved site for its kinase activation, was mutated into A599 in wt IRE1α) and CSII-EF-MCS-IRES2-Venus-IRE1αK907A-FLAG (K907, which is a key site for its RNase activity, was mutated into A907 in wt IRE1α), were generated by a site-directed mutation strategy (28). The A549 cell populations stably expressing the IRE1αK599A or IRE1αK907A mutant were sorted by flow cytometry. Restoration levels of IRE1α mutants in these cells were confirmed by Western blotting (Fig. 4C). Furthermore, the effect of IRE1αK599A on phosphorylated IRE1α was confirmed (Fig. 4D), and the effect of IRE1αK907A on the splicing of XBP1 was verified via qRT-PCR (Fig. 4E). Then, we explored the role of the kinase and RNase activities of IRE1α in ZIKV infection. The control, IRE1αKo, IRE1αRes, IRE1αK599A, and IRE1αK907A cells were infected with ZIKV at an MOI of 3 and harvested at 24 h p.i. for ZIKV replication detection. The viral RNA levels (Fig. 4F), ZIKV E protein levels (Fig. 4G), and viral yields (Fig. 4H) were significantly downregulated in IRE1αK599A and IRE1αK907A cells, which were similar to those in IRE1αKo cells.
Moreover, we investigated the effects of two IRE1α inhibitors, GSK2850163 (also named GSK163, an inhibitor of IRE1α kinase activity) and 4μ8c (an inhibitor of IRE1α RNase activity), on IRE1α proviral function (29, 30). The inhibitory effect of GSK163 (30 μM) or 4μ8c (30 μM) on IRE1α activities was confirmed by Western blotting (Fig. 4I) or by XBP1 splicing detection via qRT-PCR (Fig. 4J). A549 cells were treated with 30 μM GSK163 or 4μ8c for 1 h and then infected with ZIKV at an MOI of 3. Compared to those in the control group, GSK163 or 4μ8c treatment resulted in a marked decrease in the viral RNA levels (Fig. 4K), ZIKV E protein levels (Fig. 4L), and viral titers (Fig. 4M). Therefore, these data suggested that both the kinase and RNase activities of IRE1α are required for its proviral function.
The proviral role of IRE1α is mediated by the IRE1α-XBP1 pathway through affecting SCD1 accumulation.
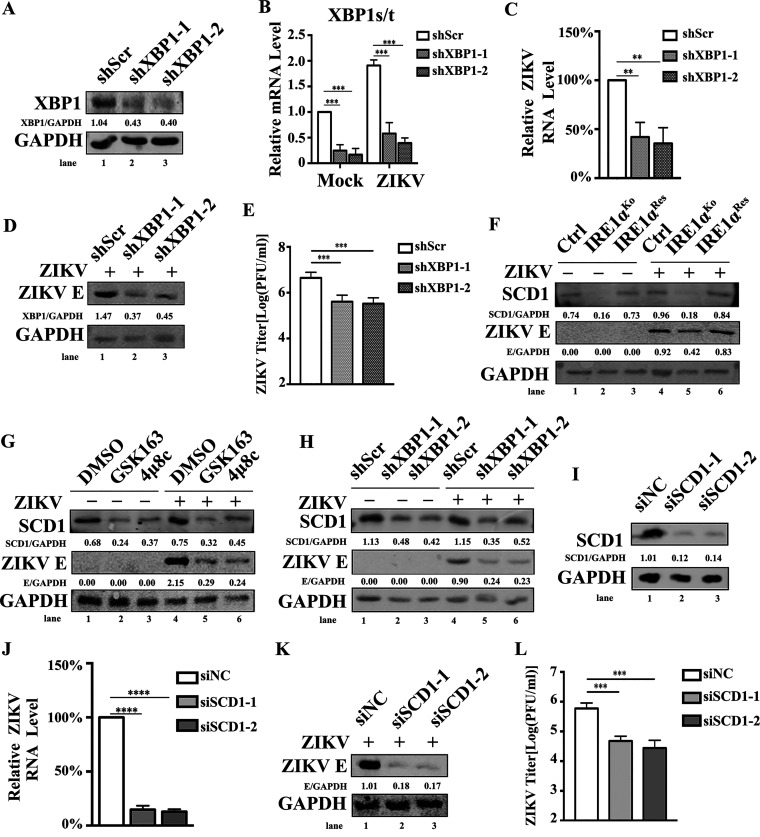
Since ZIKV replication requires both IRE1α and XBP1 in HeLa cells (23), we speculated that the proviral role of IRE1α in A549 cells also involves XBP1. To test this speculation, A549 cells were treated with scrambled short hairpin RNA (shRNA) (shScr) or two shRNAs targeting XBP1 (shXBP1-1 and shXBP1-2). Endogenous XBP1 protein level and splicing of XBP1 were significantly decreased in XPB1-specific shRNA-transduced A549 cells (Fig. 5A and B). Next, the shRNA-expressing cells were infected with ZIKV at an MOI of 3 and collected at 24 h p.i. for the examination of ZIKV replication. Viral RNA levels (Fig. 5C), ZIKV E protein levels (Fig. 5D), and viral yields (Fig. 5E) were dramatically decreased in shXBP1-transduced cells compared to those in the control cells, indicating that the IRE1α-XBP1 pathway plays an important role in the replication of ZIKV in A549 cells.
FIG 5.
The IRE1α-XBP1 signaling pathway upregulates the expression of SCD1 and promotes viral replication. (A and B) A549 cells were infected with lentivirus containing scramble shRNA (shScr), shXBP1-1, or shXBP1-2 and selected by puromycin for 1 week. Then, the cells were infected with ZIKV at an MOI of 3 and collected at 24 h p.i. to measure the expression of XBP1 (A) and the splicing of XBP1 (B). (C to E) ZIKV replication levels in XBP1 knockdown cells. shScr-, shXBP1-1-, and shXBP1-2-expressing cells were infected with ZIKV at an MOI of 3. Then, the cells and supernatants were harvested at 24 h p.i. to detect viral RNA levels (C), viral E protein levels (D), and viral yields (E) as described above. (F) SCD1 expression levels in IRE1αKo and IRE1αRes cells. Control cells, IRE1αKo cells, and IRE1αRes cells were infected with ZIKV at an MOI of 3 and were collected at 24 h p.i. for Western blotting to detect the expression levels of SCD1. (G) Impact of IRE1α inhibitors on SCD1 expression levels. A549 cells were treated with 30 μM GSK163 or 4μ8c and infected with ZIKV (MOI, 3) at 1 h posttreatment. Then, the cells were collected at 24 h p.i. for Western blotting to measure the expression levels of SCD1. (H) SCD1 expression levels in XBP1 knockdown cells. shScr-, shXBP1-1-, and shXBP1-2-expressing cells were infected with ZIKV at an MOI of 3 and harvested at 24 h p.i. for Western blotting to measure the expression levels of SCD1. (I) Knockdown efficiency of SCD1-specific siRNAs. A549 cells were transfected with siSCD1 for 48 h. Then, the cells were collected to measure the expression of SCD1 by Western blotting. (J to L) Effect of SCD1 knockdown on ZIKV replication levels. A549 cells were transfected with siSCD1, followed by ZIKV infection at an MOI of 3. Then, the cells and supernatants were harvested at 24 h p.i. to detect viral RNA levels (J), viral E protein levels (K), and viral yields (L) as described above. Human β-ACTIN mRNA level was measured as an internal control for qRT-PCR. Data are representative of those from three independent experiments. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 (two-tailed Student t test).
The literature indicated that the IRE1α-XBP1 pathway regulates the expression of lipid metabolism-associated genes, such as the SCD1 gene (31–33). SCD1 has been demonstrated to promote flavivirus replication through affecting the synthesis level of MUFA (34, 35). Therefore, we hypothesized that IRE1α might promote ZIKV replication through regulating SCD1-mediated lipid metabolism. To test this possibility, we first detected the expression levels of SCD1 in mock- or ZIKV-infected control cells, IRE1αKo cells, and IRE1αRes cells. Compared to those in the control and IRE1αRes cells, the expression levels of SCD1 in IRE1αKo cells were significantly decreased before and after viral infection (Fig. 5F), indicating that SCD1 expression is dependent on IRE1α. Moreover, the expression levels of SCD1 in both mock- and ZIKV-infected cells were also dramatically reduced by IRE1α inhibitors (GSK163 and 4μ8c) (Fig. 5G). Consistently, the knockdown of XBP1 resulted in decreased expression levels of SCD1 (Fig. 5H), implying that the IRE1α-XBP1 pathway is critical for the expression of SCD1. Importantly, in the context of IRE1α or XBP1 depletion, the expression levels of ZIKV E protein were markedly decreased accompanying the downregulation of SCD1 expression levels (Fig. 5F to H), suggesting that there is a close relationship between SCD1 and ZIKV replication. To determine the impact of SCD1 on ZIKV infection, we tested viral replication levels in SCD1 knockdown cells. A549 cells were transfected with SCD1-specific siRNAs, followed by ZIKV infection at an MOI of 3. Cells and supernatants were collected at 24 h p.i. for viral replication detection. As shown in Fig. 5I to L, SCD1 depletion led to a dramatic reduction of ZIKV RNA levels, viral E protein levels, and viral yields. Taken together, these results demonstrated that the depletion of IRE1α or blocking the IRE1α-XBP1 pathway reduces the expression of SCD1, further resulting in the decrease of ZIKV replication.
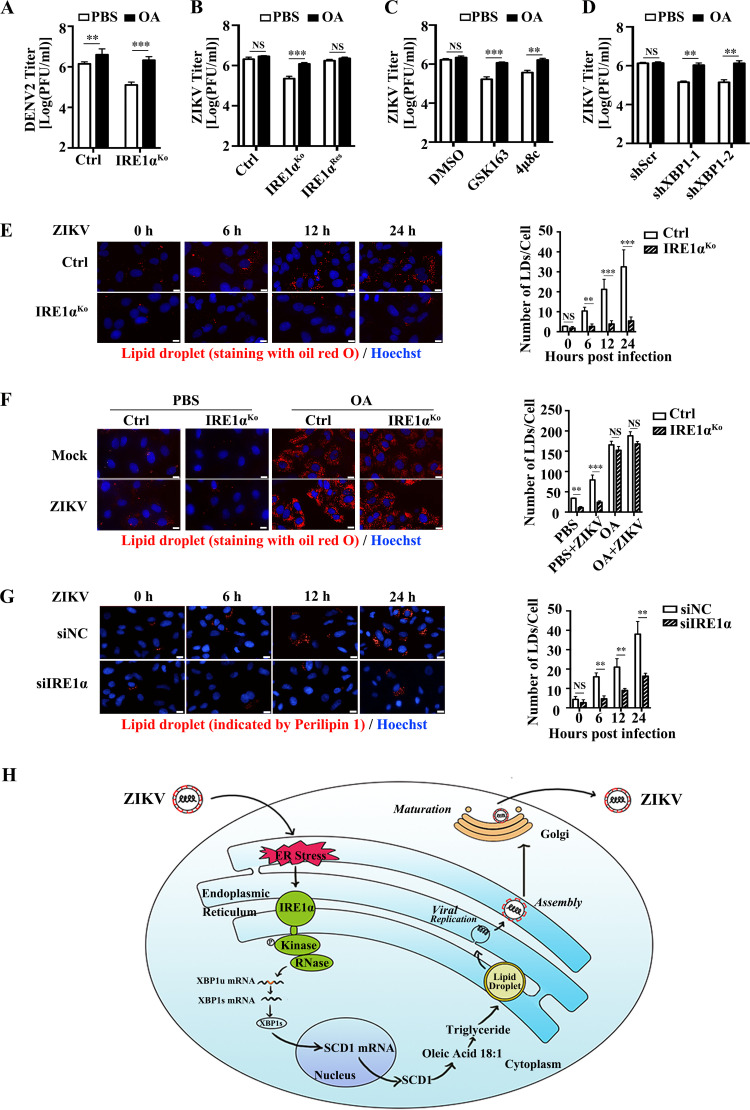
SCD1 is the only enzyme that synthesizes oleic acid (36). Oleic acid is a key component of phospholipids, cholesterol esters, and triglycerides, which are required for flavivirus infection (37–40). Therefore, we hypothesized that the addition of exogenous oleic acid might rescue the inhibitory effect of IRE1α depletion on viral replication. To test this hypothesis, the control cells, IRE1αKo cells, and IRE1αRes cells were treated with oleic acid during ZIKV or DENV2 infection. Supernatants were harvested for plaque assay at 24 h p.i. As expected, oleic acid addition significantly promoted the replication of DENV in the control A549 cells, consistent with previous studies (Fig. 6A) (34, 35). In contrast, ZIKV replication in the control A549 cells was not affected by oleic acid addition (Fig. 6B, left bars), suggesting that the requirements of oleic acid by DENV and ZIKV are different. Interestingly, addition of oleic acid successfully restored ZIKV replication in the IRE1α-deficient cells (Fig. 6B, middle bars), implying that the proviral effect of IRE1α requires oleic acid, a product of SCD1.
FIG 6.
IRE1α promotes ZIKV replication through modulating oleic acid and lipid droplet production. (A) Impact of oleic acid (OA) on DENV2 titers. Control cells and IRE1αKo cells were infected with DENV2 at an MOI of 3, followed by OA (50 μM) addition at 1 h p.i. The supernatants were collected at 24 h p.i. for plaque assay. (B) Impact of OA on ZIKV titers. Control cells, IRE1αKo cells, and IRE1αRes cells were infected with ZIKV at an MOI of 3. Then, 50 μM OA was added at 1 h p.i. The supernatants were collected at 24 h p.i. for plaque assay. (C) Effect of OA on viral titers in IRE1α inhibitor-treated cells. A549 cells were treated with 30 μM GSK163 or 4μ8c. Then, the cells were infected with ZIKV at 1 h posttreatment, followed by 50 μM OA addition at 1 h p.i. The supernatants were collected at 24 h p.i. for plaque assay. (D) Impact of OA on ZIKV titers in XBP1-deficient cells. shScr-, shXBP1-1-, and shXBP1-2-expressing cells were infected with ZIKV at an MOI of 3, followed by 50 μM OA addition. The supernatants were collected at 24 h p.i. for plaque assay. (E) Production of lipid droplets in IRE1αKo cells. Control cells and IRE1αKo cells were infected with ZIKV at an MOI of 3 and harvested at 0, 6, 12, and 24 h p.i. for staining with oil red O or Hoechst to visualize lipid droplets or the nucleus, respectively. At least 200 cells from each sample in three independent experiments were counted. The scale bar represents 20 μm. (F) Impact of OA on lipid droplet production in IRE1αKo cells. Control cells and IRE1αKo cells were infected with ZIKV (MOI 3), followed by OA (50 μM) or the same volume of PBS addition at 1 h p.i. Then, the cells were harvested at 24 h p.i. for the staining with oil red O or Hoechst to visualize lipid droplet or the nucleus, respectively. At least 200 cells from each sample in three independent experiments were counted. The scale bar represents 20 μm. (G) Production of lipid droplets in IRE1α knockdown cells. HepG2 cells were transfected with IRE1α-specific siRNAs for 48 h and then infected with ZIKV at an MOI of 3. Cells were fixed at 0, 6, 12, and 24 h p.i. and processed for immunofluorescence microscopy for detection of lipid droplet protein perilipin 1 (red) and Hoechst (blue). At least 200 cells from each sample in three independent experiments were counted. Scale bar, 20 μm. (H) A proposed model to show the mechanism of IRE1α promoting ZIKV replication. In response to ZIKV infection, IRE1α is activated and splices the XBP1 mRNA, which enhances the expression of SCD1 and subsequent lipid droplet production, ultimately promoting virus replication. Data are representative of those from three independent experiments. **, P < 0.01; ***, P < 0.001 (two-tailed Student t test).
In addition, we investigated whether oleic acid could rescue the viral replication level suppressed by IRE1α inhibitors. The ZIKV-infected cells were treated with oleic acid in the presence of IRE1α inhibitor (GSK163 or 4μ8c). In line with the result shown in Fig. 6B (left bars), the addition of oleic acid did not affect viral titers in dimethyl sulfoxide (DMSO)-treated cells (Fig. 6C, left bars). In contrast, the ZIKV replication levels were rescued by adding oleic acid in the presence of IRE1α inhibitors (Fig. 6C, middle and right bars). Moreover, we examined whether oleic acid could reverse the decrease of viral replication mediated by XBP1 knockdown. Consistently, the viral replication levels of the control cells were comparable in the absence and presence of oleic acid (Fig. 6D, left bars). But in the XBP1 knockdown cells, the addition of oleic acid largely restored the ZIKV replication level compared to that with the addition of phosphate-buffered saline (PBS) (Fig. 6D, middle and right bars). These data suggested that the proviral role of IRE1α and XBP1 in ZIKV replication is mediated by SCD1 (or oleic acid).
Considering that oleic acid is an important material of triglyceride, which is a component of lipid droplets, we examined whether the knockout of IRE1α influences the production of lipid droplet. Control cells and IRE1αKo cells were infected with ZIKV at an MOI of 3 and then stained with oil red O (a dye specific for lipid droplets) at different time points (0, 6, 12, and 24 h p.i.) (41). The numbers of lipid droplets in the control cells were increased in response to ZIKV infection, and the production of lipid droplets in the IRE1α-deficient cells was markedly reduced at each time point compared with that in the control group (Fig. 6E). Furthermore, the impact of oleic acid on lipid droplet production in IRE1αKo cells was also investigated. Mock- and ZIKV-infected cells were treated with oleic acid or PBS at 1 h p.i. and then stained with oil red O at 24 h p.i. The addition of oleic acid dramatically reversed the decrease of lipid droplet production in IRE1α knockout cells compared to that with the PBS treatment (Fig. 6F). Moreover, lipid droplet protein perilipin 1 was used to monitor the formation of lipid droplets (42, 43). Consistent with the oil red O staining results, we observed the same phenomenon in ZIKV-infected IRE1α knockdown HepG2 cells: the amounts of lipid droplets indicated by perilipin 1 were increased in the control group upon ZIKV infection, while lipid droplet production in IRE1α-deficient cells was significantly decreased compared to that in the control cells (Fig. 6G). Together, these results indicated that IRE1α is required for the regulation of SCD1 expression, which further regulates the production of oleic acid and lipid droplets and ultimately influences ZIKV replication (Fig. 6H).
IRE1α promotes ZIKV replication in neonatal mice.
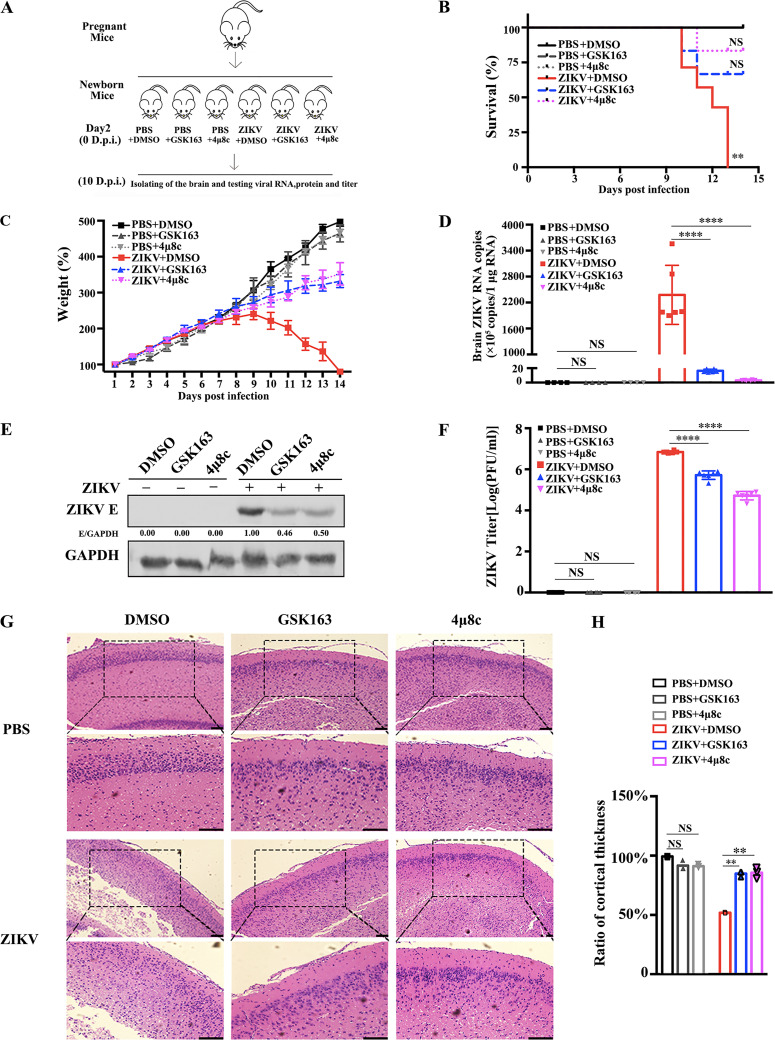
To test whether IRE1α has a proviral effect in vivo, we established a ZIKV infection mouse model and utilized two IRE1α inhibitors, GSK163 and 4μ8c, which are widely used in vitro studies (29, 30). Kunming neonatal mice, widely used for ZIKV infection (44–46), were treated with DMSO, GSK163 (1 mM), or 4μ8c (1 mM) by intracerebral inoculation. At 1 h after drug treatment, the mice were intracerebrally inoculated with a mock treatment or 4 × 104 PFU of ZIKV (Fig. 7A). The survival percentages (Fig. 7B) and weights of each group (Fig. 7C) were monitored daily after infection. In the DMSO-treated group, the survival percentages in the ZIKV-infected mice (Fig. 7B, red line) were about 50% at 10 days p.i., and all the mice died by 13 days p.i. Significant reduction in the weight of ZIKV-infected mice was observed from 10 days p.i. (Fig. 7C, red line). In contrast, the survival percentages of ZIKV-infected mice were 66.7% in GSK163-treated group and 83.3% in 4μ8c-treated group, respectively (Fig. 7B). The weights of ZIKV-infected mice treated with inhibitors were also increased significantly compared to those of the mice treated with DMSO (Fig. 7C). Moreover, the mouse brains were collected at 10 days p.i. for the detection of viral replication levels and for pathological observations by hematoxylin and eosin (H&E) staining. As expected, high ZIKV replication levels, including viral RNA copies, viral E protein, and viral loads, were readily detected in DMSO-treated mice. In contrast, the viral replication levels in the brains of GSK163- or 4μ8c-treated mice were dramatically decreased (Fig. 7D to F), indicating that IRE1α inhibitors effectively block ZIKV replication in vivo. The H&E images (Fig. 7G) showed that in the control group (PBS- or DMSO-treated group), the arrangement of cortical cells was normal and the addition of IRE1α inhibitor alone had no significant impact on the cortical layer of mouse brains. Upon ZIKV infection, the cortical thickness of mouse brain tissues was significantly reduced and the cells were not arranged in an orderly manner (Fig. 7G and H). Interestingly, the treatment with IRE1α inhibitors largely alleviated the ZIKV-mediated pathology of mouse brains: the cortical thickness and cell arrangement were similar to those in the control group, indicating that IRE1α inhibitors can effectively block ZIKV-triggered brain lesions. Taken together, our data demonstrated that IRE1α promotes ZIKV infection in vivo.
FIG 7.
IRE1α inhibitors suppress viral replication in ZIKV-infected Kunming neonatal mice. (A) An experimental scheme of the effect of IRE1α inhibitors in the ZIKV-infected neonatal mouse model. Kunming neonatal mice were treated with 1 mM GSK163 or 4μ8c (DMSO as the control) by intracerebral inoculation on day 2 after birth. Then, they were intracerebrally inoculated with 4 × 104 PFU of ZIKV suspension at 1 h posttreatment. Mock-infected controls were inoculated with the same volume of PBS. (B) Survival curves of neonatal mice injected with intracerebral inoculation under different treatment conditions (n = 6). Kaplan-Meier survival curves were analyzed by the log rank test. **, P < 0.01. (C) Changes in mouse weight were calculated daily. The weight changes were compared using one-way ANOVA. Error bars indicate standard errors of the means. (D to F) ZIKV replication levels in mouse brain tissues. The brains of neonatal mice were isolated at 10 days p.i. for the detection of viral replication levels. Viral RNA copies (D), viral protein levels (E), and viral titers (F) were determined by qRT-PCR, Western blotting, and plaque assay, respectively. Viral RNA copies were expressed as viral RNA equivalents per microgram upon comparing with a standard curve produced by gradient 10-fold dilutions of ZIKV RNA. (G) Hematoxylin and eosin (H&E) staining of mouse brain tissues was performed, and the cortical radial thickness is indicated with bars (10 μm). Coronal sections of cerebral cortex marked by dashed squares were magnified two times and shown below (scale bar, 20 μm). (H) Quantification of the above-described sample cortex thickness by ImageJ software. The thickness of the control cortex was normalized to 1. Data are representative of those from three independent experiments. **, P < 0.01; ****, P < 0.0001 (two-tailed Student t test).
DISCUSSION
IRE1α is the most evolutionarily conserved sensor of the UPR, which is a coordinated countermeasure in gene expression induced by perturbations in functions of ER, including the global arrest of new protein synthesis, enhanced expression of several chaperones, and upregulation of protein folding capability or degradation (12). The IRE1α-mediated UPR signaling pathway is activated by ER stress and plays an important role in flavivirus infection (1, 7, 20). However, the roles of the IRE1α-mediated UPR pathway in the ZIKV life cycle remain controversial. In this report, we provide strong evidence to support the notion that IRE1α functions as a ZIKV dependency factor both in vitro and in vivo. We further delineated the action mechanism by which the IRE1α-XBP1 pathway modulates the expression of SCD1 and subsequent lipid metabolism.
First, we generated two IRE1α-deficient A549 monoclonal cell lines by the CRISPR/Cas9 gene editing technique. Consistent with previous studies on DENV and ZIKV infection (1, 8), the disruption of IRE1α led to the significant reduction of ZIKV RNA levels, viral E protein levels, and viral titers. Together with data from the multistep virus growth assay, these results indicated that IRE1α contributes to both the replication and multiplication cycle of ZIKV. In addition, IRE1α-specific siRNA-mediated endogenous IRE1α knockdown in different cell lines (A549, Huh7, and SNB19 cells) resulted in dramatic downregulation of ZIKV replication, implying that the proviral function of IRE1α in ZIKV infection is not cell specific. Furthermore, restoration of IRE1α in IRE1αKo A549 cells rescued the replication levels of ZIKV, suggesting that the off-target possibility of IRE1α-specific single guide RNAs (sgRNAs) can be ruled out. Our conclusion agreed with previous work with ZIKV-infected IRE1αKd HeLa cells and DENV-infected IRE1α−/− mouse embryonic fibroblasts (MEFs) (8, 23) but disagreed with a report showing that the induction of the UPR causes a marked antiviral response depending on IRE1α in U2OS cells during flavivirus infection (DENV, ZIKV, WNV, and TBEV) (1). These contradictory conclusions may be due to the off-target effect of IRE1α siRNA or the activation of compensatory pathways induced by an activator of the UPR (tunicamycin) that influence the outcome of the experiment.
As an important sensor of the UPR, IRE1α can be autophosphorylated upon flavivirus infection and then activate its RNase activity to splice the downstream target mRNAs (7, 9, 20, 47). Therefore, we further explored the correlation between the enzyme activities and proviral effect of IRE1α. Similar to the observations in IRE1αKo cells, both the expression of IRE1α enzyme activity mutants and the treatment with two kinds of IRE1α inhibitors significantly impaired the replication levels of ZIKV, indicating that the proviral function of IRE1α relies on its kinase and RNase activities. This finding is consistent with a previous study showing that IRE1α inhibitors (KIRA6 and STF-083010) can prevent ZIKV-induced cytotoxicity (23). In 2017, Gladwyn-Ng et al. demonstrated that the UPR triggered by viral infection contributes to ZIKV-associated microcephaly and treatment with an IRE1α inhibitor (4μ8c) prevented microcephaly in mouse embryonic brains (22). Recently, Chu et al. found that an IRE1α inhibitor (KIRA6) can alleviate ZIKV-induced neurogenesis and testicular pathologies (48). In our study, we constructed a ZIKV-infected neonatal mouse model and examined the in vivo effects of two kinds of IRE1α inhibitors (GSK163 and 4μ8c). The replication levels of ZIKV in the infected mouse brains were markedly suppressed by both the kinase and RNase inhibitors of IRE1α. Moreover, the survival percentages and the weights of ZIKV-infected mice were dramatically rescued by IRE1α inhibitors. As expected, the cortical thickness and cell arrangement in virus-infected mouse brain tissues were close to normal upon the treatment with IRE1α inhibitors, which agrees with the previous conclusion reported for ZIKV-infected mouse embryonic brains (22). Our data indicated that IRE1α promotes ZIKV infection both in vitro and in vivo and its enzyme activity inhibitors can be developed as effective anti-ZIKV agents. In the future, a greater panel of IRE1α inhibitors, such as GSK163, 4μ8c, APY29, KIRA6, and STF-083010 (23, 49), and their combinations, could be further evaluated, including their antiviral effects and determination of 50% inhibitory concentrations (IC50) for the development of antiviral drugs.
Previous studies indicated that IRE1α may regulate the infection of flavivirus through different mechanisms. IRE1α increases DENV-induced autophagy via the JNK signaling pathway, thus promoting viral replication in Huh7 cells (50). We also detected the role of the IRE1α-JNK pathway in ZIKV infection. However, knockdown of JNK in A549 cells slightly reduced ZIKV replication (data not shown), suggesting that the IRE1α-JNK pathway may partially promote ZIKV replication. In addition, the IRE1α-mediated XBP1 pathway has also been reported to play a role in flavivirus replication (7, 20, 23, 48). ZIKV infection activates the IRE1α-XBP1 pathway and viral RNA levels can be decreased in IRE1α or XBP1 knockdown HeLa cells, but the underlying molecular mechanism remains unknown (23). In contrast, negative regulation of the IRE1α-XBP1 pathway in flavivirus infection has also been reported. The HCV-triggered IRE1α-XBP1 pathway results in a reduction of virus production (51). Blocking of the XBP1-mediated pathway by and IRE1α RNase inhibitor (4μ8c) in U2OS cells or knockout of XPBP1 in MEFs did not affect the infection of DENV, WNV, and TBEV (1, 8, 52). In our study, we proved the positive impact of the IRE1α-XBP1 pathway on ZIKV infection by knocking down XBP1 or using an inhibitor of IRE1α RNase activity required for XPB1 splicing. These inconsistent data indicated that different cell responses mediated by the IRE1α-XBP1 pathway could be triggered by different viruses.
Importantly, our findings established an association between the IRE1α-XBP1 pathway, lipid metabolism, and ZIKV infection. We observed that the blockade of the IRE1α-XBP1 pathway decreased the expression of SCD1 (the rate-limiting enzyme in MUFA biosynthesis), which further led to the reduction of ZIKV replication. Consistently, the addition of oleic acid, an important synthetic product of SCD1, rescued the reduction of ZIKV replication both in IRE1αKo cells, shXBP1-transduced cells, and IRE1α inhibitor-treated cells. Our results agree with the published studies showing that SCD1 is required for the replication and release of flavivirus, which controls lipid requirements during viral infection (34, 35). Hijacking host lipid metabolism (especially lipid droplet metabolism) is a characteristic of flavivirus (53, 54). The lipid droplet is an ER-localized dynamic organelle that plays a pivotal role in cellular lipid metabolism and energy storage. It regulates cellular fatty acids and neutral lipids levels and can be manipulated by flavivirus to benefit viral replication, assembly, and particle infectivity (55, 56). The size and quantity of lipid droplet in host cells can be significantly increased in response to flavivirus infection (6, 53, 57, 58). Moreover, flavivirus utilizes lipid droplets as a platform for viral assembly and encapsidation processes to facilitate viral replication (54, 59, 60). Several groups have shown the interaction between flavivirus protein (such as HCV core protein/NS5 protein or DENV C protein/NS4A protein) and lipid droplets (61–64). Disrupting the ability of HCV core protein or DENV C protein to coat lipid droplets impairs viral replication and virion formation (60, 65, 66). Recently, colocalization of ZIKV C protein and lipid droplets was observed through confocal microscopy analysis, indicating the role of lipid metabolism in ZIKV replication and pathogenesis (67, 68). As expected, we observed an increasing number of lipid droplets in response to ZIKV infection. Furthermore, the production of lipid droplets was significantly decreased in ZIKV-infected IRE1αKo cells and restored to control levels by the addition of oleic acid, implying that IRE1α is involved in SCD1-mediated lipid droplet metabolism. Our findings reveal a new mechanism in which ZIKV infection promotes cellular lipogenesis or affects dynamics of lipid droplets through the IRE1α-XBP1 pathway. Further evidence needs to be provided to elucidate the mechanism of lipid droplet formation affected by the activation of IRE1-XBP1 pathway upon ZIKV infection. Until now, research about the lipid components of flavivirus has rarely focused on its fatty acid content and saturation levels. As the requirement for MUFAs in flavivirus lipid envelope is necessary for virion maturation (35), the content or saturation level of MUFA in the envelope of ZIKV virions and the outcome of these alterations need to be explored in the future.
In conclusion, we uncovered a novel mechanism of IRE1α proviral effect by modulating lipid metabolism. Upon ZIKV infection, IRE1α is activated and splices the XBP1 mRNA, which enhances the expression of SCD1 and subsequent lipid droplet production, ultimately promoting virus replication. Moreover, the proviral effect of IRE1α on ZIKV infection is dependent on its kinase and RNase activities. Our report provides the first evidence of a close relationship between the IRE1α-mediated UPR, lipid metabolism, and ZIKV replication and indicates IRE1α inhibitors as potentially effective anti-ZIKV agents.
MATERIALS AND METHODS
Ethics statement.
The use of laboratory animals and animal-related experiments were reviewed and approved by the Sun Yat-sen University Institutional Animal Care and Use Committee (SYSU IACUC); the approval number is 2019-109. All animal-related experiments were performed in accordance with animal research reporting of in vivo experiments guidelines approved by the SYSU IACUC and were conducted in the laboratory designed to ensure biological safety.
Animal models and related experiments.
Pregnant Kunming mice were purchased and maintained under specific-pathogen-free conditions at the research animal facility of Sun Yat-sen University. Neonatal Kunming mice were breastfed by their mothers and divided into different groups as indicated in Fig. 6A. Two-day-old neonatal Kunming mice were treated with GSK2850163 (1 mM) or 4μ8c (1 mM) in a 20-μl volume by intracerebral injection. After 1 h, mice were intracerebrally inoculated with ZIKV suspension (4 × 104 PFU) in a 20-μl volume and monitored over 14 days. Control animals received an equal volume of DMSO through the same injection route. All animals were cage-bred with the mouse mothers during the experiment and observed daily until the development of symptoms. All inoculated mice were euthanized by cervical dislocation after the appearance of severe clinical signs, including lethargy and paralysis.
Viral RNA copies, viral protein levels, and viral titers in mouse brain tissues were determined. Whole-brain tissue homogenates were prepared using an automated homogenizer. Tissues were ground with PBS buffer and centrifuged at 12,000 × g for 15 min at 4°C. The supernatants were collected and stored at –80°C for later use. Total RNA was extracted using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. ZIKV copy analysis was performed using GraphPad Prism 7.0 software. Tissue specimens obtained from neonatal mouse brains were lysed with radioimmunoprecipitation assay (RIPA) lysis buffer for Western blotting. The supernatants from neonatal mouse brains were filtered through a 0.45-μm filter, and plaque assay was performed to detect the titers of ZIKV.
Cell culture.
Human lung carcinoma epithelial cells (A549), human hepatoma cells (Huh7), human glioblastoma cells (SNB19), human embryonic kidney cells (293T), baby hamster kidney cells (BHK21), and African green monkey kidney cells (Vero) were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 100 U/ml of streptomycin and penicillin (Invitrogen) and 5% or 10% fetal bovine serum (FBS; Gibco) at 37°C with 5% CO2.
Virus, virus infection, and titration.
ZIKV strain H/PF/2013 (GenBank accession number KJ776791) and DENV2 New Guinea C (NGC; GenBank accession number AF038403) were obtained from Guangzhou Centers for Disease Control. Virus was propagated in Vero cells. The supernatants were harvested when cytopathic effect appeared, and the cell debris was removed by centrifugation and filtration. Titers of virus stocks were determined and then stocks were stored at −80°C. In the single-step virus growth assay, cells were infected with ZIKV at an MOI of 3. The cells were collected for Western blotting or qRT-PCR. The supernatants were harvested at 24 h p.i. for virus titration by standard plaque or focus-forming assay (FFA) in Vero cells or BHK21 cells as previously described (69). In the multistep virus growth assay, cells were infected with ZIKV at an MOI of 0.01. The supernatants were collected at 24, 48, 72, and 96 h p.i. for virus titration.
RNA interference.
The sequences of siRNAs targeting human IRE1α mRNA and SCD1 mRNA are shown in Table 1. A control siRNA with scrambled sequence was used as a negative control (NC). Transfection was carried out with 16 nmol of siRNAs by using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer’s instructions. At 48 h posttransfection (p.t.), cells were harvested for further analysis.
TABLE 1.
Sequences for siRNAs
| Gene | Sequence (5′–3′) |
|---|---|
| siIRE1α-1-5F | GGAGAGAAGCAGCAGACUU |
| siIRE1α-1-3R | AAGUCUGCUGCUUCUCUCC |
| siIRE1α-2-5F | GCGUAAAUUCAGGACCUAU |
| siIRE1α-2-3R | AUAGGUCCUGAAUUUACGC |
| siSCD1-1-5F | UGGAAGACGACAUUCGCCCUGAUAU |
| siSCD1-1-3R | AUAUCAGGGCGAAUGUCGUCUUCCA |
| siSCD1-2-5F | CCACCUCUUCGGAUAUCGUCCUUAU |
| siSCD1-2-3R | AUAAGGACGAUAUCCGAAGAGGUGG |
Western blotting.
Cells were harvested at and treated with RIPA lysis buffer (pH 7.4) (50 mM Tris-HCl, 0.5% [vol/vol] NP-40, 1% Triton X-100, 150 mM NaCl, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride [PMSF], 1% protease inhibitor cocktails, 1 mM Na3VO4, and 1 mM NaF). Western blotting was then performed as previously described (69). The primary antibodies used in this study are as follows: anti-IRE1α (Cell Signaling Technology), anti-phospho-IRE1 (Novus Biologicals), anti-ZIKV envelope (E) (Gene Tex), anti-XBP1 (Proteintech), anti-SCD (Santa Cruz), anti-perilipin 1 (Cell Signaling Technology), and anti-glyceraldehyde-3-phosphate dehydrogenase (anti-GAPDH; Proteintech). Secondary antibodies included IRDye 800 CW-conjugated anti-rabbit IgG, IRDye 680 CW-conjugated anti-mouse IgG (LI-COR), horseradish peroxidase (HRP)-conjugated anti-mouse IgG (CST), and anti-rabbit IgG (Bio-Rad). Immunoreactive bands were analyzed with the Odyssey infrared imaging system (LI-COR). The Quantity One program (Bio-Rad) was used to quantify the Western blotting results.
qRT-PCR.
Total RNAs were extracted using TRIzol reagent (Invitrogen) and reverse transcribed using HI Script Q RT SuperMix (Vazyme) according to the manufacturer’s instructions. Quantitative real-time PCR (qRT-PCR) analysis was performed using the SYBR Premix Ex Taq (TaKaRa) on a CFX96 real-time system (Bio-Rad). Differences of gene expression in qRT-PCR data were analyzed using the change in threshold cycle (ΔCT) values as described previously (70). The primer sequences used in qRT-PCR are listed in Table 2.
TABLE 2.
Sequences of primers used in qRT-PCR
| Gene | Sequence (5′–3′) |
|---|---|
| ZIKV NS1-5F | GTCAGAGCAGCAAAGACAA |
| ZIKV NS1-3R | CAGCCTCCTTTCCCTTAACA |
| XBP1s-5F | TGCTGAGTCCGCAGCAGG |
| XBP1s-3R | GGGCTTGGTATATATGTGG |
| XBP1t-5F | CCTTGTAGTTGAGAACCAGG |
| XBP1t-3R | GGGGCTTGGTATATATGTGG |
| β-ACTIN-5F | GCTCCTCCTGAGCGCAAG |
| β-ACTIN-3R | CATCTGCTGGAAGGTGGACA |
Plasmid construction.
The oligonucleotide sequences used for generation of single guide RNAs (sgRNAs) are listed in Table 3. Pairs of forward and reverse oligonucleotides for sgRNAs were annealed and then inserted into plasmid vector LentiCRISPR v2 (Addgene). The resulting plasmids were designated pLenti-sgIRE1α-1 and pLenti-sgIRE1α-2.
TABLE 3.
Sequences of primers used in CRISPR/Cas9 gene editing for IRE1α
| Primer | Sequence (5′–3′) | Targeting region |
|---|---|---|
| sgIRE1α-1-5F | CACCGCCAAAACTACGCCTCCCCTG | Exon 4 |
| sgIRE1α-1-3R | AAACCAGGGGAGGCGTAGTTTTGGC | |
| sgIRE1α-2-5F | CACCGAGAGGACAGGCTCAATCAAA | Exon 2 |
| sgIRE1α-2-3R | AAACTTTGATTGAGCCTGTCCTCTC |
Plasmid pcDNA3.1-IRE1α-Res was mutated at synonymous sites in IRE1α-specific sgRNA-targeted sequence in order to resist the knockout by sgIRE1α. To obtain the plasmid encoding the IRE1α kinase activity mutant or IRE1α RNase activity mutant, the lysine (Lys) at amino acid (aa) 599 of IRE1α or the Lys at aa 907 of IRE1α was mutated to alanine (Ala) by site-directed mutagenesis. Briefly, IRE1α-Res, IRE1α-K599A, and IRE1α-K907A fragments were amplified from pcDNA3.1-IRE1α plasmid using the Phusion high-fidelity DNA polymerase (New England BioLabs) (the sequences of primers are listed in Table 4). The PCR products were then incubated with DpnI, which allowed for rapid circularization of the PCR product and removal of the template DNA. The digested products were transformed into Stbl3 cells. The mutations were verified by DNA sequencing. IRE1αRes, IRE1α-K599A, or IRE1α-K907A fragment was then amplified by PCR and inserted into the lentiviral vector CSII-EF-MCS-IRES2-Venus.
TABLE 4.
Sequences of oligonucleotides used in mutations
| Gene | Sequence (5′–3′) |
|---|---|
| IRE1αRes-5F | ATTTGCGGCCGCATGGCCGAGATCAAGGAGAAAAT |
| IRE1αRes-3R | CGCGGATCCCTATACTGGGCAGAGATAAAAG |
| IRE1α-K599A-5F | ATTTGCGGCCGCATGAATCCGCGGCAGGGGTATT |
| IRE1α-K599A-3R | ATTTGCGGCCGCCTATACTGGGCAGAGATAAAA |
| IRE1α-K907A-5F | GACAATTCTGCTCGAAGAAGCCAAAGCC |
| IRE1α-K907A-3R | GGCTTTGGCTTCTTCGAGCAGAATTGTC |
Generation of knockout cells by CRISPR/Cas9 gene editing.
293T cells were transfected with sgRNA vector (pLenti-sgIRE1α-1 or pLenti-sgIRE1α-2) and two packaging plasmids (psPAX2 and pMD2.G) using FuGENE HD transfection reagent (Promega). Supernatants were collected at 48 h p.t. and passed through a 0.45-μm filter. Subsequently, the lentivirus supernatants were transduced into A549 cells. Twenty-four hours later, cells were transferred to 10-cm dishes and selected by 1 μg/ml of puromycin for 10 days. Puromycin-resistant clones were sorted and confirmed by Western blotting and DNA sequencing. Genomic DNA was extracted using a cell genomic DNA extraction kit (Bioteke). Regions surrounding sgRNA target sequences were amplified by PCR. PCR products were then cloned into pMD-18T (TaKaRa) for DNA sequencing as described previously (71).
Generation of knockdown cells.
The vector pLKO.1-TRC (Addgene) was utilized to generate XBP1 shRNA or scrambled control shRNA (shSCR). XBP1 knockdown cells or shSCR cells were generated as previously described (72). The target sequences of shRNAs are listed in Table 5.
TABLE 5.
Sequences of oligonucleotides used in shRNA plasmids
| Gene | Sequence (5′–3′) |
|---|---|
| XBP1-shRNA1-5F | CCGGGACCCAGTCATGTTCTTCAAACTCGAGTTTGAAGAACATGACTGGGTCTTTTTG |
| XBP1-shRNA1-3R | AATTCAAAAAGACCCAGTCATGTTCTTCAAACTCGAGTTTGAAGAACATGACTGGGTC |
| XBP1-shRNA2-5F | CCGGAACAGCAAGTGGTAGATTTAGCTCGAGCTAAATCTACCACTTGCTGTTTTTTTG |
| XBP1-shRNA2-3R | 5-AATTCAAAAAAACAGCAAGTGGTAGATTTAGCTCGAGCTAAATCTACCACTTGCTGTT |
Flow cytometry analysis.
Control cells and IRE1αKo cells were infected with ZIKV at an MOI of 3. At 24 h p.i., the cells were suspended in PBS and incubated with anti-ZIKV E antibody (BioFront), followed by incubation with Alexa Fluor 647-conjugated anti-mouse IgG (H+L) cross-adsorbed secondary antibody (Invitrogen). Then, labeled cells were examined by flow cytometry (Beckman Coulter; CytoFLEX S).
Inhibitors and oleic acid treatments.
The inhibitors used in the experiments were GSK2850163 (Sigma) and 4μ8c (Sigma). Each inhibitor was diluted in DMSO and filtered through a 0.2-μm filter before use. Inhibitors were added to cells, followed by virus infection at 1 h posttreatment. Oleic acid was purchased from Sigma and dissolved in bovine serum albumin (BSA) at 200 mM. It was further diluted to 50 μM in 1× PBS supplemented with 1% fatty acid-free BSA (Gold Biotechnology). Diluted oleic acid was added to cells after virus attachment. A549 cells were infected with ZIKV as described above and treated with the desired treatments diluted in DMEM. The supernatants were collected at 24 h p.i. for plaque assay.
Immunofluorescence.
Control cells and IRE1αKo cells were infected with ZIKV at an MOI of 3 for 0, 6, 12, and 24 h. Cells were washed twice with PBS and fixed in 4% (vol/vol) paraformaldehyde (PFA) for 30 min. Oil red O (MP Biomedicals) was used per the manufacturer’s instructions. For the addition of oleic acid, control cells and IRE1αKo cells were infected with ZIKV at an MOI of 3 and treated with oleic acid (50 μM) at 1 h p.i. Twenty-four hours later, cells were fixed in 4% PFA for 30 min and stained with oil red O. Cells were then stained with 0.2 mg/ml of Hoechst (Invitrogen) and visualized using a Leica fluorescence microscope.
HepG2 cells were transfected with siNC or siIRE1α for 48 h. Cells were then infected with ZIKV at an MOI of 3 for 0, 6, 12, and 24 h. Cells were washed twice with cold PBS and fixed in 4% (vol/vol) paraformaldehyde for 30 min. Cells were permeabilized in 0.02% Triton X-100 for 15 min and blocked in blocking buffer (5% BSA in PBS) for 1 h, followed by incubation with primary antibodies at 4°C overnight. Then, cells were incubated with Cy3-conjugated goat anti-rabbit-IgG (Millipore) and Alexa Fluor 488-conjugated anti-mouse IgG (Invitrogen) for 1 h at room temperature. Subsequently, cells were stained with 0.2 mg/ml of Hoechst (Invitrogen) and visualized using a Leica fluorescence microscope.
Statistical analysis.
All the data are shown as means ± standard deviations (SD) from at least three independent experiments. Data were analyzed with GraphPad Prism 7.0 software. Kaplan-Meier survival curves were analyzed by the log rank test, and weight changes were compared using one-way analysis of variance (ANOVA). The statistical analysis was performed with an unpaired, two-tailed Student t test.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (no. 31970887 and 81471935), the Guangdong Science and Technology Department (no. 2018A050506029), and the Guangdong Basic and Applied Basic Research Foundation (no. 2020A1515010870, 2020A1515010570, and 2019A1515011336).
The funders had no role in study design, data collection and analysis, the decision to publish, or preparation of the manuscript.
We thank Yong Liu at Wuhan University for generously providing IRE1α-expressing plasmids.
REFERENCES
- 1.Carletti T, Zakaria MK, Faoro V, Reale L, Kazungu Y, Licastro D, Marcello A. 2019. Viral priming of cell intrinsic innate antiviral signaling by the unfolded protein response. Nat Commun 10:3889. doi: 10.1038/s41467-019-11663-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saiz JC, Vazquez-Calvo A, Blazquez AB, Merino-Ramos T, Escribano-Romero E, Martin-Acebes MA. 2016. Zika virus: the latest newcomer. Front Microbiol 7:496. doi: 10.3389/fmicb.2016.00496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weaver SC, Costa F, Garcia-Blanco MA, Ko AI, Ribeiro GS, Saade G, Shi PY, Vasilakis N. 2016. Zika virus: history, emergence, biology, and prospects for control. Antiviral Res 130:69–80. doi: 10.1016/j.antiviral.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee I, Bos S, Li G, Wang S, Gadea G, Despres P, Zhao RY. 2018. Probing molecular insights into Zika virus-host interactions. Viruses 10:233. doi: 10.3390/v10050233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paul D, Bartenschlager R. 2015. Flaviviridae replication organelles: oh, what a tangled web we weave. Annu Rev Virol 2:289–310. doi: 10.1146/annurev-virology-100114-055007. [DOI] [PubMed] [Google Scholar]
- 6.Miller S, Krijnse-Locker J. 2008. Modification of intracellular membrane structures for virus replication. Nat Rev Microbiol 6:363–374. doi: 10.1038/nrmicro1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu C-Y, Hsu Y-W, Liao C-L, Lin Y-L. 2006. Flavivirus infection activates the XBP1 pathway of the unfolded protein response to cope with endoplasmic reticulum stress. J Virol 80:11868–11890. doi: 10.1128/JVI.00879-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pena J, Harris E. 2011. Dengue virus modulates the unfolded protein response in a time-dependent manner. J Biol Chem 286:14226–14236. doi: 10.1074/jbc.M111.222703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ambrose RL, Mackenzie JM. 2011. West Nile virus differentially modulates the unfolded protein response to facilitate replication and immune evasion. J Virol 85:2723–2732. doi: 10.1128/JVI.02050-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sharma M, Bhattacharyy S, Sharma KB, Chauhan S, Asthana S, Abdin MZ, Vrati S, Kalia M. 2017. Japanese encephalitis virus activates autophagy through XBP1 and ATF6 ER stress sensors in neuronal cells. J Gen Virol 98:1027–1039. doi: 10.1099/jgv.0.000792. [DOI] [PubMed] [Google Scholar]
- 11.Tardif KD, Mori K, Kaufman RJ, Siddiqui A. 2004. Hepatitis C virus suppresses the IRE1-XBP1 pathway of the unfolded protein response. J Biol Chem 279:17158–18164. doi: 10.1074/jbc.M312144200. [DOI] [PubMed] [Google Scholar]
- 12.Hetz C, Papa FR. 2018. The unfolded protein response and cell fate control. Mol Cell 69:169–181. doi: 10.1016/j.molcel.2017.06.017. [DOI] [PubMed] [Google Scholar]
- 13.Urano F, Wang X, Bertolotti A, Zhang Y, Chung P, Harding HP, Ron D. 2000. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science 287:664–666. doi: 10.1126/science.287.5453.664. [DOI] [PubMed] [Google Scholar]
- 14.Nishitoh H, Matsuzawa A, Tobiume K, Saegusa K, Takeda K, Inoue K, Hori S, Kakizuka A, Ichijo H. 2002. ASK1 is essential for endoplasmic reticulum stress-induced neuronal cell death triggered by expanded polyglutamine repeats. Genes Dev 16:1345–1355. doi: 10.1101/gad.992302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ogata M, Hino S-i, Saito A, Morikawa K, Kondo S, Kanemoto S, Murakami T, Taniguchi M, Tanii I, Yoshinaga K, Shiosaka S, Hammarback JA, Urano F, Imaizumi K. 2006. Autophagy is activated for cell survival after ER stress. Mol Cell Biol 26:9220–9231. doi: 10.1128/MCB.01453-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hetz C, Martinon F, Rodriguez D, Glimcher LH. 2011. The unfolded protein response: integrating stress signals through the stress sensor IRE1a. Physiol Rev 91:1219–1243. doi: 10.1152/physrev.00001.2011. [DOI] [PubMed] [Google Scholar]
- 17.Neufeldt CJ, Cortese M, Acosta EG, Bartenschlager R. 2018. Rewiring cellular networks by members of the Flaviviridae family. Nat Rev Microbiol 16 :125–142. doi: 10.1038/nrmicro.2017.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hollien J, Lin JH, Li H, Stevens N, Walter P, Weissman JS. 2009. Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. J Cell Biol 186:323–331. doi: 10.1083/jcb.200903014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hollien J, Weissman JS. 2006. Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science 313:104–107. doi: 10.1126/science.1129631. [DOI] [PubMed] [Google Scholar]
- 20.Tan Z, Zhang W, Sun J, Fu Z, Ke X, Zheng C, Zhang Y, Li P, Liu Y, Hu Q, Wang H, Zheng Z. 2018. ZIKV infection activates the IRE1-XBP1 and ATF6 pathways of unfolded protein response in neural cells. J Neuroinflammation 15:275. doi: 10.1186/s12974-018-1311-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laguesse S, Creppe C, Nedialkova DD, Prevot PP, Borgs L, Huysseune S, Franco B, Duysens G, Krusy N, Lee G, Thelen N, Thiry M, Close P, Chariot A, Malgrange B, Leidel SA, Godin JD, Nguyen L. 2015. A dynamic unfolded protein response contributes to the control of cortical neurogenesis. Dev Cell 35:553–567. doi: 10.1016/j.devcel.2015.11.005. [DOI] [PubMed] [Google Scholar]
- 22.Gladwyn-Ng I, Cordón-Barris L, Alfano C, Creppe C, Couderc T, Morelli G, Thelen N, America M, Bessières B, Encha-Razavi F, Bonnière M, Suzuki IK, Flamand M, Vanderhaeghen P, Thiry M, Lecuit M, Nguyen L. 2018. Stress-induced unfolded protein response contributes to Zika virus-associated microcephaly. Nat Neurosci 21:63–71. doi: 10.1038/s41593-017-0038-4. [DOI] [PubMed] [Google Scholar]
- 23.Kolpikova EP, Tronco AR, Hartigh ABD, Jackson KJ, Iwawaki T, Fink SL. 2020. IRE1alpha promotes Zika virus infection via XBP1. Viruses 12:278–213. doi: 10.3390/v12030278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pierson TC, Diamond MS. 2018. The emergence of Zika virus and its new clinical syndromes. Nature 560:573–581. doi: 10.1038/s41586-018-0446-y. [DOI] [PubMed] [Google Scholar]
- 25.Yanagitani K, Kimata Y, Kadokura H, Kohno K. 2011. Translational pausing ensures membrane targeting and cytoplasmic splicing of XBP1u mRNA. Science 331:586–589. doi: 10.1126/science.1197142. [DOI] [PubMed] [Google Scholar]
- 26.Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, Clark SG, Ron D. 2002. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature 415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- 27.Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. 2001. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 107:881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- 28.Tomasio SM, Harding HP, Ron D, Cross BC, Bond PJ. 2013. Selective inhibition of the unfolded protein response: targeting catalytic sites for Schiff base modification. Mol Biosyst 9:2408–2416. doi: 10.1039/c3mb70234k. [DOI] [PubMed] [Google Scholar]
- 29.Concha NO, Smallwood A, Bonnette W, Totoritis R, Zhang G, Federowicz K, Yang J, Qi H, Chen S, Campobasso N, Choudhry AE, Shuster LE, Evans KA, Ralph J, Sweitzer S, Heerding DA, Buser CA, Su DS, DeYoung MP. 2015. Long-range inhibitor-induced conformational regulation of human IRE1alpha endoribonuclease activity. Mol Pharmacol 88:1011–1023. doi: 10.1124/mol.115.100917. [DOI] [PubMed] [Google Scholar]
- 30.Cross BCS, Bond PJ, Sadowski PG, Jha BK, Zak J, Goodman JM, Silverman RH, Neubert TA, Baxendale IR, Ron D, Harding HP. 2012. The molecular basis for selective inhibition of unconventional mRNA splicing by an IRE1-binding small molecule. Proc Natl Acad Sci U S A 109:E869–E878. doi: 10.1073/pnas.1115623109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xie H, Tang C-HA, Song JH, Mancuso A, Valle JRD, Cao J, Xiang Y, Dang CV, Lan R, Danielle JS, Keith B, Hu C-CA, Simon MC. 2018. IRE1a RNase-dependent lipid homeostasis promotes survival in Myc-transformed cancers. J Clin Invest 128:1300–1316. doi: 10.1172/JCI95864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Piperi C, Adamopoulos C, Papavassiliou AG. 2016. XBP1: a pivotal transcriptional regulator of glucose and lipid metabolism. Trends Endocrinol Metab 27:119–122. doi: 10.1016/j.tem.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 33.Tian S, Li B, Lei P, Yang X, Zhang X, Bao Y, Shan Y. 2018. Sulforaphane improves abnormal lipid metabolism via both ERS-dependent XBP1/ACC &SCD1 and ERS-independent SREBP/FAS pathways. Mol Nutr Food Res 62:e1700737. doi: 10.1002/mnfr.201700737. [DOI] [PubMed] [Google Scholar]
- 34.Hishiki T, Kato F, Nio Y, Watanabe S, Wen Tan NW, Yamane D, Miyazaki Y, Lin C-C, Suzuki R, Tajima S, Lim C-K, Saijo M, Hijikata M, Vasudevan SG, Takasaki T. 2019. Stearoyl-CoA desaturase-1 is required for flavivirus RNA replication. Antiviral Res 165:42–46. doi: 10.1016/j.antiviral.2019.03.002. [DOI] [PubMed] [Google Scholar]
- 35.Gullberg RC, Steel JJ, Pujari V, Rovnak J, Crick DC, Perera R. 2018. Stearoly-CoA [sic] desaturase 1 differentiates early and advanced dengue virus infections and determines virus particle infectivity. PLoS Pathog 14:e1007261. doi: 10.1371/journal.ppat.1007261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eberlin LS, Gabay M, Fan AC, Gouw AM, Tibshirani RJ, Felsher DW, Zare RN. 2014. Alteration of the lipid profile in lymphomas induced by MYC overexpression. Proc Natl Acad Sci U S A 111:10450–10455. doi: 10.1073/pnas.1409778111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lyn RK, Singaravelu R, Kargman S, O’Hara S, Chan H, Oballa R, Huang Z, Jones DM, Ridsdale A, Russell RS, Partridge AW, Pezacki JP. 2014. Stearoyl-CoA desaturase inhibition blocks formation of hepatitis C virus-induced specialized membranes. Sci Rep 4:4549. doi: 10.1038/srep04549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nguyen LN, Lim Y-S, Pham LV, Shin H-Y, Kim Y-S, Hwang SB. 2014. Stearoyl coenzyme A desaturase 1 is associated with hepatitis C virus replication complex and regulates viral replication. J Virol 88:12311–12325. doi: 10.1128/JVI.01678-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martin-Acebes MA, Vazquez-Calvo A, Saiz JC. 2016. Lipids and flaviviruses, present and future perspectives for the control of dengue, Zika, and West Nile viruses. Prog Lipid Res 64:123–137. doi: 10.1016/j.plipres.2016.09.005. [DOI] [PubMed] [Google Scholar]
- 40.Pombo JP, Sanyal S. 2018. Perturbation of intracellular cholesterol and fatty acid homeostasis during flavivirus infections. Front Immunol 9:1276. doi: 10.3389/fimmu.2018.01276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nevo-Yassaf I, Lovelle M, Nahmias Y, Hirschberg K, Sklan EH. 2017. Live cell imaging and analysis of lipid droplets biogenesis in hepatatis C virus infected cells. Methods 127:30–36. doi: 10.1016/j.ymeth.2017.05.010. [DOI] [PubMed] [Google Scholar]
- 42.Bickel PE, Tansey JT, Welte MA. 2009. PAT proteins, an ancient family of lipid droplet proteins that regulate cellular lipid stores. Biochim Biophys Acta 1791:419–440. doi: 10.1016/j.bbalip.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krahmer N, Guo Y, Farese RV, Walther TC. 2009. SnapShot: lipid droplets. Cell 139:1024–1024.e1. doi: 10.1016/j.cell.2009.11.023. [DOI] [PubMed] [Google Scholar]
- 44.Chen Y, Liu T, Zhang Z, Chen M, Rong L, Ma L, Yu B, Wu D, Zhang P, Zhu X, Huang X, Zhang H, Li YP. 2018. Novel genetically stable infectious clone for a Zika virus clinical isolate and identification of RNA elements essential for virus production. Virus Res 257:14–24. doi: 10.1016/j.virusres.2018.08.016. [DOI] [PubMed] [Google Scholar]
- 45.Yu JH, Liu XL, Ke CW, Wu QH, Lu WZ, Qin ZR, He XE, Liu YJ, Deng JL, Xu SQ, Li Y, Zhu L, Wan CS, Zhang QW, Xiao WW, Xie Q, Zhang B, Zhao W. 2017. Effective suckling C57BL/6, Kunming, and BALB/c mouse models with remarkable neurological manifestation for Zika virus infection. Viruses 9:165. doi: 10.3390/v9070165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li SX, Armstrong N, Zhao H, Hou WH, Liu J, Chen CY, Wan JK, Wang W, Zhong CL, Liu C, Zhu H, Xia NS, Cheng T, Tang QY. 2018. Zika virus fatally infects wild type neonatal mice and replicates in central nervous system. Viruses 10:49. doi: 10.3390/v10010049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bhattacharyya S, Sen U, Vrati S. 2014. Regulated IRE1-dependent decay pathway is activated during Japanese encephalitis virus-induced unfolded protein response and benefits viral replication. J Gen Virol 95:71–79. doi: 10.1099/vir.0.057265-0. [DOI] [PubMed] [Google Scholar]
- 48.Chu H, Yuen TTT, Chik KKH, Yuan S, Shuai H, Zou Z, Wang Y, Zhu Z, Yang D, Poon VKM, Chan CCS, Zhou J, Yin F, Kok K-H, Yuen K-Y, Chan JFW. 2020. Targeting the inositol-requiring enzyme-1 pathway efficiently reverts Zika virus-induced neurogenesis and spermatogenesis marker perturbations. ACS Infect Dis 6:1745–1758. doi: 10.1021/acsinfecdis.9b00526. [DOI] [PubMed] [Google Scholar]
- 49.Zhu X, Xiong T, Liu P, Guo X, Xiao L, Zhou F, Tang Y, Yao P. 2018. Quercetin ameliorates HFD-induced NAFLD by promoting hepatic VLDL assembly and lipophagy via the IRE1a/XBP1s pathway. Food Chem Toxicol 114:52–60. doi: 10.1016/j.fct.2018.02.019. [DOI] [PubMed] [Google Scholar]
- 50.Lee Y, Kuo S, Lin C, Fu P, Lin Y, Yeh T, Liu H. 2018. Dengue virus-induced ER stress is required for autophagy activation, viral replication, and pathogenesis both in vitro and in vivo. Sci Rep 8:489. doi: 10.1038/s41598-017-18909-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saeed M, Suzuki R, Watanabe N, Masaki T, Tomonaga M, Muhammad A, Kato T, Matsuura Y, Watanabe H, Wakita T, Suzuki T. 2011. Role of the endoplasmic reticulum-associated degradation (ERAD) pathway in degradation of hepatitis C virus envelope proteins and production of virus particles. J Biol Chem 286:37264–37273. doi: 10.1074/jbc.M111.259085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Medigeshi GR, Lancaster AM, Hirsch AJ, Briese T, Lipkin WI, Defilippis V, Fruh K, Mason PW, Nikolich-Zugich J, Nelson JA. 2007. West Nile virus infection activates the unfolded protein response, leading to CHOP induction and apoptosis. J Virol 81:10849–10860. doi: 10.1128/JVI.01151-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martins AS, Martins IC, Santos NC. 2018. Methods for lipid droplet biophysical characterization in Flaviviridae infections. Front Microbiol 9:1951. doi: 10.3389/fmicb.2018.01951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang J, Lan Y, Sanyal S. 2017. Modulation of lipid droplet metabolism—a potential target for therapeutic intervention in Flaviviridae infections. Front Microbiol 8:2286. doi: 10.3389/fmicb.2017.02286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hardie DG. 2011. AMP-activated protein kinase: an energy sensor that regulates all aspects of cell function. Genes Dev 25:1895–1908. doi: 10.1101/gad.17420111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Walther TC, Farese RV Jr. 2012. Lipid droplets and cellular lipid metabolism. Annu Rev Biochem 81:687–714. doi: 10.1146/annurev-biochem-061009-102430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tang WC, Lin RJ, Liao CL, Lin YL. 2014. Rab18 facilitates dengue virus infection by targeting fatty acid synthase to sites of viral replication. J Virol 88:6793–6804. doi: 10.1128/JVI.00045-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McLauchlan J. 2009. Lipid droplets and hepatitis C virus infection. Biochim Biophys Acta 1791:552–559. doi: 10.1016/j.bbalip.2008.12.012. [DOI] [PubMed] [Google Scholar]
- 59.Roingeard P, Melo RC. 2017. Lipid droplet hijacking by intracellular pathogens. Cell Microbiol 19:e12688. doi: 10.1111/cmi.12688. [DOI] [PubMed] [Google Scholar]
- 60.Samsa MM, Mondotte JA, Iglesias NG, Assuncao-Miranda I, Barbosa-Lima G, Da Poian AT, Bozza PT, Gamarnik AV. 2009. Dengue virus capsid protein usurps lipid droplets for viral particle formation. PLoS Pathog 5:e1000632. doi: 10.1371/journal.ppat.1000632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hope RG, McLauchlan J. 2000. Sequence motifs required for lipid droplet association and protein stability are unique to the hepatitis C virus core protein. J Gen Virol 81:1913–1925. doi: 10.1099/0022-1317-81-8-1913. [DOI] [PubMed] [Google Scholar]
- 62.Herker E, Harris C, Hernandez C, Carpentier A, Kaehlcke K, Rosenberg AR, Farese RV, Ott M. 2010. Efficient hepatitis C virus particle formation requires diacylglycerol acyltransferase-1. Nat Med 16:1295–1298. doi: 10.1038/nm.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Appel N, Zayas M, Miller S, Krijnse-Locker J, Schaller T, Friebe P, Kallis S, Engel U, Bartenschlager R. 2008. Essential role of domain III of nonstructural protein 5A for hepatitis C virus infectious particle assembly. PLoS Pathog 4:e1000035. doi: 10.1371/journal.ppat.1000035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang JS, Lan Y, Li MY, Lamers MM, Fusade-Boyer M, Klemm E, Thiele C, Ashour J, Sanyal S. 2018. Flaviviruses exploit the lipid droplet protein AUP1 to trigger lipophagy and drive virus production. Cell Host Microbe 23:819–831. doi: 10.1016/j.chom.2018.05.005. [DOI] [PubMed] [Google Scholar]
- 65.Boulant S, Targett-Adams P, McLauchlan J. 2007. Disrupting the association of hepatitis C virus core protein with lipid droplets correlates with a loss in production of infectious virus. J Gen Virol 88:2204–2213. doi: 10.1099/vir.0.82898-0. [DOI] [PubMed] [Google Scholar]
- 66.Boulant S, Douglas MW, Moody L, Budkowska A, Targett-Adams P, McLauchlan J. 2008. Hepatitis C virus core protein induces lipid droplet redistribution in a microtubule- and dynein-dependent manner. Traffic 9:1268–1282. doi: 10.1111/j.1600-0854.2008.00767.x. [DOI] [PubMed] [Google Scholar]
- 67.Martins A, Martins I, Faustino A, Nascimento A, Carvalho F, Santos N. 2017. Understanding the structure and function of the capsid protein of Zika, West-Nile and Dengue viruses, namely their ability to interact with host lipid systems. Protein Sci 26:150. [Google Scholar]
- 68.Shang Z, Song H, Shi Y, Qi J, Gao GF. 2018. Crystal structure of the capsid protein from Zika virus. J Mol Biol 430:948–962. doi: 10.1016/j.jmb.2018.02.006. [DOI] [PubMed] [Google Scholar]
- 69.Wang Y, Chen X, Xie J, Zhou S, Huang Y, Li YP, Li X, Liu C, He J, Zhang P. 2019. RNA helicase A is an important host factor involved in dengue virus replication. J Virol 93:e01306-18. doi: 10.1128/JVI.01306-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang P, Li Y, Xia J, He J, Pu J, Xie J, Wu S, Feng L, Huang X, Zhang P. 2014. IPS-1 plays an essential role in dsRNA-induced stress granule formation by interacting with PKR and promoting its activation. J Cell Sci 127:2471–2482. doi: 10.1242/jcs.139626. [DOI] [PubMed] [Google Scholar]
- 71.Gao H, Lin Y, He J, Zhou S, Liang M, Huang C, Li X, Liu C, Zhang P. 2019. Role of heparan sulfate in the Zika virus entry, replication, and cell death. Virology 529:91–100. doi: 10.1016/j.virol.2019.01.019. [DOI] [PubMed] [Google Scholar]
- 72.Chen C, Ma X, Hu Q, Li X, Huang F, Zhang J, Pan T, Xia J, Liu C, Zhang H. 2017. Moloney leukemia virus 10 (MOV10) inhibits the degradation of APOBEC3G through interference with the Vif-mediated ubiquitin-proteasome pathway. Retrovirology 14:56. doi: 10.1186/s12977-017-0382-1. [DOI] [PMC free article] [PubMed] [Google Scholar]