While traditional epidemiological studies are of great interest in describing the dynamics of epidemics, they struggle to fully capture the geospatial dynamics and factors driving the dispersal of pathogens like HIV as they have difficulties capturing linkages between infections. To overcome this, we used a discrete phylogeographic approach coupled to a generalized linear model extension to characterize the dynamics and drivers of the across-province spread of the five main HIV types circulating in China. Our results indicate that large urbanized areas with dense populations and developed transportation infrastructures are facilitators of HIV dispersal throughout China and highlight the need to consider harmonized country-wide public policies to control local HIV epidemics.
KEYWORDS: HIV, phylodynamics, discrete phylogeography, generalized linear model, China
ABSTRACT
The HIV epidemic in China accounts for 3% of the global HIV incidence. We compared the patterns and determinants of interprovincial spread of the five most prevalent circulating types. HIV pol sequences sampled across China were used to identify relevant transmission networks of the five most relevant HIV-1 types (B and circulating recombinant forms [CRFs] CRF01_AE, CRF07_BC, CRF08_BC, and CRF55_01B) in China. From these, the dispersal history across provinces was inferred. A generalized linear model (GLM) was used to test the association between migration rates among provinces and several measures of human mobility. A total of 10,707 sequences were collected between 2004 and 2017 across 26 provinces, among which 1,962 are newly reported here. A mean of 18 (minimum and maximum, 1 and 54) independent transmission networks involving up to 17 provinces were identified. Discrete phylogeographic analysis largely recapitulates the documented spread of the HIV types, which in turn, mirrors within-China population migration flows to a large extent. In line with the different spatiotemporal spread dynamics, the identified drivers thereof were also heterogeneous but are consistent with a central role of human mobility. The comparative analysis of the dispersal dynamics of the five main HIV types circulating in China suggests a key role of large population centers and developed transportation infrastructures as hubs of HIV dispersal. This advocates for coordinated public health efforts in addition to local targeted interventions.
IMPORTANCE While traditional epidemiological studies are of great interest in describing the dynamics of epidemics, they struggle to fully capture the geospatial dynamics and factors driving the dispersal of pathogens like HIV as they have difficulties capturing linkages between infections. To overcome this, we used a discrete phylogeographic approach coupled to a generalized linear model extension to characterize the dynamics and drivers of the across-province spread of the five main HIV types circulating in China. Our results indicate that large urbanized areas with dense populations and developed transportation infrastructures are facilitators of HIV dispersal throughout China and highlight the need to consider harmonized country-wide public policies to control local HIV epidemics.
INTRODUCTION
By the end of 2018, the number of people living with HIV (PWH) in China was close to 1.25 million (1, 2). The distribution of HIV-1 subtypes in China is diverse, with over 11 circulating genetic variants (3), each with an evolving geographical distribution, prevalence, and modes of transmission (3–6). The first nationwide molecular epidemiological survey in 1996 to 1998 showed that subtype B′/B (47.5%) and subtype C (34.3%) were the most predominant HIV types in China (7). For subtype B′, this in part resulted from its high prevalence among plasma donors in China because of unsanitary commercial plasma collection (8). Surveys conducted in 2002 to 2003 and 2006 indicated that the circulating recombinant forms (CRFs) CRF07_BC, CRF01_AE, and CRF08_BC had become the dominant HIV types in China. Founder effects show that CRF07_BC and CRF08_BC mostly circulated among injecting drug users (IDUs) in Northeastern and Southeastern China, respectively (9–12), while subtype B′ remains dominant among former plasma donors in Central China (13, 14). Meanwhile, CRF01_AE became the dominant type and replaced subtype B as the principal driver of infection among men reporting having sex with men (MSM) (3). The National Sentinel Surveillance System of China revealed that the proportion of MSM transmission increased from 14.7% in 2009 (15) to 27.6% in 2016 (16), with an increased proportion of CRF01_AE and CRF07_BC infections among MSM, while the proportion of HIV-1 subtype B decreased between 2012 and 2016 (6, 17). In addition to these predominant HIV types, CRF55_01B, generated through recombination between CRF01_AE and subtype B variants, had been first identified among MSM in the city of Shenzhen (18, 19). Circulating primarily among MSM, it has now spread throughout most provinces of China, with a prevalence ranging from 1.5% to 12.5% (20). Its prevalence has increased in the past 5 years, especially in South and East China, with higher pooled estimated rates in Guangdong Province (12.22%, 95% confidence interval [CI], 10.34 to 13.17) and Fujian Province (8.65%, 95% CI, 4.98 to 13.17) (17). It is now circulating mostly in Guangdong and neighboring provinces in China, across all risk groups (18).
The burden of HIV is also unevenly spread geographically: while HIV is present in all provinces, the top six high-prevalence provinces (Yunnan, Guangxi, Henan, Guangdong, and Xinjiang) accounted for over 60% of the national number of PWH (21). The recent upsurge of HIV among MSM in large Chinese cities, including Beijing, Chongqing, Chengdu, Guangzhou, Shanghai, and Shenyang, adds to this imbalance (22).
These multiple and diverse epidemics driven by changing risk factor patterns in part result from the inability of treatment, prevention, and control programs to halt the rapid growth of the HIV epidemics, which now account for ∼3% of the global HIV prevalence (23). The epidemic growth of ∼80,000 new infections per year (1) coincided with intense rural-to-urban migration flows (24–30) and considerable investments in land and airway transport infrastructures, expediting longer-distance human mobility (31). By the end of 2017, the migrant population, seeking better employment opportunities and living conditions in economically more developed areas, reached 244 million (32, 33), and migrant workers have become the main driver of within-country migration. Importantly, the migrant labor population is also at higher risk for HIV acquisition and transmission because of poor knowledge about self-protection and the transmission routes of HIV (34), and they have been shown to fuel local epidemics (30, 35).
While traditional epidemiological studies are of great interest in describing the dynamics of epidemics, they struggle to fully capture the geospatial dynamics and factors driving the dispersal of pathogens. By merging virus genetic, geospatial, and epidemiological data, phylodynamic models allow investigation of the migration history of pathogens and its drivers in the absence of detailed contact tracing data and when linkage among infections is not obvious (36–39). Such analyses have been widely adopted both for human (40, 41) and plant (38, 42) viruses and more recently for HIV (43, 44).
The overall goal of the present study is to characterize the dynamics and drivers of the across-province spread of the main HIV types circulating in China. For this purpose, we capitalize on a discrete phylogeographic approach coupled to a generalized linear model extension.
RESULTS
Population characteristics.
Totals of 6,800, 1,578 (822/756), 1,158, 957, and 211 available sequences were retrieved for CRF01_AE, subtype B (B/B′), CRF07_BC, CRF08_BC, and CRF55_01B, respectively. The number of provinces included in each final data set varied from 7 (CRF55_01B) to 17 (CRF01_AE and B/B′). Figure 1 shows the distribution of provinces per data set.
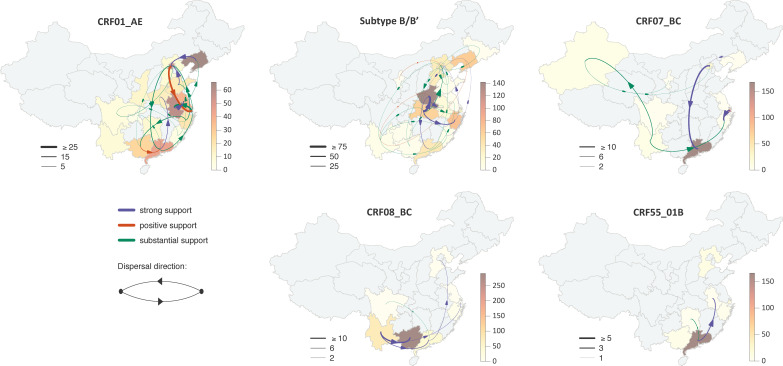
FIG 1.
Migration events between provinces in China. The thickness of the arrows corresponds to the average number of inferred migration events, their curvature indicates the migration direction, and their colors reflect the support for each link (green, orange, and purple for 3 ≤ BFadj < 10 [substantial], 10 ≤ BFadj < 20 [positive], and BFadj ≥ 20 [strong], respectively). Provinces are colored according to the number of sequences included in the clusters for each HIV type. The underlying map is from the Database of Global Administrative Areas (GADM; https://gadm.org).
Preliminary phylogenetic analysis and subsampling.
For CRF01_AE, an initial set of 6,423 HIV-1 CRF01_AE pol sequences from 53 countries across the world between 1990 and 2017 retrieved from the Los Alamos National Laboratory HIV Sequence Database (http://www.hiv.lanl.gov/) was combined with the CRF01_AE data set of 6,800 sequences to delineate clades that capture the epidemic dynamics in transmission networks that pertain to China. We identified 83 such clades (n = 1,876 sequences). To obtain data informative of interprovincial migration patterns, these were reduced to the 54 clades (sizes, 3 to 24 sequences) that included samples from at least 2 Chinese provinces (totaling 454 sequences from 17 provinces). The same rationale was used for the other data sets. Starting from totals of 1,578, 1,158, and 957 sequences for subtype B/B′, CRF07_BC, and CRF08_BC data sets, we obtained 15, 16, and 7 clades from 17, 10, and 8 provinces, respectively. For CRF55_01B, which is circulating in China only, we obtained a single clade of 197 sequences collected across 7 provinces.
Discrete phylogeographic inferences.
We used Bayesian phylogeographic inference to evaluate the dispersal history of the five main circulating types across Chinese provinces. This allows, for each subtype and CRF, identification of the significant migration events between Chinese provinces and estimation of their numbers and directionality (Fig. 1). The reconstructed patterns of spread based on the identified clades revealed strong evidence (adjusted Bayes factor [BFadj] of ≥20) of migration between provinces for all HIV populations sampled. The relative contributions of each province as source and sink of HIV dispersal throughout China are summarized in Table 1.
TABLE 1.
Relative importance of provinces in the interprovincial spread of the main HIV types in Chinaa
| HIV type | From | Mean % (95% HPD) | To | Mean % (95% HPD) |
|---|---|---|---|---|
| CRF01_AE | Beijing | 24.9 (24.8–25) | Shanghai | 25.7 (25.5–25.8) |
| Guangdong | 16.6 (16.5–16.7) | Beijing | 17.2 (17.1–17.3) | |
| Shanghai | 15.8 (15.7–15.9) | Anhui | 13.3 (13.2–13.4) | |
| Anhui | 10.6 (10.5–10.7) | Guangdong | 11.7 (11.6–11.8) | |
| Shandong | 5.6 (5.5–5.6) | Jiangsu | 10.6 (10.5–10.7) | |
| Zhejiang | 5.4 (5.4–5.5) | Henan | 7.5 (7.4–7.6) | |
| Liaoning | 5.3 (5.3–5.4) | Guangxi | 4.4 (4.4–4.5) | |
| Guangxi | 3.9 (3.8–3.9) | Liaoning | 3.1 (3.1–3.2) | |
| Sichuan | 3.3 (3.3–3.4) | Shandong | 2.4 (2.4–2.5) | |
| Jiangsu | 3.1 (3.1–3.2) | Zhejiang | 2.3 (2.3–2.3) | |
| Henan | 2.3 (2.3–2.3) | Sichuan | 1.1 (1–1.1) | |
| Fujian | 1.8 (1.8–1.8) | Chongqing | 0.5 (0.5–0.5) | |
| Hebei | 1.2 (1.2–1.2) | |||
| CRF07_BC | Beijing | 43.4 (43.1–43.8) | Guangdong | 43.4 (43.1–43.8) |
| Shanghai | 29.9 (29.5–30.3) | Zhejiang | 39.2 (38.8–39.6) | |
| Yunnan | 15.7 (15.4–16) | Beijing | 10.2 (10–10.5) | |
| Xinjiang | 7.2 (7–7.4) | Xinjiang | 6.4 (6.2–6.6) | |
| Liaoning | 3.7 (3.6–3.9) | Ningxia | 0.7 (0.7–0.8) | |
| CRF08_BC | Yunnan | 85.2 (84.8–85.7) | Guangxi | 44.2 (43.6–44.9) |
| Guangdong | 14.8 (14.3–15.2) | Guangdong | 32.4 (31.8–33) | |
| Hebei | 11.3 (10.8–11.7) | |||
| Shanghai | 8.6 (8.2–9) | |||
| Sichuan | 3.5 (3.3–3.7) | |||
| B | Hubei | 69.9 (69.7–70.1) | Henan | 55.8 (55.6–56) |
| Henan | 16.7 (16.6–16.9) | Guangdong | 10.9 (10.8–11) | |
| Liaoning | 5.4 (5.3–5.5) | Zhejiang | 9.8 (9.7–10) | |
| Zhejiang | 3 (2.9–3.1) | Hubei | 7.5 (7.4–7.6) | |
| Hebei | 2.1 (2–2.1) | Beijing | 7.4 (7.2–7.5) | |
| Beijing | 1.3 (1.3–1.4) | Anhui | 5.6 (5.5–5.7) | |
| Guangdong | 0.9 (0.9–0.9) | Shandong | 1 (0.9–1) | |
| Anhui | 0.3 (0.3–0.4) | Hebei | 0.5 (0.5–0.6) | |
| Yunnan | 0.2 (0.2–0.2) | Guangxi | 0.4 (0.3–0.4) | |
| Shandong | 0.1 (0–0.1) | Jiangsu | 0.3 (0.3–0.4) | |
| Fujian | 0 (0–0) | Shanghai | 0.3 (0.3–0.3) | |
| Jilin | 0 (0–0) | Jilin | 0.2 (0.2–0.2) | |
| Liaoning | 0.2 (0.1–0.2) | |||
| Ningxia | 0.2 (0.1–0.2) | |||
| Fujian | 0 (0–0) | |||
| Yunnan | 0 (0–0) | |||
| CRF55_01B | Guangdong | 100 (99.8–100) | Anhui | 73.9 (71.8–75.8) |
| Hunan | 26.1 (24.2–28.2) | |||
For each province or city, each percentage refers to its average proportion as the source (from) or recipient (to) of the emigration event.
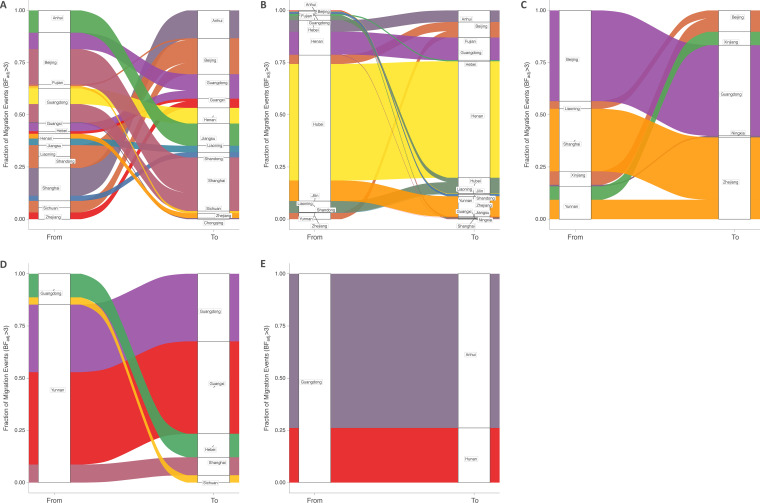
The CRF01_AE and CRF07_BC clades have become the two predominant HIV CRFs in China, with overall prevalences of 46.34% (95% CI, 40.56 to 52.17) and 19.16% (95% CI, 15.02 to 23.66), respectively (4). Here, the discrete phylogeographic analysis for CRF01_AE supported a complex migration history, with Beijing (Chinese capital, with the second highest population density), Guangdong Province (southern region, capital Guangzhou), Shanghai (the most populous urban area in China), and Anhui Province (an important part of the Yangtze River Delta and in the top four provinces of China in labor export) being the population centers most involved in the interprovincial spread of migration events, both as major sources (with 24.9% [95% CI, 24.8 to 25], 16.6% [95% CI, 16.5 to 16.7], 15.8% [95% CI, 15.7 to 15.9], and 10.6% [95% CI, 10.5 to 10.7] of the viral diffusion, respectively) and as major sinks (with 17.2% [95% CI, 17.1 to 17.3], 11.7% [95% CI, 11.6 to 11.8], 25.7% [95% CI, 25.5 to 25.8], and 13.3% [95% CI, 13.2 to 13.4] of the introduction, respectively) (Fig. 1 and 2A). For CRF07_BC, the second most prevalent type, the main sources were Beijing and Shanghai, along with Yunnan Province (northwest-central region, capital Kunming) (Fig. 1 and 2C), while for CRF08_BC, the main source was Yunnan Province. Our model also showed robust evidence of viral migration across China for HIV-1 subtype B/B′, with Hubei (capital Wuhan) being the major source of viral migration, accounting for 69.9% (95% CI, 69.7 to 70.1) of the viral dispersal and the predominant diffusion being toward Henan Province (capital Zhengzhou), with 55.8% (95% CI, 55.6 to 56) of all introduction events, acting as a sink for the B/B′ epidemic (Fig. 1 and 2B). Finally, the southern province of Guangdong, with the largest population, was the only source of migration for CRF55_01B that is supported by our data, with the dispersal directed toward Anhui Province and Hunan Province (Fig. 1 and 2E).
FIG 2.
Migration events between provinces in China. Sankey plot showing the proportions of migration events from each source province toward the recipient provinces. Left side of the plots shows the source of migration events. Right side of the plots shows the destination of migration events. Only results with adjusted Bayes factors (BFadj) of ≥3 are shown. CRF01_AE (A), subtype B/B′ (B), CRF07_BC (C), CRF08_BC (D), and CRF5501_B (E).
From a historical perspective, our analyses showed a higher density of migration events across provinces in the late 1990s and 2000s for subtype B/B′, while migration events in general are more concentrated over the past 15 years for CRF01_AE, CRF07_BC, and CRF08_BC. We also found that the historical interprovincial dispersal of CRF55_01B predominantly occurred around 2010 (data not shown).
GLM analyses.
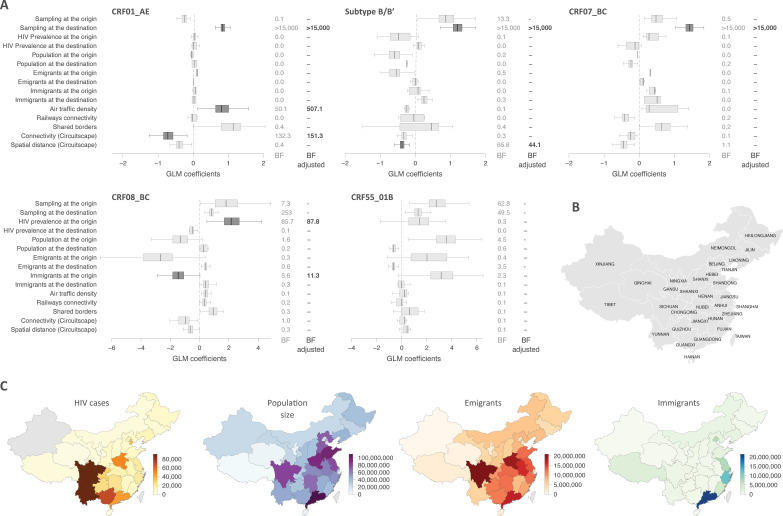
We next used the generalized linear model (GLM) extension of the phylogeographic model to evaluate the association between potential predictors and the migration frequencies among provinces (Fig. 3). For CRF01_AE, our model revealed a strong association between migration events and air traffic density, as well as connectivity among locations, associations that are robust to randomizing tip-to-location assignments (BFadj ≫ 100). The conditional effect size for connectivity between major cities over land was negative, meaning that spread between provinces that are easier to travel between over land is less frequent than between provinces that are less well connected over land. For CRF08_BC, a higher HIV prevalence in the province of origin was associated with increased HIV dispersal (BFadj = 87.8), which also associates with the number of immigrants at the origin (BFadj = 11.3). Whereas a higher HIV prevalence at the province of origin links to more frequent migration from that province, the conditional effect size for the number of immigrants at the origin was negative, implying that for CRF08_BC, more immigration toward a province links to less frequent virus migration from that province. The only other predictor that was well supported was spatial distance for subtype B/B′. For this predictor too, the conditional effect size was negative, indicating that migration is more frequent between closer locations. No other associations are well supported (i.e., BFadj ≥ 3) (Fig. 3).
FIG 3.
Predictors of transition rates among locations. (A) The boxplots report the posterior distribution of each GLM coefficient, i.e., the contribution of each predictor to the model, when included in the model (the conditional effect size). The adjusted BFs after accounting for sampling heterogeneity are reported when ≥3, and the corresponding conditional effect sizes are plotted in darker gray. (B) Map of the Chinese Provinces (Database of Global Administrative Areas [GADM; https://gadm.org]). (C) Variables tested as predictors of dispersal transition rates across locations. Data for number of HIV cases per province were obtained from the National Bureau of Statistics of China (98) and the China National Center for Disease Control and Prevention (16). Data for population size (in millions) and data for numbers of emigrants and immigrants were obtained from the National Bureau of Statistics of China (98). The numbers of sequences sampled at the origin/destination (see Fig. 1) were also included in the GLM to account for the potential impact of sampling biases within the analysis (39).
DISCUSSION
Starting from >10,000 HIV-1 pol sequences from the five main prevalent HIV-1 subtypes and CRFs collected in China between 1996 and 2017, we reconstructed the spatial diffusion of the five most prevalent HIV-1 types across provinces in China. Our reconstructions largely recapitulate their documented spread, which we discuss one by one.
CRF01_AE.
CRF01_AE has become the dominant HIV variant in most provinces (3). In line with previous epidemiological studies (3) and molecular analyses (45), our reconstructions capture that most migration events occurred recently between southern and eastern/northeastern provinces (Fig. 1 and Table 1). Specifically, we found that Beijing (the political, economic, and cultural center of China) was the main source of CRF01_AE dispersal throughout the country and that Guangdong, Shanghai, and Anhui are the other major hubs of CRF01_AE dispersal (Table 1). This largely matches the geographic scope of within-China population migration flows, which were concentrated within and between the southern and eastern main economic provinces (46, 47). It is of note that CRF01_AE is dominant among MSM (3) and that interprovincial migrants (compared to intraprovincial migrants) not only are more likely to be male but also tend to be younger and have fewer years of formal education (47), which are factors associated with higher-risk behavior (48).
The GLM analyses confirmed a strong association between the intensity of migration events and air traffic density and an inverse relation of connectivity between major cities over land with the migration intensity. Combined, our results indicate that interprovincial CRF01_AE mobility is driven predominantly by longer-distance migration, possibly MSM related, a combination that has also been noted in, e.g., regions of Canada (49).
HIV subtype B/B′.
After an initial period of dominance, the prevalence of B/B′ has declined (17). Consistent with this trend, we found that viral dispersal of HIV-1 subtype B mostly occurred in the 1990s and early 2000s (data not shown). Also in line with epidemiological surveys and with previous molecular analyses (50), we found that Hubei and Henan, both with a historically predominant circulation of B/B′ among blood donors (51–55), were the major sources of interprovincial dispersal for this HIV-1 type (Table 1 and Fig. 2).
Zhengzhou (Henan capital) is located at the junction of the major north-south Beijing-Guangzhou and east-west Lanzhou-Lianyungang railways and has evolved into a major national administrative, economic, and transportation hub (56), and Henan and Hubei are among the top five largest “migration sending areas” (47). This shows that there was ample opportunity for long-distance spread of B/B′ in relation to human mobility. In turn, the dominance of shorter-distance spread of B/B′ implies that it did not find much fertile ground in highly mobile high-risk groups, such as interprovincial migrant workers. This aspect is reflected in the results of our GLM analyses, which showed that migration events occurred more frequently among more-nearby provinces (Fig. 3).
CRF07_BC and CRF08_BC.
CRF07_BC was originally reported in Yunnan Province in the 1980s and spread quickly among IDUs. In recent years, it has been introduced in MSM populations, which drove its spread to elsewhere in China, particularly to Beijing, Shanghai, Guangdong, and Zhejiang (9, 17, 57, 58). CRF08_BC, on the other hand, was initially reported in Yunnan and Guangxi Provinces among IDUs but has rarely been reported in MSM or other risk populations.
While none of the predictors appear significantly associated with the spreading process of CRF07_BC, the origin of CRF08_BC in the southeastern part of China and its subsequent spread toward economically more developed provinces that are attraction poles for inland migration is reflected in which predictors were found to significantly associate with the migration process, as well as the direction of their effect sizes. Specifically, the migration frequency out of a province increases with increasing HIV prevalence, and the migration intensity is inversely associated with the number of immigrants in the provinces of origin, suggesting that immigration hot spots functioned as a sink for this type (Fig. 3).
CRF55_01B.
CRF55_01B was first identified among MSM in Shenzhen, Guangdong (18, 19). It has now spread across all risk groups and throughout most provinces of China (17, 20, 59), although it mostly circulates in Guangdong and neighboring provinces (18). In line with this, our results point to Guangdong as the main source of the dispersal to the western province of Anhui and, also, the adjacent province of Hunan (Table 1). As Guangdong is an economically well-developed province and attraction pole of migrant workers, it may intuitively seem at odds that it is a source rather than a destination of CRF055_01B spread. This may, however, be explained by return migration, which has become more intense over the years (60).
Prior to the 1980s, rural-urban migration in China was minimal. Since then, China has witnessed an extraordinary internal migration: rural-to-urban migrants increased the urban population by approximately 390 million. Of these rural migrants, approximately 54% were interprovincial migrants, most of which left their home province, but with many also returning after some time and many visiting their families on a regular basis (e.g., with Chinese New Year). The reconstructed patterns of interprovincial spread for the main HIV types in China support the idea that migrant workers are at least partially involved in their diffusion. Unfortunately, the lack of epidemiological metadata prevented us from more explicitly elucidating the dynamics of spread within and between relevant subpopulations by, for example, associating epidemiological characteristics with uptake in clusters of closely related viruses (49, 61–63), which can help identify on what aspects to focus screening and prevention efforts. The involvement of migrant workers can be tested more directly within the GLM framework. Regrettably, we could only dispose of the total number of immigrants/emigrants by province instead of more granular pairwise migration flow data. Nonetheless, the identified drivers of HIV dispersal in China are in line with the view that human mobility strongly impacts pathogen epidemic dynamics (64). This combines with the reconstructed patterns of spread that largely reflect within-China population migration flows (that are directed toward and between major population and economic centers) to suggest that the patterns of viral transmission for at least some of the HIV epidemics in China were driven by major population centers, which can act as gravity attractors before the virus spread to smaller populations (65, 66). This also illustrates that, in the absence of concurrent national prevention efforts with a focus on the most important drivers of ongoing transmission, local epidemics will rapidly be reseeded, challenging the long-term impact of isolated intervention efforts.
Limitations.
One major limitation of our study is that the collection of the HIV-1 pol sequences from the five main prevalent HIV-1 types in China has not been performed under a common framework, which may render our analyses prone to sampling bias. To the best of our knowledge, this drawback affects nearly every phylogeographic study of HIV-1 and other viruses. Whereas structured coalescent approaches hold promise for unbiased inferences in the face of biased sampling, inference under high-dimension state spaces and large data sets remains challenging for these models. For this reason, we relied on the computationally more efficient discrete trait analysis (67, 68). To counter this model’s sensitivity to biased sampling of subpopulations, we (i) adopted a filter based on location state randomizations and (ii) combined the geographical information from different partitions that represent different samples of the same epidemic to minimize the risk of false-positive migration linkages and associations with covariates (69, 70). Given that the reconstructed interprovincial spread largely captures the documented spread of the investigated HIV-1 types, we believe that these precautions were effective.
Several factors can explain the finding that only a few of the tested predictors associated with the spread patterns. The high-level resolution of our phylogeographic reconstructions means that potential predictors can only be evaluated against a limited number of migration events between locations, in particular for CRF07_BC, CRF08_BC, and CRF55_01B. Also, when only a few migration events are observed, the impact of imperfect representations of the location-specific diversity on the ancestral reconstructions will increase and can obfuscate the relevance of potential predictors. Furthermore, our models did not capture potential time-varying dynamics of the selected predictors over the study period. This is particularly important for longstanding epidemics, such as HIV-1 subtype B/B′. Unfortunately, we could not test this hypothesis as we did not dispose of time-variable predictors.
Conclusion.
The rapid increase of HIV-1 prevalence among migrant populations and the lack of effective intervention strategies is one of the current challenges for China (71, 72). In this study, the combined use of phylogeographic reconstructions and the generalized linear model provides insights into the spatial viral dynamics of various HIV epidemics across provinces in China. The role of large urbanized areas with dense populations and developed transportation infrastructures as facilitators of HIV dispersal throughout China illustrates the need to consider harmonized country-wide public policies to control local HIV epidemics.
MATERIALS AND METHODS
Ethics statement.
The study was approved by the ethics committee of the First Affiliated Hospital of China Medical University in Shenyang and Wuhan University of Bioengineering.
Data set compilation.
We retrieved all publicly available HIV partial pol sequences (HXB2 positions 2253 to 3554) of CRF01_AE, CRF07_BC, CRF08_BC, B/B′, and CRF55_01B with known sampling date and sampling province of China from the Los Alamos National Laboratory HIV Sequence Database (http://www.hiv.lanl.gov/).
We additionally collected 1,962 CRF01_AE and HIV-1 subtype B partial pol sequences from the NHC Key Laboratory of AIDS Immunology, China Medical University (GenBank accession numbers MT336741 to MT336811 and MT368039 to MT369927). We also retrieved publicly available HIV pol sequences from other countries, along with sampling time and related geographical information. When multiple sequences were available for one participant, only the sequence obtained closest to the estimated time of infection was kept.
Identification of Chinese clades.
The geospatial unit in all phylogeographic analyses was the Chinese province (first subnational administrative level). For all five subtypes except CRF55_01B, for which only isolates from China are available (19), we applied the step-by-step approach described below.
Step 1.
Following the approach of Cuypers et al. (73) and using an as-complete-as-possible background data set (74), we first identified clades that likely correspond to distinct HIV introductions in China. To this end, the sequences for each subtype were first complemented with the publicly available location-annotated HIV pol sequences from the same subtype and aligned to a pol reference sequence (HXB2; GenBank accession number K03455 [75]). AliView (76) was used for manually editing the alignments.
Step 2.
Next, phylogenetic trees were inferred using FastTree2 (77) under a gamma-distributed general time reversible (GTR+Γ) substitution model. These served to identify strongly supported Chinese clades, i.e., clades only including Chinese sequences and associated with a Shimodaira-Hasegawa (SH) support of at least 0.9 (78–80).
Step 3.
Within these monophyletic clades, well-supported clusters of sequences sampled from the same administrative area (province) were identified. These were downsampled by randomly selecting one sequence from each cluster. This step reduces the computational burden while preserving estimation accuracy for the migration flow quantities between Chinese provinces (40).
Time scale for the evolutionary histories.
When sequence data sets lack a clear temporal signal, it is common practice to use empirical evolutionary rate estimates for specifying a suitable prior distribution on the evolutionary rate parameter (e.g., see references 81 and 82).
(i) Subtype B/B′. To obtain plausible priors for the evolutionary rate for HIV-1 subtype B, we considered that various evolutionary rates have been reported for pol, varying from ∼0.001 to ∼0.003 substitutions/site/year (s/s/y) (83–85). For this reason, we specified a normal distribution as prior on the mean clock rate, with a mean of 0.002 s/s/y and standard deviation such that the 95% confidence interval was bounded at 0.001 s/s/y and 0.003 s/s/y.
(ii) CRF01_AE. We considered data from the literature (mean rate estimate, ∼0.0015 [84]), as well as the population-level substitution rate estimate of ∼0.0027 s/s/y (95% highest posterior density [HPD], 0.0013 to 0.0032) obtained from clade-based specific analyses of the CRF01_AE data set with a Bayesian hierarchical phylogenetic model (HPM) approach (data not shown; 85). This led us to specify a normal distribution as the prior on the mean clock rate, with a mean of 0.002 s/s/y and standard deviation of 0.0005.
(iii) CRF07_BC, CRF08_BC, and CRF55_01B. For subtypes CRF07_BC, CRF08_BC, and CRF55_01B, a normal distribution was specified as the prior on the mean clock rate of ∼0.001 s/s/y and standard deviation of 0.0005 according to clade-based estimates (data not shown).
Many of the clades that represent the HIV epidemics in China are limited in size. As this precludes reliable inference under the parameter-rich uncorrelated relaxed clock model (e.g., see reference 86), we opted to model the rate of evolutionary change in clades with ≥10 taxa with a relaxed clock model (87), while for the smaller clades, a strict clock model was assumed.
Phylogeographic inference.
Phylogeographic inference was performed using the discrete diffusion model (67, 88) implemented in the software package BEAST 1.10 (89). To promote estimation accuracy and precision of the transition rates among locations, the substitution model (GTR+Γ) and spread process were shared among clades of the same type (40, 69). A constant-size coalescent prior was assumed for all clades of CRF01_AE, subtype B, and CRF55_01B, and a nonparametric Bayesian skygrid tree prior for clades with ≥20 taxa for CRF07_BC and CRF08_BC (90, 91).
To identify the subset of transition rates that was most informative to reconstruct the dispersal history, we used a model averaging procedure (Bayesian stochastic search variable selection [BSSVS]) (67). In this procedure, the level of support depends on the a priori expected and a posteriori noted fractions of time during the Markov chain Monte Carlo (MCMC) integration that a migration link or predictor helps explain the migration history. In the default setup, however, the a priori expectation only depends on the number of locations and does not account for the relative abundance of samples by location. This can bias inference in the presence of uneven sampling. The adjusted Bayes factor (BFadj) (70) improves on this by incorporating information on the relative abundance of samples by location. It also relies on the a priori expected and a posteriori noted inclusion frequencies under BSSVS, but relative to the original test, it requires two analyses: a first analysis where the trait values remain associated with their respective taxa, and a second one during which the trait values are randomized over the tips of the tree during the MCMC sampling. The latter provides the expectation in the absence of structure in the population, akin to the date randomization test when evaluating the presence of temporal signal (42, 92). As before, support for the significance is calculated as a ratio with the posterior odds as the enumerator, but as denominator, we consider the inclusion frequency from the randomized analysis instead of the prior odds. Bayes factor (BF) support for all possible types of location exchanges was calculated with SpreaD3 (93). BF and BFadj values between 3 and 10, 10 and 20, and above 20 were considered to be substantial, positive, and strong supports, respectively, for the observed transition rates between sampled locations (94). Estimates of the posterior probability of expected number of migration events between all pairs of locations (Markov jumps) were computed through stochastic mapping techniques (95, 96).
MCMC chains were run to ensure adequate mixing. Maximum clade credibility (MCC) trees were obtained with TreeAnnotator 1.10 (89), and convergence and mixing properties were inspected using Tracer 1.7 (97).
GLM analyses.
We used the GLM extension of the discrete phylogeographic model implemented in BEAST 1.10 (39) to investigate the contribution of a series of location-associated variables to the migration rates among Chinese provinces. These variables included sociodemographic indicators (population size and numbers of emigrants and immigrants), HIV prevalence, sample size, and variables related to connectivity between locations (i.e., air traffic density, travel time by railways, the presence of shared borders, a measure of connectivity based on an accessibility model to major cities, and a proxy for spatial distance). Predictors were considered both at the origin and destination location. The population size, the numbers of emigrants and immigrants, and HIV prevalence were obtained from the National Bureau of Statistics of China (98) and the China National Center for Disease Control and Prevention (16).
The numbers of sequences sampled at the origin/destination were included in the GLM to account for the potential impact of sampling biases within the analysis (39). The air traffic density was approximated by an air passenger flux matrix that quantifies the number of passengers traveling between each pair of administrative areas (39). We used a data set provided by the OAG (Official Airline Guide; www.oag.com) and containing the annual average number of seats on scheduled commercial flights between pairs of airports between 2014 and 2016 (100), assuming that the number of seats represents a reasonable proxy for the number of passengers traveling between airports. Travel time by railways was represented by the shortest travel time between the capitals of each province obtained from the 12306 China Railway website (101).
The geospatial connectivity measures included in the GLM were the following: a binary determination of whether administrative areas share a border and the average travel time by railways between locations, as well as two measures of connectivity among administrative areas computed using an algorithm based on circuit theory and implemented in the program Circuitscape 4.0.5 (102), i.e., a measure of connectivity and a proxy of spatial distance, obtained by computing pairwise resistances on an inaccessibility grid and a uniform grid, respectively. For a given pair of locations, Circuitscape computes the pairwise electric resistance based on a geo-referenced grid (or “raster”) covering the study area and defining the local electric resistance values. To compute the proxy of spatial distance, we simply used a homogeneous raster file with cell values uniformly set to “1,” and for the pairwise connectivity measures, we used the inaccessibility raster as in reference 103 to define the local values of electric resistance. Cell values of this inaccessibility raster indicate the travel time required to reach the nearest urban center, with an urban center defined as a contiguous area with 1,500 or more inhabitants per square kilometer or a population center of at least 50,000 inhabitants (103). For computational tractability, the resolution of both the uniform and inaccessibility raster was decreased to ∼5 arcminutes (original resolution, ∼0.5 arcminutes). There are several advantages to using pairwise Circuitscape distances computed on a uniform raster alone or in complement to great-circle distances as proxies of spatial distance. First, pairwise Circuitscape distances constitute more realistic measures because the underlying path model does not assume straight-line movements and also prevents movement through inaccessible areas. Furthermore, given that the uniform raster is the homogeneous version of the inaccessibility raster, pairwise Circuitscape resistances computed on the uniform raster also represent a proper negative control (38, 104) for the inclusion in the GLM analysis of pairwise Circuitscape resistances computed on a heterogeneous raster like the inaccessibility one. Indeed, the inclusion of a GLM predictor that does not have an impact on the dispersal but for which pairwise distances have been computed using an advanced path model (like the one implemented in Circuitscape) can yield a false-positive result in the absence of an appropriate negative control (38). Circuitscape computes pairwise electric resistance between two points or between two sets of points that all have to be associated with precise geographic coordinates. Given that such precise sampling coordinates were not available for sampled sequences, we randomly assigned geographic coordinates to each sampled sequence. While this assignment was stochastic, we still used a human population density raster (resolution of ∼5 arcminutes) to define the sampling probability of all the raster cells within an administrative area. Hence, for each sequence that originated from a given administrative area, its probability of being sampled from a particular raster cell was proportional to the human population density value assigned to this cell. As this is a stochastic procedure, the sampling coordinate assignment and subsequent Circuitscape analyses were repeated 100 times. Final matrices of pairwise resistances computed on the uniform and inaccessibility rasters were obtained by averaging the 100 matrices computed after each repetition of the above procedure. Note that we used the same procedure to compute the averaged great-circle distances among locations.
To protect against a potential impact of sampling imbalances on the GLM results, support for the need for a predictor to help explain the variation in migration rates across locations was obtained after accounting for the relative abundances of the involved trait states (70).
Data availability.
HIV-1 subtype B partial pol sequences are available in GenBank under accession numbers MT336741 to MT336811 and MT368039 to MT369927.
ACKNOWLEDGMENTS
This work was supported by Mega-Projects of National Science Research for the 13th Five-Year Plan (grant number 2018ZX10721102), the Central Public-Interest Scientific Institution Basal Research Fund (grant number 2018PT31042), and The Scientific Research Funding Project of Liaoning Province Education Department in 2019, Raising Seedlings for Young Science and Technology Talents. B.V. (grant number 12U7118N) is and S.D. was supported by postdoctoral fellowships from the Fonds voor Wetenschappelijk Onderzoek (FWO, Belgium). S.D. is currently supported by the Fonds National de la Recherche Scientifique (FNRS, Belgium). A.C. acknowledges funding from the University of San Diego Center for AIDS Research (CFAR), an NIH-funded program.
We gratefully acknowledge the support of the NVIDIA Corporation with the donation of the Titan V GPU used for this research.
REFERENCES
- 1.Lyu P, Chen FF. 2019. National HIV/AIDS epidemic estimation and interpretation in China. Zhonghua Liu Xing Bing Xue Za Zhi 40:1191–1196. (In Chinese.) doi: 10.3760/cma.j.issn.0254-6450.2019.10.004. [DOI] [PubMed] [Google Scholar]
- 2.Ding Y, Ma Z, He J, Xu X, Qiao S, Xu L, Shi R, Xu X, Zhu B, Li J, Wong FY, He N. 2019. Evolving HIV epidemiology in mainland China: 2009-2018. Curr HIV/AIDS Rep 16:423–430. doi: 10.1007/s11904-019-00468-z. [DOI] [PubMed] [Google Scholar]
- 3.He X, Xing H, Ruan Y, Hong K, Cheng C, Hu Y, Xin R, Wei J, Feng Y, Hsi JH, Takebe Y, Shao Y, Group for HIV Molecular Epidemiologic Survey. 2012. A comprehensive mapping of HIV-1 genotypes in various risk groups and regions across China based on a nationwide molecular epidemiologic survey. PLoS One 7:e47289. doi: 10.1371/journal.pone.0047289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xiao P, Li J, Fu G, Zhou Y, Huan X, Yang H. 2017. Geographic distribution and temporal trends of HIV-1 subtypes through heterosexual transmission in China: a systematic review and meta-analysis. Int J Environ Res Public Health 14:830. doi: 10.3390/ijerph14070830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yuan R, Cheng H, Chen LS, Zhang X, Wang B. 2016. Prevalence of different HIV-1 subtypes in sexual transmission in China: a systematic review and meta-analysis. Epidemiol Infect 144:2144–2153. doi: 10.1017/S0950268816000212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang L, Wang YJ, Wang BX, Yan JW, Wan YN, Wang J. 2015. Prevalence of HIV-1 subtypes among men who have sex with men in China: a systematic review. Int J STD AIDS 26:291–305. doi: 10.1177/0956462414543841. [DOI] [PubMed] [Google Scholar]
- 7.Shao Y, Su L, Xing H, Shen J, Sun X, Zhang Y, Cheng H, Liu GE. 2000. HIV molecular epidemic research in China. Bull Med Res 29:19–20. [Google Scholar]
- 8.Guan Y, Chen J, Shao Y, Zhao Q, Zeng Y, Zhang J, Duan Y, Kostler J, Wolf H. 1997. [Subtype and sequence analysis of the C2-V3 region of gp120 genes among human immunodeficiency virus infected IDUs in Ruili epidemic area of Yunnan Province of China]. Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi 11(1):8–12. (In Chinese.) [PubMed] [Google Scholar]
- 9.Zhang M, Jia D, Li H, Gui T, Jia L, Wang X, Li T, Liu Y, Bao Z, Liu S, Zhuang D, Li J, Li L. 2017. Phylodynamic analysis revealed that epidemic of CRF07_BC strain in men who have sex with men drove its second spreading wave in China. AIDS Res Hum Retroviruses 33:1065–1069. doi: 10.1089/aid.2017.0091. [DOI] [PubMed] [Google Scholar]
- 10.Feng Y, Takebe Y, Wei H, He X, Hsi JH, Li Z, Xing H, Ruan Y, Yang Y, Li F, Wei J, Li X, Shao Y. 2016. Geographic origin and evolutionary history of China’s two predominant HIV-1 circulating recombinant forms, CRF07_BC and CRF08_BC. Sci Rep 6:19279. doi: 10.1038/srep19279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang R, Kusagawa S, Zhang C, Xia X, Ben K, Takebe Y. 2003. Identification and characterization of a new class of human immunodeficiency virus type 1 recombinants comprised of two circulating recombinant forms, CRF07_BC and CRF08_BC, in China. J Virol 77:685–695. doi: 10.1128/JVI.77.1.685-695.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takebe Y, Liao H, Hase S, Uenishi R, Li Y, Li XJ, Han X, Shang H, Kamarulzaman A, Yamamoto N, Pybus OG, Tee KK. 2010. Reconstructing the epidemic history of HIV-1 circulating recombinant forms CRF07_BC and CRF08_BC in East Asia: the relevance of genetic diversity and phylodynamics for vaccine strategies. Vaccine 28(Suppl 2):B39–B44. doi: 10.1016/j.vaccine.2009.07.101. [DOI] [PubMed] [Google Scholar]
- 13.Zhao CY, Li BJ, Chen SL. 2011. Molecular epidemiological investigation of HIV-1 circulating strain infected after blood receiving. Chin J Dis Control Prev 11:36–38. [Google Scholar]
- 14.Zhao CY, Zhao HR, Li BJ. 2010. Molecular epidemiology study on HIV infection among paid blood donors. Chin J Health Lab Technol 20:3136–3137. [Google Scholar]
- 15.Li D, Ge L, Wang L, Guo W, Ding Z, Li P, Cui Y. 2014. Trend on HIV prevalence and risk behaviors among men who have sex with men in China from 2010 to 2013. Zhonghua Liu Xing Bing Xue Za Zhi 35:542–546. (in Chinese.) [PubMed] [Google Scholar]
- 16.NCAIDS, NCSTD, China CDC. 2017. [Update on the AIDS/STD epidemic in China in December, 2016.] Chin J AIDS STD 23:93–94. (In Chinese.) [Google Scholar]
- 17.Yin Y, Liu Y, Zhu J, Hong X, Yuan R, Fu G, Zhou Y, Wang B. 2019. The prevalence, temporal trends, and geographical distribution of HIV-1 subtypes among men who have sex with men in China: a systematic review and meta-analysis. Epidemiol Infect 147:e83. doi: 10.1017/S0950268818003400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao J, Cai W, Zheng C, Yang Z, Xin R, Li G, Wang X, Chen L, Zhong P, Zhang C. 2014. Origin and outbreak of HIV-1 CRF55_01B among MSM in Shenzhen, China. J Acquir Immune Defic Syndr 66:e65–e67. doi: 10.1097/QAI.0000000000000144. [DOI] [PubMed] [Google Scholar]
- 19.Han X, An M, Zhang W, Cai W, Chen X, Takebe Y, Shang H. 2013. Genome sequences of a novel HIV-1 circulating recombinant form, CRF55_01B, identified in China. Genome Announc 1:e00050-12. doi: 10.1128/genomeA.00050-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wei L, Lu X, Li H, Zheng C, Li G, Yang Z, Chen L, Cheng J, Wang H, Zhao J. 2018. Impact of HIV-1 CRF55_01B infection on CD4 counts and viral load in men who have sex with men naive to antiretroviral treatment. Lancet 392:S43. doi: 10.1016/S0140-6736(18)32672-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xingyi C. 11 May 2018. A few pictures tell you the development of China’s AIDS epidemic in 2017. https://user.guancha.cn/main/content?id=16151. Accessed 15 January 2020.
- 22.Qi J, Zhang D, Fu X, Li C, Meng S, Dai M, Liu H, Sun J. 2015. High risks of HIV transmission for men who have sex with men—a comparison of risk factors of HIV infection among MSM associated with recruitment channels in 15 cities of China. PLoS One 10:e0121267. doi: 10.1371/journal.pone.0121267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.State Council AIDS Working Committee Office and UN Theme Group on HIV/AIDS. 2004. A joint assessment of HIV/AIDS prevention, treatment and care in China (2004). State Council AIDS Working Committee Office and UN Theme Group on HIV/AIDS http://www.chinaaids.cn/ddpg/lhpgbg1/zgazbyq/201312/P020131210547942626797.pdf.
- 24.Hong Y, Stanton B, Li X, Yang H, Lin D, Fang X, Wang J, Mao R. 2006. Rural-to-urban migrants and the HIV epidemic in China. AIDS Behav 10:421–430. doi: 10.1007/s10461-005-9039-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu Z, Liu H, Li X, Stanton B, Chen X. 2006. HIV-related sexual behaviour among migrants and non-migrants in a rural area of China: role of rural-to-urban migration. Public Health 120:339–345. doi: 10.1016/j.puhe.2005.10.016. [DOI] [PubMed] [Google Scholar]
- 26.Zhang T, Miao Y, Li L, Bian Y. 2019. Awareness of HIV/AIDS and its routes of transmission as well as access to health knowledge among rural residents in Western China: a cross-sectional study. BMC Public Health 19:1630. doi: 10.1186/s12889-019-7992-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mi G, Ma B, Kleinman N, Li Z, Fuller S, Bulterys M, Hladik W, Wu Z. 2016. Hidden and mobile: a web-based study of migration patterns of men who have sex with men in China. Clin Infect Dis 62:1443–1447. doi: 10.1093/cid/ciw167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zong Z, Yang W, Sun X, Mao J, Shu X, Hearst N. 2017. Migration experiences and reported sexual behavior among young, unmarried female migrants in Changzhou, China. Glob Health Sci Pract 5:516–524. doi: 10.9745/GHSP-D-17-00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dai W, Gao J, Gong J, Xia X, Yang H, Shen Y, Gu J, Wang T, Liu Y, Zhou J, Shen Z, Zhu S, Pan Z. 2015. Sexual behavior of migrant workers in Shanghai, China. BMC Public Health 15:1067. doi: 10.1186/s12889-015-2385-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Su L, Liang S, Hou X, Zhong P, Wei D, Fu Y, Ye L, Xiong L, Zeng Y, Hu Y, Yang H, Wu B, Zhang L, Li X. 2018. Impact of worker emigration on HIV epidemics in labour export areas: a molecular epidemiology investigation in Guangyuan, China. Sci Rep 8:16046. doi: 10.1038/s41598-018-33996-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hong J, Chu Z, Wang Q. 2011. Transport infrastructure and regional economic growth: evidence from China. Transportation 38:737–752. doi: 10.1007/s11116-011-9349-6. [DOI] [Google Scholar]
- 32.National Health Commission of the People's Republic of China. 2018. Report on the development of China’s floating population in 2018. Beijing, China: (In Chinese.) http://www.gov.cn/xinwen/2018-12/25/content_5352079.htm. Accessed 15 December 2019. [Google Scholar]
- 33.Lu M, Xia Y. 2016. Migration in the People’s Republic of China. Asian Development Bank Institute, Tokyo, Japan.
- 34.Yang B, Wu Z, Schimmele CM, Li S. 2015. HIV knowledge among male labor migrants in China. BMC Public Health 15:323–323. doi: 10.1186/s12889-015-1653-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang L, Chow EP, Jahn HJ, Kraemer A, Wilson DP. 2013. High HIV prevalence and risk of infection among rural-to-urban migrants in various migration stages in China: a systematic review and meta-analysis. Sex Transm Dis 40:136–147. doi: 10.1097/OLQ.0b013e318281134f. [DOI] [PubMed] [Google Scholar]
- 36.Baele G, Dellicour S, Suchard MA, Lemey P, Vrancken B. 2018. Recent advances in computational phylodynamics. Curr Opin Virol 31:24–32. doi: 10.1016/j.coviro.2018.08.009. [DOI] [PubMed] [Google Scholar]
- 37.Müller NF, Dudas G, Stadler T. 2019. Inferring time-dependent migration and coalescence patterns from genetic sequence and predictor data in structured populations. Virus Evol 5:vez030. doi: 10.1093/ve/vez030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dellicour S, Vrancken B, Trovao NS, Fargette D, Lemey P. 2018. On the importance of negative controls in viral landscape phylogeography. Virus Evol 4:vey023. doi: 10.1093/ve/vey023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lemey P, Rambaut A, Bedford T, Faria N, Bielejec F, Baele G, Russell CA, Smith DJ, Pybus OG, Brockmann D, Suchard MA. 2014. Unifying viral genetics and human transportation data to predict the global transmission dynamics of human influenza H3N2. PLoS Pathog 10:e1003932. doi: 10.1371/journal.ppat.1003932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perez AB, Vrancken B, Chueca N, Aguilera A, Reina G, Garcia-Del Toro M, Vera F, Von Wichman MA, Arenas JI, Tellez F, Pineda JA, Omar M, Bernal E, Rivero-Juarez A, Fernandez-Fuertes E, de la Iglesia A, Pascasio JM, Lemey P, Garcia F, Cuypers L. 2019. Increasing importance of European lineages in seeding the hepatitis C virus subtype 1a epidemic in Spain. Euro Surveill 24:1800227. doi: 10.2807/1560-7917.ES.2019.24.9.1800227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vrancken B, Cuypers L, Perez AB, Chueca N, Anton-Basantas J, de la Iglesia A, Fuentes J, Pineda JA, Tellez F, Bernal E, Rincon P, Von Wichman MA, Fuentes A, Vera F, Rivero-Juarez A, Jimenez M, Vandamme AM, Garcia F. 2019. Cross-country migration linked to people who inject drugs challenges the long-term impact of national HCV elimination programmes. J Hepatol 71:1270–1272. doi: 10.1016/j.jhep.2019.08.010. [DOI] [PubMed] [Google Scholar]
- 42.Trovão NS, Baele G, Vrancken B, Bielejec F, Suchard MA, Fargette D, Lemey P. 2015. Host ecology determines the dispersal patterns of a plant virus. Virus Evol 1:vev016. doi: 10.1093/ve/vev016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Graf T, Vrancken B, Maletich Junqueira D, de Medeiros RM, Suchard MA, Lemey P, Esteves de Matos Almeida S, Pinto AR. 2015. Contribution of epidemiological predictors in unraveling the phylogeographic history of HIV-1 subtype C in Brazil. J Virol 89:12341–12348. doi: 10.1128/JVI.01681-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Faria NR, Vidal N, Lourenco J, Raghwani J, Sigaloff KCE, Tatem AJ, van de Vijver DAM, Pineda-Pena AC, Rose R, Wallis CL, Ahuka-Mundeke S, Muyembe-Tamfum JJ, Muwonga J, Suchard MA, Rinke de Wit TF, Hamers RL, Ndembi N, Baele G, Peeters M, Pybus OG, Lemey P, Dellicour S. 2019. Distinct rates and patterns of spread of the major HIV-1 subtypes in Central and East Africa. PLoS Pathog 15:e1007976. doi: 10.1371/journal.ppat.1007976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang X, He X, Zhong P, Liu Y, Gui T, Jia D, Li H, Wu J, Yan J, Kang D, Han Y, Li T, Yang R, Han X, Chen L, Zhao J, Xing H, Liang S, He J, Yan Y, Xue Y, Zhang J, Zhuang X, Liang S, Bao Z, Li T, Zhuang D, Liu S, Han J, Jia L, Li J, Li L. 2017. Phylodynamics of major CRF01_AE epidemic clusters circulating in mainland of China. Sci Rep 7:6330. doi: 10.1038/s41598-017-06573-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baidu Map Eyes Big Data Team. 2015. Analysis report of big data in hometown of China. Beijing, China: (In Chinese.) https://wenku.baidu.com/view/e9320e18524de518974b7de7.html. Accessed 3 January 2020. [Google Scholar]
- 47.Su Y, Tesfazion P, Zhao Z. 2018. Where are migrants from? Inter- vs. intra-provincial rural-urban migration in China. China Economic Rev 47:142–155. doi: 10.1016/j.chieco.2017.09.004. [DOI] [Google Scholar]
- 48.Qiao Y-C, Xu Y, Jiang D-X, Wang X, Wang F, Yang J, Wei Y-S. 2019. Epidemiological analyses of regional and age differences of HIV/AIDS prevalence in China, 2004–2016. Int J Infect Dis 81:215–220. doi: 10.1016/j.ijid.2019.02.016. [DOI] [PubMed] [Google Scholar]
- 49.Vrancken B, Adachi D, Benedet M, Singh A, Read R, Shafran S, Taylor GD, Simmonds K, Sikora C, Lemey P, Charlton CL, Tang JW. 2017. The multi-faceted dynamics of HIV-1 transmission in northern Alberta: a combined analysis of virus genetic and public health data. Infect Genet Evol 52:100–105. doi: 10.1016/j.meegid.2017.04.005. [DOI] [PubMed] [Google Scholar]
- 50.Li Z, He X, Wang Z, Xing H, Li F, Yang Y, Wang Q, Takebe Y, Shao Y. 2012. Tracing the origin and history of HIV-1 subtype B′ epidemic by near full-length genome analyses. AIDS 26:877–884. doi: 10.1097/QAD.0b013e328351430d. [DOI] [PubMed] [Google Scholar]
- 51.Chu XG, Zhang XF, Zhan FX, Tang H, Chen HP, Peng TH, Gong ZJ. 2007. Study on molecular epidemiology of people infected with human immunodeficiency virus-1 in Hubei Province. Zhonghua Liu Xing Bing Xue Za Zhi 28:992–995. (In Chinese.) [PubMed] [Google Scholar]
- 52.Qian S, Guo W, Xing J, Qin Q, Ding Z, Chen F, Peng Z, Wang L. 2014. Diversity of HIV/AIDS epidemic in China: a result from hierarchical clustering analysis and spatial autocorrelation analysis. AIDS 28:1805–1813. doi: 10.1097/QAD.0000000000000323. [DOI] [PubMed] [Google Scholar]
- 53.Shan H, Wang JX, Ren FR, Zhang YZ, Zhao HY, Gao GJ, Ji Y, Ness PM. 2002. Blood banking in China. Lancet 360:1770–1775. doi: 10.1016/S0140-6736(02)11669-2. [DOI] [PubMed] [Google Scholar]
- 54.Zeng P, Wang J, Huang Y, Guo X, Li J, Wen G, Yang T, Yun Z, He M, Liu Y, Yuan Y, Schulmann J, Glynn S, Ness P, Jackson JB, Shan H, NHLBI Retrovirus Epidemiology Donor Study-II (Reds-II), International Component. 2012. The human immunodeficiency virus-1 genotype diversity and drug resistance mutations profile of volunteer blood donors from Chinese blood centers. Transfusion 52:1041–1049. doi: 10.1111/j.1537-2995.2011.03415.x. [DOI] [PubMed] [Google Scholar]
- 55.Zhang L, Chen Z, Cao Y, Yu J, Li G, Yu W, Yin N, Mei S, Li L, Balfe P, He T, Ba L, Zhang F, Lin HH, Yuen MF, Lai CL, Ho DD. 2004. Molecular characterization of human immunodeficiency virus type 1 and hepatitis C virus in paid blood donors and injection drug users in China. J Virol 78:13591–13599. doi: 10.1128/JVI.78.24.13591-13599.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.NDRC (National Development and Reform Commission). 2016. “Mid- and long-term railway network plan.” National Development and Reform Commission of the People’s Republic of China, Beijing, China: http://documents1.worldbank.org/curated/en/933411559841476316/pdf/Chinas-High-Speed-Rail-Development.pdf. Accessed January 2020. [Google Scholar]
- 57.Luo MY, Pan XH, Fan Q, Zhang JF, Ge R, Jiang J, Chen WJ. 2019. Epidemiological characteristics of molecular transmission cluster among reported HIV/AIDS cases in Jiaxing city, Zhejiang province, 2017. Zhonghua Liu Xing Bing Xue Za Zhi 40:202–206. (In Chinese.) doi: 10.3760/cma.j.issn.0254-6450.2019.02.015. [DOI] [PubMed] [Google Scholar]
- 58.Han ZG, Zhang YL, Wu H, Gao K, Zhao YT, Gu YZ, Chen YC. 2018. Prevalence of drug resistance in treatment-naive HIV infected men who have sex with men in Guangzhou, 2008–2015. Zhonghua Liu Xing Bing Xue Za Zhi 39:977–982. (In Chinese.) doi: 10.3760/cma.j.issn.0254-6450.2018.07.021. [DOI] [PubMed] [Google Scholar]
- 59.Xiao P, Zhou Y, Lu J, Yan L, Xu X, Hu H, Li J, Ding P, Qiu T, Fu G, Huan X, Yang H. 2019. HIV-1 genotype diversity and distribution characteristics among heterosexually transmitted population in Jiangsu province, China. Virol J 16:51. doi: 10.1186/s12985-019-1162-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liang Z, Li Z, Ma Z. 2014. Changing patterns of the floating population in China during 2000–2010. Popul Dev Rev 40:695–716. doi: 10.1111/j.1728-4457.2014.00007.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Poon CM, Wong NS, Kwan TH, Wong HTH, Chan KCW, Lee SS. 2018. Changes of sexual risk behaviors and sexual connections among HIV-positive men who have sex with men along their HIV care continuum. PLoS One 13:e0209008. doi: 10.1371/journal.pone.0209008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dennis AM, Volz E, Frost A, Hossain M, Poon AFY, Rebeiro PF, Vermund SH, Sterling TR, Kalish ML. 2018. HIV-1 transmission clustering and phylodynamics highlight the important role of young men who have sex with men. AIDS Res Hum Retroviruses 34:879–888. doi: 10.1089/aid.2018.0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chaillon A, Delaugerre C, Brenner B, Armero A, Capitant C, Nere ML, Leturque N, Pialoux G, Cua E, Tremblay C, Smith DM, Goujard C, Meyer L, Molina JM, Chaix ML. 2019. In-depth Sampling of High-risk Populations to Characterize HIV Transmission Epidemics Among Young MSM Using PrEP in France and Quebec. Open Forum Infect Dis 6:ofz080. doi: 10.1093/ofid/ofz080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pybus OG, Tatem AJ, Lemey P. 2015. Virus evolution and transmission in an ever more connected world. Proc R Soc B Biol Sci 282:20142878. doi: 10.1098/rspb.2014.2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Holmes EC. 2008. Evolutionary history and phylogeography of human viruses. Annu Rev Microbiol 62:307–328. doi: 10.1146/annurev.micro.62.081307.162912. [DOI] [PubMed] [Google Scholar]
- 66.Xia Y, Bjornstad ON, Grenfell BT. 2004. Measles metapopulation dynamics: a gravity model for epidemiological coupling and dynamics. Am Nat 164:267–281. doi: 10.2307/3473444. [DOI] [PubMed] [Google Scholar]
- 67.Lemey P, Rambaut A, Drummond AJ, Suchard MA. 2009. Bayesian phylogeography finds its roots. PLoS Comput Biol 5:e1000520. doi: 10.1371/journal.pcbi.1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lemey P, Rambaut A, Welch JJ, Suchard MA. 2010. Phylogeography takes a relaxed random walk in continuous space and time. Mol Biol Evol 27:1877–1885. doi: 10.1093/molbev/msq067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Faria NR, Hodges-Mameletzis I, Silva JC, Rodés B, Erasmus S, Paolucci S, Ruelle J, Pieniazek D, Taveira N, Treviño A, Gonçalves MF, Jallow S, Xu L, Camacho RJ, Soriano V, Goubau P, de Sousa JD, Vandamme A-M, Suchard MA, Lemey P. 2012. Phylogeographical footprint of colonial history in the global dispersal of human immunodeficiency virus type 2 group A. J Gen Virol 93:889–899. doi: 10.1099/vir.0.038638-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chaillon A, Gianella S, Dellicour S, Rawlings SA, Schlub TE, Faria De Oliveira M, Ignacio C, Porrachia M, Vrancken B, Smith DM. 2020. HIV persists throughout deep tissues with repopulation from multiple anatomical sources. J Clin Invest 30:1699–1712. doi: 10.1172/JCI134815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu X, Erasmus V, Wu Q, Richardus JH. 2014. Behavioral and psychosocial interventions for HIV prevention in floating populations in China over the past decade: a systematic literature review and meta-analysis. PLoS One 9:e101006. doi: 10.1371/journal.pone.0101006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li X, Gao R, Zhu K, Wei F, Fang K, Li W, Song Y, Ge Y, Ji Y, Zhong P, Wei P. 2018. Genetic transmission networks reveal the transmission patterns of HIV-1 CRF01_AE in China. Sex Transm Infect 94:111–116. doi: 10.1136/sextrans-2016-053085. [DOI] [PubMed] [Google Scholar]
- 73.Cuypers L, Vrancken B, Fabeni L, Marascio N, Cento V, Di Maio VC, Aragri M, Pineda-Pena AC, Schrooten Y, Van Laethem K, Balog D, Foca A, Torti C, Nevens F, Perno CF, Vandamme AM, Ceccherini-Silberstein F. 2017. Implications of hepatitis C virus subtype 1a migration patterns for virus genetic sequencing policies in Italy. BMC Evol Biol 17:70. doi: 10.1186/s12862-017-0913-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vrancken B, Alavian SM, Aminy A, Amini-Bavil-Olyaee S, Pourkarim MR. 2018. Why comprehensive datasets matter when inferring epidemic links or subgenotyping. Infect Genet Evol 65:350–351. doi: 10.1016/j.meegid.2018.08.012. [DOI] [PubMed] [Google Scholar]
- 75.Smith TF, Waterman MS. 1981. Identification of common molecular subsequences. J Mol Biol 147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- 76.Larsson A. 2014. AliView: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 30:3276–3278. doi: 10.1093/bioinformatics/btu531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 79.Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 80.Shimodaira H, Hasegawa M. 1999. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol Biol Evol 16:1114–1114. doi: 10.1093/oxfordjournals.molbev.a026201. [DOI] [Google Scholar]
- 81.Al-Qahtani AA, Baele G, Khalaf N, Suchard MA, Al-Anazi MR, Abdo AA, Sanai FM, Al-Ashgar HI, Khan MQ, Al-Ahdal MN, Lemey P, Vrancken B. 2017. The epidemic dynamics of hepatitis C virus subtypes 4a and 4d in Saudi Arabia. Sci Rep 7:44947. doi: 10.1038/srep44947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang Y, Vrancken B, Feng Y, Dellicour S, Yang Q, Yang W, Zhang Y, Dong L, Pybus OG, Zhang H, Tian H. 2017. Cross-border spread, lineage displacement and evolutionary rate estimation of rabies virus in Yunnan Province, China. Virol J 14:102. doi: 10.1186/s12985-017-0769-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Abecasis AB, Vandamme AM, Lemey P. 2009. Quantifying differences in the tempo of human immunodeficiency virus type 1 subtype evolution. J Virol 83:12917–12924. doi: 10.1128/JVI.01022-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Patino-Galindo JA, Gonzalez-Candelas F. 2017. The substitution rate of HIV-1 subtypes: a genomic approach. Virus Evol 3:vex029. doi: 10.1093/ve/vex029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Vrancken B, Baele G, Vandamme AM, van Laethem K, Suchard MA, Lemey P. 2015. Disentangling the impact of within-host evolution and transmission dynamics on the tempo of HIV-1 evolution. AIDS 29:1549–1556. doi: 10.1097/QAD.0000000000000731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.de Goede AL, van Deutekom HWM, Vrancken B, Schutten M, Allard SD, van Baalen CA, Osterhaus ADME, Thielemans K, Aerts JL, Keşmir C, Lemey P, Gruters RA. 2013. HIV-1 evolution in patients undergoing immunotherapy with Tat, Rev, and Nef expressing dendritic cells followed by treatment interruption. AIDS 27:2679–2689. doi: 10.1097/01.aids.0000433813.67662.92. [DOI] [PubMed] [Google Scholar]
- 87.Drummond AJ, Ho SYW, Phillips MJ, Rambaut A. 2006. Relaxed phylogenetics and dating with confidence. PLoS Biol 4:e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Edwards CJ, Suchard MA, Lemey P, Welch JJ, Barnes I, Fulton TL, Barnett R, O’Connell TC, Coxon P, Monaghan N, Valdiosera CE, Lorenzen ED, Willerslev E, Baryshnikov GF, Rambaut A, Thomas MG, Bradley DG, Shapiro B. 2011. Ancient hybridization and an Irish origin for the modern polar bear matriline. Curr Biol 21:1251–1258. doi: 10.1016/j.cub.2011.05.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Suchard MA, Lemey P, Baele G, Ayres DL, Drummond AJ, Rambaut A. 2018. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol 4:vey016. doi: 10.1093/ve/vey016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Drummond AJ, Nicholls GK, Rodrigo AG, Solomon W. 2002. Estimating mutation parameters, population history and genealogy simultaneously from temporally spaced sequence data. Genetics 161:1307–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gill MS, Lemey P, Faria NR, Rambaut A, Shapiro B, Suchard MA. 2013. Improving Bayesian population dynamics inference: a coalescent-based model for multiple loci. Mol Biol Evol 30:713–724. doi: 10.1093/molbev/mss265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Firth C, Kitchen A, Shapiro B, Suchard MA, Holmes EC, Rambaut A. 2010. Using time-structured data to estimate evolutionary rates of double-stranded DNA viruses. Mol Biol Evol 27:2038–2051. doi: 10.1093/molbev/msq088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bielejec F, Baele G, Vrancken B, Suchard MA, Rambaut A, Lemey P. 2016. SpreaD3: interactive visualization of spatiotemporal history and trait evolutionary processes. Mol Biol Evol 33:2167–2169. doi: 10.1093/molbev/msw082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kass RE, Raftery AE. 1995. Bayes factors. J Am Stat Assoc 90:773–795. doi: 10.1080/01621459.1995.10476572. [DOI] [Google Scholar]
- 95.Minin VN, Suchard MA. 2008. Counting labeled transitions in continuous-time Markov models of evolution. J Math Biol 56:391–412. doi: 10.1007/s00285-007-0120-8. [DOI] [PubMed] [Google Scholar]
- 96.Minin VN, Bloomquist EW, Suchard MA. 2008. Smooth skyride through a rough skyline: Bayesian coalescent-based inference of population dynamics. Mol Biol Evol 25:1459–1471. doi: 10.1093/molbev/msn090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA. 2018. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst Biol 67:901–904. doi: 10.1093/sysbio/syy032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.National Bureau of Statistics of China. 2016. Nationwide population census. http://www.stats.gov.cn/tjsj/pcsj/rkpc/6rp/indexch.htm. Accessed September 2019.
- 99.Reference deleted.
- 100.Woolley-Meza O, Thiemann C, Grady D, Lee JJ, Seebens H, Blasius B, Brockmann D. 2011. Complexity in human transportation networks: a comparative analysis of worldwide air transportation and global cargo-ship movements. Eur Phys J B 84:589–600. doi: 10.1140/epjb/e2011-20208-9. [DOI] [Google Scholar]
- 101.China Railway. 2019. Travel time by train in China. https://www.12306.cn/index/. Accessed September 2019.
- 102.McRae BH, Dickson BG, Keitt TH, Shah VB. 2008. Using circuit theory to model connectivity in ecology, evolution, and conservation. Ecology 89:2712–2724. doi: 10.1890/07-1861.1. [DOI] [PubMed] [Google Scholar]
- 103.Weiss DJ, Nelson A, Gibson HS, Temperley W, Peedell S, Lieber A, Hancher M, Poyart E, Belchior S, Fullman N, Mappin B, Dalrymple U, Rozier J, Lucas TCD, Howes RE, Tusting LS, Kang SY, Cameron E, Bisanzio D, Battle KE, Bhatt S, Gething PW. 2018. A global map of travel time to cities to assess inequalities in accessibility in 2015. Nature 553:333–336. doi: 10.1038/nature25181. [DOI] [PubMed] [Google Scholar]
- 104.Dellicour S, Rose R, Pybus OG. 2016. Explaining the geographic spread of emerging epidemics: a framework for comparing viral phylogenies and environmental landscape data. BMC Bioinformatics 17:82. doi: 10.1186/s12859-016-0924-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
HIV-1 subtype B partial pol sequences are available in GenBank under accession numbers MT336741 to MT336811 and MT368039 to MT369927.