Aedes mosquito-transmitted viruses such as the Zika, dengue, and chikungunya viruses have spread globally. CHIKV, similar to many other enveloped viruses, enters cells in sequential steps: step 1 involves receptor binding followed by endocytosis, and step 2 involves viral-cell membrane fusion in the endocytic vesicle. The viral envelope surface protein, E1, performs membrane fusion. E1 is triggered to undergo conformational changes by acidic pH of the maturing endosome. Different domains of E1 rearrange during the pre- to postfusion conformation change. Using in silico analysis of the E1 structure and different biochemical experiments, we explained a structural mechanism of key conformational changes in E1 triggered by acidic pH. We noted two important structural changes in E1 at acidic pH. In the first, a spring-twisted region in a loop connecting two domains (I and III) untwists, bringing a swiveling motion of domains on each other. In the second, breaking of interactions between domains I and III and domain separation are required for membrane fusion. This knowledge will help devise new therapeutic strategies to block conformation changes in E1 and thus viral entry.
KEYWORDS: acidic pH trigger, alphavirus entry, chikungunya virus, class II fusion protein, domain I and III linker, domain swiveling, fusion protein E1, prefusion conformation
ABSTRACT
Chikungunya virus (CHIKV), a mosquito-transmitted alphavirus, enters a cell through endocytosis, followed by viral and cell membrane fusion. The fusion protein, E1, undergoes an acid pH-induced pre- to postfusion conformation change during membrane fusion. As part of the conformation change, E1 dissociates from the receptor-binding protein, E2, and swivels its domains I and II over domain III to form an extended intermediate and then eventually to form a postfusion hairpin homotrimer. In this study, we tested if the domain I-III linker acts as a “hinge” for the swiveling motion of E1 domains. We found a conserved spring-twisted structure in the linker, stabilized by a salt bridge between a conserved arginine-aspartic acid pair, as a “hinge point” for domain swiveling. Molecular dynamics (MD) simulation of the CHIKV E1 or E2-E1 structure predicted that the spring-twisted region untwists at pH 5.5. Corroborating the prediction, introduction of a “cystine staple” at the hinge point, replacing the conserved arginine-aspartic acid pair with cysteine residues, resulted in loss of fusion activity of E1. MD simulation also predicted domain I-III swiveling at acidic pH. We tested if breaking the His 331-Lys 16 H bond between domains I and III, seen only in the prefusion conformation, is important for domain swiveling. When domains I and III are “stapled” by introducing a disulfide bond in between, E1 showed loss of fusion activity, implying that domain I and III dissociation is a critical acid pH-induced step in membrane fusion. However, replacement of His 331 with an acidic residue did not affect the pH threshold for fusion, suggesting His 331 is not an acid-sensing residue.
IMPORTANCE Aedes mosquito-transmitted viruses such as the Zika, dengue, and chikungunya viruses have spread globally. CHIKV, similar to many other enveloped viruses, enters cells in sequential steps: step 1 involves receptor binding followed by endocytosis, and step 2 involves viral-cell membrane fusion in the endocytic vesicle. The viral envelope surface protein, E1, performs membrane fusion. E1 is triggered to undergo conformational changes by acidic pH of the maturing endosome. Different domains of E1 rearrange during the pre- to postfusion conformation change. Using in silico analysis of the E1 structure and different biochemical experiments, we explained a structural mechanism of key conformational changes in E1 triggered by acidic pH. We noted two important structural changes in E1 at acidic pH. In the first, a spring-twisted region in a loop connecting two domains (I and III) untwists, bringing a swiveling motion of domains on each other. In the second, breaking of interactions between domains I and III and domain separation are required for membrane fusion. This knowledge will help devise new therapeutic strategies to block conformation changes in E1 and thus viral entry.
INTRODUCTION
Chikungunya virus (CHIKV) is an Aedes mosquito-transmitted alphavirus that reemerged as a global pathogen in the recent past (1, 2). CHIKV infection causes chikungunya fever. Characteristic symptoms of the infection include fever, rash, and joint and muscle pain that last for weeks to months (3).
CHIKV enters a host cell through receptor binding-induced endocytosis, followed by viral and cell membrane fusion. Membrane fusion ensues when the maturing endosome is acidified (4–6). Receptor binding and membrane fusion are achieved by the viral surface glycoproteins E2 and E1, respectively. E2 and E1 are anchored, through carboxy-terminal ends of the proteins, into the viral envelope (7). E2 and E1 are arranged on the virion surface as heterodimers. Precisely 240 E2-E1 heterodimers arrange into 80 spike-like structures, following the T = 4 icosahedral symmetry of the capsid. Each spike has three E2-E1 heterodimers winding around a vertical axis. In the spike, E2 protomers are at the center and E1 protomers are to the periphery (Fig. 1) (8). A small protein, E3, which is a proteolytic processing product of the precursor of E2, p62, is held on the trimeric spike surface through noncovalent interactions with E2 (6, 7, 9, 10).
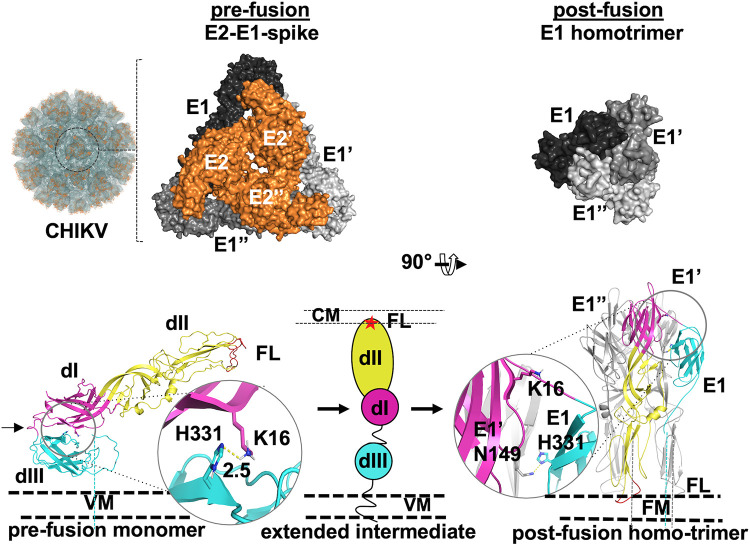
FIG 1.
CHIKV envelope proteins pre- to postfusion conformation switch. (Top, left to right) Image from 3D reconstruction of CHIKV E1-E2 into CHIKV virus-like particles (EMDB ID 5577); the surface view (labeled “CHIKV”) is shown. One trimeric spike structure is shown in the circle. An enlarged image of one E2-E1 trimeric spike is shown to the left (PDB ID 3J2W). Top views of E1 (different shades of gray) and E2 (orange) are shown in a surface representation. Three protomers of E1 and E2 are labeled E1, E′, and E′′ and E2, E2′, and E2′′, respectively. The top view of the postfusion E1 homotrimer (PDB ID 1RER) is shown to the right. (Bottom left) A single monomer of E1 (from PDB ID 3N42), in the prefusion conformation, is shown in cartoon representation with the three domains (dI, dII, and dIII) and fusion loops (FL) labeled. The dI-III linker is marked with a black arrow. The middle and right portions show a schematic of the extended intermediate (middle [dIII, dI, and dII and fusion loops depicted in cyan, magenta, and yellow ovals and a red star, respectively]) and postfusion (right) E1 homotrimer conformation. The domain I-III interface His 331-Lys 16 H bond in prefusion and the His 331-Asn 149 H bond in postfusion are shown in a cartoon and stick representation with enlarged areas in circles. VM, CM, and FM refer to viral membrane, cell membrane, and fused membrane, respectively. Dashed lines connecting protein structure figures with VM represent the stem and transmembrane region that anchor the proteins into VM or FM.
CHIKV membrane fusion protein E1, similar to other viral membrane fusion proteins, undergoes a required conformation change (from prefusion to postfusion through an intermediate extended conformation) to perform viral and cell membrane fusion (Fig. 1) (6, 11). CHIKV E1 protein (as well as those from other alphaviruses) has the following structural features of a class II viral fusion protein. (i) It has three β-sheet-rich domains: domains III, I, and II from the viral membrane proximal to distal order. (Henceforth, domains I, II, and III are referred to as dI, dII, and dIII for easy reference.) (ii) It has internal fusion loops at the tip of domain II. (iii) It forms a heterodimer with the receptor-binding protein, E2, in the prefusion state. Post-membrane fusion, E1 forms homotrimers (Fig. 1) (11, 12). During the membrane fusion process, each protomer of E1 in the trimeric spike rearranges its three domains to form an intermediate extended conformation, and then it folds back to a postfusion hairpin conformation. Also, E2 and E1 dissociate and three E1 protomers replace E2 from the center of the trimeric spike. The pre- to postfusion conformation change in the fusion protein involves rearrangements of the domains, but not secondary structure changes. The domain rearrangements in alphavirus E1 are triggered by acidic pH (5, 10, 13, 14). The optimum pH for triggering membrane fusion for CHIKV is 5.5 (15).
The crystal structure of the E2-E1 heterodimer in its prefusion conformation (crystallized in neutral-pH buffers, in a range of pH 7.1 to 7.5) (PDB identification number [ID] 3N42) (7) is presented in Fig. 1, along with the (E1-E2)3 spike structure from cryo-electron microscopy (cryo-EM) three-dimensional (3D) reconstruction of the CHIKV E2-E1 crystal structure onto electron density maps of CHIKV virus-like particles (EMDB ID 5577) (8). As revealed in these structures, in each E1 protomer, domain II (the most distal domain from the viral membrane surface) and domain I (the central domain) are arranged on a linear axis, whereas domain III folds back onto domain I. Domains I and III are held together by several weak interactions: notable of those are interactions of “acid-sensing” histidine residues with other charged residues (16). The histidine 331 (from domain III)-lysine 16 (from domain I) H bond is one such interaction that contributes to “stapling” of these two domains in the prefusion conformation (17). At the tip of domain II are fusion loops. The majority of the E1 surface area, including fusion loops, is masked by E2 in the heterodimer structure. Domains I and III are connected by a 12-residue-long linker.
The postfusion structure of CHIKV E1 is not available. However, the crystal structure of E1 from Semliki Forest virus (SFV), determined at acidic pH (pH 4.0), has been published (13) (PDB ID 1RER). In the postfusion conformation, SFV E1 is a homotrimer. In this conformation, the dI-II regions from three E1 protomers form the central core of the homotrimer, surrounding the vertical axis of the trimer. In each protomer, domain III and the rest of the C terminus of E1 are folded back onto domains I and II, to form a postfusion hairpin structure (Fig. 1, “post-fusion”). Due to the similarity of the CHIKV E1 prefusion structure to those of SFV and Sindbis virus (SINV), it is reasonable to assume that CHIKV E1 also forms a postfusion homotrimer, very similar to the SFV E1 postfusion homotrimer. In the postfusion conformation, the His 331-Lys 16 interaction is broken. Instead, His 331 forms a new interaction with Asn 149 of domain I from a neighboring protomer. (Please see the enlarged cartoon and stick representation of prefusion and postfusion structures in Fig. 1, bottom panel.) Cao et al. (18), based on cryo-EM studies of SINV at acidic pH and in the presence of liposomes, showed an extended intermediate conformation of E1. Several other biochemical studies (10, 19–21) also indicated that domains III and I separate and form an extended intermediate during early steps of membrane fusion.
In an earlier work, we showed that receptor binding leads to conformational changes in E2 that trigger E2-E1 dissociation (22). E2-E1 dissociation is completed in acidic pH of the endosome (13, 14, 18, 23, 24). Once E2 dissociates from E1, the dI-II regions from the three E1 protomers have to swivel to the center of the spike. Li et al. (14) proposed that, upon the acidic pH trigger, the dI-II region would move by 180° to the center of the spike, displacing E2. By time-resolved cryo-electron microscopy of SFV, Fuller et al. (25) also proposed that at acidic pH, E1 shows a centripetal movement by a swiveling motion toward the center of the spike to form a trimer. A similar proposal was made by Mukhopadhyay et al. (26). However, how and where swiveling of E1 domains is initiated is not known.
The domain I-III linker is the only flexible region connecting domains I and III. There is no flexible region between domain III and the C-terminal transmembrane region. As a result, domain III is anchored in position on the membrane surface, and thus swiveling of the entire ectodomain of E1 to the center of the spike is improbable. One possible way that the dI-II region of each protomer moves to the center of the spike is by a swiveling motion over the dI-III linker. We wondered if the swiveling motion of domains is brought about by conformational changes in the dI-III linker in acidic pH. Prompted by this idea, we analyzed the sequences and structures of the E1 dI-III linkers of different alphaviruses. We found a conserved spring-twisted region in the linker that is held by a salt bridge between a conserved arginine and a negatively charged residue (aspartate or glutamate) pair. Our analysis of a constant-pH molecular dynamics (MD) simulation study of the E2-E1 heterodimer and E1 alone, at the fusion-triggering pH of 5.5, shows that the conserved salt bridge in the linker is broken and the domain I-II region swivels outwards, using the spring-twisted region as a hinge point. Corroborating this, a mutant E1 protein in which the linker region conserved salt bridge region is “stapled” through an introduced cystine bond has a loss of fusion activity. Similarly, cystine stapling of domains I and III also resulted in loss of fusion activity of E1. Based on these studies, we propose that the spring-twisted region in the dI-III linker acts as a hinge point for dI-II swiveling on dIII in the pre- to postfusion conformation change of E1.
RESULTS
A conserved arginine-aspartic acid/glutamic acid pair in the alphavirus E1 dI-III linker.
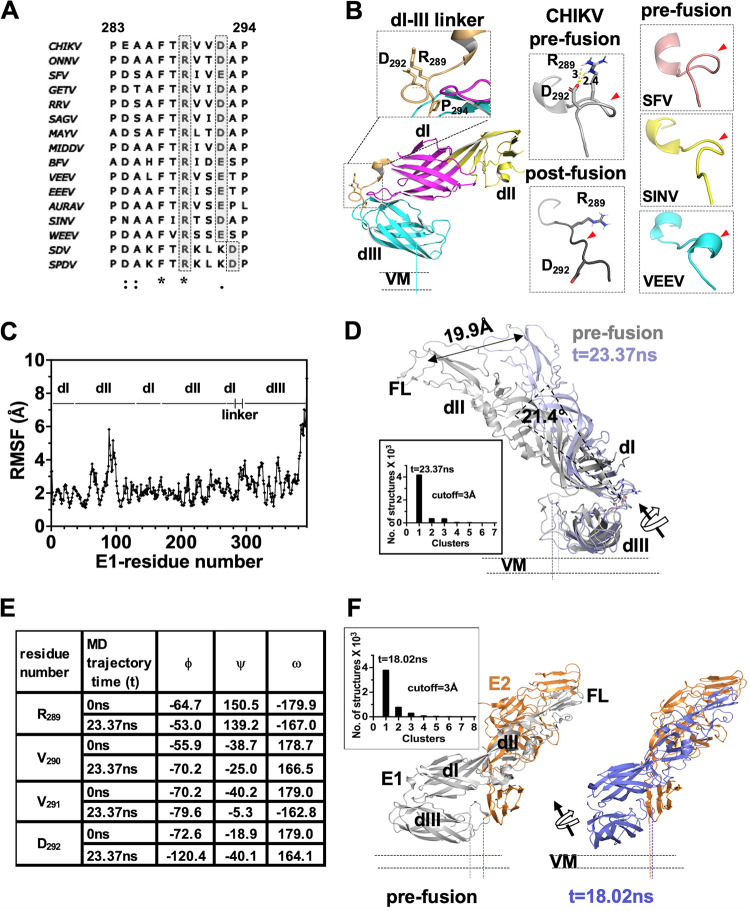
In order to understand the role of the dI-III linker in E1 domain swiveling movements during membrane fusion, we started by aligning and comparing the linker region sequences from different alphaviruses. Sequence alignment is shown in Fig. 2A. There are two conserved prolines, one on either side, as bounds of the linker in most of the sequences. Overall, no sequence conservation is noted, except for an arginine and a phenylalanine in the middle of the sequence. As per CHIKV E1 sequence numbering, the conserved arginine is at position 289. We noted that an aspartic acid or a glutamic acid residue, 2 or 3 residues in the C-terminal side of the Arg 289, is seen in 14 out of 16 sequences that we compared. Earlier, Zheng et al. (20) studied the importance of the dI-III linker in SFV E1 by creating point mutations and deletion mutants. Their observations gave insights into the importance of the linker in acid pH-induced homotrimerization of E1. They inferred that an acid-sensing conserved histidine residue in domain I, H3, forms a π-cation interaction with the conserved arginine and phenylalanine of the dI-III linker, in a postfusion homotrimer. This suggests that the conserved arginine and surrounding residues in the dI-III linker may have a critical role in acid pH-induced conformation changes in E1.
FIG 2.
Analysis of alphavirus E1 dI-III linker region in pre- and postfusion states and MD simulation analysis. (A) Multiple-sequence alignment of the alphavirus E1 dI-III linker. A conserved arginine-aspartate/glutamate pair is highlighted in gray. Sequence numbering is as per the CHIKV E1 sequence. (B) The left column shows the structure of the dI-III linker from CHIKV E1 (from PDB ID 3N42) in the prefusion conformation (cartoon representation). The conserved arginine (R289) and aspartate (D292) pair is shown in sticks. An enlarged image of the boxed region of the dI-III linker is shown in the inset above. The main chain hydrogen bond holding the spring twist region is shown by a yellow dotted line. The middle column shows the CHIKV E1 dI-III linker region in the prefusion and postfusion conformations. The distance between R289 and D292 side chain atoms is shown, and the salt bridge is shown by a yellow dotted line. The right column shows the dI-III linker regions from prefusion structures of SFV, SINV, and VEEV. In the middle and right columns, the dI-III linker is shown in a different orientation compared to that in left column, and red arrowheads point to the spring-twisted region of the linker. (C) Root mean square fluctuation (RMSF) analysis of the only E1 MD simulation trajectory. The sequence stretch is mapped to the corresponding domain (dI, dII, dIII, or dI-III linker). (D) Structural overlay of the t = 23.37-ns cluster central structure from MD simulations (slate) over the prefusion crystal structure of E1 (from PDB ID 3N42). Cluster analysis of the run is shown graphically in the inset. (E) Comparison of main chain dihedral angles (N-Cα, ϕ; Cα-C, ψ; C-N,: ω) of “RVVD” residues from the dI-dIII linker region between the t = 0-ns and t = 23.37-ns structures from E1 MD simulation studies. (F) Cluster analysis of MD simulation of E2-E1 structure. Insets show cluster analysis graphically. In panels D and F, displacement of the center of mass of dI-II regions in the MD simulation is depicted by a double-headed arrow. Rotation of the dI-II virtual axis over the dI-III linker is shown with a curved arrow. The centers of mass of dI-II regions were estimated using the center_of_mass.py script from the PyMol script repository.
A conserved spring-twisted structure in the dI-III linker is untwisted in the pre- to postfusion conformation change.
We then analyzed the structures of the dI-III linkers from different alphaviruses for which a prefusion structure of E1 is available. The linker region does not have any regular secondary structure, except for a 310 helix at the N terminus (Fig. 2B). However, two features of the dI-III linker structure caught our attention. In CHIKV E1, side chains of the conserved arginine (R289) and the conserved aspartic acid (D292) face in the same direction, away from the protein interior (Fig. 2B, left column and inset). The distance between the Arg 289 and the Asp 292 side chains (∼3.0 Å between the guanidino amino group nitrogen atoms [Nε or Nω1/ω2] on R289 and OD2 on D292; PDB ID 3N42) would allow a salt bridge formation through side-on bidentate interactions (Fig. 2B, middle column). Similarly, in SFV and Venezuelan equine encephalomyelitis virus (VEEV) as well, the Arg 289 and Glu 292 side chains are at a distance and at an orientation to form a salt bridge. Thus, the salt bridge between the conserved Arg and Asp/Glu pair in the prefusion conformation seems to be a conserved structural feature in the alphavirus E1 dI-III linker region.
Another interesting structural feature that we noted is the backbone conformation of the region between the conserved Arg 289 and Asp 292. The backbone of this region is twisted like a spring. In addition, the spring twist of the backbone seems to be stabilized by the Arg-Asp/Glu salt bridge, apart from a main chain hydrogen bond (Fig. 2B, inset in left panel). Except for VEEV E1, where this region is modeled as a 310 helix, in CHIKV, SFV, and SINV E1 this region is seen as a spring-twisted structure (Fig. 2B, panels in right column). Considering that the dI-III linker is critical for viral entry (20), we wondered if the conserved spring-twisted region of the dI-III linker acts as a “hinge” for dI-II swiveling (over dIII) during membrane fusion. If this were to be the case, then in the extended intermediate and in the postfusion conformation of E1, we would expect the spring-twisted region to be untwisted and the Arg-Asp/Glu salt bridge to be broken. The postfusion structure of CHIKV E1 is not available. Hence, we modeled the postfusion structure of CHIKV E1, using the SFV E1 postfusion homotrimer structure (PDB ID 1RER). Indeed, in the postfusion structure of SFV and in the postfusion model of CHIKV E1 (Fig. 2B, middle column, “post-fusion”), the conserved arginine side chain is on the opposite side to that of aspartic acid (glutamic acid in SFV). More importantly, the backbone of the region does not show the spring twist seen in the prefusion conformation.
Thus, our sequence and structure analyses suggest that the spring-twisted region of the E1 dI-III linker acts as a “hinge” for the domain swiveling in the pre- to postfusion conformational change of E1.
Constant-pH MD simulations of E2-E1 and E1 at fusion-triggering pH show dI-II rotation on dIII over the linker spring-twisted hinge.
In order to understand conformational changes in E1 at fusion-triggering acidic pH, we performed constant-pH (5.5) molecular dynamics simulation on E1. After titration of the E1 protein structure at pH 5.5, we compared the side chain protonation changes of acid-sensing residues. We reasoned that in acidic pH (5.5), histidine 331 gets biprotonated and the domain I-III stapling His 331-Lys 16 H bond breaks as the first step in the acidic pH trigger for domain swiveling. Untwisting of the dI-III linker spring-twisted region may happen subsequent to loss of “stapling” interactions between domains I and III. A similar change in protonation status of the acid-sensing histidine residue His 323 has been proposed as a trigger for conformational change in E protein of tick-borne encephalitis virus at fusion-triggering acidic pH (27).
Contrary to our expectation, His 331 did not show any change in protonation when the prefusion structure was titrated to pH 5.5. (The pKa of His 331 at pH 7.4 and 5.5 is 4.89.) The only change noted for the His 331 side chain was that it changed to ε-tautomer (from δ-tautomer at pH 7.4). We then used this pH 5.5-titrated E1 structure for MD simulation to predict conformational changes in the protein. The root mean square fluctuation (RMSF) deviation per residue for the trajectory is shown in Fig. 2C. As expected, there was no major change seen (less than a 3-Å RMSF deviation) in the structure of domains I, II, and III, except for the fusion loop region of domain II (RMSF deviation of ∼6 Å). Another region that showed an RMSF change above 3 Å is the dI-III linker (Fig. 2C, “linker”). Cluster analysis of the conformations of MD trajectory revealed a major cluster at t = 23.37 ns (Fig. 2D, inset). Interestingly, the His 331-Lys 16 H bond did not change in the t = 23.37-ns structure. This implies that breaking of the His 331-Lys 16 H bond is not the first acid pH-triggered structural change.
We then compared the dI-III linker region of the t = 23.37-ns cluster central structure with its position in prefusion E1 structure. We noted a rotational movement of the dI-III linker compared to the prefusion structure (Fig. 2D). Though there was no major change in the spring-twisted region of the linker, the spring twist was slightly opened up compared to the prefusion conformation. This resulted in movement of the Arg 289 side chain slightly away from Asp 292. In this orientation, the Arg 289 cannot form a salt bridge with Asp 292. We also noted that when we aligned the major cluster center structures from MD simulation over the prefusion structure, domain I-II appeared slightly rotated over the dI-III linker (by nearly 20 Å). Changes in the φ, ψ, and ω angles between R289VVD292 residues of the linker region, between prefusion and the t = 23.37-ns cluster central structure (Fig. 2E), also indicated untwisting of the spring-twisted region in acidic pH.
It is possible that when E1 is in complex with E2, the acid pH-induced changes in the E1 dI-III linker and dI-III interface may be influenced by the E2-E1 interface. To test this possibility, we also performed an MD simulation on E2-E1 heterodimer structure at pH 5.5. The conformational changes that we observed in E1 (especially the dI-III linker and dI-III interface) are very similar to that seen with E1 run alone. The spring-twisted region showed “untwisting” of the region between Arg 289 and Asp 292 side chains (Fig. 2F). Interestingly, RMSF analysis of E1 structure showed several regions with more than a 3.0-Å deviation (data not shown). However, this is not surprising as fusion-triggering acidic pH is known to destabilize the E2-E1 interaction and lead to dissociation of the complex. We ascribe the RMSF deviation seen in E1 structure to acid pH-induced changes in the E2-E1 interface. Further supporting this idea, protonation of several acid-sensing residues in the E2-E1 structure was altered (Table 1). On the other hand, protonation of the His 331 side chain did not change after titration of the E2-E1 structure to pH 5.5 (in the t = 18.02-ns cluster centric structure of the MD trajectory). We did not see any change in protonation or pKa values of the Arg 289 and Asp 292 side chains.
TABLE 1.
Comparison of protonation states of residues of E2-E1 structure from PDB ID 3N42 (at pH 7.4) or from the t = 18.02-ns structure from E2-E1 constant-pH (5.5) MD simulation studies
| Residue | Protonationa
|
|
|---|---|---|
| E2-E1 structure from PDB ID 3N42 (pH 7.4) | E2-E1 cluster 1 central structure (pH 5.5) | |
| E2 | ||
| H26 | HID | HIP |
| E35 | GLU | GLH |
| H62 | HIE | HIP |
| H73 | HID | HIP |
| H123 | HIE | HIP |
| H130 | HID | HIP |
| H147 | HID | HIP |
| H170 | HIE | HID |
| H232 | HID | HIP |
| H256 | HID | HIP |
| H291 | HIE | HIP |
| H313 | HID | HIP |
| E50 | GLU | GLH |
| E1 | ||
| H107 | HIE | HIP |
| H125 | HID | HIP |
| H152 | HIE | HIP |
| D202 | ASP | ASH |
| H375 | HID | HIP |
| H381 | HID | HIP |
| H386 | HID | HIP |
HID, proton on δ-tautomer; HIE, proton on ε-tautomer; HIP, biprotonated; GLH and ASH, protonated forms.
Based on our MD analysis, we propose that untwisting of the spring-twisted region in the dI-III linker is the first step in acidic pH-induced conformational changes of alphavirus E1. This would initiate the swiveling motion of domain I-II to the center of the spike. Breaking of the His 331-Lys 16 H bond may be a later downstream step in the mechanism. However, our analysis cannot explain how acidic pH induces breaking of the Arg 289-Asp 292 salt bridge and untwisting of the spring-twisted region in the dI-III linker. The change in E2-E1 interaction and associated conformation changes in E1 domain II may allosterically trigger conformation changes in the dI-III linker at acidic pH.
Purified E1 soluble ectodomain is functionally active in membrane association and membrane fusion assays.
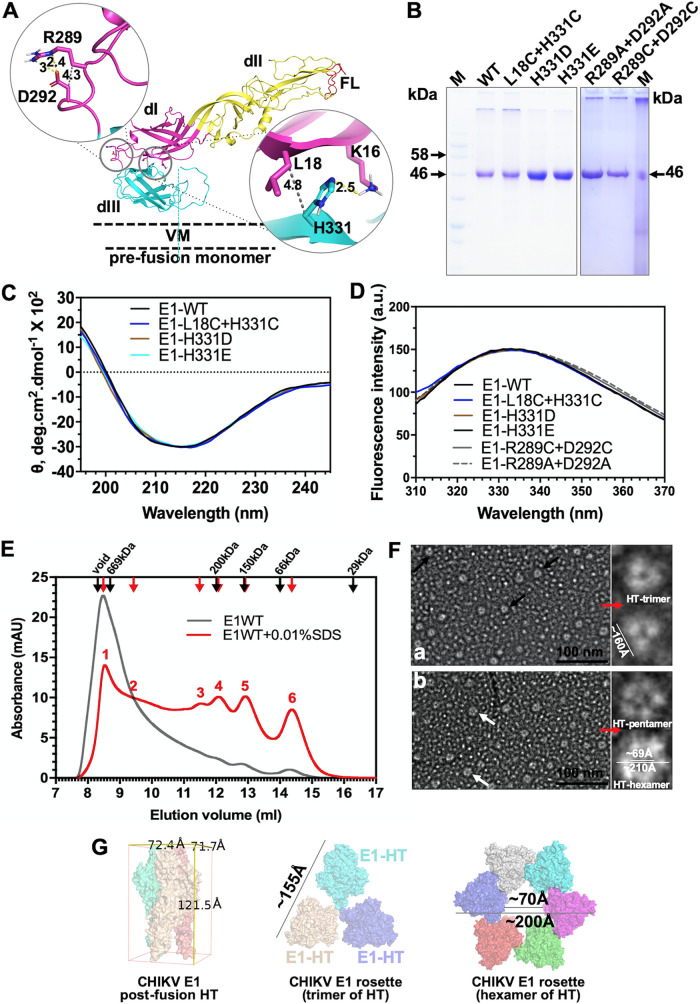
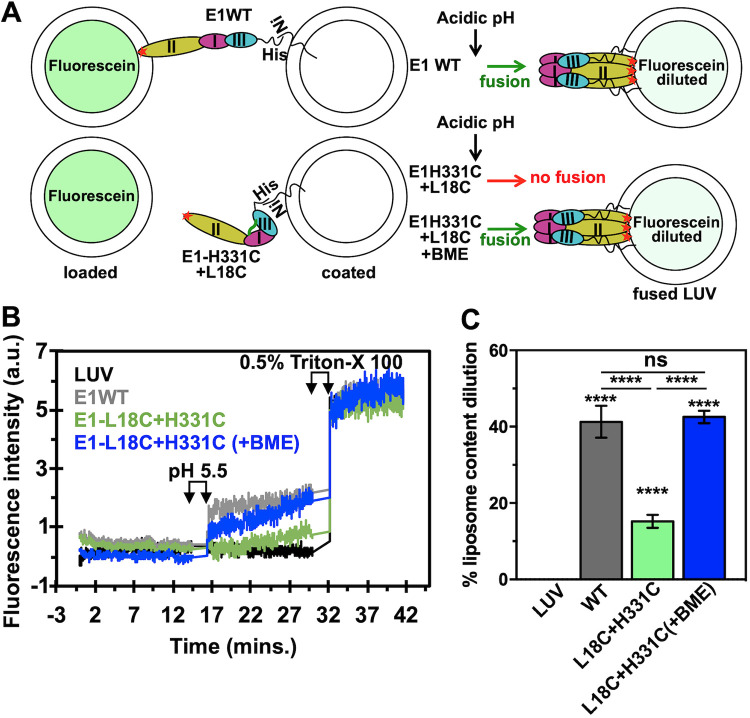
To validate our MD simulation predictions of E1 conformational changes at fusion-triggering acidic pH, we set out to purify soluble ectodomain of E1 protein and H331D/E mutants and test those in a liposome fusion assay. Our reasoning is that H331D/E mutant E1 should show a higher threshold of acidic pH (lower than pH 5.5), as the salt bridge between the introduced Asp (in place of H331) and Lys 16 should not be broken at pH 5.5. We also prepared an E1 double mutant protein in which His 331 and Leu 18 are changed to cysteines. The rationale of creating the double mutant is described in a later section. We introduced a pair of cysteines: one replacing His 331 and another in domain I, replacing Leu 18. We chose Leu 18 for introduction of cysteine in domain I, as the Cβ-Cβ distance between His 331 and Leu 18 is 4.8 Å (Fig. 3A). This Cβ-Cβ distance is taken as optimal for introducing a cysteine disulfide bond (28). We refer to this double-cysteine mutant as the “dI-III cystine staple” mutant from here onwards. Similarly, we introduced a cystine staple in the dI-III linker spring twist region, replacing the conserved Arg 289 and Asp 292 with cysteines (Fig. 3A). This mutant is referred to as the “spring twist region cystine staple.” In the cystine-stapled mutant scenario, acidic pH should be able to trigger membrane fusion between the liposomes, but only when the cystine bond is reduced by a reducing agent. In another mutant, we replaced the conserved Arg 289 and Asp 292 with alanine residues.
FIG 3.
Biochemical characterization of E1 soluble ectodomain protein and cystine staple mutants. (A) Locations of introduced cysteine residues to generate cystine staple mutants are mapped onto E1 structure (PDB ID 3N42). The left side shows the spring twist region cystine staple, and the right side shows the domain I-III cystine staple. E1 structure is shown in a cartoon representation, and the Cβ-Cβ distance is shown by a gray dotted line. Distance units are in angstroms. (B) SDS-PAGE image of the E1 WT and different mutants used in this study. “M” is the molecular weight marker. Arrows next to the marker lane mark the molecular weights of the respective bands in the gel image. (C) Far-UV CD spectra of the E1 WT and cystine staple and H331D/E mutant proteins. θ is the mean residue ellipticity. (D) Intrinsic tryptophan fluorescence spectra for E1 WT and mutant proteins. (E) Size exclusion chromatography of E. coli purified E1 protein. Elution profiles of E1 (gray line) and E1 incubated with 0.01% SDS and eluted with a buffer containing the detergent (red line) are plotted overlaid. Red arrows and numbers above the peaks (1 to 6) mark different elution peaks observed for E1 in the presence of the detergent. (F) Negative-stain single-particle electron microscopy of E1 protein. Sections of electron micrographs show possible top views of E1 trimer (black arrows in subpanel a) and pentamer or hexamer assemblies (white arrows in subpanel b). Single particles with “donut” shapes are selected and shown in enlarged insets to the right of both subpanels a and b. (G) Cryo-EM structure of homotrimers (PDB ID 3J2W) are arranged into either a trimer or hexamer assembly to compare size measurements from electron micrographs.
We expressed and purified the ectodomain of CHIKV E1 or mutant E1 proteins in Escherichia coli. Figure 3B shows an SDS-PAGE image of the purified E1 proteins. To ensure that the bacterially expressed protein is in its native conformation and functionally active, we did a thorough characterization of the purified E1 protein and its mutants. The far-UV circular dichroism (CD) spectrum of the purified E1 protein showed a minimum of mean residue ellipticity at 218 nm (Fig. 3C)—a characteristic feature for a predominantly β-sheet protein. Secondary structure content estimates from the far-UV CD spectrum showed ∼41% sheet structure for E1. (The estimate from crystal structure of PDB ID 3N42 is ∼45%.) The far-UV CD spectra of different mutant E1 proteins used in this study nearly overlapped that of the wild type (WT) (Fig. 3C). Intrinsic tryptophan fluorescence emission spectra for E1 (wild type and different mutants) overlapped (Fig. 3D) and showed an emission maximum around 335 nm, indicative of a properly folded tertiary structure. Size exclusion chromatography elution profiles of E1 in (Fig. 3E, gray line) revealed that purified E1 forms large oligomers (Fig. 3E, gray line, peak starting in the void and extending into resolution volume) that could not be resolved on a Superdex 200 column. However, in buffers containing 0.01% SDS (Fig. 3E, red line), several peaks corresponding to the trimer of trimers, tetramers, trimers and dimers could be seen. Similar size exclusion elution profiles were earlier reported for E1 protein extracted from the SFV viral surface, as well as other alphaviruses (10, 13, 29). In these studies, E1 was shown to form rosette-like structures of either pentamers or hexamers of E1 homotrimers, through interaction of exposed fusion loops or lateral interactions. In addition, these oligomers are resistant to mild detergent treatment. In the presence of detergents (e.g., 1.5% octyl-β-d-glucopyranoside [OG] or 0.01% SDS), the larger oligomer population is redistributed to smaller oligomers and the protein is functional (10, 13, 29). Further, we did negative-stain single-particle electron microscopy (Fig. 3F) of the E1 protein. As observed by others (13, 29), we noticed that the E1 homotrimer (or different assemblies of the homotrimers) preferentially “sits” on its fusion loop end, vertically orienting the assembly’s top surface view with the “head” from each homotrimer (Fig. 3F, “HT”) making the rim of the “donut” shape. We arranged the cryo-EM structure of homotrimers (PDB ID 3J2W) into either the trimer or hexamer assembly and did size measurements. Those sizes compare to measurements from our electron micrographs well. All of these observations corroborated that the protein is folded to its native conformation. We also confirmed that the introduced cysteines in the dI-III cystine staple and spring-twisted region cystine staple E1 mutant protein formed a disulfide bond, as expected. Levels of Ellman reagent reactivity, to estimate free sulfhydryl groups in the protein, were comparable between wild-type E1 and the cystine staple mutants. Levels of absorbance at 412 nm of the Ellman reagent (under identical experimental parameters) for the wild type (WT) and the cystine-staple mutants did not differ (data not shown).
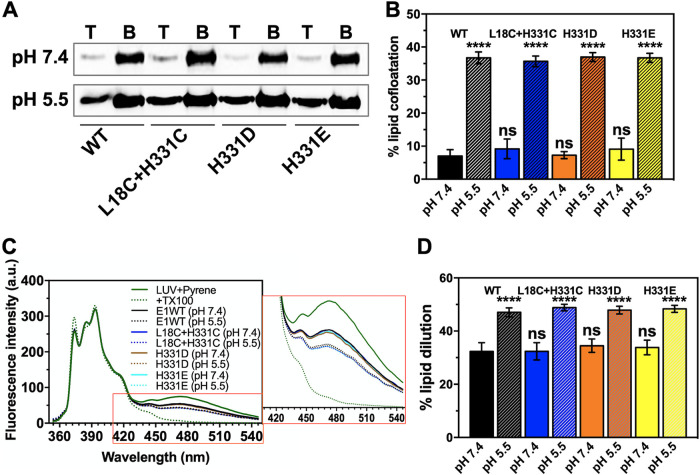
We then used the purified E1 and the mutants in a liposome coflotation assay. Earlier studies by others (10, 21, 29) have shown that E1 protein, either purified in insect cells or mammalian cells or reconstituted into lipid vesicles from viral membranes, associates with lipid membranes through exposed fusion loops in acidic pH. Nearly 4 to 8% of total E1 protein (both WT and mutants) floated along with large unilamellar vesicles (LUVs) at neutral pH (7.4) (Fig. 4A). However, when the protein and LUV mixture was incubated in a pH 5.5 buffer, membrane association increased to ∼32 to 37% (Fig. 4B). We made similar observations with the H331D, H331E, and L18C H331C mutants.
FIG 4.
Functional characterization of the E1 WT and cystine staple mutant in liposome coflotation and pyrene excimer fluorescence assays. (A) Image of Western blots for the E1 WT and dI-III cystine staple and H331D/E mutant proteins in the top (labeled “T”) and bottom (labeled “B”) fractions from the lipid flotation assay. (B) The fraction of E1 cofloating with LUVs (average of three experiments) is presented as a bar graph. (C) Pyrene emission spectrum in the presence of the E1 WT or cystine staple or H331D/E mutant proteins at pH 7.4 and 5.5. LUV plus pyrene (green solid line) acted as control (zero value of lipid dilution because of E1 FL insertion). The spectrum with pyrene-incorporated LUVs plus Triton X-100 (TX100) (green dotted line) was used to calculate the 100% value of lipid dilution. The E1 WT at pH 7.4 (solid black line) and E1 WT at pH 5.5 (dotted black line) are one comparison set, while spectra with the E1 dI-III cystine staple mutant (L18C+H331C) or H331D/E mutants at pH 7.4 (solid blue line) and 5.5 (dotted blue line) are the other comparison sets. The pyrene fluorescence emission spectrum was recorded in the 350- to 550-nm wavelength range. The excitation wavelength was set to 340 nm, and the excitation and emission slit widths were set to 5 nm. Each spectrum was an average of three readings and blank corrected. Emission spectra were normalized by setting constant the pyrene emission monomer peak at 397 nm. An inset shows an enlarged picture of the spectrum region between 420 and 540 nm. (D) The effect on lipid membrane due to E1 association as a percentage of dilution of lipids was estimated by measuring the drop in pyrene excimer peak values at ∼475 nm, in the presence of the WT or the mutant protein. Average values from three experiments are plotted in the bar graph along with error bars. One-way analysis of variance (ANOVA) was used to calculate statistical significance. ****, P ≤ 0.0001; ns, not significant.
We also characterized E1-membrane interaction at acidic pH using pyrene excimer fluorescence to record membrane destabilization activity of E1. The pyrene excimer fluorescence peak intensity decreased in the presence of E1 protein (Fig. 4C, inset). In addition, this effect was more pronounced when the protein was preexposed to fusion-triggering pH (5.5) (Fig. 4D). A similar result was seen when the dI-III cystine staple mutant or H331D/E mutant of E1 was used in the experiment (Fig. 4D). This suggests that LUV association of E1 is through insertion of its fusion loops into LUV membrane. Similar observations were made with other alphavirus fusion proteins and LUV association (30, 31).
We confirmed that the E1 and the cystine staple mutant proteins are not unfolded at the fusion-triggering pH by recording far-UV CD spectra in pH 5.5 buffer. The spectra at pH 5.5 almost overlapped with those recorded in pH 7.4 buffers (data not shown).
Taken together, the results from our structure analysis and membrane association studies establish that the CHIKV E1 protein and the mutant proteins that we expressed in E. coli are structurally and functionally similar to insect cell-expressed and viral surface-extracted E1.
Domain I and III separation and spring-twisted region unwinding are essential for fusion activity of E1.
Our MD simulations did not show any major change in the conformation surrounding His 331, at least within the time scale of the simulation study (50 ns). This suggests that at pH 5.5, the His 331-Lys 16 H bond in the dI-III interface is retained. The domain I-III separation in acidic pH might start at a place other than His 331-Lys 16 in the dI-III interface. In this scenario, replacing His 331 with another residue should not affect acidic pH triggering of E1 for fusion function. However, in the cystine staple mutant, fusion function should be seen only when the disulfide bond between domains I and III is broken by a reducing agent.
To test this idea, we used the wild type and cystine staple mutants of E1 in a liposome fusion assay. Our fusion assay is designed such that when E1 on the coated LUVs performs fusion with the fluorescein-loaded LUVs, fluorescein fluorescence is dequenched due to dilution effect (schematic in Fig. 5A). In the fusion assay, WT E1 showed 41% fusion activity (gray line in Fig. 5B and gray bar in Fig. 5C). We tested fusion activity at pH 5.0, 5.5, and 6.0. At pH 6.0, fusion activity was marginally less (but not significantly different) than that in pH 5.0 and 5.5 buffers (data not shown). When the domain I- and III-stapled (cystine staple mutant) E1 was used, only 15% fusion activity was seen (green line in Fig. 5B and green bar in Fig. 5C). In another assay with the dI-III cystine staple mutant (labeled “E1-L18C+H331C” in Fig. 5C), β-mercaptoethanol (BME) was added before acidification of the assay mixture. The fusion activity recorded in this case (blue line in Fig. 5B and blue bar in Fig. 5C) was comparable to that in the wild-type protein. We confirmed that loss of fusion activity in the cystine staple mutant (without BME) was not because of inability to insert its fusion loops into target membrane. The cystine staple E1 mutant associated with membrane vesicles and performed equal to the wild-type protein in membrane destabilization assay (Fig. 4). Based on these results, we conclude that domain I and III separation is a required conformational change step for fusion function of E1, and this event is triggered by acidic pH.
FIG 5.
Liposome fusion assay with E1 and the dI-III cystine staple mutant E1. (A) Schematic explaining the fusion assay used in this study. (Top panel) Fluorescein-loaded and E1-coated LUVs were mixed in pH 7.4 buffer, and fluorescence intensity at 520 nm (the emission maximum for fluorescein) was recorded as a time scan. The excitation wavelength was set to 490 nm. The excitation and emission slit widths were set to 5 nm. After the first 15 min of recording the fluorescence intensity, the calculated amount of 1 M sodium acetate (e.g., for a 1-ml assay mixture, 15 μl of the sodium acetate was added to bring the pH down to 5.5) was added to the assay mixture. After that, fluorescence intensity was recorded for another 15 min, before Triton X-100 was added to a concentration of 0.5% (vol/vol). After addition of Triton X-100, fluorescence intensity was recorded for another 10 min. In the bottom panel, the assay was performed exactly the same as above, except that β-mercaptoethanol either was added before the acidification step or not. (B) Fluorescence time scan in the presence of LUVs alone (black), E1 WT (gray), or the E1 dI-III cystine staple mutant (E1-L18C+H331C) (green) and with β-mercaptoethanol (+BME) added (blue). (C) The percentage of liposome content dilution was estimated as described in Materials and Methods. Average values from three experiments are plotted in the bar graph along with error bars. One-way ANOVA was used to calculate statistical significance. ****, P ≤ 0.0001; ns, not significant.
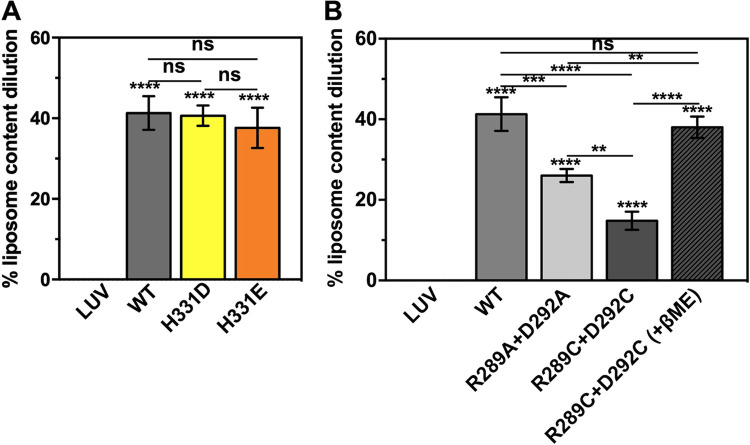
The fusion activity of H331D/E E1 was comparable to that of the wild type (Fig. 6A). If His 331 were to be an acid-sensing residue, replacing it with an acidic residue (D or E) should have increased the acidic pH threshold for fusion triggering—i.e., would have required a more acidic pH (lower than 5.5) for fusion function. This implies that acid sensing by the conserved His 331 may not play a role in domain I-III separation.
FIG 6.
Fusion activity of E1 H331D/E, spring twist region cystine staple, and double-alanine mutants. The percentage of liposome content dilution was estimated as described in Materials and Methods. Average values from three experiments are plotted in the bar graph along with error bars. (A) Fusion activity comparison between the E1 wild type and H331D or H331E mutant. (B) Fusion activity comparison between the E1 wild type and spring-twisted region cystine staple mutant. One-way ANOVA was used to calculate statistical significance. **, ***, and **** represent P ≤ 0.01, P ≤ 0.001, and P ≤ 0.0001, respectively. ns, not significant.
We also tested the fusion activity of the E1 mutants where the spring-twisted region of the dI-III linker is stapled by a cystine bond. There was significantly less fusion activity of the mutant protein compared to the wild type (Fig. 6B). When the cystine bond was reduced, the fusion function was restored (Fig. 6B). This observation corroborates our MD simulation finding that untwisting of the spring-twisted region in the dI-III linker is necessary step in acid pH-induced domain rearrangements of E1 for membrane fusion function. We also replaced the conserved salt-bridging Arg 289-Asp 292 pair with alanine residues. In membrane fusion assays, the R289A D292A mutant (Fig. 6B), surprisingly, showed significantly less activity than the wild-type E1. We expected the mutant to have fusion activity comparable to that of the wild-type protein as the spring-twist stabilizing salt bridge would not be there, and the domains are free to swivel upon acidic pH trigger. It is unclear from our studies why the mutant lost some of its activity.
DISCUSSION
Viral fusion protein undergoes a pre- to postfusion conformation change as it performs viral and cell membrane fusion. An extended conformation intermediate is formed during the conformation change. In alphaviruses and other viruses that take the endocytic pathway for cell entry, the conformation change is triggered by acidic pH of the maturing endosome. In the pre- to postfusion conformation change, two major changes are seen. In the first, E1 moves from the periphery to the center of the E1-E2 heterodimer trimeric spike and forms a homotrimer. In the second, interdomain interactions in the interface of dI and III are broken and new interactions are formed postfusion. How these conformational changes are brought about in E1, after exposure to fusion-triggering acidic pH, is addressed in this study.
A swiveling motion of domains for E1 in the pre- to postfusion conformation change was proposed earlier (7, 14, 25, 26). The only region of E1 that is relatively flexible, and thus can act as a hinge for swiveling of domains, is the linker loop connecting domains I and III. Our analysis showed that a highly conserved R-(X)2–3-D/E stretch in the linker region is twisted like a spring, stabilized by a salt bridge between R and D/E of the sequence. We reasoned that untwisting of the spring twist region in the dI-III linker would swivel the dI-II, like a hinge, on dIII. Findings from our MD simulation studies on CHIKV E1 structure at pH 5.5 corroborated our hypothesis. Corroborating our observation, Zheng et al. (20) proposed the idea that Arg 289 in the dI-III linker is a key residue for homotrimer formation of SFV E1.
Parallel to the untwisting of the spring-twisted region, the domain I-III stapling interactions should also break to allow the swiveling motion of the domains. Several charge-charge interactions between domains I and III hold both the domains in the hairpin conformation. In tick-borne encephalitis (TBE) virus E, a conserved histidine (structurally in the same context as His 331 of CHIKV) acts as an acid pH sensor and shows altered interaction between prefusion and postfusion structures (27). We expected that His 331 in CHIKV (17) E1 would play a similar role in pre- to postfusion conformation switch. Protonation changes in acidic pH on acid-sensing histidine residues have been proposed as a trigger for pre- to postfusion conformational changes for several other viral fusion proteins (32). We expected that the His 331 protonation would change when E1 structure was titrated to pH 5.5, and the H331-K16 H bond would break. Contrary to our expectation, our MD simulation study hinted that His 331 protonation would not change at acidic pH, at least in the time scale of our MD simulation. In a similar MD simulation study on CHIKV E1, Zeng et al. (17) did estimate change in pKa of His 331 from a monomeric (prefusion) state to a fusion intermediate then postfusion conformation. They also noted that in the prefusion state, His 331 has a pKa of 4.6—a value very close to the pKa that we estimated for His 331 in the prefusion and acid pH titrated state. However, in the postfusion conformation, the pKa of His 331 increased to 8.6. This implied that the change in the His 331 environment happens during an extended intermediate to postfusion conformation change.
Results of our fusion functional assays using purified E1 protein, H331D/E mutants, and cystine staple mutants validated our MD simulation predictions for His 331. Our observations with cystine staple mutant also prove that domain I-III dissociation is an essential step in the acid pH-induced conformational changes required for membrane fusion.
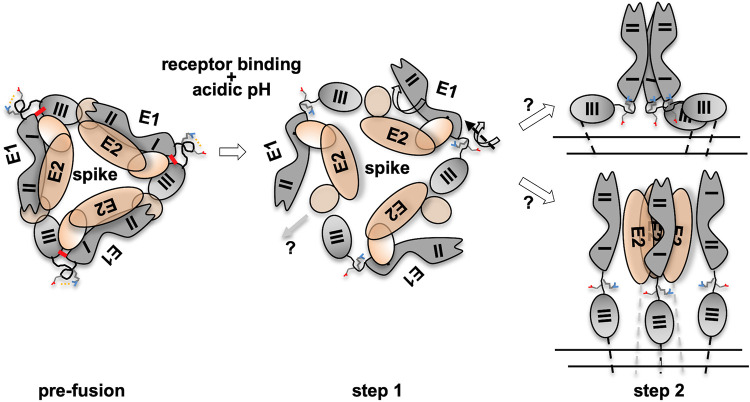
Based on interpretation of our results from this study, in the context of the universal mechanism proposed for viral fusion proteins, we present a mechanistic model to explain acidic pH-triggered conformational changes in alphavirus membrane fusion protein E1 (Fig. 7). E2-E1 dissociation is initiated after receptor binding onto E2 (22). The first conformational change that is triggered in E1 (after the endosome gets acidified to pH 5.5) is in the dI-III linker. The salt bridge is between the conserved R-(X)2–3-D/E pair in the linker region. However, it is to be noted that the salt bridge breaking is probably not because of protonation changes of Arg or Asp side chain, as at pH 5.5 protonation of either of these residues is not different. Breaking of the stapling salt bridge results in untwisting of the spring twist structure. Because of the untwisting, the dI-II region swivels outwards from the trimeric spike. Thus, the spring-twisted structure is the hinge point for the domain swiveling motion.
FIG 7.
Mechanism of acid pH-induced conformation change in E1 during viral entry. Alphavirus fusion protein E1 undergoes a pre- to postfusion conformational change, through an extended intermediate, during the membrane fusion step of viral entry. Three E2-E1 heterodimers curl around a central vertical axis to form a spike structure on the virion surface (prefusion). Step 1 involves E2-E1 dissociation and E1 domain swiveling. Receptor binding and acidic pH trigger this step. Acidic pH breaks a conserved Arg-Asp/Glu salt bridge in the dI-III linker of E1 and triggers untwisting of a spring-twisted region of the linker. This brings about the swiveling motion of the dI-II region over dIII. As a result, E1 moves away from the spike axis. The dI-III stapling interactions also break in this step. E2 may escape from the spike center at this stage. In step 2, possibility 1 (top) shows extended intermediate and E1 homotrimer formation. The swiveling-in motion of dI-II regions may bring these regions from three protomers to form the central core of the postfusion homotrimer. E1 domains may extend linearly following this. In the alternate possibility shown below, the swiveling motion will also extend the dI-III linker, to form three monomeric extended intermediate E1 protomers first. Trimerization through the dI-II region may follow at a later step. E2 may escape from the center at that stage. In prefusion and step 1, the top view of the spike is shown in the schematic. In step 2, the side view of the spike schematic is shown for clarity. E2 is shown with brown ellipses and E1 in gray shapes. The dI-III linker is shown in a hand-drawn curved shape. The spring-twisted region is depicted in a spring circled shape. The salt bridge between the conserved Arg-Asp/Glu pair in the dI-III linker is shown by yellow dotted lines. In the prefusion structure, dI-III stapling charge interactions are shown by a thick red line. In the step 2 schematic, double parallel lines below the spike shape represent viral membrane.
In parallel, acidic pH also triggers domain I-III separation. A structurally conserved histidine residue in flavivirus E protein is proposed as one of the key acid-sensing residues in flaviviruses (27). Mutation of the conserved domain III histidine (His 323 in TBE virus E) to alanine resulted in loss of fusion function. However, our observations with E1 H331D/E are in line with those of studies by Qin et al. (33) where an H331A mutant of SFV is not defective in growth properties compared to wild-type E1-containing virus. It is possible that, despite the structural similarity, alphavirus fusion protein E1 has a different mechanism of acid pH-induced pre- to postfusion conformation switch from that of flavivirus E protein. Interestingly, mutation of another highly conserved acid-sensing residue in domain I, His 3, to alanine resulted in reduced viral infection and change in the pH threshold of fusion in SFV (33). Zeng et al. (17) also suggested His 3 (along with His 331) as an acid-sensing residue in CHIKV E1. It would be interesting to see how His 3 interactions change at acidic pH and how that would trigger dI-III dissociation.
What follows after the domain swiveling motion is triggered on the spring twist region and dI-III interactions are broken? There are two possible scenarios. (i) By swiveling in, the domain I-II regions of the three E1 protomers in the trimeric spike come to the center of the spike, establish interprotomer interactions, and form the E1 homotrimer central core. (ii) Alternatively, the dI-III linker region becomes linear (as the dI-III hairpin-stapling interactions are broken), extending all three domains onto a linear axis. Homotrimerization may follow this step. Earlier studies proposed both possibilities. Sánchez-San Martin et al. (21) showed that when only the dI-II region of SFV E1 is expressed, replacing dIII and the dI-III linker with a short flexible region, acidic pH triggered formation of the dI-II region trimers. This implies that homotrimerization precedes extended intermediate formation (top in step 2 in Fig. 7). In contrast to this, Cao et al. (18) proposed a mechanism by which E1 forms an extended intermediate as a monomer, implying that homotrimerization is a later event in membrane fusion (bottom in step 2 in Fig. 7). Taken together, the findings of our study give a mechanistic insight into acid pH-triggered conformational changes in alphavirus E1 during membrane fusion.
MATERIALS AND METHODS
Sequence and structure analysis.
We used sequence corresponding to the dI-III linker region (residues 283 to 294) of CHIKV E1 (NCBI accession ID ABD95938.1) to align with the corresponding sequence region from other alphavirus E1 sequences. Altogether, 16 sequences were compared in a multiple-sequence alignment (MSA). We used the Clustal Omega program with default parameters for MSA analysis.
The crystal structure of the CHIKV E1-E2-E3 soluble ectodomain complex (PDB ID 3N42) (7) was used for analysis of the prefusion conformation of E1. The postfusion homotrimer structure of CHIKV E1 was modeled using the CHIKV S27 isolate E1 sequence (NCBI accession ID NP_690589.2) as the input and the SFV postfusion E1 structure (PDB ID 1RER) as the template. We used the SWISS-MODEL web server for homology modeling. We did Propka analysis of E1 protein in the prefusion (PDB ID 3N42) and postfusion homotrimer conformations using the PDB2PQR server available at http://nbcr-222.ucsd.edu/pdb2pqr_2.0.0/ with default parameters. PyMol was used for analysis of domain I and III interactions, in prefusion and postfusion conformations involving conserved His 331, and for preparation of structure figures.
Molecular dynamics simulations.
We performed constant-pH molecular dynamics (MD) simulation on E2-E1 heterodimer structure (PDB ID 3N42) or E1 structure (extracted from PDB ID 3N42) at pH 5.5. We used AMBER99SB force field in GROMACS 5.1.4 package. Input structure files were prepared for MD simulation by removing water molecules and HET atoms first. The “cleaned” pdb file was then used as an input at Playmolecule server available at https://playmolecule.org/ for assignment of protonation states at pH 5.5. In Gromacs, we used explicit SPC water molecules for simulation. In short, we used periodic boundary conditions to add solute molecules around the water molecules within a cubic box. Further counter ions were incorporated for system neutralization. Energy minimization was performed using the steepest-descent algorithm, and bonds were constrained using the LINCs algorithm. We performed independent coupling of solute and solvents to a modified Berendson bath at 300 K by using a solvent density of 1 atm at 300 K and a coupling time of 0.1 ps. The MD run integration time step was set to 2-fs, and 10-ps intervals were used for generation of output coordinates. We equilibrated the system twice prior to the final MD run, once each for 100 ps under NVT (constant number of atoms, volume, and temperature) and 100 ps under NPT (constant number of atoms, pressure and temperature). Production MD runs were performed at 300 K for 50 ns each.
Cloning, expression, and purification of CHIKV E1 wild-type and mutant proteins.
We PCR amplified the CHIKV E1 ectodomain region of the CHIKV LR2006_OPY1 strain. CHIKV E1 and E2 coding DNA was a kind donation from Scott C. Weaver, UTMB, Galveston, TX. The PCR amplicon of the E1 ectodomain was cloned into the pET24b E. coli expression vector. Different point mutations in E1 used in this study were created using E1-pET24b as the template by the overlapping primer extension method. (The primer sequences and description are available on request.) We sequence verified all of the clones. All of the proteins were expressed along with a C-terminal 6-histidine tag.
Overexpression of CHIKV E1 WT and mutant proteins was optimized in the BL21(DE3) strain of E. coli. Overexpressed protein formed insoluble inclusion bodies in bacteria. Protein purification was done by first solubilizing the protein in 6 M urea, followed by 6-histidine tag Ni-nitrilotriacetic acid (NTA) affinity chromatography. After purification, denaturant was removed and protein exchanged into 20 mM Tris-Cl (pH 7.4) buffer containing 150 mM NaCl and 1 mM EDTA (TNE buffer) by ultrafiltration. The concentration of the protein was estimated by taking the absorbance at 280 nm and using an estimated molar extinction coefficient of 52,800 M−1 cm−1.
Far-UV circular dichroism and intrinsic tryptophan fluorescence spectroscopy.
We used purified protein at a 0.2-mg/ml concentration for recording the far-UV circular dichroism (CD) spectrum (195 to 250 nm) and intrinsic tryptophan fluorescence measurements (excitation wavelength of 295 nm, emission wavelength range of 310 to 400 nm). We used 0.1- and 1-cm path-length quartz cuvettes for CD and fluorescence spectrum analysis, respectively. Noise removal and blank correction were applied to each spectrum. We used the K2D3 software available at http://cbdm-01.zdv.uni-mainz.de/~andrade/k2d3/ for estimation of secondary structure content from far-UV CD data.
Negative-stain single-particle electron microscopy of E1 protein.
E1 protein (100-μg/ml concentration) was adsorbed onto a 200-mesh carbon-coated Formvar copper grid. Staining was done with UranyLess stain (from Electron Microscopy Sciences, USA). Electron micrographs were collected on a Joel F200 transmission electron microscope with a 90-keV beam acceleration voltage. Single-particle images (cone/donut shapes) were picked using EMAN2 software, and size measurements were done. Electron microscopy images in the published literature (13, 29) on E1 showing similar donut shapes served as the guiding rule for identification and picking of the particles.
Size exclusion chromatography of E. coli purified CHIKV E1 protein.
A small aliquot (approximately 200 μg) of the purified E1 protein was loaded onto a Superdex 200 10/300GL analytical gel filtration column. Elution was done at a 0.5-ml/min flow rate. High-molecular-weight gel filtration standards thyroglobulin (660 kDa), alcohol dehydrogenase (200 kDa), β-amylase (150 kDa), bovine serum albumin (BSA [66 kDa]), and carbonic anhydrase (29 kDa) were used to generate a calibration curve (Kav versus log10 molecular weight plot). Blue dextran elution volume was taken as the void volume of the column.
Lipid coflotation assay.
We used large unilamellar vesicles (LUVs) prepared with purified synthetic lipids as a membrane model. LUVs were prepared by mixing 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC)–1-hexadecanoyl-2-(9Z-octadecenoyl)-sn-glycero-3-phosphoethanolamine (POPE)–palmitoyl-oleyol phosphatidic acid (POPA)–cholesterol (Avanti Polar, Ltd.) in 1:1:0.3:2 molar ratios. LUVs were prepared by a size extrusion method, using standard protocols. In each assay, LUVs (at a 0.5 mM final concentration) and 100 μg protein mixture were layered over a 40% sucrose cushion of 4 ml in an ultracentrifuge tube. Following this, 4 ml each of 20% sucrose and 5% sucrose was layered as the middle and top layers, respectively. For studying coflotation of E1 or mutants at pH 5.5, the assay was done in the same way, except that the sucrose solutions were prepared in buffer at pH 5.5. After ultracentrifugation for 3 h at 200,000 × g, fractions from the top (4 ml) and bottom (4 ml) were collected. Aliquots from the top and bottom fractions in each assay were analyzed by Western blotting using His-tagged monoclonal antibodies. The percentage of E1 floating along with liposomes was calculated by quantifying band intensities of proteins in the top fractions over band intensities of the total amount of protein (top plus bottom fractions) using ImageJ software.
Pyrene excimer fluorescence assay for studying E1-membrane association.
LUVs were prepared as described above, and then an aliquot of the liposomes was incubated with 40 μM pyrene (purchased from Sigma-Aldrich, USA). The labeled LUVs were separated from free dyes using a PD10 desalting column (purchased from GE Healthcare, USA). Pyrene partitions into LUV membrane. Pyrene-labeled LUVs and E1 protein (at a 0.2-mg/ml concentration) in TNE buffer at pH 7.4 or 5.5 (with sodium acetate at pH 4.6 added to bring the pH down to 5.5) were mixed. The assay was performed adapting the method described by Liao et al. (30). The percentage of lipid dilution calculations were performed by considering the difference between the pyrene excimer peak values of pyrene-labeled LUVs in the absence of any protein (taken as 0%) or in the absence of any protein plus 0.5% Triton X-100 (TX-100 [taken as 100%]). The effects of E1 proteins on percentage of lipid dilutions were calculated over this range. The experiment was repeated three times. The average value from three experiments is presented in the bar graphs, along with error bars.
Liposome content mixing fusion assay.
Liposome content mixing fusion assays were done adapting the protocol described by Gui and Lee (34). Briefly, a batch of LUVs was loaded with quenching concentrations (100 mM) of carboxyfluorescein (purchased from Thermo Scientific, USA). Another batch of LUVs was prepared in the same way as described above, except for inclusion of 1,2-dioleoyl-sn-glycero-3-[(N-(5-amino-1-carboxypentyl)iminodiacetic acid)succinyl] (nickel salt) [DGS-NTA-(Ni)] into the liposome at a 0.4 molar ratio. We coated DGS-NTA(Ni)-incorporated LUVs with E1 through protein 6× His tag binding to Ni-NTA. We refer to these LUVs as E1-coated LUVs.
To assess the fusion function of the E1 WT or mutant at pH 5.5, we mixed fluorescein-loaded LUVs with E1-coated LUVs. We did a fusion assay with the dI-III cystine staple mutant and dI-III linker region cystine staple mutant E1 in exactly the same manner described above, except that β-mercaptoethanol was added (to a final concentration of 5 mM), before acidification of the assay mixture with sodium acetate.
Upon acidification of the assay mixture, fluorescein fluorescence intensity was further quenched, and it showed a negative value compared to blank values. Hence, we normalized the fluorescence intensity value at the start of the time scan after acidification to the value at the end of the neutral-pH time scan. Each assay was repeated three times. An overlay of the time scan of fluorescein fluorescence intensity with E1 WT and dI-III cystine staple mutants is shown for comparison.
To quantify the fusion activity, the average of the last five data points from the only-LUV sample toward the end of the pH 5.5 reading was considered the baseline of fusion (considered 0% liposome content dilution). The average of the last five data points toward the end of the 0.5% TX-100 reading (post-acidic pH treatment) of the same sample (only-LUV sample) was considered the endpoint of the fusion assay (taken as 100%). Then fusion activity was calculated in the presence of E1 WT and mutant protein in a similar manner. Fusion activity is presented as the percentage (of liposome content dilution) of the control (only LUV) fusion activity (between 0 and 100%). The average percentage of liposome content dilution of three experiments is presented in a bar graph, along with error bars.
ACKNOWLEDGMENTS
T.K.C. is supported by an institutional intramural research grant, and N.K.G. is supported by a Ph.D. research fellowship from the Department of Atomic Energy, Government of India. B.S. is supported by a Ph.D. research fellowship from the Council of Scientific and Industrial Research, Government of India.
The authors acknowledge help from Satyamurthy Kundarapu with initial optimization of protein purification. We thank Amita Rani Sahoo, Case Western Reserve University, for help with the MD simulation runs.
T.K.C. and B.S. conceptualized and planned the work. B.S. and N.K.G. performed the experiments. T.K.C. and B.S. wrote the article.
The authors declare no conflict of interest.
REFERENCES
- 1.Weaver SC, Lecuit M. 2015. Chikungunya virus and the global spread of a mosquito-borne disease. N Engl J Med 372:1231–1239. doi: 10.1056/NEJMra1406035. [DOI] [PubMed] [Google Scholar]
- 2.Morrison TE. 2014. Reemergence of chikungunya virus. J Virol 88:11644–11647. doi: 10.1128/JVI.01432-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schwartz O, Albert ML. 2010. Biology and pathogenesis of chikungunya virus. Nat Rev Microbiol 8:491–500. doi: 10.1038/nrmicro2368. [DOI] [PubMed] [Google Scholar]
- 4.Bernard E, Solignat M, Gay B, Chazal N, Higgs S, Devaux C, Briant L. 2010. Endocytosis of chikungunya virus into mammalian cells: role of clathrin and early endosomal compartments. PLoS One 5:e11479. doi: 10.1371/journal.pone.0011479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kielian M, Chanel-Vos C, Liao M. 2010. Alphavirus entry and membrane fusion. Viruses 2:796–825. doi: 10.3390/v2040796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vaney M-C, Duquerroy S, Rey FA. 2013. Alphavirus structure: activation for entry at the target cell surface. Curr Opin Virol 3:151–158. doi: 10.1016/j.coviro.2013.04.003. [DOI] [PubMed] [Google Scholar]
- 7.Voss JE, Vaney M-C, Duquerroy S, Vonrhein C, Girard-Blanc C, Crublet E, Thompson A, Bricogne G, Rey FA. 2010. Glycoprotein organization of Chikungunya virus particles revealed by X-ray crystallography. Nature 468:709–712. doi: 10.1038/nature09555. [DOI] [PubMed] [Google Scholar]
- 8.Sun S, Xiang Y, Akahata W, Holdaway H, Pal P, Zhang X, Diamond MS, Nabel GJ, Rossmann MG. 2013. Structural analyses at pseudo atomic resolution of Chikungunya virus and antibodies show mechanisms of neutralization. eLife 2:e00435. doi: 10.7554/eLife.00435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yap ML, Klose T, Urakami A, Hasan SS, Akahata W, Rossmann MG. 2017. Structural studies of Chikungunya virus maturation. Proc Natl Acad Sci U S A 114:13703–13707. doi: 10.1073/pnas.1713166114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kielian M, Helenius A. 1985. pH-induced alterations in the fusogenic spike protein of Semliki Forest virus. J Cell Biol 101:2284–2291. doi: 10.1083/jcb.101.6.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kielian M, Rey FA. 2006. Virus membrane-fusion proteins: more than one way to make a hairpin. Nat Rev Microbiol 4:67–76. doi: 10.1038/nrmicro1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harrison SC. 2008. Viral membrane fusion. Nat Struct Mol Biol 15:690–698. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gibbons DL, Vaney M-C, Roussel A, Vigouroux A, Reilly B, Lepault J, Kielian M, Rey FA. 2004. Conformational change and protein-protein interactions of the fusion protein of Semliki Forest virus. Nature 427:320–325. doi: 10.1038/nature02239. [DOI] [PubMed] [Google Scholar]
- 14.Li L, Jose J, Xiang Y, Kuhn RJ, Rossmann MG. 2010. Structural changes of envelope proteins during alphavirus fusion. Nature 468:705–708. doi: 10.1038/nature09546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Duijl-Richter MKS, Blijleven JS, van Oijen AM, Smit JM. 2015. Chikungunya virus fusion properties elucidated by single-particle and bulk approaches. J Gen Virol 96:2122–2132. doi: 10.1099/vir.0.000144. [DOI] [PubMed] [Google Scholar]
- 16.Mueller DS, Kampmann T, Yennamalli R, Young PR, Kobe B, Mark AE. 2008. Histidine protonation and the activation of viral fusion proteins. Biochem Soc Trans 36:43–45. doi: 10.1042/BST0360043. [DOI] [PubMed] [Google Scholar]
- 17.Zeng X, Mukhopadhyay S, Brooks CL III. 2015. Residue-level resolution of alphavirus envelope protein interactions in pH-dependent fusion. Proc Natl Acad Sci U S A 112:2034–2039. doi: 10.1073/pnas.1414190112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cao S, Zhang W. 2013. Characterization of an early-stage fusion intermediate of Sindbis virus using cryoelectron microscopy. Proc Natl Acad Sci U S A 110:13362–13367. doi: 10.1073/pnas.1301911110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roman-Sosa G, Kielian M. 2011. The interaction of alphavirus E1 protein with exogenous domain III defines stages in virus-membrane fusion. J Virol 85:12271–12279. doi: 10.1128/JVI.05902-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng Y, Sanchez-San Martin C, Qin Z, Kielian M. 2011. The domain I-domain III linker plays an important role in the fusogenic conformational change of the alphavirus membrane fusion protein. J Virol 85:6334–6342. doi: 10.1128/JVI.00596-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sánchez-San Martín C, Sosa H, Kielian M. 2008. A stable prefusion intermediate of the alphavirus fusion protein reveals critical features of class II membrane fusion. Cell Host Microbe 4:600–608. doi: 10.1016/j.chom.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sahoo B, Chowdary TK. 2019. Conformational changes in Chikungunya virus E2 protein upon heparan sulfate receptor binding explain mechanism of E2-E1 dissociation during viral entry. Biosci Rep 39:BSR20191077. doi: 10.1042/BSR20191077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fields W, Kielian M. 2013. A key interaction between the alphavirus envelope proteins responsible for initial dimer dissociation during fusion. J Virol 87:3774–3781. doi: 10.1128/JVI.03310-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lescar J, Roussel A, Wien MW, Navaza J, Fuller SD, Wengler G, Wengler G, Rey FA. 2001. The fusion glycoprotein shell of Semliki Forest virus: an icosahedral assembly primed for fusogenic activation at endosomal pH. Cell 105:137–148. doi: 10.1016/s0092-8674(01)00303-8. [DOI] [PubMed] [Google Scholar]
- 25.Fuller SD, Berriman JA, Butcher SJ, Gowen BE. 1995. Low pH induces swiveling of the glycoprotein heterodimers in the Semliki Forest virus spike complex. Cell 81:715–725. doi: 10.1016/0092-8674(95)90533-2. [DOI] [PubMed] [Google Scholar]
- 26.Mukhopadhyay S, Zhang W, Gabler S, Chipman PR, Strauss EG, Strauss JH, Baker TS, Kuhn RJ, Rossmann MG. 2006. Mapping the structure and function of the E1 and E2 glycoproteins in alphaviruses. Structure 14:63–73. doi: 10.1016/j.str.2005.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fritz R, Stiasny K, Heinz FX. 2008. Identification of specific histidines as pH sensors in flavivirus membrane fusion. J Cell Biol 183:353–361. doi: 10.1083/jcb.200806081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dombkowski AA, Sultana KZ, Craig DB. 2014. Protein disulfide engineering. FEBS Lett 588:206–212. doi: 10.1016/j.febslet.2013.11.024. [DOI] [PubMed] [Google Scholar]
- 29.Gibbons DL, Erk I, Reilly B, Navaza J, Kielian M, Rey FA, Lepault J. 2003. Visualization of the target-membrane-inserted fusion protein of Semliki Forest virus by combined electron microscopy and crystallography. Cell 114:573–583. doi: 10.1016/s0092-8674(03)00683-4. [DOI] [PubMed] [Google Scholar]
- 30.Liao M, Kielian M. 2005. Domain III from class II fusion proteins functions as a dominant-negative inhibitor of virus membrane fusion. J Cell Biol 171:111–120. doi: 10.1083/jcb.200507075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smit JM, Bittman R, Wilschut J. 1999. Low-pH-dependent fusion of Sindbis virus with receptor-free cholesterol- and sphingolipid-containing liposomes. J Virol 73:8476–8484. doi: 10.1128/JVI.73.10.8476-8484.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kampmann T, Mueller DS, Mark AE, Young PR, Kobe B. 2006. The role of histidine residues in low-pH-mediated viral membrane fusion. Structure 14:1481–1487. doi: 10.1016/j.str.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 33.Qin Z-L, Zheng Y, Kielian M. 2009. Role of conserved histidine residues in the low-pH dependence of the Semliki Forest virus fusion protein. J Virol 83:4670–4677. doi: 10.1128/JVI.02646-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gui L, Lee KK. 2018. Influenza virus-liposome fusion studies using fluorescence dequenching and cryo-electron tomography. Methods Mol Biol 1836:261–279. doi: 10.1007/978-1-4939-8678-1_13. [DOI] [PMC free article] [PubMed] [Google Scholar]