Abstract
SARS-CoV-2-induced hypercytokinemia and inflammation are critically associated with COVID-19 severity. Baricitinib, a clinically approved JAK1/JAK2 inhibitor, is currently being investigated in COVID-19 clinical trials. Here, we investigated the immunologic and virologic efficacy of baricitinib in a rhesus macaque model of SARS-CoV-2 infection. Viral shedding measured from nasal and throat swabs, bronchoalveolar lavages, and tissues was not reduced with baricitinib. Type I interferon (IFN) antiviral responses and SARS-CoV-2-specific T cell responses remained similar between the two groups. Animals treated with baricitinib showed reduced inflammation, decreased lung infiltration of inflammatory cells, reduced NETosis activity, and more limited lung pathology. Importantly, baricitinib-treated animals had a rapid and remarkably potent suppression of lung macrophage production of cytokines and chemokines responsible for inflammation and neutrophil recruitment. These data support a beneficial role for, and elucidate the immunological mechanisms underlying, the use of baricitinib as a frontline treatment for inflammation induced by SARS-CoV-2 infection.
Keywords: COVID-19, SARS-CoV-2, inflammation, immune activation, baricitinib, nonhuman primate, immunology, pathogenesis
Graphical Abstract

Highlights
-
•
SARS-CoV-2-infected RMs mimic signatures of inflammation seen in COVID-19 patients
-
•
Baricitinib suppresses production of pro-inflammatory cytokines in lung macrophages
-
•
Baricitinib limits recruitment of neutrophils to the lung and NETosis
-
•
Baricitinib preserves innate antiviral and SARS-CoV-2-specific T cell responses
Using a rhesus macaque infection model, it is shown that baricitinib treatment started early after infection effectively resolves inflammatory signatures in airway macrophages, with decreased lung pathology and neutrophil infiltration.
Introduction
The rapid emergence and dissemination of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and the subsequent coronavirus disease 2019 (COVID-19) pandemic has placed an excessive burden on public and private healthcare systems with over 1,400,000 deaths worldwide. Thus, therapeutic approaches aimed at mitigating disease severity are of utmost global priority (https://www.who.int/). SARS-CoV-2 infection results in a wide spectrum of disease severity, ranging from asymptomatic individuals to critically ill patients leading to death. Severe COVID-19 presents with high-grade fever, dry cough, pneumonia, inflammation of the lungs, and infiltration of immune cells. It has been noted that individuals with co-morbidities and compromised immune systems are at higher risk for severe clinical manifestations (Guan et al., 2020).
Immunological features of COVID-19 progression includes a robust pro-inflammatory response driven by innate and adaptive immune cells, with severe cases of COVID-19 having elevated serum levels of pro-inflammatory cytokines and chemokines including: interferon (IFN)-γ, tumor necrosis factor (TNF)-α, IP-10, (granulocyte colony stimulating factor (G-CSF), interleukin-2 (IL-2), IL-6, IL-8, IL-9, IL-10, and IL-17. Therefore, the use of therapeutics targeted at Janus kinases (JAKs) have the potential to ameliorate disease severity by limiting the hypercytokinemia and cytokine release syndrome (CRS) seen in COVID-19 patients (Huang et al., 2020).
Non-human primate (NHP) models have been used extensively to study pathogenesis and potential vaccine and antiviral candidates for numerous viral diseases (Estes et al., 2018). We and others have recently used rhesus macaques (RMs) to model SARS-CoV-2 infection and pathogenesis; SARS-CoV-2-infected RMs develop transient respiratory disease and exhibit viral shedding similar to humans, recapitulating mild to moderate infection and, only in rare cases, severe disease (Chandrashekar et al., 2020; Munster et al., 2020; Rockx et al., 2020; Williamson et al., 2020; Yu et al., 2020). Baricitinib is an oral, selective inhibitor of JAKs 1 and 2 with potent anti-inflammatory activity approved for treatment of patients with moderate to severe active rheumatoid arthritis (Keystone et al., 2015). Recently, machine learning algorithms and in vitro data suggested that baricitinib could also inhibit clathrin-mediated endocytosis of SARS-CoV-2 (Cantini et al., 2020; Richardson et al., 2020; Stebbing et al., 2020; Titanji et al., 2020); thus, it could provide a dual effect of dampening inflammation and viral infection. In this study, we leveraged the ability to perform longitudinal collections, including bronchoalveolar lavages, and the availability of lung tissue for pathology, to test the immunologic and virologic effects of baricitinib treatment in SARS-CoV-2-infected RMs.
Results
Baricitinib was well-tolerated and detectable in plasma and tissues but did not limit SARS-CoV-2 replication in RMs
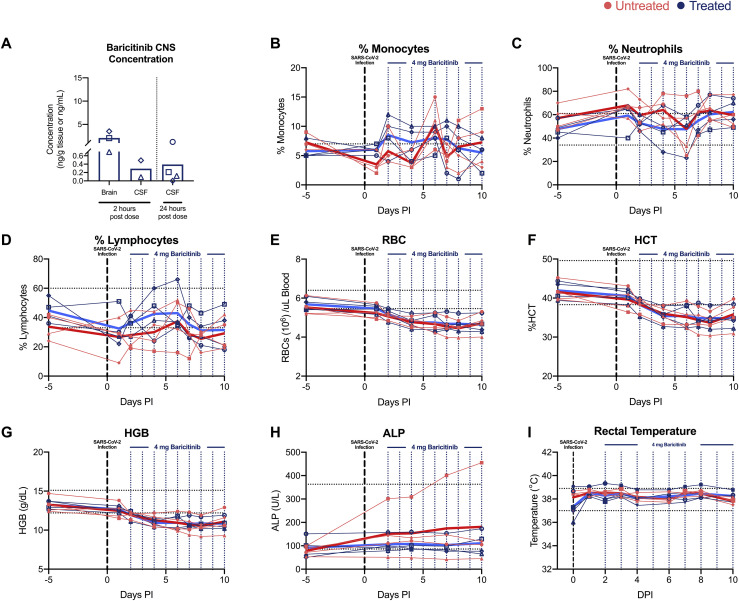
We inoculated 8 adult RMs (11–17 years old, mean = 14 years) (Table S1) with a total of 1.1 × 106 plaque-forming units (PFUs) SARS-CoV-2 (2019-nCoV/USA-WA1/2020), administered by intranasal (IN) and intratracheal (IT) routes (Yu et al., 2020). Two days after infection, 8 RMs were randomized (n = 4 treated) to receive 4 mg of oral baricitinib, daily for 8–9 days or observed without treatment until 10–11 days after infection, when all RMs were euthanized (Figure 1 A). At 24 h after dosing, baricitinib was readily detected in plasma of all treated animals (Figure 1B) (measures performed at 6 days after infection closed symbol; and 8 days after infection open symbol), achieving an average level of 2.13 ng/mL. At necropsy, baricitinib was detectable at approximately 2 h after the last dose in left/right upper and lower lung (Figure 1C) (n = 4 RMs; average of 4.41 and 4.43 ng/g, respectively), brain (n = 3 RMs; 2.09 ng/g tissue), and cerebrospinal fluid (CSF) (n = 2 RMs; 0.29 ng/mL) (Figure S1 A); we also detected baricitinib in CSF from 3 out of the 4 treated animals at 24 h after dosing on 8–9 days after the final dose (Figure S1A).
Figure 1.
Baricitinib is detectable in plasma and tissues from SARS-CoV-2-infected RMs but has no impact on viral kinetics
(A) Study design; 8 RMs were infected intranasally and intratracheally with SARS-CoV-2, and at 2 days after infection, 4 RMs began daily baricitinib administration (4 mg). Longitudinal collections performed are indicated in circles.
(B and C) Concentration of baricitinib 24 h after dosing in plasma (6 days after infection closed symbol; 8 days after infection open symbol) (B) and at necropsy in upper and lower lungs of baricitinib-treated SARS-CoV-2-infected RMs (C).
(D and E) Daily cage-side assessment and physical examination scores (D) and changes in body weight from baseline (E) in baricitinib-treated (blue symbols; n = 4) and untreated (red symbols; n = 4) SARS-CoV-2-infected RMs.
(F) Longitudinal pulse oximetry readings.
(G–J) After SARS-CoV-2 inoculation, nasal, throat, bronchoalveolar lavages (BALs), and rectal swabs were collected, and viral loads were quantified by qRT-PCR.
(K) Viral loads in tissues measured at necropsy (10–11 days after infection).
Abbreviation is as follows: Ct, cycle threshold. Different symbols represent individual RMs. Thick lines represent the average of the baricitinib-treated (blue lines) and untreated (red lines) groups. Bars in (B), (C), and (K) represent the average of the treated and untreated groups. Statistical analysis was performed using a non-parametric Mann-Whitney test.
See also Figures S1 and S2A and Tables S1, S2, and S3.
Figure S1.
Baricitinib was well-tolerated and detectable in the central nervous system in SARS-CoV-2-infected RMs, related to Figure 1
(A) Left, concentration of baricitinib 2 hours after dosing in brain and CSF and, right, 24 hours after dosing in CSF.
(B–D) Longitudinal frequency of (B) monocytes, (C) neutrophils, and (D) lymphocytes in blood of SARS-CoV-2 infected RMs.
(E–H) In (E), red blood cell counts (RBC), (F) hematocrit (HCT), (G) hemoglobin (HGB) and (H) alkaline phosphatase (ALP) levels were analyzed throughout the study.
(I) Longitudinal rectal temperatures. Different symbols represent individual animals. Bold lines represent the average of the baricitinib treated group (blue), and the untreated group (red).
A slight reduction of peripheral monocytes, neutrophils, and lymphocytes, which could be due to trafficking to the lung, as well as decreased red blood cell (RBC) counts, hematocrit (HCT), and hemoglobin (HGB) were observed starting at 2 days after infection in all RMs (Figures S1B–S1G). Blood chemistries showed elevated levels of alkaline phosphatase (ALP) in one untreated animal starting at 2 days after infection, and all other values were within the normal range (Figure S1H). Body temperature remained stable in all RMs (Figure S1I). Overall, treatment with baricitinib was well-tolerated without direct evidence of treatment-induced clinical pathology, nephrotoxicity, or hepatotoxicity when compared to untreated SARS-CoV-2-infected RMs. To further monitor response to infection and baricitinib treatment, the health status of all animals was assessed daily by veterinarians, with cage-side assessment and physical examination scored based on a standardized scoring system (modified from previous studies [Chertow et al., 2016; Huang et al., 2020]) (main parameters included in the scoring are listed on Tables S2 and S3). One day after infection, all animals exhibited changes to alertness and respiratory pattern (Figure 1D). Additional early signs of disease included changes to pulse oximetry readings, with one untreated animal dropping below 80% (Figure 1F), reduction in appetite, hunched posture, shivering, pale appearance, and agitation. Signs of disease persisted during the 10- to 11-day course of the study, without significant differences between treated and untreated animals (Figure 1D). Weight loss was observed in 4 out of 4 untreated and 3 out of 4 baricitinib-treated RMs (Figure 1E), although we cannot discriminate whether this is a result of the infection or related to frequent access for sample collection.
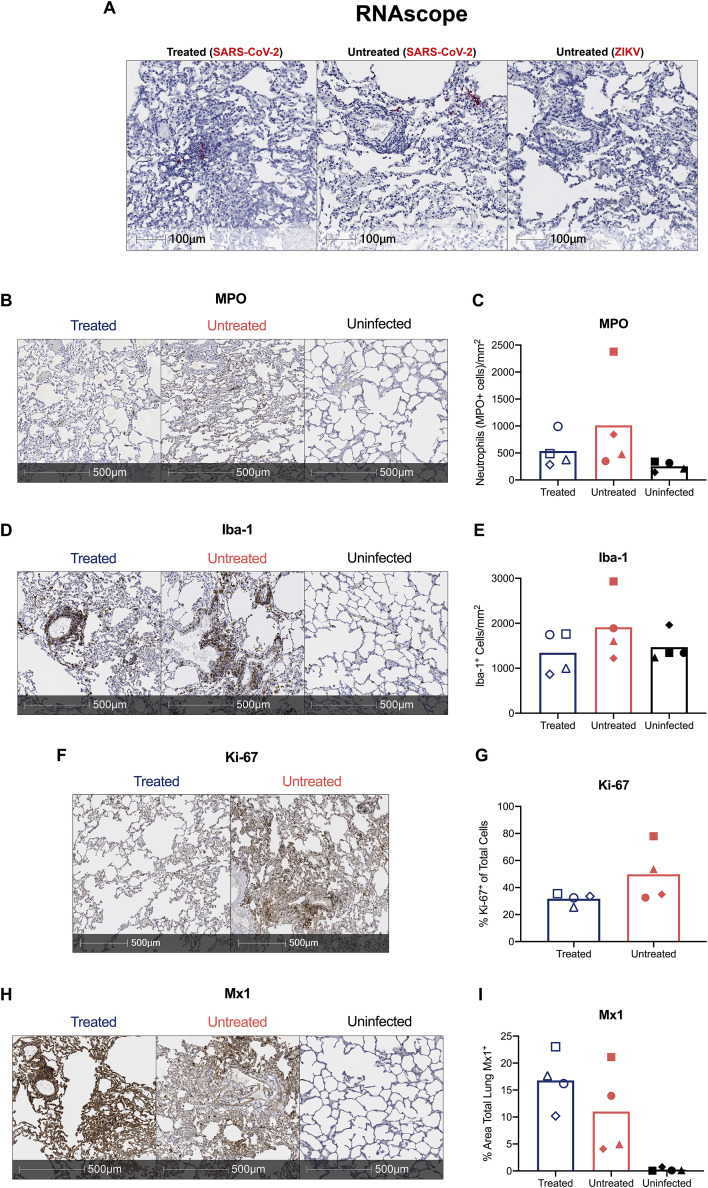
We next assessed viral RNA levels by qRT-PCR (Chandrashekar et al., 2020; Munster et al., 2020). We observed high levels of SARS-CoV-2 RNA in nasal and throat swabs, and bronchoalveolar lavages (BALs), with a peak between 2 and 4 days after infection of 1.4 × 107, 1.2 × 106, and 1.9 × 105 copies/mL, respectively (Figures 1G–1I); viral RNA then steadily decreased until 10–11 days after infection. SARS-CoV-2 RNA levels remained similar in nasal, throat, or BALs between the baricitinib-treated and the untreated groups. Virus was not detected in blood and transiently present in rectal swabs (Figure 1J). At necropsy (10–11 days after infection), viral RNA was detected for most animals in nasopharynx, lower and upper lungs, and hilar lymph nodes; viral RNA was detected in the ileum of 4 out of 4 untreated and 1 out of 4 treated RMs. Viral loads (cycle threshold value) in tissue for treated and untreated RMs were overall comparable (Figure 1K). Additionally, in situ RNA hybridization (RNAscope) targeting both positive- and negative-sense viral RNA strands identified multifocal clusters of infected cells within the lung parenchyma in both treated and untreated RMs (Figure S2 A). Thus, baricitinib treatment starting at 2 days after infection was safe and well-tolerated but did not affect the kinetics of SARS-CoV-2 replication.
Figure S2.
Baricitinib reduced lung neutrophil and macrophage infiltration, preserved IFN responses but did not reduce SARS-CoV-2 replication in RMs, related to Figures 1 and 2
(A–I) In (A), representative images of in situ RNA hybridization (RNAscope) targeting viral RNA strands identifying clusters of infected cells within the lung parenchyma in both treated and untreated SARS-CoV-2 infected RMs. Scale bars: 100 um. Representative immunohistochemistry (IHC) images of (B) neutrophils (myeloperoxidase+, MPO, cells) (D) macrophages (ionized calcium-binding adaptor molecule 1+, Iba-1, cells), (F) proliferating (Ki-67), and (H) Interferon-induced GTP-binding protein+ (Mx1), cells in lungs of baricitinib treated and untreated SARS-CoV-2 infected RMs, and uninfected RMs. Scale bars 500 μm. Quantification of (C) neutrophils (MPO+ positive cells/mm2), (E) macrophages (lba-1+ cells/mm2), (G) proliferating (Ki-67+), and (I) Interferon-induced protein Mx1 (% area total lung Mx1+) in IHC lung images of baricitinib treated, and untreated controls of SARS-CoV-2 infected RMs, and uninfected RMs.
Baricitinib reduced lung pathology and iInflammation in SARS-CoV-2-infected RMs
We then performed multiple analyses to determine the severity of SARS-CoV-2 infection in RMs and the effectiveness of baricitinib to ameliorate the pathophysiologic response. First, X-ray radiographs (RM6 X-ray) (Figure 2 A) were longitudinally (−5, 2, 4, 7, and 10 days after infection) performed (blinded scoring by a radiologist as previously reported [Munster et al., 2020; Williamson et al., 2020]). Pulmonary infiltration and ground glass opacity were observed at multiple experimental time points after infection in 2 out of 4 untreated and 0 out of 4 treated RMs (Figures 2B and 2C), with one of the untreated animals showing severe pneumonia at all time points after infection (Figures 2B and 2C). Second, we measured serum levels of several systemic inflammatory markers. Among these, ferritin (4 days after infection, p = 0.0286; and 10 days after infection, p = 0.0286) and C-reactive protein (CRP) (4 days after infection) were found to be elevated in the untreated RMs when compared with the baricitinib-treated RMs (Figures 2D and 2E). Previous reports have indicated that heightened ferritin and CRP levels are associated with increased COVID-19 severity in humans (Qin et al., 2020; Ruan et al., 2020). Finally, to assess lung damage of SARS-CoV-2 infection, all RMs were euthanized at 10 or 11 days after infection. At necropsy, multiple regions of upper, middle and lower lung lobes were taken for immunologic, virologic, and pathologic analyses. Lung pathology analyses and scoring were performed by two pathologists independently in a blinded fashion. Treated RMs showed decreased type II pneumocyte hyperplasia, peribronchiolar hyperplasia, syncytia formation, alveolar septal thickening, and inflammatory cell infiltration (Figures 2F–2K). Consistent with the pathology scoring, neutrophil (myeloperoxidase, MPO+, cells) and macrophage (ionized calcium-binding adaptor molecule, Iba-1+, cells) infiltration, as well as levels of cells expressing the proliferation marker Ki-67 appeared to be decreased in the lungs in baricitinib-treated RMs as measured by quantitative immunohistochemistry (IHC) (Figures S2B–S2G). Levels of Mx1 were similar between both groups (Figures S2H and S2I). Of note, some of the SARS-CoV-2-infected animals in both groups showed inflammatory cell infiltration levels similar to uninfected RMs, indicating a resolution of the infiltration at 10–11 days after infection, consistent with an earlier peak of pathogenesis in RMs, as previously published (Chandrashekar et al., 2020; Munster et al., 2020; Rockx et al., 2020; Williamson et al., 2020; Yu et al., 2020). The average pathology score per lobe (measuring the average severity of abnormalities per lobe, independently of how many lobes had been effected, p = 0.0286) and the total pathology score (considering severity and number of effected lobes, p = 0.0857) were lower in the baricitinib-treated group (0.99 and 22, respectively) as compared with untreated RMs (1.66 and 38.5, respectively) (Figures 2L–2N). Overall, these data support a therapeutic role of baricitinib in reducing lung pathology, infiltration of inflammatory cells in the lung, and soluble markers of inflammation associated with disease progression in humans.
Figure 2.
Reduced respiratory disease and lower levels of lung pathology in baricitinib-treated RMs
(A) Representative ventrodorsal radiograph of an untreated RM before SARS-CoV-2 infection (5 days before infection), and at 4, and 7 days after infection. Red squares indicate regions of pulmonary infiltrates and opacity.
(B and C) Daily (B) and cumulative (C) radiograph scores; ventrodorsal and lateral radiographs were scored for the presence of pulmonary infiltration by a clinical radiologist according to a standard scoring system (0: normal; 1: mild interstitial pulmonary infiltrates; 2: moderate pulmonary infiltrates with partial cardiac border effacement and small areas of pulmonary consolidation; 3: severe interstitial infiltrates, large areas of pulmonary consolidation, alveolar patterns, and air bronchograms).
(D and E) Fold change to 2 days after infection for ferritin (D) and C-reactive protein (CRP) levels (E).
(F and G) Panel (F) shows 100× magnification, and (G) shows 200× magnification (zoomed in from F), representative lung lesions in an untreated SARS-Cov-2-infected RM with focally extensive interstitial pneumonia, type 2 pneumocytes hyperplasia, alveolar septal thickening, syncytia formation (arrow), neutrophils, and macrophages infiltrations (arrowhead).
(H) 200× magnification, Thyroid Transcription Factor-1 (TTF-1) staining with prominent type 2 pneumocyte hyperplasia (brown) in a control SARS-CoV-2-infected RM.
(I and J) Panel (I) shows 100× magnification, and (J) shows 200× magnification (zoomed in from I), treatment effects of baricitinib in SARS-CoV-2-infected RMs with a reduction in pulmonary lesions, lesser inflammatory infiltrates (arrowhead), and reduced type 2 pneumocyte hyperplasia.
(K) 200× magnification, TTF-1 staining with lesser type 2 pneumocyte hyperplasia (brown) after baricitinib treatment.
(L) Average pathology score per lobe.
(M) Total pathology score.
(N) Pathology scores for individual parameters.
Scale bar, (F) and (I): 100 μM; (G), (H), (J), and (K): 50 μM. Bars in (D), (E), (L), (M), and (N) indicate mean values for baricitinib-treated (blue) and untreated (red) SARS-CoV-2-infected RMs. Each symbol represents individual animals. Statistical analysis in (D), (E), and (L)–(N) were performed using non-parametric Mann-Whitney test. Statistical analyses were performed two-sided with p ≤ 0.05 deemed significant. Ranges of significance were graphically annotated as follows: ∗p < 0.05.
See also Figures S2B–S2I.
Baricitinib treatment dampens gene signatures of macrophage inflammation and neutrophil degranulation in the BALs of SARS-CoV-2-infected RMs
To investigate the impact of baricitinib on the lower airway, we performed bulk RNA sequencing (RNA-seq) profiling of cells isolated from BALs prior to SARS-CoV-2 inoculation (5 days before infection; baseline), 2 days after virus inoculation, prior to baricitinib treatment, and 4 days after infection, and 48 h after beginning baricitinib. Relative to pre-infection, we observed a robust upregulation of differentially expressed genes (DEGs) at 2 days after infection in both the treated and untreated RMs (Figure 3 A); however, at 4 days after infection only a handful of DEGs were detected in the baricitinib-treated animals, whereas a robust transcriptional response persisted in the untreated group. To identify immunological pathways perturbed by SARS-CoV-2 infection and baricitinib treatment, we performed gene set enrichment analysis (GSEA) (Subramanian et al., 2005). To determine pathways that changed after drug administration, we directly compared gene expression profiles at 2 days after infection to 4 days after infection. Comparison of GSEA data from 2 days after infection to 4 days after infection in untreated RMs show robust, highly significant positive enrichment in pathways comprised of genes for inflammatory responses, TNF-α and IL-6 signaling, and neutrophil and granulocyte function—indicating that, in the absence of baricitinib, expression of these genes continues to increase (Figure 3B). In stark contrast, when a similar comparison of 2 days after infection versus 4 days after infection was tested in RMs receiving baricitinib, we observed negative enrichment, indicating that inflammatory genes were expressed at lower levels already after only 2 days of treatment (Figure 3B). To confirm the robustness of our enrichment analysis in detecting downregulation of inflammatory pathways with treatment, we also conducted GSEA analyses using direct cross-sectional comparisons (i.e., 4 days after infection untreated versus 4 days after infection treated); these data demonstrated that inflammatory signatures were significantly lower in animals receiving baricitinib at 4 days after infection, although equivalent when comparing 2 days after infection samples in which neither group had received the drug (Figures S3 A and S3B).
Figure 3.
Baricitinib treatment suppresses gene expression of inflammation and neutrophil degranulation in the BALs of SARS-CoV-2-infected RMs
Bulk RNA-seq profiles of BAL cell suspensions from RMs obtained at day −5 prior to SARS-CoV-2 inoculation (baseline), at 2 days after infection, prior to baricitinib treatment, and at 4 days after infection, 2 days after initiation of baricitinib.
(A) Venn diagrams indicating the number of differential expression genes (DEGs) detected at 2 or 4 days after infection relative to −5 days after infection in the untreated (red) and baricitinib-treated (blue) groups. The total DEGs for each comparison are shown in parentheses.
(B) Bar plots showing enrichment of top scoring inflammatory and immunological gene signatures from the MSIGDB (Hallmark and Canonical Pathways) and databases, and custom gene sets (interferon-stimulated genes [ISGs]; see below) ranked by GSEA comparisons of gene expression in the 4 days after infection versus 2 days after infection samples from the untreated (red bars) and baricitinib-treated (blue bars) groups. The x axis depicts the normalized enrichment score (NES); a positive enrichment score indicated higher expression at 4 days after infection relative to 2 days after infection (bars facing right); conversely, negative scores of a pathway indicate cumulatively higher expression in 2 days after infection samples relative to 4 days after infection (bars facing left). Nominal p values are indicated.
(C–F) GSEA enrichment plots depicting pairwise comparison of gene expression of 2 days after infection versus 4 days after infection samples for the untreated group and for the baricitinib-treated group. The top-scoring (i.e., leading edge) genes are indicated by solid dots. The hash plot under GSEA curves indicates individual genes and their rank in the dataset. Left-leaning curves (i.e., positive enrichment scores) indicate higher expression of pathways at 4 days after infection; right-leaning curves (negative enrichment scores) indicate higher expression at 2 days after infection. Sigmoidal curves indicate equivalent expression between the groups being compared. The NES and nominal p values testing the significance of each comparison are indicated. (C) REACTOME_ NEUTROPHIL_DEGRANULATION (MSIDB #M27620). (D) GSEA line plot of HALLMARK_TNFA_SIGNALING_VIA_NFKB pathway (MSIGDB #M5890). (E) GSEA line plot of HALLMARK_IL6_JAK_STAT3_SIGNALING (MSIGDB #M5897). (F) A custom gene set of ISGs from prior NHP studies (Nganou-Makamdop et al., 2018; Palesch et al., 2018; Sandler et al., 2014).
(G–J) Heatmaps of top-scoring (i.e., leading edge) from the untreated 4 days after infection versus 2 days after infection GSEA analyses. The color scale indicates the log2 expression relative to the median of all baseline samples.
See also Figure S3.
Figure S3.
Baricitinib Suppressed the expression of inflammatory mediators and neutrophil degranulation genes in BALs from SARS-CoV-2-infected RMs, related to Figure 3
Cross-sectional GSEA analysis comparing 4 days after infection untreated versus 4 days after infection baricitinib treated, or 2 days after infection untreated versus 2 days after infection baricitinib treated in bulk BAL from SARS-CoV-2 infected RMs.
(A–C) GSEA comparisons of 4 days after infection untreated versus 4 days after infection baricitinib treated are shown as black symbols, and comparisons of or 2 days after infection untreated versus 2 days after infection baricitinib treated are shown as gray symbols. (A) GSEA enrichment plots for the GSEA line plot of HALLMARK_IL6_JAK_STAT3_SIGNALING pathway (MSIGDB #M5897). (B) GSEA line plot of HALLMARK_TNFA_SIGNALING_VIA_NFKB pathway (MSIGDB #M5890). (C) GSEA line plot of REACTOME NEUTROPHIL DEGRANULATION gene set (REACTOME #M27620).
(D) Heatmap of leading edge genes for REACTOME NEUTROPHIL DEGRANULATION gene set based on untreated 4 days after infection versus baseline contrast. The log2 expression and the reference is the median of all baseline samples as indicated at right. The top 35 genes are shown in order of GSEA analysis of the cross-sectional 4 days after infection comparison.
(E and F) GSEA analysis for KEGG Rheumatoid Arthritis gene set (E) GSEA contrasting 4 days after infection versus 2 days after infection for untreated and treated arms. GSEA curves are colored by experimental arm. Leading edge genes are indicated by solid dots. The hash plot under GSEA curves indicate individual genes and their rank in the dataset. Left-leaning curves (i.e., positive enrichment scores) indicate enrichment at 4 days after infection, right-leaning curves (negative enrichment scores) indicate higher enrichment at 2 days after infection, and sigmoidal curves indicate a lack of enrichment, i.e., equivalent expression between the groups being compared. The normalized enrichment scores and nominal p values testing the significance of each comparison are indicated.
(F) GSEA comparisons of 4 days after infection untreated versus 4 days after infection baricitinib treated samples (black symbols); comparisons of 2 days after infection untreated versus 2 days after infection baricitinib treated samples (gray symbols).
(G) plot showing log10 average normalized counts obtained from DESeq2 for leading edge genes at 2 days after infection in untreated and treated samples, and (H) at 4 days after infection.
To explore the effect of baricitinib on the inflammatory responses induced by SARS-CoV-2 infection at the gene level, we examined several pathways in greater detail (Figures 3C–3J). One of the highest scoring pathways, neutrophil degranulation, was significantly enriched at 4 days after infection relative to 2 days after infection in the untreated group (p < 0.001) (Figure 3C). Strikingly, enrichment of this pathway was completely abrogated in the treated group (p = 0.979). When we examined individual genes that were (1) elevated by SARS-CoV-2 infection, and (2) influenced by baricitinib treatment, we observed that several genes were those encoding degradative and bactericidal enzymes present in neutrophil granules (MMP9, MMP25, BPI, and MPO), or highly expressed on polymorphonuclear neutrophils (CXCR1 and CXCR2), the alarmin S100A12, and genes for proteins that act to degrade the extracellular matrix during neutrophil extravasation: SERPINB10 and ADAM8 (Figure 3G). Of note, S100A12 (EN-RAGE), for which expression was effectively reduced by baricitinib treatment, has been associated with COVID-19 severity in humans (Arunachalam et al., 2020). These genes were highly upregulated in BAL samples of untreated RMs but substantially attenuated in treated animals, many at levels equivalent to pre-infection (Figure 3G). Collectively, these gene signatures suggest that baricitinib treatment may dampen macrophage inflammation as well as neutrophil recruitment and activity in the lower airway during acute SARS-CoV-2 infection. We also examined the enrichment of neutrophil pathway genes in cross-sectional GSEA comparisons, as shown in Figures S3C and S3D; this analysis largely mirrored our day 2 and day 4 infection observations. Additionally, we observed several alarmin proteins (S100A8 and S100A9) had lost their induction at 4 days after infection in animals receiving baricitinib, as did the MPO gene. These genes have been recently demonstrated to be highly expressed in the myeloid compartment of peripheral blood of patients exhibiting severe COVID-19 (Schulte-Schrepping et al., 2020; Silvin et al., 2020).
Baricitinib treatment also rapidly induced near complete abrogation of inflammation mediators downstream of TNF-α signaling and IL-6 signaling (Figures 3D, 3E, 3H, and 3I). Within these pathways, among the molecules suppressed by baricitinib were chemotactic factors critical for recruitment of neutrophils (CXCL6 and CXCL3) and macrophages (CCL2), inflammatory serine protease factors (SERPINB2 and TNFAIP6), and cytokines regulating inflammation and immune responses (IL-12B). Of note, genes identified as upregulated in rheumatoid arthritis (RA) were found to be significantly enriched (p = 0.0448) in untreated as compared to treated animals at 4 days after infection, despite similar gene expression at 2 days after infection (Figures S3E and S3F). In the leading-edge analysis of the RA pathway, we noted lower expression of several inflammatory mediators such as CXCL8, IL-1β, CCL5, CCL3, CCL20, IL-18, IL-6, and CXCL12 (Figures S3G and S3H). As baricitinib was developed to ameliorate inflammation in RA by inhibiting JAK1/JAK2 signaling, and consistently with the reduction in the IL-6/JAK/STAT3 signaling pathway (Figure 3I), these data confirm the effectiveness of baricitinib in the lower airway of SARS-CoV-2-infected RMs.
Several of the significantly enriched gene sets comprised genes in type I IFN signaling (Figure 3B) and multiple interferon-stimulated genes (ISGs) had elevated expression relative to baseline (Figure 3J). In both treated and untreated groups, we observed a slight reduction in expression at 4 days after infection relative to 2 days after infection (Figures 3B and 3F). However, unlike gene sets associated with inflammation, genes associated with type I IFN signaling and innate antiviral responses were unperturbed by baricitinib. Thus, baricitinib treatment potently suppressed inflammatory pathways in the lower airway of RMs infected with SARS-CoV-2 but left innate antiviral signaling largely intact.
Baricitinib treatment abolishes inflammatory cytokine and neutrophil chemoattractant expression in bronchoalveolar macrophages of SARS-CoV-2-infected RMs
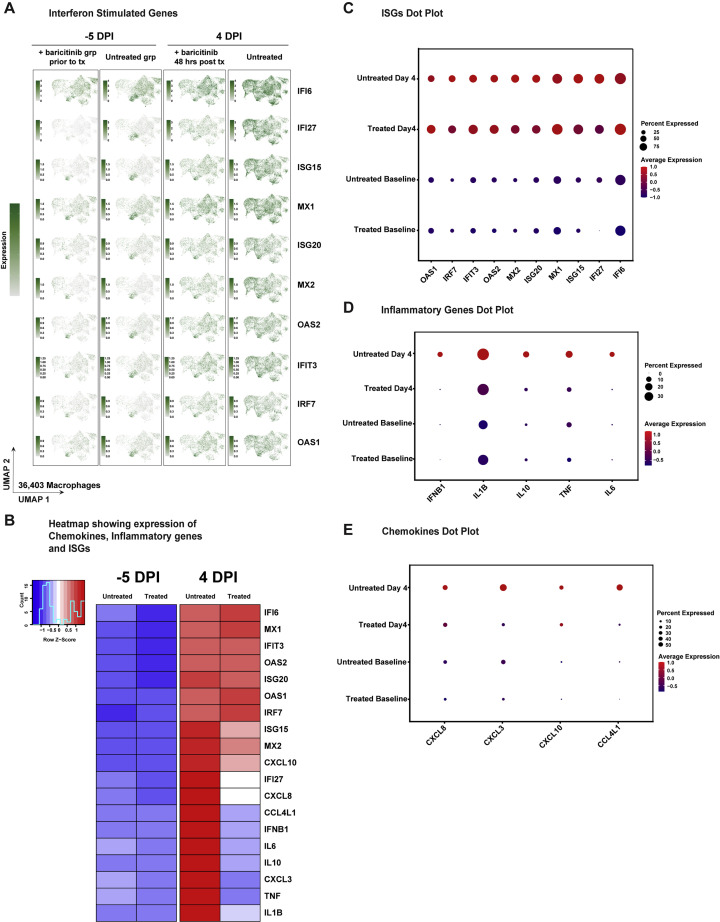
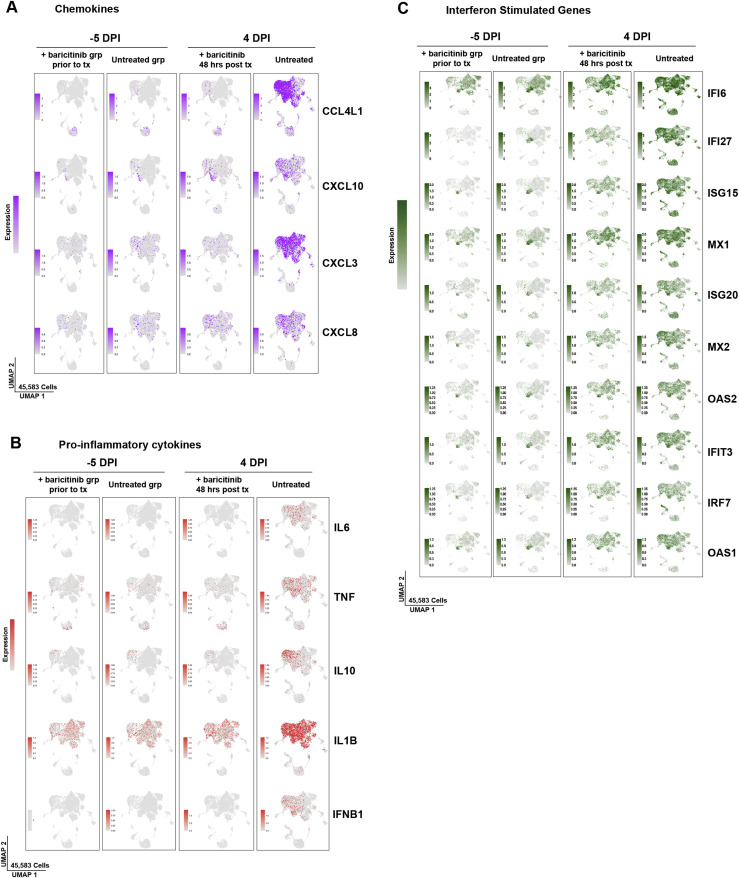
The bulk RNA-seq data indicated that gene signatures consistent with macrophage activation, neutrophil infiltration, and cytokine-driven inflammation were evident as early as 2 days after infection, and that baricitinib was capable of abrogating these pathways. To identify the cellular component orchestrating airway inflammation, we performed single-cell RNA-seq (scRNA-seq) profiling using 10× Genomics-based droplet sequencing. Single-cell suspensions of BAL samples from three untreated and two baricitinib-treated RMs prior to infection, and at 4 days after infection were subjected to 10× droplet capture within 3 h of collection. After processing to remove erythrocytes and low-quality cells, the captures yielded a cumulative 45,583 cells across all samples for analysis. The cellular distribution is summarized in the UMAP shown in Figure 4 A. Similar to observations reported in scRNA-seq data in humans infected with SARS-CoV-2 (Bost et al., 2020; Chua et al., 2020; Liao et al., 2020), the vast majority of cells in BALs were predominantly macrophage/myeloid origin (80.7%), followed by lymphocytes (CD4+ T cells, CD8+ T cells, and NK cells) (9.8%) and approximately 3.2% were identified as epithelial. Allocation of cells from the cumulative data by treatment variables (Figure 4B) demonstrated that the cellular distribution was equivalent among the experimental groups and no population was enriched due to batch or technical variation associated with individual captures. We probed the macrophage population for upstream regulators associated with the inflammatory pathways identified in the bulk RNA-seq analyses and observed elevated expression of several inflammatory mediators at 4 days after infection: IL-6, TNF-α, IL-1β, and IL-10 (Figures 4C, S4 , and S5). IFN-β was also highly expressed in the macrophage cluster; however, IFN-α transcripts were detected in a virtually negligible fraction of cells (Figures 4C, S4, and S5). Strikingly, and consistent with the bulk RNA-seq data, we observed that baricitinib treatment virtually dampened expression of TNF-α, IL-10, IFN-β, and IL-6 in pulmonary macrophages and significantly reduced expression of IL-1β (Figure 4C). We also observed a robust induction of chemokines driving neutrophil recruitment (CXCL3 and MIP2β and CXCL8 and IL-8), macrophage trafficking (CCL4L1 and MIP1β), and CXCL10 and IP10 (Figures 4D and 4E), a pleiotropic chemokine upregulated in several viral infections, and long hypothesized to be associated with pathogenesis in SARS-CoV-1 viral infection and observed in SARS-CoV-1 infection of NHPs (de Lang et al., 2007; Laing et al., 2020; Tang et al., 2005). Notably, after 48 h of baricitinib treatment, expression of these proinflammatory cytokines was reduced to basal levels (Figures 4D and 4E). Examination of the expression levels of antiviral ISGs in pulmonary macrophages yielded a much different pattern than those observed for inflammatory genes—although widespread induction of ISGs was observed after SARS-CoV-2 infection, baricitinib treatment had only a very modest effect on these pathways (Figures 4F, S4A–S4C, and S5C). Collectively, these data support a model in which baricitinib administration strongly reduces airway inflammation and neutrophil accumulation but has a minimal effect on innate antiviral immunity.
Figure 4.
Baricitinib treatment abolishes inflammatory cytokine and neutrophil chemoattractant expression in bronchoalveolar macrophages
Single-cell suspensions from BALs of SARS-CoV-2-infected RMs were subject to 10× Genomics capture and sequencing.
(A) UMAP showing major cell types in BAL samples (n = 10 samples; untreated, baseline n = 3; untreated, 4 days after infection n = 3; treated, baseline n = 2; treated, 4 days after infection n=2).
(B) UMAP showing clusters in BAL samples by treatment days (n = 10).
(C) UMAP projection of pro-inflammatory cytokines in macrophages.
(D) UMAP projection of neutrophil chemoattractant and pro-inflammatory chemokines.
(E and F) Expression of chemokines and interferon-stimulated genes (ISGs) in treated and untreated samples at baseline and 4 days after infection. The colored expression scale of expression in UMAPs is depicted on a per gene basis: the scale represents the per cell reads for each gene divided by the total reads for of that cell, scaled to the factor shown and natural log-transformed.
See also Figures S4 and S5.
Figure S4.
Baricitinib inhibited the expression of inflammatory and macrophage/neutrophil chemokine genes while preserving ISGs in lung macrophages from SARS-CoV-2-infected RMs, related to Figure 4
(A) Expression as UMAP projection of interferon stimulated genes (ISGs) in macrophages for treated and untreated samples at baseline and 4 days after infection.
(B) Heatmap showing average expression of genes of interest in macrophages for treated and untreated samples at baseline and 4 days after infection.
(C–E) Dot plots representing gene expression levels and percentage of cells expressing genes associated with inflammation, chemokine response and interferon stimulation
Figure S5.
Baricitinib reduced the expression of inflammatory and chemokine gGenes while maintaining ISGs in BALs from SARS-CoV-2-infected RMs, related to Figure 4
(A–C) Expression as UMAP projection of inflammation, chemokine and interferon stimulated genes (ISGs) across major cell types in BAL for treated and untreated samples at baseline and 4 days after infection.
Baricitinib leads to reduced BAL levels of neutrophils and neutrophil NETosis activity
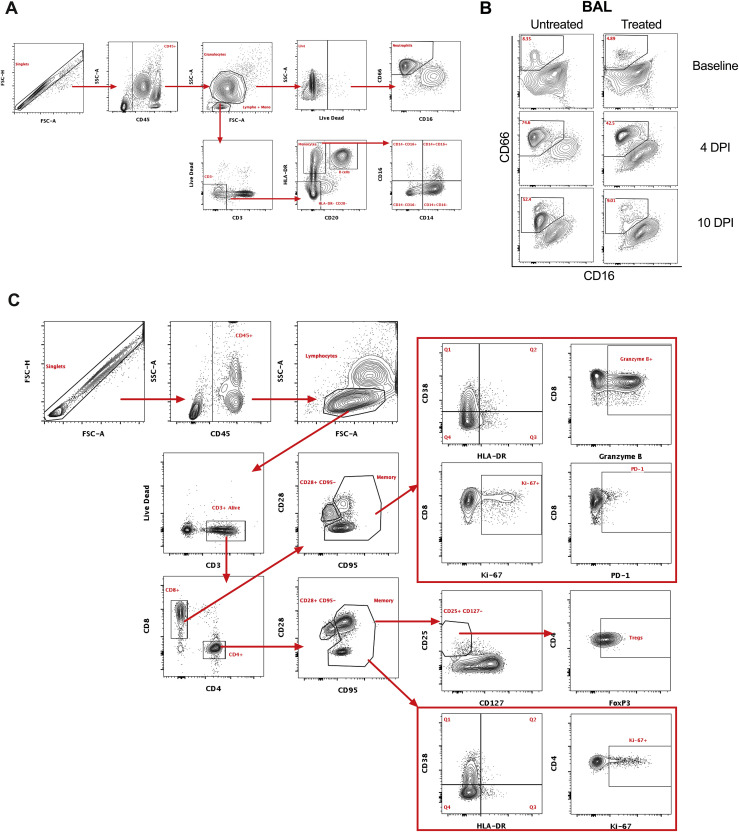
To gain insight into the immunologic effects of baricitinib treatment on cellular distribution within BALs, we applied global high-dimensional mapping of 23-parameter flow-cytometry data. As shown in the UMAP representation (Figure 5 A), untreated and baricitinib-treated RMs had different BAL cellular distribution starting from 4 days after infection, corresponding with the time point of peak inflammation and viremia, including in neutrophils. This was of interest considering the higher frequency of macrophages expressing neutrophil-attracting chemokines in untreated RMs (Figures 4D and 4E). Thus, we focused our flow-cytometry immunologic analyses in quantifying the longitudinal levels of neutrophils (CD45+CD3–CD20–CD66+ live granulocytes; representative staining in Figures S6 A and S6B). Analyses of BALs showed an early recruitment of neutrophils in the lung at 4 days after infection during the peak of viremia, particularly in the untreated RMs, which all maintained higher frequencies of neutrophils at later stages of infection (10–11 days after infection) than did baricitinib-treated RMs (Figure 5B; p = 0.0286). In blood, neutrophils (Figure 5C) remained relatively stable after infection as compared to before infection and at lower levels in untreated as compared to treated animals at the latest experimental points (p = 0.0571), consistently with a higher migration to lung in untreated RMs. The levels of CD14+CD16– (Figure 5D) and CD14+CD16+ monocytes in the BALs were, on average, slightly higher in untreated RMs at 4, 7, and 10 days after infection, with the difference due to 3 of 4 untreated RMs having levels higher than the treated animals at specific time points (Figure 5D). Given that the flow-cytometry data of BALs show a reduced migration of neutrophils to lung in baricitinib-treated RMs, we next measured neutrophil extracellular trap (NET) activity by quantification of extracellular DNA via Sytox staining, a functional readout of NETosis activity (Figures 5E and 5F) and by quantification of citrullinated H3 (Figure 5G), a systemic marker indicating a post-translational modification thought to precede DNA decondensation during NETosis. NETs have been reported as an important mechanism of inflammation and microvascular thrombosis in patients with COVID-19 (Skendros et al., 2020). Baricitinib-treated RMs showed decreased NET formation by blood neutrophils at 4 (more evident for citrullinated H3, Figure 5G; p = 0.0571) and 10 (more evident for Sytox staining, Figure 5F; p = 0.0571) days after infection when compared to untreated RMs. Finally, when the formation of NETs was examined directly in the lung by IHC staining for citrullinated H3, 3 out of 4 untreated RMs showed presence of NETs, whereas NETs were virtually absent in treated RMs (Figure 5H).
Figure 5.
Baricitinib-treated RMs have decreased infiltration of innate immune cells and lowered neutrophil NETosis
(A) UMAP analysis of BALs in baricitinib-treated (n = 4) and untreated (n = 4) SARS-CoV-2-infected RMs before infection (D −5 PI; baseline), and at 4 and 10 days after infection.
(B) Longitudinal levels of neutrophils within BAL samples depicted as a percentage of CD45+ cells
(C) Fold change to 2 days after infection of neutrophils in blood of baricitinib-treated and untreated SARS-CoV-2-infected RMs.
(D) Longitudinal levels of CD14+CD16– monocytes within BAL samples depicted as a percentage of CD45+ cells.
(E) Representative microscopy images of NETS by Sytox green assay in baricitinib-treated and untreated SARS-CoV-2-infected RMs. Scale bar, 200 μm.
(F) Quantification of NETosis activity upon staining extracellular DNA with Sytox in isolated stimulated neutrophils from blood. Fold change of Sytox levels to −5 days after infection.
(G) Quantification of citrullinated H3 in plasma.
(H) Staining of citrullinated H3 in lungs at 10–11 days after infection.
In (B)–(D), (F), and (G), each symbol represents individual animals. Thick lines represent the average of the baricitinib-treated (blue line) and untreated groups (red line). Bars in (C) and (F) represent the average of the treated and untreated groups. Statistical analysis in (B), (C), (F), and (G) was performed using a non-parametric Mann-Whitney test. Statistical analyses were performed two-sided with p ≤ 0.05 deemed significant. Ranges of significance were graphically annotated as follows: ∗p < 0.05.
See also Figures S6A and S6B.
Figure S6.
Flow cytometry gating strategy for innate and adaptive cells, related to Figures 5 and 6
Representative gating strategy of (A) neutrophils, (B) neutrophil infiltration in BAL at baseline, and 4 and 10 days after infection, and (C) T cell populations analyzed in the study.
Altogether, these data support baricitinib activity in reducing macrophage-derived inflammation and by decreasing pro-inflammatory neutrophilic levels, activity, and NETosis.
Baricitinib reduced T cell immune activation in SARS-CoV-2-infected RMs
Our transcriptomic data indicated that baricitinib reduced macrophage expression of multiple cytokines that can induce T cell immune activation. As such, we then analyzed levels of T cells and their frequency of activation and proliferation by flow cytometry (gating strategy shown in Figure S6C). CD4+ T cell levels in blood remained similar between treated and untreated animals, with 1 out of 4 baricitinib-treated and 2 out of 4 untreated RMs exhibiting a pronounced reduction in CD4+ T cell frequencies at 10 days after infection (Figure 6 A). We observed an expansion of CD4+ TRegs (CD45+CD3+CD4+CD95+CD127–CD25+FoxP3+; representative staining in Figure S6C) at 4 (p = 0.0571) and 6 days after infection in the untreated, but not in the baricitinib-treated RMs (Figure 6B). Specifically, the mean fold change in CD4+ TRegs frequency at 4 and 6 days after infection, as compared to pre-treatment baseline (2 days after infection), was of 7.43 and 4.36 in untreated and of 1.22 and 1.13 in baricitinib-treated RMs, respectively, suggesting higher levels of inflammation in the untreated group resulting in greater expansion of CD4+ TRegs (Figure 6C). Peripheral CD8+ T cells were reduced at 10 days after infection in 2 out of 4 baricitinib-treated and 2 out of 4 untreated RMs (Figure 6D). Notably, the frequency of proliferating (Ki-67+) memory CD8+ T cells in blood progressively and significantly increased in all 4 untreated animals at 7 and 10 days after infection, while significantly decreasing in all baricitinib-treated RMs already at 4 days after infection. As a result, at 10 days after infection the mean frequency of memory CD8+Ki-67+ was significantly higher in untreated RMs (24.38% versus 7.38%; p = 0.0286, Figure 6E).
Figure 6.
Decreased levels of T cell proliferation and activation in baricitinib-treated RMs
(A and B) Longitudinal levels of (A) circulating CD4+ T cells and (B) CD4+ TReg (CD45+CD3+CD4+ CD95+ CD127– CD25+ FoxP3+; representative staining in Figure S6C) cells measured by flow cytometry of baricitinib-treated (blue) and untreated (red) SARS-CoV-2-infected RMs.
(C) Fold changes to 2 days after infection of circulating CD4+ TReg cells.
(D and E) Levels of circulating CD8+ T cells (D) and proliferating (Ki-67+) memory CD8+ T cells (E).
(F and G) Levels of CD4+ T cells (F) and HLA-DR–CD38+ memory CD4+ T cells (G) in bronchoalveolar lavages (BALs) measured by flow cytometry.
(H–J) Levels of CD8+ T cells (H), proliferating (Ki-67+) memory CD8+ T cells (I), and HLA-DR–CD38+ memory CD8+ T cells (J) in BALs. Each symbol represents individual animals. Thick lines represent the average of the baricitinib-treated (blue line) and untreated groups (red line).
(K–M) Representative staining of Ki-67 and CD38 by HLA-DR. Bars in (C) represent the average of the treated and untreated groups. Statistical analysis in (C), (E), and (G) was performed using non-parametric Mann-Whitney test. Statistical analyses were performed two-sided with p ≤ 0.05 deemed significant. Ranges of significance were graphically annotated as follows: ∗p < 0.05.
See also Figures S6C and S7.
CD4+ T cells in the BALs remained relatively constant until 7 days after infection, when the majority of RMs started experiencing a reduction in their frequencies (Figure 6F). Untreated RMs showed an early (present at 4 days after infection), large (mean fold change of 3.31 at 7 days after infection versus 2 days after infection compared to 1.14 in the treated RMs), and prolonged (up to 10 days after infection) increase in the frequency of memory CD4+ T cells expressing CD38 (CD38+HLA-DR–; 4 days after infection, p = 0.0286) (Figure 6G). Remarkably, different from untreated RMs, the frequency of those activated memory CD4+ T cells decreased in baricitinib-treated animals starting at 4 days after infection and remained lower than pre-treatment until 10 days after infection (Figure 6G). Consistent with a reduced pro-inflammatory state of CD4+ T cells, baricitinib-treated RMs showed a lower frequency of CD4+ T cells that spontaneously (without stimulation) produced pro-inflammatory, Th17-related cytokines (IL-17a+; IL-17a+IL-21+; IL-17a+IL-22+) when compared to untreated RMs (Figures S7 A–S7C).
Figure S7.
Baricitinib treatment did not affect the immune T cell responses in SARS-CoV-2-infected RMs, related to Figure 6
(A–C) Frequency of circulating CD4+ T cells spontaneously (without stimulation) producing pro-inflammatory Th17 related cytokines (A) IL-17+, (B) IL-17+IL-21+, (C) IL-17+IL- 22+ at necropsy (days 10–11 after infection) in baricitinib (blue) and untreated (red) SARS-CoV-2 infected RMs.
(D) Representative flow cytometry staining of IFNγ, TNFα, IL-2, IL-4 and IL-17a in CD4+ and CD8+ T cells of a SARS-CoV-2 infected RM following stimulation with SARS-CoV-2 S peptide pool. IFNγ, Unstimulated background values were subtracted from S peptide stimulated values to determine T cell cytokine.
(E and F) IFNγ, TNFα, IL-2, IL-4 and IL-17a frequency levels in (E) CD4+ and (F) CD8+T cells following stimulation with SARS-CoV-2 S peptide pool.
(G–L) IFNγ, TNFα, IL-2, IL-4 and IL-17a frequency levels in (G) CD4+ and (H) CD8+T cells following stimulation with PMA/Ionomycin. Values from unstimulated controls were subtracted in all cases. Granzyme B and PD-1 levels in (I and J) blood and (K and L) BAL memory CD8+T cells measured by flow cytometry. Each symbol represents individual animals. Thick lines represent the average of the baricitinib treated (blue line), and untreated groups (red line). Bars represent the average of the treated and untreated groups. Statistical analysis was performed using a non-parametric Mann-Whitney Test.
As with CD4+ T cells, the reduction in CD8+ T cells was more pronounced in BAL than in blood, starting at 7 days after infection and maintained until necropsy (Figure 6H). Similarly, also in BALs, the frequency of CD8+Ki-67+ T cells increased more extensively in untreated than baricitinib-treated RMs (30.53% versus 11.53% at 7 days after infection; 39.95% versus 24.65% at 10 days after infection) (Figure 6I); as a result, the fold change (as compared to 2 days after infection, pre-treatment) in the frequency of memory CD8+Ki-67+ T cells was higher in untreated then baricitinib-treated RMs both at 7 (8.22 versus 1.02) and 10 (6.28 versus 2.48) days after infection. A similar trend was measured for activated memory CD8+ T cells, with higher frequency and fold change in untreated than baricitinib-treated RMs at 7 (FC CD38+DR–: 23.67 versus 1.62) and 10 (FC CD38+DR–: 9.81 versus 1.43) days after infection (Figure 6J). Representative staining for Ki-67 and HLA-DR by CD38 in CD4+ and CD8+ T cells are shown in Figures 6K–6M. These results corroborate the reduced frequency of Ki-67+ cells observed in baricitinib-treated compared with untreated animals in lung via quantitative IHC analysis (Figures S2F and S2G).
Finally, we assessed the ability of peripheral T cells to respond to ex vivo SARS-CoV-2 specific stimulation (with a SARS-CoV-2 S peptide pool characterized in Grifoni et al., 2020) and to non-antigen specific stimulation (with PMA/ionomycin). Importantly, the levels of SARS-CoV-2 specific CD4+ and CD8+ T cells producing IFN-γ, TNF-α, IL-2, IL-4, and IL-17a in response to S peptide pool stimulation were similar in both groups of animals (Figures S7D–S7F). Similarly, the frequency of CD4+ and CD8+ T cells producing IL-17a, IL-21, IL-22, IFN-γ, and TNF-α were similar among the two groups after PMA and Ionomycin stimulation (Figures S7G and S7H). Furthermore, levels of memory CD4+ and CD8+ T cells expressing granzyme B or PD-1 remained similar between untreated and treated RMs both in blood (Figures S7I and S7J) and BALs (Figures S7K and S7L).
Collectively, these findings indicate that baricitinib treatment lead to downstream reduction in T cell activation and proliferation, without an overall detrimental effect to antiviral function of T cells.
Discussion
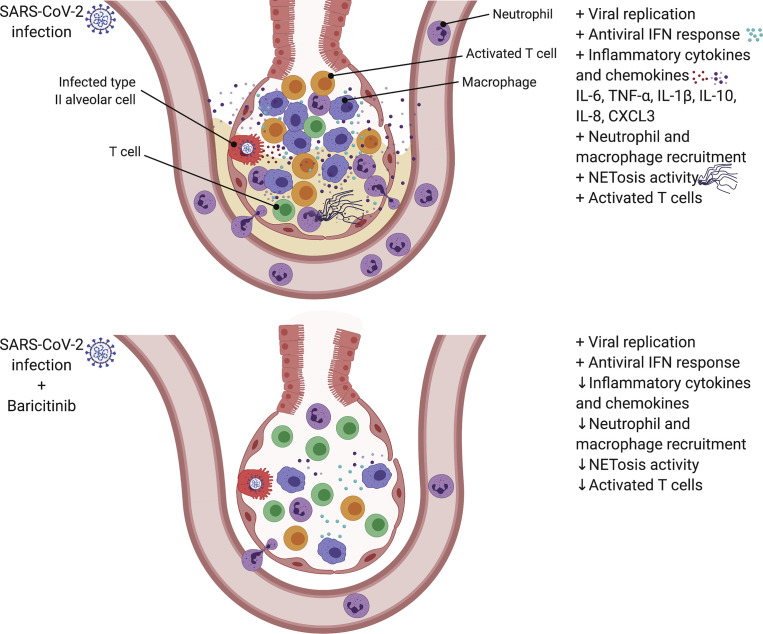
In this study, we tested baricitinib, a JAK1/JAK2 inhibitor clinically approved for rheumatoid arthritis, as a therapeutic candidate to reduce systemic inflammation caused by SARS-CoV-2 infection in RMs. Notably, baricitinib-treated RMs displayed reduced (1) lung pathology, from moderate in untreated animals to mild; (2) levels of inflammatory cytokines, chemokines, and signaling pathways associated with macrophage inflammation, neutrophil recruitment, and disease progression in SARS-CoV-2-infected humans; and (3) levels of systemic inflammation that are associated with COVID-19 severity in humans while not having an impact on type 1 IFN responses. This beneficial anti-inflammatory effect of baricitinib was confirmed by a reduced infiltration of macrophages and neutrophils into the lungs, and a reduced T cell activation in both blood and BALs as compared to untreated animals. Furthermore, we were able to observe an increased NETosis activity of neutrophils upon SARS-CoV-2 infection, previously described in serum from COVID-19 patients (Skendros et al., 2020), which was reduced in baricitinib-treated RMs. Remarkably, single-cell RNA sequencing showed reduced immune activation, neutrophil recruitment, and macrophage trafficking signatures in pulmonary macrophages from treated RMs already after two doses of baricitinib, at 4 days after infection. IL-6, TNF-α, IL-10, IL-1B, CXCL3/MIP-2β, CXCL8/IL-8, CCL4L1/MIP-1β, and CXCL10/IP-10 were all expressed at higher levels in pulmonary macrophages from untreated animals than in baricitinib-treated RMs. These data confirm very recent studies that demonstrated by RNA-seq analysis that higher levels of inflammatory cytokines in lung macrophages are associated with patients presenting with severe or critical COVID-19 cases (Liao et al., 2020). Thus, baricitinib could have clinical benefits in reducing the inflammatory response typically seen in moderate to severe cases of COVID-19 (Figure 7 ). Of note, one of the advantages of baricitinib when compared with other cytokine-specific anti-inflammatory therapies is that it can inhibit production of several cytokines involved in the cytokine storm described in severe cases of COVID-19.
Figure 7.
Effect of baricitinib treatment on the lower airway of SARS-CoV-2-infected RMs
(A) SARS-Cov-2 infection in RMs results in an accumulation of inflammatory macrophages and neutrophils in the lower airway. These airway macrophages produce high amounts of inflammatory cytokines and neutrophil-attracting chemokines and show upregulated type I interferon signaling. Neutrophil NETs and the inflammation induced by SARS-CoV-2 infection both contribute to lung pathology.
(B) Baricitinib treatment reduced the levels of macrophages producing inflammatory cytokines and neutrophil-attracting chemokines, decreased the infiltration of neutrophils into the lung, and reduced T cell activation. The Netosis activity of neutrophils was also reduced. In treated animals, the antiviral interferon response was maintained, viral replication was not impacted, and lung pathology was mild.
Clinical pathology and laboratory parameters of toxicity remained similar in the treated RMs for the 8- to 9-day treatment course at a dose comparable to humans (Bronte et al., 2020; Cantini et al., 2020; Titanji et al., 2020). Baricitinib was found distributed in lungs, a key tissue for SARS-CoV-2 replication, as well as in the central nervous system (CNS). Although several in silico modeling and in vitro studies suggested baricitinib as a possible treatment candidate to COVID-19 due to its potential antiviral activity (Cantini et al., 2020; Richardson et al., 2020; Stebbing et al., 2020; Titanji et al., 2020), we did not observe changes in viral replication dynamics in the treated animals. One of the main concerns in using a JAK inhibitor such as baricitinib is that its downstream anti-immune activation effects could limit immune responses necessary to combat SARS-CoV-2. Importantly, we did not identify reduction of SARS-CoV-2-specific and unspecific CD4+ and CD8+ T cell responses in treated animals, and baricitinib did not inhibit genes associated with type I IFN antiviral responses, indicating its mode of action in this context is primarily to dampen inflammatory responses while maintaining innate and adaptive antiviral immune responses. While ISGs can certainly be stimulated via the JAK/STAT pathways, ISGs have also been shown to be highly inducible via the STING and RIG-I pathways (Loo et al., 2008; Loo and Gale, 2011; Zevini et al., 2017), which are not affected by baricitinib. It is possible that these pathways could compensate for the reduced stimulation via the JAK/STAT pathway.
Several ongoing clinical trials are studying the effects of baricitinib in SARS-CoV-2-infected humans. Currently, the Adaptive COVID-19 Treatment Trial (ACTT-2; NCT04401579) is evaluating the effects of baricitinib in combination with remdesivir, and the COV-BARRIER trial (NCT04421027) is studying baricitinib as a monotherapy. Participants are treated with placebo, remdesivir alone or a combination of remdesivir and baricitinib. In a retrospective cohort study, 15 patients with severe COVID-19 were administered a short course of baricitinib in combination with hydroxychloroquine, and this was associated with a complete recovery in 11 out of 15 subjects (Titanji et al., 2020). In a separate pilot study, baricitinib was combined with lopinavir-ritonavir in 12 patients starting treatment 6 days after symptom onset, with all individuals showing significantly improved clinical and laboratory parameters with no treated individuals requiring ICU care (Cantini et al., 2020). Being performed in an animal model, this study has some key advantages and some important limitations. Advantages include the ability to correct for parameters that may impact clinical outcome and treatment readout, including using the same virus inoculum, dose, and route of infection and starting baricitinib at the same phase of infection in all subjects. Furthermore, the NHP model permits longitudinal collection of BALs and lung at necropsy, which in turn allows the in-depth characterization of the mechanism and impact of baricitinib on immune activation and immunologic responses, including single-cell RNA sequencing analyses of macrophages, at the foci of infection. The main limitation of the study is the small group size, with a total of eight RMs; furthermore, being that SARS-CoV-2-infected RMs are a model of mild to moderate COVID-19 with no untreated animals succumbing to infection, and with treatment initiated early after infection, we cannot determine whether the therapeutic impact of baricitinib will be the same in severe COVID-19 patients and when started at a later phase after infection. Mitigating that concern, in our study, treatment started once inflammatory signatures were already upregulated, mimicking the conditions in which baricitinib would be administered clinically. Indeed, a recent small clinical trial in 20 severe COVID-19 patients indicated that baricitinib mitigated immune dysregulation by reducing plasma levels of IL-6, IL-1β, and TNF-α and lowered time to recovery (Bronte et al., 2020). Our data provide a rationale for baricitinib treatment in COVID-19 to be given in a window where blocking immune inflammation would prevent the formation of a cytokine storm without interfering in the initial responses necessary for preventing viral dissemination and persistence.
In conclusion, this study provides rationale and mechanisms of actions for a beneficial anti-inflammatory effect of baricitinib treatment for COVID-19.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-CCR7 BB700 (3D12) | BD Biosciences | Cat#566437; RRID: AB_2744306 |
| anti-CD20 BB700 (2H7) | BD Biosciences | Cat#745889; RRID: AB_2743319 |
| anti-CD103 BV421 (Ber-ACT8) | BD Biosciences | Cat#563882; RRID: AB_2738464 |
| anti-IL-21 BV421 (3A3-N2.1) | BD Biosciences | Cat#564755; RRID: AB_2738933 |
| anti-Ki-67 BV480 (B56) | BD Biosciences | Cat#566109; RRID: AB_2739511 |
| anti-CD14 BV605 (M5E2) | BD Biosciences | Cat#564054; RRID: AB_2687593 |
| anti-CD3 BV605 (SP34-2) | BD Biosciences | Cat#562994; RRID: AB_2737938 |
| anti-CD56 BV711 (B159) | BD Biosciences | Cat#740781; RRID: AB_2740444 |
| anti-CXCR6 BV750 (13B 1E5) | BD Biosciences | Cat#747052; RRID: AB_2871813 |
| anti-CD115 BV750 (9-4D2-1E4) | BD Biosciences | Cat#747093; RRID: AB_2871846 |
| anti-CD3 BUV395 (SP34-2) | BD Biosciences | Cat#564117; RRID: AB_2738603 |
| anti-CD8 BUV496 (RPA-T8) | BD Biosciences | Cat#612942; RRID: AB_2744460 |
| anti-CD45 BUV563 (D058-1283) | BD Biosciences | Cat#741414; RRID: AB_2870903 |
| anti-CD49a BUV661 (SR84) | BD Biosciences | Cat#750628; RRID: AB_2874760 |
| anti-CCR2 BUV661 (LS132.1D9) | BD Biosciences | Cat#750472; RRID: AB_2874631 |
| anti-CD28 BUV737 (CD28.2) | BD Biosciences | Cat#612815; RRID: AB_2738808 |
| anti-CD16 BUV737 (3G8) | BD Biosciences | Cat#564434; RRID: AB_2744295 |
| anti-CD69 BUV805 (FN50) | BD Biosciences | Cat#748763; RRID: AB_2857327 |
| Fixable Viability Stain 700 | BD Biosciences | Cat#564997; RRID: AB_2869637 |
| anti-IL-2 AF488 (MQ1-17H12) | BioLegend | Cat#500314; RRID: AB_493368 |
| anti-CD8a PerCP Cy5.5 (RPA-T8) | BioLegend | Cat#301032; RRID: AB_893422 |
| anti-CD161 BV421 (HP-3G10) | BioLegend | Cat#339914; RRID: AB_2561421 |
| anti-CD4 BV421 (OKT4) | BioLegend | Cat#317434; RRID: AB_2562134 |
| anti-CD95 BV605 (DX2) | BioLegend | Cat#305628; RRID: AB_2563825 |
| anti-HLA-DR BV650 (L243) | BioLegend | Cat#307650; RRID: AB_2563828 |
| anti-IL-2 BV650 (MQ1-17H12) | BioLegend | Cat#500334; RRID: AB_2563878 |
| anti-CD25 BV711 (BC96) | BioLegend | Cat#302636; RRID: AB_2562910 |
| anti-PD-1 BV785 (EH12.2H7) | BioLegend | Cat#329930; RRID: AB_2563443 |
| anti-CD11c BV785 (3.9) | BioLegend | Cat#301644; RRID: AB_2565779 |
| anti-CD11b PE (ICRF44) | BioLegend | Cat#301306; RRID: AB_314158 |
| anti-IFNγ PE/Dazzle 594 (B27) | BioLegend | Cat#506530; RRID: AB_2566718 |
| anti-CD101 PE/Cy7 (BB27) | BioLegend | Cat#331014; RRID: AB_2716109 |
| anti-FoxP3 AF647 (150D) | BioLegend | Cat#320014; RRID: AB_439750 |
| anti-IFNγ AF647 (4S.B3) | BioLegend | Cat#502516; RRID: AB_493031 |
| anti-CD4 APC/Cy7 (OKT4) | BioLegend | Cat#317418; RRID: AB_571947 |
| anti-CD123 APC/Fire750 (6H6) | BioLegend | Cat#306042; RRID: AB_2750163 |
| anti-CD38 FITC (AT-1) | StemCell Technologies, Inc. | Cat#60131FI |
| anti-IL-17a Alexa Fluor 488 (eBio64DEC17) | Thermo Fisher Scientific | Cat#53-7179-42; RRID: AB_10548943 |
| anti-IL-22 APC (IL22JOP) | Thermo Fisher Scientific | Cat#17-7222-82; RRID: AB_10597583 |
| anti-CXCR5 PE (MU5UBEE) | Thermo Fisher Scientific | Cat#12-9185-42; RRID: AB_11219877 |
| anti-GranzymeB PE/TexasRed (GB11) | Thermo Fisher Scientific | Cat#GRB17; RRID: AB_2536540 |
| anti-IL-17a PE-efluor 610 (eBio64DEC17) | Thermo Fisher Scientific | Cat#61-7179-42; RRID: AB_2574658 |
| anti-CD127 PE/Cy5 (eBioRDR5) | Thermo Fisher Scientific | Cat#15-1278-42; RRID: AB_2043801 |
| anti-TNFα PE/Cy7 (Mab11) | Thermo Fisher Scientific | Cat#25-7349-82; RRID: AB_469686 |
| Fixable Viability Dye eFluor 780 | Thermo Fisher Scientific | Cat#65-0865-14 |
| CD66abce PE-Vio770 (TET2) | Miltenyi Biotec | Cat#130-119-849; RRID: AB_2784267 |
| anti-IL-4 PE (7A3-3) | Miltenyi Biotec | Cat#130-091-647; RRID: AB_615125 |
| CD27 PE/Cy5 (1A4CD27) | Beckman Coulter | Cat#6607107; RRID: AB_10641617 |
| anti-CD159a APC (Z199) | Beckman Coulter | Cat#A60797; RRID: AB_10643105 |
| Anti-Thyroid Transcription Factor-1 (8G7G3/1) | Agilent | Cat#M357529-2 |
| anti-MxA (M143) | EMD Millipore | Cat#MABF938 |
| anti-MPO | DAKO (Agilent) | Cat#A0398; RRID: AB_2335676 |
| anti-Iba-1 | BioCare Medical | Cat#CP290A; RRID: AB_10578940 |
| anti-Ki-67 (B56) | BD Biosciences | Cat#550609; RRID: AB_393778 |
| Histone H3 (Citrullinated R2 + R8 + R17) Monoclonal Antibody (11D3) | Cayman Chemical | Cat#17939 |
| Goat anti-Mouse IgG (H+L) Highly Cross-Adsorbed Secondary Antibody Alexa Fluor 633 | Thermo Fisher Scientific | Cat#A21052 |
| Bacterial and Virus Strains | ||
| SARS-CoV-2 (2019-nCoV/USA-WA1/2020; grown in Vero E6 cell line) | BEI Resources | Cat#NR-52281; Lot 70033175 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| FoxP3 / Transcription Factor Staining Buffer Kit | TONBO biosciences | Cat#TNB-0607 |
| Fixation/Permeabilization Solution Kit | BD Biosciences | Cat#554714 |
| Phorbol 12-myristate 13-acetate (PMA) | Sigma Aldrich | Cat#P8139 |
| Ionomycin calcium salt from Streptomyces conglobatus | Sigma Aldrich | Cat#I0634 |
| BD BD GolgiStop Protein Transport Inhibitor (containing Monensin) | BD Biosciences | Cat#51-2092KZ |
| Brefeldin A Solution (1000x) | Thermo Fisher Scientific | Cat#00-4506-51 |
| Diva Decloaker 10x | Biocare Medical | Cat#DV2004LX |
| Background Sniper | Biocare Medical | Cat#BS966L |
| Viral Transport Media | Labscoop, LLC | Cat#VTM-1L |
| Synthetic peptides | Synthetic Biomolecules (aka A&A) | www.syntheticbiomolecules.com |
| SYTOX Green Nucleic Acid Stain | Thermo Fisher Scientific | Cat#S7020 |
| Citrullinated Histone H3 (Clone 11D3) ELISA Kit | Cayman Chemical | Cat#501620 |
| Critical Commercial Assays | ||
| Ferritin | Beckman Coulter | Cat#33020 |
| C-Reactive Protein | Beckman Coulter | Cat#OSR6147 |
| Deposited Data | ||
| BAL - Bulk RNA-Seq & scRNA-Seq | Gene Expression Omnibus | GEO: GSE159214 |
| Oligonucleotides | ||
| RM-RPP30-F 5′-AGACTTGGACGTGCGAGCG-3′ | This Paper | N/A |
| RM-RPP30-R 5′-GAGCCGCTGTCTCCACAAGT-3′ | This Paper | N/A |
| RM-RPP30-Pr 5′-FAM-TTCTGACCTGAAGGCTCTGCGCG-BHQ-3′ | This Paper | N/A |
| 2019-nCoV_N2-F 5′-TTACAAACATTGGCCGCAAA-3′ | Waggoner et al., 2020 | N/A |
| 2019-nCoV_N2-R 5′-GCGCGACATTCCGAAGAA-3′ | Waggoner et al., 2020 | N/A |
| 2019-nCoV_N2-P 5′-FAM-ACAATTTGCCCCCAGCGCTTCAG-BHQ-3′ | Waggoner et al., 2020 | N/A |
| E_Sarbeco_F 5′- ACAGGTACGTTAATAGTTAATAGCGT-3′ | Waggoner et al., 2020 | N/A |
| E_Sarbeco_R 5′- ATATTGCAGCAGTACGCACACA-3′ | Waggoner et al., 2020 | N/A |
| E_Sarbeco_P 5′-CalFluor560- ACACTAGCCATCCTTACTGCGCTTCG-BHQ1-3′ | Waggoner et al., 2020 | N/A |
| RNase P – F 5′-AGATTTGGACCTGCGAGCG-3′ | Waggoner et al., 2020 | N/A |
| RNase P – R 5′-GAGCGGCTGTCTCCACAAGT-3′ | Waggoner et al., 2020 | N/A |
| RNase P – P 5′-CalFuor560-TTCTGACCTGAAGGCTCTGCGCG-BHQ1-3′ | Waggoner et al., 2020 | N/A |
| SARS-CoV2 anti-sense specific probe v-nCoV2019-S | ACD Bio | Cat#848561 |
| SARS-CoV2 anti-sense specific probe v-nCoV2019-orf1ab-sense | ACD Bio | Cat#859151 |
| Software and Algorithms | ||
| GraphPad Prism 8.0 | GraphPad Software | https://www.graphpad.com/scientific-software/prism/; RRID: SCR_002798 |
| FlowJo Software (version 10.7.1) | FlowJo, LLC | https://www.flowjo.com/solutions/flowjo/downloads; RRID: SCR_008520 |
| Olympus cellSens® Standard 2.3 digital imaging software | Olympus | https://www.olympus-lifescience.com/en/software/cellsens/; RRID: SCR_014551 |
| HALO software (version 3.0.311.405) | Indica Labs | https://indicalab.com/halo/; RRID: SCR_018350 |
| bcl2fastq v2.20.0.422 | Illumina | https://support.illumina.com/sequencing/sequencing_software/bcl2fastq-conversion-software.html |
| STAR v2.7.3a | Dobin et al., 2013 | https://github.com/alexdobin/STAR |
| DESeq2 v1.24.0 | Love et al., 2014 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| ComplexHeatMap v2.0.0 | Gu et al., 2016 | https://bioconductor.org/packages/release/bioc/html/ComplexHeatmap.html |
| VennDiagram v1.6.20 | CRAN | https://rdrr.io/cran/VennDiagram/ |
| GSEA 4.1.0 | Subramanian et al., 2005 and Mootha et al., 2003 | https://www.gsea-msigdb.org/gsea/login.jsp;jsessionid=94213B4581121AA02E710A5BE27FBE9F |
| CellRanger v3.1.0 | 10x Genomics | https://support.10xgenomics.com/single-cell-gene-expression/software/downloads/latest |
| Seurat v3.1.5 | Stuart et al., 2019 | https://satijalab.org/seurat |
| SingleR v2.0.3 | Aran et al., 2019 | https://bioconductor.org/packages/SingleR |
| DoubletFinder v2.0.3 | McGinnis et al., 2019 | https://github.com/chris-mcginnis-ucsf/DoubletFinder |
| ggplot2 | Wickham, 2016 | https://ggplot2.tidyverse.org |
| Plotly | Sievert, 2020 | https://plotly-r.com |
| Analysis scripts | This paper | https://github.com/BosingerLab/RM_Baricitinib_manuscript |
| Docker v 1.12.6 | Docker | https://www.docker.com/ |
| RStudio v1.1.453 | RStudio, Inc. | https://rstudio.com/ |
| rocker/rstudio v3.6 | Rocker Project | https://hub.docker.com/r/rocker/rstudio |
| Other | ||
| miRNeasy Micro Kit | QIAGEN | Cat#217084 |
| SMART-Seq v4 Ultra Low Input RNA Kit for Sequencing | Takara Bio | Cat#634894 |
| NexteraXT DNA Library Sample Preparation Kit | Illumina | Cat#FC-131-1096 |
| Chromium Next GEM Single Cell 5′ Library and Gel Bead Kit v1.1 | 10x Genomics | Cat#1000165 |
| QiaAmp Viral RNA mini kit | QIAGEN | Cat#52906 |
| TaqMan Fast Virus 1-Step Master Mix | Thermo Fisher Scientific | Cat#4444432 |
Resource Availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Dr. Mirko Paiardini (mirko.paiardini@emory.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
Source data supporting this work are available from the corresponding author upon reasonable request. The following sequencing data have been deposited in GenBank: SARS-CoV-2 viral stock (accession GEO: GSE162247). Data tables for expression counts for bulk and single-cell RNA-Seq for BAL are deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO accession GSE159214. Custom scripts and supporting documentation on the RNA-Seq analyses will be made available at https://github.com/BosingerLab/.
Experimental model and subject details
Study approval
YNPRC’s animal care facilities are accredited by both the U.S. Department of Agriculture (USDA) and by the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC). All animal procedures were performed in line with institutional regulations and guidelines set forth by the NIH’s Guide for the Care and Use of Laboratory Animals, 8th edition, and were conducted under anesthesia with appropriate follow-up pain management to minimize animal suffering. All animal experimentation was reviewed and approved by Emory University’s Institutional Animal Care and Use Committee (IACUC) under permit PROTO202000035.
Animal models
Eight (4 female and 4 male) specific-pathogen-free (SPF) Indian-origin rhesus macaques (RM; Macaca mulatta; Table S1) were housed at Yerkes National Primate Research Center (YNPRC) as previously described (McGary et al., 2017) in the ABSL3 facility. Animals for study assignment were requested to be greater than 11 years old without preference for gender or MHC haplotype. RMs were infected with 1.1x106 plaque forming units (PFU) SARS-CoV-2 via both the intranasal (1 mL) and intratracheal (1 mL) routes concurrently. Absent further stratification criteria, four RMs were administered 4 mg Baricitinib (Olumiant®, Eli Lilly) starting at 2 days after infection for 8-9 consecutive days. Baricitinib was supplied as a powder that was folded into food items (i.e., honey, yogurt, etc.) or distilled water, which was delivered either orally or as a gavage when animals were being anesthetically accessed, respectively. At each anesthetic access pulse oximetry was recorded and RMs were clinically scored for responsiveness and recumbency; discharges; skin condition; respiration, dyspnea, and cough; food consumption; and fecal consistency (Tables S2 and S3). At 10-11 days after infection, RMs were administered Baricitinib and subjected to necropsy after 2 hours with blood and cerebrospinal fluid (CSF) collected perimortem to assess pharmacokinetics of baricitinib. Longitudinal tissue collections of peripheral blood (PB); bronchoalveolar lavage (BAL); and nasal, and pharyngeal mucosal swabs in addition to thoracic X-rays (ventrodorsal and right lateral views) were performed immediately prior to Baricitinib administration as annotated (Figure 1A). In addition to the tissues listed above, at necropsy the following tissues were processed for mononuclear cells: hilar LN, lower lung, and upper lung. Additional necropsy tissues harvested for histology included nasopharynx.
Method details
Viral stocks
Vero E6 cell line (African Green Monkey Kidney cell line; CRL-1586, ATCC) was used in this study. Vero cells were cultured and maintained in MEM (Sigma) supplemented with 10% heat inactivated fetal bovine serum (FBS) (GIBCO) and 1 mM L-glutamine (GIBCO), 50 U/ml penicillin and 50 μg/ml streptomycin (GIBCO). The cells were kept at 37°C in the presence 5% CO2. At the time of virus inoculation and propagation, the concentration of FBS was reduced to 2%. SARS-CoV-2 (NR-52281: BEI Resources, Manassas, VA; USA-WA/2020, Lot no. 70033175) was passaged on Vero E6 cells at a MOI of 0.01 to produce the infectious viral stock. SARS-CoV-2 has been propagated and titrated by TCID50 method followed by storage of aliquots at −80°C until further use in the experiments.
Back titration of viral stocks via plaque assay was used to determine the infectious dose delivered to the RMs. The virus stock was also directly sequenced via metagenomic methods prior to inoculation to confirm the presence of the furin cleavage motif, which has been shown to be lost upon sequential passage of SARS-CoV-2 in culture (Davidson et al., 2020). Our stock contained fewer than 6% of viral genomes with a mutation that could potentially abrogate furin-mediated cleavage of S.
Determination of viral load RNA
SARS-CoV-2 genomic RNA was quantified in nasopharyngeal (NP) swabs, throat swabs, plasma, and bronchoalveolar lavages (BAL). Swabs were placed in 1mL of Viral Transport Medium (VTM-1L, Labscoop, LLC). Viral RNA was extracted from NP swabs, throat swabs, and BAL on fresh specimens, while plasma was frozen for future analysis. Viral RNA was extracted manually using the QiaAmp Viral RNA mini kit according to the manufacturer’s protocol. Quantitative PCR (qPCR) was performed on viral RNA samples using the N2 primer and probe set designed by the CDC for their diagnostic algorithm: CoV2-N2-F: 5′-TTACAAACATTGGCCGCAAA-3′, CoV2-N2-R: 5′-GCGCGACATTCCGAAGAA-3′, and CoV2-N2-Pr: 5′-FAM-ACAATTTGCCCCCAGCGCTTCAG-BHQ-3′. qPCR reactions were performed in duplicate with the TaqMan Fast Virus 1-step Master Mix using the manufacturer’s cycling conditions, 200nM of each primer, and 125nM of the probe. The limit of detection in this assay was 257 copies per mL of VTM/plasma/BAL. To verify sample quality the CDC RNase P p30 subunit qPCR was modified to account for rhesus macaque specific polymorphisms. The primer and probe sequences are RM-RPP30-F 5′-AGACTTGGACGTGCGAGCG-3′, RM-RPP30-R 5′-GAGCCGCTGTCTCCACAAGT-3′, and RPP30-Pr 5′-FAM-TTCTGACCTGAAGGCTCTGCGCG-BHQ1-3′. A single well from each extraction was run as above to verify RNA integrity and sample quality via detectable and consistent cycle threshold values.
SARS-CoV-2 quantification from necropsy samples
An approximately 0.5 cm3 sample of each tissue was collected at necropsy, placed in 500μL Nuclisens lysis buffer (Biomerieux), and stored at −80°C. Thawed samples were homogenized with a sterile pestle, treated with 50μL proteinase K (QIAGEN) for 30 minutes at 55°C, and pelleted. Total nucleic acid was extracted from 250μL of supernatant using eMAG (Biomerieux) and eluted into 50μL. RT-PCR for SARS-CoV-2 N2 was performed as previously described, and singleplex RT-PCR for RNase P was performed using primers and probes optimized for quantitation, each using 5μL of eluate (Waggoner et al., 2020). To allow for comparison of SARS-CoV-2 levels between samples that may have had subtle differences in starting material, the SARS-CoV-2 N2 Ct was normalized to the RNase P control by: 1) calculating the difference between N2 Ct and RNase P Ct for each sample, and 2) adding this to the median RNase P Ct value for the sample type. For the purposes of data visualization, samples in which SARS-CoV-2 N2 was undetected were assigned a Ct value of 40 (the assay limit of detection).
Quantification of baricitinib by LC-MS/MS in plasma, CSF and tissue
One hundred μL of plasma or CSF samples were extracted with 500 μL of methanol. For tissues like brain and lung, 0.2 to 0.5 g of tissue were homogenized and extracted with 2 mL of methanol. [2H9]-ruxolitinib dissolved in 50% methanol at 500 nM was spiked in plasma/CSF (10 μL) or tissue samples (40 μL) as internal standard before extraction. The supernatant of each extraction (50 μL) was mixed with equal amount of 0.1% formic acid and then subjected to LC-MS/MS analysis after filtration through 0.22 μm membrane with Costar Spin-X centrifuge tube filters (Corning, NY). A Vanquish Flex HPLC system (Thermo Scientific, Waltham, MA) coupled with a TSQ Quantiva triple quadrupole mass spectrometer (Thermo Scientific, Waltham, MA) with an ESI interface was used for LC-MS analysis. Analytes were separated by a Kinetex EVO-C18 column (100 × 2.1 mm, 2.6 μm; Phenomenex, Torrance, CA) at a flow rate of 300 μL/min, 35°C. Gradient elution was used for the separation with mobile phase A (0.1% formic acid) and mobile phase B (acetonitrile). The LC gradient started with 10% of mobile phase B for 0.5 min, then increased from 10% to 90% in 4 min and kept at 90% for 0.5 min before returning to the initial condition. Selected reaction monitoring in positive mode (spray voltage: 3,200 V; sheath gas: 40 Arb; auxiliary gas: 20 Arb; ion transfer tube temperature: 350°C; vaporizer temperature: 350°C) was used to detect baricitinib (372.1 → 251.1) and the internal standard [2H9]-ruxolitinib (316.2 → 186.1). Data were collected and processed by Thermo Xcalibur 3.0 software. Calibration curves were generated from standard baricitinib by serial dilutions in blank biometric samples using the same extraction method described above. For CSF, 0.5% plasma was used as surrogate to make calibration curve. The calibration curves had r2 value greater than 0.99.
All the chemicals are analytical grade or higher and were obtained commercially from Sigma-Aldrich (St. Louis, MO). [2H9]-ruxolitinib was purchased from ALSACHIM (lllkirch, Alsace, France) with purity greater than 98%.
Quantification of ferritin and CRP
Serum ferritin (Beckman Coulter; Cat# 33020) and C-Reactive protein (Beckman Coulter; Cat# OSR6147) levels were quantified by Emory Medical Laboratory using manufacturer protocols.
Histopathology and immunohistochemistry
Due to study end point, the animals were euthanized, and a complete necropsy was performed. For histopathologic examination, various tissue samples including lung, nasal turbinates, trachea, or brain, were fixed in 4% neutral-buffered paraformaldehyde for 24h at room temperature, routinely processed, paraffin-embedded, sectioned at 4 μm, and stained with hematoxylin and eosin (H& E). The H&E slides from all tissues were examined by two board certified veterinary pathologists. For each animal, all the lung lobes were used for analysis and affected microscopic fields were scored semiquantitatively as Grade 0 (None); Grade 1 (Mild); Grade 2 (Moderate) and Grade 3 (Severe). Scoring was performed based on these criteria: number of lung lobes affected, type 2 pneumocyte hyperplasia, alveolar septal thickening, fibrosis, perivascular cuffing, peribronchiolar hyperplasia, inflammatory infiltrates, hyaline membrane formation. An average lung lobe score was calculated by combining scores from each criterion. Digital images of H&E stained slides were captured at 40 × and 200 × magnification with an Olympus BX43 microscope equipped with a digital camera (DP27, Olympus) using Cellsens® Standard 2.3 digital imaging software (Olympus).
Immunohistochemical (IHC) staining of sections of lung was performed using a biotin-free polymer system. The paraffin-embedded sections were subjected to deparaffinization in xylene, rehydration in graded series of ethanol, and rinsed with double distilled water. Antigen retrieval was performed by immersing sections in DIVA Decloaker (Biocare Medical) at 125 °C for 30 s in a steam pressure decloaking chamber (Biocare Medical) followed by blocking with Background Sniper Reagent (Biocare Medical) for 10 minutes. The sections were incubated with Thyroid Transcription Factor-1 (Clone 8G7G3/1) for overnight at 4°C followed by a detection polymer system (MACH 2; Biocare Medical). Labeled antibody was visualized by development of the chromogen (DAB Chromogen Kits; Biocare Medical).
Additional IHC analysis completed by C.E.S., M.N., K.B., and J.D.E. were performed as follows: Tissues were fixed in freshly prepared 4% paraformaldehyde for 24 h, transferred to 70% ethanol, paraffin embedded within 7-10 days, and blocks sectioned at 5 μm. Slides were baked for 30-60 min at 65°C then deparaffinized in xylene and rehydrated through a series of graded ethanol to distilled water. Heat induced epitope retrieval (HIER) was performed with the antigen retrieval buffers citraconic anhydride (0.01% with 0.05% Tween; Mx1, Iba-1, and Ki-67) or citrate buffer (pH 6.0; MPO) in a Biocare NxGen Decloaking Chamber that was set to 110°C for 15 min. The slides were cooled, rinsed twice in distilled water and 1X TBS with 0.05% Tween-20 (TBS-T), blocked (TBS-T + 0.25% casein) for 30 minutes at room temperature, then incubated at room temperature with antibodies against Mx1 (EMD; Cat. No. MABF938 at 1:1000 for 1 hour), MPO (Dako; Cat. No. A0398 at 1:1000 for 1 hour), Iba-1 (BioCare; Cat. No. CP290A at 1:500 for 1 hour), and Ki67 (BD PharMingen; Cat. No. 550609 at 1:200 for 1 hour). Endogenous peroxidases were blocked with 1.5% H2O2 in TBS-T for 10 minutes. Slides were then incubated with Rabbit Polink-1 HRP (GBI Labs; Cat. No. D13-110 for MPO and Iba-1) and Mouse Polink-2 HRP (GBI Labs; Cat. No. D37-110 for Mx1 and Ki67). Slides were developed using Impact DAB (3,3′-diaminobenzidine; Vector Laboratories), washed in ddH2O, counterstained with hematoxylin, mounted in Permount (Fisher Scientific), and scanned at 20x magnification on an Aperio AT2 (Leica Biosystems). Staining for MPO, Mx1, Iba-1, and Ki67 IHC was performed as previously described using a Biocare intelliPATH autostainer.
Quantitative image analysis
Quantitative image analysis was performed using HALO software (v3.0.311.405; Indica Labs) on at least one lung lobe cross section from each animal. For MPO (neutrophil) and Iba-1 (macrophage) quantification, blood vessels (> 5mm2), bronchi, bronchioles, cartilage, and connective tissue were manually excluded; subsequently, the Multiplex IHC v2.3.4 module was used to detect MPO+ or Iba-1+ cells and is presented as a proportion of total alveolar tissue (cells/mm2). For Mx1, the Area Quantification v2 module was used to determine the percentage of Mx1 as a proportion of the total tissue area. For Ki67, the Multiplex IHC v2.3.4 module was used to quantitative the percentage of positive cells. In all instances, manual curation was performed on each sample to ensure the annotations were accurate and to correct false positives/false negatives.
RNAscope in situ hybridization
RNAscope in situ hybridization was performed as previously described (Chandrashekar et al., 2020) using SARS-CoV2 anti-sense specific probe v-nCoV2019-S (ACD Cat. No. 848561) targeting the positive-sense viral RNA and SARS-CoV2 sense specific probe v-nCoV2019-orf1ab-sense (ACD Cat. No. 859151) targeting the negative-sense genomic viral RNA. In brief, after slides were deparaffinized in xylene and rehydrated through a series of graded ethanol to distilled water, retrieval was performed for 30 min in ACD P2 retrieval buffer (ACD Cat. No. 322000) at 95-98°C, followed by treatment with protease III (ACD Cat. No. 322337) diluted 1:10 in PBS for 20 min at 40°C. Slides were then incubated with 3% H2O2 in PBS for 10 min at room temperature. Prior to hybridization, probes stocks were centrifuged at 13,000 rpm using a microcentrifuge for 10 min, then for the antisense probe diluted 1:2 in probe diluent (ACD Cat. No. 300041) to reduce probe aggregation tissue artifacts. Slides were developed using the RNAscope® 2.5 HD Detection Reagents-RED (ACD Cat. No.322360).
Tissue processing
PB was collected from the femoral vein in sodium citrate, serum separation, and EDTA tubes from which plasma was separated by centrifugation within 1 hour of phlebotomy. PB was used for complete blood counts, comprehensive serum chemistry panels, and measurement of neutrophil extracellular traps (NET) activity. From EDTA PB, peripheral blood mononuclear cells (PBMCs) were isolated using a Ficoll-Paque Premium density gradient (GE Healthcare), and washed with R-10 media. R-10 media was composed of RPMI 1640 (Corning) supplemented with 10% heat-inactivated fetal bovine serum (FBS), 100 IU/mL penicillin, 100 μg/mL streptomycin, and 200 mM L-glutamine (GeminiBio). CSF was collected by inserting a 0.75-1.5-inch, 22-25-gauge needle below the external occipital protuberance into the cisterna magna and was separated by centrifugation.
Nasopharyngeal swabs were collected under anesthesia by using a clean rayon-tipped swab (ThermoFischer Scientific, BactiSwab NPG, R12300) placed approximately 2-3cm into the nares. Oropharyneal swabs were collected under anesthesia using polyester tipped swabs (Puritan Standard Polyester Tipped applicator, polystyrene handle, 25-806 2PD, VWR International) to streak the tonsils and back of throat bilaterally (throat/pharyngeal). The swabs were dipped in 1 mL viral transport media (Viral transport Media, VTM-1L, Labscoop, LLC) and vortexed for 30 s, and the eluate was collected.
To collect BAL, a fiberoptic bronchoscope (Olympus BF-XP190 EVIS EXERA III ULTRA SLM BRNCH and BF-P190 EVIS EXERA 4.1mm) was manipulated into the trachea, directed into the primary bronchus, and secured into a distal subsegmental bronchus upon which 35-50 mL of normal saline (0.9% NaCl) was administered into the bronchus and re-aspirated to obtain a minimum of 20ml of lavage fluid. BAL was filtered through a 70 μm cell strainer.
Lung tissue was cut into small pieces, using blunt end scissors, then digested using 1.5 U/mL DNase I (Roche) and 1 mg/mL of Type I collagenase (Sigma-Aldrich) using gentleMACS C tubes and gentleMACS Dissociator (miltenyi Biotec).
Hilar LN biopsies were collected at necropsy, sectioned using blunt, micro-dissection scissors and mechanically disrupted through a 70 μm cell strainer and washed with R-10 media.
Mononuclear cells were counted for viability using a Countess II Automated Cell Counter (Thermo Fisher) with trypan blue stain and were cryo-preserved in aliquots of up to 2x107 cells in 10% DMSO in heat-inactivated FBS. Whole tissue segments (0.5 cm3) were snap frozen dry, or stored in RNAlater (QIAGEN), or Nuclisens lysis buffer (Biomerieux) for analyses of compound distribution, RNA-seq, and tissue viral quantification, respectively.
Bulk and single-cell RNA-Seq Library and sequencing from NHP BALs
Single cell suspensions from BAL were prepared in BSL3 as described above for flow cytometry; for bulk RNA-Seq, 50,000 cells were lysed directly into 700 ul of QIAzol reagent. RNA was isolated using RNeasy Mini or Micro kits (QIAGEN) with on-column DNase digestion. RNA quality was assessed using an Agilent Bioanalyzer and total RNA was used as input for cDNA synthesis using the Clontech SMART-Seq v4 Ultra Low Input RNA kit (Takara Bio) according to the manufacturer’s instructions. Amplified cDNA was fragmented and appended with dual-indexed bar codes using the NexteraXT DNA Library Preparation kit (Illumina). Libraries were validated by capillary electrophoresis on an Agilent 4200 TapeStation, pooled at equimolar concentrations, and sequenced on an Illumina NovaSeq6000 at 100SR, yielding 20-25 million reads per sample. For single-cell RNA-Seq, single-cell suspensions of 100,000 BAL-derived cells, and approximately 30,000 cells were loaded onto 10X Chromium Controller in the BSL3. Single cells were partitioned into droplets (Gel Beads in Emulsion: GEMs) using Chromium NextGEM Single Cell 5′ Library & Gel Bead kits on the 10X Chromium Controller(Zheng et al., 2017). The resulting cDNA was amplified and libraries were prepared for transcriptomic analysis according to manufacturer instructions. Gene expression libraries were sequenced as paired-end 26x91 reads on an Illumina NovaSeq6000 targeting a depth of 50,000 reads per cell in the Yerkes Genomics Core Laboratory(http://www.yerkes.emory.edu/nhp_genomics_core/). Cell Ranger software was used to perform demultiplexing of cellular transcript data, and mapping and annotation of UMIs and transcripts for downstream data analysis.
Bulk RNA-Seq analysis
The quality of reads was evaluated using FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Reads were aligned using STAR v2.7.3.(Dobin et al.). The STAR index was built by combining genome sequences for Macaca mulatta (Mmul10 Ensembl release 100), SARS-CoV2 (strain MN985325.1 - NCBI) and ERCC sequences. The gffread utility (https://github.com/gpertea/gffread) was used to convert gff3 file for SARS-CoV2 and the resulting gtf file for SARS-CoV2 was edited to include exon entries which had the same coordinates as CDS to get counts with STAR. The combined genomic and gtf files were used for generating the STAR index. Transcript abundance estimates were calculated internal to the STAR aligner using the algorithm of htseq-count(Sandler et al., 2014). The ReadsPerGene files were used to generate counts in the htseq format using a custom script that also converted the Ensembl ID to gene names using the gtf file. These files were imported in DESeq2 using the DESeqDataSetFromHTSeqCount function. DESeq2 was used for normalization (Love et al., 2014), producing both a normalized read count table and a regularized log expression table. Only the protein coding genes defined in the gtf file were used for analysis. The design used was: ∼Subject + Group where Group was a combination of Time point (baseline/2days after infection/4days after infection) and Condition (Untreated/Treated) factors. The regularized log expression values were obtained using the rlog function with the parameters blind = FALSE and filtType = “parametric.” The thresholds of padj < 0.05, fold-change > 1.5 and lfcSE < 1 were used to obtain significant differentially expressed genes. The VennDiagram R library was used to create the venn diagrams. GSEA 4.1.0 (https://www.gsea-msigdb.org/gsea/) was used for gene set enrichment analysis with the following gene sets: Hallmark and Canonical pathways (MsigDB), NHP ISGs (Sandler et al. 2014) and Rheumatoid arthritis (KEGG map05323). GSEA was run with default parameters with the permutation type set to gene_set. The input for GSEA was the regularized log expression values obtained from DESeq2 which was filtered to remove genes with mean expression ≤ 0. The regularized log expression values were also used to generate heatmaps using the Complex Heatmap R library (Gu et al., 2016).
Single-cell RNA-Seq bioinformatic analysis
Bronchoalveolar lavage (BAL) samples from five Rhesus Macaque’s were run on 2 Nova Seq 1000 lanes and the resultant bcl files were converted to counts matrices using Cell Ranger v3.1 (10X Genomics). Further, the count matrices for each sample were processed using an inhouse single-cell RNA-seq pipeline that uses Seurat v3.0 (Satija et al., 2018) to initially integrate data from SARS-CoV-2 infected and Baricitinib treated samples. The batch corrected samples were filtered for cells expressing < 250 genes, > 10% Mitochondria genes, HBB, RPS and RPL genes and any doublets were removed using DoubletFinder (McGinnis et al., 2019). After filtration, the data were normalized using scTransform normalization followed by Principal Component analysis. PCs 1-30 were chosen for clustering analysis, as there was very little additional variance observed beyond PC 30. Cells were then clustered based on PC scores using the Louvain-Jaccard method. Uniform Manifold Approximation and Projection (UMAP) (McInnes et al., 2018) method was used to visualize the single cells in 2D embedding. We used Blueprint Encode database from SingleR (Aran et al., 2019) to classify cells into different cell subtypes. Differential gene expression between the clusters was assessed by MAST (Finak et al., 2015). Heatmaps, Dot plots and Violin plots were generated using ggplot2 (Wickham, 2016) package in R.
Immunophenotyping
23-parameter flow cytometric analysis was performed on fresh PBMCs and mononuclear cells (106 cells) derived from, BAL, and lung. Immunophenotyping was performed using anti-human monoclonal antibodies (mAbs), which we (Harper et al., 2020; McGary et al., 2017; Micci et al., 2015; Pino et al., 2019) and others, including databases maintained by the NHP Reagent Resource (MassBiologics), have shown as being cross-reactive in RMs. A panel of the following mAbs was used for longitudinal T cell phenotyping in PBMCs: anti-CCR7-BB700 (clone 3D12; 2.5 μL; cat. # 566437), anti-CD103-BV421 (clone Ber-ACT8; 5 μL; cat. # 563882), anti-Ki-67-BV480 (clone B56; 5 μL; cat. # 566109), anti-CXCR6-BV750 (clone 13B 1E5; 2.5 μL; cat. # 747052), anti-CD3-BUV395 (clone SP34-2; 2.5 μL; cat. # 564117), anti-CD8-BUV496 (clone RPA-T8; 2.5 μL; cat. # 612942), anti-CD45-BUV563 (clone D058-1283; 2.5 μL; cat. # 741414), anti-CD49a-BUV661 (clone SR84; 2.5 μL; cat. # 750628), anti-CD28-BUV737 (clone CD28.2; 5 μL; cat. # 612815), anti-CD69-BUV805 (clone FN50; 2.5 μL; cat. # 748763), and Fixable Viability Stain 700 (2 μL; cat. # 564997) all from BD Biosciences; anti-CD95-BV605 (clone DX2; 5 μL; cat. # 305628), anti-HLA-DR-BV650 (clone L243; 5 μL; cat. # 307650), anti-CD25-BV711 (clone BC96; 5 μL; cat. # 302636), anti-PD-1-BV785 (clone EH12.2H7; 5 μL; cat. # 329930), anti-CD101-PE-Cy7 (clone BB27; 2.5 μL; cat. # 331014), anti-FoxP3-AF647 (clone 150D; 5 μL; cat. # 320014), and anti-CD4-APC-Cy7 (clone OKT4; 2.5 μL; cat. # 317418) all from Biolegend; anti-CD38-FITC (clone AT1; 5 μL; cat. # 60131FI) from STEMCELL Technologies; and anti-CXCR5-PE (clone MU5UBEE; 5 μL; cat. # 12-9185-42), anti-GranzymeB-PE-TexasRed (clone GB11; 2.5 μL; cat. # GRB17), and anti-CD127-PE-Cy5 (clone eBioRDR5; 5 μL; cat. # 15-1278-42) all from Thermo Fisher (Figure S6). mAbs for chemokine receptors (i.e., CCR7) were incubate at 37°C for 15 min, and cells were fixed and permeabilized for 30 min at room temperature using a FoxP3 / Transcription Factor Staining Buffer Kit (Tonbo Biosciences; cat. # TNB-0607-KIT). A panel of the following mAbs was used for the longitudinal phenotyping of innate immune cells in whole blood (500 μL), as described in (Kuri-Cervantes et al., 2020), and mononuclear cells (106 cells) derived from LN biopsies, BAL, and lung: anti-CD20-BB700 (clone 2H7; 2.5 μL; cat. # 745889), anti-Ki-67-BV480 (clone B56; 5 μL; cat. # 566109), anti-CD14-BV605 (clone M5E2; 2.5 μL; cat. # 564054), anti-CD56-BV711 (clone B159; 2.5 μL; cat. # 740781), anti-CD115-BV750 (clone 9-4D2-1E4; 2.5 μL; cat. # 747093), anti-CD3-BUV395 (clone SP34-2; 2.5 μL; cat. # 564117), anti-CD8-BUV496 (clone RPA-T8; 2.5 μL; cat. # 612942), anti-CD45-BUV563 (clone D058-1283; 2.5 μL; cat. # 741414), anti-CCR2-BUV661 (clone LS132.1D9; 2.5 μL; cat. # 750472), anti-CD16-BUV737 (clone 3G8; 2.5 μL; cat. # 564434), anti-CD69-BUV805 (clone FN50; 2.5 μL; cat. # 748763), and Fixable Viability Stain 700 (2 μL; cat. # 564997) all from BD Biosciences; anti-CD38-FITC (clone AT1; 2.5 μL; cat. # 60131FI) from STEMCELL Technologies; anti-CD161-BV421 (clone HP-3G10; 5 μL; cat. # 339914), anti-HLA-DR-BV650 (clone L243; 5 μL; cat. # 307650), anti-CD11c-BV785 (clone 3.9; 5 μL; cat. # 301644), anti-CD11b-PE (clone ICRF44; 2.5 μL; cat. # 301306), and anti-CD123-APC-Fire750 (clone 315; 2.5 μL; cat. # 306042) all from Biolegend; anti-GranzymeB-PE-TexasRed (clone GB11; 2.5 μL; cat. # GRB17) from Thermo Fisher; anti-CD66abce-PE-Vio770 (clone TET2; 1 μL; cat. # 130-119-849) from Miltenyi Biotec; and anti-CD27-PE-Cy5 (clone 1A4CD27; 2.5 μL; cat. # 6607107) and anti-NKG2A-APC (clone Z199; 5 μL; cat. # A60797) from Beckman Coulter (Figure S6). mAbs for chemokine receptors (i.e., CCR2) were incubated at 37°C for 15 min, and cells were fixed and permeabilized at room temperature for 15 min with Fixation/Permeabilization Solution Kit (BD Biosciences; cat. #554714). For each sample a minimum of 1.2x105 stopping gate events (live CD3+ T cells) were recorded. All samples were fixed with 4% paraformaldehyde and acquired within 24 hours of fixation. Acquisition of data was performed on a FACSymphony A5 (BD Biosciences) driven by FACS DiVa software and analyzed with FlowJo (version 10.7; Becton, Dickinson, and Company).
Single cells were then selected using FSC-A x FCS-H gate. A lymphocyte and granulocyte gate based on FSC-A and SSC-A was defined. Live cells were gated followed by CD45+ cells. UMAP analysis (Uniform Manifold Approximation and Projection for Dimension Reduction) was performed in live CD45+for unbiased evaluation of the distribution of the key markers. Projection of the density of cells expressing markers of interest were visualized/plotted on a 2-dimensional UMAP (https://arxiv.org/abs/1802.03426, https://github.com/lmcinnes/umap). We used the Phenograph clustering approach (https://github.com/jacoblevine/PhenoGraph)
Determination of intracellular cytokine induction following SARS-CoV-2 S peptide pool and PMA/Ionomycin stimulation
Cryo-preserved PBMCs were thawed, resuspended in RPMI medium supplemented to contain a final concentration of 10% Fetal Bovine Serum (FBS) (Corning Life Sciences/Media Tech Inc, Manassas, VA), 10mM HEPES, 1x MEM nonessential amino acids (Corning Life Sciences/Media Tech Inc, Manassas, VA), 1mM Sodium Pyruvate (Lonza, Walkersville, MD, U.S.A), 1mM Penicillin/Streptomycin containing Amphothericin B (Sigma Life Sciences, St Louis, MO, U.S.A) and 1x 2-Mercaptoethanol (GIBCO, Invitrogen, Carlsbad, CA, U.S.A). PBMCs were rested overnight at 37°C in a cell culture incubator. Cells were then stimulated for detection of cytokine production by T cells as described before (Kasturi et al., 2020). Briefly, 2 × 106 cells were cultured in 200 μL final volume in 5mL polypropylene tubes (BD Biosciences, San Diego, CA, U.S.A) in the presence of anti-CD28 (1 μg/mL) and anti-CD49d (1 μg/mL) [BD Biosciences] and the following conditions; a) negative control with DMSO only, b) S peptide pool (Grifoni et al., 2020) and c) PMA/Ionomycin in the presence of Golgi transport inhibitors - 10 μg/mL of Brefeldin A (Sigma-Aldrich). After stimulation, cells were washed and stained for cell surface antigens with two panels. Panel 1: anti-CD3 BUV395 (clone SP34-2; 2.5 μL; cat. # 564117), anti-CD8-BUV496 (clone RPA-T8; 2.5 μL; cat. # 612942), and Fixable Viability Stain 700 (2 μL; cat. # 564997) all from BD Bioscience; anti-CD4 APC/Cy7 (clone OKT4; 2.5 μL; cat. # 317418) from Biolegend; To detect intracellular expression of cytokines, mononuclear cells were fixed and permeabilized with a Cytofix/Cytoperm kit (BD Biosciences) and stained as follows: anti-IL-21 BV421 (clone 3A3-N2.1; 5 μL; cat# 564755) from BD Bioscience; anti-IL-2 BV650 (clone MQ1-17H12; 5 μL; cat# 500334) and anti-IFNγ PE/Dazzle 594 (clone B27; 5 μL; cat# 506530) both from Biolegend; anti-IL-17a Alexa Fluor 488 (clone eBio64DEC17; 5 μL; cat# 53-7179-42), anti-IL-22 APC (clone IL22JOP; 5 μL; cat# 17-7222-82), and anti-TNFα PE-Cyanine7 (clone Mab11; 0.5 μL; cat# 25-7349-82) all from Thermo Fisher Scientific; and anti-IL-4 PE (clone 7A3-3; 5 μL; cat# 130-091-647) from Miltenyl Biotech. Panel 2: anti-IL-2 Alexa Fluor 488 (clone MQ1-17H12; 3 μL; cat# 500314), anti-CD8a PerCP Cy5.5 (clone RPA-T8; 3 μL; cat# 301032), anti-CD4 BV421 (clone OKT4; 2.5 μL; cat# 317434), and anti-IFNγ Alexa 647 (clone 4S.B3; 3 μL; cat# 502516) from Biolegend; anti-CD3 BV605 (clone SP34-2; 2 μL; cat# 562994) from BD Biosciences; anti-IL-4 PE (clone 7A3-3; 5 μL; cat# 130-091-647) from Miltenyl Biotech; anti-IL-17a PE-efluor 610 (clone eBio64DEC17; 3 μL; cat# 61-7179-42), anti-TNFα PE-Cyanine7 (clone Mab11; 0.5 μL; cat# 25-7349-82), and Live Dead APC-Cy7 (1:1000; cat# 65086514) from Thermo Fisher Scientific. The frequency of SARS-CoV-2 specific CD4+ and CD8+ T cells producing single or multiple cytokines was determined after background subtraction. All samples were fixed with 4% paraformaldehyde and acquired within 24 hours of fixation. Acquisition of data was performed on a FACSymphony A5 (BD Biosciences) driven by FACS DiVa software and analyzed with FlowJo (version 10.7; Becton, Dickinson, and Company).
Isolation of Non-human primate neutrophils
Neutrophils were obtained from peripheral blood of SARS-Cov-2 infected Rhesus Macaques 5 days pre-infection and at days 4, 7, and 10 post-infection. Peripheral blood (0.5-1 ml) was collected using a citrate containing Vacutainer and the upper serum layer was removed. The red blood cell layer was lysed with 2 mL of Red Blood Cell Lysis Buffer (Cat# 11814389001, Roche) in a 15 mL tube. The tube was gently inverted for 10 minutes at room temperature and centrifuged at 500 x g for 7 minutes at room temperature. This step was repeated gently inverting for 5 minutes. Following centrifugation, the cell pellet was re-suspended in a final volume of 2 mL of 1x PBS/EDTA buffer gently. Cells were centrifuged at 500 x g for 7 mins a room temperature and the leukocyte pellet was re-suspended in 1 mL 1x PBS/EDTA buffer and carefully overlaid onto 3 mL of 65% Percoll/EDTA solution. The Percoll cell gradient was centrifuged at 400 x g for 20 mins at room temperature with the brake turned off. The neutrophil cell layer was collected, re-suspended/washed with 5 mL of 1x PBS/EDTA buffer and centrifuged at 500 x g for 10 minutes at room temperature. The neutrophil cellular pellet was re-suspended in RPMI 1640 media. Purification of the cell fragment was confirmed using flow cytometry and Wright Giemsa staining.
Quantification of extracellular DNA using SYTOX green Assay
Abundance of extracellular DNA, a surrogate of NETs, was quantified using the SYTOX green assay. Freshly, isolated non-human primate neutrophils were plated onto a 96-well plate at a density of 105 cells per well in 100 μL RPMI 1640 media then stimulated with 50 μg/mL LPS to induce NET formation. SYTOX green dye (5 μM, #S7020; Invitrogen, Carlsbad, CA) was added to each well and the fluorescence intensity was read with a filter setting at 485-nm excitation/525-nm emission using a Synergy H1 Microplate Reader and Gene5 software (Biotek, Winooski, VT). A fluorescence reading was collected every 15 mins for a total of 2 hours at 37°C. Images of the fluorescent cells were immediately taken using a fluorescent microscope (Olympus).
Staining of citH3
Paraffin-embedded lung sections were subjected to deparaffinization followed by heat induced antigen retrieval in 10 mM sodium citrate buffer (pH 6.0). Sections were blocked with 10% goat serum in 1x PBS for 1 hour. Primary antibody staining was performed for citrullinated H3 (Cayman Chemical, Cat. No. 17939, 1:50) overnight at 4°C. Slides were then incubated with Alexa Fluor 633 anti-mouse IgG secondary antibody (Thermo Fisher Scientific, Cat. No. A21052, 1:1000) for 90 mins at room temperature. Images were taken at 20x objective using a Zeiss LSM 800 Airyscan laser scanning confocal microscope.
Quantification of citrullinated H3
We quantified citrullinated histone 3 using an ELISA kit (Cayman Cat # 501620) with the antibody clone 11D3 per the manufacturer’s instructions. In short, 100 uL sample or standard was added in duplicate to a pre-coated 96-well plate and incubated for 2 hr on an orbital shaker. All steps were performed at room temperature. After 4 washes with the kit’s wash buffer, 100 uL per well horseradish peroxidase (HRP) conjugate working solution was added and the plate incubated for 1 hour on an orbital shaker. Then the plate was washed 4 times again and 100uL 3,3′,5,5′-Tetramethylbenzidine (TMB) solution was added per well then incubated for 30 minutes on an orbital shaker, followed by addition of 100 uL HRP stop solution. The plate was read at 450 nm absorbance using a microplate reader and the amount of citrullinated H3 quantified using the standards.
Quantification and statistical analysis
All statistical analyses were performed two-sided with p values ≤ 0.05 deemed significant. Ranges of significance were graphically annotated as follows: ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001. Due to the low number of animals included in our study, p values ≤ 0.1 have been indicated in the graphs. Analyses, unless otherwise noted, were performed with Prism version 8 (GraphPad).
Acknowledgments
We thank the Yerkes National Primate Research Center (YNPRC) Division of Animal Resources, especially Stephanie Ehnert, Stacey Weissman, Denise Bonenberger, John M. Wambua, Dominic M. D’Urso, Racquel Sampson-Harley, and Kalpana Patel in Research Resources for providing support in animal care. We thank Nichole Arnett for assistance with clinical pathology assays and Kevin Nguyen for additional laboratory processing. We thank Dr. Vincent Marconi for discussions and critical reading of the manuscript. Pharmaceutical-grade baricitinib was commercially obtained, provided free of charge by Dr. Schinazi, and it was found to be >99% pure by LC-MS-MS. This study was supported by an Emory University COVID-19 Molecules and Pathogens to Populations and Pandemics Initiative Seed Grant to M. Paiardini, A.P., and R.F.S.; by YNPRC Coronavirus Pilot Research Project Program grant to M. Paiardini under award P51 OD11132; and by Fast Grants Award #2144 to M. Paiardini. This work was additionally funded by the National Institute of Allergy and Infectious Diseases (NIAID, NIH) under awards R37AI141258 and R01AI116379 to M. Paiardini, R01MH116695 to R.F.S, R01AI143411, R01HL140223 to R.D.L., and R01AI149672 to J.D.E., and U24AI120134 to S.E.B. Support for this work was also provided by award NIH Office of Research Infrastructure Programs (ORIP) P51OD11132 to YNPRC, P51OD011092 to ONPRC, 1S10OD025002-01 to the Integrated Pathology Core/ONPRC, NIAID award P30 AI050409 to the Center for AIDS Research (CFAR) at Emory University, and contract Nr. 75N9301900065 (to D.W.). Next-generation sequencing services were provided by the Yerkes NHP Genomics Core, which is supported in part by NIH P51OD011132. Sequencing data were acquired on an Illumina NovaSeq6000 funded by NIH S10OD026799 to S.E.B. Pictorial illustrations were created with BioRender.com. The content of this publication does not necessarily reflect the views or policies of the U.S. Department of Health and Human Services nor does it imply endorsement of organizations or commercial products.
Author contributions
Conceptualization, T.N.H., M. Pino, J.L.H., G.S., A.P., S.E.B., R.F.S., and M. Paiardini; Methodology, T.N.H., M. Pino, A.K.B., E.G.V., N.K., K.B.-S., A.A.U., Z.S., G.K.T., K.L.P., S.G., S.K., S.T., O.M.D., K.A.C., M.N.S., L.W., P.D.F., J.W., A.P., S.P.K., C.E.S., S.W., H.A., E.A.M., M.Y.H.L., K.Z., S.T., T.R.H., E.N.B., S.P.R., and T.H.V.; Formal Analysis, T.N.H., M. Pino, Z.S., E.G.V., A.K.B., K.B.-S., A.A.U., G.K.T., S.G., S.K., S.T., P.D.F., J.W., A.P., S.P.K., S.P.R., and T.H.V; Investigation, T.N.H., M. Pino, E.G.V., J.C., S.M.J., J.S.W., F.C.-S., R.L.S., R.D.L., A.P., S.P.R., R.P.S., and T.H.V.; Resources, D.W., R.F.S., S.E.B., and M. Paiardini; Writing – Original Draft, T.N.H., M. Pino, J.L.H., and M. Paiardini; Writing – Review & Editing, T.N.H., M. Pino, J.L.H., S.E.B., and M. Paiardini; Visualization, T.N.H., M. Pino, A.K.B., A.A.U., G.K.T., Z.S., and E.G.V.; Supervision, R.F.S., S.E.B., and M. Paiardini; Funding Acquisition, A.P., S.E.B., R.F.S., and M. Paiardini.
Declaration of interests
R.F.S. served as an unpaid consultant for Eli Lilly whose drugs are being evaluated in the research described in this paper. In addition, R.F.S. owns shares in Eli Lilly. The terms of this arrangement have been reviewed and approved by Emory University in accordance with its conflict of interest policies. Eli Lilly had no role in the design of this study and did not have any role during its execution, analyses, interpretation of the data, or decision to submit results. All other authors do not have any conflicts to declare.
Published: November 10, 2021
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.cell.2020.11.007.
Supplemental Information
References
- Aran D., Looney A.P., Liu L., Wu E., Fong V., Hsu A., Chak S., Naikawadi R.P., Wolters P.J., Abate A.R. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol. 2019;20:163–172. doi: 10.1038/s41590-018-0276-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arunachalam P.S., Wimmers F., Mok C.K.P., Perera R.A.P.M., Scott M., Hagan T., Sigal N., Feng Y., Bristow L., Tak-Yin Tsang O. Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science. 2020;369:1210–1220. doi: 10.1126/science.abc6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bost P., Giladi A., Liu Y., Bendjelal Y., Xu G., David E., Blecher-Gonen R., Cohen M., Medaglia C., Li H. Host-Viral Infection Maps Reveal Signatures of Severe COVID-19 Patients. Cell. 2020;181:1475–1488. doi: 10.1016/j.cell.2020.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronte V., Ugel S., Tinazzi E., Vella A., De Sanctis F., Cane S., Batani V., Trovato R., Fiore A., Petrova V. Baricitinib restrains the immune dysregulation in severe COVID-19 patients. J. Clin. Invest. 2020 doi: 10.1172/JCI141772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantini F., Niccoli L., Matarrese D., Nicastri E., Stobbione P., Goletti D. Baricitinib therapy in COVID-19: A pilot study on safety and clinical impact. J. Infect. 2020;81:318–356. doi: 10.1016/j.jinf.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrashekar A., Liu J., Martinot A.J., McMahan K., Mercado N.B., Peter L., Tostanoski L.H., Yu J., Maliga Z., Nekorchuk M. SARS-CoV-2 infection protects against rechallenge in rhesus macaques. Science. 2020;369:812–817. doi: 10.1126/science.abc4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chertow D.S., Kindrachuk J., Sheng Z.M., Pujanauski L.M., Cooper K., Nogee D., Claire M.S., Solomon J., Perry D., Sayre P. Influenza A and methicillin-resistant Staphylococcus aureus co-infection in rhesus macaques - A model of severe pneumonia. Antiviral Res. 2016;129:120–129. doi: 10.1016/j.antiviral.2016.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua R.L., Lukassen S., Trump S., Hennig B.P., Wendisch D., Pott F., Debnath O., Thürmann L., Kurth F., Völker M.T. COVID-19 severity correlates with airway epithelium-immune cell interactions identified by single-cell analysis. Nat. Biotechnol. 2020;38:970–979. doi: 10.1038/s41587-020-0602-4. [DOI] [PubMed] [Google Scholar]
- Coronavirus Disease . 2019. Situation report number 148. Accessed August 18, 2020.https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200616-covid-19-sitrep-148-draft.pdf?sfvrsn=9b2015e9_2 [Google Scholar]
- Davidson A.D., Williamson M.K., Lewis S., Shoemark D., Carroll M.W., Heesom K.J., Zambon M., Ellis J., Lewis P.A., Hiscox J.A., Matthews D.A. Characterisation of the transcriptome and proteome of SARS-CoV-2 reveals a cell passage induced in-frame deletion of the furin-like cleavage site from the spike glycoprotein. Genome Med. 2020;12:68. doi: 10.1186/s13073-020-00763-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lang A., Baas T., Teal T., Leijten L.M., Rain B., Osterhaus A.D., Haagmans B.L., Katze M.G. Functional genomics highlights differential induction of antiviral pathways in the lungs of SARS-CoV-infected macaques. PLoS Pathog. 2007;3:e112. doi: 10.1371/journal.ppat.0030112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estes J.D., Wong S.W., Brenchley J.M. Nonhuman primate models of human viral infections. Nat. Rev. Immunol. 2018;18:390–404. doi: 10.1038/s41577-018-0005-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finak G., McDavid A., Yajima M., Deng J., Gersuk V., Shalek A.K., Slichter C.K., Miller H.W., McElrath M.J., Prlic M. MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 2015;16:278. doi: 10.1186/s13059-015-0844-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., Rawlings S.A., Sutherland A., Premkumar L., Jadi R.S. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell. 2020;181:1489–1501. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z., Eils R., Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- Guan W.J., Ni Z.Y., Hu Y., Liang W.H., Ou C.Q., He J.X., Liu L., Shan H., Lei C.L., Hui D.S.C., China Medical Treatment Expert Group for Covid-19 Clinical Characteristics of Coronavirus Disease 2019 in China. N. Engl. J. Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper J., Gordon S., Chan C.N., Wang H., Lindemuth E., Galardi C., Falcinelli S.D., Raines S.L.M., Read J.L., Nguyen K. CTLA-4 and PD-1 dual blockade induces SIV reactivation without control of rebound after antiretroviral therapy interruption. Nat. Med. 2020;26:519–528. doi: 10.1038/s41591-020-0782-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasturi S.P., Rasheed M.A.U., Havenar-Daughton C., Pham M., Legere T., Sher Z.J., Kovalenkov Y., Gumber S., Huang J.Y., Gottardo R. 3M-052, a synthetic TLR-7/8 agonist, induces durable HIV-1 envelope-specific plasma cells and humoral immunity in nonhuman primates. Sci. Immunol. 2020;5 doi: 10.1126/sciimmunol.abb1025. Published online June 19, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keystone E.C., Taylor P.C., Drescher E., Schlichting D.E., Beattie S.D., Berclaz P.Y., Lee C.H., Fidelus-Gort R.K., Luchi M.E., Rooney T.P. Safety and efficacy of baricitinib at 24 weeks in patients with rheumatoid arthritis who have had an inadequate response to methotrexate. Ann. Rheum. Dis. 2015;74:333–340. doi: 10.1136/annrheumdis-2014-206478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuri-Cervantes L., Pampena M.B., Meng W., Rosenfeld A.M., Ittner C.A.G., Weisman A.R., Agyekum R.S., Mathew D., Baxter A.E., Vella L.A. Comprehensive mapping of immune perturbations associated with severe COVID-19. Sci. Immunol. 2020;5:eabd7114. doi: 10.1126/sciimmunol.abd7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laing A.G., Lorenc A., Del Molino Del Barrio I., Das A., Fish M., Monin L., Muñoz-Ruiz M., McKenzie D.R., Hayday T.S., Francos-Quijorna I. A dynamic COVID-19 immune signature includes associations with poor prognosis. Nat. Med. 2020;26:1623–1635. doi: 10.1038/s41591-020-1038-6. [DOI] [PubMed] [Google Scholar]
- Liao M., Liu Y., Yuan J., Wen Y., Xu G., Zhao J., Cheng L., Li J., Wang X., Wang F. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 2020;26:842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- Loo Y.M., Gale M., Jr. Immune signaling by RIG-I-like receptors. Immunity. 2011;34:680–692. doi: 10.1016/j.immuni.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo Y.M., Fornek J., Crochet N., Bajwa G., Perwitasari O., Martinez-Sobrido L., Akira S., Gill M.A., García-Sastre A., Katze M.G., Gale M., Jr. Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J. Virol. 2008;82:335–345. doi: 10.1128/JVI.01080-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGary C.S., Deleage C., Harper J., Micci L., Ribeiro S.P., Paganini S., Kuri-Cervantes L., Benne C., Ryan E.S., Balderas R. CTLA-4(+)PD-1(-) Memory CD4(+) T Cells Critically Contribute to Viral Persistence in Antiretroviral Therapy-Suppressed, SIV-Infected Rhesus Macaques. Immunity. 2017;47:776–788. doi: 10.1016/j.immuni.2017.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis C.S., Murrow L.M., Gartner Z.J. DoubletFinder: doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst. 2019;8:329–337. doi: 10.1016/j.cels.2019.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInnes L., Healy J., Melville J. Umap: Uniform manifold approximation and projection for dimension reduction. arXiv. 2018:180203426. [Google Scholar]
- Micci L., Ryan E.S., Fromentin R., Bosinger S.E., Harper J.L., He T., Paganini S., Easley K.A., Chahroudi A., Benne C. Interleukin-21 combined with ART reduces inflammation and viral reservoir in SIV-infected macaques. J. Clin. Invest. 2015;125:4497–4513. doi: 10.1172/JCI81400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha V.K., Lindgren C.M., Eriksson K.F., Subramanian A., Sihag S., Lehar J., Puigserver P., Carlsson E., Ridderstrale M., Laurila E. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nature genetics. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- Munster V.J., Feldmann F., Williamson B.N., van Doremalen N., Pérez-Pérez L., Schulz J., Meade-White K., Okumura A., Callison J., Brumbaugh B. Respiratory disease in rhesus macaques inoculated with SARS-CoV-2. Nature. 2020;585:268–272. doi: 10.1038/s41586-020-2324-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nganou-Makamdop K., Billingsley J.M., Yaffe Z., O’Connor G., Tharp G.K., Ransier A., Laboune F., Matus-Nicodemos R., Lerner A., Gharu L. Type I IFN signaling blockade by a PASylated antagonist during chronic SIV infection suppresses specific inflammatory pathways but does not alter T cell activation or virus replication. PLoS Pathog. 2018;14:e1007246. doi: 10.1371/journal.ppat.1007246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palesch D., Bosinger S.E., Tharp G.K., Vanderford T.H., Paiardini M., Chahroudi A., Johnson Z.P., Kirchhoff F., Hahn B.H., Norgren R.B. Sooty mangabey genome sequence provides insight into AIDS resistance in a natural SIV host. Nature. 2018;553:77–81. doi: 10.1038/nature25140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pino M., Paganini S., Deleage C., Padhan K., Harper J.L., King C.T., Micci L., Cervasi B., Mudd J.C., Gill K.P. Fingolimod retains cytolytic T cells and limits T follicular helper cell infection in lymphoid sites of SIV persistence. PLoS Pathog. 2019;15:e1008081. doi: 10.1371/journal.ppat.1008081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin C., Zhou L., Hu Z., Zhang S., Yang S., Tao Y., Xie C., Ma K., Shang K., Wang W., Tian D.S. Dysregulation of Immune Response in Patients With Coronavirus 2019 (COVID-19) in Wuhan, China. Clin. Infect. Dis. 2020;71:762–768. doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson P., Griffin I., Tucker C., Smith D., Oechsle O., Phelan A., Rawling M., Savory E., Stebbing J. Baricitinib as potential treatment for 2019-nCoV acute respiratory disease. Lancet. 2020;395:e30–e31. doi: 10.1016/S0140-6736(20)30304-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockx B., Kuiken T., Herfst S., Bestebroer T., Lamers M.M., Oude Munnink B.B., de Meulder D., van Amerongen G., van den Brand J., Okba N.M.A. Comparative pathogenesis of COVID-19, MERS, and SARS in a nonhuman primate model. Science. 2020;368:1012–1015. doi: 10.1126/science.abb7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Q., Yang K., Wang W., Jiang L., Song J. Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from Wuhan, China. Intensive Care Med. 2020;46:846–848. doi: 10.1007/s00134-020-05991-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandler N.G., Bosinger S.E., Estes J.D., Zhu R.T., Tharp G.K., Boritz E., Levin D., Wijeyesinghe S., Makamdop K.N., del Prete G.Q. Type I interferon responses in rhesus macaques prevent SIV infection and slow disease progression. Nature. 2014;511:601–605. doi: 10.1038/nature13554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satija R., Butler A., Hoffman P. 2018. Seurat: Tools for Single Cell Genomics. R package version 2. [Google Scholar]
- Schulte-Schrepping J., Reusch N., Paclik D., Baßler K., Schlickeiser S., Zhang B., Krämer B., Krammer T., Brumhard S., Bonaguro L., Deutsche COVID-19 OMICS Initiative (DeCOI) Severe COVID-19 Is Marked by a Dysregulated Myeloid Cell Compartment. Cell. 2020;182:1419–1440. doi: 10.1016/j.cell.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievert C. Interactive web-based data visualization with R, plotly, and shiny. CRC Press, Taylor and Francis Group; Boca Raton, FL: 2020. [Google Scholar]
- Silvin A., Chapuis N., Dunsmore G., Goubet A.G., Dubuisson A., Derosa L., Almire C., Hénon C., Kosmider O., Droin N. Elevated Calprotectin and Abnormal Myeloid Cell Subsets Discriminate Severe from Mild COVID-19. Cell. 2020;182:1401–1418. doi: 10.1016/j.cell.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skendros P., Mitsios A., Chrysanthopoulou A., Mastellos D.C., Metallidis S., Rafailidis P., Ntinopoulou M., Sertaridou E., Tsironidou V., Tsigalou C. Complement and tissue factor-enriched neutrophil extracellular traps are key drivers in COVID-19 immunothrombosis. J. Clin. Invest. 2020;130:6151–6157. doi: 10.1172/JCI141374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbing J., Krishnan V., de Bono S., Ottaviani S., Casalini G., Richardson P.J., Monteil V., Lauschke V.M., Mirazimi A., Youhanna S., Sacco Baricitinib Study Group Mechanism of baricitinib supports artificial intelligence-predicted testing in COVID-19 patients. EMBO Mol. Med. 2020;12:e12697. doi: 10.15252/emmm.202012697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., 3rd, Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive Integration of Single-Cell Data. Cell. 2019;177:1888–1902.e1821. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang N.L., Chan P.K., Wong C.K., To K.F., Wu A.K., Sung Y.M., Hui D.S., Sung J.J., Lam C.W. Early enhanced expression of interferon-inducible protein-10 (CXCL-10) and other chemokines predicts adverse outcome in severe acute respiratory syndrome. Clin. Chem. 2005;51:2333–2340. doi: 10.1373/clinchem.2005.054460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titanji B.K., Farley M.M., Mehta A., Connor-Schuler R., Moanna A., Cribbs S.K., O’Shea J., DeSilva K., Chan B., Edwards A. Use of Baricitinib in Patients With Moderate to Severe Coronavirus Disease 2019. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa879. Published online June 29, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waggoner J.J., Stittleburg V., Pond R., Saklawi Y., Sahoo M.K., Babiker A., Hussaini L., Kraft C.S., Pinsky B.A., Anderson E.J., Rouphael N. Triplex Real-Time RT-PCR for Severe Acute Respiratory Syndrome Coronavirus 2. Emerg. Infect. Dis. 2020;26:1633–1635. doi: 10.3201/eid2607.201285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. 2016. ggplot2: elegant graphics for data analysis (springer) [Google Scholar]
- Williamson B.N., Feldmann F., Schwarz B., Meade-White K., Porter D.P., Schulz J., van Doremalen N., Leighton I., Yinda C.K., Pérez-Pérez L. Clinical benefit of remdesivir in rhesus macaques infected with SARS-CoV-2. Nature. 2020;585:273–276. doi: 10.1038/s41586-020-2423-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Tostanoski L.H., Peter L., Mercado N.B., McMahan K., Mahrokhian S.H., Nkolola J.P., Liu J., Li Z., Chandrashekar A. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science. 2020;369:806–811. doi: 10.1126/science.abc6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zevini A., Olagnier D., Hiscott J. Crosstalk between Cytoplasmic RIG-I and STING Sensing Pathways. Trends Immunol. 2017;38:194–205. doi: 10.1016/j.it.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G.X., Terry J.M., Belgrader P., Ryvkin P., Bent Z.W., Wilson R., Ziraldo S.B., Wheeler T.D., McDermott G.P., Zhu J. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 2017;8:14049. doi: 10.1038/ncomms14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Source data supporting this work are available from the corresponding author upon reasonable request. The following sequencing data have been deposited in GenBank: SARS-CoV-2 viral stock (accession GEO: GSE162247). Data tables for expression counts for bulk and single-cell RNA-Seq for BAL are deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO accession GSE159214. Custom scripts and supporting documentation on the RNA-Seq analyses will be made available at https://github.com/BosingerLab/.