Abstract
The sudden outbreak of novel coronavirus 2019 (COVID-19) increased the diagnostic burden of radiologists. In the time of an epidemic crisis, we hope artificial intelligence (AI) to reduce physician workload in regions with the outbreak, and improve the diagnosis accuracy for physicians before they could acquire enough experience with the new disease. In this paper, we present our experience in building and deploying an AI system that automatically analyzes CT images and provides the probability of infection to rapidly detect COVID-19 pneumonia. The proposed system which consists of classification and segmentation will save about 30%–40% of the detection time for physicians and promote the performance of COVID-19 detection. Specifically, working in an interdisciplinary team of over 30 people with medical and/or AI background, geographically distributed in Beijing and Wuhan, we are able to overcome a series of challenges (e.g. data discrepancy, testing time-effectiveness of model, data security, etc.) in this particular situation and deploy the system in four weeks. In addition, since the proposed AI system provides the priority of each CT image with probability of infection, the physicians can confirm and segregate the infected patients in time. Using 1,136 training cases (723 positives for COVID-19) from five hospitals, we are able to achieve a sensitivity of 0.974 and specificity of 0.922 on the test dataset, which included a variety of pulmonary diseases.
Keywords: COVID-19, Deep learning, Neural network, Medical assistance system, Classification, Segmentation
Highlights
-
•
We build a system for detecting COVID-19 infection regions.
-
•
We build a dataset of COVID-19.
-
•
It is easy to distinguish pneumonia from healthy cases.
1. Introduction
COVID-19 started to spread in January 2020. Up to early March 2020, it has infected over 100,000 people worldwide [1]. The virus is harbored most commonly with little or no symptoms, but can also lead to a rapidly progressive and often fatal pneumonia in 2%–8% of those infected. COVID-19 causes acute respiratory distress syndrome on patients [2], [3]. Laboratory confirmation of SARS-CoV-2 is performed with a virus-specific RT-PCR, but it has several challenges, including high false negative rates, delays in processing, variabilities in test techniques, and sensitivity sometimes reported as low as 60%–70%.
The CT image can show the characteristics of each stage of disease detection and evolution. Although many challenges are still existed with rapid diagnosis of COVID-19, there are some characteristic with typical features. The preliminary prospective analysis by Huang et al. [2] showed that all 41 patients in the study had abnormal chest CT, with bilateral ground-glass shape lung opacities in subpleural areas of the lungs. Many recent studies [4], [5], [6], [7], [8] also viewed chest CT as a low-cost, accurate and efficient method for novel coronavirus pneumonia diagnosis. The official guidelines for COVID-19 diagnosis and treatment (7th edition) by China’s National Health Commission [9] also listed chest CT result as one of the main clinical features. CT evaluation has been an important approach to evaluate the patients with suspected or confirmed COVID-19 in multiple centers in Wuhan China and northern Italy.
The sudden outbreak of COVID-19 overwhelmed health care facilities in the Wuhan area. Hospitals in Wuhan had to invest significant resources to screen suspected patients, further increasing the burden of radiologists. As Ji et al. [10] pointed out, there was a significant positive correlation between COVID-19 mortality and health-care burden. It was essential to reduce the workload of clinicians and radiologists and enable patients to get early diagnoses and timely treatments. In a large country like China, it is nearly impossible to train such a large number of experienced physicians in time to screen this novel disease, especially in regions without an outbreak yet. To handle this dilemma, in this research, we present our experience in developing and deploying an artificial intelligence (AI) based method to assist novel coronavirus pneumonia screening using CT imaging.
At present, the physicians often obtain a ID from the Hospital Information System (HIS), then assess the results of CT images from the Picture archiving and communication systems (PACS), and give a conclusion of CT images to HIS. Due to the rapid increase in number of new and suspected COVID-19 cases, we expect to detect the CT images in order of importance (i.e. high risk patient). However, since the IDs from HIS are assigned with capturing time, the existing the AI system can only perform with these IDs on PACS. Therefore, it still spends a large amount of time to detect COVID-19 patients, and affects the treatment time of severely infected COVID-19 patients. In this paper, we introduce a automatically AI system that can provide the probability of infection and the ranked IDs. Specifically, the proposed system which consists of classification and segmentation will save about 30%–40% of the detection time for physicians and promote the performance of COVID-19 detection. The classification subsystem tries to give the probability of getting COVID-19 for each sample, the segmentation subsystem will highlight the position of the suspected area.
In addition, training AI models requires a lot of samples. However, at the beginning of a new epidemic, there were not many positive cases confirmed by nucleic acid test (NAT). To build a dataset for detect COVID-19, we collect 877 samples from 5 hospitals. All imaging data come from the COVID-19 patients confirmed by NAT who underwent lung CT scans. This requirement also ensured that the image data had the diagnostic characteristics. Based on these samples, we employ experienced annotators to annotate all the samples. While it is easy to distinguish pneumonia from healthy cases, it is non-trivial for the model to distinguish COVID-19 from other pulmonary diseases, which is the top clinical requirement. Thus, we add other pulmonary diseases in the proposed dataset. Using the dataset, we train and evaluate several deep learning based models to detect and segment the COVID-19 regions. Finally, the construction of the AI model included four stages: (1) Data collection; (2) Data annotation; (3) Model training and evaluation, and (4) Model deployment.
The contributions of this paper can be summarized as follows:
-
•
In this paper, we present our experience in building and deploying an AI system that automatically analyzes CT images to rapidly detect COVID-19 pneumonia.
-
•
We build a new dataset on top of real images which labeled the contour and infection regions to promote the development of COVID-19 detection.
-
•
The proposed AI system can reduce physician workload in regions with the outbreak based on the disease priority, and improve the diagnosis accuracy for physicians.
-
•
The proposed AI systems have deployed in 16 hospitals and can provide professional deployment service on-premise of the hospitals.
Starting from the introduction in Section 1, the paper is organized as follows: In Section 2, we give related work on data acquisition, lung segmentation and AI-assisted diagnosis. In Section 3 describes the details of the proposed method. The experimental results and ablation studies are presented in Section 4. In Section 5 describes the advantages and disadvantages of the method. The paper is concluded in Section 6.
2. Related work
In this section we introduce some related studies on AI-assisted diagnosis techniques towards COVID-19, including data collection, medical images segmentation, and diagnosis.
2.1. Data acquisition
The very first step of building an AI-assisted diagnosis system for COVID-19 is image acquisition, in which Chest X-ray and CT images are most widely used. And there are more applications using CT images for COVID-19 diagnosis [11], [12], [13], [14], since the analysis and segmentation of CT images are always more precise and efficient than X-ray images.
Recently, there has been some progress on COVID-19 dataset construction. Zhao et al. [15] build a COVID-CT dataset which includes 288 CT slices of confirmed COVID-19 patients, collected from about 700 COVID-19 related publications on medRxiv and bioRxiv. The Coronacases Initiative releases some CT images of 10 confirmed COVID-19 patients on its website [16]. COVID-19 CT segmentation dataset [17] is also a publicly available dataset. It contains 100 axial CT slices of 60 confirmed COVID-19 patients, and all the CT slices are manually annotated with segmentation labels. Besides, Cohen et al. [18] collect 123 frontal view X-rays from some publications and websites and build the COVID-19 Image Data Collection.
Some efforts have been made on contactless data acquisition to reduce the risk of infection during COVID-19 pandemic [19], [20], [21]. For example, an automated scanning workflow equipped with mobile CT platform is built [19], in which the mobile CT platform has more flexible access to patients. During CT data acquisition, the positioning and scanning of patients are operated remotely by a technician.
2.2. Lung segmentation
The medical image segmentation and deep neural network [22], [23], [24], [25], [26], [27] plays an important role in AI-assisted COVID-19 analysis in many works. It highlights the regions of interest (ROIs) in CT or X-ray images for further examination. The segmentation tasks in COVID-19 applications can be divided into two groups: Lung region segmentation and lung lesion segmentation. During lung region segmentation, the whole lung region is separated from background, while in lung lesion segmentation tasks the lesion areas are distinguished from other lung areas. Lung region segmentation is often executed as a preprocessing step in CT segmentation tasks [28], [29], [30], in order to decrease the difficulty of lesion segmentation.
Several widely-used segmentation models are applied in COVID-19 diagnosis systems such as U-Net [31], V-Net [14] and U-Net++ [32]. Among them U-Net is a fully convolutional network in which skip connection is employed to fuse information of multi-resolution layers. V-Net adopts a volumetric, fully convolutional neural network and achieves 3D image segmentation. VB-Net [33] replaces the conventional convolutional layers inside down block and up block with the bottleneck, to achieve promising and efficient segmentation results. U-Net++ is composed of deeply-supervised encoder and decoder sub-networks. Nested skip connections are equipped to connect the two sub-networks, which could increase the segmentation performance.
In AI-assisted COVID-19 analysis applications, Li et al. [12] develop a U-Net based segmentation system to distinguish COVID-19 from community-acquired pneumonia on CT images. Qi et al. [34] also build a U-Net based segmentation model to separate lung lesions and extract the radiologic characteristics in order to predict the hospital stay of a patient. Shan et al. [35] propose a VB-Net based segmentation system to segment lung, lung lobes and lung lesion regions. And the segmentation results can also provide accurate quantification data for further study towards COVID-19. Chen et al. [36] train a U-Net++ based segmentation model to provide COVID-19 related lesions.
2.3. AI-assisted diagnosis
Medical imaging AI systems such as disease classification and segmentation are increasingly inspired and transformed from computer vision based AI systems. Morteza et al. [37] propose a data-driven model that recommends the necessary set of diagnostic procedures based on the patients’ most recent clinical record extracted from the Electronic Health Record (EHR). This has the potential to enable health systems expand timely access to initial medical specialty diagnostic workups for patients. Gu et al. [38] propose a series of collaborative techniques to engage human pathologists with AI given AI’s capabilities and limitations, based on which they prototype Impetus - a tool where an AI takes various degrees of initiatives to provide various forms of assistance to a pathologist in detecting tumors from histological slides. Samaniego et al. [39] propose a blockchain-based solution to enable distributed data access management in Computer-Aided Diagnosis (CAD) systems. This solution has been developed as a distributed application (DApp) using Ethereum in a consortium network. Li et al. [40] develop a visual analytics system that compares multiple models’ prediction criteria and evaluates their consistency. With this system, users can generate knowledge on different models’ inner criteria and how confidently we can rely on each model’s prediction for a certain patient.
AI-assisted COVID-19 diagnosis based on CT and X-ray images could accelerate the diagnosis and decrease the burden of radiologists, thus is highly desired in COVID-19 pandemic. And a series of models which can distinguish COVID-19 from other pneumonia and diseases have been widely explored.
The AI-assisted diagnosis systems can be grouped into two categories, i.e., X-ray based screening COVID-19 systems and CT based screening COVID-19 systems. Among X-ray based AI-assisted systems, Ghoshal et al. [41] develop a Bayesian Convolutional Neural network to measure the diagnosis uncertainty of COVID-19 prediction. Narin et al. [42] develop three widely-used models, i.e., ResNet-50 [43], Inception-V3 [44], and Inception-ResNet-V2 [45], to detect COVID-19 lesion in X-ray images and among them ResNet-50 achieves the best classification performance. And Zhang et al. [46] present a ResNet based model to detect COVID-19 lesions from X-ray images. This model can provide an anomaly score to help optimize the classification between COVID-19 and non-COVID-19.
As X-ray images are always the typical common-used imaging modality in pulmonary diseases diagnosis, they are usually not as sensitive as 3D CT images. Besides, the positive COVID-19 X-ray data in these studies is mainly from one online datasets [18] which only contains insufficient X-ray images from confirmed COVID-19 patients. And the lack of data could affect the generalization property of the diagnosis systems.
As for CT based AI-assisted diagnosis, a series of approaches with different frameworks are proposed. Some approaches employ a single model to determine the presence of COVID-19 disease or certain other diseases in CT images. Ying et al. [11] propose DeepPneumonia, which is a ResNet-50 based CT diagnosis system, to distinguish the COVID-19 patients from bacteria pneumonia patients and healthy people. Jin et al. [47] build a 2D CNN based model to segment the lung and then identify slices of COVID-19 cases. Li et al. [12] propose COVNet, which is a ResNet-50 based model employed on 2D slices with shared weights, to discriminate COVID-19 from community-acquired pneumonia and non-pneumonia. Shi et al. [13] apply VB-Net [33] to segment the CT images into the left or right lung, 5 lung lobes, and 18 pulmonary segments, then select hand-crafted features to train a random forest model to make diagnosis.
Some other works follow a segmentation and classification mechanism. Taking some approaches for instance, Xu et al. [48] build a deep learning model based on V-Net [14] and ResNet-18 to classify COVID-19 patients, Influenza-A patients, and healthy people. In this model, lung lesion region in CT image is extracted using V-Net first, then the type of lesion region is determined via ResNet-18. Zheng et al. [49] propose DeCoVNet, which is a combination of a U-Net [31] model and a 3D CNN model. The U-Net model is used for lung segmentation, then the segmentation results are input into the 3D CNN model for predicting the probability of existence of COVID-19.
3. Methods
3.1. Overview
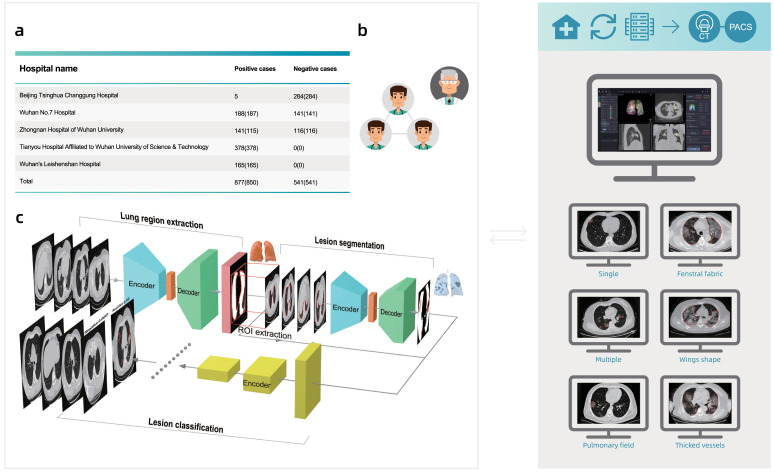
As shown in Fig. 1, the construction of the AI model included four stages: (1) Data collection; (2) Data annotation; (3) Model training and evaluation, and (4) Model deployment. As we accumulated data, we iterated through the stages to continuously improve model performance.
Fig. 1.
The framework of the proposed system.
3.2. Data collection
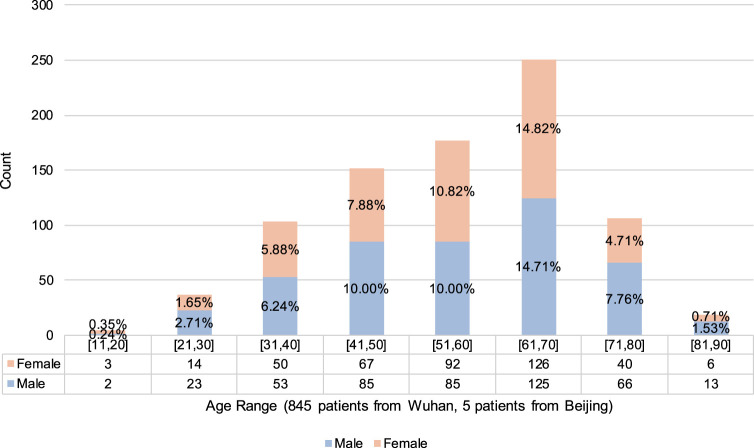
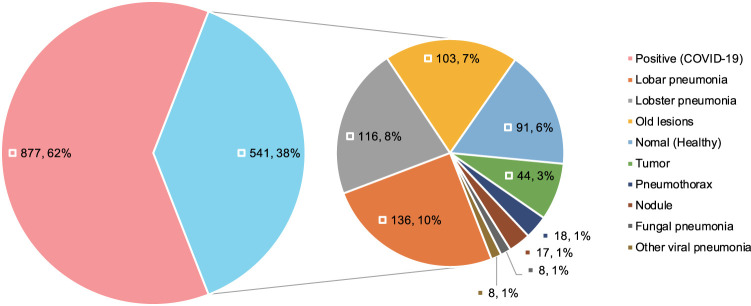
Our dataset was obtained from 5 hospitals (see Table 1). Most of the 877 positive cases were from hospitals in Wuhan, while half of the 541 negative cases were from hospitals in Beijing. Our positive samples were all collected from confirmed patients, following China’s national diagnostic and treatment guidelines at the time of the diagnosis, which required positive results in NAT. The positive cases offered a good sample of confirmed cases in Wuhan, covering different age and gender groups (see Fig. 4). We collect many CT images with other types of pneumonias, e.g. common pneumonia patients, viral pneumonia patients, fungal pneumonia patients, tumor patients, emphysema patients, lung lesions patients. To choose the reasonable negative cases, we employ several senior physicians to manually confirm each case. Based on the experiences of senior physicians, they choose the negative cases which have similar characteristics with COVID-19. Finally, we also had 450 cases with other known lung diseases with CT imaging features similar to COVID-19 to some extent (see Fig. 5).
Table 1.
Case-level data source distribution of the dataset by hospital.
| Hospital name | Positive cases (#Patients) | Negative cases (#Patients) |
|---|---|---|
| Beijing Tsinghua Changgung Hospital | 5 (5) | 284 (284) |
| Wuhan No.7 Hospital | 188 (187) | 141 (141) |
| Zhongnan Hospital of Wuhan University | 141 (115) | 116 (116) |
| Tianyou Hospital Affiliated to Wuhan University of Science & Technology | 378 (378) | 0 (0) |
| Wuhan’s Leishenshan Hospital | 165 (165) | 0 (0) |
| Total | 877 (850) | 541 (541) |
Fig. 4.
Patient-level age–gender distribution of positive cases.
Fig. 5.
Case-level clinical typing distribution of the dataset.
The hospitals used different models of CT equipment from different manufacturers (see Table 2). Due to the shortage of CT scanners for hospitals in Wuhan, slice thicknesses varied from 0.625 mm to 10 mm, with a majority (81%) under 2 mm. We believed this variety helped to improve the generalizability of our model in real deployment. In addition, we removed the personally identifiable information (PII) from all CT scans for patients’ privacy. We randomly divided the whole dataset into a training set and a test set for each model training (see Table 4).
Table 2.
Case-level data source distribution of the dataset by CT scanner models.
| Equipment model | Positive cases (#Patients) | Negative cases (#Patients) |
|---|---|---|
| UIH uCT 760 | 13 (10) | 195 (195) |
| UIH uCT 530 | 253 (252) | 0 (0) |
| GE Optima CT660 | 378 (378) | 27 (27) |
| GE Discovery CT750 HD | 0 (0) | 86 (86) |
| GE Discovery CT | 192 (176) | 0 (0) |
| GE BrightSpeed | 0 (0) | 3 (3) |
| SIEMENS SOMATOM Definition | 22 (19) | 0 (0) |
| SIEMENS Sensation Open | 2 (2) | 0 (0) |
| Philips iCT 256 | 0 (0) | 228 (228) |
| Philips Ingenuity CT | 17 (13) | 0 (0) |
| Philips Brilliance Big Bore | 0 (0) | 2 (2) |
| Total | 877 (850) | 541 (541) |
Table 4.
Case-level dataset division for each model training task.
| Training set |
Testing set |
|||||
|---|---|---|---|---|---|---|
| Positive cases (#Patients) | Negative (#Patients) |
Positive (#Patients) | Negative (#Patients) |
|||
| Healthy | Other diseases | Healthy | Other diseases | |||
| Lung region extraction | 361 (360) | 9 (9) | 27 (27) | 93 (93) | 0 (0) | 1 (1) |
| Lesion segmentation | 704 (680) | 7 (7) | 21 (21) | 168 (168) | 2 (2) | 5 (5) |
| Lesion classification | 723 (696) | 70 (70) | 343 (343) | 154 (154) | 21 (21) | 107 (107) |
3.3. Data annotation
To train the models, a team of six data annotators annotated the lesion regions (if there are any), lung boundaries, and the parts of the lungs for transverse section layers in all CT samples.
Saving time for radiologists was essential during the epidemic outbreak, so our data annotators performed most of the tasks, and we relied on a three-step quality inspection process to achieve reasonable accuracy for annotation. All of the annotators had radiology background, and we conducted a four-day hands-on training led by a senior radiologist with clinical experience of COVID-19 before they performed annotations.
Our three-step quality inspection process was the key to obtain high-quality annotations. We divided the six-annotator team into a group of four (Group A) and a group of two (Group B).
- Step 1
Group A made all the initial annotations, and Group B performed a back-to-back quality check, i.e., each of the two members in Group B checked all the annotations independently and then compared their results. The pass rate for this initial inspection was 80%. The cases that failed to pass mainly had minor errors in small lesion region missing or the inexact boundary shape.
- Step 2
Group A revised the annotations, and then Group B rechecked the annotations. This process continued until all of them passed the back-to-back quality test within the two-people group.
- Step 3
When a batch of data was annotated and passed the first two steps, senior radiologists randomly checked 30% of the revised annotations for each batch. We observed a pass rate of 100% in this step, showing reasonable annotation quality.
Of course, there might still be errors remaining, and we relied on the model training process to tolerate these random errors.
3.4. Pre-processing
We performed the following preprocessing steps before we used them for training and testing. (1) Since different samples had different resolutions and slice thicknesses, we first normalized them to (1, 1, 2.5) mm using standard interpolation algorithms (e.g. nearest neighbor interpolation, bilinear interpolation and cubic interpolation [50], [51]). Note that we use cubic interpolation to obtain better image effect. (2) We adjusted the window width (WW) and window level (WL) for each model, generating three image sets, each with a specific window setting. For brevity, we used the [min, max] interval format in programming for WW and WL. Specifically, we set them to [−150, 350] for the lung region segmentation model, and [−1024, 350] for both of the lesion segmentation and classification models. (3) We first ran the lung segmentation model to extract areas of the lungs from each image and used only the extraction results in the subsequent steps. (4) We normalized all the values to the range of [0, 1]. (5) We applied typical data augmentation techniques [52], [53] to increase the diversity of data. For example, we randomly flipped, panned, and zoomed images for more variety, which had been shown to improve the generalization ability for the trained model.
3.5. Model library
Our model was a combination of a segmentation model and a classification model. Specifically, we used the segmentation model to obtain the lung lesion regions, and then the classification model to determine whether it was COVID-19-like for each lesion region. We selected both models empirically by training and testing all models in our previously-developed model library.
For the segmentation task, we considered several widely-used segmentation models such as fully convolutional networks (FCN-8s) [54], U-Net [31], V-Net [14] and 3D U-Net++ [32].
FCN-8s [54] was a “fully convolutional” network in which all the fully connected layers were replaced by convolution layers. Thus, the input of FCN-8s could have arbitrary size. FCN-8s introduced a novel skip architecture to fuse information of multi-resolution layers. Specifically, upsampled feature maps from higher layers were combined with feature maps skipped from the encoder, to improve the spatial precision of the segmentation details.
Similar to FCN-8s, U-Net [31] was a variant of encoder– decoder architecture and employed skip connection as well. The encoder of U-Net employed multi-stage convolutions to capture context features, and the decoder used multi-stage convolutions to fuse the features. Skip connection was applied in every decoder stage to help recover the full spatial resolution of the network output, making U-Net more precise, and thus suitable for biomedical image segmentation.
V-Net [14] was a 3D image segmentation approach, where volumetric convolutions were applied instead of processing the input volumes slice-wise. V-Net adopted a volumetric, fully convolutional neural network and could be trained end-to-end. Based on the Dice coefficient between the predicted segmentation and the ground truth annotation, a novel objective function was introduced to cope with the imbalance between the number of foregrounds and background voxels.
3D U-Net++ [32] was an effective segmentation architecture, composed of deeply-supervised encoder and decoder sub- networks. Concretely, a series of nested, dense re-designed skip pathways connected the two sub-networks, which could reduce the semantic gap between the feature maps of the encoder and the decoder. Integrate the multi-scale information, the 3D U-Net++ model could simultaneously utilize the semantic information and the texture information to make the correct predictions. Besides, deep supervision enabled more accurate segmentation, particularly for lesion regions. Both re-designed skip pathways and deep supervision distinguished U-Net++ from U-Net, and assisted U-Net++ to effectively recover the fine details of the target objects in biomedical images. Also, allowing 3D inputs could capture inter-slice features and generate dense volumetric segmentation.
For all the segmentation models, we used patch size (i.e., the input image size to the model) of (256, 256, 128). The positive data for the segmentation models were those images with arbitrary lung lesion regions, regardless of whether the lesions were COVID-19 or not. Then the model made per-pixel predictions of whether the pixel was within the lung lesion region.
In the classification task, we evaluated some state-of-the-art classification models such as ResNet-50 [43], Inception networks [44], [45], [55], DPN-92 [56], and Attention ResNet-50 [57].
Residual network (ResNet) [43] was a widely-used deep learning model that introduced a deep residual learning framework. ResNet was composed of a number of residual blocks, the shortcut connections element-wisely combined the input features with the output of the same block. These connections could assist the higher layers to access information from distant bottom layers and effectively alleviated the gradient vanishing problem, since they backpropagated the gradient to the bottom gradient without diminishing magnitude. For this reason, ResNet was able to be deeper and more accurate. Here, we used a 50-layer model ResNet-50.
Inception families [44], [45], [55] had evolved a lot over time, while there was an inherent property among them, which was a split-transform-merge strategy. The input of the inception model was split into a few lower-dimensional embeddings, transformed by a set of specialized filters, and merged by concatenation. This split-transform-merge behavior of inception module was expected to approach the representational power of large and dense layers but at a considerably lower computational complexity.
Dual path network (DPN-92) [56] was a modularized classification network that presented a new topology of connections internally. Specifically, DPN-92 shared common features and maintained the flexibility of exploring new features via dual path architectures, and realized effective feature reuse and exploration. Compared with some other advanced classification models such as ResNet-50, DPN-92 had higher parameter efficiency and was easy to optimize.
Residual attention network (Attention ResNet) [57] was a classification model which adopted attention mechanism. Attention ResNet could generate adaptive attention-aware features by stacking attention modules. In order to extract valuable features, the attention-aware features from different attention modules change adaptively when layers going deeper. In that way, the meaningful areas in the images could be enhanced while the invalid information could be suppressed. We used Attention ResNet-50 in the residual attention network.
All the classification models took the input of dual-channel information, i.e., the lesion regions and their corresponding segmentation masks (obtained from the previous segmentation models) were simultaneously sent into the classification models, then gave the classification results (positive or negative).
3.6. Model training
For neural network training, we trained all models from scratch with random initial parameters. Table 4 described the training and test data distribution of both segmentation and classification tasks. We trained the models on a server with eight NVIDIA TITAN RTX GPUs using the PyTorch [58] framework. We used Adam optimizer with an initial learning rate of and learning rate decay of .
3.7. Deployment
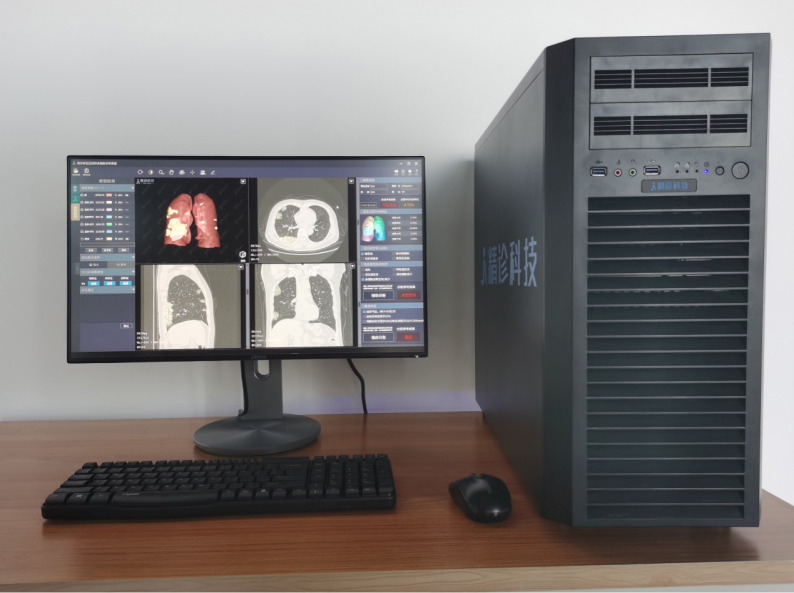
We deployed the trained models on workstations that we deployed on premise of the hospitals. A typical workstation contained an Intel Xeon E5-2680 CPU, an Intel I210 NIC, two TITAN X GPUs, and 64GB RAM (see Fig. 6). The server imported images from the hospital’s Picture Archiving and Communication Systems (PACS), and displayed the results iteratively. The server automatically checked for model/software updates and installed them so we could update the models remotely.
Fig. 6.
Demonstration of the deployment workstation.
3.8. Evaluation metrics
We used the Dice coefficient to evaluate the performance of the segmentation tasks and the area under the curve (AUC) to evaluate the performance of the classification tasks. Besides, we also analyzed the selected best classification model with sensitivity and specificity.
Concretely, the Dice coefficient was the double area of overlap divided by the total number of pixels in both images, which was widely used to measure the ability of the segmentation algorithm in medical image segmentation tasks. AUC denoted “area under the ROC curve”, in which ROC stood for “receiver operating characteristic”. ROC curve was drawn by plotting the true positive rate versus the false positive rate under different classification thresholds. Then AUC calculated the two-dimensional area under the entire ROC curve from (0, 0) to (1, 1), which could provide an aggregate measure of the classifier performance across varied discrimination thresholds. Sensitivity/specificity was also known as the true positive/negative rate, measured the fraction of positives/negatives that were correctly identified as positive/negative.
3.9. Reader study
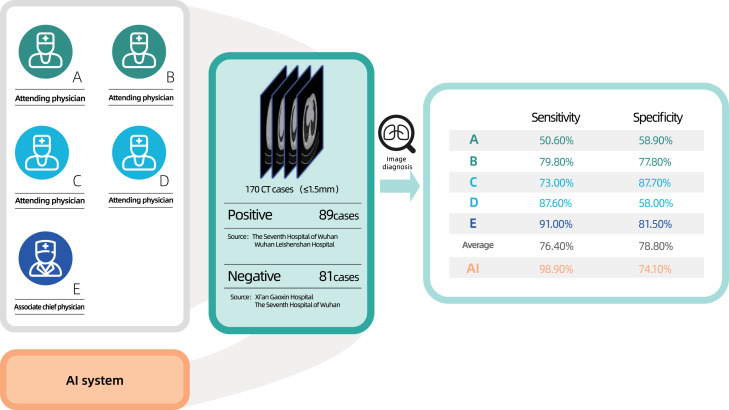
Five qualified physicians (three from hospitals in Wuhan, two from hospitals in Beijing) participated in this reader study. Four of them were attending physicians with average working years of five, while the last one was an associate chief physician with working years of eighteen. For this reader study, we generated a new dataset consisting of 170 cases. Among them, 89 were positive cases provided by The Seventh Hospital of Wuhan and Wuhan Leishenshan Hospital. The other 81 negative cases were chest CT scans with other lung diseases provided by Xi’an Gaoxin Hospital and The Seventh Hospital of Wuhan. Both the physicians and the AI system performed the diagnosis purely based on CT images.
4. Results
We proposed a combined “segmentation - classification” model pipeline, which highlighted the lesion regions in addition to the screening result. The model pipeline was divided into two stages: 3D segmentation and classification. The pipeline leveraged the model library we had previously developed. This library contained the state-of-the-art segmentation models such as fully convolutional network (FCN-8s) [54], U-Net [31], V-Net [14], and 3D U-Net++ [32], as well as classification models like dual path network (DPN-92) [56] , Inception-v3 [44], residual network (ResNet-50) [59], and Attention ResNet-50 [57]. We selected the best diagnosis model by empirically training and evaluating the models within the library.
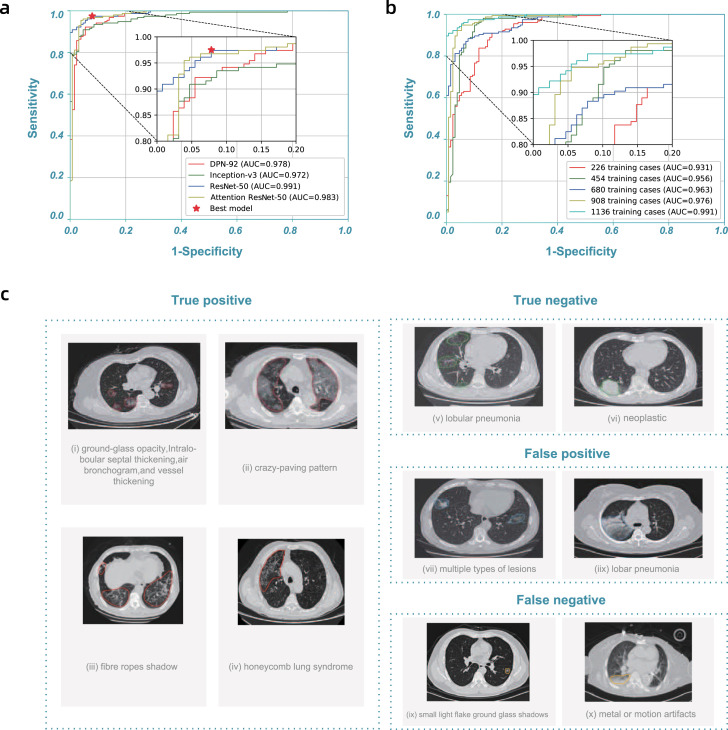
The latest segmentation model was trained on 732 cases (704 contained inflammation or tumors). The 3D U-Net++ model obtained the highest Dice coefficient of 0.754, and Table 3 showed the detailed segmentation model performance. By fixing the segmentation model as 3D U-Net++, we used 1136 (723 were positive)/282 cases (154 were positive) to train/test the classification/combined model, the detailed data distribution was given in Table 4, Table 5, Table 6. Fig. 2(a) showed the receiver operating characteristic (ROC) curves of these four combined models. The “3D Unet++ - ResNet-50” combined model achieved the best area under the curve (AUC) of 0.991. Fig. 2(a) plotted the best model by a star, which achieved a sensitivity of 0.974 and specificity of 0.922.
Table 3.
Dice coefficients of segmentation models.
| Segmentation model | Dice coefficient |
|---|---|
| FCN-8s | 0.681 |
| V-Net | 0.739 |
| U-Net | 0.742 |
| 3D U-Net++ | 0.754 |
Table 5.
Case-level data source distribution of the classification dataset by hospital.
| Hospital name | Training set |
Testing set |
||||
|---|---|---|---|---|---|---|
| Positive cases | Negative cases |
Positive cases | Negative cases |
|||
| Healthy | Other diseases | Healthy | Other diseases | |||
| Beijing Tsinghua Changgung Hospital | 4 | 43 | 183 | 1 | 10 | 48 |
| Wuhan No.7 Hospital | 150 | 15 | 88 | 38 | 6 | 32 |
| Zhongnan Hospital of Wuhan University | 113 | 12 | 72 | 28 | 5 | 27 |
| Tianyou Hospital | 319 | 0 | 0 | 59 | 0 | 0 |
| Wuhan’s Leishenshan Hospital | 132 | 0 | 0 | 33 | 0 | 0 |
| Total | 718 | 70 | 343 | 159 | 21 | 107 |
Table 6.
Case-level data source distribution of the classification dataset by CT scanner models.
| Equipment model | Training set |
Testing set |
||||
|---|---|---|---|---|---|---|
| Positive cases | Negative cases |
Positive cases | Negative cases |
|||
| Healthy | Other diseases | Healthy | Other diseases | |||
| UIH uCT 760 | 12 | 30 | 127 | 1 | 5 | 33 |
| UIH uCT 530 | 201 | 0 | 0 | 52 | 0 | 0 |
| GE Optima CT660 | 319 | 3 | 17 | 59 | 0 | 7 |
| GE Discovery CT750 HD | 0 | 13 | 54 | 0 | 5 | 14 |
| GE Discovery CT | 156 | 0 | 0 | 36 | 0 | 0 |
| GE BrightSpeed | 0 | 0 | 2 | 0 | 0 | 1 |
| SIEMENS SOMATOM Definition | 20 | 0 | 0 | 2 | 0 | 0 |
| SIEMENS Sensation Open | 1 | 0 | 0 | 1 | 0 | 0 |
| Philips iCT 256 | 0 | 24 | 141 | 0 | 11 | 52 |
| Philips Ingenuity CT | 14 | 0 | 0 | 3 | 0 | 0 |
| Philips Brilliance Big Bore | 0 | 0 | 2 | 0 | 0 | 0 |
| Total | 723 | 70 | 343 | 154 | 21 | 107 |
Fig. 2.
Model performance and highlights of model predictions.a, Receiver operating characteristic (ROC) curves of DPN-92, Inception-v3, ResNet-50, and Attention ResNet-50 with 3D U-Net++, respectively. b, ROC curves of 3D U-Net++ - ResNet-50 trained with different numbers of training cases. c, Typical predictions of the segmentation model.
The performance of the model improved steadily as the training data accumulated. In practice, the model was continually retrained in multiple stages (the average time between stages was about three days). Table 7 showed the training datasets we used in each stage. Fig. 2(b) showed the improvement of the ROC curves at each stage. At the first stage, the AUC reached 0.931 using 226 training cases. The model performance at the last stage, AUC reached 0.991 with 1136 training cases which was sufficient for clinical applications.
Table 7.
Training sets distribution in multiple training stages.
| Collection dates | Positive | Negative | Total |
|---|---|---|---|
| 2020.02.07 | 144 | 82 | 226 |
| 2020.02.10 | 289 | 165 | 454 |
| 2020.02.14 | 433 | 247 | 680 |
| 2020.02.17 | 578 | 330 | 908 |
| 2020.02.20 | 723 | 413 | 1136 |
With the model prediction, physicians could acquire insightful information from highlighted lesion regions in the user interface. Fig. 2(c) showed some examples. The model identified typical lesion characteristics of COVID-19 pneumonia, including ground-glass opacity, intralobular septal thickening, air bronchogram sign, vessel thickening, crazy-paving pattern, fiber stripes, and honeycomb lung syndrome. The model also picked out abnormal regions for cases with negative classification, such as lobular pneumonia and neoplastic lesion. These highlights helped physicians to quickly locate the slides and regions for detailed examination, improving their diagnosis efficiency.
It was necessary to study the false positive and false negative predictions, given in Fig. 2(c). Most notably, the model sometimes missed positive cases for patchy ground glass opacities with diameters less than 1 cm. The model might also introduce false positives with other types of viral pneumonia, for instance, lobar pneumonia, with similar CT features. Also, the model did not perform well when there were multiple types of lesions, or with significant metal or motion artifacts. We plan to obtain more cases with these features for training as our next steps.
Since the AI has powerful ability to locate the lesions in seconds, it can extremely reduce the workload of physicians which carefully search and decide the lesions from hundreds of CT images one by one. Thanks to this system, the physicians only need to examine the estimated results of artificial intelligence. To verify the efficiency of the proposed system, we employ 5 senior physicians to detect the COVID-19 infection regions, as shown in Fig. 3. We found the system to be effective in reducing the rate of missed diagnosis. By only using the CT scans, for 170 cases (89 were positive) randomly selected from the test set, five radiologists achieved an average sensitivity of 0.764 and specificity of 0.788, while the deep learning model obtained a sensitivity of 0.989 and specificity of 0.741. On 100 cases misclassified by at least one of the radiologists, the model sensitivity and specificity were 0.981 and 0.646, respectively. The radiologists showed a very low average sensitivity of 0.2 on the cases misclassified by the model, and 82.8% (18/22) of the cases were also misdiagnosed by at least one of the radiologists.
Fig. 3.
Illustration of the reader study. Five qualified physicians participated in this reader study. A total of 170 cases (89 were positive) were randomly selected from the test set.
At the time of writing, we had deployed the system in 16 hospitals, including Zhongnan Hospital of Wuhan University, Wuhan’s Leishenshan Hospital, Beijing Tsinghua Changgung Hospital, and Xi’an Gaoxin Hospital, etc. Physicians first ran the system once automatically, which took 0.8 s on average. The model prediction would be checked in the next step. Regardless of whether the classification was positive or negative, the physicians would check the segmentation results to quickly locate the suspected legions and examine if there were missing ones. Finally, physicians confirmed the screening result.
5. Discussion
In this section, we will discuss the benefits and drawbacks of the proposed system. As mentioned above, the deployed system is an effective to reduce the rate of missed diagnosis, and can distinguish the COVID-19 pneumonia and common pneumonia. In addition, the proposed system will produce the results of classification and segmentation, simultaneously. This combination facilitates doctors to make a definite diagnosis further. Furthermore, our system had deployed in 16 hospitals, which took 0.8 s on average and made the crucial contributions to cope the COVID-19 in practice.
Although the proposed system have achieved significant effects, it still has some failed cases. First, it does not perform well when there were multiple types of lesions, or with significant metal or motion artifacts. How to enhance the generalization ability of the system is our study in the future. Second, to train the network in the proposed system, we need a large annotated CT images which consist of lung contour, lesions and classification. Thus, the another flaw of our system is too dependent on fully annotated CT images.
6. Conclusion
The system would help the heavily affected areas, where enough radiologists were unavailable, by giving out the preliminary CT results to speed up the filtering process of COVID-19 suspected patients. For the less affected area, it could help less-experienced radiologists, who faced a challenge in distinguishing COVID-19 from normal pneumonia, to better detect the highly-indicative features of the presence of COVID-19.
While it was not currently possible to build a general AI that could automatically diagnose every new disease, we could have a generally applicable methodology that allowed us to quickly construct a model targeting a specific one, like COVID-19. The methodology not only included a library of models and training tools, but also the process for data collection, annotation, testing, user interaction design, and clinical deployment. Based on this methodology, we were able to produce the first usable model in 7 days after we received the first batch of data, and conducted additional four iterations in the model in the next 13 days while deploying it in 16 hospitals. The model was performing more than 1300 screenings per day at the time of writing.
Being able to take in more data continuously was an essential feature for epidemic response. The performance could be quickly improved by updating the model with continuous data taken in. To further improve the detection accuracy, we need to focus on adding training samples with complicated cases, such as cases with multiple lesion types. Besides, CT is only one of the factors for the diagnosis. We are building a multi-modal model allowing other clinical data inputs, such as patient profiles, symptoms, and lab test results, to produce a better screening result.
CRediT authorship contribution statement
Bo Wang: Conceptualization, Methodology. Shuo Jin: Conceptualization, Methodology. Qingsen Yan: Methodology, Writing - original draft. Haibo Xu: Conceptualization, Methodology. Chuan Luo: Conceptualization, Methodology. Lai Wei: Conceptualization, Methodology. Wei Zhao: Conceptualization, Methodology. Xuexue Hou: Methodology, Writing - original draft. Wenshuo Ma: Methodology, Writing - original draft. Zhengqing Xu: Methodology, Writing - original draft. Zhuozhao Zheng: Methodology, Writing - original draft. Wenbo Sun: Methodology, Writing - original draft. Lan Lan: Methodology, Writing - original draft. Wei Zhang: Methodology, Writing - original draft. Xiangdong Mu: Visualization, Investigation. Chenxi Shi: Visualization, Investigation. Zhongxiao Wang: Visualization, Investigation. Jihae Lee: Visualization, Investigation. Zijian Jin: Formal analysis. Minggui Lin: Formal analysis. Hongbo Jin: Formal analysis. Liang Zhang: Writing - review & editing. Jun Guo: Writing - review & editing. Benqi Zhao: Software, Validation. Zhizhong Ren: Software, Validation. Shuhao Wang: Software, Validation. Wei Xu: Supervision. Xinghuan Wang: Supervision. Jianming Wang: Supervision. Zheng You: Supervision. Jiahong Dong: Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work is supported by National Key Research and Development Program of China No. 2020YFC0845500, National Natural Science Foundation of China (NSFC) No. 61532001, Tsinghua Initiative Research Program Grant No. 20151080475, Application for Independent Research Project of Tsinghua University (Project Against SARI) .
References
- 1.World Health Organization (WHO) 2020. Coronavirus disease 2019 (COVID-19) situation report - 43.https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200303-sitrep-43-covid-19.pdf [Google Scholar]
- 2.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Graham R.L., Donaldson E.F., Baric R.S. A decade after SARS: strategies for controlling emerging coronaviruses. Nat. Rev. Microbiol. 2013;11(12):836–848. doi: 10.1038/nrmicro3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen H., Guo J., Wang C., Luo F., Yu X., Zhang W., Li J., Zhao D., Xu D., Gong Q. Clinical characteristics and intrauterine vertical transmission potential of COVID-19 infection in nine pregnant women: a retrospective review of medical records. Lancet. 2020 doi: 10.1016/S0140-6736(20)30360-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chung M., Bernheim A., Mei X., Zhang N., Huang M., Zeng X., Cui J., Xu W., Yang Y., Fayad Z. CT imaging features of 2019 novel coronavirus (2019-nCoV) Radiology. 2020 doi: 10.1148/radiol.2020200230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pan F., Ye T., Sun P., Gui S., Liang B., Li L., Zheng D., Wang J., Hesketh R.L., Yang L. Time course of lung changes on chest CT during recovery from 2019 novel coronavirus (COVID-19) pneumonia. Radiology. 2020 doi: 10.1148/radiol.2020200370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ai T., Yang Z., Hou H., Zhan C., Chen C., Lv W., Tao Q., Sun Z., Xia L. Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology. 2020 doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee E.Y., Ng M.-Y., Khong P.-L. COVID-19 pneumonia: what has CT taught us? Lancet Infect. Dis. 2020 doi: 10.1016/S1473-3099(20)30134-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.National Health Commission of the People’s Republic of China E.Y. seventh ed. 2020. The Notice of Launching Guideline on Diagnosis and Treatment of the Novel Coronavirus Pneumonia.http://www.nhc.gov.cn/yzygj/s7653p/202003/46c9294a7dfe4cef80dc7f5912eb1989.shtml [Google Scholar]
- 10.Ji Y., Ma Z., Peppelenbosch M.P., Pan Q. Potential association between COVID-19 mortality and health-care resource availability. Lancet Glob. Health. 2020 doi: 10.1016/S2214-109X(20)30068-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Song Y., Zheng S., Li L., Zhang X., Zhang X., Huang Z., Chen J., Zhao H., Jie Y., Wang R. Cold Spring Harbor Laboratory Press; 2020. Deep Learning Enables Accurate Diagnosis of Novel Coronavirus (COVID-19) with CT Images. medRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li L., Qin L., Xu Z., Yin Y., Wang X., Kong B., Bai J., Lu Y., Fang Z., Song Q. Artificial intelligence distinguishes covid-19 from community acquired pneumonia on chest ct. Radiology. 2020 doi: 10.1148/radiol.2020200905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi F., Xia L., Shan F., Wu D., Wei Y., Yuan H., Jiang H., Gao Y., Sui H., Shen D. 2020. Large-scale screening of COVID-19 from community acquired pneumonia using infection size-aware classification. arXiv preprint arXiv:2003.09860. [DOI] [PubMed] [Google Scholar]
- 14.Milletari F., Navab N., Ahmadi S.-A. 2016 Fourth International Conference on 3D Vision. IEEE; 2016. V-Net: Fully convolutional neural networks for volumetric medical image segmentation; pp. 565–571. [Google Scholar]
- 15.Zhao J., Zhang Y., He X., Xie P. 2020. Covid-ct-dataset: a ct scan dataset about covid-19. arXiv preprint arXiv:2003.13865. [Google Scholar]
- 16.Coronacases Initiative J. 2020. Helping radiologists to help people in more than 100 countries.https://coronacases.org/ [Google Scholar]
- 17.MedSeg J. 2020. COVID-19 CT segmentation dataset.http://medicalsegmentation.com/covid19/ [Google Scholar]
- 18.Cohen J.P., Morrison P., Dao L. 2020. COVID-19 image data collection. arXiv preprint arXiv:2003.11597. [Google Scholar]
- 19.United imaging’s emergency radiology departments support mobile cabin hospitals J.P. 2020. Facilitate 5G remote diagnosis.https://www.prnewswire.com/news-releases/united-imagings-emergency-radiology-departments-support-mobile-cabin-hospitals-facilitate-5g-remote-diagnosis-301010528.html [Google Scholar]
- 20.Li R., Cai C., Georgakis G., Karanam S., Chen T., Wu Z. 2019. Towards robust RGB-D human mesh recovery. arXiv preprint arXiv:1911.07383. [Google Scholar]
- 21.Wang Y., Lu X., Zhang Y., Zhang X., Wang K., Liu J., Li X., Hu R., Meng X., Dou S. Precise pulmonary scanning and reducing medical radiation exposure by developing a clinically applicable intelligent CT system: Toward improving patient care. EBioMedicine. 2020;54 doi: 10.1016/j.ebiom.2020.102724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yan Q., Gong D., Zhang Y. Two-stream convolutional networks for blind image quality assessment. IEEE Trans. Image Process. 2019;28(5):2200–2211. doi: 10.1109/TIP.2018.2883741. [DOI] [PubMed] [Google Scholar]
- 23.Yan Q., Zhang L., Liu Y., Zhu Y., Sun J., Shi Q., Zhang Y. Deep HDR imaging via a non-local network. IEEE Trans. Image Process. 2020;29:4308–4322. doi: 10.1109/TIP.2020.2971346. [DOI] [PubMed] [Google Scholar]
- 24.Yan Q., Wang B., Li P., Li X., Zhang A., Shi Q., You Z., Zhu Y., Sun J., Zhang Y. Ghost removal via channel attention in exposure fusion. Comput. Vis. Image Underst. 2020;201 doi: 10.1016/j.cviu.2020.103079. [DOI] [Google Scholar]
- 25.Yan Q., Wang B., Gong D., Luo C., Zhao W., Shen J., Shi Q., Jin S., Zhang L., You Z. 2020. COVID-19 chest CT image segmentation – a deep convolutional neural network solution. arXiv preprint arXiv:2004.10987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Q. Yan, D. Gong, P. Zhang, Q. Shi, J. Sun, I. Reid, Y. Zhang, Multi-scale dense networks for deep high dynamic range imaging, in: IEEE Winter Conference on Applications of Computer Vision, 2019, pp. 41–50.
- 27.Yan Q., Gong D., Shi Q., Hengel A.v.d., Shen C., Reid I., Zhang Y. 2019. Attention-guided network for ghost-free high dynamic range imaging. arXiv preprint arXiv:1904.10293. [Google Scholar]
- 28.Cao Y., Xu Z., Feng J., Jin C., Han X., Wu H., Shi H. Longitudinal assessment of COVID-19 using a deep learning–based quantitative CT pipeline: Illustration of two cases. Radiol.: Cardiothorac. Imaging. 2020;2(2) doi: 10.1148/ryct.2020200082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gozes O., Frid-Adar M., Greenspan H., Browning P.D., Zhang H., Ji W., Bernheim A., Siegel E. 2020. Rapid ai development cycle for the coronavirus (covid-19) pandemic: Initial results for automated detection & patient monitoring using deep learning ct image analysis. arXiv preprint arXiv:2003.05037. [Google Scholar]
- 30.Huang L., Han R., Ai T., Yu P., Kang H., Tao Q., Xia L. Serial quantitative chest CT assessment of COVID-19: Deep-learning approach. Radiol.: Cardiothorac. Imaging. 2020;2(2) doi: 10.1148/ryct.2020200075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ronneberger O., Fischer P., Brox T. International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer; 2015. U-Net: convolutional networks for biomedical image segmentation; pp. 234–241. [Google Scholar]
- 32.Zhou Z., Siddiquee M.M.R., Tajbakhsh N., Liang J. Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support. Springer; 2018. UNet++: A nested U-Net architecture for medical image segmentation; pp. 3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mu G., Lin Z., Han M., Yao G., Gao Y. University of Minnesota Libraries Publishing; 2019. Segmentation of Kidney Tumor by Multi-Resolution VB-Nets. [Google Scholar]
- 34.Qi X., Jiang Z., Yu Q., Shao C., Zhang H., Yue H., Ma B., Wang Y., Liu C., Meng X. Cold Spring Harbor Laboratory Press; 2020. Machine Learning-Based CT Radiomics Model for Predicting Hospital Stay in Patients with Pneumonia Associated with SARS-Cov-2 Infection: a Multicenter Study. medRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shan F., Gao+ Y., Wang J., Shi W., Shi N., Han M., Xue Z., Shen D., Shi Y. 2020. Lung infection quantification of covid-19 in ct images with deep learning. arXiv preprint arXiv:2003.04655. [Google Scholar]
- 36.Chen J., Wu L., Zhang J., Zhang L., Gong D., Zhao Y., Hu S., Wang Y., Hu X., Zheng B. Cold Spring Harbor Laboratory Press; 2020. Deep Learning-Based Model for Detecting 2019 Novel Coronavirus Pneumonia on High-Resolution Computed Tomography: a Prospective Study. medRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Noshad M., Jankovic I., Chen J.H. 2020. Clinical recommender system: Predicting medical specialty diagnostic choices with neural network ensembles. arXiv preprint arXiv:2007.12161. [Google Scholar]
- 38.Gu H., Huang J., Hung L., Chen X.A. 2020. Lessons learned from designing an AI-enabled diagnosis tool for pathologists. arXiv preprint arXiv:2006.12695. [Google Scholar]
- 39.Samaniego M., Kassani S.H., Espana C., Deters R. 2020. Access control management for computer-aided diagnosis systems using blockchain. arXiv preprint arXiv:2006.11522. [Google Scholar]
- 40.Li Y., Fujiwara T., Choi Y.K., Kim K.K., Ma K.-L. 2020. A visual analytics system for multi-model comparison on clinical data predictions. arXiv preprint arXiv:2002.10998. [Google Scholar]
- 41.Ghoshal B., Tucker A. 2020. Estimating uncertainty and interpretability in deep learning for coronavirus (COVID-19) detection. arXiv preprint arXiv:2003.10769. [Google Scholar]
- 42.Narin A., Kaya C., Pamuk Z. 2020. Automatic detection of coronavirus disease (COVID-19) using X-ray images and deep convolutional neural networks. arXiv preprint arXiv:2003.10849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.He K., Zhang X., Ren S. Deep residual learning for image recognition. Comput. Sci. 2015 [Google Scholar]
- 44.C. Szegedy, V. Vanhoucke, S. Ioffe, J. Shlens, Z. Wojna, Rethinking the Inception architecture for computer vision, in: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2016, pp. 2818–2826.
- 45.C. Szegedy, S. Ioffe, V. Vanhoucke, A.A. Alemi, Inception-v4, Inception-ResNet and the impact of residual connections on learning, in: Thirty-First AAAI Conference on Artificial Intelligence, 2017.
- 46.Zhang J., Xie Y., Li Y., Shen C., Xia Y. 2020. COVID-19 screening on chest X-ray images using deep learning based anomaly detection. arXiv preprint arXiv:2003.12338. [Google Scholar]
- 47.Jin C., Chen W., Cao Y., Xu Z., Zhang X., Deng L., Zheng C., Zhou J., Shi H., Feng J. Cold Spring Harbor Laboratory Press; 2020. Development and Evaluation of an AI System for COVID-19 Diagnosis. medRxiv. [Google Scholar]
- 48.Xu X., Jiang X., Ma C., Du P., Li X., Lv S., Yu L., Chen Y., Su J., Lang G. 2020. Deep learning system to screen coronavirus disease 2019 pneumonia. arXiv preprint arXiv:2002.09334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zheng C., Deng X., Fu Q., Zhou Q., Feng J., Ma H., Liu W., Wang X. Cold Spring Harbor Laboratory Press; 2020. Deep Learning-based Detection for COVID-19 from Chest CT using Weak Label. medRxiv. [Google Scholar]
- 50.Lehmann T.M., Gonner C., Spitzer K. Survey: Interpolation methods in medical image processing. IEEE Trans. Med. Imaging. 1999;18(11):1049–1075. doi: 10.1109/42.816070. [DOI] [PubMed] [Google Scholar]
- 51.Isensee F., Petersen J., Kohl S.A., Jäger P.F., Maier-Hein K.H. 2019. nnu-net: Breaking the spell on successful medical image segmentation, 1; pp. 1–8. arXiv preprint arXiv:1904.08128. [Google Scholar]
- 52.Eaton-Rosen Z., Bragman F., Ourselin S., Cardoso M.J. 2018. Improving data augmentation for medical image segmentation. [Google Scholar]
- 53.Hussain Z., Gimenez F., Yi D., Rubin D. AMIA Annual Symposium Proceedings, Vol. 17. American Medical Informatics Association; 2017. Differential data augmentation techniques for medical imaging classification tasks; pp. 9–19. [PMC free article] [PubMed] [Google Scholar]
- 54.J. Long, E. Shelhamer, T. Darrell, Fully convolutional networks for semantic segmentation, in: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2015, pp. 3431–3440. [DOI] [PubMed]
- 55.C. Szegedy, W. Liu, Y. Jia, P. Sermanet, S. Reed, D. Anguelov, D. Erhan, V. Vanhoucke, A. Rabinovich, Going deeper with convolutions, in: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2015, pp. 1–9.
- 56.Chen Y., Li J., Xiao H., Jin X., Yan S., Feng J. Advances in Neural Information Processing Systems. 2017. Dual path networks; pp. 4467–4475. [Google Scholar]
- 57.F. Wang, M. Jiang, C. Qian, S. Yang, C. Li, H. Zhang, X. Wang, X. Tang, Residual attention network for image classification, in: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2017, pp. 3156–3164.
- 58.A. Paszke, S. Gross, S. Chintala, G. Chanan, E. Yang, Z. DeVito, Z. Lin, A. Desmaison, L. Antiga, A. Lerer, Automatic differentiation in PyTorch, in: NIPS Workshop, 2017.
- 59.K. He, X. Zhang, S. Ren, J. Sun, Deep residual learning for image recognition, in: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2016, pp. 770–778.